Abstract
Yeast isolates from raw and processed poultry products were characterized using PCR amplification of the internally transcribed spacer (ITS) 5.8S ribosomal DNA region (ITS-PCR), restriction analysis of amplified products, randomly amplified polymorphic DNA (RAPD) analysis, and pulsed-field gel electrophoresis (PFGE). ITS-PCR resulted in single fragments of 350 and 650 bp, respectively, from eight strains of Yarrowia lipolytica and seven strains of Candida zeylanoides. Digestion of amplicons with HinfI and HaeIII produced two fragments of 200 and 150 bp from Y. lipolytica and three fragments of 350, 150, and 100 bp from C. zeylanoides, respectively. Although these fragments showed species-specific patterns and confirmed species identification, characterization did not enable intraspecies typing. Contour-clamped heterogeneous electric field PFGE separated chromosomal DNA of Y. lipolytica into three to five bands, most larger than 2 Mbp, whereas six to eight bands in the range of 750 to 2,200 bp were obtained from C. zeylanoides. Karyotypes of both yeasts showed different polymorphic patterns among strains. RAPD analysis, using enterobacterial repetitive intergenic sequences as primers, discriminated between strains within the same species. Cluster analysis of patterns formed groups that correlated with the source of isolation. For ITS-PCR, extraction of DNA by boiling yeast cells was successfully used.
Contamination and spoilage of raw and processed poultry meat with bacteria are well documented. Much less is known about the presence and contribution of yeasts to spoilage of poultry, although yeasts are a stable part of the microbiota on raw red meat, poultry, and fish (11). The few surveys done to determine the number of yeasts on raw poultry have revealed log10 values of 4.8 to 6.0 CFU/g of chicken and turkey (6), 3.1 CFU/g of chicken carcass (19), 2.2 to 4.1 CFU/cm2 of chicken carcass (2), and 2.5 CFU/cm2 of skin on freshly slaughtered chicken (18). Viljoen et al. (56) described a study designed to identify yeasts on slaughtered poultry and correlate the presence of specific species with spoilage. They concluded that yeasts are substantially represented in the total microbial ecology of spoilage of raw poultry.
In a recent study (25), we determined the populations of yeasts associated with 50 samples of raw, marinated, smoked, or roasted chicken and turkey products. The predominant yeasts were identified, and lipolytic and proteolytic properties were characterized. Yarrowia lipolytica and Candida zeylanoides constituted 39 and 26%, respectively, of the isolates. The same species have been observed to predominate on frozen chicken (13). Identification of yeasts from poultry analyzed in our study (25) was achieved using traditional physiological and biochemical tests that allow species assignment but do not reveal molecular differences between strains of a given species or their potential contributions to spoilage. Molecular characterization of Y. lipolytica and C. zeylanoides may enable correlation with relative ability to produce lipases and/or proteases, thus providing a tool to predict the extent to which they contribute to spoilage of raw poultry and poultry products.
Various molecular techniques have been developed which permit species identification and typing of food-borne microorganisms, among them yeasts. These include pulsed-field gel electrophoresis (PFGE; karyotyping), restriction enzyme analysis, PCR-based techniques, and sequencing (for reviews, see references 12, 42, 44, and 51). Some of these techniques have been applied successfully to characterize yeasts isolated from various food products; however, most are too sophisticated or cumbersome for use in routine industrial practice. In recent years, PCR-based techniques targeting ribosomal RNA genes that can be performed with relative ease have emerged. Of these, restriction analysis of variable internal transcribed spacer (ITS) sequences framing the more conservative 5.8S rRNA gene (rDNA) has proven most useful, allowing both species identification and typing of isolates (17, 21, 49). Based on an extensive database, this technique has been proposed for rapid, routine identification for yeasts (16). Randomly amplified polymorphic DNA (RAPD) analysis can also be used to reliably type yeast strains (4, 5, 35).
The aim of this study was to further characterize two predominant yeast species isolated from poultry meat by restriction analysis of PCR-amplified ITS-5.8S rDNA (ITS-PCR) and to typify strains of these species using RAPD and PFGE analysis in order to assess their biodiversity and potential contribution to spoilage.
MATERIALS AND METHODS
Yeasts strains.
Yeasts (152 strains) isolated from commercial raw and processed chicken and turkey products were identified as belonging to 12 different species using traditional morphological, physiological, and biochemical tests (25). Y. lipolytica and C. zeylanoides were predominant, making up 39 and 26%, respectively, of isolates. Eight strains of Y. lipolytica and seven strains of C. zeylanoides isolated from a range of poultry products were subjected to molecular characterization (Table 1). Laboratory stock strains of Y. lipolytica and Candida species were also examined for comparison.
TABLE 1.
