Abstract
An understanding of the factors influencing colonization of the rhizosphere is essential for improved establishment of biocontrol agents. The aim of this study was to determine the origin and composition of bacterial communities in the developing barley (Hordeum vulgare) phytosphere, using denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA genes amplified from extracted DNA. Discrete community compositions were identified in the endorhizosphere, rhizoplane, and rhizosphere soil of plants grown in an agricultural soil for up to 36 days. Cluster analysis revealed that DGGE profiles of the rhizoplane more closely resembled those in the soil than the profiles found in the root tissue or on the seed, suggesting that rhizoplane bacteria primarily originated from the surrounding soil. No change in bacterial community composition was observed in relation to plant age. Pregermination of the seeds for up to 6 days improved the survival of seed-associated bacteria on roots grown in soil, but only in the upper, nongrowing part of the rhizoplane. The potential occurrence of skewed PCR amplification was examined, and only minor cases of PCR bias for mixtures of two different DNA samples were observed, even when one of the samples contained plant DNA. The results demonstrate the application of culture-independent, molecular techniques in assessment of rhizosphere bacterial populations and the importance of the indigenous soil population in colonization of the rhizosphere.
The use of antagonistic bacteria for the protection of crops against soilborne pathogens provides a promising environment-friendly alternative to chemical pesticides. However, the root colonization efficiency of introduced biocontrol strains is often limited, potentially reducing the effectiveness of protection (8, 25, 32). The selection and use of biocontrol strains therefore depend heavily on our knowledge of survival of the inoculant and its potential activity in the rhizosphere ecosystem of a particular plant and soil. Consequently, successful biological control with inoculated strains requires an understanding of the dynamics and composition of the bacterial communities colonizing the rhizosphere.
Previous studies, employing cultivation-based, laboratory methods or microscopy, have shown that different bacterial populations are present or active at different stages of root development and that rhizosphere communities are distinct from those found in bulk soil (1, 14, 17, 19, 21, 27, 30). However, recent molecular studies involving PCR amplification of 16S rRNA genes (rDNA), question some of these results. Duineveld et al. (9) reported that the bacterial communities of the Chrysanthemum rhizosphere, as measured by denaturing gradient gel electrophoresis (DGGE) of 16S rDNA PCR products, changed very little with plant age and were similar to those of bulk soil. In contrast, different bacterial communities were identified in soil and in the root tissue of white clover and ryegrass by cluster analysis of a 16S rDNA clone library (16). Furthermore, DGGE analysis has revealed different bacterial communities along barley roots (33). Hence, it seems that 16S rDNA-based techniques, in particular DGGE, give rise to contradictory results, and additional, more-detailed studies involving DGGE are needed to enable interpretation and validation of this approach to rhizosphere community studies.
In DGGE and temperature gradient gel electrophoresis (TGGE), PCR-amplified 16S rDNA products with the same length but with different sequence can be separated on a gel, resulting in unique fingerprints of environmental DNA samples. DGGE or TGGE analysis does not require laboratory cultivation of bacteria and consequently enables assessment of the diversity of total bacterial populations, including nonculturable organisms that may constitute 90 to 99% of the total rhizosphere bacteria (4). Molecular methods, including PCR amplification, may, however, introduce bias from a number of sources which will influence assessment and interpretation of true bacterial biodiversity (10, 12, 22, 29).
The efficiency of bacterial seed inoculants can be considerably improved by optimizing the presowing conditions, such as the formulation of the seed coating, the number and viability of bacteria applied, and the germination stage of the seed. Entrapment of bacteria in granular peat or polymer gels (32), optimization of the inoculum density (11), and pregermination of seeds (5) have been shown to improve root colonization by seed-associated bacteria. However, previous studies examining the importance of the presowing state have focused on individual strains, and knowledge of the root colonization capacity of broad bacterial groups is lacking.
The aim of this study was to analyze the composition and origin of barley-colonizing bacterial communities by DGGE analysis. Separate habitats were studied, defined as the endorhizosphere, the rhizoplane, and the rhizosphere soil, and the importance of presowing conditions was examined by germinating the barley seeds for up to 6 days prior to sowing. Unique bands on the DGGE gels were identified and sequenced to obtain phylogenetic information. To verify the validity of the results, the potential occurrence of skewed PCR amplification between samples was assessed.
MATERIALS AND METHODS
Growing of barley plants.
