Abstract
A strategy to measure bacterial functional redundancy was developed and tested with soils collected along a soil reclamation gradient by determining the richness and diversity of bacterial groups capable of in situ growth on selected carbon substrates. Soil cores were collected from four sites along a transect from the Jamari tin mine site in the Jamari National Forest, Rondonia, RO, Brazil: denuded mine spoil, soil from below the canopy of invading pioneer trees, revegetated soil under new growth on the forest edge, and the forest floor of an adjacent preserved forest. Bacterial population responses were analyzed by amending these soil samples with individual carbon substrates in the presence of bromodeoxyuridine (BrdU). BrdU-labeled DNA was then subjected to a 16S-23S rRNA intergenic analysis to depict the actively growing bacteria from each site. The number and diversity of bacterial groups responding to four carbon substrates (l-serine, l-threonine, sodium citrate, and α-lactose hydrate) increased along the reclamation-vegetation gradient such that the preserved forest soil samples contained the highest functional redundancy for each substrate. These data suggest that bacterial functional redundancy increases in relation to the regrowth of plant communities and may therefore represent an important aspect of the restoration of soil biological functionality to reclaimed mine spoils. They also suggest that bacterial functional redundancy may be a useful indicator of soil quality and ecosystem functioning.
Identification of microbial community characteristics that influence terrestrial ecosystem performance is of considerable importance to ecology and conservation biology. One long-standing assumption has been that ecosystem stability and productivity are influenced by biodiversity (7, 16). This concept has been particularly well supported by empirical studies on plant communities (8, 15, 23, 31). For example, a long-term study of grasslands showed that more diverse plant communities were more resistant to a major drought (24). Recently, this concept has been extended to below-ground diversity, in which species loss at various trophic levels was shown to negatively influence several parameters such as respiration and organic-matter decomposition (19). When assessing biodiversity, meaningful characterization must consider not only the number and distribution of species but also functional diversity and redundancy. Functional diversity can be defined as the number of distinct processes or functions that are carried out by a community, whereas functional redundancy is a measure of the number of different species within the various functional groups or guilds (10).
One of the best community-based approaches for indexing the functional diversity of microbial communities has been the use of substrate or metabolic fingerprinting. Here, microbial communities are typically screened for their ability to utilize selected carbon substrates by using MicroPlates (Biolog, Hayward, Calif.) (9, 33). A potential limitation of this strategy is that the substrate utilization patterns are examined in liquid media, which may introduce a culture bias that can vary depending on the medium used (20, 22). This bias may lead to a depiction of the functional diversity that does not reflect the natural microbial community, since the majority of microorganisms cannot be ready cultured in standard media. An alternative technique that avoids this bias characterizes substrate utilization patterns by measuring CO2 respiration after selected carbon substrates are added to soil (5).
Although potentially an important ecosystem performance parameter, functional redundancy of microbial communities has not been well studied. Culture-based methods have been used to study the impact of heavy metals on the numbers of different species that are capable of using selected rare substrates (28). This work showed that elevated heavy metal levels resulted in a decline in the number of different microorganisms that used these substrates. It also provided the basis for concern about decreased reliability in the degradation of possibly toxic compounds that might eventually accumulate to undesirable levels in soils. Similar concern could also be expressed about any activity that causes a disturbance or decline in functional redundancy.
This report presents a culture-independent strategy to examine bacterial functional redundancy and tests its use with soils collected along a vegetation gradient on reclaimed mine spoils. This study focused on a tin mine spoil in Rondonia, Brazil, at a location that has proved to be extraordinarily difficult to revegetate. On this site, natural revegetation follows a pattern in which a few pioneer tree species eventually become established in spot locations and begin to deposit organic matter on the soil surface. Revegetation also proceeds from the forest edge, where there is dense plant growth due to the availability of light and seeds and from organic-matter deposition. As the forest is reestablished, the canopy closes between the pioneer community and the old forest growth. Introduction of organic matter along the vegetation gradient should increase the range of different niches that can be occupied by soil microorganisms. It was thus hypothesized that functional redundancy of the soil bacterial community on this mine spoil would increase in relation to the revegetation pattern and plant succession. The site sampled within the preserved forest had been previously characterized as one that was unusually rich in organic matter, a consequence of its prior prehistoric use as an Amazon Indian settlement. These anthropogenic soils (terra preta) resulted from hunting, gardening, and agroforestry and are distinguished by deep organic-rich soil profiles (6). Other survey evidence in this area suggests that the Indian group Karitiana, described as a small remnant population from Northern Amazonas (4), probably developed the site. The soil transect across the revegetation gradient thus provided a particularly interesting sample set for testing of the functional-redundancy analysis.