Yeast species and strains examined and their sources
| Species | Strain(s) | Source |
|---|---|---|
| Candida zeylanoides | Type, NRRL Y-6360 | Georgia State Universitya |
| 103, 130, 155 | Smoked turkey drumstickb | |
| 157, 217 | Turkey sausageb | |
| 121, 128 | Roasted chicken breastb | |
| C. guilliermondii | NRRL 2075 | Georgia State Universitya |
| C. krusei (Issatchenkia orientalis) | NRRL 7179 | Georgia State Universitya |
| C. famata (Debaryomyces hansenii) | CBS 767 | Georgia State Universitya |
| C. parapsilosis | ATCC 22019 | Georgia State Universitya |
| C. sake | CBS 159 | Georgia State Universitya |
| C. tropicalis | CDC-59 | Georgia State Universitya |
| C. lipolytica | 90-019095 | Georgia State Universitya |
| Yarrowia lipolytica | 142, 160, 218 | Chicken liverb |
| 138, 253 | Chicken liver IIb | |
| 173, 175, 246 | Roasted chicken breastb |
Courtesy of Sally Meyer.
From Ismail et al. (25).
DNA extraction for PCR.
Yeasts were cultured for 16 to 18 h at 25°C on tryptone-glucose-yeast extract (TGY) broth, which contains, per liter of deionized water, tryptone (Difco, Detroit, Mich.), 5 g; glucose, 10 g; and yeast extract, 5 g. Cells were collected by centrifugation at 16,000 × g for 2 min and then washed and resuspended in 100 μl of sterile distilled water. After washing again, the suspension was boiled for 10 min and centrifuged at 16,000 × g for 5 min. An aliquot of the supernatant was used for PCR analysis.
DNA extraction for PFGE.
Yeasts were grown in TGY broth at 25°C for 24 h on a gyratory shaker (150 rpm). Cells were centrifuged (16,000 × g, 2 min) and washed in LET buffer (500 mM EDTA, 10 mM Tris [pH 7.5]); 6 × 108 cells/ml were embedded in 100-μl plugs of 0.75% low-melting-point agarose and incubated overnight at 37°C in LET buffer containing 25 mg of Lyticase per ml and 7.5% 2-mercaptoethanol. The plugs were washed and incubated in NDS buffer (500 mM EDTA, 10 mM Tris [pH 7.5], 1% laurylsarcosine, 2 mg of proteinase K per ml) at 50°C for 24 h, then thoroughly washed four times with 50 mM EDTA (pH 8.0), and kept at 4°C until used. All enzymes and reagents were obtained from Bio-Rad (Hercules, Calif.).
ITS-PCR.
Two primers, ITS1 (5′TCCGTAGGTGAACCTGCGG3′) and ITS4 (5′TCCTCCGCTTATTGATATGC3′), custom synthesized by Gibco Life Technologies (Grand Island, N.Y.), were used as described by White et al. (55). The amplification reaction was performed with a Thermocycler 480 (Perkin-Elmer Corp., Norwalk, Conn.) using a mixture containing 2 U of Taq DNA polymerase (1 U/μl), 2 μl of deoxynucleoside triphosphate (dNTP) mix (10 mM each dNTP), 10 μl of 10× PCR buffer with MgCl2 (all Boehringer reagents; Roche Diagnostics, Indianapolis, Ind.), 2 μl of each primer (0.1 μg/μl), and 10 μl of boiled cell extract as a template. The volume was made up to 100 μl with sterile distilled water. After an initial denaturation at 95°C for 5 min, amplification was made through 35 cycles, each consisting of 1 min at 95°C, 2 min at 55°C, and 2 min at 72°C, followed by a final extension step of 10 min at 72°C.
Restriction analysis.
HinfI and HaeIII restriction endonucleases (Boehringer) were used separately to digest the amplification products of ITS-PCR. The digestion mixture consisted of 16 μl of amplicon, 2 μl of restriction enzyme (10 U/μl), and 2 μl of each of the buffers provided by Roche Diagnostics. The mixture was incubated 16 to 18 h at 37°C.
RAPD analysis.
Enterobacterial repetitive intergenic consensus (ERIC) primers were used singly or in combination as described by Metzgar et al. (31). ERIC1R (5′ATGTAAGCTCCTGGGGATTCAC3′) and ERIC2 (5′AAGTAAGTGACTGGGGTGAGCG3′) were custom made by Gibco Life Technologies. The same thermocycler and Boehringer reagents as described above were used. The PCR mix (50 μl) contained 20 μl of boiled cell extract, 5 μl of 10× PCR buffer with MgCl2, 6 μl of dNTP mix, 1 μl of Taq DNA polymerase, and 2 μl of primer. This mixture was heated at 94°C for 5 min and then subjected to 40 cycles at 92°C for 45 s, 25°C for 1 min, and 68°C for 10 min, followed by a final extension at 72°C for 20 min.
Gel electrophoresis.