Barley seeds (Hordeum vulgare cv. Pastoral) were pregerminated on moist filter paper for 2 days. Individual seedlings with primary roots 5 to 10 mm long were planted in pots containing approximately 130 g of a sandy loam soil (Insch, Scotland). The soil had a water content of 28% (wt/wt), a pH of 6.6 (measured in water), and an organic matter content of 3.6% (wt/wt). Prior to use, the soil was sieved (mesh size, 3 mm). The pots were incubated in a growth chamber at 20 to 22°C and 60% relative humidity with a 12-h light, 12-h darkness cycle. On alternate days, 10 to 12 ml of tap water was added to the top-soil to maintain water content at 28% (wt/wt).
Sampling of barley microcosms.
Triplicate barley microcosms were sampled 6, 12, 18 and 36 days after sowing. Bulk soil samples (0.3 g) were obtained from subsurface soil not associated with the roots, and control soil samples (0.3 g) were taken from pots containing no plant. Each soil sample was transferred to a 2-ml Ribolyser tube containing 0.5 g of a mixture of ceramic and silica beads (Hybaid Ltd., Ashford, United Kingdom), 300 μl of NaPO4 buffer (0.12 M; pH 8.0), and 200 μl of Tris-HCl (1.0 M; pH 8.0).
Root samples were divided into rhizosphere soil, rhizoplane, and endorhizosphere fractions. The rhizosphere soil, defined as the soil firmly adhering to the roots (0.2 to 0.5 g of soil per sample), was removed manually and transferred to a Ribolyser tube with the chemicals described above. The roots were separated from the seed and transferred to a glass test tube with 10 ml of sterile MilliQ water. The samples were vortexed at full speed for 60 s, sonicated for 5 min in an ultrasonic bath, and vortexed for an additional 60 s. The root material was then removed, and the remaining extract was centrifuged at 15,000 × g for 10 min. The pellet (the rhizoplane fraction) was resuspended in 300 μl of NaPO4 buffer and 200 μl of Tris-HCl and transferred to a Ribolyser tube. The root material (the endorhizosphere fraction) was frozen by the addition of liquid nitrogen and ground in a porcelain mortar (8), resuspended in 300 μl of NaPO4 buffer and 200 μl of Tris-HCl, and transferred to a Ribolyser tube.
Leaf and seed samples were each divided into two fractions to provide phylloplane, endophyllosphere, spermoplane, and endospermosphere fractions. The fractions were obtained as described above for the rhizoplane and endorhizosphere fractions, except that sterile-filtered Tween 20 was added to the glass tubes with the leaf samples at a concentration of 1% (wt/wt) to improve the extraction of bacteria from the hydrophobic leaf material.
To study the impact of establishment of bacterial communities on barley seedlings prior to sowing, seeds were pregerminated on moist filter paper for up to 6 days. The seedlings were then grown in soil for 6 days (triplicate samples) and sampled as described above, but with the root divided into upper (near the seed) and lower regions of equal length.
Extraction and purification of DNA.
Water-saturated phenol (500 μl; pH 8.0) was added to each sample in a Ribolyser tube. Cells were lysed mechanically in a Hybaid Ribolyser cell disrupter at speed 4 twice for 10 s. Further phenol-chloroform purification, Microcon dialysis, and low-melting-point agarose gel purification of DNA were performed as described by Stephen et al. (28). Gel bands containing DNA of more than approximately 12 kb were excised, and DNA was finally purified with the Hybaid recovery DNA purification kit II.
PCR amplification of 16S rDNA.
PCR amplifications were performed with the universal eubacterial primers F341 (GC clamped) and R534 (18). Each PCR mixture contained 10 to 50 ng of DNA template, a 0.4 μM concentration of each primer, a 250 μM concentration of each deoxynucleoside triphosphate, 5 μl of reaction buffer (10×; pH 8.8; Bioline, London, United Kingdom), 1.5 mM MgCl2, 0.4 mg of bovine serum albumin ml−1, 1 U of BIOTAQ polymerase (Bioline), and sterile MilliQ water to 50 μl. Thirty amplification cycles were carried out as follows: one cycle consisting of an initial denaturation at 95°C for 5 min followed by primer annealing at 50°C for 30 s and primer extension at 72°C for 30 s; 29 cycles of 94°C (92°C for the final 15 cycles) for 30 s, 50°C for 30 s, and 72°C for 45 s; and a final step consisting of 10 min of incubation at 72°C. The DNA content was quantified by running 1 μl of each PCR mixture on a 1.2% (wt/vol) agarose gel stained with ethidium bromide.
