Abstract
All sexually fertile strains in the Gibberella fujikuroi species complex are heterothallic, with individual mating types conferred by the broadly conserved ascomycete idiomorphs MAT-1 and MAT-2. We sequenced both alleles from all eight mating populations, developed a multiplex PCR technique to distinguish these idiomorphs, and tested it with representative strains from all eight biological species and 22 additional species or phylogenetic lineages from this species complex. In most cases, either an ∼800-bp fragment from MAT-2 or an ∼200-bp fragment from MAT-1 is amplified. The amplified fragments cosegregate with mating type, as defined by sexual cross-fertility, in a cross of Fusarium moniliforme (Fusarium verticillioides). Neither of the primer pairs amplify fragments from Fusarium species such as Fusarium graminearum, Fusarium pseudograminearum, and Fusarium culmorum, which have, or are expected to have, Gibberella sexual stages but are thought to be relatively distant from the species in the G. fujikuroi species complex. Our results suggest that MAT allele sequences are useful indicators of phylogenetic relatedness in these and other Fusarium species.
Fusarium isolates in the Gibberella fujikuroi species complex include important fungal pathogens of agricultural crops and trees and may be divided into at least eight different biological species and 32 additional asexual species or phylogenetic lineages (4, 10, 13, 15, 16). All sexually fertile species in the G. fujikuroi species complex are dimictic, i.e., two isolates are cross-fertile if they carry the different mating-type idiomorphs MAT-1 and MAT-2 (4, 6, 9, 13), which share no sequence similarity with respect to either DNA sequence or the proteins encoded (5). The MAT-2 idiomorphs thus far characterized have a conserved HMG (high-mobility-group) domain (3, 5, 6, 9). The MAT-1 idiomorphs have a conserved α-domain (5, 25), but no MAT-1 alleles have yet been described from any members of the G. fujikuroi species complex.
Our objectives in this study were (i) to sequence conserved portions of the MAT-1 and MAT-2 alleles from the eight known G. fujikuroi mating populations, (ii) to develop a multiplex PCR reaction to be used for the identification of both mating-type idiomorphs within the defined biological species of the G. fujikuroi species complex, (iii) to determine the range of Fusarium species which have Gibberella teleomorphs to which this technique can be successfully applied, and (iv) to test the use of the sequence of the MAT idiomorphs for phylogenetic analyses. This technique eases the identification of strains to be used in crosses to identify new biological species and eliminates the need for sexual crosses to score this trait. MAT sequence variations may be used as phylogenetic characters, providing another marker that can be used to test phylogenies for robustness.
(Portions of this work have been published in abstract form [E. T. Steenkamp, B. D. Wingfield, T. A. Coutinho, M. J. Wingfield, W. F. O. Marasas, and J. F. Leslie, Abstr., Phytopathology 89:S75, 1999].)
MATERIALS AND METHODS
Fungal isolates.
We examined 16 standard mating-type testers (4, 10, 13) from the eight described mating populations in the G. fujikuroi species complex, 128 progeny from the mapping population described by Xu and Leslie (23), and all of the strains examined by Kérenyi et al. (9). We also examined 37 additional strains representing 29 additional species or phylogenetic lineages; names indicated with an * are invalid (1). These strains were [species, strain number(s)] as follows: F. acuminatum MRC 7681, KSU X-05020, and FRC R-6666; F. acutatum MRC 7544, KSU X-10679, and BBA 69580; F. annulatum MRC 2577, KSU X-03831, FRC M-1220, and BBA 63629; F. anthophilum MRC 2578, KSU X-03818, FRC M-0854, and BBA 63270; F. avenaceum MRC 7680, KSU X-05017, and FRC R-6550; F. begonieae* MRC 7542, KSU X-10767, and BBA 67781; F. beomiforme MRC 4602, KSU X-05013, and FRC M-1088; F. brevicatenulatum* MRC 7531, KSU X-10756, and BBA 69197; F. bulbicola MRC 7534, KSU X-10759, and BBA 63628; F. concentricum* MRC 7540, KSU X-10765, and BBA 64354; F. crookwellense MRC 2878 and KSU X-04833; F. culmorum MRC 7682, KSU X-06576, and FRC R-5626; F. denticulatum* MRC 