We read with interest the study by Kayisoglu et al1 comparing the gene expression between embryo-derived and adult-derived intestinal organoids. Some innate immune system genes were differentially expressed between the two organoid types, suggesting a potential role of exposure to the environment, including gut microbiota, in shaping the intestinal gene expression. In extremely preterm infants (<32 weeks gestation), microbial–host interaction at the epithelial surface has been associated with various morbidities including late onset sepsis and necrotising enterocolitis.2 Thus, preterm intestinal organoids may provide a specific and robust model for this population.
We expand on the work carried by Kayisoglu et al1 by comparing whole transcriptome sequencing (RNA-sequencing) analysis performed on four preterm (23–32 weeks gestation) and five adult (30–60 years of age)3 intestinal organoid lines exposed to Enterococcus faecalis, a prevalent gut pathobiont.4 Human intestinal organoid lines were generated from preterm (surgical resection) and adult (biopsy) following the protocol described by Stewart et al.5 All specimens were from the ileum. Intestinal organoids were exposed to Enterococcus faecalis in form of monolayer using the organoid-anaerobe co-culture (OACC) model.6 All data are available at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), Accession: GSE161953.
We first compared transcriptomic profiles from preterm and adult organoid monolayers following 24 hours of culture in the OACC system, finding significant separation of preterm and adult lines (2000 permutations, pR2Y=0.047, pQ2=0.007; figure 1A). A total of 490 differentially expressed genes (DEGs) were found when comparing all preterm and adult monolayers (all adj. p<0.05; figure 1B). These included immune response associated genes such as eight human leucocyte antigen system genes, interleukin 7 (IL7), IL1 receptor antagonist and IL10 receptor subunit betaKyoto Encyclopedia of Genes and Genomes (KEGG)pathways significantly over-represented included ‘Antigen processing and presentation’ and ‘Intestinal immune network for IgA production’ (all adj. p<0.05). To further explore the response to bacteria within preterm compared with adult lines, we next analysed the two exposure conditions (media only or bacteria) separately. While 137 DEGS were found between preterm and adult monolayers exposed to media only, the presence of bacteria increased the number of DEGs to 1204 (figure 1C). This suggests that preterm tissue derived organoids respond differently to specific bacteria (E. faecalis) when compared with ileal organoids established from adults.
