Abstract
Plastic pollution is a growing environmental problem, in part due to the extremely stable and durable nature of this polymer. As recycling does not provide a complete solution, research has been focusing on alternative ways of degrading plastic. Fungi provide a wide array of enzymes specialized in the degradation of recalcitrant substances and are very promising candidates in the field of plastic degradation. This review examines the present literature for different fungal enzymes involved in plastic degradation, describing their characteristics, efficacy and biotechnological applications. Fungal laccases and peroxidases, generally used by fungi to degrade lignin, show good results in degrading polyethylene (PE) and polyvinyl chloride (PVC), while esterases such as cutinases and lipases were successfully used to degrade polyethylene terephthalate (PET) and polyurethane (PUR). Good results were also obtained on PUR by fungal proteases and ureases. All these enzymes were isolated from many different fungi, from both Basidiomycetes and Ascomycetes, and have shown remarkable efficiency in plastic biodegradation under laboratory conditions. Therefore, future research should focus on the interactions between the genes, proteins, metabolites and environmental conditions involved in the processes. Further steps such as the improvement in catalytic efficiency and genetic engineering could lead these enzymes to become biotechnological applications in the field of plastic degradation.
Keywords: plastic, biodegradation, enzymes, fungi, bioremediation, biotechnology
1. Introduction
The word plastic derives from the Greek “plastikos”, meaning “able to be modeled” [1]. Today, the term plastic refers to a range of synthetic long-chain polymeric molecules which, in the 1950s, started to be substituted for natural materials across a range of different sectors and in everyday applications [2]. The rapid development of plastics can be attributed to their combination of lightness, durability and other intrinsic properties, along with their easy and low-cost production [3]. As a result of their versatility, plastic materials have been increasingly used, reaching global production of almost 370 million tonnes in 2020 [4]. Almost 55 million tonnes of plastic were produced in Europe in 2020 [4]. The most commonly used plastics are polyethylene (PE; 30.3%), polypropylene (PP; 19.7%), polyvinyl chloride (PVC; 9.6%), polyethylene terephthalate (PET; 8.4%), polyurethane (PUR; 7.8%) and polystyrene (PS; 6.1%) [4,5].
1.1. Plastic Pollution
The mass of plastics in municipal solid waste in high-income and developing countries increased from less than 1% in 1960 to more than 10% in 2005 [6]. In Europe, 6.9 million tonnes of plastics were dumped in landfills in 2020 [4]. One of the problems of plastics accumulated in landfills or released into the environment, is the long time they take to decay. This long decay time derives from the inherent characteristics of plastic, especially the high molecular weight, crystallinity and hydrophobicity [3,7], and from the fact that its monomers, such as ethylene and propylene, originate from fossil hydrocarbons [8]. This results in the accumulation and persistence of plastic in land, freshwater, and oceans for many decades [9,10]. Moreover, supplementary chemicals and additives are often added to plastic polymers to increase the quality of the final products [11]. These additives such as endocrine disrupting chemicals (bisphenol A, bisphenol S, octylphenol and nonylphenol) [12], dioxin-like compounds [13] and heavy metals [14,15] can cause negative effects on organisms. Reproductive abnormalities, disruption of the endocrine system, diabetes and obesity could be linked to additives in plastics [16].
A further problem associated with plastic pollution is the formation of small particles called microplastics, which originate from plastic fragmentation. Microplastics and bigger plastic fragments can enter the food chain and be transferred to higher trophic level organisms where they accumulate [17]. Microplastics can also be the carrier for toxic chemicals and pathogens, facilitating their dispersion in the environment and threatening ecosystems [3,5].
In 2020, 12.4 million tonnes of plastics were used for energy recovery [4] and plastic incineration plays an important role in the management of municipal solid waste [18]. However, energy recovery by incineration can lead to harmful and toxic emissions, such as dioxins, furans, heavy metals and sulphides, contributing to environmental pollution [19,20,21]. Recycling is a better alternative for plastic waste management, but it is not the ultimate solution to the plastic problem. For example, the mechanical properties of recycled PET are reduced with each reuse and the thermal degradation of PE by pyrolysis leads to the random breaking of C–C bonds with the consequent drastic change in its mechanical properties [22]. Moreover, the extremely low percentage of PP recycling (<1%) is alarming since in many cases it is not found as the only polymer forming an object. In addition, the tertiary carbon in PP is susceptible to photo-oxidative and thermo-oxidative degradation, requiring a stabiliser to be added in the production stage, contributing to the deterioration of recycled PP properties [22].
1.2. Plastic Biodegradation
Biodegradation is a complex process of physico-chemical transformation of polymers into smaller units mediated by microorganisms [23,24]. Microorganisms, including fungi, are able to biochemically degrade, assimilate and metabolise complex organic compounds, xenobiotics and recalcitrant substances for their energy needs [25,26]. Several organisms and different mechanisms are being investigated at the moment to improve and promote the biodegradation of complex and polluting polymers. For example, the addition of bacteria with specific engineered plasmids in polluted sites could transfer the catabolic genes in the plasmids to the indigenous bacterial population increasing their capacity for xenobiotic degradation [27]. Furthermore, it is possible to insert mutations in bacterial genes that increase the biodegradative capacity of their enzymes. Thanks to this technique, Lu et al. [28] created an algorithm for an engineered and robust PET hydrolase that can be active in a wide range of temperatures and pHs. Another interesting line of research is to exploit extremophile microorganisms, so that they can be used in the bioremediation of extremely polluted sites. For example, some fungi, such as strains belonging to Fusarium, Verticillium, Penicillium and Aspergillus, are able to produce metal nanoparticles that allow them to tolerate and remove heavy metals from heavily polluted water or soil [29].
The first step required for the biodegradation of high molecular weight and long-chain polymers, such as plastics, is the weakening of the polymers’ structure. Many different factors can influence plastic biodegradation, for example, the hydrophobicity of the exposed area as well as the chemical structure, crystallinity grade and structure, glass transition, melting temperature and elasticity [30,31,32]. Environmental factors, such as UV exposure or temperature, can decrease the hydrophobicity of plastics or introduce carbonyl/carboxyl/hydroxyl groups, increasing their biodegradability [33,34,35]. These environmental factors can lead to surface roughness, cracks and molecular changes in plastics [36]. Similarly, the growth of microorganisms on plastic surfaces can modify the physical properties of the plastic by creating cracks and enlarging the pore size. Microorganisms can also chemically deteriorate plastics, for example, by changing the pH of the surrounding microenvironment [37].
The second step in plastic biodegradation is the depolymerisation into shorter chains. The microbial exoenzymes involved in this process create intermediates with modified properties, that increase their cellular assimilation [38]. After intermediates are created and assimilated, they are used by cells as carbon sources and broken down into water and carbon dioxide or methane to complete the mineralization process [5,24].
The complexity of plastic biodegradation is due to their chemical and physical characteristics, such as their high molecular weight, hydrophobicity and insolubility [39]. The use of filamentous fungi for the bioremediation process of plastics can overcome this problem. Indeed, filamentous fungi present a typical hyphal apical growth form that allows them to extend their mycelial networks into different kinds of materials [40]. The penetrative abilities of fungal hyphae are associated with their secretion of exoenzymes and hydrophobins, increasing their adhesion to hydrophobic substrates [41]. Moreover, the non-specificity of fungal exoenzymes allows them to break down different plastic polymers [42]. For example, fungal hydrolases (lipases, carboxylesterases, cutinases and proteases) can modify the plastic surface, increasing its hydrophilicity [43]. These enzymes are also involved in PET and PUR biodegradation due to the presence of hydrolysable chemical bonds in the polymer structures [44,45,46]. On the other hand, oxidoreductases (laccases and peroxidases) are involved in plastic degradation into smaller molecules such as oligomers, dimers and monomers [47,48]. Due to their highly stable carbon–carbon (C–C) bonds, plastic polymers such as PE, PS, PP and PVC require oxidation before the depolymerisation process [39,49].
In this context, this review will examine the different fungal enzymes involved in the degradation processes of the primary petroleum-based plastic polymers, describing their main characteristics, their efficacy and their possible biotechnological applications. Therefore, the aim of this review is to provide an extensive and reliable assessment of all the present knowledge on fungal enzymes in plastic biodegradation as a start base for new bioremediation applications.
2. Fungal Enzymes Involved in Plastic Biodegradation
The main classes of enzymes involved in plastic biodegradation are hydrolases and oxidoreductases (Table 1). These enzymes have been extensively studied due to their involvement both in natural and industrial processes. For example, in nature and industry they are essential in lignocellulose biodegradation [50], fungal pathogenesis [51] and the hydrolysis of triacylglycerol to fatty acids [52]. These enzymes also have applications in the food and textile industries [53,54,55], and in bioremediation processes [56,57].
Table 1.
Reaction schemes of the main fungal enzymes involved in plastic degradation.
| Enzyme | Enzyme Commission (EC) Number | Activity | Reaction Scheme |
|---|---|---|---|
| Laccases | 1.10.3.2 | Oxidoreductases |
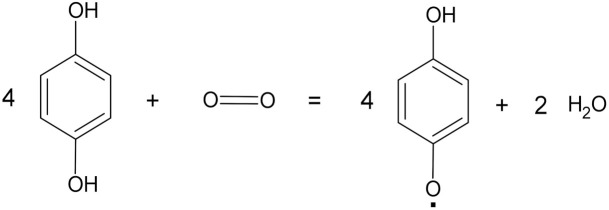 4 benzenediol + O2 = 4 benzosemiquinone + 2 water |
| Manganese peroxidases | 1.11.1.13 | Oxidoreductases |
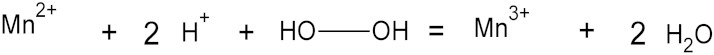
|
| Lignin peroxidases | 1.11.1.14 | Oxidoreductases |
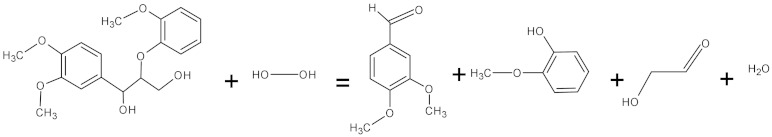 1-(3,4-dimethoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol + hydrogen peroxide = 3,4-dimethoxybenzaldehyde + 2-methoxyphenol + glycolaldehyde + water |
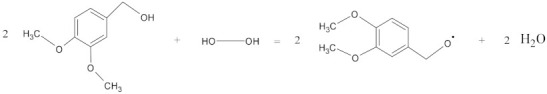 2 (3,4-dimethoxyphenyl)methanol + hydrogen peroxide = 2 (3,4-dimethoxyphenyl)methanol radical + 2 water |
|||
| Versatile peroxidase | 1.11.1.16 | Oxidoreductases |
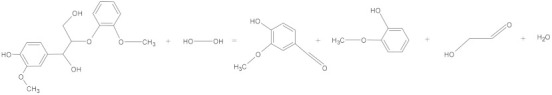 1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol + hydrogen peroxide = 4-hydroxy-3-methoxybenzaldehyde + 2-methoxyphenol + glycolaldehyde + water |
| Versatile peroxidase | 1.11.1.16 | Oxidoreductases |
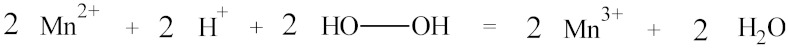
|
| Lipases | 3.1.1.3 | Hydrolases |
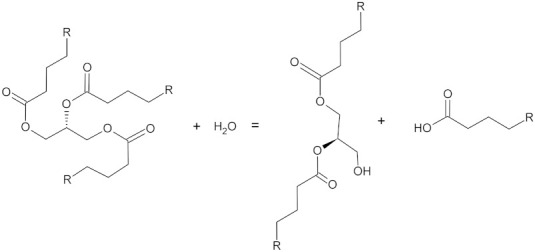 Triacylglycerol + water = diacylglycerol + a carboxylate |
| Cutinases | 3.1.1.74 | Hydrolases |
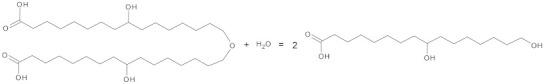 Cutin + water = 2 cutin monomers |
2.1. Laccases (EC 1.10.3.2)
Laccases (EC 1.10.3.2) are a class of enzyme belonging to the blue copper oxidases and are multicopper monomeric glycoproteins [58]. They use oxygen as an electron acceptor to oxidize phenolic and non-phenolic compounds, and they are involved in the reduction of molecular oxygen to water [59,60,61].
Laccases were first discovered in the plant species Rhus vernicifera in 1883 [62]. In 1896 they were identified in fungi for the first time by Bertrand and Laborde [63,64]. In the following years, laccases were discovered in many species of fungi belonging to Ascomycetes, Basidiomycetes and Deuteromycetes [65]. White-rot fungi are the most studied group in relation to their ability to produce laccases to degrade lignin [63]. Indeed, in nature, fungal laccases are involved in lignin degradation and in the removal of toxic phenols produced during this process [66]. Moreover, they play a role in the synthesis of dihydroxynaphthalene melanins, compounds that are useful for protection against environmental stress [67]. Thanks to their characteristics, laccases are used in a number of industrial applications such as delignification, pulp bleaching and bioremediation processes removing toxic compounds through oxidative enzymatic coupling [64].
2.2. Peroxidases (EC 1.11.1)
Peroxidases (EC 1.11.1) are a group of haem containing oxidoreductases, which catalyse the oxidation of organic and inorganic compounds and the reduction of hydrogen peroxide [68]. Peroxidases are divided into three classes: class-I are intracellular peroxidases that are found in most living organisms, except animals; class-II are extracellular fungal peroxidases; class-III are extracellular plant peroxidases [69]. The main fungal peroxidases are manganese peroxidases (MnP; EC 1.11.1.13), lignin peroxidases (LiP; EC 1.11.1.14), versatile peroxidases (EC 1.11.1.16) and dye decolorizing peroxidases (EC 1.11.1.19), depending on what they use for the reducing substrate [70].
The most well-known group of peroxidase-producing fungi are the ligninolytic fungi such as white-rot fungi Phanerochaete chrysosporium, Trametes versicolor, Pleurotus spp., Phlebia radiata, Bjerkandera adusta, Ceriporiopsis subvermispora and Dichomitus squalens [68,70,71]. LiP and MnP were first discovered and purified in the extracellular medium of a Phanerochaete chrysosporium culture in the 1980s [72,73,74,75]. Since then, studies have continued to better understand the functioning of these enzymes, and in 2010 the structure of the MnP of P. chrysosporium was refined at 0.93 Å resolution [76].
The main characteristics of peroxidases are their non-specificity and their ability to oxidise substrates with high redox potential [70,77]. These properties have led to these enzymes being used in a large number of applications. Peroxidases are involved in the production of biofuels and paper [78], in waste treatment [68] and in the bioremediation of industrial pollutants such as synthetic dyes and polycyclic aromatic hydrocarbons (PAHs) [40,79,80,81,82,83,84,85].
2.3. Cutinases (E.C. 3.1.1.74)
Cutinases (E.C. 3.1.1.74) are extracellular serine esterases and are divided into two fungal subfamilies and one bacterial subfamily [86]. These subfamilies have a different primary structure according to whether they are eukaryotic or prokaryotic [87]. Cutinases have an α/β fold, and a central β-sheet composed of five parallel strands covered by two or three helices on either side of the sheet [88]. Their active site is uncovered and consists of a catalytic triad of Ser-His-Asp/Glu [89]. One exception is the active site of Trichoderma reesei cutinase, which has a covered active site, similar to a lipase [90]. In this case the imidazole of the histidine removes a proton from the serine hydroxyl group and the serine oxygen makes a nucleophilic attack on the substrate acyl carbonyl carbon [91]. Then, by a transacylation reaction with the serine, the substrate becomes an acyl-enzyme intermediate, which is then hydrolysed to release the product. The development of a negative charge during the formation of the acyl-enzyme intermediate is stabilized by an oxyanion hole of cutinase [88,92].
In nature, because they can degrade ester bonds, cutinases are involved in fungal pathogenesis [88]. Cutinases are also multifunctional enzymes with many industrial applications due to their ability to catalyse hydrolysis reactions, esterifications and transesterifications [93]. These properties make them suitable for the degradation of high molecular weight polyesters [94] and useful in synthetic fibre modification [95].
2.4. Lipases (EC 3.1.1.3)
Fungal lipases (EC 3.1.1.3) are extracellular triacylglycerol acyl hydrolases that can hydrolyse ester bonds from insoluble substrates of tri-, di- and mono-glycerides into free fatty acids and glycerol [96,97]. Therefore, lipases are involved in lipid metabolism in processes such as digestion, absorption and reconstitution [98].
