Abstract
Two PCR primer pairs were designed to amplify rRNA genes (rDNA) from all four major phyla of fungi: Ascomycota, Basidiomycota, Chytridomycota, and Zygomycota. PCRs performed with these primers showed that both pairs amplify DNA from organisms representing the major taxonomic groups of fungi but not from nonfungal sources. To test the ability of the primers to amplify fungal rDNA from environment samples, clone libraries from two avocado grove soils were constructed and analyzed. These soils possess different abilities to inhibit avocado root rot caused by Phythophthora cinnamomi. Analysis of the two rDNA clone libraries revealed differences in the two fungal communities. It also revealed a markedly different depiction of the soil fungal community than that generated by a culture-based analysis, confirming the value of rDNA-based approaches for identifying organisms that may not readily grow on agar media. Additional evidence of the usefulness of the primers was obtained by identifying fungi associated with avocado leaves. In both the soil and leaf analyses, no nonfungal rDNA sequences were identified, illustrating the selectivity of these PCR primers. This work demonstrates the ability of two newly developed PCR primer sets to amplify fungal rDNA from soil and plant tissue, thereby providing unique tools to examine this vast and mostly undescribed community of organisms.
Fungi play complex and diverse roles in ecosystems and human society. They comprise the majority of the biomass in soil, decompose organic material, provide nutrients to plants, and act as indicators of ecosystem health (2, 7). In agriculture, fungi can both devastate crop yields and provide a means to control plant pests, including other fungi (29). In humans, fungi can cause a range of conditions from psoriasis to meningitis. They have also proven to be effective curative agents; for example, Saccharomyces boulardii can prevent Clostridium difficile toxicity and other intestinal disturbances caused by antibiotic usage (6). In the biotechnology arena, fungi produce numerous secondary metabolites that have valuable pharmaceutical properties (24). Their importance to this industry and other bioprospecting endeavors is enhanced by the vast diversity of extant fungi (10).
Although considerable knowledge about fungi has been amassed, most of these organisms remain uncharacterized. Estimates suggest that there are 1.5 million species of fungi on Earth; however, only approximately 70,000 species have been described, leaving 95% of the species undescribed (16). The reasons for this deficiency include habitats that have not been well investigated, organisms that are difficult to culture axenically, such as obligately associated fungi, and inaccurate identification of catalogued samples (16). Strategies used to rectify this deficit should include the use of comparative sequence analysis of rRNA and rRNA genes (rDNA), which has led to the discovery of many new bacterial and archaeal phylotypes in environments such as Yellowstone hot springs, soil, and rock (3, 22).
Several PCR primers that amplify fungal rDNA from a wide range of taxonomic groups have been described (31), but few of these were designed for use with environmental samples. Such a tool must have high specificity, as fungal DNA may be rare compared to DNA from other sources, such as bacteria, plants, or other eukaryotes (14). The ITS1-F and ITS4-B primers have been used to amplify basidiomycete ITS1, ITS2, and 5.8S rDNA sequences from plant tissues containing fungi (12). Similarly, the VANS1 primer has been used in combination with other primers to amplify rDNA from vesicular-arbuscular endomycorrhizal fungi (27). To identify disease-causing fungi, PCR primers have been designed to specifically amplify both human (4, 18, 21) and plant (17) pathogens. In addition, three PCR primer pairs described by Smit et al. were recently used to amplify fungal rDNA from wheat rhizosphere samples (28). In this report, we describe two new PCR primer pairs designed to amplify rDNA from all major taxonomic groups of fungi, and in this study we demonstrated the use of these primer pairs by examining the fungal communities of two avocado grove soils.
MATERIALS AND METHODS
Primer design.