7538, KSU X-10763, and BBA 67772; F. dlamini MRC 3023, KSU X-05009, and FRC M-1557; F. graminearum (Gibberella zeae) MRC 7677, and KSU Z-03639; F. guttiforme* MRC 7539, KSU X-10764, and BBA 69661; F. lactis MRC 7532, KSU X-10757, and BBA 68590; F. napiforme MRC 3105, KSU X-05015, and FRC M-1646; F. nisikadoi MRC 7533, KSU X-10758, and BBA 69015; F. oxysporum f. sp. cubense MRC 7671 and KSU O-02332; F. oxysporum f. sp. chrysanthemi MRC 7672, KSU O-02523, and FRC O-734; F. oxysporum f. sp. niveum MRC 7673, KSU O-02529, and FRC O-1087; F. oxysporum f. sp. radicis-lycopersici MRC 7674, KSU O-02530, and FRC O-1090; F. oxysporum f. sp. vasinfectum MRC 7675, KSU O-02533, and FRC O-1139; F. phyllophilum MRC 2576, KSU X-03829, FRC M-1218, and BBA 62262, and MRC 7543, KSU X-10768, and BBA 63625; F. pseudoanthophilum* MRC 7530, KSU X-10755, and BBA 69002; F. pseudocircinatum* MRC 7678 and KSU X-04379, and MRC 7536, KSU X-10761, and BBA 69636; F. pseudograminearum (Gibberella coronicola) MRC 7670, KSU X-00629, and FRC R-5210; F. pseudonygamai* MRC 7537, KSU X-10762, and BBA 69552; F. ramigenum* MRC 7535, KSU X-10760, and BBA 68592; F. solani MRC 7676 and KSU X-03198; F. subglutinans (mango) MRC 7679 and KSU X-04706; and F. succisae MRC 2579, KSU X-03832, FRC M-1221, and BBA 63627. Strains were from the Medical Research Council (MRC), Tygerberg, South Africa; Kansas State University (KSU), Manhattan; the Fusarium Research Center (FRC), The Pennsylvania State University, University Park, Pennsylvania; and the Biologische Bundesanstalt für Land und Forstwirtschaft (BBA), Berlin, Germany.
DNA isolation and manipulation.
DNA manipulations and general molecular biology protocols followed those of Sambrook et al. (19). Fungal tissue was harvested from liquid cultures and ground to a powder in the presence of liquid nitrogen. DNA was isolated as previously described (9, 21), resuspended in deionized water or Tris-EDTA, and stored at −20°C.
PCR amplification of MAT-2.
We examined the conserved HMG domain and 3′-idiomorph flank of MAT-2, including the variable sequence between these regions. We initially used a previously described primer pair, NcHMG1 and NcHMG2 (3), to amplify the HMG domain from MRC 6213. This PCR reaction mixture contained 1 ng of DNA per μl, a 1 mM concentration of deoxynucleoside triphosphates (dNTPs) (0.25 mM concentrations of each), 2.5 mM MgCl2, 2 μM concentrations of each primer, and 0.05 U of Super-Therm DNA polymerase and reaction buffer (Southern Cross Biotechnology, Cape Town, South Africa) per μl. Reaction mixtures were overlaid with mineral oil to prevent evaporation. The initial denaturation at 92°C for 1 min was followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. A final extension was performed at 72°C for 10 min. Fragments were resolved and sized on a 2% agarose gel in 0.5× Tris-borate-EDTA. A 300-bp fragment was excised from the gel and purified with the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), after which it was cloned into the pCR-Script Amp SK(+) vector (Stratagene Cloning Systems, La Jolla, Calif.). Plasmids were harvested by alkaline lysis, and inserts were sequenced using M13 forward and reverse primers. Based on this sequence, we designed a Fusarium-specific primer for the 5′ end of the HMG domain, Gfmat2c (5′-AGCGTCATTATTCGATCAAG-3′).
To amplify the HMG domain and a portion of the conserved 3′-idiomorph flank, we performed PCR with the primers Gfmat2c and Fo14. Fo14 (25) is part of the conserved 3′-idiomorph flank from F. oxysporum (GenBank accession no. AB011378). The PCR conditions were the same as those described for the degenerate PCR, except that 0.1 μl of the primers Gfmat2c and Fo14 were used. The ∼900-bp PCR products from each of the eight G. fujikuroi MAT-2 tester strains were sequenced, and these sequences were used to design a second Fusarium-specific MAT-2 primer GFmat2d (5′-CTACGTTGAGAGCTGTACAG-3′). Gfmat2c and Gfmat2d can be used to amplify an 800-bp fragment that includes part of the conserved HMG domain and the 3′-idiomorph flank, as well as a variable sequence between these regions. We also analyzed some strains using the GfHMG1 and GfHMG2 primers and the PCR amplification conditions of Kerényi et al. (9).