DGGE and sequencing.
Approximately 200 to 400 ng of DNA of selected PCR products was loaded on 8% (wt/vol) polyacrylamide gels containing a top-to-bottom linear denaturant gradient of 30 to 70% (where 100% corresponds to 40% [vol/vol] formamide and 7 M urea). Electrophoresis was performed by running the gels at 60°C and 200 V for 5 h in a DGGE chamber (Bio-Rad Laboratories, Hercules, Calif.) containing approximately 7 liters of TAE buffer (40 mM Tris, 20 mM acetate, 1 mM EDTA). Gels were silver stained (24) and photographed with a digital camera. Bands were detected, aligned, and linked with the Dendron 2.2 software (Solltech Inc., Oakdale, Iowa) and dendrograms were constructed by simple matching (SM) of unweighted pair groups with mathematical averages using the NT-SYS program (Exeter Software, New York, N.Y.). Banding patterns with a level of similarity of 70% or more (SSM ≥ 0.7) were grouped into clusters. The goodness of fit for each cluster analysis was interpreted by comparing the cophenetic value matrix with the SM matrix (cophenetic correlation, r).
Additional gels were stained with ethidium bromide, and unique bands were excised and sequenced to obtain phylogenetic information and to verify bands that were suspected to contain plant DNA. The DNA was eluted overnight at 5°C in sterile MilliQ water, reamplified with the eubacterial primer set, and purified by Microcon dialysis. Sequencing of the PCR products was performed using the BigDye Terminator cycle sequencing kit (PE Biosystems, Warrington, United Kingdom), and sequencing products were analyzed with a model ABI 377 automated sequencer (PE Biosystems) using the R534 primer. The DNA sequences obtained were compared to sequences in nucleotide databases using the BLAST search program (http://www.ncbi.nlm.nih.gov).
Control for differential PCR amplification.
To determine whether the DNA templates of a particular sample type were preferentially amplified, DNA from different samples was mixed at concentration ratios of 1:100, 1:10, 1:1, 10:1, and 100:1 prior to PCR amplification. The PCR products obtained were analyzed by DGGE, and the banding patterns were identified with the Dendron 2.2 software.
RESULTS
Bacterial community composition in barley microcosms.
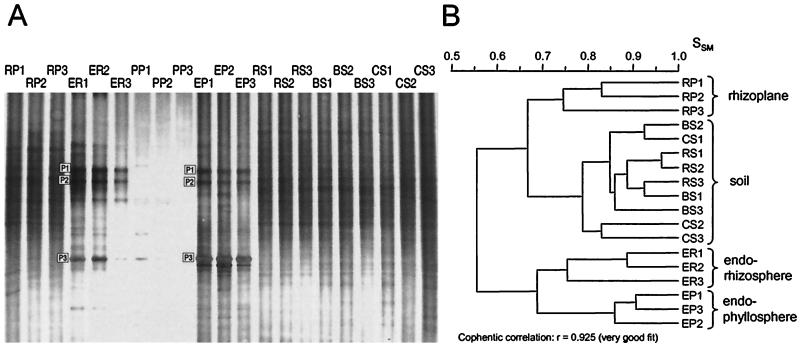
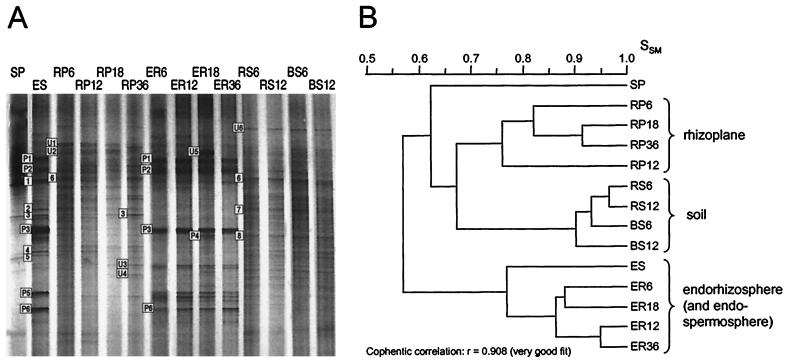
Bacterial community composition was investigated by PCR amplification and DGGE analysis of 16S rRNA genes from DNA extracted from plant and soil samples from microcosms containing 6- to 36-day-old barley plants. A comparison of the DGGE banding patterns of replicate samples from day 6 revealed a relatively high degree of reproducibility for samples from each habitat (Fig. 1A), with similarity coefficients, SSM, ranging from 0.75 to 0.96 in triplicate samples (Fig. 1B). In general, therefore, patterns from triplicates formed discrete clusters, with the exception of the soil samples (rhizosphere, bulk, and control) which formed a single cluster, reflecting a high degree of similarity between the different soil habitats (Fig. 1B). Similar clustering profiles and SSM values were obtained on the remaining sampling days (data not shown). To facilitate comparison of DGGE profiles from different sampling days on the same gel, the PCR products of triplicate samples were pooled prior to DGGE analysis. This approach