ITS-PCR- and RAPD-PCR-amplified products and restriction digests were separated by agarose gel electrophoresis using a horizontal submarine gel system (E-C Apparatus Corp., Holbrook, N.Y.). Agarose (Gibco BRL Life Technologies) at a concentration of 0.8% (wt/vol) was used to separate ITS-PCR products and RAPD products, whereas a concentration of 1.4% (wt/vol) agarose was used to separate restriction fragments. Electrophoresis was conducted in 0.5× TBE buffer (5.4 g of Tris base, 2.75 g of boric acid, and 2 ml of 0.5 M EDTA [pH 8.0] in 1 liter of distilled water) at 10 V/cm for various times, depending on the size of the gel unit; DNA size markers (Boehringer XII and XIV) were used as standards. DNA bands were stained with ethidium bromide and then visualized and photographed under UV light using a GelDoc2000 Transilluminator (Bio-Rad).
PFGE.
A CHEF-DR II apparatus (Bio-Rad) was used for karyotyping of yeasts. C. zeylanoides chromosomal DNA prepared in agarose plugs was separated in 1% agarose at 100 V for 15 h with a pulse time of 120 s followed with a 240-s pulse time for 33 h at 14°C in 0.5× TBE buffer. For separation of much larger DNA molecules of Y. lipolytica, PFGE conditions were modified as follows: 50 V and 1,200-s pulse time for 36 h followed with an 1,800-s pulse time for another 36 h. Saccharomyces cerevisiae chromosomal DNA size standards (Bio-Rad) were used as markers. After electrophoresis, bands were visualized as described above.
Cluster analysis.
Genetic relationships and divergence between ERIC-RAPD patterns of strains isolated from poultry were calculated from the Pearson coefficient using Bio-Rad Molecular Analyst software (3) and are illustrated in a dendrogram constructed using the unweighted pair-group method with arithmetic averaging (UPGMA) and single linkage.
RESULTS
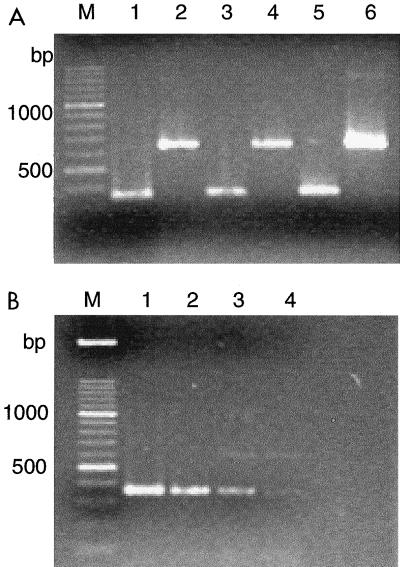
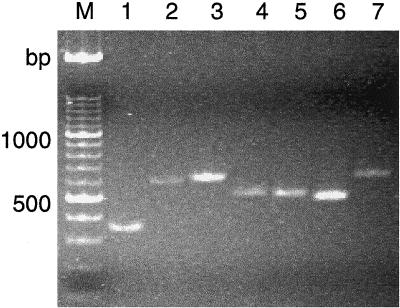
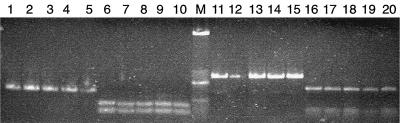
Direct extraction of DNA from washed, boiled yeast cells enabled PCR amplification using ITS1 and ITS4 primers. Similar patterns were obtained with cells from overnight TGY broth cultures and 24- to 36-h colonies formed on TGY agar; however, working with broth cultures gave more uniform results (Fig. 1). Cell populations up to 3 × 108 CFU/ml did not interfere with PCR, whereas less than 3 × 104 CFU/ml did not result in amplified product. Each yeast species produced a single amplified ITS-5.8S rDNA fragment (Fig. 2). The fragment in Y. lipolytica was shorter than that in C. zeylanoides (about 350 and 650 bp, respectively), and all strains belonging to these species gave a single, uniform band (Fig. 3). Digestion of PCR products with restriction endonuclease HinfI resulted in two fragments of 200 and 150 bp from Y. lipolytica strains, whereas three fragments of 350, 150, and 100 bp were obtained from C. zeylanoides, using restriction endonuclease HaeIII (Fig. 3). Again, all strains of both species produced uniform digestion patterns.
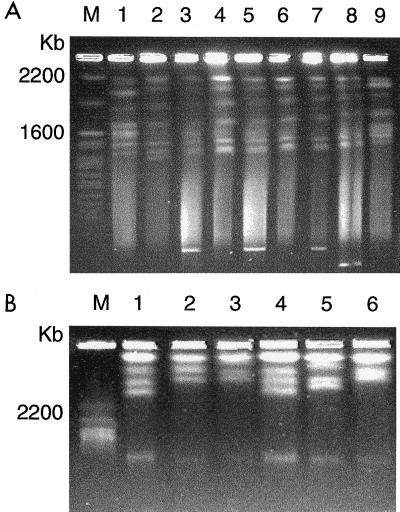
FIG. 1.