PCR amplification of MAT-1.
We used the Falpha1 and Falpha2 degenerate primers (25) to PCR amplify the MAT-1 α-domain from the eight mating-type tester strains that were not MAT-2. The ∼-200-bp PCR products from each of the mating-type tester strains were sequenced. Based on these sequences we constructed two specific primers, GFmat1a (5′-GTTCATCAAAGGGCAAGCG-3′) and GFmat1b (5′-TAAGCGCCCTCTTAACGCCTTC-3′) that can be used to amplify an ∼200-bp portion of the relatively conserved G. fujikuroi MAT-1 α domain.
DNA sequencing.
MAT-1 and MAT-2 fragments were sequenced in both directions using either primers Gfmat2c and Fo14 or primers Falpha1 and Falpha2. PCR products were purified with a QIAquick PCR Purification Kit (Qiagen) and sequenced by using an ABI PRISM 377 automated DNA sequencer and an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Warrington, United Kingdom).
We translated the DNA sequence and analyzed the inferred amino acid sequence with Sequence Navigator version 1.0.1 (Applied Biosystems). DNA sequences were manually aligned by inserting gaps and then analyzed with PAUP (Phylogenetic Analysis Using Parsimony, version 4.0*; Sinauer Associates, Sunderland, Mass.
In these analyses gaps were treated as fifth characters (newstate) in heuristic searches, with tree-bisection-reconnection branch swapping. We also performed bootstrap analyses to estimate the confidence of branching points. Trees generated in this way were rooted to F. oxysporum MAT-1 and MAT-2 sequences (GenBank accession nos. AB011378 and AB011379).
Diagnostic multiplex PCR for MAT-1 and MAT-2.
The multiplex PCR included the four primers GFmat1a, GFmat1b, GFmat2c, and GFmat2d. We optimized the reaction conditions by varying the Mg2+ concentrations (1.5, 2.0, 2.5, and 3.0 mM), the Taq polymerase concentrations (0.35, 0.40, and 0.45 U per reaction), the target DNA concentrations (∼100 to 20 and ∼10 to 2 ng/μl), the annealing temperatures (61, 63, 65, and 67°C), and the annealing times (30 or 60 s). After optimization, we used the following reaction conditions (10 μl, final volume): 1× PCR buffer (Sigma, St. Louis, Mo.), 2.5 mM MgCl2, 0.2 mM concentrations of each dNTP, a 0.1 μM concentration of each of the four primers, and 0.4 U of Taq DNA polymerase (Sigma). We amplified PCR products according to the following program: an initial denaturation at 94°C for 1 min, followed by 35 cycles of 30 s at 92°C, 30 s at 67°C, and 30 s at 72°C. After the last cycle there was a final elongation step for 5 min at 70°C.
Blind test verification of diagnostic multiplex PCR.
We examined 60 strains from G. fujikuroi mating population H (4) and 80 strains from G. fujikuroi mating populations A to F (24). To demonstrate Mendelian segregation and cosegregation of the molecular markers with their corresponding mating phenotype, we examined 128 progeny of a cross between two F. verticillioides isolates from the A mating population (23). Prior to these analyses, all isolates were renumbered, and the tests were done blind.
RESULTS
Analysis of MAT-2.
We amplified and sequenced the MAT-2 HMG-domain, the 3′-idiomorph flank, and the variable sequence flanked by these conserved regions from one mating-type tester representing each of the eight described G. fujikuroi mating populations (GenBank accession no. AF236765 to AF236772). The sequenced portion of the HMG domain and the 3′-idiomorph flank were highly similar (>92% nucleotide sequence similarity), whereas the variable sequences flanked by these conserved regions were relatively heterogeneous (<88% sequence similarity). N. crassa a (GenBank accession no. M54787), Cochliobolus heterostrophus MAT-2 (GenBank accession no. X68398), and Podospora anserina mat+ (GenBank accession no. X64195) and all eight MAT-2 alleles from the G. fujikuroi mating-type testers have an intron at a conserved position within the HMG domain (data not shown). Although there were some differences in the sequence (<90% sequence similarity) of this intron among the Fusarium strains, there was no significant similarity (<30% sequence similarity) to the intron from the other three ascomycetes. The ∼800-bp MAT-2 fragment was amplified from the “+” mating-type tester strains from mating populations A, B, D, E, and H and from the “−” mating-type tester strains from mating populations C, F, and G.