Lipase-producing fungi have been isolated from different habitats such as contaminated soils, wastes and deteriorated food [98,99,100]. The main genera of lipase-producing fungi are Aspergillus, Acremonium, Alternaria, Beauveria, Candida, Eremothecium, Fusarium, Geotrichum, Humicola, Mucor, Ophiostoma, Penicillium, Rhizomucor, Rhizopus and Trichoderma [96,101].
Fungal lipases are used in a number of industrial applications such as in the food, textile and manufacturing industries and in the production of detergents, cosmetics and pharmaceuticals [102]. Moreover, lipases can degrade fatty wastes [103] and polyurethane (PUR) [104].
3. Types of Plastics and Their Biodegradation by Fungi
3.1. Polyethylene (PE)
Polyethylene (PE) is one of the most abundant and most widely commercialized synthetic, petroleum-based, thermoplastic materials [105,106]. PE consists of long linear chains of ethylene monomers (C2H4)n [107] and can be divided into high-density polyethylene (HDPE) and low-density polyethylene (LDPE), according to the number of carbon atoms and the molecular weight [108] (Table 2). HDPE, so called because it has a molecular weight between 100,000–250,000 Daltons [109], is used in bottles for cleaning products, houseware items and for toy production, making up 12.9% of the total plastic demand in Europe [4]. On the other hand, LDPE has a molecular weight of 40,000 Daltons [109] and is mainly used in single-use plastics such as bags and food packaging films, composing 17.4% of the total plastic demand in Europe [4]. Due to the short time period for the use of PE products, they are rapidly accumulated in the environment in large numbers [110,111]. Over the years, this large abundance of PE in the environment has led to the investigation of alternative disposal methods, including microbial biodegradation.
Table 2.
European plastic demand, molecular weight (Daltons) and backbone structural formula of polyethylene, high-density polyethylene and low-density polyethylene.
| Name | Plastic Demand in Europe | Molecular Weight (Daltons) | Structure |
|---|---|---|---|
| High-density polyethylene (HDPE) | 12.9% | 100,000–250,000 |
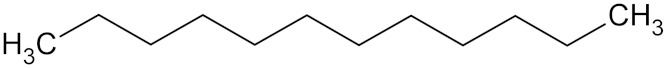
|
| Low-density polyethylene (LDPE) | 17.4% | 40,000 |
|
One of the main problems of using microbes to degrade PE is its high molecular weight, which limits the number of possible enzymatic reactions. Usually, a microorganism’s enzymatic systems use substrates with 10–50 carbons. Therefore, in order to degrade PE using microbes, a reduction in its molecular weight is necessary. Moreover, the transport of molecules through the cell membrane also requires a lower molecular weight [49]. After the decrease in molecular weight, enzymes must oxidise the polymer to transform it into a carboxylic acid that can be metabolized by β-oxidation and the Krebs cycle [112]. The reduction in molecular weight can be performed using abiotic factors such as UV light and heat, or by microbial enzymes [49]. Most studies investigating this enzymatic reduction have focused on bacteria that use laccase and alkane hydroxylase from the AlkB family [113,114]. However, the advantage of fungal biodegradation is in the production of exoenzymes. Hydrophobins, fungal surface hydrophobic proteins that facilitate hyphae adhesion to a plastic surface, and exoenzymes, such as peroxidases and oxidases, promote oxidation or hydrolysis [38,39,41,115,116]. The most studied fungal genera belong to Ascomycota and are Aspergillus, Penicillium, Trichoderma and Fusarium [117,118,119,120,121,122,123], whereas Basidiomycota or Mucoromycota are less investigated. In addition, the involvement of fungi such as Phanerochaete chrysosporium [124], Bjerkandera adusta [125], Trametes versicolor [115] and Rhizopus oryzae [126] in PE biodegradation has also been reported.
3.2. Fungal Enzymes Involved in PE Biodegradation
The main fungal enzymes involved in polyethylene biodegradation are the lignolitic enzymes laccases (Lac, EC 1.10.3.2.) and peroxidases (EC 1.11.1.7) [116,117,120,127].
The effect of these enzymes on PE has been studied extensively in Basidiomycota, but they are also present in Ascomycota. For example, the ascomycete Trichoderma harzianum is able to produce both laccase (Mw 88 kDa) and peroxidase (Mw 55 kDa) when involved in PE biodegradation [120]. Indeed, the treatment of PE with 0.01071 IU/mL of its laccase caused a reduction in mass of 0.5% after 10 days incubation, while the treatment of PE with 0.01080 IU/mL of its peroxidase caused a 0.6% loss of mass. As a result of the enzymatic treatment, carboxylic acids, aldehydes, aromatics, alcohols, esters, ethers and alkyl halides groups were formed and detected by Fourier-transform infrared spectroscopy (FTIR) analysis.
A particularly interesting ascomycete involved in HDPE microplastic biodegradation (density 0.955 g/cm3, size below 200 μm) is Aspergillus flavus PEDX3, which was isolated from the gut of the wax moth Galleria mellonella [38]. This strain was able to depolymerize HDPE long chains and produce lower molecular weight fragments after 28 days of incubation. A. flavus PEDX3 action could be attributed to its capacity to produce laccases and laccase-like multicopper oxidases (LMCOs). Gene sequencing analysis using RT-PCR led to the identification of two genes (AFLA_006190 and AFLA_053930) that may encode potential degrading LMCOs [38].
In the Basidiomycota, partially purified manganese peroxidase (MnP, EC 1.11.1.13) from Phanerochaete chrysosporium ME-446 caused significant PE degradation when 0.1% Tween 80 was present in the growing medium, reducing tensile strength and elongation [115]. Moreover, after the addition of 0.1 mM manganese sulfate (MnSO4), PE molecular weight (Mw) decreased from 716,000 to 89,500 Daltons and the relative elongation changed from 100% to 0%. Although exogenous H2O2 supply is not necessary for polyethylene degradation, it is essential for the MnP reaction system [115].
Fujisawa et al. [128] investigated the effects of a laccase-mediator system (LMS) from Trametes versicolor IFO 6482 in PE biodegradation. LMS (500 nkat) was able to reduce the PE elongation by 20% in 3 days, while the addition of 0.2 mM 1-hydroxybenzotriazole (HBT) to the medium caused no elongation and relative tensile strength decreased by 60%. Moreover, Mw changed from 242,000 to 28,300 Daltons after 3 days of LMS with HBT mediator treatment at 30 °C.
Another Basidiomycete involved in PE biodegradation is Pleurotus ostreatus, which can hydrolyse C–C bonds by producing extracellular ligninolytic enzymes including lignin peroxidase (LiP), manganese peroxidase (MnP) and laccases (Lac). During growth on semisolid Radha modified medium in the presence of LDPE sheets, high enzyme production was detected. Specifically, the highest Lac and LiP activities were 2.817 U/g and 70.755 U/g after 30 days and 90 days, respectively, while the highest MnP production was observed at day 120 (1.097 U/g) [48].
A recent study reports computational molecular simulations between PE (dodecane, 170.3 Daltons) and different enzymes known to degrade it. Santacruz-Juárez et al. [116] studied the interactions between MnP (manganese peroxidase from Phanerochaete chrysosprium), LiP (lignin peroxidase from Trametes cervine), Lac (laccase from Trametes versicolor), UnP (unspecific peroxygenase from Agrocybe aegerita) or Cut (cutinase from Fusarium solani, as a negative control) and PE. They measured the binding affinity, i.e., the strength of the binding interaction between the enzyme and its ligand (PE), and found affinities were UnP (34.34 μM) > Lac (40.11 μM) > LiP (66.93 μM) > MnP (82.16 μM) > Cut (5590 μM). The high area (659.920 Å2), volume (367.243 Å3) and hydrophobicity of the UnP catalytic cavity was suggested to be the reason for the high interaction with PE [129,130,131]. The hydrophobicity is caused by the presence of phenylalanine residues in the UnP active site [116]. The binding affinity mirrored the binding energy scores, which resulted in −6.09, −6.00, −5.69, −5.57 and −3.07 Kcal/mol for UnP–PE, Lac–PE, LiP–PE, MnP–PE and Cut–PE complexes, respectively. Indeed, the lower the required binding energy, the easier the bonds are created. These computational observations showed that peroxidases can play an important role in PE biodegradation, and that the non-specific UnP enzymes can be used in practical applications due to their distinctive cavity composed of Val244, Phe121, Phe191, Phe199, Phe274, Ala77, Thr192, Gly195, Glu196, Ser123, Cys33, haem propionate, 1H-imidazol-5-yl methanol (Mzo354) and two water molecules [116].
Hypothetical biodegradation pathways involving the ligninolytic enzymes (Lac, LiP and MnP) and PE have been proposed, using as the biosurfactant a fungal hydrophobin from class-II [41,116]. Specifically, Bertrand et al. [132] hypothesized a PE degradation pathway that uses Lac from Trametes versicolor; Miki et al. [133] suggested using LiP from Trametes cervine; Sánchez [41], in a computational study, proposed using MnP from Phanerochaete chrysosporium. In order to perform their degradative activity, both MnP and LiP require the addition of H2O2 to the culture medium [116] and acidic conditions [41]. The involvement of H2O2 is to act as an electron accepting co-substrate in the oxidation-reduction reactions promoted by MnP and LiP [41]. Alternatively, Lac causes the transfer of electrons from organic substrates to molecular oxygen. Therefore, the main difference in PE biodegradation pathways between laccases and haem peroxidases (LiP and MnP) is based on the different methods of electron transfer [41].
3.3. Polyethylene Terephthalate (PET)
Polyethylene terephthalate (PET) is another important synthetic, petroleum-based, thermoplastic polymer commonly used in everyday life [134,135]. PET is a saturated polyester consisting of terephthalic acid (TPA) and ethylene glycol (C10H8O4)n with an average molecular weight ranging from 20,000 to 50,000 Daltons depending on the field of application [136] (Table 3). PET is widely used in beverage bottles, synthetic textile fibres, films and resins, and it has a significant market share of plastic fibres thanks to its versatile performance characteristics [137,138]. PET fibres are employed in cloth, technical and medical textiles, and in furnishings [137]. Nowadays, PET represents 8.4% of the plastics demand in Europe [4].
Table 3.
European plastic demand, molecular weight (Daltons) and backbone structural formula of polyethylene terephthalate.
| Name | Plastics Demand in Europe | Molecular Weight (Daltons) | Structure |
|---|---|---|---|
| Polyethylene terephthalate (PET) | 8.4% | 20,000–50,000 |
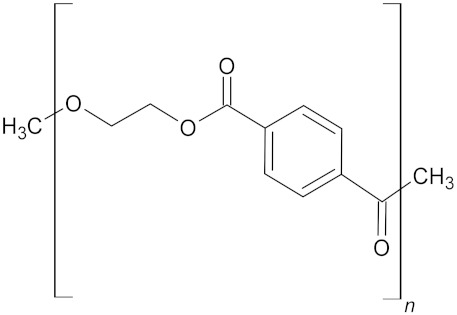
|
PET depolymerisation takes place when it is converted to terephthalic or isophthalic acid, ethylene glycol and small oligomers such as bis(2-hydroxyethyl) terephthalate (BHET) and mono(2-hydroxyethyl) terephthalate (MHET) [139]. These polymers are less harmful to the environment than PET [140]. Although PET is a polyester (compounds that are considered to degrade more easily), it is recalcitrant to biodegradation [141]. However, fungal genera such as Aspergillus [142,143], Fusarium [144,145] and Penicillium [146,147] have been reported to be involved in PET biodegradation.
3.4. Fungal Enzymes Involved in PET Biodegradation
In recent times, several studies have looked for enzymes involved in PET biodegradation. However, most studies have focused on bacterial enzymes [148,149,150] with fungal enzymes being less investigated.
The main fungal enzymes involved in PET biodegradation are hydrolytic enzymes acting on ester bonds (esterases; EC 3.1.1), such as cutinases (EC 3.1.1.74), lipases (EC 3.1.1.3) and carboxylesterases (EC 3.1.1.1) [138,149,151].
3.4.1. Cutinases Involved in PET Biodegradation
Specific cutinases able to degrade PET were identified from Humicola insolens (HiC) [148,152], Fusarium solani pisi (FsC) [148,152,153,154,155] and Fusarium oxysporum (FoCut5a) [94].
The most studied enzymes are the cutinases HiC and FsC. HiC has good thermostability with a temperature range from 30 to 85 °C, an optimum at 80 °C, and maximum initial activity from 70 to 80 °C. On the other hand, FsC has a lower temperature range of 30–60 °C with the best performance at 50 °C. Ronkvist et al. [152] tested the biodegradation capacity of HiC and FsC. They found that the hydrolysis rate constant k2 was 7-fold higher for HiC at 70 °C than FsC at 40 °C (0.62 μmol/cm2/h compared to 0.09 μmol/cm2/h) [152]. Moreover, the results showed a 97 ± 3% weight loss when low-crystallinity PET was incubated with HiC for 96 h at 70 °C, while there was only a 5% decrease after 96 h of incubation with FsC at 40 °C.
A few studies noted that the activity of cutinases was higher for an amorphous PET polymer compared to that of a highly crystalline substrate [43,154]. Indeed, these enzymes are sensitive to chain distribution and length [156]. An increase in the PET crystallinity rate from 7% to 35% caused a decrease in the initial enzymatic activities up to 25-fold for HiC and 6-fold for FsC [152]. The enzymes’ preference for amorphous regions of PET led to an increase in the biodegradation of these regions and an increase in the crystallinity rate of the biodegraded polymer [152,153]. Moreover, a high presence of aromatic rings lowers the rate of hydrolysis. On the other hand, HiC preferably hydrolysed both internal (terephthalic acid-1,4-butanediol) and external (benzoic acid-1,4 butanediol) ester bonds, and more rapidly hydrolysed substrates with longer terminal alcohols but shorter chain length acids [157].
Several studies have focused on trying to explain the biodegradation pathways and functioning of HiC and FsC. For example, the ability of cutinases to cleave ester bonds of dissolved materials [158] was confirmed by Eberl et al. [153], who studied the PET monomer bis(2-hydroxyethyl) terephthalate (BHET). They observed that a cutinase from F. solani was able to completely hydrolyse BHET in only 30 min. Terephthalic acid (TPA) formation started after the complete hydrolysis of BHET to the monoester mono (2-hydroxyl ethyl) terephthalate (MHET). After the 96 h incubation of low-crystallinity PET with FsC in 1 M Tris-HCl (pH 8) at 40 °C, or with HiC at 70 °C, MHET was reduced to terephthalic acid (TPA) and ethylene glycol [152,159]. Nevertheless, after FsC-catalysed PET hydrolysis, Vertommen et al. [43] observed a predominant production of MHET, some TPA and small traces of BHET. These differences could be ascribed to changes in the ratio of substrate—enzyme, or changes in the incubation conditions, which could lead to the incomplete conversion of water-soluble PET degradation products into TPA.
The active site of a cutinase from Fusarium solani pisi (PDB code1CEX) was genetically modified to improve its activity towards PET fibres [160]. Previous studies proposed a 3D structure of this enzyme [88] and the native cutinase gene sequence that can be PCR-amplified with the primers CutFor (5′-CGGGATCCCATGAAACAAAGCACTATTGCACTG-3′) and CutRev (5′-CGAGCTCGCAGCAGAACCACGGACAGCC-3′) from the vector pDrFST [161]. This information allowed Araújo et al. [157] to carry out computational studies showing that the increase in the size of the active site corresponds with a 4–5-fold enhancement in activity. Indeed, the large PET polymer can fit better in the active site after the amplification. Furthermore, it was shown that the mutations L81A, N84A, L182A, V184A and L189A result in a better stabilization of the tetrahedral intermediate of the model substrates [160] due to the enlargement of the active site.
Another cutinase with potential in PET bioremediation is produced by Fusarium oxysporum, and is called FoCut5a [94,144]. It is highly homologous to F. solani pisi cutinase (FsC), but the hydrophobic residues Ala62 and Phe63 present in F. solani are replaced by Lys63 and Tyr64 polar amino acids in FoCut5a at the end of helix a2. Due to these and other small but significant differences, FoCut5a seems slightly more thermostable than FsC, underlining a possible important role in industrial applications [94]. The optimized parameters for PET hydrolysis are 40 °C, pH 8 and 1.92 mg FoCut5a per gram of fabric [144]. FoCut5a efficacy was confirmed by superficial changes observable by Fourier-transform infrared spectroscopy (FTIR) ATR analysis, X-ray photoelectron spectroscopy (XPS), Scanning Electron Microscope (SEM), as well as through dyeability tests using reactive dyes [144].
