Abstract
A new series of (S)-flurbiprofen derivatives 4a–4p and 5a–5n were synthesized with different aromatic or aliphatic aldehydes and ketones to produce Schiff’s bases and their structures were confirmed through HR-ESI-MS, 1H, and 13C-NMR spectroscopy. The α-glucosidase inhibitory activities of the newly synthesized compounds were scrutinized, in which six compounds 5k, 4h, 5h, 4d, 4b, and 5i showed potent inhibition in the range of 0.93 to 10.26 µM, respectively, whereas fifteen compounds 4c, 4g, 4i, 4j, 4l, 4m, 4o, 4p, 5c, 5d, 5j, 5l, 5m, 5n and 1 exhibited significant inhibitory activity with IC50 in range of = 11.42 to 48.39 µM. In addition, compounds 5g, 5f, 4k, 4n, and 4f displayed moderate-to-low activities. The modes of binding of all the active compounds were determined through the molecular docking approach, which revealed that two residues, specifically Glu277 and His351 are important in the stabilization of the active compounds in the active site of α-glucosidase. Furthermore, these compounds block the active site with high binding energies (−7.51 to −3.36 kcal/mol) thereby inhibiting the function of the enzyme.
Keywords: (S)-flurbiprofen, Schiff’s bases, α-glucosidase inhibition, molecular docking
1. Introduction
Compounds containing at least one carbon–nitrogen double bond with the general formula R1R2C=NR3 are named Schiff’s base [1]. Hydrazones are the bimolecular condensation product of an alkyl or aryl hydrazine and a carbonyl compound which may be an aldehyde or a ketone. Due to their easy synthesis and active pharmacophore group (C=N moiety), hydrazones mostly display good biological and catalytic activities [2]. The Schiff base moiety is a valuable group which is found several bioactive compounds including antioxidant [3,4], antibacterial [5,6], anti-fungal, antiviral [6,7], anti-diabetic [2], anticancer [8], antitumor [9], xanthine oxidase and α-glucosidase inhibitors [10,11,12], anti-inflammatory [13], and anti-convulsant molecules [14]. Khan et al. synthesized and reported the flurbiprofen derivatives as novel α-amylase inhibitors [15], whereas studies have also shown the potent inhibitory potential of Schiff’s bases for cyclooxygenase enzymes (COX-I and COX-II) [15,16].
The non-steroidal anti-inflammatory drugs (NSAIDs) are the most common drugs used in the world to reduce pain, fever, and inflammation due to their analgesic, antipyretic and anti-inflammatory properties [17]. Flurbiprofen is a well-known NSAID, known by the trade name Ansaid, a derivative of phenyl alkanoic acid [18]. This drug is a well-known anti-inflammatory agent because it exerts its effect by blocking prostaglandin synthesis through inhibition of cyclooxygenase enzymes COX-I and COX-II [19]. Being an important class of NSAIDs family, flurbiprofen is extensively used to treat pain, inflammation, acute gout, migraine [20], osteoarthritis [21], soft tissue injuries [22], rheumatoid arthritis [23], postoperative ocular inflammation, herpetic stromal keratitis and periodontal surgery [24]. The hydrazide and hydrazones of flurbiprofen derivatives demonstrated a large number of pharmacological and pharmaceutical activities including anti-microbial, anti-inflammatory, and antioxidant properties, and also inhibit a wide range of bacterial infections [25,26]. In search of biologically active compounds with α-glucosidase inhibitory potential, we have synthesized a new series of flurbiprofen Schiff’s base derivatives.
Diabetes mellitus (DM) is a challenging endocrine disease globally which affects the metabolism of fats, proteins, and carbohydrates [27]. It is a group of metabolic diseases represented by hyperglycemia stemming from either the lack of insulin action or insulin secretion or both [28]. Chronic hyperglycemia is related to dysfunction, long-lasting damage, and malfunction of many organs, such as the eyes, heart, kidneys, blood vessels, and nerves. Primarily, it is sorted into type-I and type-II DM. Type-I DM is caused due to pancreatic cell injury, and sufficient insulin cannot be produced, while type-II DM is caused through the resistance of insulin or less insulin action in the body [29,30]. The inhibition of α-glucosidase slow the digestion of polysaccharides into core monomers, thereby lowering blood sugar levels [31]. In contrast to other hypoglycemic agents that regulate certain biochemical processes, the α-glucosidase inhibitors act locally in the human intestine [32]. However, the clinical uses of these inhibitors are associated with side effects such as abdominal discomfort, diarrhea, and flatulence. Therefore, designing new α-glucosidase inhibitors with no or minimal side effects is of dire need [33,34,35].
The main objective of this study was to synthesize various derivatives of the marketed available drug flurbiprofen through simple transformation and to screen the synthesized compounds for their various biological activities. In the past, many derivatives of different drugs have been synthesized and evaluated for different biological activities which showed potent enzyme inhibitory activities, for example, derivatives of secnidazole showed excellent carbonic anhydrase and α-glucosidase inhibitory potential [36], while metronidazole showed potent α-amylase and β-glucuronidase inhibition. Similarly, atenolol derivatives show potent inhibition against the urease enzyme, and piroxicam derivatives were screened for their anti-nociceptive activity with excellent results [37]. Likewise, NSAIDS (ibuprofen and naproxen) are also evaluated for their enzyme inhibitory activities, which showed promising results [36].
