Abstract
Human caliciviruses (HuCVs) cause waterborne outbreaks of gastroenteritis. Standard indicators of a safe water supply do not adequately predict contamination of water by viruses, including HuCVs. We developed a method to concentrate and detect HuCVs in water samples by using a cultivable primate calicivirus (Pan-1) as a model. Viable Pan-1 was seeded in different types of water and then filtered with a 1MDS filter, eluted with beef extract (BE), and reconcentrated by polyethylene glycol (PEG) precipitation. The viruses in the final samples were tested by plaque assay or by reverse transcription (RT)-PCR following extraction of the RNA with Trizol. Pan-1 was more sensitive to high-pH treatment than poliovirus was; a pH 9.0 BE solution was found to recover 35% more viable Pan-1 than a pH 9.5 BE solution recovered. Pan-1 was recovered from small volumes of deionized, finished, ground, and surface waters at efficiencies of 94, 73, 67, and 64%, respectively, when samples were assayed after elution without further concentration. When larger volumes of water (up to 40 liters) were tested after elution and concentration with PEG, 38, 19, and 14% of the seeded Pan-1 were recovered from finished, ground, and surface waters, respectively. The limit of detection of Pan-1 by RT-PCR was estimated to be 0.75 to 1.5 PFU in 40 liters of finished water. This method may be adapted for monitoring HuCVs in drinking water and other types of water for public health safety.
Diarrhea remains an important disease in both developed and developing countries. Among the different gastroenteritis viruses, Norwalk virus and Norwalk-related viruses, now classified as caliciviruses (CVs), play a major role in nonbacterial outbreaks of acute gastroenteritis (23). Large outbreaks of waterborne acute gastroenteritis caused by human CVs (HuCVs) have been documented; in these outbreaks fecal contamination of drinking water or indirect contamination of water or water products occurred (15, 16, 18, 20, 25, 26).
Unlike many bacterial pathogens, which have been controlled largely by modern water and wastewater treatment practices, the incidence of water-related viral diseases, including gastroenteritis and hepatitis, has remained virtually unchanged over the past several decades (23). Use of bacterial pathogens as indicators of clean and unpolluted water does not predict the safety of water with respect to viral pathogens (8, 17, 21, 24, 27). Therefore, development of sensitive methods to monitor enteric viruses in water is necessary.
Monitoring water quality by direct detection of human enteric viruses has been difficult because only a few infectious units are required for most human enteric viruses to cause infection. Detection of such low concentrations of viruses in environmental samples usually requires concentration of virus from large volumes of water. Even with viruses highly concentrated from water samples, the methods routinely used to detect enteric viruses in clinical specimens, such as enzyme immune assays and cell culture, still are not sensitive enough. In addition, detection of viruses by cell culture is applicable to certain virus families but many enteric viruses cannot be replicated in cell culture.
The recent development of molecular methods, such as PCR, reverse transcription-PCR (RT-PCR), and nucleotide hybridization (10, 13), provides hope for rapid and sensitive detection of pathogenic enteric viruses in water at levels that could predict water safety. Such techniques have been developed for detection of enteroviruses (poliovirus, coxsackieviruses, and echoviruses) and hepatitis A and E viruses in water and water products (1, 6, 7, 11, 12, 14, 19, 22, 29, 30, 35, 36, 40). Methods for detection of HuCVs by RT-PCR in sewage, oysters, and other food products have been reported (2–4, 20, 32–34, 40). Methods for detection of CVs in large volumes of drinking water, surface water, and groundwater are lacking.
In this study, we developed a method for concentration and detection of CVs in large-volume water samples by RT-PCR. Because HuCVs have not been cultivated in cell culture, we used a cultivable CV, the primate CV Pan-1 strain, as a model in seeding experiments to study CV recovery and detection in different types of water.
MATERIALS AND METHODS
Viruses and cell cultures.
Pan-1 originally was isolated from a pygmy chimpanzee (37). The three-dimensional structure of the Pan-1 virion and the sequence of the Pan-1 genome are known (28, 31). Pan-1 grows to a high titer and causes cytopathic effects in monkey cell lines. Pan-1 was grown in LLC-MK2 cells (American Type Culture Collection), which were maintained in medium 199 (Gibco BRL, Grand Island, N.Y.) supplemented with 5% fetal bovine serum. Plaque-purified Pan-1 was used to infect cell monolayers in T-150 flasks at a multiplicity of infection of 1. Infected cells were harvested 16 to 24 h postinfection. The cells were frozen and thawed three times and then were clarified by centrifugation for 15 min at 11,300 × g to remove debris. The supernatant was divided into aliquots, titrated by a plaque assay (PA) and RT-PCR, and stored at −70°C as a virus stock. For each experiment, a fresh aliquot from the freezer was used to avoid repeated freeze-thaw cycles.
PA.