was employed to assess changes in the community composition in relation to plant age (Fig. 2A), and the emergence or disappearance of bands, as a function of sampling day, was observed in a few cases. At least two bands (U3 and U4) emerged in the rhizoplane at day 36, and two unique bands (U5 and P4) appeared in the endorhizosphere sample at day 18 (Fig. 2A; Table 1). However, based on these observations and the cluster analysis (Fig. 2B), the differences were not sufficiently significant to assign any relationship between plant age and bacterial community composition.
FIG. 1.
(A) DGGE analysis of 16S rDNA fragments from soil and barley samples obtained 6 days after sowing. The gel shown is the original photograph before straightening of lanes and alignment of bands. P1 to P3 refer to bands with barley rDNA (see Table 1). (B) Dendrogram showing the relatedness of the DGGE banding patterns. Bands with barley DNA and the phylloplane samples, which contained no bacterial bands, were not considered in the cluster analysis. Abbreviations: RP, rhizoplane; ER, endorhizosphere; PP, phylloplane; EP, endophyllosphere; RS, rhizosphere soil; BS, bulk soil; CS, control soil. The number after each abbreviation indicates replicate number.
FIG. 2.
(A) DGGE analysis of 16S rDNA fragments from soil and barley samples obtained 6, 12, 18, and 36 days after sowing. The gel shown is the original photograph before straightening of lanes and alignment of bands. PCR products of triplicate samples were pooled prior to the DGGE analysis. Sequenced bands are labeled; 1 to 8 refer to bacterial bands, and P1 to P6 refer to bands with barley rDNA (see Table 1). U1 to U6 refer to bands that could not be sequenced. (B) Dendrogram showing the relatedness of the DGGE banding patterns. Bands with barley DNA were not considered in the cluster analysis. Abbreviations: SP, spermoplane; ES, endospermosphere; RP, rhizoplane; ER, endorhizosphere; RS, rhizosphere soil; BS, bulk soil. The number after each abbreviation indicates sampling day. For the SP and ES samples, the RP and ER of the 5- to 10-mm-long primary roots are included.
TABLE 1.
Results of sequence analysis of selected DGGE bands labeled in Fig. 2
| Band no. | Habitat(s)a | Most similar sequence (accession no.) | Homology (%) |
|---|---|---|---|
| 1 | WT, SP, ES, RPUP6+6, ER | Acinetobacter sp. DSM590 (X81659) | 97 |
| 2 | SP6, ES | Burkholderia sp. (U96927 and more) | 99 |
| 3 | SP, ES, RP | Uncultured soil bacterium C026 (AF128717) | 96 |
| 4 | SP6, ES | Pantoea agglomerans (Z96082 and 2 more) | 100 |
| 5 | SP, ES | Pantoea agglomerans (AF157694) | 99 |
| 6 | RP, RS, BS | Bacillus sp. (AF071856) | 96 |
| 7 | RS, BS | Bacillus megaterium (Y15052) | 76 |
| 8 | RS, BS | Burkholderia sp. (U96927 and more) | 83 |
| 9 | WT, SP6, RPUP6+6 | Pseudomonas sp. (AF184918 and more) | 100 |
| P1 | ES, ER, EP | Zea mays 18S rRNA (AF168884)b | 86 |
| P2 | ES, ER, EP | Zea mays 18S rRNA (AF168884)b | 84 |
| P3 | ES, ER, EP | Triticum aestivum chloroplast 16S rRNA (AJ239003)b | 98 |
| P4 | ER 18 | Triticum aestivum chloroplast 16S rRNA (AJ239003)b | 94 |
| P5 | ES, EP | Triticum aestivum mitochondrion 18S rRNA (Z14060)b | 89 |
| P6 | ES, ER, EP | Cytinus ruber mitochondrion 18S rRNA (U82639)b | 81 |
Abbreviations as in Fig. 3.
rDNA sequences for barley do not appear in the nucleotide databases.
