Abstract
Bovine viral diarrhea virus (BVDV), also known as Pestivirus A, causes severe infection mostly in cattle, but also in pigs, sheep and goats, causing huge economical losses on agricultural farms every year. The infections are actually controlled by isolation of persistently infected animals and vaccination, but no antivirals are currently available to control the spread of BVDV on farms. BVDV binds the host cell using envelope protein E2, which has only recently been targeted in the research of a potent and efficient antiviral. In contrast, RdRp has been successfully inhibited by several classes of compounds in the last few decades. As a part of an enduring antiviral research agenda, we designed a new series of derivatives that emerged from an isosteric substitution of the main scaffold in previously reported anti-BVDV compounds. Here, the new compounds were characterized and tested, where several turned out to be potent and selectively active against BVDV. The mechanism of action was thoroughly studied using a time-of-drug-addition assay and the results were validated using docking simulations.
Keywords: benzimidazole-2-phenyl-carboxamides, BVDV, dual target, antivirals, entry inhibition, replication inhibition
1. Introduction
Bovine viral diarrhea virus (BVDV) is a single-stranded positive-sense RNA virus that belongs to the Pestivirus genus of the Flaviviridae family, recently renamed Pestivirus A in the 2017 report from the International Committee on Taxonomy of Viruses [1]. The Pestivirus genus also contains more animal pathogens, such as the border disease virus (BDV) and the classical swine fever virus (CSFV) [2]. Not only can cows be the host of BVDV, but pigs, sheep, goats and other wild and domestic ruminants can also be infected [3]. BVDV causes teratogenesis, abortion, early embryonic death, immune system and respiratory disorders in cows, resulting in acute infections of immunocompetent cattle, giving rise to a mortality rate ranging from approximately 20 to 30% [4] and an estimated economical loss of approximately 10 and 40 million dollars per million calvings. Newborns can be infected through the placenta, leading to the presence of cattle persistently infected (PI) with BVDV on farms [5]. BVDV was also detected as a troublesome pollutant in commercially available lots of fetal bovine serum and cell lines commonly used in the laboratories [6], and therefore, was revealed as a contaminant in interferons and vaccines for medical use [7,8].
No antivirals are currently available for controlling BVDV infections in laboratories or on farms; the vaccination protocols and the isolation of PI animals is the only validated strategy currently in use on farms to limit the transmission and reduce the economic loss [9]. BVDV continues to cause agronomical trouble; therefore, the identification of molecules that are capable of binding and inhibiting the virus replication and transmission remains a major antiviral research goal.
BVDV single-stranded RNA is translated by the host cell into a polyprotein, which is processed by viral and host proteases and cleaved into four structural (Protein C, Erns, E1 and E2) and eight functional proteins [10]. E2 glycoprotein binds the cell-surface receptor (CD46 or CD81) and causes the membrane fusion to begin, which allows the BVDV single-stranded RNA to be released in the host cytoplasm [2,11]. The non-structural viral proteins have been the most inhibited in drug discovery in the last few decades, such as NS5B RNA-dependent RNA-polymerase (RdRp), NS4a protease and NS3 helicase, while only recently, the structural proteins were targeted in order to inhibit the early stages of virus infection [12].
In recent years, many selective anti-BVDV compounds were reported, with a virus-targeting or host-targeting approach, inter alia viral polymerase ligands [13,14,15,16], protease inhibitors [17] and human cellular enzymes targeting compounds [18,19,20,21]. The antiviral research against Pestivirus genus pathogens is still a challenge. Among others, benzimidazole derivatives were demonstrated to be active against RNA viruses in the last few decades [22,23,24,25,26,27].
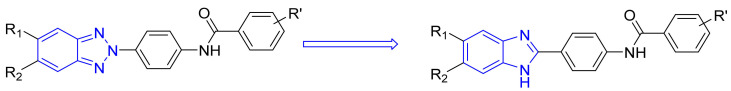
As a part of our enduring antiviral research agenda, a series of angular and linear N-polycyclic derivatives active against BVDV, HCV and other related viruses were designed and synthetized [24,28,29]. The viral target for BVDV inhibition was ascertained in the RNA-dependent RNA-polymerase (RdRp) [30,31]. Aiming to select the best scaffold endowed with antiviral activity, we synthesized several polycyclic heteroaromatic derivatives, in turn, improving selectivity and potency [29,32,33] The most promising anti-BVDV compounds that emerged possess the halo-benzotriazole-2-phenyl carboxamide scaffold. Based on the bioisostere theory and the literature on benzimidazole scaffold, we designed the isosteric substitution of the N2 of benzotriazole derivatives depicted in Figure 1 with a carbon atom, obtaining benzimidazole derivatives. This variance limited the free rotation of the phenyl moiety bound to the atom in position 2 and transformed the nitrogen in position 1 from an H-bond acceptor to an H-bond donor. This resulted in a series of benzimidazole derivatives (1–7n, 1–7o, 1–7p) wherein substitution based on benzimidazole moiety was evaluated; meanwhile, the substitution of R’ = 4-Cl, 4-NO2 or 3,4,5-trimethoxy was conserved.
Figure 1.
Isosteric substitution of benzotriazole scaffold with benzimidazole scaffold.
2. Materials and Methods
2.1. Chemistry
2.1.1. General Synthetic Strategies
All starting materials were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany), Across Organics (Geel, Belgium) and Carlo Erba (Milan, Italy) producers. Nitrogen group reduction to amine derivatives was performed in ethanol using three different routes as follows: chlorine derivatives reductions were made with methylhydrazine as a bland reducing agent in ethanol using the autoclave at 100 °C. All the other reductions were conducted in ethanol with Pd/C as a catalyst and H2 at room temperature, or with Pd/C and hydrazine at 80 °C. Benzimidazole ring closure was carried out as described by Bahrami et al. [34] by mixing o-phenylenediamines and aldehydes in a ratio of 1:1 in acetonitrile as the solvent, with H2O2 30% (ratio 1:7) and HCl 37% (ratio 1:3.5) added at room temperature for the proper number of hours. Amide groups were obtained from the reaction of aniline derivatives with benzoyl chloride derivatives in a ratio of 1:1.2 in DMF at 80 °C till the reaction was completed. Products were purified via crystallization from ethanol or methanol, or via flash chromatography using proper elution by mixing suitable chosen solvents among petroleum ether (PE), diethyl ether (DE), ethyl acetate (EA), chloroform and methanol.
2.1.2. Chemical Characterization
Nuclear magnetic resonance (NMR) spectra were recorded with a Bruker Avance III 400 NanoBay (400 MHz) instrument from Bruker (Billerica, MA, US) from sample solutions in deuterated solvents, Acetone or DMSO. 1H NMR chemical shifts are reported in parts per million (ppm) downfield from tetramethylsilane (TMS) used as an internal standard. Chemical shift values are reported in ppm (δ) and coupling constants (J) are reported in hertz (Hz). Signal multiplicities are represented as s (singlet), ws (wide singlet), d (doublet), dd (doublet of doublets), ddd (doublet of doublet of doublets), t (triplet), td (triplet of doublets), q (quadruplet) and m (multiplet). The assignment of exchangeable protons (OH and NH) was confirmed via the addition of D2O. 13C NMR chemical shifts are reported downfield from tetramethylsilane (TMS) used as an internal standard. A suitable method among the APT (attached proton test) and jmod (J-modulated spin-echo for X-nuclei coupled to H-1 to determine the number of attached protons) was selected for each compound. Two-dimensional NMR experiments HSQC (heteronuclear single quantum coherence) and HMBC (heteronuclear multiple bond correlation) were performed to correctly assign the peaks. The solutions for ESI-MS measurements were prepared at a concentration of 1.0–2.0 ppm by dissolving and serially diluting the solid compounds in HPLC acetonitrile. Mass spectra in the positive-ion mode were obtained on a Q Exactive Plus Hybrid Quadrupole-Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer. The solutions were infused at a flow rate of 5.00 μL/min into the ESI chamber. The spectra were recorded in the m/z range of 150–800 at a resolution of 140,000 and accumulated for at least 2 min to increase the signal-to-noise ratio. The instrumental conditions used for the measurements were as follows: spray voltage 2300 V, capillary temperature 250 °C, sheath gas 10 (arbitrary units), auxiliary gas 3 (arbitrary units), sweep gas 0 (arbitrary units) and probe heater temperature 50 °C. ESI-MS spectra were analyzed by using Thermo Xcalibur 3.0.63 software (Thermo Fisher Scientific, Waltham, MA, USA), and the average deconvoluted monoisotopic masses were obtained through the Xtract tool integrated with the software. Each compound melting point (m.p.) was taken in open capillaries in a Digital Electrothermal melting point apparatus and was uncorrected. Retention factors (Rf) were measured via thin-layer chromatography (TLC) using Merck F-254 commercial plates (Merck KGaA, Darmstadt, Germany).
2.2. Biology
2.2.1. Cell Lines and Viruses
Cell lines were purchased from the American Type Culture Collection (ATCC). The absence of mycoplasma contamination was periodically checked using the Hoechst staining method. The cell line supporting the multiplication of BVDV was the following: Madin-Darby Bovine Kidney (MDBK) (ATCC CCL 22 (NBL-1) Bos Taurus). The virus was purchased from American Type Culture Collection (ATCC), Bovine Viral Diarrhoea Virus (BVDV) (strain NADL (ATCC VR-534)).
2.2.2. Cytotoxicity Assay
Cytotoxicity assays were run in parallel with antiviral assays. MDBK cells were seeded at an initial density of 6 × 105 cells/cm3 in 96-well plates in culture medium (Minimum Essential Medium with Earle′s salts (MEM-E) with L-glutamine, supplemented with 10% horse serum and 1 mM sodium pyruvate, and 0.025 g/L kanamycin). Cell cultures were then incubated at 37 °C in a humidified, 5% CO2 atmosphere in the absence or presence of serial dilutions of test compounds. The cell viability was determined after 48–96 h at 37 °C using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method [35]. The MTT data were processed using the statistical method of linear regression.
2.2.3. Antiviral Assay
Compound activity against BVDV was based on the inhibition of virus-induced cytopathogenicity in MDBK cells acutely infected with an m.o.i. of 0.01. Briefly, MDBK cells were seeded in 96-well plates at a density of 3 × 104 cells/well and were allowed to form confluent monolayers by incubating overnight in a growth medium at 37 °C in a humidified CO2 (5%) atmosphere. Cell monolayers were then infected with 50 mL of virus dilution in a maintenance medium (MEM-E with L-glutamine, supplemented with 0.5% inactivated FBS, 1 mM sodium pyruvate and 0.025 g/L kanamycin) to give an m.o.i. of 0.01. After 2 h, 50 mLof maintenance medium, without or with serial dilutions of test compounds, was added. After 3 days of incubation at 37 °C, cell viability was determined using the MTT method. Linear regression analysis: viral and cell growth at each drug concentration is expressed as a percentage of untreated controls and concentrations resulting in 50% (EC50 and CC50) growth inhibition. NM 108 (2′-C-methylguanosine) and ribavirin were used as positive controls. The selectivity index (S.I.) was used, which is a parameter of preferential antiviral activity of a compound related to its cytotoxicity (CC50/EC50). Each experiment was conducted in duplicate (two wells in parallel) and the experiments were performed in three copies.
2.2.4. Time of Drug Addition Assay
A time-of-addition experiment was carried out with MDBK cells. The confluent monolayers of MDBK cells, seeded in 24-well tissue culture plates, were infected for 1 h at room temperature with 6650 PFU of BVDV, corresponding to a multiplicity of infection of 3 PFU/cell. After adsorption for 60 min, the monolayers were washed two times with a maintenance medium in the presence of HS inactivated and incubated with the same medium at 5% CO2 and 37 °C. The test medium containing 10 × EC50 compound concentration was added at –1 to 0 (pretreatment), 0 to 2 (during infection), 2 to 4, 4 to 6, 6 to 8, 8 to 10, 10 to 12, 12 to 14 and 14 to 16. After each incubation period, the monolayers were washed two times with a maintenance medium and incubated with a fresh medium until 12 h post-infection. Then, the monolayers were frozen at −80 °C and the viral titers were determined using a plaque reduction assay.
2.2.5. Virucidal activity
A suspension of viral particles was directly exposed to each test compound and incubated for 1 h at 4 °C. Then, the mixtures were diluted in series and used to infect the cells; the infectious titers were recorded and compared with those obtained with an untreated viral suspension.