A PA was performed with LLC-MK2 cell monolayers in six-well tissue culture plates (Falcon, Becton Dickinson and Company, Franklin Lakes, N.J.) for both Pan-1 and poliovirus. Triplicates of serial dilutions of each sample were inoculated onto 90 to 100% confluent cell monolayers. The viruses were adsorbed to the monolayers for 2 h at 37°C. The inoculum then was removed, and an agarose overlay (0.6% Seakem LE agarose [FMC BioProducts, Rockland, Maine] in medium 199 with 5% fetal bovine serum) was added to each well. After incubation for 12 to 16 h at 37°C, a second agarose medium overlay containing 0.05% neutral red was added. Pan-1 plaques were counted after 4 h of incubation at 37°C. Poliovirus plaques were counted 24 to 36 h postinfection. Wells containing 5 to 60 plaques per well were counted. PFU were calculated by using the average number of plaques in duplicate samples and multiplying by the dilution factor for the wells.
RT-PCR.
Viral RNA was extracted from seeded water samples by the Trizol method (Gibco BRL) and was detected by RT-PCR (9). The primers used for RT-PCR included Pan-1 36 (5′-ATCCAAGTTGGCATCAACA; nucleotides 4727 to 4745 of the Pan-1 genome) and Pan-1 35 (5′-CGGGTCGGTTTCAGACCAAAC; nucleotides 5220 to 5200), which were designed based upon the RNA polymerase sequence of Pan-1 (GenBank accession number AF091736). RT was performed in 50 μl of RT reaction mixture that contained 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 50 mM KCl), each deoxynucleoside triphosphate at a concentration of 400 μM, 0.2 μg (30 to 50 pmol) of negative-strand primer, 10 U of RNasin, and 7.5 U of avian myeloblastosis virus reverse transcriptase and was incubated for 1 h at 42°C. For PCR, 50 μl of 1× PCR buffer containing 10 U of Taq polymerase and 0.2 μg of positive-sense primer was added. The thermocycling program included 2 min at 94°C, 40 cycles of 30 s at 94°C, 1 min at 49°C, and 1 min at 72°C, and a final extension for 10 min at 72°C. RT-PCR products were analyzed by agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light.
To quantify Pan-1 by RT-PCR, serial 10-fold dilutions of each sample were tested to determine the end point of detection for the assay, which represented 1 RT-PCR detection unit. The total number of RT-PCR detection units in a sample was calculated from the dilution factor and the original volume of the sample tested.
Internal RNA control.
RNA transcripts were generated by in vitro transcription using DNA templates cloned into the pGEM-T vector (Promega, Madison, Wis.). Each DNA template contained the Pan-1 36 primer sequence at one end and the Pan-1 35 primer sequence at the other end. Two such DNA templates (700 and 400 bp) were generated. In vitro transcription was performed by using the cloned plasmid DNA as the template and T7 polymerase (Promega) under conditions described by the manufacturer. After transcription, the template DNA was removed by treatment of the sample with DNase. Removal of DNA was confirmed by the lack of reaction of the final preparation containing the RNA transcripts in a PCR without RT. Detection of the RNA transcripts, as determined by RT-PCR performed with serial dilutions, was used to monitor inhibitors in the field studies.
Seeding and filtration of CVs.
Seeding experiments were performed to evaluate individual steps, including concentration, reconcentration, and detection of CVs in water samples. Different volumes of finished water (tap water), surface water (source water), and groundwater were seeded with predetermined amounts of Pan-1 and then processed to concentrate, reconcentrate, and detect the virus. Tap water was collected in the laboratory, and the water was dechlorinated by addition of 52 mg of sodium thiosulfate per liter before seeding. Surface water was collected from Lake Wright and Lake West at outlets in the Norfolk City Utilities Laboratory. Groundwater was collected from a well in a residential area of Virginia Beach, Va. The groundwater was acidic (pH 5.2) and was adjusted to pH 7.0 to 7.5 before seeding with Pan-1. The pH of the surface water was around 7.0, and this water was used without pretreatment. After the virus was added, each water sample was held for 30 min at room temperature before it was processed to determine the concentration. Forty liters of a water sample was seeded with Pan-1, and a double layer of 90-mm-diameter 1MDS disk filters (Cuno, Inc., Meriden, Conn.) was used to adsorb the virus. The flow rate was controlled at 1.0 to 1.2 ml/min per cm2 of filter surface by using a regulated air gas cylinder as the source of positive pressure. All 40 liters of finished water or ground water was passed through the filter, but only 2 liters of surface water was passed through due to the high turbidity of the water. After filtration, the virus on the filter was eluted and reconcentrated as described below.
Elution and reconcentration of CVs.
To elute virus from a 1MDS filter, the filter was flushed with variable eluents with positive pressure from an air gas cylinder. The filter was allowed to soak in the eluent for 1 to 2 min before the eluent was washed off the filter. The elution step was repeated once. The eluents used for comparison included different concentrations of beef extract V (BE) (Becton Dickinson and Company, Cockeysville, Md.), 0.4 M NaCl, and 0.05 M glycine. To evaluate the efficiencies of individual eluents for recovery of CVs, the eluates were assayed directly or after further concentration steps.