A total of 213 fungal small-subunit rDNA sequences of representatives of all major phylogenetic groups were obtained from GenBank (National Center for Biotechnology Information [NCBI]) and were aligned with PILEUP (Genetics Computer Group, Madison, Wis.). Conserved sequences within this group were identified with PRETTY (Genetics Computer Group). The specificity of these sequences was examined by comparison to the nonredundant nucleotide database at GenBank by using BLAST (NCBI). The PCR primers identified through this process were nu-SSU-0817-5′ (TTAGCATGGAATAATRRAATAGGA), nu-SSU-1196-3′ (TCTGGACCTGGTGAGTTTCC), and nu-SSU-1536-3′ (ATTGCAATGCYCTATCCCCA).
DNA extraction.
DNA were extracted from pure cultures of fungi, dried fungal samples, and avocado leaves by using a FastDNA Kit as described by the manufacturer (Bio 101, Vista, Calif.). DNA were extracted from two avocado grove soils, collected at the Vanoni and Powell ranches, by using a FastDNA Kit for Soil as described by the manufacturer (Bio 101) (5). DNA that were not amplified in PCRs containing universal rDNA primers 530F (GTGCCAGCMGCCGCGG) and 1392R (ACGGGCGGTGTGTRC) (19) were further purified by electrophoresis on 1% agarose gels and isolated with a QIAquick gel extraction kit (Qiagen, Valencia, Calif.).
PCR parameters.
DNA from fungi and other sources were amplified in 10-μl PCR mixtures containing the following final concentrations or total amounts: 3 to 8 ng of DNA, 50 mM Tris (pH 8.3), 500 μg of bovine serum albumin per ml, 2.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 250 μM, 400 nM forward primer nu-SSU-0817-5′, 400 nM reverse primer nu-SSU-1196-3′ or nu-SSU-1536-3′, and 0.5 U of Taq DNA polymerase. All reagents were combined and heated at 94°C for 2 min. Thirty-five cycles of PCR were then performed by using 94°C for 0 s, 56°C for 10 s, and 72°C for 30 s, followed by 72°C for 2 min. PCRs were performed in glass capillary tubes with a model 1002 Rapidcycler (Idaho Technologies, Idaho Falls, Idaho). PCRs which used primers EF4 and EF3, primers EF4 and fung5, and primers EF4 and NS3 were performed as previously described by Smit et al. (28) by using both an MJ Research PTC-200 thermocycler and an Idaho Technologies model 1002 Rapidcycler.
Small-subunit rDNA clone library construction.
DNAs isolated from soil and avocado leaves were amplified by PCR as described above. rDNA libraries were produced by gel isolating the amplified genes, ligating them into the pGEM-T vector (Promega, Madison, Wis.), and transforming the plasmids into competent JM109 cells. Bacterial colonies containing plasmids with rDNA inserts were identified by α-complementation (26).
Analysis of rDNA clone libraries.
Plasmid DNA were isolated from randomly selected rDNA clones. To sort the clones into groups or operational taxonomic units (OTUs), the rDNA inserts were amplified by PCR, digested individually with several DNA restriction endonuclease treatments (HpaI, MseI, ApaI-SacI, and RsaI), and resolved on 2% agarose gels. Approximately 430 bases of nucleotide sequence were obtained from one representative clone of each OTU. Similarities of the rDNA clones to sequences in the GenBank database were determined by using BLAST (NCBI) and Gap (Genetics Computer Group).
Nucleotide sequence accession numbers.
rDNA sequences AL10, AL15, AL3, JK1-1, JK1-14, and JK1-16 have been deposited in the GenBank database under accession no. AF183384, AF183386, AF183388, AF247743, AF247744, and AF247745, respectively. rDNA sequences JK2-2, JK2-4, JK2-9, JK2-17, JK2-22, JK3-21, JK3-25, JK3-30, JK3-18, JKX-3, and JK3-26 have been deposited in the GenBank database under accession no. AF246619 through AF246629, respectively.