MATERIALS AND METHODS
Soil collection.
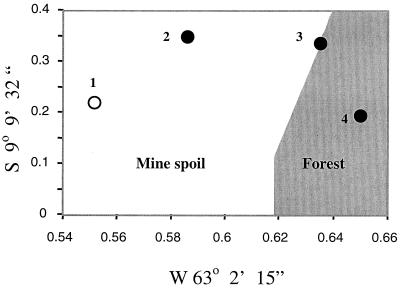
Four intact soil cores measuring 10 cm deep and 8 cm wide were collected using individual sterile soil-coring devices that encase the soil in a stainless steel sleeve with a removable ring on both the top and bottom of the core. The sleeved soil samples were then excavated, and the top and bottom rings were removed so that the interfacing soil at either end could be aseptically removed. The cores were then wrapped in aluminum foil and placed in boxes for transport to the laboratory. The soil cores were shipped to the laboratory at the University of California, Riverside, which required 3 days, and were then frozen at −70°C until used in the experiment. Soil cores were collected from denuded mine spoil approximately 100 m from the forest edge (soil site 1); from underneath a pioneer tree, Cecropia sciadophylla, in the same location (soil site 2); from the forest edge in dense vegetation (soil site 3); and from a prehistoric Amazon Indian archaeological site in the adjacent, mature preserved forest (soil site 4) (Fig. 1). The first three soils consisted of a coarse mine spoil material consisting largely of crushed rock and sand. The soil from the archeological site was characterized by a deep, organic matter-enriched A soil horizon that was approximately 1 m thick and overlaid a clay ultisol in the B horizon. In the laboratory, soil samples that had been stored at −70°C were taken from the freezer, removed from the cores, sieved to pass a 1.0-mm screen, and then air dried at 30°C.
FIG. 1.
Soil collection sites for samples analyzed in this study. The x and y axes designate the map coordinates for this site: 1, bare mine spoil approximately 100 m from the forest edge; 2, mine spoil from underneath a pioneer tree, Cecropia sciadophylla; 3, mine spoil from the forest edge in dense vegetation; and 4, undisturbed soil from a prehistoric Amazon Indian archaeological site in mature preserved forest.
Soil incubation and DNA extraction.
Soil samples (0.5 g) were amended with 5 mg of a single carbon substrate (either l-serine, l-threonine, sodium citrate, or α-lactose hydrate), 5 μl of 100 mM bromodeoxyuridine (BrdU), and 60 μl of H2O and incubated in petri dishes (60 by 15 mm) at room temperature in the dark. After 24 h, DNA was extracted from the soil samples with the FastDNA Kit for Soil as described by the manufacturer (Bio 101, Vista, Calif.).
Immunocapture of BrdU-labeled DNA.
Immunocapture of BrdU-labeled DNA was performed by a modification of previously described methods (2, 27). A 9-μl volume of herring sperm DNA (0.63 mg per ml in phosphate-buffered saline [PBS]) was heated at 95°C for 5 min, quickly cooled on ice for 5 min, mixed with 1 μl of anti-BrdU antibody (Boehringer Mannheim, Indianapolis, Ind.), and rotated at room temperature in the dark for 30 min. An 8-μl volume of BrdU-labeled DNA was combined with 2 μl of PBS, heated at 95°C for 5 min, quickly cooled on ice for 5 min, mixed with the herring sperm DNA–anti-BrdU antibody mixture, and rotated at room temperature for 30 min in the dark. Dynabeads (M-450) coated with sheep anti-mouse immunoglobulin G (3.13 μl) were washed once with 50 μl of PBS containing 0.1% bovine serum albumin per ml (PBS-BSA) as described by the manufacturer (Dynal, Lake Success, N.Y.), resuspended in 3.13 μl of PBS-BSA, and added to the BrdU-labeled DNA–anti-BrdU antibody mixture. This was rotated at room temperature for 30 min in the dark and washed three times each with 100 μl of PBS-BSA using magnetic separation. The pellets were resuspended in 10 μl of 1.7 mM BrdU in PBS-BSA and rotated at room temperature for 30 min in the dark, and the BrdU-labeled DNA fraction (supernatant) was collected by magnetic separation.