Two methods for reconcentration of CVs from eluates were compared: polyethelene glycol (PEG) precipitation and organic flocculation (OF). For PEG precipitation, eluates from filters were adjusted to pH 7.0 to 7.5, and PEG 8000 (Sigma Chemical Co., St. Louis, Mo.) was added to the eluates at a final concentration of 8%. In our early experiments NaCl was added in this step, but in our final protocol it was added in the BE eluent as described above. Viruses were precipitated by stirring the eluates for 1.5 to 2 h at room temperature or overnight at 4°C, and precipitated viruses were collected by centrifugation for 20 min at 10,000 × g. The resulting pellets were either processed for extraction of viral RNA or suspended in Hanks balanced salt solution (HBSS) and treated with antibiotics for PA. For OF, eluates were adjusted to pH 3.5 with 1 N HCl, stirred for 30 min to allow flocculation, and then centrifuged for 20 min at 10,000 × g. The pellet was treated in the same way that the PEG-precipitated samples were treated.
Extraction of viral RNA from water concentrates.
The cetyltrimethylammonium bromide (CTAB) (hexadecyl, trimethylammonium bromide; Sigma catalog no. H5882) method for extraction of HuCV RNA from stool specimens (10) was used directly for extraction of Pan-1 RNA in our initial comparison of the methods. After comparison with the Trizol method, the CTAB method was excluded from the seeding experiments. Different Trizol method conditions for extraction and detection of Pan-1 RNA in water concentrates were compared; the volume of Trizol used for extraction and the times of Trizol-chloroform extraction were varied according to the volume and turbidity of the sample tested. Briefly, 0.5 to 1 ml of Trizol was added to each PEG-concentrated virus pellet. After vortexing for 1 min and incubation for 5 min at room temperature, the samples were extracted with 200 μl of chloroform. If a thick interphase between the water and organic phases was seen, the sample was reextracted with Trizol and chloroform until the interphase cleared. Viral RNA in the aqueous phase then was precipitated with 1 volume of isopropanol, and the final RNA pellets were resuspended in water and the RNA was directly detected by RT-PCR.
Southern blot hybridization.
The identities of RT-PCR products were confirmed by Southern blot hybridization by using an internal oligonucleotide probe (5′-TCGGCATGTGCAGCACTCAAA; nucleotides 4910 to 4930 of the Pan-1 genome). Digoxigenin (DIG)-labeled oligonucleotide probes were prepared by using a DIG oligonucleotide 3′-end labeling kit (Boehringer GmbH, Mannheim, Germany). PCR products were denatured and electrophoretically transferred from the agarose gel to a positively charged nylon membrane (Nytran plus; Schleicher & Schuell, Keene, N.H.). The transferred DNA then was cross-linked to the membrane by UV light. The membrane was prehybridized for at least 1 h in a hybridization solution and then hybridized overnight in the presence of 20 pmol of labeled oligonucleotide probe per ml. The membrane then was washed twice with 50 ml of 2× SSC containing 1% sodium dodecyl sulfate per 100 cm2 and twice with 0.1× SSC containing 0.5% sodium dodecyl sulfate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). A hybridization signal was detected with a DIG nucleic acid detection kit (Boehringer).
RESULTS
Outline of methods used for concentration and detection of CVs in large volumes of water.
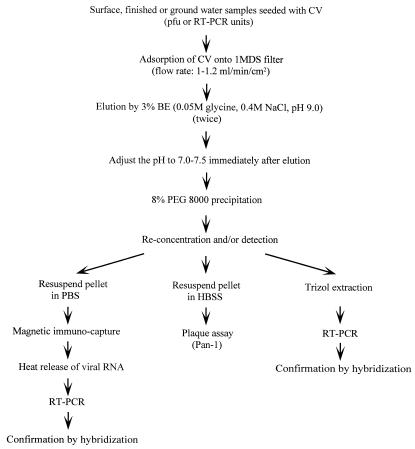
Figure 1 shows the steps used for concentration and detection of CVs in different types of water samples. Each step was evaluated by Pan-1 seeding experiments, which are described below.
FIG. 1.
Outline of the steps used for concentration and detection of CVs in different water samples by RT-PCR. HBSS, Hanks balanced salt solution.
Titration of Pan-1 by PA and RT-PCR.
The Pan-1 stock used for the seeding experiments was titrated for viable viruses by PA and for total viral RNA by RT-PCR. These analyses yielded 2.2 × 107 PFU per ml and 5.0 × 108 RT-PCR units per ml. Therefore, 1 PFU of Pan-1 was estimated to represent 23 RT-PCR units. This ratio remained consistent when preparations were retested during seeding experiments.
Utilization of internal RNA controls to monitor inhibitors.
Both RNA transcripts were useful for monitoring inhibitors of RT-PCR (Fig. 2), but the larger transcript (700 bp) was preferred because it had a less competitive effect on the viral RNA (data not shown). For best results, a minimal detectable amount of RNA (2 μl, 0.14 to 1.4 pg/μl) was used. To ensure reproducible results, this amount of RNA was prepared in small aliquots and stored at −70°C. For some experiments, the control RNAs were tested separately from Pan-1 with duplicate samples.