RESULTS AND DISCUSSION
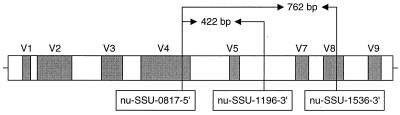
Two PCR primer pairs were designed to amplify rDNA from all four major phyla of fungi: Ascomycota, Basidiomycota, Chytridomycota, and Zygomycota. The primers were named nu-SSU-0817-5′, nu-SSU-1196-3′, and nu-SSU-1536-3′ by using the nomenclature convention described by Gargas and DePriest (13). nu-SSU-0817-5′ and nu-SSU-1196-3′ amplify a 422-bp region of the Saccharomyces cereviseae small-subunit rDNA molecule (GenBank accession no. J01353) and contain the V4 (partial) and V5 variable regions (Fig. 1) (30). nu-SSU-0817-5′ and nu-SSU-1536-3′ amplify a 762-bp region and contain the V4 (partial), V5, V7, and V8 (partial) variable regions (Fig. 1).
FIG. 1.
Diagram of the eukaryotic small-subunit rDNA with the variable regions highlighted in gray. The numerical positions of the primers and the PCR product sizes were obtained by using S. cereviseae (GenBank accession no. J01353) as the reference template.
Both primer pairs show strong specificity for fungal rDNA sequences. Using BLAST (NCBI), the percentages of identical matches of nu-SSU-0817-5′, nu-SSU-1196-3′, and nu-SSU-1536-3′ with fungal rDNA sequences in the GenBank database (NCBI) were determined to be 83, 85, and 99.8%, respectively (Table 1). PCRs with these primers showed that both pairs amplify DNA from representatives of all major taxonomic groups of fungi but not from representative nonfungal groups, including the oomycete Phytophthora infestans (Fig. 2 and Table 2). These results were obtained when annealing temperatures ranging from 50 to 58°C and from 52 to 58°C were used with the nu-SSU-0817-5′–nu-SSU-1196-3′ and nu-SSU-0817-5′–nu-SSU-1536-3′ primer pairs, respectively (data not shown). Similar results were obtained with two different thermocyclers (data not shown).
TABLE 1.
Specificity of the PCR primers for fungal rDNA sequences in the GenBank database
| Taxon | Identical matches witha:
|
||
|---|---|---|---|
| Primer nu-SSU-0817-5′ | Primer nu-SSU-1196-3′ | Primer nu-SSU-1536-3′ | |
| Fungi | 83 (1,089) | 85 (793) | 99.8 (848) |
| Rhodophyta | 12 (155) | ||
| Alveolata | 2 (32) | 10 (94) | |
| Viridiplantae | 2.4 (22) | ||
| Metazoa | 0.9 (12) | 1.7 (16) | 0.2 (2) |
| Choanoflagellida | 0.3 (4) | 0.4 (4) | |
| Glaucocystophyceae | 0.2 (2) | ||
| Cryptophyta | 1.7 (23) | 0.1 (1) | |
| Myxozoa | 0.5 (5) | ||
| Stramenopiles | 0.2 (3) | ||
| Total | 100 (1,320) | 100 (935) | 100 (850) |
These data are the percentages (numbers) of identical matches between the PCR primers described in this study and sequences in the GenBank (NCBI) nonredundant nucleotide database. These data were obtained by using BLAST (NCBI) during the period from December 1998 to April 1999.
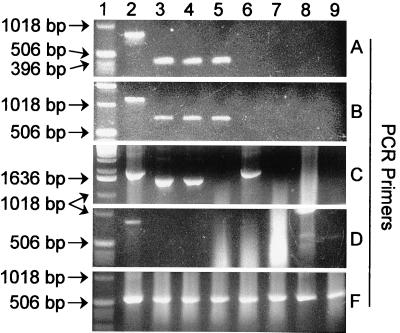
FIG. 2.