Community structure analysis.
Bacterial 16S-23S rRNA intergenic fragments were amplified in 10 μl of PCR mixture containing the following final concentrations or amounts: 1 μl of immunocaptured DNA or 1 μl of 1:40-diluted uncaptured soil DNA, 50 mM Tris (pH 8.3), 2.5 mM MgCl2, 500 μg of BSA per ml, 250 μM concentrations of each deoxynucleoside triphosphate, 2 μM forward primer 1406F (TGYACACACCGCCCGT) (11), 4 μM reverse primer 23SR (GGGTTBCCCCATTCRG) (3), and 5 U of Taq DNA polymerase. All reagents were combined and heated at 94°C for 2 min. Thirty-five cycles of PCR were performed at 94°C for 0 s, 52°C for 10 s, and 72°C for 30 s, followed by one episode at 72°C for 2 min. PCRs were performed in 10-μl glass capillary tubes with a model 1002 Rapidcycler (Idaho Technologies, Idaho Falls, Idaho).
Functional-redundancy analysis.
The 16S-23S rRNA intergenic-region PCR products were resolved by electrophoresis on 2% agarose gels and photographed. The DNA-banding profiles obtained from the photographs were converted into computer digital images using an image scanner. A peak analysis was performed to resolve the individual peaks and quantify the band intensities using Scion Image (Scion Corp., Frederick, Md.). The number and diversity of bacteria capable of utilizing each substrate were estimated by using the number of bands as a representation of the number of different organisms (richness) and the intensity of the bands as the number of individuals within an organism type. The diversity index used was Shannon's index: H = −Σ (ni/N) ln (ni/N), where ni is the area of each peak and N is the sum of all peak area.
RESULTS AND DISCUSSION
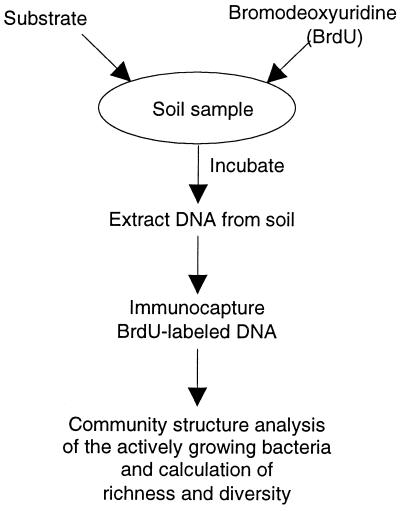
A culture-independent strategy to measure bacterial functional redundancy in environmental samples was developed. This approach is an extension of the aforementioned Biolog (9) and CO2 respiration (5) methods combined with a technique that permits the identification of actively growing bacteria (2). A carbon substrate and the thymidine analog BrdU are added to soil (Fig. 2). After incubation, the organisms that grow in response to the amendment are identified by a community structure analysis of the BrdU-labeled DNA, which is isolated by immunocapture. In this study, the bacterial community analyses were performed by resolution of PCR amplified 16S-23S rRNA intergenic-region fragments on 2% agarose gels, where the size heterogeneity of these fragments provides a simple method to depict bacterial community structure (3). The number of DNA bands provides a measure of the number of different organisms (richness), while the intensity of each band represents the number of individuals within an organism type. Since functional redundancy can be defined as the number of different species that perform a specified function, the richness of bacteria capable of utilizing a selected substrate can be used as a measure of this parameter. An alternative measure of functional redundancy can be obtained by measuring the bacterial diversity instead of the richness, providing an accounting of both the number of different organisms that perform a specified function and their distribution.
FIG. 2.
Strategy to measure bacterial functional redundancy in soil.