(A) Direct ITS-PCR amplification from broth cultures and colonies. Lanes: 1, 3, and 5, Y. lipolytica strain 160; 2, 4, and 6, C. zeylanoides strain 155; 1 and 2: 36-h TGY broth culture; 3 and 4, 12-h TGY broth culture; 5 and 6, 36-h colony on TGY agar plate; M, DNA size marker. (B) Effect of cell concentration on amplification by direct ITS-PCR from 48-h TGY broth culture of Y. lipolytica strain 160. Cell populations (CFU per milliliter): lane 1, 3 × 108; lane 2, 3 × 106; lane 3, 3 × 104; lane 4, 3 × 103.
FIG. 2.
Direct ITS-PCR amplification products of several yeast species. Template DNA was extracted from washed overnight TGY broth cultures by boiling. Lanes: 1, Y. lipolytica; 2, C. guilliermondii; 3, C. sake; 4, C. parapsilosis; 5, C. tropicalis; 6, C. krusei; 7, C. famata; M, DNA size marker.
FIG. 3.
Direct ITS-PCR amplification products and restriction fragments from Y. lipolytica and C. zeylanoides. Lanes 1 to 10, Y. lipolytica strains 160 (lanes 1 and 6), 138 (lanes 2 and 7), 173 (lanes 3 and 8), 246 (lanes 4 and 9), 175 (lanes 5 and 10); lanes 11 to 20, C. zeylanoides strains 155 (lanes 11 and 16), 130 (lanes 12 and 17), 103 (lanes 13 and 18), 121 (lanes 14 and 19), 128 (lanes 15 and 20). ITS-PCR amplicon, lanes 1 to 5 and 11 to 15; HinfI digest, lanes 6 to 10; HaeIII digest, lanes 16 to 20; M, DNA size marker.
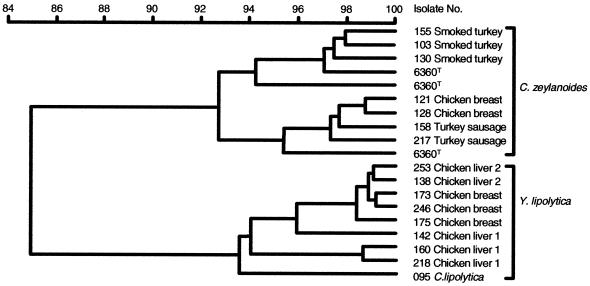
RAPD amplification with commercial decamer primers of crude DNA extract did not result in amplified products. Using the much longer ERIC primers enabled typing of strains by producing patterns consisting of several different bands. Cluster analysis was used to determine if strains with different patterns can be grouped according to their source of isolation (Fig. 4). The dendogram also shows that isolates of Y. lipolytica and C. zeylanoides are clearly distinct and separated. Within each branch, strains exhibit a high degree of similarity, and with a few exceptions, groups correlate well with the source of isolation.
FIG. 4.
Cluster analysis of RAPD-ERIC2 patterns of Y. lipolytica and C. zeylanoides showing relations with the source of isolation. Dendogram was derived using UPGMA and Pearson product method. Reference strain, C. zeylanoidesT 6360. Other strains: C. zeylanoides 103, 158, 121, 128, 130, 155, and 217; Y. lipolytica 138, 142, 160, 173, 175, 218, 246, and 253; C. lipolytica 095.
Chromosomal DNA molecules were separated successfully by contour-clamped electric field (CHEF) PFGE. From C. zeylanoides, we obtained six to eight bands in the range of 750 to 2,200 kbp; chromosomal DNA molecules in Y. lipolytica were much larger, mostly over 2 Mbp, and separated into three to five bands (Fig. 5). Karyotypes of both yeast species showed polymorphic patterns differing between strains.
FIG. 5.
(A) Separation of chromosomal DNA from C. zeylanoides by CHEF PFGE. Lanes 1 and 9, C. zeylanoides type strain; lanes 2 and 8, isolates 217 and 158, respectively, from turkey sausage; lanes 3 and 5, isolates 128 and 121, respectively, from chicken breast; lanes 4, 6, and 7, isolates 130, 103, and 155, respectively, from smoked turkey; M, S. cerevisiae DNA size marker. Electrophoretic conditions: 100 V for 15 h with 120-s pulse time, followed by 180-s pulse time for 33 h at 14°C in 1% agarose gel. (B) Separation of chromosomal DNA from Y. lipolytica by CHEF PFGE. Lanes 1 and 4, isolates 218 and 160, respectively, from chicken liver 1; lanes 2, 3, and 6, isolates 246, 173, and 175, respectively, from chicken breast; lane 5, isolate 138 from chicken liver 2; M, S. cerevisiae DNA size marker. Electrophoretic conditions: 50 V for 36 h with 1,200-s pulse time, followed with 1,800-s pulse time for 36 h at 14°C in 0.75% agarose gel.