The samples grouped into four main clusters, representing the rhizoplane, soil, endorhizosphere, and endophyllosphere (Fig. 1B and 2B). The rhizoplane samples showed a higher similarity to the soil samples (SSM, 0.67) than to the endorhizosphere samples (SSM, 0.55 to 0.57) (Fig. 1B and 2B). The spermoplane sample of ground 2-day-old seedlings was distinct from the other samples, with the highest resemblance to the rhizoplane and soil samples (SSM, 0.62) (Fig. 2B). Hence, the composition of the bacterial communities on the rhizoplane was comparable to that found in the surrounding soil (rhizosphere and bulk) but not to the composition seen in the root tissue or the seedlings. Finally, the DGGE profiles of the endophyllosphere revealed the closest resemblance to those of the endorhizosphere and the endospermosphere (SSM, 0.68 to 0.77) (Fig. 1B; gel with pooled endophyllosphere samples not shown).
Improved survival of seed-borne bacteria.
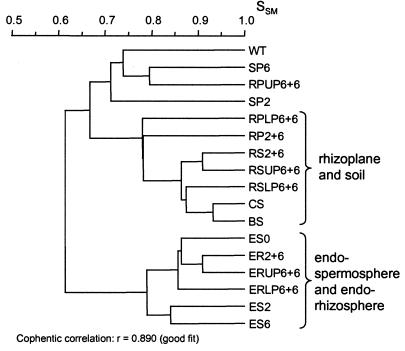
To determine whether it is possible to improve the capacity of seed-associated bacteria to colonize the root, barley seeds were pregerminated for 6 days prior to sowing, rather than 2 days. The seedlings were then grown for 6 days in soil and samples were obtained for DGGE analysis as previously described, but with the root divided into upper and lower regions. As the 6-day-old seedings, on average, doubled in length from 6 to 12 cm during the 6-day-incubation period in soil, the upper part of the root system closely resembled the length of the original seedling while the lower part was a result of root extension after sowing.
The bacteria colonizing a seedling presumably originate from the nongerminated seed or the water source (tap water). No PCR product was obtained from the spermoplane samples of nongerminated seeds, indicating a very low number of bacteria on the seed surface. The water sample gave rise to six DGGE bands, of which three also appeared on the surface of the 2- and 6-day-old seedlings (SP2 and SP6) and in the upper part of the rhizoplane of plants grown in soil (RPUP6+6). Two of the bands contained sequences similar to Acinetobacter sp. and Pseudomonas sp. (bands 1 and 9 in Table 1).
Cluster analysis of DGGE patterns revealed a soil-rhizoplane cluster and an endorhizosphere-endospermosphere cluster (Fig. 3). The DGGE profile of the lower part of the rhizoplane (RPLP6+6) had the highest resemblance to the profiles of the soil samples and the rhizoplane sample originating from 2-day-old seedlings (SSM, 0.78) (Fig. 3). However, the DGGE profile of the upper part of the rhizoplane (RPUP6+6) was more similar to the profiles of the 2- and 6-day-old seedlings and the water sample (SSM, 0.71, 0.80, and 0.74, respectively) than to the soil-rhizoplane cluster (SSM, 0.67) (Fig. 3).
FIG. 3.
Dendrogram for DGGE banding patterns of barley samples showing the impact of seed pregermination. Abbreviations: WT, water source; SP, spermoplane; ES, endospermosphere; RP, rhizoplane; ER, endorhizosphere; RS, rhizosphere soil; BS, bulk soil; CS, control soil; UP, upper part of root; LP, lower part of root; 2+6 and 6+6, number of days of seed pregermination + number of days of growth in soil. For the SP and ES samples, the RP and ER of the 5- to 10-mm-long primary roots are included.
Bacterial identities in relation to habitat.