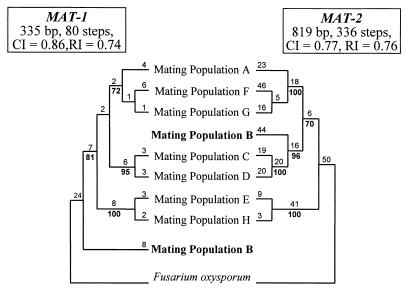
Phylogenetic analysis based on the MAT-2 sequence data resulted in a single most parsimonious tree with three distinct clades (Fig. 1). The first clade included the isolates from mating populations A, F, and G; the second clade included the isolates from mating populations B, C, and D; the third clade included isolates from mating populations E and H.
FIG. 1.
Phylograms generated from partial MAT-1 (left) and MAT-2 (right) DNA sequence data for the eight identified mating populations (A to H) in the G. fujikuroi species complex. The trees were generated with PAUP and rooted to F. oxysporum. Bootstrap values are based on 1,000 replications and are indicated as percentages in boldface below the branches. Branch lengths, character state (nucleotide) changes, are indicated above the branches.
Analysis of MAT-1.
We designed a pair of primers, GFmat1a and GFmat1b, that are specific for the MAT-1 alleles in G. fujikuroi. These alleles shared more than 94% nucleotide sequence similarity in their α-domains (GenBank accession nos. AF236757 to AF236764). The G. fujikuroi MAT-1 α-domain contained an intron at a position similar to the intron in N. crassa A (GenBank accession no. M33876), C. heterostrophus MAT-1 (GenBank accession no. X68399), and P. anserina mat− (GenBank accession no. 64194). Although there was a significant amount of variation in the G. fujikuroi α-domain intron sequences (>73% sequence similarity), these sequences shared little similarity (<40% sequence similarity) with those from the other three ascomycetes. The ∼200-bp MAT-1 fragment was amplified from the “−” mating-type tester strains from mating populations A, B, D, E, and H and from the “+” mating-type tester strains from mating populations C, F, and G.
Phylogenetic analysis of the MAT-1 α-domain sequences resulted in a tree with three clades (Fig. 1). The compositions of these clades were similar to those obtained from the MAT-2 sequence data in that mating populations A, F, and G, mating populations C and D, and mating populations E and H remained grouped. The sequence of the MAT-1 α-domain from the B-mating population shared significant similarities with the α-domains in all of the other mating populations and could be basal to the other seven mating populations (Fig. 1).
Diagnostic PCR for MAT-1 and MAT-2.
PCR reactions containing primers GFmat1a, GFmat1b, GFmat2c, and GFmat2d resulted in amplification of either the ∼200-bp MAT-1 or the ∼800-bp MAT-2 fragment (Fig. 2). We obtained better results when either primers GFmat1a and GFmat1b or primers GFmat2c and GFmat2d were used as pairs rather than as a multiplex reaction. The amount of DNA was an important variable. Results were more reproducible and there was less background with the 1:100 (∼2 to 10 ng of DNA/μl) dilutions of initial DNA preparations than with the 1:10 (∼20 to 100 ng of DNA/μl) dilutions. Annealing temperature also was an important variable. If only the MAT-1 primers (GFmat1a and GFmat1b) were used, then a single ∼200-bp product was detected at all four temperatures tested. If only the MAT-2 primers (GFmat2c and GFmat2d) were used, then clear amplification of a single ∼800-bp product was observed only at 65 and 67°C. Increasing the annealing time from 30 to 60 s resulted in more degenerate amplification products.
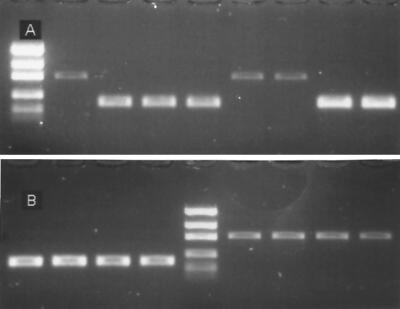
FIG. 2.