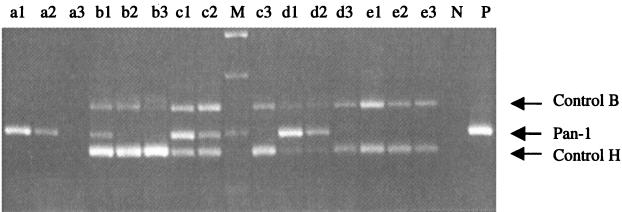
FIG. 2.
Titration of internal control RNAs by RT-PCR in the presence of Pan-1 RNA. Serial dilutions of internal RNA controls (10−6 to 10−8) and Pan-1 RNA (10−5 to 10−7) were tested separately (lanes a1 through a3, Pan-1 RNA diluted 10−5, 10−6, and 10−7, respectively; lanes e1 through e3, control RNAs H and B diluted 10−6, 10−7, and 10−8, respectively) or with combinations of different dilutions of RNAs. Lanes b1 through b3 contained 10−6 dilutions of control RNAs H and B and 10−5, 10−6, and 10−7 dilutions of Pan-1 RNA, respectively; lanes c1 through c3 contained 10−7 dilutions of control RNAs H and B and 10−5, 10−6, and 10−7 dilutions of Pan-1 RNA, respectively; and lanes d1 through d3 contained 10−8 dilutions of control RNAs H and B and 10−5, 10−6, and 10−7 dilutions of Pan-1 RNA, respectively. Lane M contained a 1-kb DNA marker. Lanes N and P contained negative and positive controls for RT-PCR.
Adsorption of Pan-1 by the 1MDS filter.
The first step used to concentrate viruses from water samples was to adsorb the viruses by filtration. High levels of adsorption (86 to 99%) of Pan-1 from seeded water samples by the 1MDS filter were observed (Table 1). The level of adsorption of Pan-1 from seeded surface water was lower than the levels of adsorption of Pan-1 from other types of water. The surface water had higher turbidity (∼9 nephelometric turbidity units) than the other types of water. The pHs of the finished water (∼7.0), surface water (∼7.0), and groundwater (∼5.2) were within the range (pH 3.5 to 8.0) for adsorption of enteric viruses, as suggested by the manufacturer, based on studies of poliovirus (38, 39). Adsorption was significantly decreased when Pan-1 was seeded in 0.1 M phosphate-buffered saline (data not shown), suggesting that the presence of Cl− and PO43− anions might affect binding of Pan-1 to the filter.
TABLE 1.
Adsorption and elution of Pan-1 seeded in different types of water followed by passage through a 1MDS filtera
| Type of water | % Adsorbed (range) | % Eluted (range) | % Recovery (range) |
|---|---|---|---|
| Deionized | >99 | 95 (85–104) | 94 (84–103) |
| Finished | >99 | 74 (74–75) | 73 (73–74) |
| Ground | >99 | 68 (62–73) | 67 (62–72) |
| Surface | 86 (85–87) | 74 (66–82) | 64 (57–71) |
Pan-1 was seeded in different types of water at a concentration of 220 PFU/ml. Adsorption of Pan-1 to a 1MDS filter was determined by comparison of Pan-1 titers in the water, as determined by PA, before and after passage through the filter. The recovery rate was determined by dividing the Pan-1 titer in the eluate by the titer seeded. The elution rate was calculated based on the Pan-1 titer in the eluate relative to the estimated amount of Pan-1 adsorbed to the filter. In the case of deionized, finished, and ground water samples, the adsorption rates were estimated to be 99%. Viruses were eluted with 3% BE (pH 9.0).
Elution and reconcentration of Pan-1 from the 1MDS filter.
The next steps were to elute the viruses from the filters and to concentrate the viruses. We compared two concentrations (1.5 and 3.0%) of BE for elution of Pan-1 from the 1MDS filter; 3% BE resulted in a higher rate of recovery of Pan-1 than 1.5% BE (data not shown). A BE concentration of 3% also resulted in larger pellets when the samples were processed by the OF method but not when they were processed by the PEG method (data not shown). Therefore, 3% BE and PEG were used. The PEG-derived samples also were suitable for extraction of viral RNA by the Trizol method. In repeated seeding experiments, up to 40 liters of tap water processed with 200 ml of 3% BE followed by PEG precipitation resulted in no inhibition of RT-PCR, as determined by using seeded internal control RNA (data not shown).
In the elution and reconcentration experiments, we observed significant inactivation of Pan-1 by high pH (pH 9.5 for elution) and low pH (pH 3.5 for precipitation by OF) (Table 2). Similar pH treatments did not decrease the infectivity of polioviruses (Table 2). The rates of recovery of viable Pan-1 were highest when a pH 9.0 eluent was used (Table 3). Therefore, pH 9.0 was used to elute Pan-1, and this step was performed as quickly as possible (within 15 min).
TABLE 2.