2.3. Docking
AutoDock Vina [36] was selected as the docking program. As a starting point, the crystal structure of BVDV1 envelope glycoprotein E2 was retrieved from the RCSB Protein Data Bank (rcsb.org, PDB code 2YQ2) [22]. NAG (2-acetamido-2-deoxy-β-d-glucopyranose) and water molecules were removed from the co-crystallized structure.
Compounds 2c, 2d, 2e, 6c and 7c, were first minimized through Avogadro [37] version 1.1.1 and then saved as PDB files. Protein and ligand structures were processed through AutoDockTools (ADT) [38] version 1.5.6, generating the PDBQT files and obtaining the coordinate and volume information for the docking grid box: 30–30–30 Å; x = –17.095, y = 35.857 and z = –38.325. For the ligands′ structure, all rotatable bonds were made flexible, including the amide bonds. Docking was carried out by making some amino acids flexible, viz., Ser57, Thr60, Arg61, Gln87, Glu89, Asp91, Pro105, Val153 and Arg154. These amino acids were exposed in the target binding pocket of the protein and shown to stabilize the structural conformation of different literature compounds that docked in that precise site [12]. As docking parameters, the exhaustiveness value was set to 64 and the number of poses to 10 for each docking prediction. The affinity energy range was set to be ≤ 2 kcal·mol−1 above that of the best binding pose for each ligand. For the evaluation of the affinity prediction, the best-docked pose of each investigated ligand was taken and visualized in PyMOL 2.3.4 [39]. The docking studies were conducted on a PC with an Intel® Core™ i7–9705H CPU @ 2.60 GHz with 8 GB RAM running Ubuntu 16.04 as an operating system. The RdRp crystal structure was retrieved by RSCB PDB (PDB code 1S48). The grid box was set to 18–18–22 Å (spacing 1 Å) around the coordinating point with x/y/z values of –28.322/157.375/22.518.
3. Results and Discussion
3.1. Chemistry
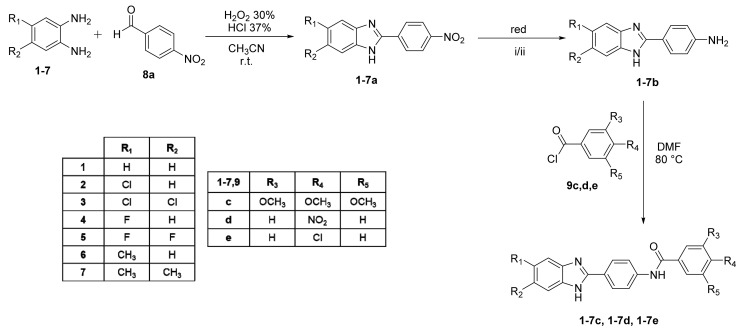
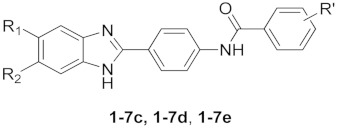
Derivatives 1–7c, 1–7d and 1–7e were obtained as described in Scheme 1. Intermediate o-phenylenediamines 1–7 (commercially purchased) were mixed with 4-nitrobenzaldehyde 8a in a ratio of 1:1 to obtain nitro-derivatives 1–7a that were appropriately reduced to aniline derivatives 1–7b. Derivatives 1–7c, 1–7d and 1–7e were gained via the reaction of aniline derivatives 1–7b with the corresponding benzoyl chloride derivative (9c,d,e) in a ratio of 1:1.2 in DMF at 80 °C till reaction was completed.
Scheme 1.
Synthetic route for derivatives 1–7c, 1–7d and 1–7e. i: when R1 or both R1 and R2 = Cl, reaction conditions: CH3NHNH2/EtOH in an autoclave at 100 °C for 48 h; ii: all the other derivative reaction conditions: NH2NH2 and Pd/C in EtOH at 80 °C for 1 h.
3.2. Anti-BVDV Activity
Concerning the benzimidazole derivatives (1–7c, 1–7d, 1–7e), which were synthesized and evaluated for activity and selectivity against BVDV, the results are reported in Table 1. It clearly shows that when R’ is a chlorine atom (1–7e), the corresponding compounds had no relevant antiviral activity and the EC50 values were always higher than CC50 ones. All trimethoxy-derivatives (1–7c) were found to be highly potent BVDV inhibitors with EC50 values ranging between 0.09 and 41 µM, whereas only some nitro-derivatives (2–5d) showed interesting EC50 values: 1.9, 53, 7.9 and 3.6 µM, respectively. Selecting the most active trimethoxy-derivatives (1–7c), preliminary SARs highlighted that the substitution on the benzimidazole scaffold in position 5 or both 5 and 6 generally increased the activity, and the presence of one (2c) or two chlorine atoms (3c) strengthened the activity but was coupled with cytotoxicity, with CC50 values of 45 and 28 µM, respectively. The presence of one or two smaller atoms, such as fluorine (4c and 5c), augmented the activity (EC50: 41 and 1.4 µM) with cytotoxicity values always higher than 100 µM. A valuable anti-BVDV activity was obtained when a methyl group was in position 5 or both 5 and 6 on the benzimidazole main scaffold, resulting in derivatives 6c and 7c, with EC50 values of 0.23 and 0.3 µM, respectively. This SAR analysis was confirmed by the selectivity index (S.I.) values, also reported in Table 1. The three most potent compounds, namely, 2c, 6c and 7c, also showed the highest S.I. values. Several new compounds showed a better score than the positive control compounds, namely, NM108 (S.I. 66.7) and the gold-standard reference ribavirin, for which the S.I. was 5.6.
Table 1.
Antiviral activity and cytotoxicity of benzimidazole derivatives (1–7c, 1–7d, 1–7e); NM108 and ribavirin were used as positive controls.
| ||||||
|---|---|---|---|---|---|---|
| Compound | R1 | R2 | R’ | MDBK CC50 a |
BVDV EC50 b |
S.I. |
| 1c | H | H | 3,4,5-tri-OCH3 | 28.5 | 2.2 | 12.9 |
| 1d | H | H | 4-NO2 | >100 | >100 | - |
| 1e | H | H | 4-Cl | >100 | >100 | - |
| 2c | Cl | H | 3,4,5-tri-OCH3 | 45 | 0.09 | 500 |
| 2d | Cl | H | 4-NO2 | >100 | 1.9 | >52.6 |
| 2e | Cl | H | 4-Cl | >100 | >100 | - |
| 3c | Cl | Cl | 3,4,5-tri-OCH3 | 28 | 1.3 | 21.5 |
| 3d | Cl | Cl | 4-NO2 | >100 | 53 | >1.9 |
| 3e | Cl | Cl | 4-Cl | 60 | >60 | - |
| 4c | F | H | 3,4,5-tri-OCH3 | >100 | 41 | >2.4 |
| 4d | F | H | 4-NO2 | >100 | 7.9 | >12.7 |
| 4e | F | H | 4-Cl | >100 | >100 | - |
| 5c | F | F | 3,4,5-tri-OCH3 | >100 | 1.4 | >71.4 |
| 5d | F | F | 4-NO2 | >100 | 3.6 | >27.8 |
| 5e | F | F | 4-Cl | >100 | >100 | - |
| 6c | CH3 | H | 3,4,5-tri-OCH3 | >100 | 0.23 | >434.8 |
| 6d | CH3 | H | 4-NO2 | >100 | >100 | - |
| 6e | CH3 | H | 4-Cl | >100 | >100 | - |
| 7c | CH3 | CH3 | 3,4,5-tri-OCH3 | >100 | 0.3 | >333.3 |
| 7d | CH3 | CH3 | 4-NO2 | >100 | >100 | - |
| 7e | CH3 | CH3 | 4-Cl | >100 | >100 | - |
| NM 108 | >100 | 1.5 | >66.7 | |||
| Ribavirin | >100 | 18 | >5.6 | |||
a Compound concentration (µM) required to reduce the viability of mock-infected MDBK cells by 50%, as determined using the MTT method. b Compound concentration (µM) required to achieve 50% protection of MDBK cells from the BVDV-induced cytopathogenicity, as determined using the MTT method. In bold relevant EC50 values and significant S. I.
3.3. Time of Drug Addition
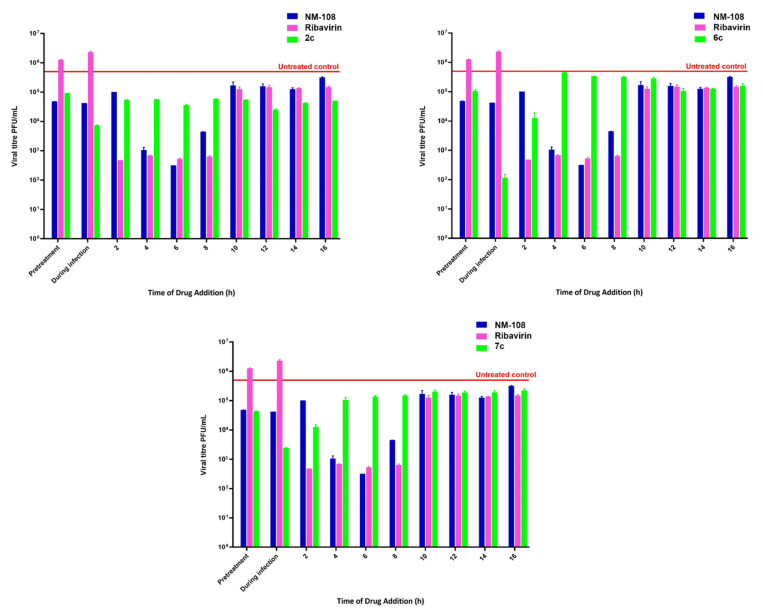
According to the discussed results, the three most promising compounds, namely, 2c, 6c and 7c, were selected to deepen the study of the mechanism of virus inhibition for this new class of compounds. A time of drug addition assay was performed to identify the time point in the virus replication cycle where the new compounds exerted the highest antiviral activity, and the results are depicted in Figure 2.
Figure 2.
Effect of the time of drug addition on antiviral activity with SD of derivatives 2c (0.09 µM), 6c (0.23 µM) and 7c (0.3 µM) (green). Parallel experiments were performed using reference compounds NM 108 (1.5 µM) (blue) and Ribavirin (16 µM) (purple) for comparison. We show the untreated control in red.
A time of drug addition assay indicates the time at which the compound reaches the maximum antiviral activity; thus, knowing the virus life cycle, the virus replication step inhibited by the analyzed compound can be identified. The results showed that the pretreatment with the three tested compounds resulted in a slight reduction in virus replication; the same poor activity was detected between 10- and 16-h post-infection. The best score was detected when cells were treated at the infection moment. The poor activity detected in pretreated cells and the highest potency when the cells were treated simultaneously with compounds and BVDV suggested that benzimidazole compounds inhibited the early stage of the virus infection of the host cell. Moreover, these data let us suppose that the target lay on the virus envelope rather than on the cells since no cell protection during pretreatment was detected.
Chlorinated compound 2c showed a consistent antiviral activity along the viral replication cycle more distinctly than derivatives 6c and 7c. This suggested that a non-structural protein was also targeted by the present series of compounds; indeed, they were designed and synthesized to target and inhibit the viral RdRp.
The time of drug addition output showed that the three tested compounds turned out to be the most active in the early stage of infection. The host-targeting effect was ruled out by the poor results of the cell pretreatment. The viral-targeting effect may result in virucidal or virustatic activity. The former was evaluated.