PCR amplification of representative DNA templates with five different fungal rDNA primer sets. PCR products were resolved on agarose gels and stained with ethidium bromide. (A) Primers nu-SSU-0817-5′ and nu-SSU-1196-3′; (B) primers nu-SSU-0817-5′ and nu-SSU-1536-3′; (C) primers EF4 and EF3; (D) primers EF4 and NS3; (E) primers EF4 and fung5. Lane 1, 1-kb ladder (Gibco BRL, Grand Island, N.Y.); lane 2, Monilinia fructicola; lane 3, Tilletia caries; lane 4, Allomyces javanicus; lane 5, Glomus deserticola; lane 6, Escherichia coli; lane 7, Caenorhabditis elegans; lane 8, Cucumis melo; lane 9, Phytophthora infestans. The larger sizes of the M. fructicola bands (lane 2) are likely due to intron insertion. The results of these experiments and a more extensive analysis are summarized in Table 2.
TABLE 2.
Amplification of fungal and nonfungal templates with five fungal rDNA primer pairsa
| Taxon | Division | Class | Species | Primers
|
||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | ||||
| Fungi | Ascomycota | Archiascomycetes | Schizosaccharomyces octosporus | + | + | + | − | + |
| Taphrina deformans | + | + | + | − | + | |||
| Euascomycetes | Monilinia fructicola | + | + | + | − | + | ||
| Erysiphe graminis | + | + | + | − | + | |||
| Penicillium digitatum | + | + | + | − | + | |||
| Sordaria sp. | + | + | + | − | + | |||
| Hemiascomycetes | Saccharomyces cerevisiae | + | + | + | − | + | ||
| Saccharomyces ludwigii | + | + | + | − | + | |||
| Saccharomyces octosporus | + | + | + | − | + | |||
| Basidiomycota | Gasteromycetes | Calvatia subcretacea | + | + | + | − | + | |
| Pisolithus tinctorius | + | + | + | − | + | |||
| Scleroderma arenicola | + | + | + | − | + | |||
| Hymenomycetes | Armillaria sp. | + | + | + | − | + | ||
| Sclerotium rolfsii | + | + | + | − | + | |||
| Urediniomycetes | Tranzschelia discolor | + | + | + | − | + | ||
| Ustilaginomycetes | Tilletia caries | + | + | + | − | + | ||
| Chytridiomycota | Chytridiomycetes | Allomyces javanicus | + | + | + | − | + | |
| Spizellomyces punctatus | + | + | − | − | − | |||
| Zygomycota | Trichomycetes | Amoebidium parasiticum | + | + | − | + | − | |
| Smittium sp. | + | + | + | − | − | |||
| Zygomycetes | Glomus deserticola | + | + | − | − | + | ||
| Mucor rouxii | + | + | + | + | + | |||
| Phycomyces blakesleeanus | + | + | − | − | + | |||
| Proteobacteria | Escherichia coli | − | − | + | − | + | ||
| Metazoa | Caenorhabditis elegans | − | − | − | − | + | ||
| Streptophyta | Cucumis melo | − | − | − | − | + | ||
| Oomycetes | Phytophthora infestans | − | − | − | − | + | ||
| Water | − | − | − | − | − | |||
Summary of the results obtained with PCR mixtures containing pairs of fungal rDNA primers and various DNA templates. PCRs that produced an amplification product of the expected size are indicated by a plus sign. The following primer pairs were used: A, nu-SSU-0817-5′ and nu-SSU-1196-3′; B, nu-SSU-0817-5′ and nu-SSU-1536-3′; C, EF4 and EF3; D, EF4 and NS3; and E, EF4 and fung5. All DNA templates were determined to be of sufficient purity for PCR amplification by successful amplification with universal rDNA primers 530F (GTGCCAGCMGCCGCGG) and 1392R (ACGGGCGGTGTGTRC) (19) (data not shown).