DISCUSSION
Karyotyping is a reliable and discriminative technique that has been used commonly for typing yeast isolates from food samples and clinical specimens (36, 54). In agreement with another report (38), Y. lipolytica has few but large chromosomal DNA molecules. To our knowledge, this is the first study reporting the karyotype of C. zeylanoides. The number and size of its chromosomal DNA molecules are grossly similar to those of several other Candida species (14).
Characterization of polymorphic karyotypes permitted discrimination of strains and enabled correlation with the source of isolation. However, the greatest drawback of karyotyping is the long time necessary to obtain results. Preparation of chromosomal DNA takes at least 2 days, and PFGE proceeds for another 2 to 3 days to obtain good separation of chromosomal bands.
The direct PCR technique, i.e., extracting DNA by boiling yeast cell suspensions, has been used by other researchers. A survey of the literature revealed more than 20 publications using this method since 1992. The 1997 Cold Spring Harbor laboratory course manual (1) describes a yeast colony PCR protocol. Experience has shown that for successful amplification, the density of the cell suspension is crucial. However, a detailed study to optimize amplification as influenced by cell density has not been published. Yeast densities between 105 and 108 cells/ml have been used. Pearson and McKee (40) added an ice-cold suspension of yeast cells directly to the PCR mix, which was then heated at 92°C for 2 min before being subjected to amplification. This method was used in subsequent studies (26). Hopfer et al. (22) obtained DNA for PCR amplification from yeast cells boiled in EDTA, but Howell et al. (23) reported that RAPD amplification from boiled cells was not sufficiently reproducible. Steffan et al. (45), using colony lysates, optimized conditions of RAPD for the identification of Candida species of clinical significance. Their toothpick protocol involved picking up a barely visible amount of cells from a single colony and suspending it in a buffered Zymolase solution. The direct addition of intact cells to the PCR mix resulted in successful amplification in a less reproducible manner. In contrast, several workers have observed that boiled extracts of yeast cells can be used reliably for PCR amplification (15, 28, 29, 39, 46). Lachance et al. (27), using whole yeast cell extracts, obtained amplification products with quality equivalent or better than that of purified DNA. This simple and rapid method gave reproducible patterns in a study with a broad range of yeasts (16). In our study, the ITS-PCR method was robust enough to produce amplification products in a reproducible way. The RAPD technique is influenced more by experimental conditions, and in agreement with Power (41), the use of a crude DNA preparation cannot be recommended.
In food and beverage industry laboratories, of great significance is the time saved by analytical methods. Standard methods for DNA extraction and purification may require 24 h or more, compared with extraction by boiling, followed by centrifugation, all within 15 min. The time required for identification of an isolate can be as short as 8 h, which includes DNA extraction, amplification, restriction digestion, and electrophoretic analysis. Additional typing by RAPD can be finished the following morning.
Numerous studies have confirmed the efficiency of ITS-PCR for identifying yeast species. The region of the rDNA amplified by ITS1 and ITS4 primers includes variable ITSs and the less variable 5.8S rRNA gene (55), allowing differentiation of more closely related species than can be achieved using conservative 18S and 25S rDNA sequences. ITS-PCR has been used for the taxonomic study and delineation of Candida (30), Saccharomyces (37), Kluyveromyces (7), and Metschnikowia (49) species and for rapid identification and population analysis of yeasts in wine (10, 17, 21), kefyr (58), and other foods (5). The technique has also been used for taxonomy, diagnosis, and epidemiology of yeasts of clinical importance, in particular, Candida albicans and other Candida species (9, 30, 33, 57). These studies demonstrated that the amplicon length alone can sometimes differentiate species (33, 49). Unequivocal identification, however, requires more precise determination. This can be achieved in various ways, e.g., using inner primers in nested PCR (9), separating PCR products by capillary electrophoresis (47), or hybridization with specific DNA probes (8). However, a simple but sufficiently sensitive method is restriction analysis of amplified fragments that has been widely used (16). The amplicon lengths obtained from Y. lipolytica and C. zeylanoides in our study agree well with those described for one strain of each species investigated by Esteve-Zarzoso et al. (16). Digestion patterns, however, differed slightly in that HinfI produced two unequal fragments from the amplicon of Y. lipolytica, whereas HaeIII resulted in three rather than two fragments of the C. zeylanoides amplicon. In our study, eight strains of Y. lipolytica and seven strains of C. zeylanoides gave uniform digestion patterns. Guillamón et al. (21) tested the same strain of C. zeylanoides used by Esteve-Zarzoso et al. (16) and obtained similar results. In an earlier investigation (52), two size classes found in rDNA units of Y. lipolytica were attributed to differences in the nontranscribed spacer.