Effect of pH on the infectivity of Pan-1 and poliovirus as determined by PA
| Treatmenta | Pan-1
|
Poliovirus
|
||
|---|---|---|---|---|
| Concn (PFU/ml) | % Reduction | Concn (PFU/ml) | % Reduction | |
| Untreated | 2.8 × 107 | NAc | 4.5 × 108 | NA |
| pH 9.5 | 1.7 × 107 | 39 | 3.6 × 108 | 20 |
| pH 9.5 and then pH 3.5b | 9.0 × 104 | 99 | 4.1 × 108 | 9 |
Samples were not treated or were treated for 30 min at different pHs and then assayed for infectious viruses by the PA. The reductions in virus titers were determined based on the numbers of PFU in the untreated samples compared with those in the treated samples.
pH 9.5 and 3.5 were recommended for elution and reconcentration, respectively, of enteric viruses from water samples by the OF method.
NA, not applicable.
TABLE 3.
Elution of Pan-1 from 1MDS filters with 3% BE at different pH values
| pH | Mean % recovery (range)a |
|---|---|
| 8.5 | 68 (52–84) |
| 9.0 | 87 (83–90) |
| 9.5 | 52 (35–69) |
| 10.0 | <1.0 |
The recovery rates were based on PA results of three replicate experiments.
Comparison of RNA extraction methods for RT-PCR.
After reconcentration, viruses in the water samples were detected by cell culture or by RT-PCR. We compared different methods, including the CTAB method and different versions of Trizol methods, for extraction of viral RNA from the water concentrates for RT-PCR. The Trizol methods were comparable to, or more sensitive than, the CTAB method for detection of Pan-1 and involved fewer steps than the CTAB method (data not shown). The Trizol methods were particularly useful for highly turbid samples because they permitted multiple extractions of a single sample.
Confirmation of RT-PCR results by Southern blot hybridization.
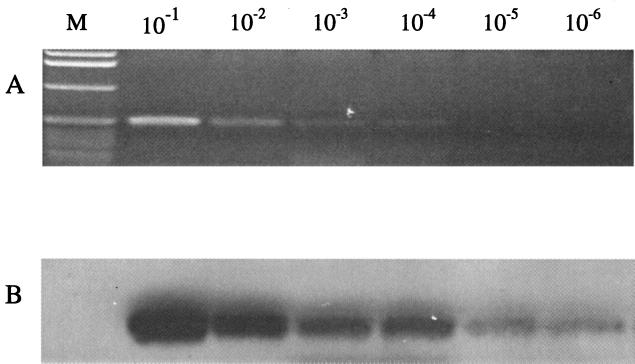
The last step was to confirm the results by hybridization. Direct comparison of RT-PCR results by agarose gel electrophoresis and by Southern blot hybridization showed that hybridization increased the sensitivity 10- to 100-fold with radiolabeled (data not shown) or nonradiolabeled probes compared with ethidium bromide-stained gels (Fig. 3). The nonradiolabeled probes resulted in less biohazard waste.
FIG. 3.
Detection of Pan-1 RNA by RT-PCR followed by ethidium bromide staining or Southern blot hybridization with DIG-labeled oligonucleotide probes. Serial 10-fold dilutions of Pan-1 RT-PCR products were electrophoresed in an agarose gel. The gel then was stained with ethidium bromide (A) and subsequently transferred to a Nytran membrane for hybridization (B) by using conditions described in Materials and Methods, and the hybridized signals were detected by chemoluminescence with a DIG nucleic acid detection kit (Boehringer). Lane M contained a 1-kb marker (Gibco BRL).
Recovery of Pan-1 in seeded field water samples.
After evaluation of the individual steps, the entire procedure was further evaluated to determine the rates of recovery of Pan-1 from different types of water in two sets of seeding experiments. In the first set, small volumes (200 ml) of water samples and a high concentration of Pan-1 (220 PFU/ml) were used. Pan-1 from the 1MDS filter was tested with the PA without further concentration. Pan-1 was recovered from deionized, finished, ground, and surface waters at efficiencies of 94, 73, 67, and 64%, respectively (Table 1).
In the second set of seeding experiments, larger volumes (up to 40 liters) of water samples were seeded with Pan-1 at concentrations of 0.375 to 1.5 PFU/ml. After concentration and elution from a 1MDS filter, the samples were processed by performing PEG reconcentration followed by the PA. The rates of recovery of seeded Pan-1 were 38, 19, and 14% for finished, ground, and surface waters, respectively, and the rates of recovery of poliovirus (a pH 9.5 eluent was used) in finished water were 51 to 55% (Table 4). Aliquots consisting of one-third of the final concentrates of finished water samples were processed and tested by RT-PCR. The end point for detection of Pan-1 by this method was at dilutions of the final extracted RNA of 1:500 to 1:1,000 (data not shown). On the basis of these data, the limit of detection by RT-PCR was estimated to be 0.75 to 1.5 PFU in 40 liters.
TABLE 4.
Recovery of Pan-1 and poliovirus seeded in different water samples followed by concentration and detection with PA
| Water type | Sample pH | Vol (liter) | Turbidity (NTU)a | % Recovery (range)b |
|---|---|---|---|---|
| Pan-1 | ||||
| Surface | 7.1 | 1.9–2.3 | 8.9 | 14 (12–16) |
| Ground | 5.2c | 40 | 0.5 | 19 (18–20) |
| Finished | 6.9 | 40 | NTd | 38 (32–44) |
| Poliovirus | ||||
| Finished | 6.9 | 40 | NT | 53 (51–55) |
NTU, nephelometric turbidity units.