3.4.2. Lipases Involved in PET Biodegradation
Lipases are another class of enzymes involved in PET biodegradation [149]. The most studied that are involved in PET biodegradation are produced by Aspergillus oryzae CCUG 33812 [143] and by the yeasts Candida antarctica (CALB) [140,162] and Pichia pastoris [163].
Aspergillus oryzae CCUG 33812 can produce a lipase able to catalyse PET hydrolysis using 0.1 g/L bis(2-hydroxyethyl) terephthalate (BHT) as an inducer [143]. An increase in hydrophilicity and antistatic ability, as well as a 0.74% weight loss and a decrease in both the water contact angle and static half decay time, were observed after 24 h at 55 °C [143].
The lipase triacylglycerol hydrolase produced by the yeast Pichia pastoris was able to modify the surface morphology of polyester fibres at 60 °C and at pH 7.5–8. Moreover, 7 h treatment with the combination of 10 g/L P. pastoris lipase and 0.5 g/L non-ionic surfactant JFC (a fatty alcohol polyoxyethylene ether) at 60 °C and pH 7.5 changed the surface morphology of the PET fibres and increased the number of hydrophilic groups [163].
3.4.3. Polyesterases Involved in PET Biodegradation
Extracellular polyesterases involved in PET hydrolyzation are secreted by Beauveria brongniartii [164] and Penicillium citrinum grown on a medium containing cutin with molecular weight 14.1 kDa, temperature optimum 36 °C and pH 8.2 [165].
Polyesterase from B. brongniartii released TPA during treatment of PET [164], while P. citrinum enzymatic activity liberates only low amounts of TPA in favour of BHET and MHET [165].
3.4.4. Synergic Action of Cutinase HiC and Lipase CALB
An important PET depolymerization enzyme that can act synergically with HiC is lipase B from Candida antarctica (CALB) [140,162].
HiC and CALB present two different activity profiles at the final stage of PET depolymerization. Indeed, TPA was the predominant molecule after 24 h of CALB action, while HiC very quickly converted BHET into MHET, but then TPA formation was slow [140]. Despite this, when the two enzymes are used alone, HiC is more efficient than CALB in degrading PET [149], while a synergic action between HiC and CALB led to a more intense MHET consumption and TPA formation [159]. Better results were obtained by de Castro et al. [140] using HiC and CALB sequentially. This method led to an initial release of MHET (HiC action at 60 °C), which was rapidly converted to TPA after CALB addition (37 °C), resulting in a degradation 141-times higher than when using the two enzymes at the same temperature [140].
3.4.5. PET Hydrophobicity Modification after Fungal Enzymes Action
An interesting reaction observable with most enzymes involved in PET hydrolysation is the increase in hydrophilicity. The action of hydrolases from Fusarium oxysporum LCH1 [145] leads to a higher PET hydrophilicity than cutinase from F. solani pisi [137,166,167,168]. Indeed, the water adsorption of PET fabrics treated with 80 U of cutinase and hydrolase was 36 mm/10 min and 57 mm/10 min, respectively [145]. An increase in hydrophilicity was also observed after the action of polyesterase obtained from Penicillium citrinum [165]. Comparing the measure of rising height, the enzyme preparation of P. citrinum caused the largest increase in hydrophilicity on PET (5.1 cm) [165], followed by Aspergillus sp. (4.3 cm), F. solani (2.3 cm) and Beauveria sp. (1.2 cm) [155]. The increase in hydrophilicity could be caused by the introduction of polar groups onto the polymer surface [167,168]. This was confirmed by the increase in hydrophilicity associated with the gain of hydroxylic and carboxylic acid groups on the PET surface after hydrolysis by an enzyme preparation from Beauveria brongniartii (0.5 nKat/mL) [164].
3.5. Polyurethane (PUR)
Polyurethane (PUR) is a polymeric, synthetic, petroleum-based, thermosets polymer composed of repeating units containing ethyl carbamate (H2NCO2C2H5) and urethane bonds with urea, ether, ester and aromatic groups [169]. Depending on the polyols used in the condensation reaction to produce PUR, polyurethane can be classified as a polyester (esters were used) or a polyether (utilisation of ethers) [47] (Table 4). PUR is a very versatile material, which can be used as a foam in building insulation, in pillows and mattresses, in semirigid plastics, as an elastomer, and in paint, adhesive and textile fibres [170,171]. As with PET, polyurethane made up 7.8% of European plastic demand in 2020 [4].
Table 4.
Backbone structural formula of polyurethane (PU), ether-PU and ester-PU.
| Name | Structure |
|---|---|
| Polyurethane (PU) |
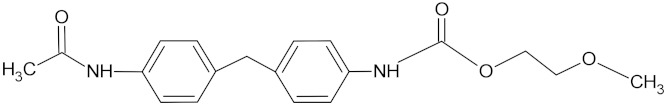
|
| Ether-PU |
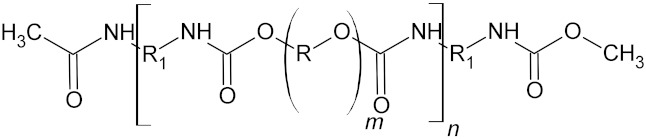
|
| Ester-PU |
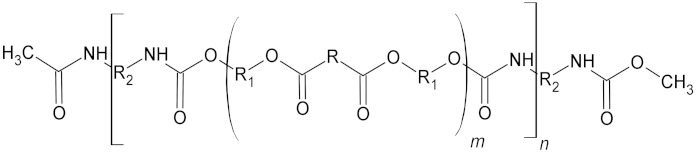
|
Today the disposal of polyurethane is a major problem as its degradation takes hundreds of years [172,173]. Moreover, incineration generates toxic emissions, and the material becomes useless after a few rounds of recycling [174].
The effectiveness of fungal biodegradation of PUR depends on the polymer molecular orientation, cross-linking and crystallinity [175]. It is easier to attack polyurethane in the amorphous regions rather than in the crystalline regions [169]. Moreover, the availability depends on the absence of ramifications or on the presence of long repetitive units that decrease the formation of highly crystalline areas [176,177]. In addition, the presence of methylene groups encourages fungal degradation [177].
Depending on its composition, polyurethane can be classified as a polyester or a polyether polyurethane, which can be more or less targeted by fungi [177]. The ester types are more susceptible to microbial degradation than ether types [178]. Indeed, during biodegradation, polyesters are subjected to hydrolysis, a reaction catalysed by both acids and bases and which follows the three-centre mechanism. The result of the hydrolysis of ester bonds is the liberation of a free acid, making this reaction autocatalytic [173]. On the other hand, ether bonds are very stable and as a result ether-PU products are a serious problem in environmental pollution [179]. Biodeterioration of polyether polyurethane can be carried out by oxidation rather than hydrolysis, and it involves the abstraction of α-hydrogen adjacent to oxygen [180,181]. Adding metal ions such as cobalt can accelerate this type of degradation [173].
Fungal biodegradation of ether-PU appears to be more promising than bacterial, especially in foams. This could be due to the ability of fungal hyphae to penetrate into foam pores, cracking the material mechanically [182] and thereby increasing the accessibility of chemical bonds for fungal exoenzymes. On the other hand, ester-PU is a substrate that is more susceptible to fungal enzymes than ether-PU, and several ester-PU degrading fungi are reported in the literature [183].
3.6. Fungal Enzymes Involved in PUR Biodegradation
Fungal hydrolases such as proteases, esterases, ureases and lipase are involved in the polyurethane biodegradation process. Ester bonds are cleaved by all these enzymes, while amide and urethane bonds are only hydrolysed by proteases, and urea is only targeted by ureases. Urease activity can be detected by phenolic compounds released into culture media, and this activity is exploited to estimate the amount of biodegradation [177]. However, the action of degradative enzymes is susceptible to the distance between urethane bonds, which can interfere negatively in the biodegradation [183]. There are also specific enzymes able to degrade polyurethane, and they are known as polyurethanases (PUase) [39,184].
3.6.1. Esterases Involved in Polyester-PUR Biodegradation
The mechanism of polyester-PUR degradation by esterases was proposed for the first time by Wales and Sagar in 1988 [185]. They suggested that extracellular esterases act by hydrolysis on polymers containing ester and urethane links. Boubendir [186] reported the correlation between the increase in esterase activity and the addition of liquid polyester PUR in cultural medium, suggesting the induction of enzymes with esterase and urethane hydrolase activities in Chaetomium globosum and Aspergillus terreus. Another extracellular enzyme-like factor with esterase properties was secreted by Curvularia senegalensis [185]. This enzyme was found to be stable at 100 °C for 10 min, displayed a band of 28 kDa in SDS PAGE analysis and was inhibited by phenylmethylsulphonyl fluoride (PMSF). Its involvement in polyurethane degradation was observed through the formation of clear zones in Impranil agar plates [185]. Impranil® DLN (Impranil, Leverkusen, Germany) is a class of plastics belonging to the polyurethane family [187], and it is commonly used to determine the ability of a microorganism or a protein to degrade PUR [188].
The esterase ability of PU biodegradation was confirmed by the 1500-fold increase in esterase production by Cladosporium pseudocladosporioides T1.PL.1 using Impranil as the sole carbon source [47], and by the increase in Aspergillus fumigatus strain S45’s esterase activity from 0.1440 µM/Min/mg to 2.687 µM/Min/mg after 15 days of incubation with polyester PUR as the sole carbon source [189].
Another fungal enzyme able to biodegrade Impranil DLN is produced by Pestalotiopsis microspore E2712A. This observation suggests the involvement of the 21 kDa serine hydrolase-like enzyme in PU biodegradation [190].
3.6.2. Lipases Involved in Polyester-PUR Biodegradation
Lipases involved in the hydrolytic degradation of polyester polyurethane are secreted mainly by yeasts belonging to the Candida genus. Kinetics and mathematical models of PUR degradation by Candida rugosa were proposed by Gautam et al. [191]. In this study, the maximum degradation rate of 2.5 g PUR/L was reached using 70 μg lipase/mL at pH 7 and 35 °C. Moreover, a linear increase in the PUR degradation product, diethylene glycol, was observed during PUR biodegradation [191].
Candida antarctica lipase represents another potential candidate for PUR biodegradation [104]. Indeed, oligomers with a molecular weight lower than 500 Daltons were produced after the activity of 20 mg of lipase in toluene (10 mL) at 60 °C for 24 h [104].
The Ascomycete Aspergillus tubingenesis is able to degrade polyester PU, producing both esterases and lipases [192]. The optimum pH required for A. tubingenesis esterase maximum activity was pH 7, while lipase has an optimum at pH 5. For both enzymes, the optimum temperature was 37 °C. An interesting observation made by the authors of this study is that lipase production augments in the first two weeks of culture when the surfactants Tween 20 or Tween 80 were added to the culture medium, then the lipase concentration decreased by the end of the month. On the other hand, esterase activities reached the maximum activity after two weeks in the presence of Tween 80 and continued to increase slowly until the fourth week in the presence of Tween 20. The gradual decrease in enzyme production was hypothesized to be due to the surfactants’ repression effect on the gene responsible for enzyme production [192].
3.6.3. Cutinase Involved in Polyester-PUR Biodegradation
A cutinase demonstrating the ability to degrade PUR was secreted by the thermophilic fungus Thielavia terrestris CAU709 [193]. The highest production of this enzyme (TtcutA) was observed when T. terrestris CAU709 grew using 2% (w/v) wheat bran as the sole carbon source amended with 0.1% (w/v) Tween 80. The highest enzyme activity (90.4 U/mL) was observed after 96 h of cultivation. The molecular mass of the purified TtcutA was between 25.3 and 22.8 kDa with optimum conditions at pH 4.0 and a temperature of 50 °C [193]. PU biodegradation was estimated using 0.6% emulsified PU [193].
3.6.4. Fungal Hydrolyses Involved in Polyether-PUR Biodegradation
As previously reported, enzymatic biodegradation of polyether PUR is more complex than polyester PUR and only a few studies have reported its degradation by fungal hydrolyses [151,169,194]. Urethane-bond-degrading enzymes and urea-bond-degrading enzymes were found in a culture supernatant of Alternaria sp. PURDK2. Polyamines (4,4′-methylenedianiline) and polyols were released after the hydrolysation of urethane bonds, while 4,4′-methylenedianiline and n-polyamines were generated after the degradation of urea bonds by enzymes of Alternaria sp. PURDK2 [179]. However, an initial mechanical disruption by fungal hyphae was required for polyether-PUR biodegradation [179].
3.7. Polyvinyl Chloride (PVC)
Polyvinyl chloride (PVC) is a commonly used synthetic, thermoplastic, petroleum-based material [195]. Pure PVC consists of long chains of ethylene monomers containing 56.77% (w/w) chlorine element (C2H3Cl)n [193] (Table 5). It is a white, brittle, solid polymer, is highly hydrophobic, soluble in tetrahydrofuran (THF) and resilient to chemical abrasion [24,196].
Table 5.
European plastic demand and backbone structural formula of polyvinylchloride.
| Name | Plastics Demand in Europe | Structure |
|---|---|---|
| Polyvinylchloride (PVC) | 9.6% |
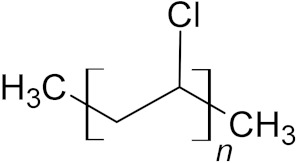
|
PVC represents 9.6% of the total European plastic use [4]. It is used in its rigid or flexible form in pipes and electrical wire insulation, profiles of windows or doors, coverings for floors and walls, and in textiles such as synthetic leather products, shoe soles, packaging and credit cards [24,170,196].
Some PVC products have a long lifespan so there is a long time between PVC production and waste, but in some products, such as packaging, PVC has a short life. In any case, long- and short-term PVC products are disposed of at the end of their use [195]. Depositing in landfill and incineration are the most widely used disposal methods for PVC [196]. However, during incineration a large amount of hydrogen chloride and tetrachlorodibenzo-p-dioxin are produced, causing secondary pollution [195,196,197,198]. Therefore, PVC biodegradation has become a topic of particular interest.
The biodegradation of PVC involves three main reactions: chain depolymerization; oxidation; mineralisation of formed intermediates [196,197]. Fungal PVC biodegradation is a very difficult process due to the hydrophobicity of the material, its resistance to abrasion and the persistence of its structure [199]. Indeed, modification of the molecular weight of PVC by microorganisms has been reported only in a small number of studies [196]. For example, fungal involvement in PVC biodegradation has been found in white-rot fungi [200] such as Pleurotus sp., Polyporus versicolor, Phanerochaete chrysosporium and Lentinus tigrinus [201,202]; or by Ascomycetes belonging to the genera Aspergillus [202,203,204,205,206,207,208], Cochliobolus [209], Chaetomium [203,210,211], Fusarium [203,205], Mucor [203,212] and Penicillium [203,205,206,212,213].
3.8. Fungal Enzymes Involved in PVC Biodegradation
The fungal enzymes involved in PVC biodegradation are currently not well known [178,214,215,216].
It has been reported that PVC biodegradation is related to the ability to degrade lignin [196,217], as demonstrated by the modification of the PVC structure by fungal lignin peroxidase (EC 1.11.1.14) from Phanerocheate chrysosporium [218]. P. chrysosporium lignin peroxidase had a molecular weight of 46 kDa and reached its maximum production after 4 weeks at 25 °C and pH 5. The weight of PVC films decreased by 31% when this partially purified enzyme was used and a stretch of alkenyl C–H in the PVC structure was observed by FTIR analysis (peak at 2943 cm−1) [218].
Another ligninolytic enzyme involved in PVC biodegradation is laccase from Cochliobolus sp. [219]. The higher laccase production (1.793 nKat/mL) was observed after 6 days of incubation at 30 °C and at pH 6.5. Modifications in FTIR spectra, such as the shifting of the CH-stretching mode (from 2912 cm−1 to 2915 cm−1) and the appearance of new peaks corresponding to a carbonyl group, suggested there was activity of Cochliobolus sp. laccase on the PVC structure. Changes in the PVC structure and surface were confirmed by GC-MS analysis and SEM photography [219].
Other studies have reported the involvement of fungal ligninolytic enzymes in PVC biodegradation [201]; however, no attempt was made to quantify the enzyme production or to demonstrate substrate degradation by purified enzymes [220].