Using boiled extract as template DNA, we did not obtain amplicons with the RAPD method using five commercially available decamer primers (A-01 to A-05; Operon Technology, Alameda, Calif.). This may be due to the small number of primers tested, as previous studies showed that sometimes 40 or more primers need to be screened for successful RAPD amplification (20, 34, 43). For RAPD amplification, oligonucleotide primers can be of variable length, from five to 24 bp, often used singularly but sometimes in pairs, and can be synthesized arbitrarily or selected from natural sequences, e.g., microsatellite and repetitive sequences which are also used in PCR-fingerprinting (32). ERIC sequences are frequently used primers in PCR-based molecular typing of bacteria (53) and, under the nonstringent conditions of RAPD, may also anneal to eukaryotic DNA. This allowed their use in typing dermatophytes and clinical yeast strains (24, 31, 50). In our study, both ERIC1 and ERIC2 produced several polymorphic fragments resulting in band patterns differing between strains within a given species. Cluster analysis of patterns made it possible to trace the source of isolation of strains. Thus, ERIC-PCR can be used as a tool to study the ecological diversity of yeasts.
REFERENCES
- 1.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 2.Al-Mohizea S, Mashhadi A, Fawwal A, Al-Shalhat A. Microbiological and shelf life assessment of chilled eviscerated whole chicken broilers in Saudia Arabia. Br Poultry Sci. 1994;35:519–526. doi: 10.1080/00071669408417717. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Molecular analyst software, Fingerprinting Plus and Fingerprinting DST software, version 1.6, instruction manual. Hercules, Calif: Bio-Rad Laboratories, Life Science Group; 1999. [Google Scholar]
- 4.Baleiras Couto M M, Eijsma B, Hofstra H, Huis in't Veld J H J, van der Vossen J M B M. Evaluation of molecular typing techniques to assign genetic diversity among Saccharomyces cerevisiae strains. Appl Environ Microbiol. 1996;62:41–46. doi: 10.1128/aem.62.1.41-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baleiras Couto M M, Vogels J T W E, Hofstra H, Huis in't Veld J H J, van der Vossen J M B M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food-borne yeasts. J Appl Bacteriol. 1995;79:525–535. doi: 10.1111/j.1365-2672.1995.tb03173.x. [DOI] [PubMed] [Google Scholar]
- 6.Banks J G, Board R G. Some factors influencing the recovery of yeast and moulds from chilled foods. Int J Food Microbiol. 1987;4:197–206. [Google Scholar]
- 7.Belloch C, Barrio E, Dolores Garcia M, Querol A. Phylogenetic reconstruction of the yeast genus Kluyveromyces: restriction map analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Syst Appl Microbiol. 1998;21:266–273. doi: 10.1016/S0723-2020(98)80032-5. [DOI] [PubMed] [Google Scholar]
- 8.Botelho A R, Planta R J. Specific identification of Candida albicans by hybridization with oligonucleotides derived from ribosomal DNA internal spacers. Yeast. 1994;10:709–710. doi: 10.1002/yea.320100603. [DOI] [PubMed] [Google Scholar]
- 9.Bougnoux E, Dupont C, Mateo J, Saulnier P, Faivre V, Payen D, Nicolas-Chanoine M-H. Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR. J Clin Microbiol. 1999;37:925–930. doi: 10.1128/jcm.37.4.925-930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Constanti M, Reguant C, Poblet M, Zamora F, Mas A, Guillamón J M. Molecular analysis of yeast population dynamics: effect of sulphur dioxide and inoculum on must fermentation. Int J Food Microbiol. 1998;41:169–175. doi: 10.1016/s0168-1605(98)00041-5. [DOI] [PubMed] [Google Scholar]
- 11.Deák T, Beuchat L R. Handbook of food spoilage yeasts. Boca Raton, Fla: CRC Press; 1996. [Google Scholar]
- 12.Deák T. Methods for the rapid detection and identification of yeasts in foods. Trends Food Sci Technol. 1995;6:287–292. [Google Scholar]
- 13.Diriye F U, Scorzetti G, Martini N. Methods for the separation of yeast cells from the surfaces of processed frozen foods. Int J Food Microbiol. 1993;19:27–37. doi: 10.1016/0168-1605(93)90121-v. [DOI] [PubMed] [Google Scholar]
- 14.Doi M, Homma M, Chindamporn A, Tanaka K. Estimation of chromosome number and size by pulsed-field gel electrophoresis (PFGE) in medically important Candida species. J Gen Microbiol. 1992;138:2243–2251. doi: 10.1099/00221287-138-10-2243. [DOI] [PubMed] [Google Scholar]
- 15.Donnelly S M, Sullivan D J, Shanley D B, Coleman D C. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–1882. doi: 10.1099/13500872-145-8-1871. [DOI] [PubMed] [Google Scholar]