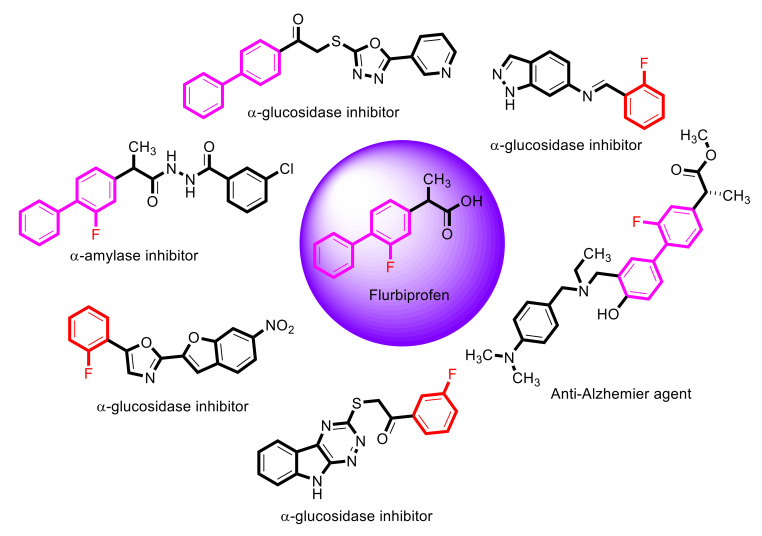
Recently, Momin khan et al. reported oxadiazole-based derivatives of flurbiprofen and screened them for their in vitro α-amylase inhibitory potential with excellent results [15]. Similarly, compounds containing biphenyl rings were checked for their anti-Alzheimer and α-glucosidase inhibitory activities and considered as lead molecules for diabetes mellitus type-II (Figure 1) [37]. Likewise, Bushra et al. and Taha et al. reported a number of compounds with fluorine atoms attached to the benzene ring with excellent α-glucosidase inhibitory potential and confirmed that the fluorine group is responsive to the strength of compounds [38]. Flurbiprofen also has a fluorine group attached to the benzene ring; therefore, we planned to synthesize derivatives of flurbiprofen for their α-glucosidase inhibitory activity.
Figure 1.
2. Results and Discussion
2.1. Chemistry
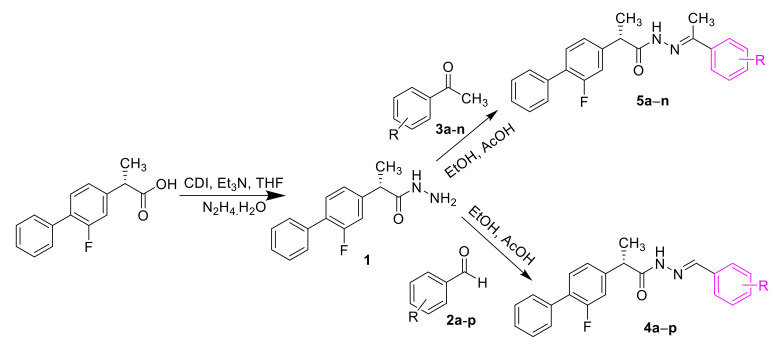
In continuation of our efforts to discover pharmacologically potent α-glucosidase inhibitors, thirty Schiff base derivatives of flurbiprofen were successfully synthesized through multi-step reactions. Flurbiprofen was reacted with hydrazine hydrate in the presence of CDI (1, 1′-carbonyldiimidazole) and triethylamine (Et3N) in THF solvent. The hydrazide was further reacted with various substituted aliphatic/aromatic aldehydes or ketones in the presence of acid using ethanol solvent to give the desired Schiff’s base derivatives of flurbiprofen (Scheme 1).
Scheme 1.
Schiff’s base derivatives of flurbiprofen.
2.2. In Vitro α-Glucosidase Inhibitory Activity
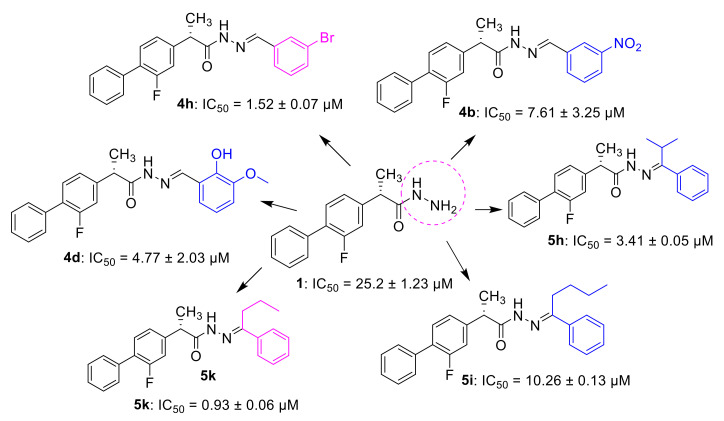
The synthesized compounds were tested for in vitro α-glucosidase inhibitory potential and compared with standard drug acarbose. All the compounds showed α-glucosidase inhibitory activity except five compounds (4a, 4e, 5a, 5b, and 5e). In this series, compounds 4b, 4d, 4h, 5k, 5h and 5i were found potent against α-glucosidase as compared to the standard inhibitor, acarbose (IC50 = 875.75 ± 1.24 µM). Compound 5k (IC50 = 0.93 µM), was the most potent inhibitor followed by 4h (IC50 = 1.52 µM), 5h (IC50 = 3.41 µM), 4d (IC50 = 4.77 µM), 4b (IC50 = 7.61 µM), and 5i (IC50 = 10.26 µM), (Figure 1, Table 1). Furthermore, compounds 4g, 5m, 4i, 5n, 5l, 4l, 4p, 4j, 4m, 4o, 5j, 1, 5c, 5d, and 4c also demonstrated significant inhibitory activity (IC50 value ranging from 11.42 to 48.39 µM) against α-glucosidase, while five compounds [5g, (IC50 = 68.75 µM), 5f (IC50 = 136.36 µM), 4k (IC50 = 146.78 µM), 4n (IC50 = 147.26 µM), and 4f (IC50 = 426.82 µM)] showed moderate to least inhibitory activities (Table 1).
Table 1.
α-Glucosidase inhibitory potential of Schiff’s bases of flurbiprofen.