3.9. Polypropylene (PP)
Polypropylene (PP) is a linear, thermoplastic hydrocarbon synthetic polymer, where a methyl-group replaces one hydrogen for every carbon of PE structure (C3H6)n [35,221]. PP is an inert, semicrystalline material, slightly harder and more resistant to heat and chemical reactions than PE [222]. It has a high hydrophobicity [223] and a molecular weight varying from 10,000 to 40,000 Daltons [214].
Polypropylene is the most used plastic polymer in Europe, accounting for 19.7% of the plastic demand [4]. It is used in many industries such as packaging for food and materials, pipes, automotive parts and diapers [32] (Table 6).
Table 6.
European plastic demand, molecular weight (Daltons) and backbone structural formula of polypropylene.
| Name | Plastics demand in Europe | Molecular Weight (Daltons) | Structure |
|---|---|---|---|
| Polypropylene (PP) | 19.7% | 10,000–40,000 |
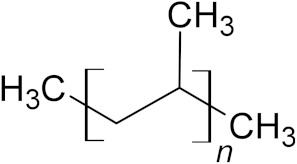
|
Due to the short-term use of these packaging products and the massive and rapid accumulation of PP in the environment, the study of alternative disposal methods, such as fungal biodegradation, is an important field of research.
Polypropylene proprieties such as hydrophobicity and high molecular weight make it particularly resistant to microbial attack and biodegradation [214]. Moreover, the addition of stabilizers and antioxidants to preserve PP from oxidation by atmospheric agents increase its biodegradation resistance even more [224]. Despite these problems, fungi such as Aspergillus niger were found to be involved in PP biodegradation [225,226,227]. Only a few other fungal species such as Lasiodiplodia theobromae [228], Bjerkandera adusta [229] and Engyodontium album [228] were reported as being able to attack PP. Due to its high resistance and recalcitrance, most studies used pretreatments such as UV or γ-irradiation to make PP more susceptible to degradation by fungi [226,228,230].
3.10. Fungal Enzymes Involved in PP Biodegradation
Although some studies reported the involvement of fungi in polypropylene biodegradation, so far none have investigated the use of fungal enzymes in the biodegradation of PP [101,178,214,216,231,232]. Investigating which microorganisms are able to degrade PP and the identification of their enzymes would be an interesting and useful area of further research.
3.11. Polystyrene (PS)
Polystyrene (PS) is a very stable, synthetic, thermoplastic polymer formed by an aromatic styrene monomer (C8H8)n [216,233]. PS has a large molecular weight and high hydrophobicity, which contribute to its rigidity and hardness [214]. The low cost and ease of production, as well as its lightweight, rigid and transparent properties make it ideal for the packaging industry [219,233]. PS is also used in electrical and electronic equipment, and in construction applications [170,234], composing 6.1% of the plastics demand in Europe [4] (Table 7).
Table 7.
European plastic demand, molecular weight (Daltons) and backbone structural formula of polypropylene.
| Name | Plastics Demand in Europe | Molecular Weight (Daltons) | Structure |
|---|---|---|---|
| Polystyrene (PS) | 6.1% | 150,000–400,000 |
|
Due to the high resistance of the C–C backbone [235], only a few studies have reported fungi involved in PS biodegradation. The main fungal genera involved in PS modification are the Ascomycetes Rhizopus [236], Aspergillus [236], Penicillium [237] and Curvularia [238]; the Basidiomycetes white-rot fungi Phanerochaete chrysosporium [236,239], Trametes versicolor [239] and Pleurotus ostreatus [239]; and the brown-rot fungus Gloeophyllum trabeum [44].
3.12. Fungal Enzymes Involved in PS Biodegradation
Although some studies have reported fungi involved in PS biodegradation, research describing fungal enzymes involved in this process are scarce [25,215]. Only an esterase produced by Lentinus tigrinus has been reported to biodegrade PS film into non-toxic smaller molecules [240]. The optimum activity of this purified hydrolytic enzyme was observed at 45 °C and pH 9. Moreover, the maximum production reached was 38.62 U/mL when L. tigrinus was grown in the presence of urea and yeast extract [240].
4. Conclusions and Perspectives for Future Research
Plastic waste is an increasingly urgent environmental problem, and a large amount of scientific research has been focused on using microbes for its biodegradation. In this context, the use of fungal enzymes as bioremediation tools is being increasingly studied. This review has reported the main fungal enzymes involved in plastic degradation, describing their characteristics and efficiency (Table 8). Fungal ligninolytic enzymes (laccases and peroxidases), already used in several biotechnological applications, were found to be successful in partially degrading PE and PVC. Moreover, fungal esterases, such as cutinase and lipase, were able to degrade PET and PUR. In addition, protease and urease can also contribute to PUR degradation. On the other hand, there is an absence of studies on fungal enzymes able to act on PP and PS, which opens the door for further research in this area.
Table 8.
Fungal enzymes documented as able to degrade plastics and the relative producing fungal species.
| Enzymes | Fungal Species | Plastic Polymers | References |
|---|---|---|---|
| Cutinase HiC | Humicola insolens | PET | [148,152,157] |
| Cutinase FoCut5a | Fusarium oxysporum | PET | [94,144] |
| Cutinase FsC | Fusarium solani pisi | PET | [43,148,151,152,153,154,155,159,160] |
| Cutinase TtcutA | Thielavia terrestris CAU709 | PUR | [193] |
| Esterases | Aspergillus fumigatus S45 | PUR | [189] |
| Aspergillus terreus | PUR | [186] | |
| Aspergillus tubingenesis | PUR | [192] | |
| Chaetomium globosum | PUR | [186] | |
| Cladosporium pseudocladosporioides T1.PL.1 | PUR | [47] | |
| Curvularia senegalensis | PUR | [185] | |
| Laccases | Aspergillus flavus PEDX3 | PE | [38] |
| Pleurotus ostreatus | PE | [48] | |
| Trichoderma harzianum | PE | [120] | |
| Laccase-like multicopper oxidases (LMCOs) | Aspergillus flavus PEDX3 | PE | [38] |
| Laccase-mediator system (LMS) | Trametes versicolor IFO 6482 | PE | [128] |
| Lipases | Aspergillus oryzae CCUG 33812 | PET | [143] |
| Aspergillus tubingenesis | PUR | [192] | |
| Candida antarctica (CALB) | PET | [140,162] | |
| Candida antarctica | PUR | [104] | |
| Candida rugosa | PUR | [191] | |
| Pichia pastoris | PET | [163] | |
| Peroxidases | Trichoderma harzianum | PE | [120] |
| Lignin peroxidases | Pleurotus ostreatus | PE | [48] |
| Manganese peroxidases | Phanerochaete chrysosporium ME-446 | PE | [115] |
| Pleurotus ostreatus | PE | [48] | |
| Polyesterases | Beauveria brongniartii | PET | [164] |
| Penicillium citrinum | PET | [165] | |
| Serine hydrolase-like enzyme | Pestalotiopsis microspora E2712A | PUR | [190] |
| Urethane hydrolases | Aspergillus terreus | PUR | [186] |
| Chaetomium globosum | PUR | [186] |
Biochemical methods and tools can be developed for controlling and improving enzyme catalytic activities and could be applied in several sectors including green chemistry [241]. For example, enzyme immobilization is a powerful tool that allows the enzyme of interest to be confined in a matrix, usually consisting of inert polymers and inorganic materials [241]. This technique enhances catalytic activity and increases the stability, specificity and selectivity of the immobilized enzymes, thus significantly reducing production costs [242]. A number of studies have reported that the immobilisation of PET-degrading bacterial enzymes enhances their catalytic activity and thermal stability [243,244,245], but few studies have focused on fungi [246]. Su et al. [247] immobilised cutinase from Aspergillus oryzae, Humicola insolens and Thielavia terrestris on the macroporous support Lewatit VP OC 1600 obtaining immobilization yields higher than 98%. Enzyme immobilisation is currently used in the treatment of dye-based wastewater from industries [248], anthracene degradation [249] and decomposition of the insecticide methyl parathion [250].
A recently emerged research approach to plastic biodegradation by fungal enzymes is the genetic engineering of fungi involving recombinant DNA technology, gene cloning, manipulation and modification [251]. Such techniques can be used to increase yields and improve the kinetics of the enzymes produced [252,253]. In the case of bacterial enzymes, improvement in the PET degradation rate was accomplished by genetic engineering, which enhanced the enzyme thermostability [141,254,255]. Such results were obtained by reinforcing the binding of the substrate to the active site [256,257,258], or by improving the substrate/enzyme surface interaction [259,260]. If similar manipulations could be carried out of genes coding for key plastic-degrading enzymes, improvements in fungi biodegradation potential could be achieved. For example, the mutations L81A, N84A, L182A, V184A and L189A enlarged the active site of the cutinase from Fusarium solani pisi leading to an improved fitting of the large polymer chains of PET. This modification resulted in a fivefold increase in this cutinase activity [170]. Janatunaim and Fibriani [261] were able to express the genes of the bacterium Ideonella sakaiensis 201-F6 coding for the enzyme monohydroxyethyl terephthalate hydrolase, which is involved in PET biodegradation, in Escherichia coli BL21. A similar genetic engineering process was reported for the laccase gene FoLacc5 from Fusarium oxysporum cDNA. After its expression in Pichia pastoris X33, this methylotroph yeast was able to easily degrade synthetic blue dyes [262]. Other fungal laccase genes, for example lac-En3–1 in Ganoderma sp. [263] or genes for Lac in Aspergillus sp. [264], were expressed in Pichia pastoris GS115 and then used in dye degradation. Although this technique for plastic degradation is still at the pioneer stage, it appears to produce promising results and has very high potential [251].
A further step towards biotechnological use of fungal enzymes for plastic degradation could be achieved using gene editing tools [251]. The manipulation of specific, desired genes by genome editing allows the loss or gain of a function of interest [265]. However, very few studies of this kind have been carried out. One example increased the PETase activity by the inclusion of two conserved residues of the gene coding cutinases from Thermobifida fusca at the polyesterase active site of the bacterium Ideonella sakaiensis [256]. Therefore, it appears there is a very high potential of this tool to open new avenues for future research in the area of the genetic editing of fungal enzymes for plastic degradation.
In conclusion, fungal enzymes show high potential in the fight against plastic waste thanks to their broad biodegradation ability. Currently, most studies are still performed in vitro under laboratory conditions. Further research is required to better understand the mechanisms of action of these enzymes and the genetics behind them. These interdisciplinary investigations could lead to optimised fungal cell factories with high degradation efficiency and vast industrial applications to help reduce plastic waste pollution and improve the environment.
Acknowledgments
We acknowledge Scuola di Alta Formazione Dottorale (SAFD) of the University of Pavia for giving the opportunity for this work.
Author Contributions
M.E.E.T. conceptualization, writing original draft and data curation, L.N. writing review, editing and supervision, E.N. editing and supervision, S.T. supervision and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fried J.R. Polymer Science and Technology. Prentice Hall; Upper Saddle River, NJ, USA: 1995. pp. 4–9. [Google Scholar]
- 2.Millet H., Vangheluwe P., Block C., Sevenster A., Garcia L., Antonopoulos R. The nature of plastics and their societal usage. In: Hester R.E., editor. Plastic and the Environment. Royal Society of Chemistry’s; London, UK: 2018. pp. 1–20. [Google Scholar]
- 3.Matjašič T., Simčič T., Medvešček N., Bajt O., Dreo T., Mori N. Critical evaluation of biodegradation studies on synthetic plastics through a systematic literature review. Sci. Total Environ. 2021;752:141959. doi: 10.1016/j.scitotenv.2020.141959. [DOI] [PubMed] [Google Scholar]
- 4.Plastic Europe Plastics–The Facts 2021: An Analysis of European Plastics Production, Demand and Waste Data. 2021. [(accessed on 19 January 2021)]. Available online: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/
- 5.Bahl S., Dolma J., Singh J.J., Sehgal S. Biodegradation of plastics: A state of the art review. Mater. Today Proc. 2021;39:31–34. doi: 10.1016/j.matpr.2020.06.096. [DOI] [Google Scholar]
- 6.Jambeck J.R., Geyer R., Wilcox C., Siegler T.R., Perryman M., Andrady A., Narayan R., Law K.L. Plastic waste inputs from land into the ocean. Science. 2015;347:768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 7.Devi R.S., Kannan V.R., Natarajan K., Nivas D., Kannan K., Chandru S., Antony A.R. The role of microbes in plastic degradation. Environ. Waste Manag. 2016;341:341–370. [Google Scholar]
- 8.Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen M., Huang W., Chen M., Song B., Zeng G., Zhang Y. (Micro) plastic crisis: Un-ignorable contribution to global greenhouse gas emissions and climate change. J. Clean Prod. 2020;254:120138. doi: 10.1016/j.jclepro.2020.120138. [DOI] [Google Scholar]
- 10.Chamas A., Moon H., Zheng J., Qiu Y., Tabassum T., Jang J.H., Suh S. Degradation rates of plastics in the environment. ACS Sustain. Chem. Eng. 2020;8:3494–3511. doi: 10.1021/acssuschemeng.9b06635. [DOI] [Google Scholar]
- 11.Kumar M., Chen H., Sarsaiya S., Qin S., Liu H., Awasthi M.K., Taherzadeh M.J. Current research trends on micro-and nano-plastics as an emerging threat to global environment: A review. J. Hazard. Mater. 2021;409:124967. doi: 10.1016/j.jhazmat.2020.124967. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q., Allgeier A., Yin D., Hollert H. Leaching of endocrine disrupting chemicals from marine microplastics and mesoplastics under common life stress conditions. Environ. Int. 2019;130:104938. doi: 10.1016/j.envint.2019.104938. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Zhang H., Allgeier A., Zhou Q., Ouellet J.D., Crawford S.E., Hollert H. Marine microplastics bound dioxin-like chemicals: Model explanation and risk assessment. J. Hazard. Mater. 2019;364:82–90. doi: 10.1016/j.jhazmat.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Turner A., Filella M. Hazardous metal additives in plastics and their environmental impacts. Environ. Int. 2021;156:106622. doi: 10.1016/j.envint.2021.106622. [DOI] [PubMed] [Google Scholar]
- 15.Mohsen M., Lin C., Tu C., Zhang C., Xu S., Yang H. Association of heavy metals with plastics used in aquaculture. Mar. Pollut. Bull. 2022;174:113312. doi: 10.1016/j.marpolbul.2021.113312. [DOI] [PubMed] [Google Scholar]