The recovery rates were based on the results of three replicate experiments. One-third of each final sample was tested by PA at dilutions of 1:50 and 1:100.
The original pH of groundwater was 5.2, and the pH was adjusted to 7.0 to 7.5 before the virus was added.
NT, not tested.
DISCUSSION
We developed a method to concentrate and detect CVs in water samples by using a cultivable animal CV as a model. Pan-1 seeded in water samples was measured by both plaque and RT-PCR assays after each concentration step. Pan-1 was efficiently adsorbed to and eluted from 1MDS filters and was effectively recovered in the subsequent reconcentration steps. RT-PCR was significantly more sensitive than the PA for detection of Pan-1 in the water concentrates. In seeding experiments in which up to 40 liters of finished water was used, the limit of detection of Pan-1 by RT-PCR was estimated to be 0.75 to 1.5 PFU. This level of sensitivity may allow monitoring of HuCVs in drinking water for public health safety. The use of a cultivable virus as a model in the study also provided ways of estimating effects of the methods used on viable CVs in water.
When we compared our method for CVs with methods described in the literature for other viruses, several differences were noted. First, the pH of the solution used to elute viruses from the filter in our protocol was slightly lower. Most previously described methods used solutions with a higher pH (pH 9.5) for elution of enteric viruses. However, we found that Pan-1's infectivity was reduced at high pH more than the infectivity of poliovirus was reduced. A slightly lower pH of BE (pH 9.0 instead of pH 9.5) resulted in significantly decreased inactivation of Pan-1. Whether the lower pH is suitable for recovery of other CVs and enteric viruses remains to be determined.
Second, a high concentration of BE (3%) was used in our protocol to elute Pan-1 from the filters. One concern about the high concentration of BE is that BE may coprecipitate with viral RNA and interfere with the RT-PCR assay. This influence was encountered when the samples were concentrated by the OF method but not when they were concentrated by the PEG method. The PEG-treated RNA pellets were significantly smaller than the OF-treated samples, while detection of Pan-1 in the PEG-treated samples was more sensitive than detection in the OF-treated samples. The use of PEG also avoided potential inactivation of Pan-1 at a low pH (pH 3.5), which is required for the OF method.
Third, the BE eluent in our protocol included a high concentration of salts. A high salt concentration is required for reconcentration of viruses from water samples by PEG precipitation. In our experiments, we observed that a high salt concentration facilitated Pan-1 elution from 1MDS filters. In standard protocols for PEG methods, salts usually are added together with PEG. We added the salt directly to the eluent before PEG was added, and this modification increased the rates of recovery of Pan-1. In a direct comparison of elution of Pan-1 by 3% BE with and without NaCl, BE with 0.4 M NaCl recovered 14 to 16% more virus (data not shown).
Last, in the previous studies, internal control RNAs smaller than the target RNA usually were used (3, 34). We selected an RNA transcript larger than the viral target RNA, because a larger RNA has less interference, is less easily detected, and, therefore, is more sensitive to inhibitors than a smaller RNA. Our results showed that the competition between internal and viral RNAs was dose dependent, and a minimum detectable amount of the control RNA is recommended. Furthermore, because the amounts of target viral RNA in environmental samples are expected to be low, parallel testing for viral and control RNAs in separate tubes is recommended.
During development of our methods, we also noticed some problems. First, the rates of recovery of Pan-1 varied for different water types and were lower than those of poliovirus. This could be related to the unique sensitivity of CVs to high pH values. Second, our method has a limited ability to remove inhibitors in heavily contaminated and large-volume surface water samples. Further modification of our method, such as incorporation of the silica membrane method or high-salt precipitation to remove polysaccharide and proteoglycan from water samples, should be tried (5). Third, because HuCVs still cannot be cultivated in cell culture, alternative methods should be developed to measure infectious viruses in water samples. One approach currently being assessed in our laboratory is an immune capture RT-PCR. This method may be more specific and more sensitive than conventional RT-PCR. It also detects capsid-associated viral RNA, which is likely to be more infectious than naked RNA. A panel of hyperimmune and monoclonal antibodies against baculovirus-expressed HuCV capsid antigens from different genogroups or genetic clusters of HuCVs is now available, which should facilitate development of this method in the near future. Finally, although Pan-1 has been well studied and its genetic and morphological features have revealed many similarities with Norwalk virus, Pan-1 is an animal CV, and it is not known whether it replicates in the gastrointestinal tract like most HuCVs. Therefore, Pan-1 may not provide a perfect model for HuCV transmission and survival in the environment. Further studies to characterize Pan-1 and to search for other candidate model strains for HuCVs are necessary.
ACKNOWLEDGMENTS
This study was supported by the American Water Works Association Research Foundation (AWWARF project 345).
We thank the advisory committee members, Shay Fout, Dennis R. Lang, Bruce Roll, and Marleen Wekell, and the program manager, Kathryn Martin, for their critical comments and help in the study.