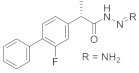 1 | |||||
|---|---|---|---|---|---|
| Compounds | R | IC50 ± µM (S.E.M) |
Compounds | R | IC50 ± µM (S.E.M) |
| 4a |
|
N/A | 5a |
|
N/A |
| 4b |
|
7.16 ± 3.25 | 5b |
|
N/A |
| 4c |
|
48.39 ± 3.75 | 5c |
|
35.52 ± 2.04 |
| 4d |
|
4.77 ± 2.03 | 5d |
|
42.03 ± 3.51 |
| 4e |
|
N/A | 5e |
|
N/A |
| 4f |
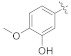
|
426.82 ± 4.63 | 5f |
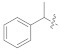
|
136.36 ± 2.57 |
| 4g |
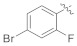
|
11.42 ± 1.09 | 5g |
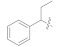
|
68.75 ± 1.42 |
| 4h |
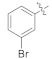
|
1.52 ± 0.07 | 5h |
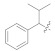
|
3.41 ± 0.05 |
| 4i |
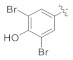
|
13.20 ± 2.47 | 5i |
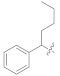
|
10.26 ± 0.13 |
| 4j |
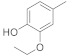
|
18.71 ± 0.25 | 5j |
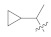
|
29.31 ± 0.60 |
| 4k |

|
146.78 ± 0.86 | 5k |
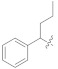
|
0.93 ± 0.06 |
| 4l |

|
17.36 ± 1.68 | 5l |
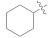
|
14.73 ± 0.27 |
| 4m |
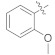
|
19.84 ± 1.89 | 5m |
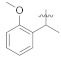
|
11.83 ± 0.18 |
| 4n |
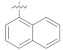
|
147.26 ± 2.78 | 5n |
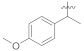
|
14.24 ± 0.16 |
| 4o |

|
22.72 ± 1.46 | Acarbose | 875.75 ± 1.24 | |
| 4p |
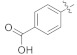
|
18.61 ± 0.14 | |||
NA = Not active; S.E.M = standard error mean.
The structure–activity relationship (SAR), for better understanding, was discussed by analyzing the changes in position and nature of attached substituents (R) on the benzene ring. In the case of the aldehydes series, compound 4h was the most active inhibitor of α-glucosidase having IC50 value of 1.52 ± 0.07 μM (Figure 2), ~580 times stronger than acarbose as the standard inhibitor (Figure 2). The most potent activity of 4h could be due to the presence of bromine substituent at the meta position of the benzene ring. The decrease in α-glucosidase inhibition of 4g (IC50 = 11.42 ± 1.09 μM) may be likely due to the presence of bromine at the para substitution and additional substituent (fluorine) at the ortho position of the benzene ring. Similarly, a further slight decline in the inhibition of 4i (IC50 = 13.20 ± 2.47 μM) could be due to the presence of additional bromine at the ortho substituent and hydroxyl group at the para position of the benzene ring. Comparing 4d (IC50 = 4.77 ± 2.03 μM) with 4j (IC50 = 18.71 ± 0.25 μM) and 4m (IC50 = 19.84 ± 1.89), the higher activity of the 4d might be due to the presence of hydroxyl and methoxy groups at ortho and meta positions, respectively, attached to the benzene ring. The compounds (4j and 4m) attributed almost the same inhibition, while the lower activity of the compound 4f (IC50 = 426.82 ± 4.63 μM) suggested that the –OH group at ortho position and –OCH3 at ortho and meta substitution on the benzene ring may interact well with the α-glucosidase enzyme compared to para (Table 1). The slight decline in activity of 4c (IC50 = 48.39 ± 3.75 μM) is likely due to the substitution of second –OH at para position, indicating the interaction of –OH at para substitution with the screen enzyme as well. Comparing straight-chain alkanes, compounds 4l (IC50 = 17.36 ± 1.68 μM), 4o (IC50 = 22.72 ± 1.46 μM), displayed outstanding inhibition than 4k (IC50 =146.78 ± 0.86 μM), indicating that n-hexyl and octyl groups might have better interaction with the enzyme compared to butyl in the aldehyde series. Compound 4p revealed promising activity with IC50 value of 18.61 ± 0.14 μM having a carboxylic group at the para substitution of the benzene ring, while 4n (IC50 = 147.26 ± 2.78 μM, naphthalene substituent), determined good inhibition compared to the standard.
Figure 2.
Substitution influence on the Schiff’s base derivatives of flurbiprofen.
Similarly, in the case of ketone series, 5k attributed excellent inhibitory activity ((IC50 = 0.93 ± 0.06 μM), ~940 times stronger) compared to the standard, which could be attributed due to the presence of benzene ring on one side and propyl group on the other side of the attached moiety.
Comparing the most active compound 5k with 5h and 5i, a decrease in inhibition of the can be attributed to the replacement of propyl with isopropyl (5h, IC50 = 3.41 ± 0.05 μM) and butyl (5i, IC50 = 10.26 ± 0.13 μM) on one side of the attached moiety. On the other side, comparing compounds 5h and 5i, the higher inhibition of 5i is likely due to the isomerization of the propyl group (Figure 2). Comparing compounds 5f and 5g with the most active compound (5k), a further decrease in inhibition of the 5g (IC50 = 68.75 ± 1.42 μM) and 5f (IC50 = 136.36 ± 2.57 μM) may be possible due to the replacement of ethyl and methyl groups on one side of the attached moiety, respectively. The lower inhibition activity of 5f than 5g could be possibly due to the replacing of ethyl with a methyl group on one side of the attached substituent. Overall, it can be concluded that the propyl group on one side of the substituent, among the ketone series having straight-chain alkanes on one side and benzene ring on the other side, may have a beneficial effect on the interaction with the α-glucosidase enzyme. Compound 5a and 5b (having methyl group on one side and electron-withdrawing substituent on the para position of a benzene ring on the other side) was found inactive, indicating that methyl with para hydroxyl and methyl with para nitro benzene groups had no beneficial effect on the interaction with the α-glucosidase enzyme. Similarly, compound 5e having cyclohexane on one side and benzene on the other side was found to be inactive. The higher inhibition of 5m (IC50 = 11.83 ± 0.18 μM) than 5n (IC50 = 14.24 ± 0.16 μM) might be possibly due to the presence of methoxy group at ortho substation on benzene ring on one side in addition to having the same functional group on the other side. Compounds 5j (IC50 = 29.31 ± 0.60 μM), 5c (IC50 = 35.52 ± 2.04 μM), and 5d (IC50 = 42.03 ± 3.51 μM) displayed promising inhibition in the series as well (Table 1).