- 16.Kumar P. Role of plastics on human health. Indian J. Pediatr. 2018;85:384–389. doi: 10.1007/s12098-017-2595-7. [DOI] [PubMed] [Google Scholar]
- 17.Yang D., Shi H., Li L., Li J., Jabeen K., Kolandhasamy P. Microplastic pollution in table salts from China. Environ. Sci. Technol. 2015;49:13622–13627. doi: 10.1021/acs.est.5b03163. [DOI] [PubMed] [Google Scholar]
- 18.Istrate I.R., Medina-Martos E., Galvez-Martos J.L., Dufour J. Assessment of the energy recovery potential of municipal solid waste under future scenarios. Appl. Energy. 2021;293:116915. doi: 10.1016/j.apenergy.2021.116915. [DOI] [Google Scholar]
- 19.Kunwar B., Cheng H.N., Chandrashekaran S.R., Sharma B.K. Plastics to fuel: A review. Renew. Sust. Energ. Rev. 2016;54:421–428. doi: 10.1016/j.rser.2015.10.015. [DOI] [Google Scholar]
- 20.Verma R., Vinoda K.S., Papireddy M., Gowda A.N.S. Toxic pollutants from plastic waste—A review. Procedia Environ. Sci. 2016;35:701–708. doi: 10.1016/j.proenv.2016.07.069. [DOI] [Google Scholar]
- 21.Aguado J., Serrano D.P., Escola J.M. Fuels from waste plastics by thermal and catalytic processes: A review. Ind. Eng. Chem. Res. 2008;47:7982–7992. doi: 10.1021/ie800393w. [DOI] [Google Scholar]
- 22.Thiounn T., Smith R.C. Advances and approaches for chemical recycling of plastic waste. J. Polym. Sci. 2020;58:1347–1364. doi: 10.1002/pol.20190261. [DOI] [Google Scholar]
- 23.Chiellini E., Solaro R. Biodegradable polymeric materials. Adv. Mater. 1996;8:305–313. doi: 10.1002/adma.19960080406. [DOI] [Google Scholar]
- 24.Shah A.A., Hasan F., Hameed A., Ahmed S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008;26:246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Amobonye A., Bhagwat P., Singh S., Pillai S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021;759:143536. doi: 10.1016/j.scitotenv.2020.143536. [DOI] [PubMed] [Google Scholar]
- 26.Harms H., Schlosser D., Wick L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011;9:177–192. doi: 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- 27.Bhatt P., Bhandari G., Bhatt K., Maithani D., Mishra S., Gangola S., Bhatt R., Huang Y., Chen S. Plasmid-mediated catabolism for the removal of xenobiotics from the environment. J. Hazard. Mater. 2021;420:126618. doi: 10.1016/j.jhazmat.2021.126618. [DOI] [PubMed] [Google Scholar]
- 28.Lu H., Diaz D.J., Czarnecki N.J., Zhu C., Kim W., Shroff R., Alper H.S. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature. 2022;604:662–667. doi: 10.1038/s41586-022-04599-z. [DOI] [PubMed] [Google Scholar]
- 29.Bhatt P., Pandey S.C., Joshi S., Chaudhary P., Pathak V.M., Huang Y., Chen S. Nanobioremediation: A sustainable approach for the removal of toxic pollutants from the environment. J. Hazard. Mater. 2022;427:128033. doi: 10.1016/j.jhazmat.2021.128033. [DOI] [PubMed] [Google Scholar]
- 30.Zara Z., Mishra D., Pandey S.K., Csefalvay E., Fadaei F., Minofar B., Řeha D. Surface Interaction of Ionic Liquids: Stabilization of Polyethylene Terephthalate-Degrading Enzymes in Solution. Molecules. 2022;27:119. doi: 10.3390/molecules27010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace N.E., Adams M.C., Chafin A.C., Jones D.D., Tsui C.L., Gruber T.D. The highly crystalline PET found in plastic water bottles does not support the growth of the PETase-producing bacterium Ideonella sakaiensis. Environ. Microbiol. Rep. 2020;12:578–582. doi: 10.1111/1758-2229.12878. [DOI] [PubMed] [Google Scholar]
- 32.Tokiwa Y., Calabia B.P., Ugwu C.U., Aiba S. Biodegradability of plastics. Int. J. Mol. Sci. 2009;10:3722–3742. doi: 10.3390/ijms10093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taghavi N., Zhuang W.Q., Baroutian S. Enhanced biodegradation of non-biodegradable plastics by UV radiation: Part 1. J. Environ. Chem. Eng. 2021;9:106464. doi: 10.1016/j.jece.2021.106464. [DOI] [Google Scholar]
- 34.Vedrtnam A., Kumar S., Chaturvedi S. Experimental study on mechanical behavior, biodegradability, and resistance to natural weathering and ultraviolet radiation of wood-plastic composites. Compos. B. Eng. 2019;176:107282. doi: 10.1016/j.compositesb.2019.107282. [DOI] [Google Scholar]
- 35.Arutchelvi J., Sudhakar M., Arkatkar A., Doble M., Bhaduri S., Uppara P.V. Biodegradation of polyethylene and polypropylene. IJBT. 2008;7:9–22. [Google Scholar]
- 36.Jacquin J., Cheng J., Odobel C., Pandin C., Conan P., Pujo-Pay M., Ghiglione J.F. Microbial ecotoxicology of marine plastic debris: A review on colonization and biodegradation by the “Plastisphere”. Front. Microbiol. 2019;10:865. doi: 10.3389/fmicb.2019.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dussud C., Ghiglione J.F. Bacterial degradation of synthetic plastics. In: Briand F., editor. Marine Litter in the Mediterranean and Black Seas. Volume 46. CIESM; Paris, France: 2014. pp. 49–54. [Google Scholar]
- 38.Zhang J., Gao D., Li Q., Zhao Y., Li L., Lin H., Zhao Y. Biodegradation of polyethylene microplastic particles by the fungus Aspergillus flavus from the guts of wax moth Galleria mellonella. Sci. Total Environ. 2020;704:135931. doi: 10.1016/j.scitotenv.2019.135931. [DOI] [PubMed] [Google Scholar]
- 39.Wei R., Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017;10:1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daccò C., Girometta C., Asemoloye M.D., Carpani G., Picco A.M., Tosi S. Key fungal degradation patterns, enzymes and their applications for the removal of aliphatic hydrocarbons in polluted soils: A review. Int. Biodeterior. Biodegrad. 2020;147:104866. doi: 10.1016/j.ibiod.2019.104866. [DOI] [Google Scholar]
- 41.Sánchez C. Fungal potential for the degradation of petroleum-based polymers: An overview of macro-and microplastics biodegradation. Biotechnol. Adv. 2020;40:107501. doi: 10.1016/j.biotechadv.2019.107501. [DOI] [PubMed] [Google Scholar]
- 42.Da Luz J.M.R., da Silva M.D.C.S., dos Santos L.F., Kasuya M.C.M. Plastics polymers degradation by fungi. In: Blumenberg M., editor. Microorganisms. IntechOpen; London, UK: 2019. pp. 261–270. [Google Scholar]
- 43.Vertommen M.A.M.E., Nierstrasz V.A., van der Veer M., Warmoeskerken M.M.C.G. Enzymatic surface modification of poly (ethylene terephthalate) J. Biotechnol. 2005;120:376–386. doi: 10.1016/j.jbiotec.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Krueger M.C., Hofmann U., Moeder M., Schlosser D. Potential of wood-rotting fungi to attack polystyrene sulfonate and its depolymerisation by Gloeophyllum trabeum via hydroquinone-driven Fenton chemistry. PLoS ONE. 2015;10:e0131773. doi: 10.1371/journal.pone.0131773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb H., Arnott J., Crawford R., Ivanova E. Plastic degradation and its environmental implicationswith special reference to poly(ethylene terephthalate) Polymers. 2013;5:1. doi: 10.3390/polym5010001. [DOI] [Google Scholar]
- 46.Cregut M., Bedas M., Durand M.J., Thouand G. New insights into polyurethane biodegradation andrealistic prospects for the development of a sustainablewaste recycling process. Biotechnol. Adv. 2013;31:1634–1647. doi: 10.1016/j.biotechadv.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Álvarez-Barragán J., Domínguez-Malfavón L., Vargas-Suárez M., González-Hernández R., Aguilar-Osorio G., Loza-Tavera H. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 2016;82:5225–5235. doi: 10.1128/AEM.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gómez-Méndez L.D., Moreno-Bayona D.A., Poutou-Piñales R.A., Salcedo-Reyes J.C., Pedroza-Rodríguez A.M., Vargas A., Bogoya J.M. Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PLoS ONE. 2018;13:e0203786. doi: 10.1371/journal.pone.0203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Restrepo-Florez J.M., Bassi A., Thompson M.R. Microbial degradation and deterioration of polyethylene—A review. Int. Biodeterior. Biodegrad. 2014;88:83–90. doi: 10.1016/j.ibiod.2013.12.014. [DOI] [Google Scholar]
- 50.Daly P., Cai F., Kubicek C.P., Jiang S., Grujic M., Rahimi M.J., Druzhinina I.S. From lignocellulose to plastics: Knowledge transfer on the degradation approaches by fungi. Biotechnol. Adv. 2021;50:107770. doi: 10.1016/j.biotechadv.2021.107770. [DOI] [PubMed] [Google Scholar]
- 51.Arya G.C., Cohen H. The Multifaceted Roles of Fungal Cutinases during Infection. J. Fungi. 2022;8:199. doi: 10.3390/jof8020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehta A., Bodh U., Gupta R. Fungal lipases: A review. J. Biotech Res. 2017;8:58–77. [Google Scholar]
- 53.Da Silva Santiago S.R.S., Santiago P.A.L., de Oliveira M.R., de Souza Rodrigues R., Barbosa A.N., da Silva G.F., de Souza A.Q.L. Evaluation of enzymatic production of hydrolases and oxyredutases by Fusarium pseudocircinatum and Corynespora torulosa isolated from caesarweed (Urena lobata L., 1753) Res. Soc. Dev. 2022;11:e13211225325. doi: 10.33448/rsd-v11i2.25325. [DOI] [Google Scholar]
- 54.Kabir S.M.M., Koh J. Sustainable textile processing by enzyme applications. In: Mendes K.F., de Sousa R.O., Mielke K.C., editors. Biodegradation Technology of Organic and Inorganic Pollutants. IntechOpen; London, UK: 2021. [DOI] [Google Scholar]
- 55.Drozłowska E. The use of enzymatic fungal activity in the food industry-review. World Sci. News. 2019;116:222–229. [Google Scholar]
- 56.Behbudi G., Yousefi K., Sadeghipour Y. Microbial Enzymes Based Technologies for Bioremediation of Pollutions. J. Environ. Treat. Tech. 2021;9:463–469. [Google Scholar]
- 57.Siddiqui N.M., Dahiya P. Enzyme-based biodegradation of toxic environmental pollutants. In: Rodriguez-Couto S., Shah M.P., editors. Development in Wastewater Treatment Research and Processes. Elsevier; Amsterdam, The Netherlands: 2022. pp. 311–333. [Google Scholar]
- 58.Bilal M., Rasheed T., Nabeel F., Iqbal H.M., Zhao Y. Hazardous contaminants in the environment and their laccase-assisted degradation—A review. J. Environ. Manag. 2019;234:253–264. doi: 10.1016/j.jenvman.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 59.Demarche P., Junghanns C., Nair R.R., Agathos S.N. Harnessing the power of enzymes for environmental stewardship. Biotechnol. Adv. 2012;30:933–953. doi: 10.1016/j.biotechadv.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Surwase S.V., Patil S.A., Srinivas S., Jadhav J.P. Interaction of small molecules with fungal laccase: A surface plasmon resonance based study. Enzym. Microb. Technol. 2016;82:110–114. doi: 10.1016/j.enzmictec.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Desai S.S., Nityanand C. Microbial laccases and their applications: A review. Asian J. Biotechnol. 2011;3:98–124. doi: 10.3923/ajbkr.2011.98.124. [DOI] [Google Scholar]
- 62.Giardina P., Faraco V., Pezzella C., Piscitelli A., Vanhulle S., Sannia G. Laccases: A never-ending story. Cell Mol. Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madhavi V., Lele S.S. Laccase: Properties and applications. BioResources. 2009;4:1694–1717. [Google Scholar]
- 64.Kunamneni A., Ballesteros A., Plou F.J., Alcalde M. Fungal laccase—A versatile enzyme for biotechnological applications. In: Méndez-Vilas A., editor. Communicating Current Research and Educational Topics and Trends in Applied Microbiology. Formatex; Guadalajara, Mexico: 2007. pp. 233–245. [Google Scholar]
- 65.Rivera-Hoyos C.M., Morales-Álvarez E.D., Poutou-Pinales R.A., Pedroza-Rodríguez A.M., Rodriguez-Vazquez R., Delgado-Boada J.M. Fungal laccases. Fungal. Biol. Rev. 2013;27:67–82. doi: 10.1016/j.fbr.2013.07.001. [DOI] [Google Scholar]
- 66.Thurston C.F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. doi: 10.1099/13500872-140-1-19. [DOI] [Google Scholar]
- 67.Henson J.M., Butler M.J., Day A.W. The dark side of the mycelium: Melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 1999;37:447–471. doi: 10.1146/annurev.phyto.37.1.447. [DOI] [PubMed] [Google Scholar]
- 68.Ikehata K., Buchanan I.D., Smith D.W. Recent developments in the production of extracellular fungal peroxidases and laccases for waste treatment. J. Environ. Eng. Sci. 2004;3:1–19. doi: 10.1139/s03-077. [DOI] [Google Scholar]
- 69.Welinder K.G. Structure and evolution of peroxidase. In: Welinder K.G., Rasmussen S.K., Penel C., Greppin H., editors. Plant Peroxidases, Biochemistry and Physiology. 1st ed. University of Geneva; Geneva, Switzerland: 1993. pp. 35–42. [Google Scholar]
- 70.Conesa A., Punt P.J., van den Hondel C.A. Fungal peroxidases: Molecular aspects and applications. J. Biotechnol. 2002;93:143–158. doi: 10.1016/S0168-1656(01)00394-7. [DOI] [PubMed] [Google Scholar]
- 71.Ayuso-Fernández I., Martínez A.T., Ruiz-Dueñas F.J. Experimental recreation of the evolution of lignin-degrading enzymes from the Jurassic to date. Biotechnol. Biofuels. 2017;10:67. doi: 10.1186/s13068-017-0744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tien M., Kirk T.K. Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium Burds. Science. 1983;221:661–663. doi: 10.1126/science.221.4611.661. [DOI] [PubMed] [Google Scholar]
- 73.Tien M., Kirk T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. USA. 1984;81:2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huynh V.B., Crawford R.L. Novel extracellular enzymes (ligninases) of Phanerochaete chrysosporium. FEMS Microbiol. Lett. 1985;28:119–123. doi: 10.1111/j.1574-6968.1985.tb00776.x. [DOI] [Google Scholar]
- 75.Paszczynski A., Huynh V.B., Crawford R. Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol. Lett. 1985;29:37–41. doi: 10.1111/j.1574-6968.1985.tb00831.x. [DOI] [Google Scholar]
- 76.Sundaramoorthy M., Gold M.H., Poulos T.L. Ultrahigh (0.93Å) resolution structure of manganese peroxidase from Phanerochaete chrysosporium: Implications for the catalytic mechanism. J. Inorg. Biochem. 2010;104:683–690. doi: 10.1016/j.jinorgbio.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maciel M.J.M., Ribeiro H.C.T. Industrial and biotechnological applications of ligninolytic enzymes of the basidiomycota: A review. Electron. J. Biotechnol. 2010;13:14–15. doi: 10.2225/vol13-issue6-fulltext-2. [DOI] [Google Scholar]
- 78.Li C., Zhao X., Wang A., Huber G.W., Zhang T. Catalytic transformation of lignin for the production of chemicals and fuels. Chem. Rev. 2015;115:11559–11624. doi: 10.1021/acs.chemrev.5b00155. [DOI] [PubMed] [Google Scholar]
- 79.Sung H.J., Khan M.F., Kim Y.H. Recombinant lignin peroxidase-catalyzed decolorization of melanin using in-situ generated H2O2 for application in whitening cosmetics. Int. J. Biol. Macromol. 2019;136:20–26. doi: 10.1016/j.ijbiomac.2019.06.026. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H., Zhang J., Zhang X., Geng A. Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process Biochem. 2018;66:222–229. doi: 10.1016/j.procbio.2017.12.011. [DOI] [Google Scholar]
- 81.Chen W., Zheng L., Jia R., Wang N. Cloning and expression of a new manganese peroxidase from Irpex lacteus F17 and its application in decolorization of reactive black 5. Process Biochem. 2015;50:1748–1759. doi: 10.1016/j.procbio.2015.07.009. [DOI] [Google Scholar]