- 16.Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol. 1999;49:329–337. doi: 10.1099/00207713-49-1-329. [DOI] [PubMed] [Google Scholar]
- 17.Fernández M, Ubeda J F, Briones A I. Comparative study of non-Saccharomyces microflora of musts in fermentation, by physiological and molecular methods. FEMS Microbiol Lett. 1999a;173:223–229. [Google Scholar]
- 18.Gallo L, Schmitt R E, Schmidt-Lorenz W. Microbial spoilage of refrigerated fresh broilers. I. Bacterial flora and growth during storage. Lebensm Wiss Technol. 1998;21:216–223. [Google Scholar]
- 19.Geornaras I, Dykes G A, von Holy A. Microbial populations associated with refrigerated poultry. S Afr J Sci. 1994;90:570–582. [Google Scholar]
- 20.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillamón J M, Sabate J, Barrio E, Cano J, Querol A. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcribed spacer (ITS) region. Arch Microbiol. 1998;169:387–392. doi: 10.1007/s002030050587. [DOI] [PubMed] [Google Scholar]
- 22.Hopfer R L, Walden P, Setterquist S, Highsmith W E. Detection and differentiation of fungi in clinical specimens using polymerase chain reaction (PCR) amplification and restriction enzyme analysis. J Med Vet Mycol. 1993;31:65–75. doi: 10.1080/02681219380000071. [DOI] [PubMed] [Google Scholar]
- 23.Howell S A, Anthony R M, Power E. Application of RAPD and restriction enzyme analysis to the study of oral carriage of Candida albicans. Lett Appl Microbiol. 1996;22:125–128. doi: 10.1111/j.1472-765x.1996.tb01124.x. [DOI] [PubMed] [Google Scholar]
- 24.Howell S A, Barnard R J, Humphreys F. Application of molecular typing methods to dermatophyte species that cause skin and nail infections. J Med Microbiol. 1999;48:33–40. doi: 10.1099/00222615-48-1-33. [DOI] [PubMed] [Google Scholar]
- 25.Ismail, S. A. S., T. Deak, H. A. Abd-Rahman, M. A. M. Zassien, and L. R. Beuchat. Presence and changes in populations of yeasts associated with raw and processed poultry products stored at refrigeration temperature. Int. J. Food Microbiol., in press. [DOI] [PubMed]
- 26.James S A, Cai J, Roberts I N, Collins M D. A phylogenetic analysis of the genus Saccharomyces based on 18S rRNA gene sequences: description of Saccharomyces kunashirensis sp. nov. and Saccharomyces martinae sp. nov. Int J Syst Bacteriol. 1997;47:453–460. doi: 10.1099/00207713-47-2-453. [DOI] [PubMed] [Google Scholar]
- 27.Lachance M-A, Bowles J M, Starmer W T, Barker J S F. Kodamaea kakaduensis and Candida tolerans, two new ascomycetous yeast species from Australian Hibiscus flowers. Can J Microbiol. 1999;45:172–177. doi: 10.1139/w98-225. [DOI] [PubMed] [Google Scholar]
- 28.Maiwald M, Kappe R, Sonntag H-G. Rapid presumptive identification of medically relevant yeasts to the species level by polymerase chain reaction and restriction enzyme analysis. J Med Vet Mycol. 1994;32:115–122. doi: 10.1080/02681219480000161. [DOI] [PubMed] [Google Scholar]
- 29.Masneuf I, Aigle M, Dubourdieu D. Development of a polymerase chain reaction/restriction fragment length polymorphism method for Saccharomyces cerevisiae and Saccharomyces bayanus identification in enology. FEMS Microbiol Lett. 1996;138:239–244. doi: 10.1111/j.1574-6968.1996.tb08164.x. [DOI] [PubMed] [Google Scholar]
- 30.McCullough M J, Clemons K V, Stevens D A. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37:417–421. doi: 10.1128/jcm.37.2.417-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzgar D, van Belkum A, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–2313. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meunier J-R, Grimont P A D. Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol. 1993;144:373–379. doi: 10.1016/0923-2508(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell T G, Freedman E Z, White T J, Taylor J W. Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J Clin Microbiol. 1994;32:253–255. doi: 10.1128/jcm.32.1.253-255.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitrakul C M, Henick-Kling T, Egli C M. Discrimination of Brettanomyces/Dekkera yeast isolates from wine by using various DNA fingerprinting methods. Food Microbiol. 1999;16:3–14. [Google Scholar]
- 35.Molnár O, Messner R, Prillinger H, Stahl U, Slavikova E. Genotypic identification of Saccharomyces species using random amplified polymorphic DNA analysis. Syst Appl Microbiol. 1995;18:136–145. [Google Scholar]
- 36.Monod M, Porchet S, Baudraz-Rosselet F B, Frank E. The identification of pathogenic yeast strains by electrophoretic analysis of their chromosomes. J Med Microbiol. 1990;32:123–129. doi: 10.1099/00222615-32-2-123. [DOI] [PubMed] [Google Scholar]