2.3. Molecular Docking Studies
The mode of binding of all the active compounds was studied in the active site of the α-glucosidase enzyme by molecular docking. The binding orientation of the parent compound (1) indicates that the 2-fluoro-substituted-biphenyl moiety of the compound lies near the entrance of the active site where Tyr158 provides hydrophobic interaction to the biphenyl ring of the compound 1, while its hydrazide moiety mediated H-bonding (H-bond) with the side chains of Asp69 and Arg442. However, the substitution of the R group at the terminal amino group of 1 was responsible to tilt the other compounds towards the hydrophobic pocket in the active site constituted by Trp58, Phe301, Phe303, and Tyr347 and the R group resided in that pocket.
Among compounds 4a–4p, 4h was found most active inhibitor with an IC50 value of 1.52 µM, followed by 4d and 4b which exhibited an IC50 value of 4.77 µM and 7.16 µM, respectively. The binding orientation of compound 4h, which is the second most active compound in this series, was like the docked conformation of 5k. The addition of isobutyl-benzene at the R position of compound 4h made this molecule tilt more towards the Phe303 which stabilized the 4h by π-π interaction. Due to the tilted position, the hydrazide moiety of 4h got far away from Glu277 and His351. A similar trend of binding was followed by compounds 4d and 4b. The hydrazide moiety of 4d mediated H-bonding with the side chains of Asp215 and His351, while the hydrazide group of 4b did not form any significant interaction; however, its nitro group interacted with the side chain of Arg213 through H-bond.
The compounds 4g, 4i, 4l, 4p, 4j, 4m, and 4o showed moderate inhibitory activities with IC50 in the range of 11.42 to 22.72 µM. The docked orientation of 4g reflects that the substituted bromo–flouro–benzene ring is responsible to decrease the activity of this compound since the bulky group is not favorable for binding at this position. The addition of dibromophenol ring at the R position in 4i further reduced the inhibitory activity of 4i. The compound 4i further slipped towards the entrance of the active site, thereby its carbonyl only interacted with the side chain of Arg213. The docked orientations of compounds 4g, 4i, 4j, 4l, 4m, and 4p are quite similar; however, the hydrazide moiety of 4g, 4l, and 4m interacted with the Glu277 and His351 like compound 4h, while the hydrazide group of 4p mediated H-bonding with Asp215 and His351. The hydrazide carbonyl group of 4j was stabilized by His351, whereas the ethoxy group of 4j and hydrazide carbonyl of 4o accepted H-bond with the side chain of Tyr347. One of the −OH groups of 4c (IC50 = 48.39 µM) exhibited H-bonding with Asp352, while its hydrazide group interacted with Asp215 and His351. The compounds 4k, 4n, and 4f exhibited the least activity with IC50 values of 146.78, 147.26, and 426.82 µM, respectively. The compound 4k slipped more towards the outside of the active site while 4n adopt a bent position, the carbonyl of 4k was stabilized by the side chain of Arg442 and the hydrazide of 4n mediated H-bonding with Asp215 and Arg213. The docked orientation of 4f was twisted and its biphenyl ring acquired a position in opposite direction, due to the reason this compound did not form any significant interaction in the active site, which may be the reason for the least activity in this compound.
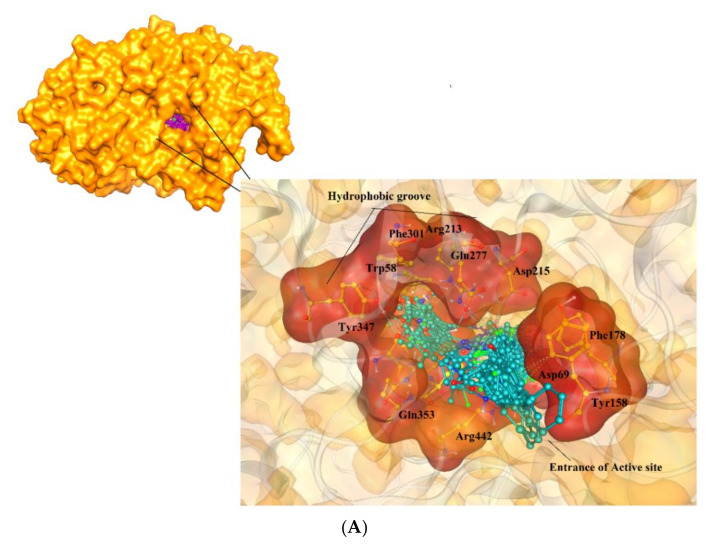
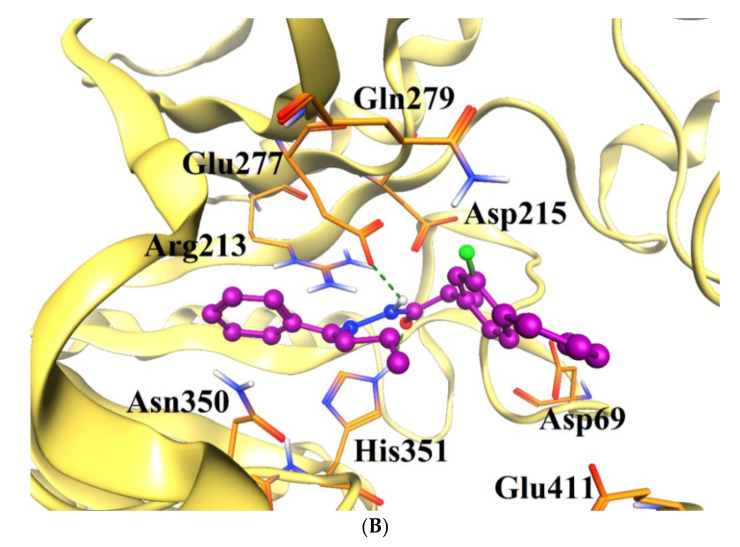
In this series, compound 5k was identified as the most active inhibitor against α-glucosidase. Like 4h, the hydrazide group of 5k mediated H-bonds with the side chains of Glu277 and His351, and the side chains of Trp58, and Phe301 provide hydrophobic interaction to the R group of 5k, and the biphenyl ring of the compound formed hydrophobic interaction with the side chain of Phe178 at the entrance of the active site of the enzyme. The docked mode of 5h was found more twisted, therefore the carbonyl group of 5h interacted with the side chain of Arg213. The addition of pentyl-benzene moiety at the R position of 5i made the compound more flexible. Due to high flexibility, 5i moved more towards the entrance of the active site, however, the hydrazide group of 5i retained its H-bonding with Glu277 and His351, and π-π interaction with Phe303. The hydrazide moiety of 5l, 5n, 5j, 5c, and 5f interacted with the Glu277 and His351-like compounds 5k and 4h. The biphenyl ring of 5m was tilted towards Arg69, His112, and Phe178, and its hydrazide group mediated H-bond with the Asn350 and Glu277. Like 4f, the biphenyl ring of 5d was tilted in opposite direction, therefore 5d could only make hydrophobic interaction with Glu277 and Phe303, as a result, its activity was decreased. Similarly, the biphenyl ring of 5g slipped more outside the active site, while its R-group acquired the position near the biphenyl ring of other compounds, instead of residing in the hydrophobic groove. Due to this conformational change, the compound showed the least inhibitory activity. The docking results are tabulated in Table 2. In docking studies, it was observed that Glu277 and HIS351 play a crucial role in the stabilization of binding of those compounds in the active site of the enzyme (Figure 3). The docking scores of the compounds are in the range of −7.51 to −3.36 kcal/mol, which indicates that these compounds are good binders of protein, therefore exhibit excellent inhibitory potential for α-glucosidase.