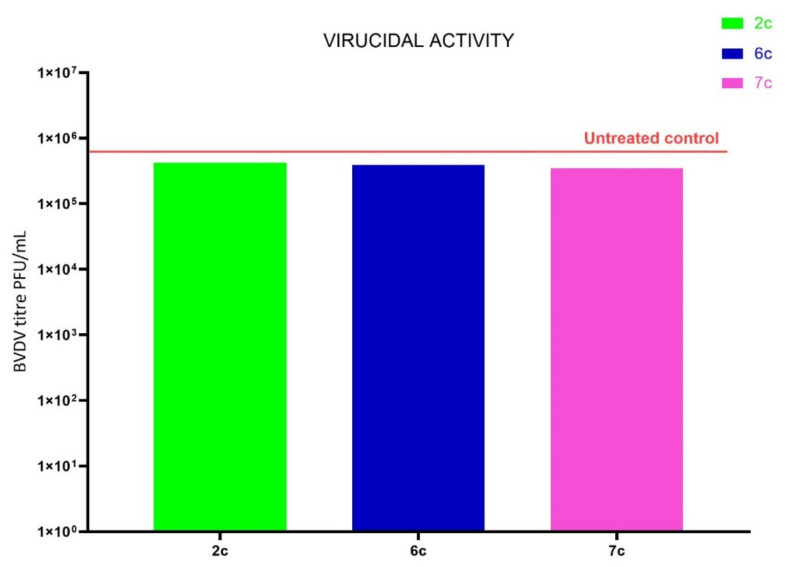
3.4. Virucidal Activity
To detect the antiviral mechanism of action, the virucidal activity of the three selected compounds, namely, 2c, 6c and 7c, was investigated. BVDV was incubated at 4 °C for 1 h; the mixture was then used to infect cells. It resulted in infected cultures of host cells that were almost comparable to the untreated but infected ones, as shown in Figure 3. The single treatment of virus or cell pretreatment did not result in the highest antiviral activity, while it was demonstrated that when the three components, namely, virus, cell and ligand, were mixed at the same time, the compounds inhibited the receptor binding.
Figure 3.
Virucidal activity of the 2c, 6c and 7c compounds. BVDV suspensions were incubated with the title compound at 4 °C for 1 h, and then the remaining infectivity was determined. Results are expressed as a titer reduction in compound-treated samples compared to non-treated ones.
3.5. Molecular Docking
3.5.1. Trimethoxy-Substituted Compounds 2c, 6c and 7c Docked in a Strategic Binding Pocket in E2 Protein
BVDV E2 glycoprotein was demonstrated to have a crucial role in cell entry [40]. Docking predictions were carried out to evaluate the correlation between the biological results and the predicted mode of binding of the different ligands. Considering the biological assessment, compounds 2c, 6c and 7c were reported as the hit compounds endowed with the best antiviral activity of the present series of compounds. The docking predictions provided a value of affinity energy for each ligand targeting the same protein binding pocket. The tri-methoxy derivatives 2c, 6c and 7c showed comparable affinity energies between −6.4 and −6.9 kcal·mol−1, as reported in Table 2.
Table 2.
Affinity energy values for compounds 2c, 6c and 7c from Vina docking predictions.
| Affinity Energy | ||
|---|---|---|
| Compound | 2c | −6.4 kcal·mol−1 |
| 6c | −6.9 kcal·mol−1 | |
| 7c | −6.8 kcal·mol−1 |
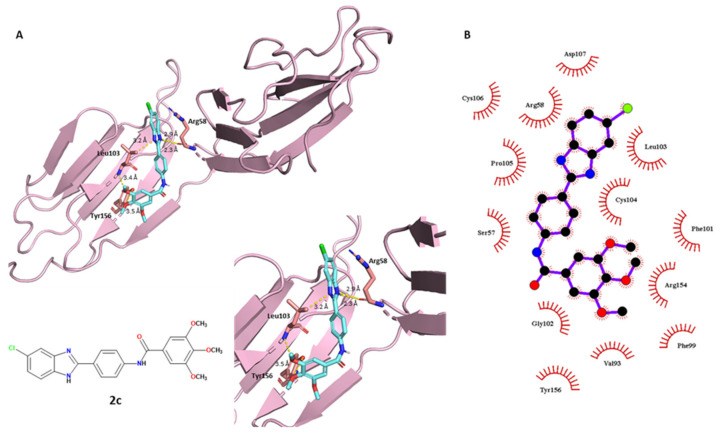
More in-depth analyses of the protein–ligand interactions are reported in the following discussion, involving the polar and non-polar contacts between the ligands and a specific portion of the BVDV glycoprotein E2 (PDB ID: 2YQ2). Starting with compound 2c reported in Figure 4, its best-predicted pose showed that the molecule establishing different polar contacts with some exposed amino acids of the binding pocket of E2 glycoprotein. The docking of the ligands to the glycoprotein E2 was performed on a specific region located in the DB domain (residues 88–164) of E2 glycoprotein, one of the most distal portions from the virus membrane, and hence, one of the most exposed on the viral surface. This specific portion is often targeted by anti-BVDV compounds [40]. The methoxy groups have a crucial role: the oxygen of OCH3 in the C–3 position accepts a hydrogen bond with the intrinsic NH of Leu103. The side-chain isopropyl CH3 of Leu103 has a polarized C–H bond, which can interact with the benzimidazole nitrogen atom of compound 2c (3.2 Å length). The phenolic hydroxyl group of Tyr156 contacts the C–4 methoxy group of the ligand. Moreover, the best-predicted conformation of the benzimidazole scaffold played a crucial role, with two polar contacts (2.3 and 2.9 Å length) within the acceptor carbonyl oxygen of Arg58. This showed how the polar term was fundamental for E2–2c affinity, suggesting a strong binding of compound 2c and explaining its good experimental antiviral profile. Regarding the hydrophobic interactions involving compound 2c, the tri-methoxy phenyl moiety was the most-involved portion: it was surrounded by different amino acids: Gly102, Tyr156, Val93, Phe99, Arg154 and Phe101. The middle phenyl-amide moiety interacted with Gly102, but also Ser57, Cys104 and Pro105. Furthermore, the 5-chlorobenzimidazole scaffold had nonpolar contacts with Pro105, Cys106, Arg58, Asp107, Leu103 and Cys104.
Figure 4.
Conformational representation of the best-docked pose of compound 2c. (A) Protein–ligand polar interactions between 2c and the exposed amino acids of E2, investigated in PyMOL. Protein is depicted in a light-pink tint, compound 2c is in blue marine. Polar contacts are reported as yellow dashes. (B) Non-polar interactions between 2c and E2, generated through LigPlot+. Colors: black (carbon), blue (nitrogen) and red (oxygen).
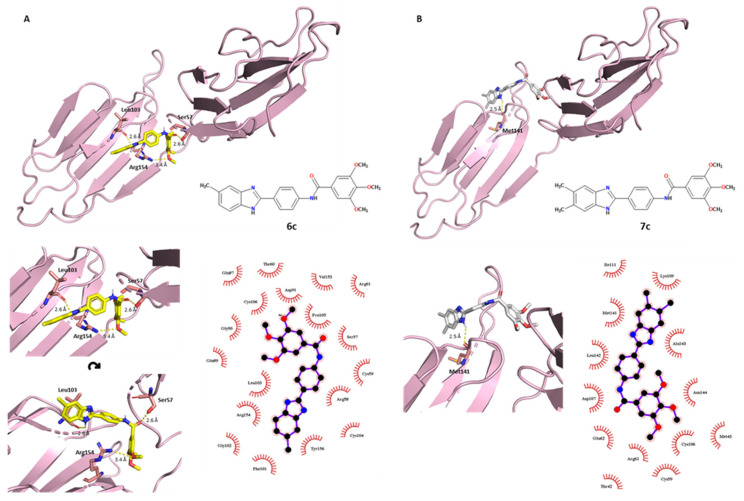
Compound 6c showed a characteristic affinity with the target, where the polar protein–ligand interactions involved each moiety of the ligand. The OCH3 in the C–3 position established an H-bond with the guanidine NH2 group of Arg154. As previously discussed for 2c, compound 6c interacted with Leu103, involving a polar interaction between the benzimidazole NH of the ligand and the carbonyl oxygen of the leucine residue (2.6 Å bond length). In addition, the carbonyl oxygen of the amide linker accepted a hydrogen bond 2.6 Å in length from the side-chain hydroxyl group of Ser57. The great antiviral activity of compound 6c was shown through the biological assessment and it was confirmed here by this docking prediction (Figure 5A), where the electrostatic term seemed crucial for the binding. Different hydrophobic interactions were established within neighboring amino acids: of particular interest were Tyr156, Arg154 and Leu103 on the benzimidazole scaffold side; Arg58, Ser57 and Pro105 for the phenyl-amide middle portion; and Leu103, Pro105, Asp91 and Cys106 for the tri-methoxy-phenyl moiety.
Figure 5.
PyMOL (3D) and LigPlot+ (2D) representations showing polar and nonpolar interactions between the E2 protein and (A) compounds 6c and (B) 7c. Compounds 6c and 7c are colored in yellow and white, respectively.
To complete the series of tri-methoxy-derived compounds, derivative 7c resulted in a good antiviral agent but was slightly less potent than compounds 2c and 6c. The docking results showed that derivative 7c had a unique H-bond with a methionine amid the benzimidazole NH of the ligand and the carbonyl oxygen of Met141 (2.5 Å length) of the E2 target. The same amino acid also established nonpolar interactions with the benzimidazole scaffold, as was the case for Ala143 and Lys109. As can be seen in Figure 5B, the hydrophobic term was crucial for the protein–ligand relationship. Furthermore, Leu142 and Asp107 set hydrophobic interactions with the phenyl-amide portion, while Arg61, Cys106 and Asn144 contacted the tri-methoxy-phenyl moiety.
3.5.2. Trimethoxy-Substituted Compounds 2c, 6c and 7c Docked in a Strategic Binding Pocket in E2 Protein
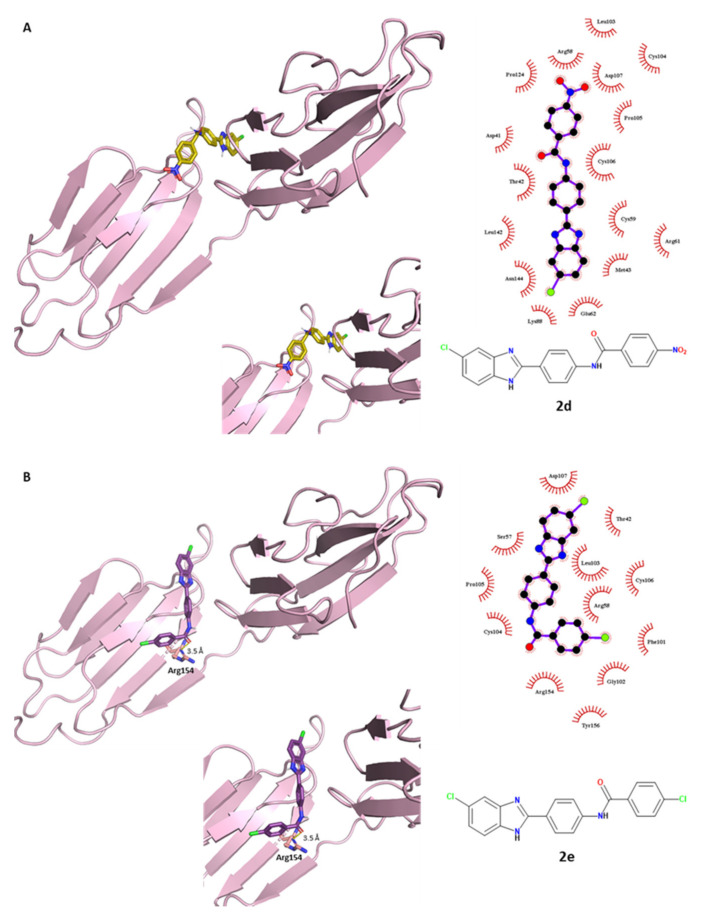
The 5-chloro derivatives 2c (tri-methoxy), 2d (p-NO2) and 2e (p-Cl) were used for deeper docking simulations in order to compare the prediction outputs with the experimental biological results and to better understand the different interactions of the R3, R4 and R5 substituents within the E2 glycoprotein target. The binding profile of compound 2c was previously described. The high number of polar–nonpolar contacts gave a tight binding of the molecule to the binding pocket surface. As for the p-nitro derivative 2d, it interacted with E2 protein through some nonpolar interactions, while it did not show hydrogen bonds with neighboring polar amino acids (Figure 6A). Regarding compound 2e, depicted in its best-binding pose in Figure 6B, it established a unique H-bond: the amidic oxygen of the ligand interacts as an acceptor with the side-chain NH of Arg154. Few hydrophobic interactions contributed to its interaction with E2 protein. In general, we could assume that the weaker antiviral effect of compounds 2d and 2e in comparison to 2c could be predictively confirmed by these docking studies. In general, analyzing the polar interactions between this class of molecules and the E2 binding pocket, Leu103 and Arg154 amino acids were predicted to be crucial for the ligand binding and were important for their conformational stabilization of their protein-bound state.
Figure 6.