- 82.Lee H., Jang Y., Choi Y.S., Kim M.J., Lee J., Lee H., Hong J.-H., Lee Y.M., Kim G.H., Kim J.J. Biotechnological procedures to select white rot fungi for the degradation of PAHs. J. Microbiol. Methods. 2014;97:56–62. doi: 10.1016/j.mimet.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Torres-Farradá G., Manzano León A.M., Rineau F., Ledo Alonso L.L., Sánchez-López M.I., Thijs S., Colpaert J., Ramos-Leal M., Guerra G., Vangronsveld J. Diversity of ligninolytic enzymes and their genes in strains of the genus Ganoderma: Applicable for biodegradation of xenobiotic compounds? Front. Microbiol. 2017;8:898. doi: 10.3389/fmicb.2017.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pozdnyakova N.N. Involvement of the ligninolytic system of white-rot and litter-decomposing fungi in the degradation of polycyclic aromatic hydrocarbons. Biotechnol. Res. Int. 2012;2012:243217. doi: 10.1155/2012/243217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qayyum H., Maroof H., Yasha K. Remediation and treatment of organopollutants mediated by peroxidases: A review. Crit. Rev. Biotechnol. 2009;29:94–119. doi: 10.1080/07388550802685306. [DOI] [PubMed] [Google Scholar]
- 86.Skamnioti P., Furlong R.F., Gurr S.J. Evolutionary history of the ancient cutinase family in five filamentous Ascomycetes reveals differential gene duplications and losses and in Magnaporthe grisea shows evidence of sub- and neo-functionalization. New Phytol. 2008;180:711–721. doi: 10.1111/j.1469-8137.2008.02598.x. [DOI] [PubMed] [Google Scholar]
- 87.Chen S., Tong X., Woodard R.W., Du G., Wu J., Chen J. Identification and characterization of bacterial cutinase. J. Biol. Chem. 2008;283:25854–25862. doi: 10.1074/jbc.M800848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Longhi S., Czjzek M., Nicolas A., Cambillau C. Atomic resolution (1.0 Å) crystalstructure of Fusarium solani cutinase: Stereochemical analysis. J. Mol. Biol. 1997;268:779–799. doi: 10.1006/jmbi.1997.1000. [DOI] [PubMed] [Google Scholar]
- 89.Martinez C., de Geus P., Lauwereys M., Matthyssens G., Cambillau C. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 90.Roussel A., Amara S., Nyyssola A., Mateos-Diaz E., Blangy S., Kontkanen H., Westerholm-Pantinen A., Carriere F., Cambillau C. A cutinase from Trichoderma reesei with a lid-covered active site and kinetic properties of true lipases. J. Mol. Biol. 2014;426:3757–3772. doi: 10.1016/j.jmb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 91.Lau E.Y., Bruice T.C. Consequences of breaking the Asp-His hydrogen bond of the catalytic triad: Effects on the structure and dynamics of the serine esterase cutinase. Biophys. J. 1999;77:85–98. doi: 10.1016/S0006-3495(99)76874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Longhi S., Nicolas A., Creveld L., Egmond M., Verrips C.T., Vlieg J., Martinez C., Cambillau C. Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variant. Protein. 1996;26:442–458. doi: 10.1002/(SICI)1097-0134(199612)26:4<442::AID-PROT5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 93.Chen S., Su L., Chen J., Wu J. Cutinase: Characteristics, preparation, and application. Biotechnol. Adv. 2013;31:1754–1767. doi: 10.1016/j.biotechadv.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 94.Dimarogona M., Nikolaivits E., Kanelli M., Christakopoulos P., Sandgren M., Topakas E. Structural and functional studies of a Fusarium oxysporum cutinase with polyethylene terephthalate modification potential. Biochim. Biophys. Acta Gen. Subj. 2015;1850:2308–2317. doi: 10.1016/j.bbagen.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 95.Silva C.M., Carneiro F., O’Neill A., Fonseca L.P., Cabral J.S., Guebitz G., Cavaco-Paulo A. Cutinase-a new tool for biomodification of synthetic fibers. J. Polym. Sci. A Polym. Chem. 2005;43:2448–2450. doi: 10.1002/pola.20684. [DOI] [Google Scholar]
- 96.Singh A.K., Mukhopadhyay M. Overview of fungal lipase: A review. Appl. Biochem. Biotechnol. 2012;166:486–520. doi: 10.1007/s12010-011-9444-3. [DOI] [PubMed] [Google Scholar]
- 97.Ramos-Sánchez L.B., Cujilema-Quitio M.C., Julian-Ricardo M.C., Cordova J., Fickers P. Fungal lipase production by solid-state fermentation. J. Bioprocess. Biotech. 2015;5:1. doi: 10.4172/2155-9821.1000203. [DOI] [Google Scholar]
- 98.Sharma R., Chisti Y., Banerjee U.C. Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 2001;19:627–662. doi: 10.1016/S0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- 99.Sztajer H., Maliszewska I., Wieczorek J. Production of exogenous lipases by bacteria, fungi, and actinomycetes. Enzym. Microb. Technol. 1988;10:492–497. doi: 10.1016/0141-0229(88)90027-0. [DOI] [Google Scholar]
- 100.Ko W.H., Wang I.T., Ann P.J. A simple method for detection of lipolytic microorganisms in soils. Soil Biol. Biochem. 2005;37:597–599. doi: 10.1016/j.soilbio.2004.09.006. [DOI] [Google Scholar]
- 101.Chandra P., Singh R., Arora P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories. 2020;19:169. doi: 10.1186/s12934-020-01428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Houde A., Kademi A., Leblanc D. Lipases and their industrial applications. Appl. Biochem. Biotechnol. 2004;118:155–170. doi: 10.1385/ABAB:118:1-3:155. [DOI] [PubMed] [Google Scholar]
- 103.Masse L., Kennedy K.J., Chou S.P. The effect of an enzymatic pretreatment on the hydrolysis and size reduction of fat particles in slaughterhouse wastewater. J. Chem. Technol. Biotechnol. 2001;76:629–635. doi: 10.1002/jctb.428. [DOI] [Google Scholar]
- 104.Takamoto T., Shirasaka H., Uyama H., Kobayashi S. Lipase-catalyzed hydrolytic degradation of polyurethane in organic solvent. Chem. Lett. 2001;30:492–493. doi: 10.1246/cl.2001.492. [DOI] [Google Scholar]
- 105.Montazer Z., Habibi Najafi M.B., Levin D.B. Challenges with verifying microbial degradation of polyethylene. Polymers. 2020;12:123. doi: 10.3390/polym12010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mierzwa-Hersztek M., Gondek K., Kopeć M. Degradation of polyethylene and biocomponent-derived polymer materials: An overview. J. Polym. Environ. 2019;27:600–611. doi: 10.1007/s10924-019-01368-4. [DOI] [Google Scholar]
- 107.Gajendiran A., Krishnamoorthy S., Abraham J. Microbial degradation of low-density polyethylene (LDPE) by Aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech. 2016;6:52. doi: 10.1007/s13205-016-0394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bardají D.K.R., Moretto J.A.S., Furlan J.P.R., Stehling E.G. A mini-review: Current advances in polyethylene biodegradation. World J. Microbiol. Biotechnol. 2020;36:32. doi: 10.1007/s11274-020-2808-5. [DOI] [PubMed] [Google Scholar]
- 109.Kurtz S., Manley M. Cross-linked polyethylene. In: Hozack W., editor. Surgical Treatment of Hip Arthritis. Reconstruction, Replacement, and Revision. Saunders (W.B.) Co., Ltd.; Philadelphia, PA, USA: 2009. pp. 456–467. [Google Scholar]
- 110.Ragaert K., Delva L., van Geem K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017;69:24–58. doi: 10.1016/j.wasman.2017.07.044. [DOI] [PubMed] [Google Scholar]
- 111.Harshvardhan K., Jha B. Biodegradation of low-density polyethylene by marine bacteria from pelagic waters, Arabian Sea, India. Mar. Pollut. Bull. 2013;77:100–106. doi: 10.1016/j.marpolbul.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 112.Albertsson A.C., Andersson S.O., Karlsson S. The mechanism of biodegradation of polyethylene. Polym. Degrad. Stab. 1987;18:73–87. doi: 10.1016/0141-3910(87)90084-X. [DOI] [Google Scholar]
- 113.Santo M., Weitsman R., Sivan A. The role of the copper-binding enzyme e laccase e in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2012;208:1.e7. doi: 10.1016/j.ibiod.2012.03.001. [DOI] [Google Scholar]
- 114.Yoon M.G., Jeon J.H., Kim M.N. Biodegradation of polyethylene by a soil bacterium and AlkB cloned recombinant cell. J. Bioremed. Biodegr. 2012;3:145. doi: 10.4172/2155-6199.1000145. [DOI] [Google Scholar]
- 115.Iiyoshi Y., Tsutsumi Y., Nishida T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 1998;44:222–229. doi: 10.1007/BF00521967. [DOI] [Google Scholar]
- 116.Santacruz-Juárez E., Buendia-Corona R.E., Ramírez R.E., Sánchez C. Fungal enzymes for the degradation of polyethylene: Molecular docking simulation and biodegradation pathway proposal. J. Hazard. Mater. 2021;411:125118. doi: 10.1016/j.jhazmat.2021.125118. [DOI] [PubMed] [Google Scholar]
- 117.Malachová K., Novotný Č., Adamus G., Lotti N., Rybková Z., Soccio M., Šlosarčíková P., Verney V., Fava F. Ability of Trichoderma hamatum Isolated from Plastics-Polluted Environments to Attack Petroleum-Based, Synthetic Polymer Films. Processes. 2020;8:467. doi: 10.3390/pr8040467. [DOI] [Google Scholar]
- 118.Ojha N., Pradhan N., Singh S., Barla A., Shrivastava A., Khatua P., Bose S. Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Sci. Rep. 2017;7:39515. doi: 10.1038/srep39515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sáenz M., Borodulina T., Diaz L., Banchon C. Minimal Conditions to Degrade Low Density Polyethylene by Aspergillus terreus and niger. J. Ecol. Eng. 2019;20:44–51. doi: 10.12911/22998993/108699. [DOI] [Google Scholar]
- 120.Sowmya H.V., Krishnappa M., Thippeswamy B. Degradation of polyethylene by Trichoderma harzianum—SEM, FTIR, and NMR analyses. Environ. Monit. Assess. 2014;186:6577–6586. doi: 10.1007/s10661-014-3875-6. [DOI] [PubMed] [Google Scholar]
- 121.Sowmya H.V., Krishnappa M., Thippeswamy B. Degradation of polyethylene by Penicillium simplicissimum isolated from local dumpsite of Shivamogga district. Environ. Dev. Sustain. 2015;17:731–745. doi: 10.1007/s10668-014-9571-4. [DOI] [Google Scholar]
- 122.Shi K., Jing J., Song L., Su T., Wang Z. Enzymatic hydrolysis of polyester: Degradation of poly (ε-caprolactone) by Candida antarctica lipase and Fusarium solani cutinase. Int. J. Biol. Macromol. 2020;144:183–189. doi: 10.1016/j.ijbiomac.2019.12.105. [DOI] [PubMed] [Google Scholar]
- 123.Spina F., Tummino M.L., Poli A., Prigione V., Ilieva V., Cocconcelli P., Puglisi E., Bracco P., Zanetti M., Varese G.C. Low density polyethylene degradation by filamentous fungi. Environ. Pollut. 2021;274:116548. doi: 10.1016/j.envpol.2021.116548. [DOI] [PubMed] [Google Scholar]
- 124.Corti A., Sudhakar M., Chiellini E. Assessment of the whole environmental degradation of oxo-biodegradable linear low density polyethylene (LLDPE) films designed for mulching applications. J. Polym. Environ. 2012;20:1007–1018. doi: 10.1007/s10924-012-0493-7. [DOI] [Google Scholar]
- 125.Kang B.R., Kim S.B., Song H.A., Lee T.K. Accelerating the biodegradation of high-density polyethylene (HDPE) using Bjerkandera adusta TBB-03 and lignocellulose substrates. Microorganisms. 2019;7:304. doi: 10.3390/microorganisms7090304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Awasthi S., Srivastava N., Singh T., Tiwary D., Mishra P.K. Biodegradation of thermally treated low density polyethylene by fungus Rhizopus oryzae NS 5. 3 Biotech. 2017;7:73. doi: 10.1007/s13205-017-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shimao M. Biodegradation of plastics. Curr. Opin. Biotechnol. 2001;12:242–247. doi: 10.1016/S0958-1669(00)00206-8. [DOI] [PubMed] [Google Scholar]
- 128.Fujisawa M., Hirai H., Nishida T. Degradation of polyethylene and nylon-66 by the laccase-mediator system. J. Polym. Environ. 2001;9:103–108. doi: 10.1023/A:1020472426516. [DOI] [Google Scholar]
- 129.Volkamer A., Griewel A., Grombacher T., Rarey M. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010;50:2041–2052. doi: 10.1021/ci100241y. [DOI] [PubMed] [Google Scholar]
- 130.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 131.Isvoran A. Web-based computational tools used in protein surface analysis and characterization. Applications for protein–protein and protein–ligand interactions. In: Putz M.V., editor. Exotic Properties of Carbon Nanomatter. Carbon Materials: Chemistry and Physics. Volume 8. Springer; Dordrecht, The Netherlands: 2015. pp. 203–227. [Google Scholar]
- 132.Bertrand T., Jolivalt C., Briozzo P., Caminade E., Joly N., Madzak C., Mougin C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41:7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- 133.Miki Y., Calviño F.R., Pogni R., Giansanti S., Ruiz-Dueñas F.J., Martínez M.J., Basosi R., Romero A., Martínez A.T. Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine. J. Biol. Chem. 2011;286:15525–15534. doi: 10.1074/jbc.M111.220996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sang T., Wallis C.J., Hill G., Britovsek G.J. Polyethylene terephthalate degradation under natural and accelerated weathering conditions. Eur. Polym. J. 2020;136:109873. doi: 10.1016/j.eurpolymj.2020.109873. [DOI] [Google Scholar]
- 135.Raheem A.B., Noor Z.Z., Hassan A., Abd Hamid M.K., Samsudin S.A., Sabeen A.H. Current developments in chemical recycling of post-consumer polyethylene terephthalate wastes for new materials production: A review. J. Clean Prod. 2019;225:1052–1064. doi: 10.1016/j.jclepro.2019.04.019. [DOI] [Google Scholar]
- 136.Pang J., Zheng M., Sun R., Wang A., Wang X., Zhang T. Synthesis of ethylene glycol and terephthalic acid from biomass for producing PET. Green Chem. 2016;18:342–359. doi: 10.1039/C5GC01771H. [DOI] [Google Scholar]
- 137.Zimmermann W., Billig S. Enzymes for the biofunctionalization of poly (ethylene terephthalate) In: Nyanhongo G.S., editor. Biofunctionalization of Polymers and Their Applications. Springer; Berlin/Heidelberg, Germany: 2010. pp. 97–120. [Google Scholar]
- 138.Fischer-Colbrie G., Heumann S., Liebminger S., Almansa E., Cavaco-Paulo A., Guebitz G.M. New enzymes with potential for PET surface modification. Biocatal. Biotransform. 2004;22:341–346. doi: 10.1080/10242420400024565. [DOI] [Google Scholar]
- 139.Malafatti-Picca L., de Barros Chaves M.R., de Castro A.M., Valoni É., de Oliveira V.M., Marsaioli A.J., de Franceschi de Angelis D., Attili-Angelis D. Hydrocarbon-associated substrates reveal promising fungi for poly (ethylene terephthalate) (PET) depolymerization. Braz. J. Microbiol. 2019;50:633–648. doi: 10.1007/s42770-019-00093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Castro A.M., Carniel A., Nicomedes Junior J., da Conceição Gomes A., Valoni É. Screening of commercial enzymes for poly (ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J. Ind. Microbiol. Biotechnol. 2017;44:835–844. doi: 10.1007/s10295-017-1942-z. [DOI] [PubMed] [Google Scholar]
- 141.Kawai F., Kawabata T., Oda M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019;103:4253–4268. doi: 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarkhel R., Sengupta S., Das P., Bhowal A. Comparative biodegradation study of polymer from plastic bottle waste using novel isolated bacteria and fungi from marine source. J. Polym. Res. 2020;27:16. doi: 10.1007/s10965-019-1973-4. [DOI] [Google Scholar]