Table 2.
Molecular Docking results of the Active α-glucosidase Inhibitors.
| Compounds | Docking Score (kcal/mol) |
Protein-Ligand Interaction | |||
|---|---|---|---|---|---|
| Ligand Atom | Receptor Atom | Interaction | Distance (Å) | ||
| 5k | −7.51 | N9 O8 6-ring |
OE2-GLU277 NE2-HIS351 CE1-PHE178 |
HBD HBA π-H |
1.97 1.74 3.21 |
| 4h | −6.98 | N9 O8 6-ring |
OE2-GLU277 NE2-HIS351 CD2-TYR158 |
HBD HBA π-H |
1.84 1.69 2.72 |
| 5h | −6.71 | O8 6-ring |
NH2-ARG213 6-ring-PHE303 |
HBA π-π |
2.65 2.49 |
| 4d | −6.41 | N9 N11 |
OD1-ASP215 NE2-HIS351 |
HBD HBA |
1.80 1.95 |
| 4b | −6.04 |
O26 6-ring |
NE-ARG213 6-ring-PHE303 |
HBA π-π |
3.09 3.75 |
| 5i | −5.70 | N9 O8 6-ring |
OE1-GLU277 NE2-HIS351 6-ring-PHE303 |
HBD HBA π-π |
2.53 2.26 2.52 |
| 4g | −5.36 |
N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
2.62 2.67 |
| 5m | −5.70 | N9 O8 6-ring 6-ring 6-ring |
OE2-GLU277 ND2-ASN350 6-ring-TYR347 6-ring-PHE301 6-ring-PHE178 |
HBD HBA π-π π-π π-π |
2.82 2.98 3.75 3.27 3.59 |
| 4i | −5.69 | O8 | NH2-ARG213 | HBA | 2.67 |
| 5n | −5.24 | N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
1.84 1.70 |
| 5l | −5.70 | N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
1.82 1.71 |
| 4l | −5.59 |
N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
2.09 2.59 |
| 4p | −5.59 | N18 N17 |
OD1-ASP215 NE2-HIS351 |
HBD HBA |
1.71 1.95 |
| 4j | −5.36 | O8 O46 |
NE2-HIS351 OH-TYR347 |
HBA HBA |
2.28 2.09 |
| 4m | −5.44 |
N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
1.95 1.73 |
| 4o | −2.12 | O8 | OH-TYR347 | HBA | 2.05 |
| 1 | −5.11 | N11 O8 6-ring |
OD2-ASP69 NH1-ARG442 CD2-TYR158 |
HBD HBA π-H |
1.96 1.93 3.64 |
| 5j | −4.77 | N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
1.82 1.69 |
| 5c | −4.27 |
N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
1.85 1.70 |
| 5d | −4.24 |
6-ring 6-ring |
CG-GLU277 6-ring-PHE303 |
π-H π-π |
3.03 2.58 |
| 4c | −4.21 |
N9 O46 O8 |
OD1-ASP215 OD2-ASP352 NE2-HIS351 |
HBD HBD HBA |
1.80 1.83 2.62 |
| 5g | −3.48 | 6-ring | 6-ring TYR72 | π-π | 2.80 |
| 5f | −3.12 |
N9 O8 |
OE2-GLU277 NE2-HIS351 |
HBD HBA |
2.08 2.44 |
| 4k | −3.50 | O8 | NH1-ARG442 | HBA | 2.46 |
| 4n | −3.64 | N9 | OD1-ASP215 | HBD | 2.73 |
| 4f | −3.36 | 6-ring | 6-ring-PHE303 | π-π | 3.69 |
HBA = Hydrogen bond acceptor, HBD = Hydrogen bond donor.
Figure 3.
(A) The docked orientations of all the active compounds are shown in the active site of α-glucosidase enzyme. The entrance of the active site and the hydrophobic pocket is displayed in the surface model, the compounds are presented in cyan ball and stick model. (B) The binding mode of the most active compound (5k, shown in magenta ball and stick model) is displayed in the active site of α-glucosidase. The active site residues are presented in orange stick model, and the protein is shown in gold ribbon. The H-bonds are displayed in green dotted line.