Figure 1.
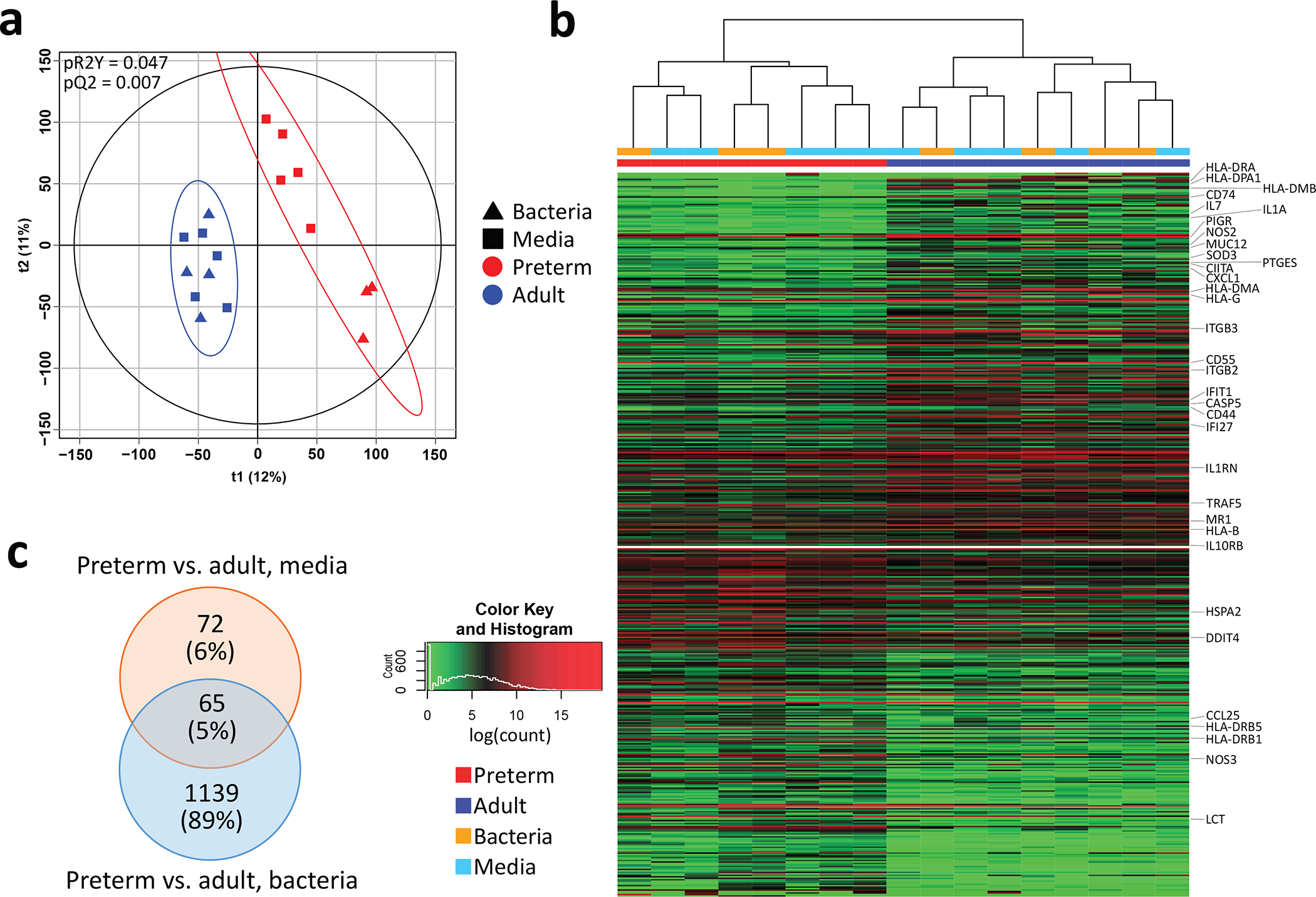
Comparison of preterm and adult ileal organoid lines following 24 hours of culture. Bacterial treatment was with Enterococcus faecalis. (A) Partial least squares discriminant analysis of preterm and adult intestinal organoid lines. Plot coloured by line and shaped by exposure (media only or bacteria). P values were calculated based on 2000 permutations. (B) Heatmap of genes differentially expressed between preterm and adult organoids (all adj. p<0.05). (C) Venn diagram of significantly differentially expressed genes between preterm and adult lines within media only or bacterial exposure conditions (all adj. p<0.05).
Within the preterm lines, we next investigated the influence of culture time on the transcriptome by comparing monolayers maintained in the OACC model for 8hours and 24 hours. Transcriptomic profiles were significantly different depending on time of culture (2000 permutations, pR2Y=0.006, pQ2 <0.001; figure 2A). A total of 598 DEGs were identified when comparing the two time points without separating for exposure condition (all adj. p<0.05; figure 2B), including genes associated to cell cycle, such as M-phase inducer phosphatase 3 (CDC25C), and immune response, including IL6 receptor and IL18 binding protein. Comparing between 8hours and 24 hours within media only cultures showed 112 DEGs, but comparing between the two time points following bacterial co-culture showed 595 DEGs (figure 2C).
Figure 2.
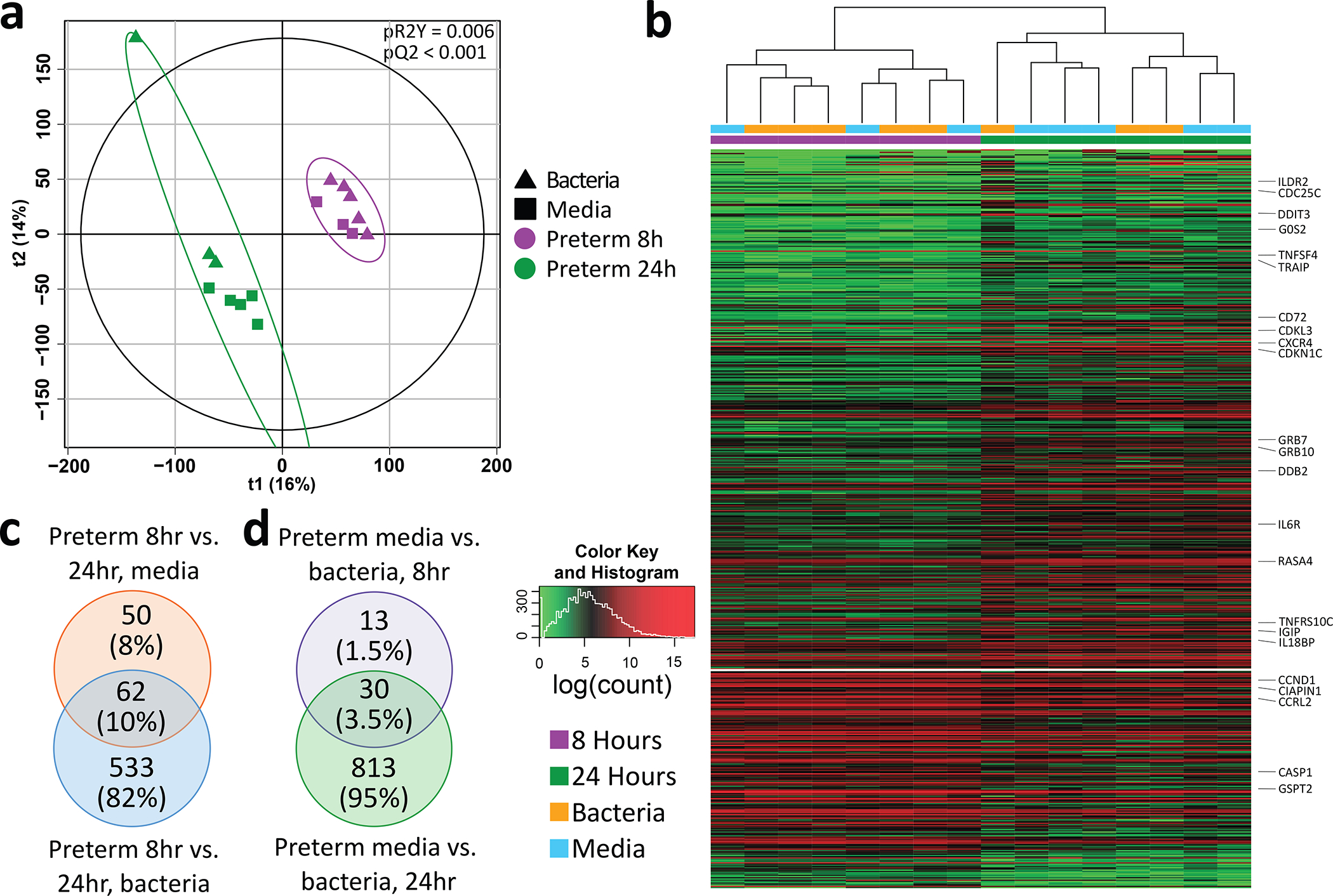
Comparison of preterm lines cultured for 8 hours and 24 hours. Bacterial treatment was with Enterococcus faecalis. (A) partial least squares discriminant analysis of preterm monolayers cultured for 8 and 24. Plot coloured by time point and shaped by exposure (media only or bacteria). P values were calculated based on 2000 permutations. (B) Heatmap of genes differentially expressed between preterm organoids cultured for 8 and 24 hours (all adj. p<0.05). log transformed normalised counts were used. (C) Venn diagram of significantly differentially expressed genes associated to comparison of preterm organoids cultured for 8 and 24 hours separating by exposure condition (all adj. p<0.05). (D) Venn diagram of significantly differentially expressed genes between preterm organoids exposed to media and bacteria by time point (8 and 24 hours) (all adj. p<0.05).