- 143.Wang X., Lu D., Jönsson L.J., Hong F. Preparation of a PET-hydrolyzing lipase from Aspergillus oryzae by the addition of bis(2-hydroxyethyl)terephthalate to the culture medium and enzymatic modification of PET fabrics. Eng. Life Sci. 2008;8:268–276. doi: 10.1002/elsc.200700058. [DOI] [Google Scholar]
- 144.Kanelli M., Vasilakos S., Nikolaivits E., Ladas S., Christakopoulos P., Topakas E. Surface modification of poly (ethylene terephthalate) (PET) fibers by a cutinase from Fusarium oxysporum. Process Biochem. 2015;50:1885–1892. doi: 10.1016/j.procbio.2015.08.013. [DOI] [Google Scholar]
- 145.Nimchua T., Punnapayak H., Zimmermann W. Comparison of the hydrolysis of polyethylene terephthalate fibers by a hydrolase from Fusarium oxysporum LCH I and Fusarium solani f. sp. pisi. Biotechnol. J. Healthc. Nutr. Technol. 2007;2:361–364. doi: 10.1002/biot.200600095. [DOI] [PubMed] [Google Scholar]
- 146.Sepperumal U., Markandan M., Palraja I. Micromorphological and chemical changes during biodegradation of polyethylene terephthalate (PET) by Penicillium sp. J. Microbiol. Biotechnol. Res. 2013;3:47–53. [Google Scholar]
- 147.Nowak B., Pająk J., Łabużek S., Rymarz G., Talik E. Biodegradation of poly (ethylene terephthalate) modified with polyester “Bionolle®” by Penicillium funiculosum. Polimery. 2011;56:35–44. doi: 10.14314/polimery.2011.035. [DOI] [Google Scholar]
- 148.Taniguchi I., Yoshida S., Hiraga K., Miyamoto K., Kimura Y., Oda K. Biodegradation of PET: Current status and application aspects. ACS Catal. 2019;9:4089–4105. doi: 10.1021/acscatal.8b05171. [DOI] [Google Scholar]
- 149.Carr C.M., Clarke D.J., Dobson A.D.W. Microbial Polyethylene Terephthalate Hydrolases: Current and Future Perspectives. Front. Microbiol. 2020;11:571265. doi: 10.3389/fmicb.2020.571265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao R., Pan H., Lian J. Recent advances in the discovery, characterization, and engineering of poly (ethylene terephthalate) (PET) hydrolases. Enzym. Microb. Technol. 2021;150:109868. doi: 10.1016/j.enzmictec.2021.109868. [DOI] [PubMed] [Google Scholar]
- 151.Danso D., Chow J., Streit W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019;85:e01095-19. doi: 10.1128/AEM.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ronkvist A.M., Xie W.C., Lu W.H., Gross R.A. Cutinase catalyzed hydrolysis of poly(ethylene terephthalate) Macromolecules. 2009;42:5128–5138. doi: 10.1021/ma9005318. [DOI] [Google Scholar]
- 153.Eberl A., Heumann S., Bruckner T., Araujo R., Cavaco- Paulo A., Kaufmann F., Kroutil W., Guebitz G.M. Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis- (benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J. Biotechnol. 2009;143:207–212. doi: 10.1016/j.jbiotec.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 154.Donelli I., Freddi G., Nierstrasz V.A., Taddei P. Surface structure and properties of poly-(ethylene terephthalate) hydrolyzed by alkali and cutinase. Polym. Degrad. Stab. 2010;95:1542–1550. doi: 10.1016/j.polymdegradstab.2010.06.011. [DOI] [Google Scholar]
- 155.Heumann S., Eberl A., Pobeheim H., Liebminger S., Fischer-Colbrie G., Almansa E., Cavaco-Paulo A., Gübitz G.M. New model substrates for enzymes hydrolysing polyethyleneterephthalate and polyamide fibres. J. Biochem. Biophys. Methods. 2006;69:89–99. doi: 10.1016/j.jbbm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 156.Ping L.F., Chen X.Y., Yuan X.L., Zhang M., Chai Y.J., Shan S.D. Application and comparison in biosynthesis and biodegradation by Fusarium solani and Aspergillus fumigatus cutinases. Int. J. Biol. Macromol. 2017;104:1238–1245. doi: 10.1016/j.ijbiomac.2017.06.118. [DOI] [PubMed] [Google Scholar]
- 157.Perz V., Bleymaier K., Sinkel C., Kueper U., Bonnekessel M., Ribitsch D., Guebitz G.M. Substrate specificities of cutinases on aliphatic–aromatic polyesters and on their model substrates. New Biotechnol. 2016;33:295–304. doi: 10.1016/j.nbt.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 158.Müller R.J., Schrader H., Profe J., Dresler K., Deckwer W.D. Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005;26:1400–1405. doi: 10.1002/marc.200500410. [DOI] [Google Scholar]
- 159.De Castro A.M., Carniel A., Stahelin D., Junior L.S.C., de Angeli Honorato H., de Menezes S.M.C. High-fold improvement of assorted post-consumer poly (ethylene terephthalate) (PET) packages hydrolysis using Humicola insolens cutinase as a single biocatalyst. Process Biochem. 2019;81:85–91. doi: 10.1016/j.procbio.2019.03.006. [DOI] [Google Scholar]
- 160.Araújo R., Silva C., O’Neill A., Micaelo N., Guebitz G.M., Soares C., Casal M., Cavaco-Paulo A. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J. Biotechnol. 2007;128:849–857. doi: 10.1016/j.jbiotec.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 161.Griswold K., Mahmood N.A., Iverson B.L., Georgiou G. Effects of codon usage versus putative 5_-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Prot. Expr. Purif. 2003;27:134–142. doi: 10.1016/S1046-5928(02)00578-8. [DOI] [PubMed] [Google Scholar]
- 162.Carniel A., Valoni É., Junior J.N., da Conceição Gomes A., de Castro A.M. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 2017;59:84–90. doi: 10.1016/j.procbio.2016.07.023. [DOI] [Google Scholar]
- 163.Gao A., Shen H., Zhang H., Feng G., Xie K. Hydrophilic modification of polyester fabric by synergetic effect of biological enzymolysis and non-ionic surfactant, and applications in cleaner production. J. Clean Prod. 2017;164:277–287. doi: 10.1016/j.jclepro.2017.06.214. [DOI] [Google Scholar]
- 164.Almansa E., Heumann S., Eberl A., Fischer-Colbrie G., Martinkova L., Marek J., Cavaco-Paulo A., Guebitz G.M. Enzymatic surface hydrolysis of PET enhances bonding in PVC coating. Biocatal. Biotransform. 2008;26:365–370. doi: 10.1080/10242420802357613. [DOI] [Google Scholar]
- 165.Liebminger S., Eberl A., Sousa F., Heumann S., Fischer-Colbrie G., Cavaco-Paulo A., Guebitz G.M. Hydrolysis of PET and bis-(benzoyloxyethyl) terephthalate with a new polyesterase from Penicillium citrinum. Biocatal. Biotransform. 2007;25:171–177. doi: 10.1080/10242420701379734. [DOI] [Google Scholar]
- 166.Alisch-Mark M., Herrmann A., Zimmermann W. Increase of the hydrophilicity of polyethylene terephthalate fibres by hydrolases from Thermomonospora fusca and Fusarium solani f. sp. pisi. Biotechnol. Lett. 2006;28:681–685. doi: 10.1007/s10529-006-9041-7. [DOI] [PubMed] [Google Scholar]
- 167.Yoon M.Y., Kellis J., Poulouse A.J. Enzymatic modification of polyester. AATCC Rev. 2002;2:3336. [Google Scholar]
- 168.Gouda M.K., Kleeberg I., van den Heuvel J., Müller R.J., Deckwer W.D. Production of a polyester degrading extracellular hydrolase from Thermomonospora fusca. Biotechnol. Progr. 2002;18:927934. doi: 10.1021/bp020048b. [DOI] [PubMed] [Google Scholar]
- 169.Howard G.T. Biodegradation of polyurethane: A review. Int. Biodeterior. Biodegrad. 2002;49:245–252. doi: 10.1016/S0964-8305(02)00051-3. [DOI] [Google Scholar]
- 170.Plastic Europe Plastics—The Facts 2020: An Analysis of European Plastics Production, Demand and Waste Data. 2020. [(accessed on 19 January 2021)]. Available online: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2020/
- 171.Nakajima-Kambe T., Shigeno-Akutsu Y., Nomura N., Onuma F., Nakahara T. Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes. Appl. Microbiol. Biotechnol. 1999;51:134–140. doi: 10.1007/s002530051373. [DOI] [PubMed] [Google Scholar]
- 172.Gautam R., Bassi A.S., Yanful E.K. A review of biodegradation of synthetic plastic and foams. Appl. Biochem. Biotechnol. 2007;141:85–108. doi: 10.1007/s12010-007-9212-6. [DOI] [PubMed] [Google Scholar]
- 173.Mahajan N., Gupta P. New insights into the microbial degradation of polyurethanes. RSC Adv. 2015;5:41839–41854. doi: 10.1039/C5RA04589D. [DOI] [Google Scholar]
- 174.Ignatyev I.A., Thielemans W., Vander Beke B. Recycling of polymers: A review. Chem. Sus. Chem. 2014;7:1579–1593. doi: 10.1002/cssc.201300898. [DOI] [PubMed] [Google Scholar]
- 175.Pathirana R.A., Seal K.J. Gliocladium roseum (Bainier), a potential biodeteriogen of polyester polyurethane elastomers. In: Oxley T.A., editor. Biodeterioration 5: Papers Presented at the 5th International Biodeterioration Symposium, Aberdeen, September, 1981. Wiley; Chichester, UK: 1983. p. c1983. [Google Scholar]
- 176.Howard G.T., Blake R.C. Growth of Pseudomonas fluorescens on a polyester–polyurethane and the purification and characterization of a polyurethanase–protease enzyme. Int. Biodeterior. Biodegrad. 1998;42:213–220. doi: 10.1016/S0964-8305(98)00051-1. [DOI] [Google Scholar]
- 177.Loredo-Treviño A., García G., Velasco-Téllez A., Rodríguez-Herrera R., Aguilar C.N. Polyurethane foam as substrate for fungal strains. Adv. Biosci. Biotechnol. 2011;2:52. doi: 10.4236/abb.2011.22009. [DOI] [Google Scholar]
- 178.Ru J., Huo Y., Yang Y. Microbial degradation and valorization of plastic wastes. Front. Microbiol. 2020;11:442. doi: 10.3389/fmicb.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Matsumiya Y., Murata N., Tanabe E., Kubota K., Kubo M. Isolation and characterization of an ether-type polyurethane-degrading micro-organism and analysis of degradation mechanism by Alternaria sp. J. Appl. Microbiol. 2010;108:1946–1953. doi: 10.1111/j.1365-2672.2009.04600.x. [DOI] [PubMed] [Google Scholar]
- 180.Stachelek S.J., Alferiev I., Choi H., Chan C.W., Zubiate B., Sacks M., Composto R., Chen I.W., Levy R.J. Prevention of oxidative degradation of polyurethane by covalent attachment of di-tert-butylphenol residues. J. Biomed. Mater. Res. 2006;78:653–661. doi: 10.1002/jbm.a.30828. [DOI] [PubMed] [Google Scholar]
- 181.Christenson E.M., Anderson J.M., Hiltner A. Oxidative mechanisms of poly (carbonate urethane) and poly (ether urethane) biodegradation: In vivo and in vitro correlations. J. Biomed. Mater. Res. A. 2004;70:245–255. doi: 10.1002/jbm.a.30067. [DOI] [PubMed] [Google Scholar]
- 182.Kemona A., Piotrowska M. Polyurethane recycling and disposal: Methods and prospects. Polymers. 2020;12:1752. doi: 10.3390/polym12081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Loredo-Treviño A., Gutiérrez-Sánchez G., Rodríguez-Herrera R., Aguilar C.N. Microbial enzymes involved in polyurethane biodegradation: A review. J. Polym. Environ. 2012;20:258–265. doi: 10.1007/s10924-011-0390-5. [DOI] [Google Scholar]
- 184.Howard G.T. Polyurethane biodegradation. In: Singh S.N., editor. Microbial Degradation of Xenobiotics. Springer; Heidelberg, Germany: 2012. pp. 371–394. [Google Scholar]
- 185.Crabbe J.R., Campbell J.R., Thompson L., Walz S.L., Schultz W.W. Biodegradation of a colloidal ester-based polyurethane by soil fungi. Int. Biodeterior. Biodegrad. 1994;33:103–113. doi: 10.1016/0964-8305(94)90030-2. [DOI] [Google Scholar]
- 186.Boubendir A. Purification and biochemical evaluation of polyurethane degrading enzymes of fungal origin. Diss. Abstr. Int. 1993;53:4632. [Google Scholar]
- 187.Saffarzadeh N., Moghimi H. The study of impranil (DLN) polymer biodegradation by fungus Sarocladium kiliense. Nova Biol. Reper. 2019;6:20–29. doi: 10.29252/nbr.6.1.20. [DOI] [Google Scholar]
- 188.Biffinger J.C., Barlow D.E., Cockrell A.L., Cusick K.D., Hervey W.J., Fitzgerald L.A., Nadeau L.J., Hung C.S., Crookes-Goodson W.J., Russell J.N., Jr. The applicability of Impranil® DLN for gauging the biodegradation of polyurethanes. Polym. Degrad. Stab. 2015;120:178–185. doi: 10.1016/j.polymdegradstab.2015.06.020. [DOI] [Google Scholar]
- 189.Osman M., Satti S.M., Luqman A., Hasan F., Shah Z., Shah A.A. Degradation of polyester polyurethane by Aspergillus sp. strain S45 isolated from soil. J. Polym. Environ. 2018;26:301–310. doi: 10.1007/s10924-017-0954-0. [DOI] [Google Scholar]
- 190.Russell J.R., Huang J., Anand P., Kucera K., Sandoval A.G., Dantzler K.W., Strobel S.A. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011;77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gautam R., Bassi A.S., Yanful E.K. Candida rugosa lipase-catalyzed polyurethane degradation in aqueous medium. Biotechnol. Lett. 2007;29:1081–1086. doi: 10.1007/s10529-007-9354-1. [DOI] [PubMed] [Google Scholar]
- 192.Khan S., Nadir S., Shah Z.U., Shah A.A., Karunarathna S.C., Xu J., Khan A., Munir S., Hasan F. Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environ. Pollut. 2017;225:469–480. doi: 10.1016/j.envpol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 193.Yang S., Xu H., Yan Q., Liu Y., Zhou P., Jiang Z. A low molecular mass cutinase of Thielavia terrestris efficiently hydrolyzes poly (esters) J. Ind. Microbiol. Biotechnol. 2013;40:217–226. doi: 10.1007/s10295-012-1222-x. [DOI] [PubMed] [Google Scholar]
- 194.Owen S., Otani T., Masaoka S., Ohe T. The biodegradation of low-molecular-weight urethane compounds by a strain of Exophiala jeanselmei. Biosci. Biotechnol. Biochem. 1996;60:244–248. doi: 10.1271/bbb.60.244. [DOI] [PubMed] [Google Scholar]
- 195.Sadat-Shojai M., Bakhshandeh G.R. Recycling of PVC wastes. Polym. Degrad. Stab. 2011;96:404–415. doi: 10.1016/j.polymdegradstab.2010.12.001. [DOI] [Google Scholar]
- 196.Peng B.Y., Chen Z., Chen J., Yu H., Zhou X., Criddle C.S., Zhang Y. Biodegradation of polyvinyl chloride (PVC) in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Environ. Int. 2020;145:106106. doi: 10.1016/j.envint.2020.106106. [DOI] [PubMed] [Google Scholar]
- 197.Winkler D.E. Mechanism of polyvinyl chloride degradation and stabilization. J. Polym. Sci. 1959;35:3–16. doi: 10.1002/pol.1959.1203512802. [DOI] [Google Scholar]
- 198.Pospíšil J., Horák Z., Kruliš Z., Nešpůrek S., Kuroda S.I. Degradation and aging of polymer blends I. Thermomechanical and thermal degradation. Polym. Degrad. Stab. 1999;65:405–414. doi: 10.1016/S0141-3910(99)00029-4. [DOI] [Google Scholar]
- 199.Amass W., Amass A., Tighe B. A review of biodegradable polymers: Uses, current developments in the synthesis and characterization of biodegradable polyesters, blends of biodegradable polymers and recent advances in biodegradation studies. Polym. Int. 1998;47:89–144. doi: 10.1002/(SICI)1097-0126(1998100)47:2<89::AID-PI86>3.0.CO;2-F. [DOI] [Google Scholar]
- 200.Roberts W.T., Davidson P.M. Growth characteristics of selected fungi on polyvinyl chloride film. Appl. Environ. Microbiol. 1986;51:673–676. doi: 10.1128/aem.51.4.673-676.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Klrbas Z., Keskin N., Güner A. Biodegradation of polyvinylchloride (PVC) by white rot fungi. Bull. Environ. Contam. Toxicol. 1999;63:335–342. doi: 10.1007/s001289900985. [DOI] [PubMed] [Google Scholar]
- 202.Ali M.I., Ahmed S., Robson G., Javed I., Ali N., Atiq N., Hameed A. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbiol. 2014;54:18–27. doi: 10.1002/jobm.201200496. [DOI] [PubMed] [Google Scholar]
- 203.Sakhalkar S., Mishra R.L. Screening and identification of soil fungi with potential of plastic degrading ability. Indian J. Appl. Res. 2013;3:3. doi: 10.15373/2249555X/DEC2013/16. [DOI] [Google Scholar]