Electrostatic and hydrophobic contacts between E2 protein and compounds (A) 2d and (B) 2e, colored in gold and purple, respectively.
3.5.3. Docking of Compound 2c to BVDV RNA-Dependent RNA Polymerase (RdRp)
In order to collect in silico data regarding its mechanism of action, the binding to the envelope glycoprotein E2 or the RNA-dependent RNA polymerase (RdRp) of the benzimidazole-based lead compound 2c was compared. The favorable binding of compound 2c to the binding site in the envelope protein E2 is reported below. The binding to the RdRp of BVDV was evaluated, considering the most important amino acids for the docking to this protein, which was previously reported by us and other researchers in the literature [29,30,31].
The affinity score of the binding between the best-docked pose of derivative 2c and the RdRp binding site was −5.9 kcal·mol−1, with average energy between the first 10 most-ranked poses of ~ −5.3 kcal·mol−1. The binding of compound 2c was better predicted to occur at the E2 active site in comparison to that at RdRp superficial site, considering the affinity scores.
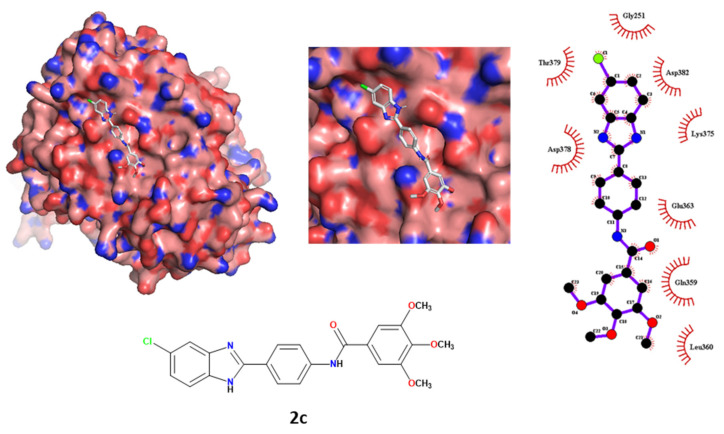
Moreover, regarding the protein–ligand interactions, the importance of the polar and nonpolar contacts between compound 2c and the envelope E2 protein were previously-reported. In contrast, the contact between ligand 2c and the superficial site of RdRp was predicted not to create polar contacts, not showing any H-bond with the exposed amino acids. The ligands had different hydrophobic interactions with the RdRp site. The 5-chlorobenzimidazole scaffold interacted with Asp378, Thr379, Gly251, Asp382 and Lys375. The remaining moieties of the ligand interacted with Glu363, Gln359 and Leu360. The results are depicted in Figure 7.
Figure 7.
Tri- and bidimensional representation of the spatial conformation of compound 2c docked at the RdRp surface binding site.
We concluded that ligand 2c, in its best-predicted pose, showed an interesting pattern of electrostatic and hydrophobic interactions with the exposed amino acids of the envelope E2 protein. In contrast, when the best-docked pose of ligand 2c approached the superficial pocket of RdRp, its binding profile appeared weaker in terms of the affinity energy score and polar/nonpolar interactions.
4. Conclusions
We undertook a bioisosteric substitution of benzotriazole scaffold of our previously reported anti-BVDV compounds with benzimidazole scaffold. The resulting 21 compounds 1–7c, 1–7d and 1–7e were tested against BVDV and MDBK cell lines and selectivity indexes were also calculated. Three of them were found to be active in the low micromolar range, and 2c showed the highest EC50 value of 90 nM. It was found to be active and potent in the early stages of infection, inhibiting the entry of the virus into the host cell and interfering with the receptor binding, whilst no direct virucidal activity was detected. Compound 2c showed detectable activity, also between 2 and 14 h post-infection, when viral replication is ongoing. The key protein in the cell surface recognition was the viral E2 glycoprotein and our hit compound 2c was shown to bind the target pocket with an affinity energy value of −6.4 kcal·mol−1. We concluded that ligand 2c, in its best-predicted pose, showed an interesting pattern of electrostatic and hydrophobic interactions with the exposed amino acids of the envelope E2 protein. In contrast, when the best-docked pose of ligand 2c approached the superficial pocket of RdRp, its binding profile appeared weaker in terms of affinity energy score and polar/nonpolar interactions. These results appeared in agreement with the biological outputs. Further studies should be carried out to validate these results.
5. Experimental Characterization
5.1. Characterization of Intermediates 1–7a and 1–7b
5.1.1. 2-(4-Nitrophenyl)-1H-benzo[d]imidazole (1a)
Compound 1a (C13H9N3O2, MW 239.229) was obtained with a total yield of 44%; m.p. 214–215 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.72. 1H-NMR (DMSO-d6): δ 8.49 (4H, s, H-2′,3′,5′,6′), 7.76 (2H, m, H-4,7), 7.41 (2H, m, H-5,6). 13C-NMR (jmod, DMSO-d6): δ 148.45 (C), 148.24 (C), 136.98 (2C), 133.63 (C), 128.10 (2CH), 124.41 (2CH), 124.18 (2CH), 115.18 (2CH). ESI-MS (m/z): calcd. for C13H9N3O2 240.077, found 240.077 [M+H]+.
5.1.2. 5-Chloro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (2a)
Compound 2a (C13H8ClN3O2, MW 273.674) was obtained with a total yield of 97%; m.p. 256–258 °C; TLC (PE/ES 7/3): Rf 0.44. 1H-NMR (DMSO-d6): δ 8.43 (4H, s, H-2′,3′,5′,6′), 7.73 (1H, d, J = 1.6 Hz, H-4), 7.69 (1H, d, J = 8.4 Hz, H-7), 7.308 (1H, dd, 1J = 10.4 Hz, 2J = 2 Hz, H-6), 3.68 (1H, s, NH). 13C-NMR (DMSO-d6): δ 150.30 (C), 148.07 (C), 140.27 (C), 137.78 (C), 135.25 (C), 127.62 (2CH), 127.36 (C), 124.31 (2CH), 123.39 (CH), 116.70 (CH), 115.27 (CH). ESI-MS (m/z): calcd. for C13H8ClN3O2 274.038, found 274.038 [M+H]+.
5.1.3. 5,6-Dichloro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (3a)
Compound 3a (C13H7Cl2N3O2, MW 308.120) was obtained with a total yield of 69%; m.p. 226 °C; TLC (PE/EA 7/3): Rf 0.58. 1H-NMR (DMSO-d6): δ 8.40 (4H, ws, H-2′,3′,5′,6′), 7.91 (2H, s, H-4,7). 13C-NMR (jmod, DMSO-d6): δ 151.50 (C), 148.25 (C), 188.80 (2C), 134.88 (C), 127.42 (2CH), 125.47 (2C), 124.32 (2CH), 116.83 (2CH). ESI-MS (m/z): calcd. for C13H7Cl2N3O2 307.999, found 307.999 [M+H]+.
5.1.4. 5-Fluoro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (4a)
Compound 4a (C13H8FN3O2, MW 257.220) was obtained with a total yield of 87%; m.p. 226.3 °C; TLC (PE/EA 7/3): Rf 0.45. 1H-NMR (DMSO-d6): δ 8.45 (4H, d, J = 1.6 Hz, H-2′,3′,5′,6′), 7.74 (1H, m, H-7′), 7.45 (1H, m, H-4′), 7.23 (1H, m, H-6′). 13C-NMR (jmod, DMSO-d6): δ 160.54 (C), 158.18 (C), 149.72 (C), 148.35 (C), 137.91 (C), 133.97 (C), 127.95 (2CH), 124.36 (2CH), 116.44 (CH), 112.26 (CH), 101.29 (CH). ESI-MS (m/z): calcd. for C13H8FN3O2 258.067, found 258.067 [M+H]+.
5.1.5. 5,6-Difluoro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (5a)
Compound 5a (C13H7F2N3O2, MW 275.210) was obtained with a total yield of 52%; m.p. 292 °C; TLC (PE/EA 7/3): Rf 0.40. 1H-NMR (DMSO-d6): δ 8.39 (4H, m, H-2′,3′,5′,6′), 7.715 (2H, t, H-4,7). 13C-NMR (jmod, DMSO-d6): δ 149.70 (C), 148.88 (C), 148.39 (2C, dd, 1JC-F = 245 Hz, 2JC-F = 19 Hz, 2C-F), 131.84 (C, m), 131.66 (C), 131.25 (C, m), 128.35 (2CH), 124.28 (2CH), 102.90 (2C, dd, 2JC-F = 15 Hz, 3JC-F = 8 Hz, CH-4,7). ESI-MS (m/z): calcd. for C13H7F2N3O2 276.058, found 276.058 [M+H]+.
5.1.6. 5-Methyl-2-(4-nitrophenyl)-1H-benzo[d]imidazole (6a)
Compound 6a (C14H11N3O2, MW 253.256) was obtained with a total yield of 42%; m.p. 94–95 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.74. 1H-NMR (DMSO-d6): δ 8.57 (2H, d, J = 9.2 Hz, H-3′,5′), 8.52 (2H, d, J = 8.8 Hz, H-2′,6′), 7.73 (1H, d, J = 8.4 Hz, H-7), 7.63 (1H, s, H-4), 7.37 (1H, d, J = 8.4 Hz), 2.50 (3H, s, CH3). 13C-NMR (jmod, DMSO-d6): δ 149.13 (C), 146.57 (C), 135.75 (C), 133.82 (C), 131.98 (C), 130.62 (C), 128.91 (2CH), 127.32 (CH), 124.48 (2CH), 114.28 (CH), 113.76 (CH), 21.23 (CH3). ESI-MS (m/z): calcd. for C14H11N3O2 254.092, found 254.092 [M+H]+.
5.1.7. 5,6-Dimethyl-2-(4-nitrophenyl)-1H-benzo[d]imidazole (7a)
Compound 7a (C15H13N3O2, MW 267.283) was obtained with a total yield of 62%; m.p. 260–262 °C; TLC (PE/EA 7/3): Rf 0.43. 1H-NMR (DMSO-d6): δ 8.38 (4H, s, H-2′,3′,5′,6′), 7.42 (2H, s, H-4,7), 2.34 (6H, s, 2CH3). 13C-NMR (DMSO-d6): δ 147.92 (C), 147.52 (C), 137.75 (C), 135.79 (2C), 132.07 (2C), 128.85 (2CH), 127.69 (2CH), 124.19 (2CH), 19.96 (2CH3). ESI-MS (m/z): calcd. for C15H13N3O2 268.108, found 268.108 [M+H]+.
5.1.8. 4-(1H-Benzo[d]imidazol-2-yl)aniline (1b)
Compound 1b (C13H11N3, MW 209.247) was obtained with a total yield of 63%; m.p. >300 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.38. 1H-NMR (DMSO-d6): δ 7.86 (2H, d, J = 8.4 Hz, H-2′,6′), 7.50 (2H, m, H-4,7), 7.14 (2H, m, H-5,6), 6.68 (2H, d, J = 8.4 Hz, H-3′,5′). 13C-NMR (jmod, DMSO-d6): δ 152.38 (C), 150.76 (2C), 138.88 (C), 127.85 (2CH), 121.46 (2CH), 116.63 (C), 113.54 (2CH), 113.22 (2CH). ESI-MS (m/z): calcd. for C13H11N3 210.103, found 210.103 [M+H]+.
5.1.9. 4-(5-Chloro-1H-benzo[d]imidazol-2-yl)aniline (2b)
Compound 2b (C13H10ClN3, MW 243.692) was obtained with a total yield of 86%; m.p. 133–135 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.29. 1H-NMR (DMSO-d6): δ 7.83 (2H, d, J = 8.4 Hz, H-2′,6′), 7.54 (1H, m, H-4), 7.43 (1H, ws, H-7), 7.13 (1H, d, J = 8.4 Hz, H-6), 6.66 (2H, d, J = 8.4 Hz), 5.66 (2H, s, NH2). 13C-NMR (APT, DMSO-d6+TFA-d): δ 153.63 (C), 150.93 (C), 132.17 (C), 130.24 (C), 129.65 (C), 129.60 (2CH), 125.52 (CH), 114.42 (CH), 114.13 (2CH), 112.76 (CH), 107.87 (C). ESI-MS (m/z): calcd. for C13H10ClN3 244.064, found 244.064 [M+H]+.