Between 10 and 27 bacterial bands of various intensities were detected in all samples, except for the water and spermoplane samples, where fewer than 10 bands were detected, and the phylloplane samples, where no bacterial band appeared. Seven different bacterial species were identified by comparing the sequences obtained to nucleotides in databases (Table 1). Mitochondrial, chloroplast, and 18S plant rDNA were detected in the plant tissue samples and commonly resulted in strong bands, indicating large amounts of plant DNA in the original sample. Six bands gave rise to nonsense or poor quality sequences (U1 to U6 in Fig. 2A), possibly due to the occurrence of two or more different PCR products in the same band.
Four bands (1, 2, 4, and 5) representing species of the genera Acinetobacter and Burkholderia and the species Pantoea agglomerans were specific to the barley seedlings, and one band (3), representing an unknown 16S rDNA sequence, was found on both the seedlings and in the rhizoplane (Table 1 and Fig. 2A). Band 6, representing Bacillus sp., was common to the rhizoplane and soil samples, whereas bands 7 and 8, representing Bacillus megaterium and Burkholderia sp., only appeared in the soil samples (Table 1 and Fig. 2A). Furthermore, Acinetobacter sp. and Pseudomonas sp. were detected in the water source as described in the previous subsection.
Control for differential PCR amplification.
In four instances two different DNA samples were mixed at different concentrations prior to PCR amplification to test whether the DNA templates of one sample were preferentially amplified. When mixed at the concentrations 10:1 and 100:1, the banding pattern of the sample with the higher concentration completely dominated (data not shown). However, differential PCR amplification was observed to some extent when DNA samples were mixed 1:1. From 9 to 19% of the DGGE-bands of the two individual samples were not present in the lane of bands based on a 1:1 mixture (Table 2). However, in all cases the repressed bands appeared as faint bands when the samples were analyzed separately.
TABLE 2.
Band repression measured after DGGE of PCR-amplified 1:1 DNA sample mixtures
| Sample 1a | Sample 2a | No. of bands absent, in 1:1 mixture
|
Total no. of bands detected | % of bands, absent | |
|---|---|---|---|---|---|
| Sample 1 | Sample 2 | ||||
| ER18 | BS18 | 0 | 3 | 33 | 9.1 |
| SP6 | CS6 | 5 | 1 | 32 | 18.8 |
| ER6 | RS6 | 2 | 3 | 31 | 16.1 |
| RPUP6+6 | RSUP6+6 | 2 | 2 | 32 | 12.5 |
Abbreviations as in Fig. 3
DISCUSSION
The results obtained by DGGE of 16S rDNA indicated that different locations of the barley phytosphere harbored distinct bacterial communities. The DGGE banding profiles for samples representing the same phytosphere or soil habitat were reproducible but with occasional variations between replicates. Yang and Crowley (33) observed comparable variations in the barley rhizosphere and suggested that these may result from stochastic events during plant growth, such as the colonization of the root by various microorganisms as the root elongates through soil and organic matter particles. Also, the heterogeneity of soil with respect to pore structure, humidity, and organic matter content may lead to variations in the bacterial populations. In our study, differences due to natural variability were reduced, but not eliminated, by pooling triplicate samples prior to DGGE analysis. For example, the emergence of two unique DGGE bands in the 18-day-old endorhizosphere most probably was a result of a random event, as they were present in a single replicate only (data not shown).
The physical presence of a root surface and the release of organic root exudates, such as amino acids and sugars, are believed to influence the composition of bacterial communities in the rhizosphere (7, 23, 31). Our study indicates that the influence of root exudates on bacterial community composition did not extend further than 1 mm from the barley root surface, as no difference could be detected between the DGGE banding profiles of rhizosphere soil and bulk soil. The banding patterns of the rhizoplane, however, were different from those found in soil. Nevertheless, they revealed a higher resemblance to those in the soil than to the patterns associated with the root tissue and the seedlings. This suggests that rhizoplane bacteria primarily originated from the surrounding soil. Marilley and Aragno (16) studied rhizosphere bacterial communities, by sequence analysis of a 16S rDNA clone library, and found that communities in the rhizoplane-endorhizosphere of Trifolium repens and Lolium perenne differed from those of rhizosphere soil and bulk soil. In contrast, Duineveld et al. (9), using DGGE, found no apparent rhizosphere effect, with bacterial communities in the Chrysanthemum rhizosphere and in bulk soil being indistinguishable. Different definitions of root habitats may explain the discrepancies in the observations, although factors such as soil type and plant species also may influence the results. We suggest that the occurrence of soil particles in rhizosphere samples will lead to a bacterial community composition similar to that of soil, but by distinguishing between endorhizosphere, rhizoplane, and rhizosphere this hindrance can be overcome.