PCR amplification of the MAT region by using a multiplex PCR. The larger differential band (∼800 bp) is from amplification of the MAT-2 region. The smaller differential band (∼200 bp) is from amplification of the MAT-1 region. The constant band beneath both of the amplified bands is unincorporated primer. (A) Segregation of MAT-1 and MAT-2 in 8 of the progeny from the mapping cross of F. verticillioides (23). Lane 1, size markers (200, 400, 800, 1,200, and 2,000 bp); lanes 2 to 9, progeny from A-0015 × A-4643. (B) MAT-1 and MAT-2 alleles from the mating-type tester strains for G. fujikuroi mating populations A, B, D, and H. Lane 5, size markers; lanes 1 to 4, MAT-1 tester strains; lanes 6 to 9, MAT-2 tester strains.
Blind tests.
We tested the multiplex PCR amplification on 102 strains from mating populations A to F (24) and 60 strains from mating population H (4) and found that the amplification products detected were the same as those predicted based on the results of sexual crosses. The amplified MAT DNA fragments cosegregated with mating type in a genetic mapping cross (9, 24). Thus, the amplified fragments map with 95% certainty to a 2.3-map unit region that includes MAT and are unlikely to map more than one map unit from MAT, if they are not coincident with it.
MAT alleles in other Fusarium species and phylogenetic lineages.
We observed no amplification of either MAT-1 or MAT-2 fragments from the seven strains from species outside the Liseola or Elegans sections of the genus: F. acuminatum, F. avenaceum, F. crookwellense, F. culmorum, F. graminearum, F. pseudograminearum, and F. solani. Of the five F. oxysporum strains, two were MAT-1 (KSU O-02523 and O-02529) and three were MAT-2 (KSU O-02332, O-02530, and O-02533). The MAT-1 results were clear in the multiplex reaction. No amplification of the MAT-2 allele was detected in the multiplex reaction, but clear bands were observed from all three strains following PCR amplification using the Kerényi et al. (9) primers. The 23 strains from the G. fujikuroi species complex represented 21 species or phylogenetic lineages other than the eight identified G. fujikuroi mating populations. We tested two isolates each of F. phyllophilum and F. pseudocircinatum. Both F. phyllophilum isolates were MAT-1, while one F. pseudocircinatum isolate (KSU X-04379) was MAT-1 and the other (KSU X-10761) was MAT-2. Of the remaining 19 species, 9 were represented by a strain from which the MAT-1 fragment could be amplified: F. annulatum, F. anthophilum, F. begonieae, F. bulbicola, F. concentricum, F. lactis, F. napiforme, F. ramigenum, and F. succisae. The other 10 species were represented by a strain from which a MAT-2 fragment could be amplified: F. acutatum, F. beomiforme, F. brevicatenulatum, F. denticulatum, F. dlamini, F. guttiforme, F. nisikadoi, F. pseudoanthophilum, F. pseudonygamai, and F. subglutinans (mango). Representatives from all 10 of these species also yielded a fragment when the Kerényi et al. (9) primers were used.
DISCUSSION
Kerényi et al. (9) described a primer pair that could be used to prime a PCR reaction that amplified the MAT-2 idiomorph, and they standardized the terminology for mating type in G. fujikuroi mating populations A to G. Covert et al. (6) identified a MAT-2 allele in Gibberella circinata (an invalid name for G. fujikuroi mating population H) and adopted the Kerényi et al. (9) terminology. In this report we extend their results by developing primers for the α box of the MAT-1 idiomorph and by identifying limits on the diversity of species in which the primers will function. We developed a multiplex PCR reaction in which both MAT-1 and MAT-2 can be diagnosed as the positive outcome of a PCR amplification reaction without the worry that a lack of amplification, as with the Kerényi et al. (9) or Covert et al. (6) protocols, might have more than one meaning, i.e., no MAT sequence to amplify or MAT-1 allele present. It is not possible to use the Kerényi et al. (9) MAT-2 primers in a multiplex reaction with our MAT-1 primers because the annealing temperatures for the PCR reactions differ (61 and 67°C) and because the fragments amplified are similar enough in size to be difficult to distinguish easily on an agarose gel.