REFERENCES
- 1.Abbaszadegan M, Huber M S, Gerba C P, Pepper I L. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:1318–1324. doi: 10.1128/aem.59.5.1318-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar R L, Metcalf T G, Neill F H, Estes M K. Detection of enteric viruses in oysters by using the polymerase chain reaction. Appl Environ Microbiol. 1993;59:631–635. doi: 10.1128/aem.59.2.631-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar R L, Neill F H, Romalde J L, Le Guyader F, Woodley C M, Metcalf T G, Estes M K. Detection of Norwalk virus and hepatitis A virus in shellfish tissues with the PCR. Appl Environ Microbiol. 1995;61:3014–3018. doi: 10.1128/aem.61.8.3014-3018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmar R L, Neill F H, Woodley C M, Manger R, Fout G S, Burkhardt W, Leja L, McGovern E R, Le Guyader F, Metcalf T G, Estes M K. Collaborative evaluation of a method for the detection of Norwalk virus in shellfish tissues by PCR. Appl Environ Microbiol. 1996;62:254–258. doi: 10.1128/aem.62.1.254-258.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski P, Mackey K. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. BioTechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- 6.Chung H, Jaykus L A, Sobsey M D. Detection of human enteric viruses in oysters by in vivo and in vitro amplification of nucleic acids. Appl Environ Microbiol. 1996;62:3772–3778. doi: 10.1128/aem.62.10.3772-3778.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng M Y, Day S P, Cliver D O. Detection of hepatitis A virus in environmental samples by antigen-capture PCR. Appl Environ Microbiol. 1994;60:1927–1933. doi: 10.1128/aem.60.6.1927-1933.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerba C P, Goyal S M, LaBelle R L, Cech I, Bodgan G F. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am J Public Health. 1979;69:1116–1119. doi: 10.2105/ajph.69.11.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Matson D O, Cubitt W D, Estes M K. Genetic and antigenic diversity of human caliciviruses (HuCVs) using RT-PCR and new EIAs. Arch Virol Suppl. 1996;12:251–262. doi: 10.1007/978-3-7091-6553-9_27. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, Wang J, Graham D Y, Estes M K. Detection of Norwalk virus in stool by polymerase chain reaction. J Clin Microbiol. 1992;30:2529–2534. doi: 10.1128/jcm.30.10.2529-2534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jothikumar N, Aparna K, Kamatchiammal S, Paulmurugan R, Saravanadevi S, Khanna P. Detection of hepatitis E virus in raw and treated wastewater with the polymerase chain reaction. Appl Environ Microbiol. 1993;59:2558–2562. doi: 10.1128/aem.59.8.2558-2562.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jothikumar N, Cliver D O, Mariam T W. Immunomagnetic capture PCR for rapid concentration and detection of hepatitis A virus from environmental samples. Appl Environ Microbiol. 1998;64:504–508. doi: 10.1128/aem.64.2.504-508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jothikumar N, Khanna P, Kamatchiammal S, Murugan R P. Rapid detection of waterborne viruses using the polymerase chain reaction and a gene probe. Intervirology. 1992;34:184–191. doi: 10.1159/000150281. [DOI] [PubMed] [Google Scholar]
- 14.Jothikumar N, Khanna P, Paulmurugan R, Kamatchiammal S, Padmanabhan P. A simple device for the concentration and detection of enterovirus, hepatitis E virus and rotavirus from water samples by reverse transcription-polymerase chain reaction. J Virol Methods. 1995;55:401–415. doi: 10.1016/0166-0934(95)00089-9. [DOI] [PubMed] [Google Scholar]
- 15.Kayashima N, Abe S, Akanuma M, Arai K, Takei K, Mitani K, Oniwa K, Aoki H, Yasuda H, Shirahama T, Natori K. Outbreak of diarrhea due to SRSV infection. Nippon Naika Gakkai Zasshi. 1998;87:2504–2506. [PubMed] [Google Scholar]
- 16.Khan A S, Moe C L, Glass R I, Monroe S S, Estes M K, Chapman L E, Jiang X, Humphrey C, Pon E, Iskander J K. Norwalk virus-associated gastroenteritis traced to ice consumption aboard a cruise ship in Hawaii: comparison and application of molecular method-based assays. J Clin Microbiol. 1994;32:318–322. doi: 10.1128/jcm.32.2.318-322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaBelle R L, Gerba C P, Goyal S M, Melnick J L, Cech I, Bogdan G F. Relationships between environmental factors, bacterial indicators, and the occurrence of enteric viruses in estuarine sediments. Appl Environ Microbiol. 1980;39:588–596. doi: 10.1128/aem.39.3.588-596.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefkowitz A, Fout G S, Losonsky G, Wasserman S S, Israel E, Morris J G., Jr A serosurvey of pathogens associated with shellfish: prevalence of antibodies to Vibrio species and Norwalk virus in the Chesapeake Bay region. Am J Epidemiol. 1992;135:369–380. doi: 10.1093/oxfordjournals.aje.a116298. [DOI] [PubMed] [Google Scholar]