- 37.Montrocher R, Verner M-C, Briolay J, Gautier C, Marmeisse R. Phylogenetic analysis of the Saccharomyces cerevisiae group based on polymorphisms of rDNA spacer sequences. Int J Syst Bacteriol. 1998;48:295–303. doi: 10.1099/00207713-48-1-295. [DOI] [PubMed] [Google Scholar]
- 38.Naumova E, Naumov G, Fournier P, Nguyen H-V, Gaillardin C. Chromosomal polymorphism of the yeast Yarrowia lipolytica and related species: electrophoretic karyotyping and hybridization with cloned genes. Curr Genet. 1993;23:450–454. doi: 10.1007/BF00312633. [DOI] [PubMed] [Google Scholar]
- 39.Okhravi N, Adamson P, Mant R, Matheson M M, Midgley G, Towler H M A, Lightman S. Polymerase chain reaction and restriction fragment length polymorphism mediated detection and specification of Candida spp causing intraocular infection. Investig Ophthalmol Vis Sci. 1998;39:859–866. [PubMed] [Google Scholar]
- 40.Pearson B M, McKee R A. Rapid identification of Saccharomyces cerevisiae, Zygosaccharomyces bailii, and Zygosaccharomyces rouxii. Int J Food Microbiol. 1992;16:63–67. doi: 10.1016/0168-1605(92)90126-n. [DOI] [PubMed] [Google Scholar]
- 41.Power E G M. RAPD typing in microbiology—a technical review. J Hosp Infect. 1996;34:247–265. doi: 10.1016/s0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- 42.Querol A, Ramón D. The application of molecular techniques in wine microbiology. Trends Food Sci Technol. 1996;7:73–78. [Google Scholar]
- 43.Quesada M P, Cenis J L. Use of random amplified polymorphic DNA (RAPD-PCR) in the characterization of wine yeasts. Am J Enol Vitic. 1995;46:204–208. [Google Scholar]
- 44.Smole Mozina S, Raspor P. Molecular techniques for yeast identification in food processing. Food Technol Biotechnol. 1997;35:55–61. [Google Scholar]
- 45.Steffan P, Vazquez J A, Boikov D, Xu C, Sobel J D, Akins R A. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J Clin Microbiol. 1997;35:2031–2039. doi: 10.1128/jcm.35.8.2031-2039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Török T, Mortimer R K, Romano P, Suzzi G, Polsinelli M. Quest for wine yeasts—an old story revisited. J Indust Microbiol. 1996;17:303–313. [Google Scholar]
- 47.Turenne C, Sanche S E, Hoban D J, Karlowsky J A, Kabani A M. Rapid identification of fungi using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37:1846–1851. doi: 10.1128/jcm.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valente P, Gouveia F C, Lemos G A, Pimentel D, Elsas J D, Mendonca-Hagler L C, Hagler A N. PCR amplification of the rDNA internal transcribed spacer region for differentiation of Saccharomyces cultures. FEMS Microbiol Lett. 1996;137:253–256. doi: 10.1111/j.1574-6968.1996.tb08114.x. [DOI] [PubMed] [Google Scholar]
- 49.Valente P, Gouveia F C, Lemos G A, Pimentel D, Mendonca-Hagler L C, Hagler A N. PCR-amplified ITS length variation within the yeast genus Metschnikowia. J Gen Appl Microbiol. 1997;43:179–181. doi: 10.2323/jgam.43.179. [DOI] [PubMed] [Google Scholar]
- 50.Van Belkum A, Beached T, Bosom R. Monitoring spread of Malassezia infections in a neonatal intensive care unit by PCR-mediated genetic typing. J Clin Microbiol. 1994;32:2528–2532. doi: 10.1128/jcm.32.10.2528-2532.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Vossen J M B M, Hofstra H. DNA based typing, identification and detection systems for food spoilage microorganisms: development and implementation. Int J Food Microbiol. 1996;33:35–49. doi: 10.1016/0168-1605(96)01136-1. [DOI] [PubMed] [Google Scholar]
- 52.Van Heerikhuizen H, Ykema A, Klootwijk J, Gaillardin C, Ballas C, Fournier P. Heterogeneity in the ribosomal RNA genes of the yeast Yarrowia lipolytica; cloning and analysis of two size classes of repeats. Gene. 1985;39:213–222. doi: 10.1016/0378-1119(85)90315-4. [DOI] [PubMed] [Google Scholar]
- 53.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acid Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vezinhet F, Blondin B, Hallet J N. Chromosomal DNA patterns and mitochondrial DNA polymorphism as tools for identification of enological strains of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1990;32:568–571. [Google Scholar]
- 55.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 56.Viljoen B C, Gernaras I, Lamprecht A, von Holy A. Yeast populations associated with processed poultry. Int J Food Microbiol. 1998;15:113–117. [Google Scholar]
- 57.Williams D W, Wilson M J, Lewis M A O, Potts A J C. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wyder M-T, Puhan Z. A rapid method for identification of yeasts from kefyr at species level. Milchwissenschaft. 1997;52:327–330. [Google Scholar]