To examine the ability of these primers to selectively amplify fungal rDNA from environmental samples, the fungal communities of two avocado grove soils were analyzed by performing PCRs with mixtures containing nu-SSU-0817-5′ and nu-SSU-1536-3′ (Fig. 3). The Vanoni soil possesses the ability to inhibit avocado root rot caused by Phytophthora cinnamomi and is therefore considered a disease-suppressive soil. Conversely, the Powell soil is classified as a disease-conducive soil because it does not inhibit avocado root rot. Numerous suppressive soils have been described, many of which possess biological components that may contribute to this phenomenon (8). In the past, identification of these components has been challenging because the majority of microorganisms do not readily grow on agar media (1), which leads to analyses that may not accurately reflect the true fungal community in a soil. Alternative approaches that avoid this culture bias include analysis of rRNA genes isolated from soil. In this study, we examined two avocado grove soils by sorting 62 fungal rDNA clones into 10 different clone types or OTUs, 4 of which were found only in the suppressive soil (Table 3). In addition, the dominant genera in the Vanoni soil identified by the rDNA analysis (Tritirachium, Aspergillus, Pleospora, Petriella, Monilinia, and Exophiala) (Table 3) were markedly different from those identified by a traditional culture-based approach (Aspergillus, Penicillium, Sporothrix, Phoma, Trichoderma, and Fusarium) (9). While PCR can also introduce errors (11, 20, 25), these results show the potential of the rDNA-based approach for biological control research as this approach revealed a very different depiction of the soil fungal community than that provided by the culture-based analysis. Future strategies to obtain biological control organisms could include culture-independent rDNA analysis followed by isolation of specific organisms on selective media.
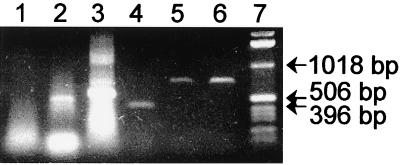
FIG. 3.
PCR amplification of DNA extracted from two soils with five different fungal rDNA primer pairs. PCR products were resolved on agarose gels and stained with ethidium bromide. Lane 1, primers EF4 and EF3; lane 2, primers EF4 and fung5; lane 3, primers EF4 and NS3; lane 4, primers nu-SSU-0817-5′ and nu-SSU-1196-3′; lanes 5 and 6, primers nu-SSU-0817-5′ and nu-SSU-1536-3′; lanes 1 through 5, Vanoni soil DNA; lane 6, Powell soil DNA; lane 7, 1-kb ladder (Gibco BRL).
TABLE 3.
Fungal community analysis for two avocado grove soils
| Identity of OTU (GenBank accession no.)a | Cloneb | No. of clones identified in:
|
|
|---|---|---|---|
| Powell soil | Vanoni soil | ||
| Tritirachium sp. (AB003951) | JK2-2 (99) | 11 | 13 |
| Pleospora rudis (U00975) | JK2-4 (97) | 9 | 2 |
| Petriella setifera (U43908) | JK3-18 (98) | 6 | 2 |
| Aspergillus versicolor (AB008411) | JK2-9 (99) | 2 | 5 |
| Chromocleista cinnabarina (AB006747) | JK2-22 (99) | 3 | 1 |
| Monilinia laxa (Y14210) | JK2-17 (96) | 1 | 2 |
| Exophiala jeanselmei (X80705) | JK3-21 (96) | 0 | 2 |
| Nectria cinnabarina (AB003949) | JK3-36 (98) | 0 | 1 |
| Verticillium dahliae (U33637) | JK3-30 (99) | 0 | 1 |
| Coccodinium bartschii (U77668) | JK3-25 (97) | 0 | 1 |
Fungal rDNA clone libraries from each soil were constructed by performing PCRs with the nu-SSU-0817-5′ and nu-SSU-1536-3′ primers. rDNA clones from each soil were randomly selected and sorted into OTUs by restriction enzyme analysis. The nucleotide sequence of one representative clone from each OTU was determined and analyzed by using BLAST (NCBI) and Gap (Genetics Computer Group) to identify the database sequence most similar to the clone and to obtain a phylogenetic identity for the OTU.
The numbers in parentheses are the percentages of similarity of the rDNA clones to the OTU sequences.