3. Experimental Part
3.1. General Instrumentation
All the chemicals, solvents, and reagents used in this research work were analytical grade and purchased from Sigma-Aldrich, MO, USA. For thin-layer chromatography (TLC), pre-coated aluminum sheets (silica gel 60F-254, Merck, Darmstadt, Hesse, Germany) were used and visualized with the help of UV light at 254 nm. LC-HRESIMS (liquid chromatography high-resolution electrospray ionization mass spectrometry) spectra were recorded on a mass spectrometer (Waters Quattro Premier XE, Waters, Milford, MA, USA). Melting point apparatus Stuart® SMP10 was used for the determination of melting points of the synthesized derivatives. The 1H- and 13C NMR spectra were recorded on a nuclear magnetic resonance (NMR) spectrometer (Bruker, Zürich, Switzerland) operating at 600 MHz (150 MHz for 13C; 600 MHz for 1H; chemical shift (δ) = ppm; coupling constants (J) = Hz) using the solvent peaks as internal references (CDCl3, δH: 7.26; δC: 77.0). Column chromatography was carried out by using silica gel of the selected particle size of 100–200 mesh.
3.2. 2-(2-Fluorobiphenyl-4-yl)propanehydrazide (1)
Coupling reagent CDI (1,1′-carbonyldiimidazole) was used for the synthesis of hydrazide by reacting an appropriate amount of flurbiprofen with hydrazine hydrate in the presence of triethyl amine in THF solvent. In this reaction, flurbiprofen (0.01 M) and CDI (0.012 M) were taken in a round bottom flask and dissolved in minimum amount of tetrahydrofuran (THF). The reaction mixture was stirred for 30–40 min to activate the carboxylic group of flurbiprofen. Finally, hydrazine hydrates (4 mL) were added and reflux it for 5 h, with continuous stirring. The completion of the reaction was checked through TLC (4:6 = ethyl acetate: hexane) from time to time. After completion the reaction mixture was cooled to room temperature (RT) and poured onto crushed ice (100 mL), precipitates were formed, filtered, and washed with excess distilled water to obtain pure hydrazide for further reactions, and the ppt. were dried, weighed and recrystallize with ethanol earlier reported by Momin khan et al. [15].
3.3. General Procedure for the Synthesis of Substituted Schiff’s Bases of Flurbiprofen (4a–p and 5a–n)
Acid hydrazide of flurbiprofen (0.0005 mole) and corresponding substituted aromatic/aliphatic aldehydes and ketones (0.0005 mole) were added to a 100 mL RB flask and dissolved in 15 mL ethanol in the presence of 4–5 drops glacial acetic acid as a catalyst. The reaction mixture was refluxed for 3–4 h with constant stirring. The reaction progress was checked through TLC (3:7 = ethyl acetate:hexane). After completion of reaction, it was poured into ice-cold distilled water, precipitates were formed, filtered, and washed with distilled water and hot hexane to remove unreacted aldehydes/ketones. The ppt. was dried under air, recrystallized with ethanol, and weighed. The desired products were further confirmed by different spectroscopic techniques like HRMS, and NMR (Figures S1–S30, Supplementary Materials).
3.4. Spectral Interpretation of the Synthesized Compounds
3.4.1. 2-(2-Fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (1)
Solid; yield: 90%; M.P.: 120–121 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.12 (s, 1H, NH), 7.42 (m, 7H, H-5, H-6, H-8, H-9, H-10, H-11, H-12), 7.22 (s, 1H, H-2), 4.24 (s, 2H, NH2), 3.60 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.35 (d, J = 7.2 Hz, 3H, -CH3-CH-), 13C-NMR (150 MHz, CDCl3 δ (ppm): 172.3 (C-14), 157.8 (C-3), 137.0 (C-1), 134.9 (C-7), 130.1 (C-5), 129.0 (C-9, C-11), 128.0 (C-4, C-8, C-12), 127.5 (C-10), 125.5 (C-6), 118.2 (C-2), 42.7 (C-13), 18.1 (C-15). LC-HRMS (ESI+): Found [M + H]+: 258.2176; C15H15FN2O.
3.4.2. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(3,4,5-trimethoxybenzylidene)propanehydrazide (4a)
Solid; yield: 88%; M.P.: 139–140 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.68 (s, 1H, -CH=N-), 7.66 (s, 1H, -NH), 7.46 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.39 (t, J = 7.2 Hz, 2H, Ar-H, H-9, H-11), 7.33 (m, 4H, Ar-H, H-2, H-5, H-6, H-10), 6.85 (s, 2H, Ar-H, H-2′, H-6′), 4.70 (q, J = 6.6 Hz, 1H, -CH-CH3), 3.91 (s, 6H, 2-OCH3), 3.86 (s, 3H, -OCH3), 1.57 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C NMR 150 MHz, CDCl3 δ (ppm): 175.7 (C-14), 160.4 (C-3), 158.8 (C-3′, C-5′), 153.5 (C-1′′), 143.7 (C-4′), 139.9 (C-1), 135.4 (C-7), 130.7 (C-5), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 123.5 (C-1′), 115.7 (C-10), 115.5 (C-2), 104.2 (C-2′, C-6′), 60.9 (C-3′′), 56.1 (C-2′′, C-4′′), 41.6 (C-13), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 437.1862; C22H25FN2O4.
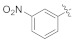
3.4.3. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(3-nitrobenzylidene)propanehydrazide (4b)
Solid; yield: 70%; M.P.: 145–147 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.64 (s, 1H, -CH=N-), 8.52 (s, 1H, -NH), 8.26 (d, J = 8.4 Hz, 1H, Ar-H, H-5), 7.95 (d, J = 7.8 Hz, 1H, Ar-H, H-4′), 7.83 (s, 1H, H-2′), 7.61 (t, J = 7.2 Hz, 1H, Ar-H, H-5′), 7.52 (d, J = 7.2 Hz,, 2H, Ar-H, H-8, H-12), 7.41 (m, 2H, H-9, H-11), 7.36 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 7.31 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 7.30 (s, 1H, Ar-H, H-2), 7.26 (d, J = 7.2 Hz, 1H, H-6′), 3.84 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.66 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 176.0 (C-14), 158.9 (C-3), 148.6 (C-3′), 140.8 (C-1′′), 135.5 (C-1), 135.4 (C-7), 132.5 (C-6′), 130.8 (C-5), 130.8 (C-5′), 128.9 (C-9, C-11), 128.4 (C-8), 127.6 (C-4), 127.6 (C-10), 127.6 (C-1′), 124.4 (C-6), 123.9 (C-4′), 121.6 (C-2′), 115.4 (C-2), 41.6 (C-13), 18.3 (C-15). LC-HRMS (ESI+): Found [M + H]+: 392.1387; C22H18FN3O3.