Finally, we sought to analyse the impact of exposure (media only or bacteria) within each time point in preterm lines. At 8hours, 43 DEGs were found between media only and bacteria, whereas at 24 hours 843 DEGs were found, of which 30 were shared between 8 and 24 hours (figure 2D).
Taken together, the results show that preterm intestinal organoids are inherently different and respond differently to bacterial exposure when compared with adult lines. This provides evidence for the utility of preterm intestinal organoids for modelling the role of microbe–host interaction in this population.
Funding
CS received funding from the Wellcome Institutional Strategic Support Fund (ISSF) and Newcastle University Academic Track scheme.
Disclaimer The funders had no role in the design and conduct of the study.
Footnotes
Competing interests TYF, JMA, RAB, MKE, JFP, CJS have filled a patent for the enteroid anaerobic co-culture system. CJS has received honorarium from Danone Nutricia.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication Not applicable.
Ethics approval This study was approved by Institutional Review Board for Baylor College of Medicine and Affiliated Hospitals (H-38591).
Provenance and peer review Not commissioned; internally peer reviewed.
REFERENCES
- 1.Kayisoglu O, Weiss F, Niklas C, et al. Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium. Gut 2021;70:687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranowski JR, Claud EC. Necrotizing enterocolitis and the preterm infant microbiome. Adv Exp Med Biol 2019;1125:25–36. [DOI] [PubMed] [Google Scholar]
- 3.Rajan A, Vela L, Zeng X-L. Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. MBio 2018;9:e02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black CG, Tavares L, Stachel A, et al. Distribution of late-onset neonatal sepsis pathogens differs in inpatient and outpatient settings. Am J Perinatol 2019;36:1136–41. [DOI] [PubMed] [Google Scholar]
- 5.Stewart CJ, Estes MK, Ramani S. Establishing human intestinal Enteroid/Organoid lines from preterm infant and adult tissue. Methods Mol Biol 2020;2121:185–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fofanova TY et al. A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia. bioRxiv 2019:555755. [Google Scholar]