- 19.Le Guyader F, Dubois E, Menard D, Pommepuy M. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl Environ Microbiol. 1994;60:3665–3671. doi: 10.1128/aem.60.10.3665-3671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Guyader F, Neill F H, Estes M K, Monroe S S, Ando T, Atmar R L. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl Environ Microbiol. 1996;62:4268–4272. doi: 10.1128/aem.62.11.4268-4272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Pila J M, Dizer H, Dorau W. Wastewater treatment and elimination of pathogens: new prospects for an old problem. Microbiologia. 1996;12:525–536. [PubMed] [Google Scholar]
- 22.Ma J F, Gerba C P, Pepper I L. Increased sensitivity of poliovirus detection in tap water concentrates by reverse transcriptase-polymerase chain reaction. J Virol Methods. 1995;55:295–302. doi: 10.1016/0166-0934(95)00065-6. [DOI] [PubMed] [Google Scholar]
- 23.Metcalf T G, Melnick J L, Estes M K. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu Rev Microbiol. 1995;49:461–487. doi: 10.1146/annurev.mi.49.100195.002333. [DOI] [PubMed] [Google Scholar]
- 24.Moe C L, Sobsey M D, Samsa G P, Mesolo V. Bacterial indicators of risk of diarrhoeal disease from drinking-water in the Philippines. Bull W H O. 1991;69:305–317. [PMC free article] [PubMed] [Google Scholar]
- 25.O'Ryan M L, Vial P A, Mamani N, Jiang X, Estes M K, Ferrecio C, Lakkis H, Matson D O. Seroprevalence of Norwalk virus and Mexico virus in Chilean individuals: assessment of independent risk factors for antibody acquisition. Clin Infect Dis. 1998;27:789–795. doi: 10.1086/514949. [DOI] [PubMed] [Google Scholar]
- 26.Payment P, Franco E, Fout G S. Incidence of Norwalk virus infections during a prospective epidemiological study of drinking water-related gastrointestinal illness. Can J Microbiol. 1994;40:805–809. doi: 10.1139/m94-128. [DOI] [PubMed] [Google Scholar]
- 27.Payment P, Gamache F, Paquette G. Microbiological and virological analysis of water from two water filtration plants and their distribution systems. Can J Microbiol. 1988;34:1304–1309. doi: 10.1139/m88-228. [DOI] [PubMed] [Google Scholar]
- 28.Prasad B V, Rothnagel R, Jiang X, Estes M K. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol. 1994;68:5117–5125. doi: 10.1128/jvi.68.8.5117-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds K A, Gerba C P, Pepper I L. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl Environ Microbiol. 1996;62:1424–1427. doi: 10.1128/aem.62.4.1424-1427.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds K A, Roll K, Fujioka R S, Gerba C P, Pepper I L. Incidence of enteroviruses in Mamala Bay, Hawaii using cell culture and direct polymerase chain reaction methodologies. Can J Microbiol. 1998;44:598–604. [PubMed] [Google Scholar]
- 31.Rinehart-Kim J E, Zhong W M, Jiang X, Smith A W, Matson D O. Complete nucleotide sequence and genomic organization of a primate calicivirus, Pan-1. Arch Virol. 1999;144:199–208. doi: 10.1007/s007050050497. [DOI] [PubMed] [Google Scholar]
- 32.Schwab K J, De Leon R, Sobsey M D. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:531–537. doi: 10.1128/aem.61.2.531-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwab K J, De Leon R, Sobsey M D. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl Environ Microbiol. 1996;62:2086–2094. doi: 10.1128/aem.62.6.2086-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab K J, Neill F H, Estes M K, Metcalf T G, Atmar R L. Distribution of Norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT-PCR. J Food Prot. 1998;61:1674–1680. doi: 10.4315/0362-028x-61.12.1674. [DOI] [PubMed] [Google Scholar]
- 35.Schweiger B, Schreier E, Bothig B, Lopez-Pila J M. Differentiation of vaccine and wild-type polioviruses using polymerase chain reaction and restriction enzyme analysis. Arch Virol. 1994;134:39–50. doi: 10.1007/BF01379105. [DOI] [PubMed] [Google Scholar]
- 36.Severini G M, Mestroni L, Falaschi A, Camerini F, Giacca M. Nested polymerase chain reaction for high-sensitivity detection of enteroviral RNA in biological samples. J Clin Microbiol. 1993;31:1345–1349. doi: 10.1128/jcm.31.5.1345-1349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A W, Skilling D E, Ensley P K, Benirschke K, Lester T L. Calicivirus isolation and persistence in a pygmy chimpanzee (Pan paniscus) Science. 1983;221:79–81. doi: 10.1126/science.6304880. [DOI] [PubMed] [Google Scholar]
- 38.Sobsey M D, Glass J S. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl Environ Microbiol. 1980;40:201–210. doi: 10.1128/aem.40.2.201-210.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobsey M D, Jones B L. Concentration of poliovirus from tap water using positively charged microporous filters. Appl Environ Microbiol. 1979;37:588–595. doi: 10.1128/aem.37.3.588-595.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai Y L, Sobsey M D, Sangermano L R, Palmer C J. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl Environ Microbiol. 1993;59:3488–3491. doi: 10.1128/aem.59.10.3488-3491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]