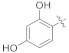
3.4.4. (E)-N′-(2,4-Dihydroxybenzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4c)
Solid; yield: 67%; M.P.: 133–135 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 10.16 (s, 1H, -CH=N-), 8.84 (s, 1H, -OH), 8.14 (s, 1H, -OH), 7.77 (s, 1H, -NH), 7.53 (m, 6H, Ar-H, H-5, H-6, H-9, H-10, H-11, H-6′), 7.05 (d, J = 7.8 Hz, 1H, Ar-H, H-5′), 6.47 (s, 1H, H-3′), 6.43 (d, J = 8.4 Hz, 2H, Ar-H, H-8, H-12), 6.33 (s, 1H, Ar-H, H-2), 4.38 (q, J = 6.6 Hz, 1H, -CH-CH3), 1.69 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 173.7 (C-14), 169.1 (C-2′), 160.7 (C-4′), 158.6 (C-3), 154.8 (C-1′′), 150.3 (C-1), 147.5 (C-7), 142.8 (C-6′), 135.3 (C-5), 130.6 (C-9, C-12), 128.7 (C-4, C-8, C-12), 127.4 (C-10), 123.6 (C-6), 115.2 (C-2), 110.2 (C-1′), 107.6 (C-5′), 103.3 (C-3′), 29.4 (C-13), 18.8 (C-15). LC-HRMS (ESI+): Found [M + H]+: 379.1430; C22H19FN3O3.
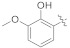
3.4.5. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(2-hydroxy-3-methoxybenzylidene)propanehydrazide (4d)
Solid; yield: 80%; M.P.: 154–155 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.94 (s, 1H, -CH=N-), 9.81 (s, 1H, -NH), 7.98 (s, 1H, Ar-H, H-2), 7.54 (m, 3H, Ar-H, H-6, H-8, H-12), 7.28 (m, 3H, Ar-H, H-9, H-10, H-11), 7.03 (m, 3H, Ar-H, H-4′, H-5′, H-6′), 4.49 (q, J = 6.6 Hz, 1H, -CH-CH3), 3.95 (d, J = 7.2 Hz, 3H, CH3-CH-), 1.69 (s, 3H, -OCH3), 13C-NMR 150 MHz, CDCl3 δ (ppm): 196.6 (C-14), 160.6 (C-3), 151.6 (C-2′), 148.0 (C-3′), 147.1 (C-1′′), 141.6 (C-1), 135.4 (C-7), 131.0 (C-5), 128.9 (C-9, C-11), 128.3 (C-4, C-8, C-12), 127.5 (C-10), 124.5 (C-6), 123.9 (C-6′), 122.2 (C-1′), 119.6 (C-2), 117.9 (C-5′), 115.2 (C-4′), 56.2 (C-2′′), 41.6 (C-13), 18.8 (C-15). LC-HRMS (ESI+): Found [M + H]+: 393.1653; C23H21FN3O3.
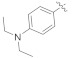
3.4.6. (E)-N′-(4-(Diethylamino)benzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4e)
Solid; yield: 84%; M.P.: 144–145 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.81 (s, 1H, -NH), 7.68 (s, 1H, -CH=N-), 7.57 (s, 1H, Ar-H, H-2), 7.49 (m, 4H, Ar-H, H-8, H-9, H-11, H-12), 7.43 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 7.35 (m, 2H, Ar-H, H-5, H-6), 7.31 (d, J = 7.2 Hz, 2H, Ar-H, H-2′, H-6′), 6.66 (d, J = 8.4 Hz, 2H, Ar-H, H-3′, H-5′), 4.75 (q, J = 6.6 Hz, 1H, -CH-CH3), 4.22 (m, 4H, 2 x -CH2-CH3), 1.67 (d, J = 6.6 Hz, CH3-CH-), 1.18 (m, 6H, 2 x CH3-CH2), 13C-NMR 150 MHz, CDCl3 δ (ppm): 190.0 (C-14), 175.1 (C-3), 167.7 (C-4′), 160.4 (C-1′′), 158.8 (C-1), 143.0 (C-7), 135.7 (C-5), 128.9 (C-9, C-11), 128.3 (C-2′, C-6′), 128.4 (C-4, C-8, C-12), 127.4 (C-3′, C-5′), 123.9 (C-10), 115.7 (C-6), 111.2 (C-1′), 110.5 (C-2), 68.1 (C-2′′, C-4′′), 44.6 (C-13), 18.1 (C-15), 10.9 (C-3′, C-5′′). LC-HRMS (ESI+): Found [M + H]+: 418.2281; C26H28FN3O.
3.4.7. (E)-2-(2-Fluoro-[1, 1′-biphenyl]-4-yl)-N′-(3-hydroxy-4-methoxybenzylidene)propanehydrazide (4f)
Solid; yield: 74%; M.P.: 126–127 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.42 (s, 1H, -NH), 7.63 (s, 1H, -CH=N-), 7.49 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.39 (t, J = 9.0 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.32 (m, 2H, Ar-H, H-5, H-6), 7.27 (d, J = 8.4 Hz, 1H, Ar-H, H-5′), 6.84 (d, J = 8.4 Hz, 1H, H-6′), 5.73 (s, 1H, -OH), 4.76 (q, J = 7.2 Hz, 1H, -CH-CH3), 3.90 (s, 3H, -OCH3), 1.57 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.8 (C-14), 160.4 (C-3), 158.8 (C-4′), 148.5 (C-3′), 146.0 (C-1′′), 143.7 (C-1), 142.7 (C-7), 135.6 (C-1′), 130.6 (C-9, C-11), 128.9 (C-4, C-8, C-12), 127.4 (C-10), 124.0 (C-5), 120.8 (C-6′), 115.6 (C-2), 111.9 (C-2′), 110.4 (C-5′), 56.0 (C-2′′), 41.0 (C-13), 18.1 (C-15). LC-HRMS (ESI+): Found [M + H]+: 393.1625; C23H21FN2O3.