We examined both of the strains from the B mating population (MRC 6524 and MRC 6525) that Britz et al. (4) identified as, at least occasionally, homothallic. Both of these strains clearly yield only a single product in the multiplex PCR amplification reaction; MRC 6524 is MAT-2 and MRC 6525 is MAT-1. Thus, the basis for homothallism in these strains cannot be due to mating-type switching, as has been observed in some yeasts and a few filamentous fungi (7, 8, 21).
The fragment amplified by our MAT-2 primers is larger than that of Kerényi et al. (9), ∼800 and ∼260 bp, respectively, and includes a ∼560-bp region that is not a part of the conserved HMG box. The Kerényi et al. (9) primers and our primers do not differ significantly in their ability to amplify fragments from the eight identified mating populations of G. fujikuroi, but they do differ in their abilities to prime PCR reactions with DNA from strains of more distantly related species. For example, the Kerényi et al. (9) primers can be used to amplify a fragment from strains of F. beomiforme and F. nisikadoi. These two species are not closely related to the other species in the group based on sequences from the 28S ribosomal DNA, the mitochondrial small-subunit ribosomal DNA and β-tubulin (17).
The conserved nature of the MAT alleles has led some to suggest their possible use in phylogenetic and taxonomic studies (21). The phylogenetic trees generated from the partial sequences of both MAT-1 and MAT-2 (Fig. 1) are similar to those of O'Donnell et al. (17) and Steenkamp et al. (20), with the exception of the placement of mating population B. The B mating population groups with the isolates from the C and D mating populations based on MAT-2 (Fig. 1), histone (21), and β-tubulin (17) DNA sequences. The partial MAT-1 sequences, however, suggest that mating population B is approximately equally distant from the seven other mating populations (Fig. 1). Thus, the B mating population α-domain could have resulted from a hybridization event between the α-domains of strains from the other mating populations, or it could be the basal progenitor of the α-domain in the other mating populations. To resolve this problem, a larger, and perhaps more variable, portion of the MAT-1 idiomorph from more strains and species will have to be sequenced and analyzed.
Molecular scoring of mating type will reduce the amount of effort required to screen field populations for sexual fertility and should increase the efficiency of the process through which new mating populations are identified. Molecular diagnosis of the mating type of strains assigned to a known mating population can reduce the number of crosses needed in two ways. First, only crosses with the tester of the opposite mating type need to be made, thereby reducing the number of crosses by one-half. Second, if the initial crosses are successful, then the crosses need not be repeated to confirm fertility since the molecular diagnosis provides this confirmation.
For the identification of a new mating population, each putative member of the new mating population must be used as both a male and a female parent in crosses with all of the other putative members of that mating population to identify female-fertile strains. If a set of 60 strains is used, then 3,600 crosses (602) are needed to test the 60 strains for the presence of female fertility at the 5% frequency level with 95% confidence. If mating type is scored molecularly, then the number of crosses that need to be made is significantly reduced. For example, if a 40:20 split at mating type is detected following PCR amplification, then only 1,660 crosses are needed.
The availability of molecular diagnostics for mating type also may enable the analysis of purportedly asexual fungi, e.g., F. oxysporum, and 12 of the 13 recently described Fusarium taxa (15, 16). There is circumstantial evidence in F. oxysporum for sexual reproduction in the form of high levels of diversity with respect to the multilocus vegetative compatibility trait (see, for example, references 8, 11, 12, and 22), especially in populations of putatively nonpathogenic strains. Sexual reproduction need not be frequent to still play an important role in the maintenance and generation of genotypic diversity within field populations of these fungi (14), and the availability of mating-type data should make it easier to identify potentially cross-fertile strains that can be used to test these hypotheses.
In conclusion, we developed a multiplex PCR reaction for scoring mating type within the existing mating populations of G. fujikuroi that will speed the analysis of natural populations of these fungi. MAT-1 and MAT-2 sequences may be useful in taxonomic and phylogenetic studies of this group of fungi, but sequences from more strains and species still need to be analyzed.
ACKNOWLEDGMENTS
This work was supported in part by the National Research Foundation, the Tree Pathology Co-operative Programme, and the Kansas Agricultural Experiment Station.
Footnotes
Contribution 00-322-J from the Kansas Agricultural Experiment Station, Manhattan.