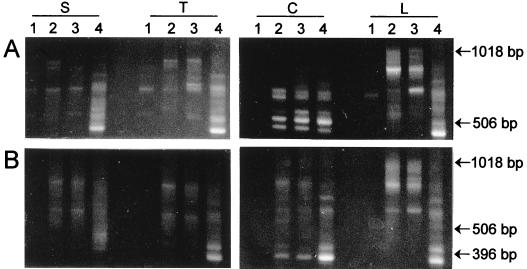
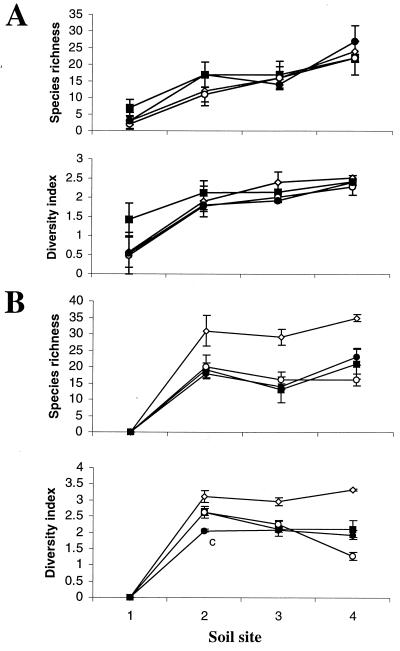
A test of the functional-redundancy analysis was done on four soils taken along a vegetation gradient from a tin mine site in Brazil which had been subjected to strip mining of the overburden (Fig. 1). With the exception of the preserved forest soil adjacent to the mine, the soil at this location was severely disturbed and depleted of organic matter since all of the overburden was processed by crushing, sieving, and washing of the gravel, which was then replaced over the mine spoil with no further amendments or topsoil. Several attempts to revegetate this site by tree plantings had failed, although after several years a few pioneer species had become established. For the functional-redundancy analysis, four different carbon substrates were selected: two amino acids, l-serine and l-threonine; one carboxylic acid, sodium citrate; and one carbohydrate, α-lactose hydrate (Fig. 3A). The selected substrates are representatives of different chemical groups commonly used in Biolog MicroPlates. The community structure banding patterns show that the soils capable of supporting more plant life also contain a greater variety of bacteria that responded to the carbon substrate amendments. The richness and diversity values, calculated from these data, confirm this visual observation for all substrates (Fig. 4A).
FIG. 3.
Bacterial community structure analysis of four soils collected along a mine reclamation gradient in Rondonia, Brazil, were performed on immunocaptured (A) and noncaptured (B) DNA. Lanes: 1, soil site 1; 2, soil site 2; 3, soil site 3; 4, soil site 4. The carbon substrates used were l-serine (S), l-threonine (T), sodium citrate (C), and α-lactose (L).
FIG. 4.
Bacterial species richness and Shannon diversity indices from four soils taken along a mine reclamation gradient were calculated by using the community structure data from Fig. 3. Four substrate amendments were used: l-serine (◊), l-threonine (■), sodium citrate (○), and α-lactose (●). Values were obtained from immunocaptured (A) and noncaptured (B) DNA. Error bars represent standard error measurements from three replicate PCR experiments.
For comparison, we also measured the total bacterial diversity of the four soils by analyzing the uncaptured DNA isolated in the substrate amendment experiments described in the previous paragraph. As expected, the uncaptured DNA showed higher diversity and richness values (Fig. 4B) than did immunocaptured samples (Fig. 4A). The rDNA fingerprints also showed that the four different carbon amendments had little impact on the total bacterial communities within each soil type (Fig. 3B) while revealing significant differences in the populations that grew in response to the substrates (Fig. 3A). In addition, these data showed that changes in soil quality, as evaluated by the ability to support plant growth along the reclamation gradient, correlated positively with bacterial functional redundancy whereas total bacterial richness and diversity were relatively constant for all soils except the denuded mine spoil (soil site 1). This result suggests that bacterial functional redundancy increases in relation to the regrowth of plant communities and may represent an important aspect of the restoration of soil biological functionality to reclaimed mine spoils in the Amazon forest.