5.1.10. 4-(5,6-Dichloro-1H-benzo[d]imidazol-2-yl)aniline (3b)
Compound 3b (C13H9Cl2N3, MW 278.137) was obtained with a total yield of 97%; m.p. 222–223 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.28. 1H-NMR (DMSO-d6): δ 7.83 (2H, d, J = 7.6 Hz, H-2′6′), 7.78 (1H, s, H-4), 7.61 (1H, s, H-7), 6.67 (2H, d, J = 7.6 Hz, H-3′5′), 5.72 (2H, s, NH2). 13C-NMR (jmod, DMSO-d6): δ 155.21 (C), 151.22 (C), 143.89 (C), 134.56 (C), 128.12 (2CH), 123.46 (C), 123.28 (C), 118.78 (CH), 113.48 (2CH), 116.05 (C), 111.76 (CH). ESI-MS (m/z): calcd. for C13H9Cl2N3 278.025, found 278.025 [M+H]+.
5.1.11. 4-(5-Fluoro-1H-benzo[d]imidazol-2-yl)aniline (4b)
Compound 4b (C13H10FN3, MW 227.237) was obtained with a total yield of 63%; m.p. 271 °C; TLC (PE/EA 7/3): Rf 0.12. 1H-NMR (DMSO-d6): δ 8.05 (2H, d, J = 8.8 Hz, H-2′,6′), 7.72 (1H, m, H-7), 7.54 (1H, m, H-4), 7.40 (1H, m, H-6), 6.78 (2H, d, J = 8.8 Hz, H-3′,5′). 13C-NMR (jmod, DMSO-d6): δ 160.81 (C), 158.42 (C), 153.90 (C), 151.15 (C), 132.42 (C), 129.67 (2CH), 128.48 (C), 114.43 (CH), 113.66 (2CH), 113.02 (CH), 99.95 (CH). ESI-MS (m/z): calcd. for C13H10FN3 228.093, found 228.093 [M+H]+.
5.1.12. 4-(5,6-Difluoro-1H-benzo[d]imidazol-2-yl)aniline (5b)
Compound 5b (C13H7F2N3O2, MW 245.227) was obtained with a total yield of 87%; m.p. 246–247 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.40. 1H-NMR (DMSO-d6): δ 7.81 (2H, d, J = 8.4 Hz, H-2′,6′), 7.54 (1H, s, H-4), 7.448 (1H, s, H-7), 6.67 (2H, d, J = 8.8 Hz, H-3′,5′), 5.64 (2H, s, NH2). 13C-NMR (jmod, DMSO-d6): δ 154.62 (C), 150.85 (2C), 146.21 (2C, dd, 1JC-F = 236 Hz, 2JC-F = 16 Hz, C-F), 127.75 (2CH), 116.53 (2C), 113.51 (2CH), 102.45-101.30 (2C, m, CH-F). ESI-MS (m/z): calcd. for C13H9F2N3 246.083, found 246.083 [M+H]+.
5.1.13. 4-(5-Methyl-1H-benzo[d]imidazol-2-yl)aniline (6b)
Compound 6b (C14H13N3, MW 223.273) was obtained with a total yield of 31%; m.p. 110–112 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.26. 1H-NMR (acetone-d6): δ 7.94 (2H, d, J = 8.8 Hz, H-2′,6′), 7.39 (1H, d, J = 8 Hz, H-7), 7.31 (1H, s, H-4), 6.97 (1H, d, J = 8 Hz, H-6), 6.78 (2H, d, J = 8.4 Hz, H-3′,5′), 5.09 (2H, s, NH2), 2.42 (3H, s, CH3). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 153.69 (C), 149.49 (C), 135.10 (C), 131.54 (C), 129.33 (2CH), 129.11 (C), 126.45 (CH), 114.07 (2CH), 113.42 (2CH), 108.04 (C), 20.97 (CH3). ESI-MS (m/z): calcd. for C14H13N3 224.118, found 224.118 [M+H]+.
5.1.14. 4-(5,6-Dimethyl-1H-benzo[d]imidazol-2-yl)aniline (7b)
Compound 7b (C15H15N3, MW 237.299) was obtained with a total yield of 88%; m.p. 109–111 °C; TLC (CHCl3/CH3OH 95/5): Rf 0.17. 1H-NMR (DMSO-d6): δ 7.80 (2H, d, J = 8.4 Hz, H-2′,6′), 7.31 (1H, s, H-7), 7.18 (1H, s, H-4), 6.65 (2H, d, J = 8.8 Hz, H-3′,5′), 5.55 (2H, s, NH2), 2.29 (6H, d, J = 7.2 Hz). 13C-NMR (jmod, DMSO-d6): δ 151.62 (C), 150.21 (C), 137.32 (2C), 130.01 (2C), 127.52 (2CH), 117.13 (2C), 114.37 (C), 113.78 (2CH), 19.84 (2CH3). ESI-MS (m/z): calcd. for C15H15N3 238.134, found 238.134 [M+H]+.
5.2. Synthesis, Purification and Characterization of Benzimidazole Derivatives
5.2.1. N-(4-(1H-Benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (1c), N-(4-(1H-Benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (1d) and N-(4-(1H-Benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (1e)
A mixture of 230 mg of 4-(1H-benzo[d]imidazol-2-yl)aniline (1.09 mmol) (1b) and 276 mg of 3,4,5-trimethoxybenzoyl chloride (1.20 mmol) (9c), or 222 mg of 4-nitrobenzoyl chloride (1.20 mmol) (9d) or 210 mg of 4-nitrobenzoyl chloride (1.20 mmol) (9e) (ratio 1:1.1) in 10 mL of DMF was stirred at 80 °C to obtain 1c, 1d and 1e, respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in an oven overnight. The pure products (1c, 1d and 1e) were obtained via crystallization using ethanol.
Compound 1c (C23H21N3O4, MW 403.43) was obtained with a total yield of 20%; m.p. 164 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.58. 1H-NMR (DMSO-d6): δ 10.61 (1H, s, NH), 8.30 (2H, d, J = 8.8 Hz, H-2′,6′), 8.09 (2H, d, J = 8.8 Hz, H-3′,5′), 7.75 (2H, m, H-4,7), 7.43 (2H, m, H-5,6), 8.09 (2H, d, J = 8.8, H-3′,5′), 7.35 (2H, s, H-2″,6″), 3.90 (6H, s, 2CH3-C3″,5″), 3.75 (3H, s, CH3-C4″). 13C-NMR (jmod, DMSO-d6): δ 165.34 (C), 152.64 (2C), 149.52 (C), 142.59 (C), 140.59 (C), 134.57 (C), 129.52 (2C), 128.15 (2CH), 124.35 (2CH), 120.52 (2CH), 120.39 (C), 114.18 (2CH), 105.54 (2CH), 60.12 (CH3), 56.16 (2CH3). ESI-MS (m/z): calcd. for C23H21N3O4 404.160, found 404.161 [M+H]+.
Compound 1d (C20H14N4O3, MW 358.35) was obtained with a total yield of 56%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.49. 1H-NMR (DMSO-d6): δ 10.79 (1H, s, NH), 8.41 (2H, d, J = 8.8 Hz, H-3″,5″), 8.21 (2H, d, J = 2.8 Hz, H-2″,6″), 8.20 (2H, d, J = 8.8 Hz, H-3′,5′), 7.98 (2H, d, J = 8.8 Hz, H-2′,6′), 7.65 (1H, s, H-7), 7.53 (1H, s, H-4), 7.20 (2H, d, J = 5.2 Hz, H-5.6). 13C-NMR (jmod, DMSO-d6): δ 164.09 (C=O), 150.98 (C), 149.22 (C), 143.82 (C), 140.39 (C), 140.11 (C), 135.07 (C), 129.26 (2CH), 127.00 (2CH), 125.80 (C), 123.58 (2CH), 122.35 (CH), 121.59 (CH), 120.42 (2CH), 118.65 (CH), 111.15 (CH). ESI-MS (m/z): calcd. for C20H14N4O3 359.114, found 359.114 [M+H]+.
Compound 1e (C20H14ClN3O, MW 347.79) was obtained with a total yield of 62%; m.p. 170.5 °C; TLC (PE/EA 6/4): Rf 0.60. 1H-NMR (DMSO-d6): δ 8.36 (1H, s, NH), 8.20 (2H, d, J = 8.8 Hz, H-2′,6′), 8.02 (2H, m, H-3′,5′), 7.94 (2H, d, J = 8.4 Hz, H-2″,6″), 7.63 (2H, m, H-4,7), 7.56 (2H, d, J = 8.4 Hz, H-3″,5″), 7.24 (1H, m, H-5,6). 13C-NMR (jmod, DMSO-d6): δ 166.42 (C), 164.63 (C), 150.72 (C), 140.90 (C), 137.76 (C), 136.61 (C), 133.35 (C), 131.10 (2CH), 129.69 (2CH), 129.61 (C), 129.35 (CH), 129.24 (CH), 128.69 (CH), 128.49 (2CH), 127.23 (CH), 122.49 (CH), 120.33 (CH). ESI-MS (m/z): calcd. for C20H14ClN3O 348.090, found 348.090 [M+H]+.
5.2.2. N-(4-(5-Chloro-1H-benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (2c), N-(4-(5-Chloro-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (2d) and N-(4-(5-Chloro-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (2e)
A mixture of 400 mg of 4-(5-chloro-1H-benzo[d]imidazol-2-yl)aniline (1.64 mmol) (2b) and 416 mg of 3,4,5-trimethoxybenzoyl chloride (1.80 mmol) (9c), or 334 mg of 4-nitrobenzoyl chloride (1.80 mmol) (9d) or 315 mg of 4-nitrobenzoyl chloride (1.80 mmol) (9e) (ratio 1:1.1) in 10 mL of DMF was stirred at 80 °C to obtain 2c, 2d and 2e, respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in an oven overnight. The crudes obtained were all purified using flash chromatography (CHCl3/CH3OH in a ratio of 96/4).
Compound 2c (C23H20ClN3O4, MW 437.88) was obtained with a total yield of 15%; m.p. 286–287 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.53. 1H-NMR (DMSO-d6): δ 10.35 (1H, s, NH), 8.17 (2H, d, J = 8 Hz, H-2′,6′), 7.95 (2H, d, J = 8.4 Hz, H-3′,5′), 7.70-7.64 (1H, m, H-6), 7.53 (1H, d, J = 8.4 Hz, H-7), 7.31 (2H, s, H-2″,6″), 7.24-7.19 (1H, m, H-4), 3.89 (6H, s, 2OCH3), 3.75 (3H, s, OCH3). 13C-NMR (APT, DMSO-d6+TFA-d): δ 165.66 (C=O), 152.63 (2C), 149.98 (C), 143.85 (C), 140.68 (C), 132.57 (C), 130.66 (C), 130.25 (C), 129.24 (C), 128.81 (2CH), 126.17 (CH), 120.64 (2CH), 117.19 (C), 115.22 (CH), 113.48 (CH), 105.38 (2CH), 60.03 (OCH3), 55.96 (2OCH3). ESI-MS (m/z): calcd. for C23H20ClN3O4 438.121, found 438.122 [M+H]+.
Compound 2d (C20H13ClN4O3, MW 392.80) was obtained with a total yield of 25%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.45. 1H-NMR (DMSO-d6): δ 10.81 (1H, s, NH), 8.40 (2H, d, J = 8.8 Hz, H-3″,5″), 8.22 (2H, d, J = 8.8 Hz, H-2″,6″), 8.18 (2H, d, J = 8 Hz, H-3′,5′), 7.99 (2H, d, J = 8.8 Hz, H-2′,6′), 7.71-7.65 (1H, m, H-7), 7.54 (1H, d, J = 8.8 Hz, H-4), 7.25-7.20 (1H, m, H-6). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 164.67 (C=O), 149.88 (C), 149.31 (C), 143.37 (C), 139.81 (C), 132.50 (C), 130.70 (C), 130.33 (C), 129.24 (2CH), 128.88 (2CH), 126.19 (CH), 123.49 (2CH), 120.64 (2CH), 117.61 (C), 115.25 (CH), 113.50 (CH). ESI-MS (m/z): calcd. for C20H13ClN4O3 393.075, found 393.075 [M+H]+.