Several root-associated bacterial species were identified by sequence analysis but none matched species identified in another recent DGGE study of the barley rhizosphere (33). However, all species have previously been observed in the rhizosphere environment (2, 3, 6, 13, 16). In particular, Acinetobacter, Burkholderia, Pantoea agglomerans, and Pseudomonas species appeared in a 16S rDNA clone library obtained from L. perenne and T. repens roots (16).
No changes in banding profiles as a function of plant age could be observed for any of the examined habitats. Similarly, Duineveld et al. (9) observed no clear temporal or spatial changes in the bacterial DGGE profiles derived from the rhizosphere of 2- to 10-week-old Chrysanthemum plants. In contrast, differences in bacterial communities between old and young (root tip) regions of wheat and barley roots have been observed in at least two culture-dependent studies (15, 26) and in one culture-independent study involving DGGE (33). However, a comparison of the results of the studies described above and our own leads us to suggest that the community composition on young root tips may be unique, but that nongrowing regions of the root will instantaneously develop a constant community composition which is, as shown in this study, comparable to that of the surrounding soil.
We were able to recover PCR-amplifiable 16S rDNA from all types of samples, except for the phylloplane samples. Possibly, the highly hydrophobic barley leaves effectively restricted any microbial colonization of the leaf surface, although bacterial DNA was detected in the interior of the leaves. The DGGE profiles of the endophyllosphere most closely resembled the profiles of the endospermosphere and endorhizosphere, indicating related community compositions in different parts of the interior of the barley plants. However, large amounts of barley DNA, released by the grinding procedure, may have repressed the PCR amplification of less-dominant bacterial DNA in the endophytosphere samples, since the primer set used also recognized plant rRNA sequences. Yang and Crowley (33) also observed chloroplast DNA on a DGGE gel of barley root samples. However, we only observed minor cases of skewed PCR amplification for mixtures of two different DNA samples, even when one of the samples contained plant DNA. Thus, the analysis of banding patterns of different sample types appeared reliable, although we cannot completely exclude differential amplification within individual samples (as specified by Heuer and Smalla [12]).
When using seed-borne biocontrol agents, it is essential that the presowing conditions be optimal to enable colonization of the roots by the introduced strain(s). By pregerminating the barley seeds for up to 6 days, we examined whether seed-associated bacteria can be stimulated to colonize plant roots grown in soil. The survival on the roots of bacteria originating from the seed or the water source did improve when the seeds were pregerminated, but only in the upper, nongrowing part of the rhizoplane. In the growing part of the rhizoplane none of the original predominant DGGE bands of the seedlings was recovered. This implies that emerging plant roots are colonized mainly by soilborne and not by seed- or root-borne bacteria, and it underlines a main obstacle using seed-borne bacteria as biocontrol agents: the insufficient colonization of the roots. Previous studies have been aimed at studying the fate of single seed-inoculated strains. A general approach has not, to our knowledge been attempted, but we suggest that studying the root colonization of broader bacterial groups, for example using group-specific primers or probes in DGGE analysis, will improve the knowledge considerably on how to select and use seed-inoculated biocontrol agents.
ACKNOWLEDGMENTS
B.N. was supported by a grant from the Danish Ministry of Environment and Energy.
We thank Gordon Webster and Katie Cuschieri for skillful assistance with the experimental work and Ole Nybroe for important comments on the manuscript.