Investigators in various disciplines such as agriculture, conservation, and reclamation science have attempted to measure soil quality. Abiotic measures of soil quality have included analyses of physical and chemical parameters, whereas biotic indicators have included measurements of enzyme activities, microbial biomass, and keystone species and enumerations of culturable microorganisms on agar media. Given the positive relationships between biodiversity and ecosystem performance identified in other studies (see the introduction for references), it may prove fruitful to use species diversity (26) and functional redundancy (this report) as indicators of soil quality. The molecular approach of Torsvik et al. (26) has been used to show that chemical pollutants decrease the diversity of bacteria in soil (1) and marine sediment (25). In our work, there was a clear positive correlation between the bacterial functional redundancy of soils and the regrowth of plant communities along a mine reclamation gradient. When evaluating biological diversity, it is important to distinguish between these two parameters, since changes in microbial diversity do not always correspond to changes in functional redundancy (1) (see above). Total bacterial diversity values provide a broad measure of biological diversity, whereas functional diversity determinations yield targeted assessments of the diversity within a functional group.
The extent and role of functional redundancy in microbial communities are unknown. Its purpose and importance has been debated, with some viewing it as an unnecessary luxury and others suggesting that it promotes reliability and is therefore an essential component of ecosystem productivity (12, 18, 31, 32). Determining the role of redundancy in ecosystem functioning is of significant importance and may provide the basis for more rational decisions concerning the value of biological diversity (32). The work described in this report should advance microbial ecology investigations by providing a new approach to examine bacterial functional redundancy in soil. To optimize this strategy toward a comprehensive functional-redundancy measurement, substrate selection could encompass a wide range of chemical groups; for example, the Eco MicroPlates (Biolog) use 31 different carbon substrates. Alternatively, this strategy could be used for highly focused investigations where only specific and narrowly defined substrates (not necessarily carbon) are used (30).
One current limitation of this functional-redundancy method is the resolving power of the community structure analysis. In this study, the community data were obtained using ribosomal intergenic spacer analysis (3). Other methods such as denaturing gradient gel electrophoresis (17) and terminal restriction fragment length polymorphism (13) could also be used. Unfortunately, none of these methods can accurately depict the true diversity of bacteria in a soil sample, since communities that may contain thousands of different species are resolved into approximately 10 to 50 groups. Thorough examination of rDNA clone libraries by extensive nucleotide sequence analysis, amplified rDNA restriction analysis (ARDRA) (14, 29), or future breakthroughs in DNA microarray approaches may significantly increase the resolution of this functional-redundancy analysis. Another potential limitation of this method is the breadth of organisms that incorporate BrdU. A definitive determination of this range will not be obtained without a comprehensive examination of numerous bacteria from all taxonomic groups. Thus far, with a few notable exceptions, it appears that the majority of bacteria incorporate [3H]thymidine (21). Since BrdU has been successfully used as a thymidine analog in numerous applications, it is likely that most organisms will also take up and incorporate BrdU. In addition, although the purpose of this strategy is to identify the organisms that respond to specified amendments, any organism that is growing at the time of the analysis will also be identified. Finally, in this research we attempted to preserve the soils as closely as possible by taking intact cores and then minimizing storage and contamination problems by freezing them. Ideally, samples such as these should be processed immediately. Studies examining functional redundancy and community structures will need to consider the best methods available for preserving samples prior to their analysis.
ACKNOWLEDGMENT
We thank FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) for financial support.