To test the primers further, nu-SSU-0817-5′ and nu-SSU-1536-3′ were used to examine fungi associated with avocado leaves. After epidemics of P. cinnamomi moved through several southern California avocado groves, the soils were determined to be more disease suppressive and were also covered with avocado leaves coated with fungi (unpublished observations). To identify these fungi, three randomly selected clones were analyzed, all of whose rDNA sequences showed similarity to previously described fungal rDNA sequences. Specifically, clones AL3, AL10, and AL15 showed similarity to Panellus serotinus (96%), Arthrobotrys dactyloides (98%), and Monilinia fructicola (99%), respectively. To test the nu-SSU-0817-5′–nu-SSU-1196-3′ primer pair, an rDNA clone library from the aforementioned Vanoni soil DNA was constructed. Three randomly selected clones from this library, JK1-1, JK1-14, and JK1-16, were analyzed and shown to have similarity to Fusarium oxysporum (98%), Echinosporangium transversale (98%), and Pseudallescheria ellipsoidea (100%), respectively. In these analyses and the other rDNA analyses performed with the primers developed in this study, no nonfungal rDNA sequences were identified, demonstrating the selectivity of these newly developed tools.
To compare our PCR primers with others designed for similar purposes, we also examined three recently developed fungal rDNA primer pairs described by Smit et al. (28). In our laboratory, two of these pairs (primers EF4 and EF3 and primers EF4 and fung5) amplified most of our fungal templates and some of our nonfungal templates (Fig. 2 and Table 2). The third primer pair, primers EF4 and NS3, produced no amplification products with any of the templates except Amoebidium parasiticum and Mucor rouxii (Fig. 2 and Table 2). For all three of these primer pairs, similar results were obtained with two different thermocyclers (data not shown). Except for the inability of primers EF4 and NS3 to amplify most of the fungal rDNA templates, these results were similar to those described by Smit et al. (28). To test the ability of these primers to amplify fungal rDNA from environmental samples, the primers were used in PCRs with DNA extracted from soil (Fig. 3). The resulting PCR products were then gel isolated and cloned. For the EF4-fung5 primer pair, 30 clones were sorted into three OTUs. A nucleotide sequence analysis of one representative clone from each OTU identified two sequences that do not have significant similarity to any rDNA or other database sequence and one sequence (JKX-3) that is similar to the fungus Opegrapha varia sequence (92%). For the EF4-NS3 primer pair, 29 clones were sorted into six OTUs. A nucleotide sequence analysis of these clones identified six sequences that do not have significant similarity to any rDNA or other database sequence. For the EF4-EF3 primer pair, a band of the correct size was not obtained and therefore there was no further analysis. These results show the variability that can occur with PCR-based techniques, as our soil analysis produced results that were significantly different from those obtained in a wheat rhizosphere analysis in which the same primers were used (28). They also reinforce the idea that PCR-based community structure studies should include some nucleotide sequence analysis as the resulting amplification products may not always be comprised of the intended DNA.
The results described in this report provide evidence that PCR primers nu-SSU-0817-5′, nu-SSU-1196-3′, and nu-SSU-1536-3′ will be useful tools for identifying fungi in environmental samples, such as soil and plant tissues. In addition, the primer sequences will likely be useful in the construction of fungal rDNA libraries from other environmental samples, for denaturation gradient gel electrophoresis analysis, or as fungus-specific hybridization probes. Fungi play important roles in agriculture, medicine, and ecosystems and are also considered critical components of human civilization and evolution (15). Indeed, fungi may have been essential in the evolution of land plants (15, 23). The scope of their functional roles and the extent of their diversity have yet to be understood, as most fungi remain undescribed (16). The PCR primers described in this report provide unique tools to further characterize this important group of organisms.