- 204.Muthukumar A., Veerappapillai S. Biodegradation of plastics: A brief review. Int. J. Pharm. Sci. Rev. Res. 2015;31:204–209. [Google Scholar]
- 205.Pradeep S., Benjamin S. Mycelial fungi completely remediate di (2-ethylhexyl) phthalate, the hazardous plasticizer in PVC blood storage bag. J. Hazard. Mater. 2012;235:69–77. doi: 10.1016/j.jhazmat.2012.06.064. [DOI] [PubMed] [Google Scholar]
- 206.Webb J.S., Nixon M., Eastwood I.M., Greenhalgh M., Robson G.D., Handley P.S. Fungal colonization and biodeterioration of plasticized polyvinyl chloride. Appl. Environ. Microbiol. 2000;66:3194–3200. doi: 10.1128/AEM.66.8.3194-3200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gumargalieva K.Z., Zaikov G.E., Semenov S.A., Zhdanova O.A. The influence of biodegradation on the loss of a plasticiser from poly (vinyl chloride) Polym. Degrad. Stab. 1999;63:111–112. doi: 10.1016/S0141-3910(98)00071-8. [DOI] [Google Scholar]
- 208.Mogil’nitskii G.M., Sagatelyan R.T., Kutishcheva T.N., Zhukova S.V., Kerimov S.I., Parfenova T.B. Disruption of the protective properties of the polyvinyl chloride coating under the effect of microorganisms. Prot. Met. 1987;23:173–175. [Google Scholar]
- 209.Sumathi T., Viswanath B., Sri Lakshmi A., SaiGopal D.V.R. Production of laccase by Cochliobolus sp. isolated from plastic dumped soils and their ability to degrade low molecular weight PVC. Biochem Res Int. 2016;2016:9519527. doi: 10.1155/2016/9519527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Vivi V.K., Martins-Franchetti S.M., Attili-Angelis D. Biodegradation of PCL and PVC: Chaetomium globosum (ATCC 16021) activity. Folia Microbiol. 2019;64:1–7. doi: 10.1007/s12223-018-0621-4. [DOI] [PubMed] [Google Scholar]
- 211.Kaczmarek H., Bajer K. Biodegradation of plasticized poly (vinyl chloride) containing cellulose. J. Polym. Sci. B Polym. Phys. 2007;45:903–919. doi: 10.1002/polb.21100. [DOI] [Google Scholar]
- 212.Pardo-Rodríguez M.L., Zorro-Mateus P.J.P. Biodegradation of polyvinyl chloride by Mucor sp. and Penicillium sp. isolated from soil. Rev. Investig. Desarro. Innovación. 2021;11:387–400. doi: 10.19053/20278306.v11.n2.2021.12763. [DOI] [Google Scholar]
- 213.Sabev H.A., Handley P.S., Robson G.D. Fungal colonization of soil-buried plasticized polyvinyl chloride (pPVC) and the impact of incorporated biocides. Microbiology. 2006;152:1731–1739. doi: 10.1099/mic.0.28569-0. [DOI] [PubMed] [Google Scholar]
- 214.Mohanan N., Montazer Z., Sharma P.K., Levin D.B. Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 2020;11:2837. doi: 10.3389/fmicb.2020.580709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cobongela S.Z.Z. Enzymes involved in plastic degradation. In: Inamuddin, editor. Degradation of Plastics. Materials Research Foundations. Volume 99. Materials Research Forum LLC; Millersville, PA, USA: 2021. pp. 95–110. [Google Scholar]
- 216.Othman A.R., Hasan H.A., Muhamad M.H., Ismail N.I., Abdullah S.R.S. Microbial degradation of microplastics by enzymatic processes: A review. Environ. Chem. Lett. 2021;19:3057–3073. doi: 10.1007/s10311-021-01197-9. [DOI] [Google Scholar]
- 217.Yang S.S., Kang J.H., Xie T.R., He L., Xing D.F., Ren N.Q., Ho S.H., Wu W.M. Generation of high-efficient biochar for dye adsorption using frass of yellow mealworms (larvae of Tenebrio molitor Linnaeus) fed with wheat straw for insect biomass production. J. Clean Prod. 2019;227:33–47. doi: 10.1016/j.jclepro.2019.04.005. [DOI] [Google Scholar]
- 218.Khatoon N., Jamal A., Ali M.I. Lignin peroxidase isoenzyme: A novel approach to biodegrade the toxic synthetic polymer waste. Environ. Technol. 2019;40:1366–1375. doi: 10.1080/09593330.2017.1422550. [DOI] [PubMed] [Google Scholar]
- 219.Chaudhary A.K., Vijayakumar R.P. Studies on biological degradation of polystyrene by pure fungal cultures. Environ. Dev. Sustain. 2020;22:4495–4508. doi: 10.1007/s10668-019-00394-5. [DOI] [Google Scholar]
- 220.Pointing S. Feasibility of bioremediation by white-rot fungi. Appl. Microbiol. Biotechnol. 2001;57:20–33. doi: 10.1007/s002530100745. [DOI] [PubMed] [Google Scholar]
- 221.Baker M.A.M., Mead J. Thermoplastics. In: Harper C.A., editor. Handbook of Plastics, Elastomers and Composites. 4th ed. McGraw-Hill; New York, NY, USA: 2002. pp. 1–90. [Google Scholar]
- 222.Karian H. Handbook of Polypropylene and Polypropylene Composites, Revised and Expanded. CRC Press; Boca Raton, FL, USA: 2003. [Google Scholar]
- 223.Khoironi A., Hadiyanto H., Anggoro S., Sudarno S. Evaluation of polypropylene plastic degradation and microplastic identification in sediments at Tambak Lorok coastal area, Semarang. Indones. Mar. Pollut. Bull. 2020;151:110868. doi: 10.1016/j.marpolbul.2019.110868. [DOI] [PubMed] [Google Scholar]
- 224.Zheng Y., Yanful E.K. A review of plastic waste degradation. Crit. Rev. Biotechnol. 2005;25:243–250. doi: 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]
- 225.Alariqi S.A.S., Kumar A.P., Rao B.S.M., Singh R.P. Biodegradation of γ-sterilised biomedical polyolefins under composting and fungal culture environments. Polym. Degrad. Stab. 2006;91:1105–1116. doi: 10.1016/j.polymdegradstab.2005.07.004. [DOI] [Google Scholar]
- 226.Pandey J.K., Singh R.P. UV-irradiated biodegradability of ethylene-propylene copolymers, LDPE, and I-PP in composting culture environments. Biomacromolecules. 2001;2:880–885. doi: 10.1021/bm010047s. [DOI] [PubMed] [Google Scholar]
- 227.Cacciari I., Quatrini P., Zirletta G., Mincione E., Vinciguerra V., Lupattelli P., Sermanni G.G. Isotactic polypropylene biodegradation by a microbial community: Physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 1993;59:3695–3700. doi: 10.1128/aem.59.11.3695-3700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jeyakumar D., Chirsteen Doble M. Synergistic effects of pretreatment and blending on fungi mediated biodegradation of polypropylenes. Bioresour. Technol. 2013;148:78–85. doi: 10.1016/j.biortech.2013.08.074. [DOI] [PubMed] [Google Scholar]
- 229.Butnaru E., Darie-Niţă R.N., Zaharescu T., Balaeş T., Tănase C., Hitruc G., Doroftei F., Vasile C. Gamma irradiation assisted fungal degradation of the polypropylene/biomass composites. Radiat. Phys. Chem. 2016;125:134–144. doi: 10.1016/j.radphyschem.2016.04.003. [DOI] [Google Scholar]
- 230.Sheik S., Chandrashekar K.R., Swaroop K., Somashekarappa H.M. Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int. Biodeterior. Biodegrad. 2015;105:21–29. doi: 10.1016/j.ibiod.2015.08.006. [DOI] [Google Scholar]
- 231.Anjana K., Hinduja M., Sujitha K., Dharani G. Review on plastic wastes in marine environment: Biodegradation and biotechnological solutions. Mar. Pollut. Bull. 2020;150:110733. doi: 10.1016/j.marpolbul.2019.110733. [DOI] [PubMed] [Google Scholar]
- 232.Pires J.P., Miranda G.M., de Souza G.L., Fraga F., da Silva Ramos A., de Araújo G.E., de Lima J.E.A. Investigation of degradation of polypropylene in soil using an enzymatic additive. Iran Polym. J. 2019;28:1045–1055. doi: 10.1007/s13726-019-00766-8. [DOI] [Google Scholar]
- 233.Ho B.T., Roberts T.K., Lucas S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2018;38:308–320. doi: 10.1080/07388551.2017.1355293. [DOI] [PubMed] [Google Scholar]
- 234.Krueger M.C., Seiwert B., Prager A., Zhang S., Abel B., Harms H., Schlosser D. Degradation of polystyrene and selected analogues by biological Fenton chemistry approaches: Opportunities and limitations. Chemosphere. 2017;173:520–528. doi: 10.1016/j.chemosphere.2017.01.089. [DOI] [PubMed] [Google Scholar]
- 235.Goldman A.S. Carbon–carbon bonds get a break. Nature. 2010;463:435–436. doi: 10.1038/463435a. [DOI] [PubMed] [Google Scholar]
- 236.Atiq N. Ph.D. Thesis. Quaid-i-Azam University; Islamabad, Pakistan: 2011. Biodegradability of Synthetic Plastics Polystyrene and Styrofoam by Fungal Isolates. [Google Scholar]
- 237.Tian L., Kolvenbach B., Corvini N., Wang S., Tavanaie N., Wang L., Ma Y., Scheu S., Corvini P.F.X., Ji R. Mineralisation of 14C-labelled polystyrene plastics by Penicillium variabile after ozonation pre-treatment. New Biotechnol. 2017;38:101–105. doi: 10.1016/j.nbt.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 238.Motta O., Proto A., de Carlo F., De Caro F., Santoro E., Brunetti L., Capunzo M. Utilization of chemically oxidized polystyrene as co-substrate by filamentous fungi. Int. J. Hyg. Environ. Health. 2009;212:61–66. doi: 10.1016/j.ijheh.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 239.Milstein O., Gersonde R., Huttermann A., Chen M.J., Meister J.J. Fungal biodegradation of lignopolystyrene graft copolymers. Appl. Environ. Microbiol. 1992;58:3225–3232. doi: 10.1128/aem.58.10.3225-3232.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tahir L., Ali M.I., Zia M., Atiq N., Hasan F., Ahmed S. Production and Characterization of Esterase in Lantinus tigrinus for Degradation of Polystyrene. Pol. J. Microbiol. 2013;62:101–108. doi: 10.33073/pjm-2013-015. [DOI] [PubMed] [Google Scholar]
- 241.Datta S., Christena L.R., Rajaram Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech. 2013;3:1–9. doi: 10.1007/s13205-012-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Samak N.A., Jia Y., Sharshar M.M., Mu T., Yang M., Peh S., Xing J. Recent advances in biocatalysts engineering for polyethylene terephthalate plastic waste green recycling. Environ. Int. 2020;145:106144. doi: 10.1016/j.envint.2020.106144. [DOI] [PubMed] [Google Scholar]
- 243.Hegde K., Veeranki V.D. Studies on immobilization of cutinases from Thermobifida fusca on glutaraldehyde activated chitosan beads. Biotechnol. J. Int. 2014;4:1049–1063. doi: 10.9734/BBJ/2014/12558. [DOI] [Google Scholar]
- 244.Barth M., Honak A., Oeser T., Wei R., Belisário-Ferrari M.R., Then J., Schmidt J., Zimmermann W. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol. J. 2016;11:1082–1087. doi: 10.1002/biot.201600008. [DOI] [PubMed] [Google Scholar]
- 245.Pellis A., Vastano M., Quartinello F., Herrero Acero E., Guebitz G.M. His-Tag Immobilization of Cutinase 1 From Thermobifida cellulosilytica for Solvent-Free Synthesis of Polyesters. Biotechnol. J. 2017;12:1700322. doi: 10.1002/biot.201700322. [DOI] [PubMed] [Google Scholar]
- 246.Ahmaditabatabaei S., Kyazze G., Iqbal H., Keshavarz T. Fungal Enzymes as Catalytic Tools for Polyethylene Terephthalate (PET) Degradation. J. Fungi. 2021;7:931. doi: 10.3390/jof7110931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Su A., Shirke A., Baik J., Zou Y., Gross R. Immobilized cutinases: Preparation, solvent tolerance and thermal stability. Enzym. Microb. Technol. 2018;116:33–40. doi: 10.1016/j.enzmictec.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 248.Aslam S., Asgher M., Khan N.A., Bilal M. Immobilization of Pleurotus nebrodensis WC 850 laccase on glutaraldehyde cross-linked chitosan beads for enhanced biocatalytic degradation of textile dyes. J. Water Process Eng. 2021;40:101971. doi: 10.1016/j.jwpe.2021.101971. [DOI] [Google Scholar]
- 249.Imam A., Suman S.K., Singh R., Vempatapu B.P., Ray A., Kanaujia P.K. Application of laccase immobilized rice straw biochar for anthracene degradation. Environ. Pollut. 2021;268:115827. doi: 10.1016/j.envpol.2020.115827. [DOI] [PubMed] [Google Scholar]
- 250.Gao Y., Truong Y.B., Cacioli P., Butler P., Kyratzis I.L. Bioremediation of pesticide contaminated water using an organophosphate degrading enzyme immobilized on nonwoven polyester textiles. Enzym. Microb. Technol. 2014;54:38–44. doi: 10.1016/j.enzmictec.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 251.Jaiswal S., Sharma B., Shukla P. Integrated approaches in microbial degradation of plastics. Environ. Technol. Innov. 2020;17:100567. doi: 10.1016/j.eti.2019.100567. [DOI] [Google Scholar]
- 252.Saravanan A., Kumar P.S., Vo D.V.N., Jeevanantham S., Karishma S., Yaashikaa P.R. A review on catalytic-enzyme degradation of toxic environmental pollutants: Microbial enzymes. J. Hazard. Mater. 2021;419:126451. doi: 10.1016/j.jhazmat.2021.126451. [DOI] [PubMed] [Google Scholar]
- 253.Zhu B., Wang D., Wei N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022;40:22–37. doi: 10.1016/j.tibtech.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 254.Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- 255.Son H.F., Cho I.J., Joo S., Seo H., Sagong H.Y., Choi S.Y., Lee S.Y., Kim K.J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019;9:3519–3526. doi: 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- 256.Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., Silveira R.L., Pollard B.C., Dominick G., Duman R., El Omari K., et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA. 2018;115:E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ma Y., Yao M., Li B., Ding M., He B., Chen S., Zhou X., Yuan Y. Enhanced poly (ethylene terephthalate) hydrolase activity by protein engineering. Engineering. 2018;4:888–893. doi: 10.1016/j.eng.2018.09.007. [DOI] [Google Scholar]
- 258.Silva C., Da S., Silva N., Matamá T., Araújo R., Martins M., Chen S., Chen J., Wu J., Casal M., et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnol. J. 2011;6:230–1239. doi: 10.1002/biot.201000391. [DOI] [PubMed] [Google Scholar]
- 259.Herrero Acero E., Ribitsch D., Dellacher A., Zitzenbacher S., Marold A., Steinkellner G., Gruber K., Schwab H., Guebitz G.M. Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol. Bioeng. 2013;110:2581–2590. doi: 10.1002/bit.24930. [DOI] [PubMed] [Google Scholar]
- 260.Biundo A., Ribitsch D., Steinkellner G., Gruber K., Guebitz G.M. Polyester hydrolysis is enhanced by a truncated esterase: Less is more. Biotechnol. J. 2017;12:1600450. doi: 10.1002/biot.201600450. [DOI] [PubMed] [Google Scholar]
- 261.Janatunaim R.Z., Fibriani A. Construction and Cloning of Plastic-degrading Recombinant Enzymes (MHETase) Recent Pat. Biotechnol. 2020;14:229–234. doi: 10.2174/1872208314666200311104541. [DOI] [PubMed] [Google Scholar]
- 262.Huy N.D., Le N.T.M., Chew K.W., Park S.M., Show P.L. Characterization of a recombinant laccase from Fusarium oxysporum HUIB02 for biochemical application on dyes removal. Biochem. Eng. J. 2021;168:107958. doi: 10.1016/j.bej.2021.107958. [DOI] [Google Scholar]
- 263.Zhuo R., He F., Zhang X., Yang Y. Characterization of a yeast recombinant laccase rLAC-EN3-1 and its application in decolorizing synthetic dye with the coexistence of metal ions and organic solvents. Biochem. Eng. J. 2015;93:63–72. doi: 10.1016/j.bej.2014.09.004. [DOI] [Google Scholar]
- 264.Liu S., Xu X., Kang Y., Xiao Y., Liu H. Degradation and detoxification of azo dyes with recombinant ligninolytic enzymes from Aspergillus sp. with secretory overexpression in Pichia pastoris. Royal. Soc. Open Sci. 2020;7:200688. doi: 10.1098/rsos.200688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang S., Chen H., Tang X., Zhang H., Chen W., Chen Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017;101:8063–8075. doi: 10.1007/s00253-017-8486-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