Compound 2e (C20H13Cl2N3O, MW 382.24) was obtained with a total yield of 34%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.62. 1H-NMR (APT, DMSO-d6+ TFA-d): δ 10.55 (1H, s, NH), 8.16 (2H, d, J = 8.8 Hz, H-3′,5′), 8.02 (2H, d, J = 8.8 Hz, H-2″,6″), 7.97 (2H, d, J = 8.8 Hz, H-2′,6′), 7.65 (2H, d, J = 8.4 Hz, H-3″,5″), 7.59 (2H, ws, H-4,7), 7.22 (1H, dd, Jortho= 8.6 Hz, Jmeta= 2 Hz, H-6). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 165.16 (C=O), 149.93 (C), 143.81 (C), 136.97 (C), 132.88 (C), 132.50 (C), 130.67 (C), 130.30 (C), 129.67 (2CH), 128.88 (2CH), 128.49 (CH), 126.16 (CH), 120.47 (2CH), 119.25 (CH), 116.38 (C), 115.24 (CH), 113.54 (CH). ESI-MS (m/z): calcd. for C20H13Cl2N3O 382.051, found 382.051 [M+H]+.
5.2.3. N-(4-(5,6-Dichloro-1H-benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (3c), N-(4-(5,6-Dichloro-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (3d), N-(4-(5,6-Dichloro-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (3e)
A mixture of 300 mg of 4-(5,6-dichloro-1H-benzo[d]imidazol-2-yl)aniline (1.08 mmol) (3b) and 274 mg of 3,4,5-trimethoxybenzoyl chloride (1.19 mmol) (9c), or 220 mg of 4-nitrobenzoyl chloride (1.19 mmol) (9d), or 208 mg of 4-nitrobenzoyl chloride (1.19 mmol) (9e) [ratio 1:1.1] in 10 ml of DMF was stirred at 80 °C, to obtain 3c, 3d, 3e respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in oven overnight. The pure products were obtained by crystallisation from ethanol (3c and 3e) or from methanol (3d).
Compound 3c (C23H19Cl2N3O4, MW 472.32) was obtained in total yield 32%; m.p. 150.6 °C; TLC (PS/EA 6/4): Rf 0.34. 1H-NMR (DMSO-d6): δ 10.43 (1H, s, NH), 8.13 (2H, d, J = 8.8 Hz), 7.95 (2H, d, J = 8.4 Hz), 7.83 (2H, s, H-4,7), 7.29 (2H, s, H-2′,6′), 3.87 (3H, s, CH3), 3.81 (6H, ws, 2CH3). 13C-NMR (jmod, DMSO-d6): δ 166.94 (C), 165.29 (C), 153.61 (C), 152.62 (C), 152.59 (C), 141.32 (C), 141.20 (C), 140.45 (C), 129.63 (C), 127.32 (2CH), 125.81 (C), 124.50 (C), 124.00 (C), 120.58 (2CH), 113.75 (2CH), 105.30 (2CH), 60.10 (CH3), 55.88 (2CH3). ESI-MS (m/z): calcd for C23H19Cl2N3O4 472.083, found 472.082 [M+H]+.
Compound 3d (C20H12Cl2N4O3, MW 427.24) was obtained in total yield 10%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.53. 1H-NMR (DMSO-d6): δ 10.83 (1H, s, NH), 8.41 (2H, d, J = 8.8 Hz, H-3″,5″), 8.22 (2H, d, J = 8.8 Hz, H-2″,6″), 8.19 (2H, d, J = 8.8 Hz, H-3′,5′), 8.00 (2H, d, J = 8.8 Hz, H-2′,6′), 7.93 (1H, s, H-4), 7.76 (1H, s, H-7). 13C-NMR (APT, DMSO-d6, TFA-d): δ 164.57 (C=O), 153.64 (C), 151.60 (C), 149.32 (C), 142.96 (C), 139.94 (C), 133.20 (C), 131.48 (C), 129.92 (CH), 129.29 (2CH), 128.73 (CH), 127.62 (C), 123.53 (2CH), 120.56 (CH), 119.21 (C), 115.54 (CH), 114.56 (CH), 114.11 (CH). ESI-MS (m/z): calcd for C20H12Cl2N4O3 427.036, found 427.036 [M+H]+.
Compound 3e (C20H12Cl3N3O, MW 416.69) was obtained in total yield 23%; m.p. 161–162 °C; TLC (PS/EA 6/4): Rf 0.69. 1H-NMR (DMSO-d6): δ 10.63 (1H, s, NH), 8.17 (2H, d, J = 8.4 Hz, H-2′,6′), 7.86 (2H, s, H-4,7), 7.81 (2H, d, J = 8.4 Hz, H-3′,5′), 7.62 (2H, d, J = 8.4 Hz, H-2″-6″), 7.55 (2H, d, J = 8.4 Hz, H-3″,5″). 13C-NMR (jmod, DMSO-d6): δ 166.47 (C), 164.82 (C), 153.22 (C), 141.47 (C), 137.78 (2C), 136.68 (C), 133.20 (C), 131.10 (2CH), 129.66 (CH), 129.53 (C), 128.69 (2CH), 128.50 (CH), 127.56 (CH), 124.97 (C), 120.34 (CH), 115.94 (CH), 113.90 (CH). ESI-MS (m/z): calcd for C20H12Cl3N3O 416.012, found 416.012 [M+H]+.
5.2.4. N-(4-(5-Fluoro-1H-benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (4c), N-(4-(5-Fluoro-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (4d), N-(4-(5-Fluoro-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (4e)
A mixture of 500 mg of 4-(5-fluoro-1H-benzo[d]imidazol-2-yl)aniline (2.20 mmol) (4b) and 558 mg of 3,4,5-trimethoxybenzoyl chloride (2.42 mmol) (9c), or 449 mg of 4-nitrobenzoyl chloride (2.42 mmol) (9d), or 424 mg of 4-nitrobenzoyl chloride (2.42 mmol) (9e) [ratio 1:1.1] in 10 mL of DMF was stirred at 80 °C, to obtain 4c, 4d, 4e respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in oven overnight. The crudes obtained were purified by flash chromatography (CHCl3/CH3OH in ratio 95/5) for 4d. Pure 4c and 4e were obtained by crystallisation from ethanol.
Compound 4c (C23H20FN3O4, MW 421.42) was obtained in total yield 13%; m.p. 171.8 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.46. 1H-NMR (DMSO-d6): δ 10.60 (1H, s, NH), 8.37 (2H, s, H-2″,6″), 8.16 (2H, d, J = 8.4 Hz, H-2′6′), 7.80 (2H, d, J = 8.4, H-3′,5′), 7.63 (1H, m, H-6), 7.43 (1H, m, H-4), 7.14 (1H, m, H-7). 13C-NMR (jmod, DMSO-d6): δ 165.19 (C), 158.94 (1C, 1JC-F = 235 Hz, C-F), 151.67 (2C), 140.91 (C), 140.52 (2C), 137.86 (C), 133.78 (C), 128.21 (CH), 127.75 (2CH), 122.96 (2C), 122.80 (C), 120.48 (2CH), 117.20 (CH), 115.29 (1C, 3JC-F = 10 Hz, CH), 111.05 (1C, 2JC-F = 25 Hz, CH), 100.85 (1C, 2JC-F = 26 Hz, CH), 60.10 (CH3)¸ 56.13 (2CH3). ESI-MS (m/z): calcd for C23H20FN3O4 422.151, found 422.151 [M+H]+.
Compound 4d (C20H13FN4O3, MW 376.34) was obtained in total yield 29%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.35. 1H-NMR (DMSO-d6): δ 10.79 (1H, s, NH), 8.4 (2H, d, J = 8.8 Hz, H-3″,5″), 8.22 (2H, d, J = 8.8 Hz, H-2″,6″), 8.17 (2H, d, J = 8.4 Hz, H-3′,5′), 7.98 (2H, d, J = 8.8 Hz, H-2′,6′), 7.58 (1H, ws, H-7), 7.38 (1H, ws, H-4), 7.06 (1H, m, H-6). 13C-NMR (APT, DMSO-d6+TFA-d): δ 164.59 (C=O), 160.11 (C, d, 1J = 240 Hz, C-F), 149.93 (C), 149.32 (C), 143.29 (C), 139.84 (C), 132.23 (C, m, C-F), 129.28 (2CH), 128.79 (2CH), 128.36 (C), 123.52 (2CH), 120.60 (2CH), 115.34 (C, d, 3JC-F = 10 Hz, CH-7), 114.41 (C, d, 2JC-F= 25 Hz, CH-6), 100.47 (C, d, 2JC-F = 29 Hz, CH-4). ESI-MS (m/z): calcd for C20H13FN4O3 377.104, found 377.104 [M+H]+.
Compound 4e (C20H13ClFN3O, MW 365.79) was obtained in total yield 37%; m.p. 138.6 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.66. 1H-NMR (DMSO-d6): δ 10.55 (1H, s, NH), 8.15 (1H, m, H-6), 7.98-7.93 (4H, m, H-3’,5’,3″,5″), 7.65-7.56 (4H, m, H-2′,6′,2″,6″), 7.39 (1H, m, H-4), 7.07 (1H, m, H-7). 13C-NMR (jmod, DMSO-d6): δ 165.55 (C), 159.49 (1C, d, 1JC-F = 234 Hz, C-F), 153.39 (C), 141.51 (2C), 137.49 (2C), 134.31 (C), 132.02 (CH), 130.58 (2CH), 129.62 (2CH), 129.41 (2CH), 126.00 (C), 121.23 (2CH), 110.88 (1C, d, 2JC-F = 25 Hz, CH). ESI-MS (m/z): calcd for C20H13ClFN3O 366.080, found 366.081 [M+H]+.
5.2.5. N-(4-(5,6-Difluoro-1H-benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (5c), N-(4-(5,6-Difluoro-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (5d), N-(4-(5,6-Difluoro-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (5e)
A mixture of 300 mg of 4-(5,6-difluoro-1H-benzo[d]imidazol-2-yl)aniline (1.22 mmol) (5b) and 309 mg of 3,4,5-trimethoxybenzoyl chloride (1.34 mmol) (9c), or 249 mg of 4-nitrobenzoyl chloride (1.34 mmol) (9d), or 235 mg of 4-nitrobenzoyl chloride (1.34 mmol) (9e) [ratio 1:1.1] in 10 mL of DMF was stirred at 80 °C, to obtain 5c, 5d, 5e respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in oven overnight. The crudes obtained were purified by flash chromatography (CHCl3/CH3OH in ratio 98/2) for 5d and 5e. Pure 5c was obtained by crystallisation from methanol.
Compound 5c (C23H19F2N3O4, MW 439.41) was obtained in total yield 28%; m.p. 285–287 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.50. 1H-NMR (DMSO-d6): δ 10.34 (1H, s, NH), 8.14 (2H, d, J = 8.4 Hz, H-2′,6′), 7.94 (2H, d, J = 8.4 Hz, H-3′,5′), 7.69 (1H, m, H-7), 7.55 (1H, m, H-4), 7.30 (2H, s, H-2″,6″), 3.89 (6H, s, C-3″,5″-OCH3), 3.75 (3H, s, C-4″-OCH3). 13C-NMR (jmod, DMSO-d6): δ 165.10 (C=O), 153.17 (C), 152.63 (2C), 146.64 (2C, dd, 1JC-F = 251 Hz, 2JC-F = 14 Hz, C-F), 140.78 (C), 140.45 (C), 139.28 (C, d, JC-F = 12 Hz, C), 130.34 (C, d, JC-F = 12 Hz, C), 129.78 (C), 126.94 (2CH), 124.76 (C), 120.47 (2CH), 105.87 (C, d, JC-F = 19 Hz, CH-F), 105.37 (2CH), 99.17 (C, d, JC-F = 22 Hz, CH-F), 60.12 (CH3), 56.11 (2CH3). ESI-MS (m/z): calcd for C23H19F2N3O4 440.142, found 440.142 [M+H]+.
Compound 5d (C20H12F2N4O3, MW 394.33) was obtained in total yield 48%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.37. 1H-NMR (DMSO-d6): δ 10.79 (1H, s, NH), 8.40 (2H, d, J = 8.8 Hz, H-3″,5″), 8.22 (2H, d, J = 8.8 Hz, H-2″,6″), 8.16 (2H, d, J = 8.8 Hz, H-3′,5′), 7.98 (2H, d, J = 8.8 Hz, H-2′,6′), 7.64 (2H, ws, H-4,7). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 164.55 (C=O), 150.62 (C), 149.34 (C), 148.71 (2C, dd, 1JC-F = 245 Hz, 2JC-F = 17 Hz, C-F), 143.34 (C), 139.88 (C), 138.42 (C), 129.25 (2CH), 128.78 (2CH), 127.76 (C, m, C), 123.44 (2CH), 120.59 (2CH), 117.87 (C, m, C), 102.50 (2C, dd, 2J = 15.5 Hz, 3J = 7 Hz, CH). ESI-MS (m/z): calcd for C20H12F2N4O3 395.095, found 395.095 [M+H]+.