REFERENCES
- 1.Anonymous. Index of fungi. Index fungi. 1999;6:979–980. [Google Scholar]
- 2.Appel D J, Gordon T R. Local and regional variation in populations of Fusarium oxysporum from agricultural field soils. Phytopathology. 1994;84:786–791. [Google Scholar]
- 3.Arie T, Christiansen S K, Yoder O C, Turgeon B G. Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol. 1997;21:118–130. [PubMed] [Google Scholar]
- 4.Britz H, Coutinho T A, Wingfield M J, Marasas W F O, Gordon T R, Leslie J F. Fusarium subglutinans f. sp. pini represents a distinct mating population in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:1198–1201. doi: 10.1128/aem.65.3.1198-1201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppin E, Debuchy R, Arnaise S, Picard M. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 1997;61:411–428. doi: 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covert S F, Briley A, Wallace M M, McKinney V T. Partial MAT-2 gene structure and the influence of temperature on mating success in Gibberella circinata. Fungal Genet Biol. 1999;28:43–54. doi: 10.1006/fgbi.1999.1161. [DOI] [PubMed] [Google Scholar]
- 7.Gutz H, Schmidt H. The genetic basis of homothallism and heterothallism in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Semin Dev Biol. 1990;1:169–176. [Google Scholar]
- 8.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerényi Z, Zeller K A, Hornok L, Leslie J F. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:4071–4076. doi: 10.1128/aem.65.9.4071-4076.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaasen J A, Nelson P E. Identification of a mating population, Gibberella nygamai sp. nov., within the Fusarium nygamai anamorph. Mycologia. 1996;88:965–969. [Google Scholar]
- 11.Kondo N, Kodama F, Ogoshi A. Vegetative compatibility groups of Fusarium oxysporum f. sp. adzukicola and nonpathogenic Fusarium oxysporum on adzuki bean isolated from adzuki bean fields in Hokkaido. Ann Phytopathol Soc Jpn. 1997;63:8–12. [Google Scholar]
- 12.Leslie J F. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–150. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 13.Leslie J F. Gibberella fujikuroi: available populations and variable traits. Can J Bot. 1995;73:S282–S291. [Google Scholar]
- 14.Leslie J F, Klein K K. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics. 1996;144:557–567. doi: 10.1093/genetics/144.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nirenberg H I, O'Donnell K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. [Google Scholar]
- 16.Nirenberg H I, O'Donnell K, Kroschel J, Andrianaivo A P, Frank J M, Mubatanhema W. Two new species of Fusarium: Fusarium brevicatenulatum from the noxious weed Striga asiatica in Madagascar and Fusarium pseudoanthophilum from Zea mays in Zimbabwe. Mycologia. 1998;90:459–464. [Google Scholar]
- 17.O'Donnell K, Cigelnik E, Nirenberg H I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 18.Perkins D D. Mating-type switching in filamentous ascomycetes. Genetics. 1987;115:215–216. doi: 10.1093/genetics/115.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J E, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Steenkamp E T, Wingfield B D, Coutinho T A, Wingfield M J, Marasas W F O. Differentiation of Fusarium subglutinans f. sp. pini by histone gene sequence data. Appl Environ Microbiol. 1999;65:3401–3406. doi: 10.1128/aem.65.8.3401-3406.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turgeon B G. Application of mating type gene technology to problems in fungal biology. Annu Rev Phytopathol. 1998;36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 22.Woudt L P, Neuvel A, Sikkema A, van Grinsven M Q J M, de Milliano W A J, Campbell C L, Leslie J F. Genetic variation in Fusarium oxysporum from cyclamen. Phytopathology. 1995;85:1348–1355. [Google Scholar]
- 23.Xu J-R, Leslie J F. A genetic map of Fusarium moniliforme (G. fujikuroi mating population A) Genetics. 1996;143:175–189. doi: 10.1017/s0016672300034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan K, Dickman M B, Xu J-R, Leslie J F. Sensitivity of field strains of Gibberella fujikuroi (Fusarium section Liseola) to benomyl and hygromycin B. Mycologia. 1993;85:206–213. [Google Scholar]
- 25.Yoshida T, Arie T, Nomura Y, Yoder O C, Turgeon B G, Yamaguchi I. Proceedings of the VIIth International Congress on Plant Pathology. Edinburgh, Scotland: International Congress of Plant Pathology; 1998. Cloning mating type genes of the asexual ascomycete Fusarium oxysporum; p. 1.10.7. [Google Scholar]