REFERENCES
- 1.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence J R, Hartmann A. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg G. Rhizobacteria of oilseed rape antagonistic to Verticillium dahliae var. longisporum STARK. Zeitschr Pflanzenkrank Pflanzenschutz. 1996;103:20–30. [Google Scholar]
- 3.Berg G, Ballin G. Bacterial antagonists to Verticillium dahliae Kleb. J Phytopathol. 1994;141:99–110. [Google Scholar]
- 4.Campbell R, Greaves M P. Anatomy and community structure of the rhizosphere. In: Lynch J M, editor. The rhizosphere. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 11–35. [Google Scholar]
- 5.Creus C M, Sueldo R J, Barassi C A. Azospirillum inoculation in pregerminating wheat seeds. Can J Microbiol. 1996;42:83–86. [Google Scholar]
- 6.De Freitas J R, Banerjee M R, Germida J J. Phosphate-solubizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.) Biol Fertil Soils. 1997;24:358–364. [Google Scholar]
- 7.Dellaporta S L, Wood J, Hicks J B. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 8.De Weger L A, van der Bij A J, Dekkers L C, Simons M, Wijffelman C A, Lugtenberg B J J. Colonization of the rhizosphere of crop plants by plant-beneficial pseudomonads. FEMS Microbiol Ecol. 1995;17:221–228. [Google Scholar]
- 9.Duineveld B M, Rosado A S, Van Elsas J D, van Veen J A. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol. 1998;64:4950–4957. doi: 10.1128/aem.64.12.4950-4957.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen M C, Tolker-Nielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 11.Hatzinger P B, Alexander M. Relationship between the number of bacteria added to soil or seeds and their abundance and distribution in the rhizosphere of alfalfa. Plant Soil. 1994;158:211–222. [Google Scholar]
- 12.Heuer H, Smalla K. Application of denaturing gradient gel electrophoresis and temperature gradient gel electrophoresis for studying soil microbial communities. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 353–373. [Google Scholar]
- 13.Kremer R J, Begonia M-FT, Stanley L, Lanham E T. Characterization of rhizobacteria associated with weed seedlings. Appl Environ Microbiol. 1990;56:1649–1655. doi: 10.1128/aem.56.6.1649-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J-M, Alabouvette C. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol. 1995;61:1004–1012. doi: 10.1128/aem.61.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liljeroth E, Burgers S L G E, van Veen J A. Changes in bacterial populations along roots of wheat (Triticum aestivum L.) seedlings. Biol Fertil Soils. 1991;10:276–280. [Google Scholar]
- 16.Marilley L, Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. [Google Scholar]
- 17.Mavingui P, Laguerre G, Berge O, Heulin T. Genetic and phenotypic diversity of Bacillus polymyxa in soil and in the wheat rhizosphere. Appl Environ Microbiol. 1992;58:1894–1903. doi: 10.1128/aem.58.6.1894-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muyzer G, Dewaal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijhuis E H, Maat M J, Zeegers I W E, Aalwijk C, van Veen J A. Selection of bacteria suitable for introduction into the rhizosphere of grass. Soil Biol Biochem. 1993;25:885–895. [Google Scholar]
- 20.Normander B, Hendriksen N B, Nybroe O. Green fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere. Appl Environ Microbiol. 1999;65:4646–4651. doi: 10.1128/aem.65.10.4646-4651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsson S, Persson P. The composition of bacterial populations in soil fractions differing in their degree of adherence to barley roots. Appl Soil Ecol. 1999;12:205–215. [Google Scholar]
- 22.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rovira A D. Interactions between plant roots and soil microorganisms. Annu Rev Microbiol. 1965;19:241–266. doi: 10.1146/annurev.mi.19.100165.001325. [DOI] [PubMed] [Google Scholar]
- 24.Sanguinetti C J, Neto E D, Simpson A J G. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques. 1994;17:915–919. [PubMed] [Google Scholar]
- 25.Schippers B, Bakker A W, Bakker P A H M. Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu Rev Phytopathol. 1987;25:339–358. [Google Scholar]
- 26.Semenov A M, van Bruggen A H C, Zelenev V V. Moving waves of bacterial populations and total organic carbon along roots of wheat. Microb Ecol. 1999;37:116–128. doi: 10.1007/s002489900136. [DOI] [PubMed] [Google Scholar]
- 27.Söderberg K H, Bååth E. Bacterial activity along a young barley root measured by the thymidine and leucine incorporation techniques. Soil Biol Biochem. 1998;30:1239–1268. [Google Scholar]
- 28.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Peer R, Punte H L M, de Weger L A, Schippers B. Characterization of root surface and endorhizosphere pseudomonads in relation to their colonization of roots. Appl Environ Microbiol. 1990;56:2462–2470. doi: 10.1128/aem.56.8.2462-2470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 33.Yang C-H, Crowley D E. Rhizosphere microbial community structure in relation to root location and plant iron status. Appl Environ Microbiol. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]