Compound 5e (C20H12ClF2N3O, MW 383.78 °C) was obtained in total yield 47%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.41. 1H-NMR (DMSO-d6): δ 10.54(1H, s, NH), 8.14 (1H, d, J = 8.8 Hz, H-7), 8.02 (2H, d, J = 8.4 Hz, H-2′,6′), 7.954 (2H, m, H-4, 2″,6″), 7.64 (2H, d, J = 8.4 Hz, H-3′,5′), 7.57 (2H, d, J = 8.4 Hz, H-3″,5″). 13C-NMR (jmod, DMSO-d6): δ 166.49 (C=O), 164.70 (C) 153.18 (C), 146.76 (2C, dd, 1JC-F = 237 Hz, 2JC-F = 14 Hz, C-F), 140.72 (C), 137.77 (C), 136.63 (C), 133.43 (C), 131.15 (2CH), 129.73 (2CH), 128.74 (2CH), 128.55 (2CH), 127.01 (CH), 124.93 (C), 120.37 (CH). ESI-MS (m/z): calcd for C20H12ClF2N3O 384.071, found 384.071 [M+H]+.
5.2.6. N-(4-(5-Methyl-1H-benzo[d]imidazol-2-yl)phenyl)- 3,4,5-trimethoxybenzamide (6c), N-(4-(5-Methyl-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (6d), N-(4-(5-Methyl-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (6e)
A mixture of 350 mg of 4-(5-methyl-1H-benzo[d]imidazol-2-yl)aniline (1.57 mmol) (6b) and 399 mg of 3,4,5-trimethoxybenzoyl chloride (1.73 mmol) (9c), or 321 mg of 4-nitrobenzoyl chloride (1.73 mmol) (9d), or 303 mg of 4-nitrobenzoyl chloride (1.73 mmol) (9e) [ratio 1:1.1] in 10 mL of DMF was stirred at 80 °C, to obtain 6c, 6d, 6e respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in oven overnight. The solid obtained were pure for derivatives 6d and 6e. Crude of 6c was purified by flash chromatography (CHCl3/CH3OH in ratio 95/5).
Compound 6c (C24H23N3O4, MW 417.46) was obtained in total yield 35%; m.p. 287-279 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.50. 1H-NMR (DMSO-d6): δ 10.32 (1H, s, NH), 8.15 (2H, d, J = 8.8 Hz, H-3′,5′), 7.93 (2H, d, J = 8.8 Hz, H-2′,6′), 7.52-7.50 (1H, m, H-7), 7.43-7.39 (1H, m, H-4), 7.31 (2H, s, H-2″,6″), 7.02-7.00 (1H, m, H-6), 3.90 (6H, s, 2OCH3), 3.75 (3H, s, OCH3), 2.44 (3H, s, CH3). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 165.49 (C=O), 152.65 (2C), 148.24 (C), 143.68 (C), 140.73 (C), 136.11 (C), 131.69 (C), 129.50 (C), 129.28 (C), 128.62 (2CH), 127.40 (CH), 120.57 (2CH), 120.47 (CH), 117.28 (C), 113.06 (CH), 105.44 (2CH), 59.99 (OCH3), 55.97 (2OCH3), 20.94 (CH3). ESI-MS (m/z): calcd for C24H23N3O4 418.176, found 418.176 [M+H]+.
Compound 6d (C21H16N4O3, MW 372.38) was obtained in total yield 80%; m.p. 297–298 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.45. 1H-NMR (DMSO-d6): δ 10.77 (1H, s, NH), 8.40 (2H, d, J = 8.8 Hz, H-3″,5″), 8.22 (2H, d, J = 8.8 Hz, H-2″,6″), 8.17 (2H, d, J = 8.8 Hz, H-3′,5′), 7.97 (2H, d, J = 8.8 Hz, H-2′,6′), 7.47 (1H, d, J = 8 Hz, H-7), 7.37 (1H, s, H-4), 7.03 (1H, d, J = 8 Hz, H-6), 2.44 (3H, s, CH3). 13C-NMR (APT, DMSO-d6+ TFA-d): δ 164.48 (C=O), 149.35 (C), 148.11 (C), 143.23 (C), 139.87 (C), 136.15 (C), 131.69 (C), 129.50 (C), 129.29 (2CH), 128.72 (2CH), 127.44 (CH), 123.49 (2CH), 120.58 (2CH), 117.80 (C), 113.29 (CH), 113.10 (CH), 20.93 (CH3). ESI-MS (m/z): calcd for C21H16N4O3 373.130, found 373.130 [M+H]+.
Compound 6e (C21H16ClN3O, MW 361.82) was obtained in total yield 60%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.82. 1H-NMR (DMSO-d6): δ 10.52 (1H, s, NH), 8.14 (2H, d, J = 8.4 Hz, H-2″,6″), 8.02 (2H, d, J = 8 Hz, H-2′,6′), 7.95 (2H, d, J = 8.4 Hz, H-3′,5′), 7.64 (2H, d, J = 8.4 Hz, H-3″,5″), 7.46 (1H, d, J = 7.6 Hz, H-7), 7.36 (1H, s, H-4), 7.02 (1H, d, J = 8 Hz, H-6), 2.43 (3H, s, CH3). 13C-NMR (APT, DMSO-d6+TFA-d): δ 165.08 (C=O), 148.21 (C), 143.52 (C), 136.94 (C), 136.11 (C), 132.93 (C), 131.71 (C), 129.68 (2CH), 129.52 (C), 128.65 (2CH), 128.49 (2CH), 127.41 (CH), 120.46 (CH), 117.44 (C), 113.27 (CH), 113.08 (CH), 20.95 (CH3). ESI-MS (m/z): calcd for C21H16ClN3O 362.105, found 362.105 [M+H]+.
5.2.7. N-(4-(5,6-Dimethyl-1H-benzo[d]imidazol-2-yl)phenyl)-3,4,5-trimethoxybenzamide (7c), N-(4-(5,6-Dimethyl-1H-benzo[d]imidazol-2-yl)phenyl)-4-nitrobenzamide (7d), N-(4-(5,6-Dimethyl-1H-benzo[d]imidazol-2-yl)phenyl)-4-chlorobenzamide (7e)
A mixture of 300 mg of 4-(5,6-dimethyl-1H-benzo[d]imidazol-2-yl)aniline (1.26 mmol) (7b) and 321 mg of 3,4,5-trimethoxybenzoyl chloride (1.39 mmol) (9c), or 258 mg of 4-nitrobenzoyl chloride (1.39 mmol) (9d), or 243 mg of 4-nitrobenzoyl chloride (1.39 mmol) (9e) [ratio 1:1.1] in 10 mL of DMF was stirred at 80 °C, to obtain 7c, 7d, 7e respectively. The reactions were completed after 24 h. The reaction mixtures were poured into cold water, obtaining precipitation of the solid compounds. The precipitates were filtered off, washed with water and dried in oven overnight. The pure solid products 7c, 7d, 7e were obtained by washing the crudes with few millilitres of diethyl ether.
Compound 7c (C25H25N3O4, MW 431.48) was obtained in total yield 40%; m.p. 272–274 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.47. 1H-NMR (DMSO-d6): δ 10.43 (1H, s, NH), 8.16 (2H, d, J = 8.4 Hz, H-2′,6′), 8.00 (2H, d, J = 8.4 Hz, H-3′,5′), 7.46 (2H, s, H-4,7), 7.32 (2H, s, H-2″,6″), 3.89 (6H, s, 2OCH3), 3.75 (3H, s, OCH3), 2.37 (6H, s,2CH3). 13C-NMR (jmod, DMSO-d6): δ 165.20 (C=O), 152.65 (2C), 149.05 (C), 141.66 (C), 140.56 (C), 134.42 (C), 132.63 (2C), 129.62 (2C), 127.51 (2CH), 122.04 (C), 120.50 (2CH), 114.24 (2CH), 105.42 (2CH), 60.13 (OCH3), 56.13 (2OCH3), 19.94 (2CH3). ESI-MS (m/z): calcd for C25H25N3O4 432.192, found 432.192 [M+H]+.
Compound 7d (C22H18N4O3, MW 386.40) was obtained in total yield 18%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.40. 1H-NMR (DMSO-d6): δ 10.77 (1H, s, NH), 8.40 (2H, d, J = 8.8 Hz, H-3″,5″), 8.34 (2H, d, J = 2 Hz, H-2″,6″), 8.23-8.14 (4H, m, H-2′,3′,5′,6′), 7.96 (1H, d, J = 8.8 Hz, H-7), 7.35 (1H, s, H-4), 2.33 (6H, s, 2CH3). 13C-NMR (jmod, DMSO-d6): δ 165.77 (C), 164.03 (C), 150.05 (C), 149.21 (C), 140.41 (C), 139.84 (C), 136.36 (C), 132.77 (C), 130.67 (2CH), 130.48 (C), 129.25 (2CH), 126.77 (CH), 125.97 (C), 123.71 (2CH), 123.57 (2CH), 120.38 (C), 19.99 (2CH3). ESI-MS (m/z): calcd for C22H18N4O3 387.145, found 387.145 [M+H]+.
Compound 7e (C22H18ClN3O, MW 375.85) was obtained in total yield 19%; m.p. >300 °C; TLC (CHCl3/CH3OH 9.5/0.5): Rf 0.32. 1H-NMR (DMSO-d6): δ 10.54 (1H, s, NH), 8.14-7.93 (6H, m, H-2′, 3′, 5′, 6′, 2″, 6″), 7.65-7.56 (3H, m, H-3″, 5″, 7), 7.37 (1H, s, H-4), 2.34 (6H, s, 2CH3). 13C-NMR (jmod, DMSO-d6): δ 166.43 (C), 164.63 (C), 149.89 (C), 140.43 (C), 137.76 (C), 136.58 (C), 133.39 (C), 131.11 (2CH), 130.92 (C), 129.66 (2CH), 129.59 (C), 128.72 (2CH), 128.51 (2CH), 126.88 (CH), 124.86 (C), 120.30 (CH), 19.97 (2CH3). ESI-MS (m/z): calcd for C22H18ClN3O 376.121, found 376.121 [M+H]+.
Acknowledgments
The authors would like to thank Fondazione di Sardegna and the University of Sassari for supporting this research.