ACKNOWLEDGMENTS
We thank James Adaskaveg, Salomon Bartnicki-Garcia, Howard Judelson, John Menge, Elinor Pond, Sam Roberts, Karl Steddom, John Taylor, and Barbara Waaland for kindly providing tissue or DNA samples for this study.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apinis A E. Facts and problems. Mycopathol Mycol Appl. 1972;48:93–109. doi: 10.1007/BF02051773. [DOI] [PubMed] [Google Scholar]
- 3.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock M, Maiwald M, Kappe R, Nickel P, Naeher H. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses. 1994;37:79–84. doi: 10.1111/j.1439-0507.1994.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 5.Borneman J, Skroch P W, O'Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buts J P, Bernasconi P, Vaerman J P, Dive C. Stimulation of secretory IgA and secretory component of immunoglobulins in small intestine of rats treated with Saccharomyces boulardii. Dig Dis Sci. 1990;35:251–256. doi: 10.1007/BF01536771. [DOI] [PubMed] [Google Scholar]
- 7.Carroll G C, Wicklow D T. The fungal community: its organization and role in the ecosystem. 2nd ed. New York, N.Y: M. Dekker; 1992. [Google Scholar]
- 8.Cook R J, Baker K F. The nature and practice of biological control of plant pathogens. St. Paul, Minn: APS Press; 1983. [Google Scholar]
- 9.Downer A J. Ph.D. thesis. Riverside: University of California; 1998. [Google Scholar]
- 10.Dreyfuss M M, Chapela I H. Potential of fungi in the discovery of novel, low-molecular weight pharmaceuticals. In: Gullo V P, editor. Biotechnology series. 26. The discovery of natural products with therapeutic potential. Newton, Mass: Butterworth-Heinemann; 1994. pp. 49–80. [DOI] [PubMed] [Google Scholar]
- 11.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardes M, Bruns T D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 13.Gargas A, DePriest P T. A nomenclature for fungal PCR primers with examples from intron-containing SSU rDNA. Mycologia. 1996;88:745–748. [Google Scholar]
- 14.Harris D. Analyses of DNA extracted from microbial communities. In: Ritz K, Dighton J, Giller K E, editors. Beyond the biomass. Chichester, United Kingdom: John Wiley & Sons; 1994. pp. 111–118. [Google Scholar]
- 15.Hawksworth D L. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res. 1991;95:641–655. [Google Scholar]
- 16.Hawksworth D L, Rossman A Y. Where are all the undescribed fungi? Phytopathology. 1997;87:888–891. doi: 10.1094/PHYTO.1997.87.9.888. [DOI] [PubMed] [Google Scholar]
- 17.Henson J M, French R. The polymerase chain reaction and plant disease diagnosis. Annu Rev Phytopathol. 1993;31:81–109. doi: 10.1146/annurev.py.31.090193.000501. [DOI] [PubMed] [Google Scholar]
- 18.Kappe R, Fauser C, Okeke C N, Maiwald M. Universal fungus-specific primer systems and group-specific hybridization oligonucleotides for 18S rDNA. Mycoses. 1996;39:25–30. doi: 10.1111/j.1439-0507.1996.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 19.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: Wiley; 1991. pp. 115–175. [Google Scholar]
- 20.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 21.Makimura K, Murayama S Y, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 22.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 23.Pirozynski K A, Malloch D W. The origin of land plants: a matter of mycotrophism. BioSystems. 1975;6:153–164. doi: 10.1016/0303-2647(75)90023-4. [DOI] [PubMed] [Google Scholar]
- 24.Porter N, Fox F M. Diversity of microbial products: discovery and application. Pestic Sci. 1993;39:161–168. [Google Scholar]
- 25.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Simon L, Lalonde M, Bruns T D. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol. 1992;58:291–295. doi: 10.1128/aem.58.1.291-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smit E, Leeflang P, Glandorf B, van Elsas J D, Wernars K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–2621. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Driesche R G, Bellows T S. Biological control. New York, N.Y: Chapman & Hall; 1996. [Google Scholar]
- 30.Verweij P E, Meis J F G M, Van Den Hurk P, Zoll J, Samson R A, Melchers W J G. Phylogenetic relationships of five species of Aspergillus and related taxa as deduced by comparison of sequences of small subunit ribosomal RNA. J Med Vet Mycol. 1995;33:185–190. doi: 10.1080/02681219580000381. [DOI] [PubMed] [Google Scholar]
- 31.White T J, Bruns T, Lee S, Taylor J W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]