REFERENCES
- 1.Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microb Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 2.Borneman J. Culture-independent identification of microorganisms that respond to specified stimuli. Appl Environ Microbiol. 1999;65:3398–3400. doi: 10.1128/aem.65.8.3398-3400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callegarijacques S M, Salzano F M, Wwimer T A, Hutz T A, Black F F, Santos S E B, Guerreiro J F, Mestriner M A, Pandy J P. Further blood genetic-studies on Amazonian diversity—data from 4 Indian groups. Ann Hum Biol. 1994;21:465–481. doi: 10.1080/03014469400003482. [DOI] [PubMed] [Google Scholar]
- 5.Degens B P, Harris J A. Development of a physiological approach to measuring the catabolic diversity of soil microbial communities. Soil Biol Biochem. 1997;29:1309–1320. [Google Scholar]
- 6.Denevan W M. A bluff model of riverine settlement in prehistoric Amazonia. Ann Assoc Am Geogr. 1996;86:654–681. [Google Scholar]
- 7.Ehrlich P R, Ehrlich A H. Extinction: the causes and consequences of the disappearance of species. New York, N.Y: Random House; 1981. [Google Scholar]
- 8.Ewel J J, Mazzarino M J, Berish C W. Tropical soil fertility changes under monocultures and successional communities of different structure. Ecol Appl. 1991;1:289–302. doi: 10.2307/1941758. [DOI] [PubMed] [Google Scholar]
- 9.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaston K J. Biodiversity: a biology of numbers and difference. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1996. [Google Scholar]
- 11.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 12.Lawton J H, Brown V K. Redundancy in ecosystems. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem function. New York, N.Y: Springer Verlag; 1993. pp. 255–270. [Google Scholar]
- 13.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massol-Deya A A. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–8. [Google Scholar]
- 15.McNaughton S J. Biodiversity and function of grazing ecosystems. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem functioning. New York, N.Y: Springer Verlag; 1993. pp. 361–384. [Google Scholar]
- 16.McNaughton S J. Diversity and stability of ecological communities: a comment on the role of empiricism in ecology. Am Nat. 1977;111:515–525. [Google Scholar]
- 17.Muyzer G, Waal E C D, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naeem S. Species redundancy and ecosystem reliability. Conserv Biol. 1998;12:39–45. [Google Scholar]
- 19.Naeem S, Thompson L J, Lawler S P, Lawton J H, Woodfin R M. Declining biodiversity can alter the performance of ecosystems. Nature (London) 1994;368:734–737. [Google Scholar]
- 20.Ovreas L, Torsvik V. Microbial diversity and community structure in two different agricultural soil communities. Microb Ecol. 1998;36:303–315. doi: 10.1007/s002489900117. [DOI] [PubMed] [Google Scholar]
- 21.Robarts R D, Zohary T. Fact or fiction—bacterial growth rates and production as determined by (methyl-3H)-thymidine? Adv Microb Ecol. 1993;13:371–425. [Google Scholar]
- 22.Smalla K, Wachtendorf U, Heuer H, Liu W-T, Forney L. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–1225. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swift M J, Anderson J M. Biodiversity and ecosystem function in agricultural systems. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem function. New York, N.Y: Springer Verlag; 1993. pp. 15–41. [Google Scholar]
- 24.Tilman D, Downing J A. Biodiversity and stability in grasslands. Nature (London) 1994;367:363–365. [Google Scholar]
- 25.Torsvik V, Daae F L, Sandaa R A, Ovreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 26.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urbach E, Vergin K L, Giovannoni S J. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl Environ Microbiol. 1999;65:1207–1213. doi: 10.1128/aem.65.3.1207-1213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Beelen P, Doelman P. Significance and application of microbial toxicity tests in assessing ecotoxicological risks of contaminants in soil and sediment. Chemosphere. 1997;34:455–499. [Google Scholar]
- 29.Vaneechoutte M, Rossau R, Devos P, Gillis M, Janssens D, Paepe N, Derouck A, Fiers T, Claeys G, Kersters K. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 30.Victorio L, Gilbride K A, Allen D G, Liss S N. Phenotypic fingerprinting of microbial communities in wastewater treatment systems. Water Res. 1996;30:1077–1086. [Google Scholar]
- 31.Vitousek P M, Hooper D U. Biological diversity and terrestrial ecosystem biogeochemistry. In: Schulze E D, Mooney H A, editors. Biodiversity and ecosystem functioning. New York, N.Y: Springer Verlag; 1993. pp. 3–14. [Google Scholar]
- 32.Walker B H. Biodiversity and ecological redundancy. Conserv Biol. 1992;6:18–23. [Google Scholar]
- 33.Zak J C, Willig M R, Moorhead D L, Wildman H G. Functional diversity of microbial communities: a quantitative approach. Soil Biol Biochem. 1994;26:1101–1108. [Google Scholar]