Author Contributions
Conceptualization, R.I., F.R. and I.D.; methodology, R.I., F.R., I.D. and I.L.; investigation, R.I., F.R., I.D., I.L. and G.C.; data curation, R.I. and F.R.; writing—original draft preparation, R.I. and F.R.; writing—review and editing, all the authors.; visualization, R.I. and F.R.; supervision, S.P., R.L. and A.C.; funding acquisition, S.P. and A.C. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Fondazione di Sardegna through the grant “Bando Fondazione di Sardegna 2022 e 2023”.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Smith D.B., Meyers G., Bukh J., Gould E.A., Monath T., Scott Muerhoff A., Pletnev A., Rico-Hesse R., Stapleton J.T., Simmonds P., et al. Proposed Revision to the Taxonomy of the Genus Pestivirus, Family Flaviviridae. J. Gen. Virol. 2017;98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierson T.C., Kielian M. Flaviviruses: Braking the Entering. Curr. Opin. Virol. 2013;3:3. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newcomer B.W. 75 Years of Bovine Viral Diarrhea Virus: Current Status and Future Applications of the Use of Directed Antivirals. Antivir. Res. 2021;196:105205. doi: 10.1016/j.antiviral.2021.105205. [DOI] [PubMed] [Google Scholar]
- 4.Ellis J.A., West K.H., Cortese V.S., Myers S.L., Carman S., Martin K.M., Haines D.M. Lesions and Distribution of Viral Antigen Following an Experimental Infection of Young Seronegative Calves with Virulent Bovine Virus Diarrhea Virus-Type II. Can. J. Vet. Res. Rev. Can. Rech. Vet. 1998;62:161–169. [PMC free article] [PubMed] [Google Scholar]
- 5.Hessman B.E., Fulton R.W., Sjeklocha D.B., Murphy T.A., Ridpath J.F., Payton M.E. Evaluation of Economic Effects and the Health and Performance of the General Cattle Population after Exposure to Cattle Persistently Infected with Bovine Viral Diarrhea Virus in a Starter Feedlot. Am. J. Vet. Res. 2009;70:73–85. doi: 10.2460/ajvr.70.1.73. [DOI] [PubMed] [Google Scholar]
- 6.Harasawa R., Mizusawa H. Demonstration and Genotyping of Pestivirus RNA from Mammalian Cell Lines. Microbiol. Immunol. 1995;39:979–985. doi: 10.1111/j.1348-0421.1995.tb03301.x. [DOI] [PubMed] [Google Scholar]
- 7.Harasawa R., Sasaki T. Sequence Analysis of the 5’ Untranslated Region of Pestivirus RNA Demonstrated in Interferons for Human Use. Biologicals. 1995;23:263–269. doi: 10.1006/biol.1995.0044. [DOI] [PubMed] [Google Scholar]
- 8.Harasawa R., Tomiyama T. Evidence of Pestivirus RNA in Human Virus Vaccines. J. Clin. Microbiol. 1994;32:1604–1605. doi: 10.1128/jcm.32.6.1604-1605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newcomer B.W., Marley M.S., Galik P.K., Walz P.H., Zhang Y., Riddell K.P., Dykstra C.C., Boykin D.W., Kumar A., Cruz-Espindola C., et al. Antiviral Treatment of Calves Persistently Infected with Bovine Viral Diarrhoea Virus. Antivir. Chem. Chemother. 2012;22:171–179. doi: 10.3851/IMP1903. [DOI] [PubMed] [Google Scholar]
- 10.Al-Kubati A.A.G., Hussen J., Kandeel M., Al-Mubarak A.I.A., Hemida M.G. Recent Advances on the Bovine Viral Diarrhea Virus Molecular Pathogenesis, Immune Response, and Vaccines Development. Front. Vet. Sci. 2021;8:475. doi: 10.3389/fvets.2021.665128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D., Sainz I.F., Ansari I.H., Gil L.H.V.G., Vassilev V., Donis R.O. The Envelope Glycoprotein E2 Is a Determinant of Cell Culture Tropism in Ruminant Pestiviruses. J. Gen. Virol. 2003;84:1269–1274. doi: 10.1099/vir.0.18557-0. [DOI] [PubMed] [Google Scholar]
- 12.Bollini M., Leal E.S., Adler N.S., Aucar M.G., Fernández G.A., Pascual M.J., Merwaiss F., Alvarez D.E., Cavasotto C.N. Discovery of Novel Bovine Viral Diarrhea Inhibitors Using Structure-Based Virtual Screening on the Envelope Protein E2. Front. Chem. 2018;6:79. doi: 10.3389/fchem.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baginski S.G., Pevear D.C., Seipel M., Sun S.C., Benetatos C.A., Chunduru S.K., Rice C.M., Collett M.S. Mechanism of Action of a Pestivirus Antiviral Compound. Proc. Natl. Acad. Sci. USA. 2000;97:7981–7986. doi: 10.1073/pnas.140220397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King R.W., Scarnati H.T., Priestley E.S., De Lucca I., Bansal A., Williams J.K. Selection of a Thiazole Urea-Resistant Variant of Bovine Viral Diarrhoea Virus That Maps to the RNA-Dependent RNA Polymerase. Antivir. Chem. Chemother. 2002;13:315–323. doi: 10.1177/095632020201300507. [DOI] [PubMed] [Google Scholar]
- 15.Sun J.-H., Lemm J.A., O’Boyle D.R., Racela J., Colonno R., Gao M. Specific Inhibition of Bovine Viral Diarrhea Virus Replicase. J. Virol. 2003;77:6753–6760. doi: 10.1128/JVI.77.12.6753-6760.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabarrini O., Manfroni G., Fravolini A., Cecchetti V., Sabatini S., De Clercq E., Rozenski J., Canard B., Dutartre H., Paeshuyse J., et al. Synthesis and Anti-BVDV Activity of Acridones as New Potential Antiviral Agents. J. Med. Chem. 2006;49:2621–2627. doi: 10.1021/jm051250z. [DOI] [PubMed] [Google Scholar]
- 17.Bukhtiyarova M., Rizzo C.J., Kettner C.A., Korant B.D., Scarnati H.T., King R.W. Inhibition of the Bovine Viral Diarrhoea Virus NS3 Serine Protease by a Boron-Modified Peptidyl Mimetic of Its Natural Substrate. Antivir. Chem. Chemother. 2001;12:367–373. doi: 10.1177/095632020101200607. [DOI] [PubMed] [Google Scholar]
- 18.Branza-Nichita N., Durantel D., Carrouée-Durantel S., Dwek R.A., Zitzmann N. Antiviral Effect of N-Butyldeoxynojirimycin against Bovine Viral Diarrhea Virus Correlates with Misfolding of E2 Envelope Proteins and Impairment of Their Association into E1-E2 Heterodimers. J. Virol. 2001;75:3527–3536. doi: 10.1128/JVI.75.8.3527-3536.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durantel D., Branza-Nichita N., Carrouée-Durantel S., Butters T.D., Dwek R.A., Zitzmann N. Study of the Mechanism of Antiviral Action of Iminosugar Derivatives against Bovine Viral Diarrhea Virus. J. Virol. 2001;75:8987–8998. doi: 10.1128/JVI.75.19.8987-8998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zitzmann N., Mehta A.S., Carrouée S., Butters T.D., Platt F.M., McCauley J., Blumberg B.S., Dwek R.A., Block T.M. Imino Sugars Inhibit the Formation and Secretion of Bovine Viral Diarrhea Virus, a Pestivirus Model of Hepatitis C Virus: Implications for the Development of Broad Spectrum Anti-Hepatitis Virus Agents. Proc. Natl. Acad. Sci. USA. 1999;96:11878–11882. doi: 10.1073/pnas.96.21.11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markland W., McQuaid T.J., Jain J., Kwong A.D. Broad-Spectrum Antiviral Activity of the IMP Dehydrogenase Inhibitor VX-497: A Comparison with Ribavirin and Demonstration of Antiviral Additivity with Alpha Interferon. Antimicrob. Agents Chemother. 2000;44:859–866. doi: 10.1128/AAC.44.4.859-866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musiu S., Pürstinger G., Stallinger S., Vrancken R., Haegeman A., Koenen F., Leyssen P., Froeyen M., Neyts J., Paeshuyse J. Substituted 2,6-Bis(Benzimidazol-2-Yl)Pyridines: A Novel Chemical Class of Pestivirus Inhibitors That Targets a Hot Spot for Inhibition of Pestivirus Replication in the RNA-Dependent RNA Polymerase. Antivir. Res. 2014;106:71–79. doi: 10.1016/j.antiviral.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Pascual M.J., Merwaiss F., Leal E., Quintana M.E., Capozzo A.V., Cavasotto C.N., Bollini M., Alvarez D.E. Structure-Based Drug Design for Envelope Protein E2 Uncovers a New Class of Bovine Viral Diarrhea Inhibitors That Block Virus Entry. Antivir. Res. 2018;149:179–190. doi: 10.1016/j.antiviral.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Vitale G., Corona P., Loriga M., Carta A., Paglietti G., Giliberti G., Sanna G., Farci P., Marongiu M.E., la Colla P. 5-Acetyl-2-Arylbenzimidazoles as Antiviral Agents. Part 4. Eur. J. Med. Chem. 2012;53:83–97. doi: 10.1016/j.ejmech.2012.03.038. [DOI] [PubMed] [Google Scholar]
- 25.Tonelli M., Simone M., Tasso B., Novelli F., Boido V., Sparatore F., Paglietti G., Pricl S., Giliberti G., Blois S., et al. Antiviral Activity of Benzimidazole Derivatives. II. Antiviral Activity of 2-Phenylbenzimidazole Derivatives. Bioorganic. Med. Chem. 2010;18:2937–2953. doi: 10.1016/j.bmc.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 26.Tonelli M., Novelli F., Tasso B., Vazzana I., Sparatore A., Boido V., Sparatore F., la Colla P., Sanna G., Giliberti G., et al. Antiviral Activity of Benzimidazole Derivatives. III. Novel Anti-CVB-5, Anti-RSV and Anti-Sb-1 Agents. Bioorg. Med. Chem. 2014;22:4893–4909. doi: 10.1016/j.bmc.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Yang Y., Li Y., Wang Y. Computational Study Exploring the Interaction Mechanism of Benzimidazole Derivatives as Potent Cattle Bovine Viral Diarrhea Virus Inhibitors. J. Agric. Food Chem. 2016;64:5941–5950. doi: 10.1021/acs.jafc.6b01067. [DOI] [PubMed] [Google Scholar]
- 28.Carta A., Loriga M., Paglietti G., Ferrone M., Fermeglia M., Pricl S., Sanna T., Ibba C., La Colla P., Loddo R. Design, Synthesis, and Preliminary in Vitro and in Silico Antiviral Activity of [4,7]Phenantrolines and 1-Oxo-1,4-Dihydro-[4,7]Phenantrolines against Single-Stranded Positive-Sense RNA Genome Viruses. Bioorganic Med. Chem. 2007;15:1914–1927. doi: 10.1016/j.bmc.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Carta A., Briguglio I., Piras S., Corona P., Boatto G., Nieddu M., Giunchedi P., Marongiu M.E., Giliberti G., Iuliano F., et al. Quinoline Tricyclic Derivatives. Design, Synthesis and Evaluation of the Antiviral Activity of Three New Classes of RNA-Dependent RNA Polymerase Inhibitors. Bioorganic Med. Chem. 2011;19:7070–7084. doi: 10.1016/j.bmc.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Asthana S., Shukla S., Ruggerone P., Vargiu A.V. Molecular Mechanism of Viral Resistance to a Potent Non-Nucleoside Inhibitor Unveiled by Molecular Simulations. Biochemistry. 2014;53:6941–6953. doi: 10.1021/bi500490z. [DOI] [PubMed] [Google Scholar]
- 31.Asthana S., Shukla S., Vargiu A.V., Ceccarelli M., Ruggerone P., Paglietti G., Marongiu M.E., Blois S., Giliberti G., La Colla P. Different Molecular Mechanisms of Inhibition of Bovine Viral Diarrhea Virus and Hepatitis C Virus RNA-Dependent RNA Polymerases by a Novel Benzimidazole. Biochemistry. 2013;52:3752–3764. doi: 10.1021/bi400107h. [DOI] [PubMed] [Google Scholar]
- 32.Carta A., Briguglio I., Piras S., Corona P., Ibba R., Laurini E., Fermeglia M., Pricl S., Desideri N., Atzori E.M.M., et al. A Combined in Silico/in Vitro Approach Unveils Common Molecular Requirements for Efficient BVDV RdRp Binding of Linear Aromatic N-Polycyclic Systems. Eur. J. Med. Chem. 2016;117:321–334. doi: 10.1016/j.ejmech.2016.03.080. [DOI] [PubMed] [Google Scholar]
- 33.Ibba R., Corona P., Carta A., Giunchedi P., Loddo R., Sanna G., Delogu I., Piras S. Antiviral Activities of 5-Chlorobenzotriazole Derivatives. Mon. Fur. Chem. 2018;149:1247–1256. doi: 10.1007/s00706-018-2234-7. [DOI] [Google Scholar]
- 34.Bahrami K., Khodaei M.M., Kavianinia I. A Simple and Efficient One-Pot Synthesis of 2-Substituted Benzimidazoles. Synthesis. 2007;4:547–550. doi: 10.1055/s-2007-965878. [DOI] [Google Scholar]
- 35.Pauwels R., Balzarini J., Baba M., Snoeck R., Schols D., Herdewijn P., Desmyter J., de Clercq E. Rapid and Automated Tetrazolium-Based Colorimetric Assay for the Detection of Anti-HIV Compounds. J. Virol. Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 36.Trott O., Olson A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. Adv. Math. 2014;262:476–483. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. [(accessed on 30 March 2022)]. Available online: https://pymol.org/2/support.html.
- 40.El Omari K., Iourin O., Harlos K., Grimes J.M., Stuart D.I. Structure of a Pestivirus Envelope Glycoprotein E2 Clarifies Its Role in Cell Entry. Cell Rep. 2013;3:30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]