3.4.8. (E)-N′-(4-Bromo-2-fluorobenzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4g)
Solid; yield: 85%; M.P.: 166–168 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.34 (s, 1H, -NH), 8.25 (s, 1H, -CH=N-), 7.89 (s, 1H, Ar-H, H-3′), 7.73 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 7.51 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.42 (t, J = 7.8 Hz, 2H, Ar-H, H-5, H-6), 7.28 (d, J = 9.6 Hz, 1H, Ar-H, H-5′), 7.21 (m, 2H, Ar-H, H-2, H-6′), 4.69 (q, J = 6.6 Hz, 1H, -CH-CH3), 1.57 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.6 (C-14), 161.7 (2′), 160.5 (C-3), 160.0 (1′′), 158.8 (C-1), 142.4 (C-7), 136.1 (C-6′), 135.5 (C-5), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 127.6 (C-10), 127.5 (C-4′), 123.7 (C-5′), 119.7 (C-6), 119.6 (C-2′), 115.6 (C-1′), 41.4 (C-13), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 445.0526; C22H17BrF2N2O
3.4.9. (E)-N′-(3-Bromobenzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4h)
Solid; yield: 70%; M.P.: 147–148 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.04 (s, 1H, -NH), 7.78 (s, 1H, -CH=N-), 7.63 (s, 1H, Ar-H, H-2′), 7.51 (m, 5H, Ar-H, H-4′, H-5, H-6, H-8, H-12), 7.40 (t, J = 7.2 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.33 (t, J = 7.2 Hz, 3H, 1H, Ar-H, H-5′), 7.27 (d, J = 7.8 Hz, 1H, Ar-H, H-6′), 7.20 (s, 1H, Ar-H, H-2), 4.71 (q, J = 6.6 Hz, 1H, -CH-CH3), 1.57 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 190.7 (C-14), 175.6 (C-3), 160.5 (C-1′′), 158.8 (C-1), 141.7 (C-7), 137.3 (C-4′), 135.6 (C-2′) 133.0 (C-5), 132.3 (C-5′), 130.7 (C-3′), 130.3 (C-6′), 129.7 (C-4), 128.9 (C-9, C-11), 128.3 (C-8, C-12), 127.5 (C-6), 125.8 (C-2), 123.8 (C-10), 123.0 (C-1′), 41.5 (C-13), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 425.0662; C22H18BrFN2O.
3.4.10. (E)-N′-(3,5-Dibromo-4-hydroxybenzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4- yl)propane hydrazide (4i)
Solid; yield: 95%; M.P.: 161–162 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.19 (s, 1H, -NH), 7.98 (s, 1H, -CH=N-), 7.78 (s, 1H, -OH), 7.69 (s, 2H, Ar-H, H-2′, H-6′), 7.54 (s, 1H, Ar-H, H-2), 7.49 (d, J = 7.8 Hz, 2H, H-8, H-12), 7.40 (m, 4H, Ar-H, H-5, H-6, H-9, H-11), 7.33 (t, J = 7.2 Hz, 1H, Ar-H, H-10), 4.67 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.56 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 188.0 (C-14), 175.6 (C-3), 160.5 (C-4′), 158.8 (C-1′′), 135.5 (C-1) 133.6 (C-7), 132.3 (C-2′, C-6′), 128.9 (C-4, C-8, C-12), 128.4 (C-9, C-11), 127.5 (C-3′, C-5′), 123.8 (C-1′), 123.0 (C-5), 110.7 (C-10), 41.5 (C-13), 18.2 (C-15). HR-EIMS (ESI+): Found [M + H]+: 520.9690; C22H17Br2FN2O2.
3.4.11. (E)-N′-(3-Ethoxy-4-hydroxybenzylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4j)
Solid; yield: 90%; M.P.: 173–174 °C; 1H-NMR (600 MHz, MeOD ; ⸹, ppm): 8.01 (s, 1H, -NH), 7.60 (s, 1H, -CH=N-), 7.53 (d, J = 7.8 Hz, 2H, Ar-H, H-8, H-12), 7.45 (t, J = 8.4 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.38 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.31 (m, 2H, Ar-H, H-5′, H-6′), 7.05 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 6.84 (m, 1H, Ar-H, H-2′) 4.22 (m, 2H, -CH2-CH3), 3.79 (q, J = 6.6 Hz, 1H, -CH-CH3), 1.58 (d, J = 7.2 Hz, 3H, CH3-CH-), 1.47 (m, 3H, CH3-CH2-) 13C-NMR 150 MHz, MeOD δ (ppm): 177.2 (C-14), 172.7 (C-3), 161.8 (C-4′), 160.1 (C-3′), 150.8 (C-1′′), 150.3 (C-1), 148.7 (C-7), 146.0 (C-1′), 144.3 (C-5) 136.8 (C-10), 131.9 (C-6), 129.9 (C-9, C-11), 129.4 (C-8, C-12), 128.7 (C-4), 127.2 (C-6′), 124.8 (C-5′), 124.3 (C-2′), 123.2 (C-2), 65.4 (C-2′′), 49.1 (C-13), 19.0 (C-15), 15.0 (C-3′′). LC-HRMS (ESI+): Found [M + H]+: 407.1765; C24H23FN2O3.
3.4.12. (E)-N′-Butylidene-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (4k)
Solid; yield: 87%; M.P.: 168–169 °C; 1H-NMR (600 MHz, MeOD ; ⸹, ppm): 7.53 (t, J = 7.8 Hz, 2H, Ar-H, H-9, H-11), 7.43 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.40 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.38 (t, J = 7.2 Hz, 1H, Ar-H, H-10), 7.28 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 7.23 (s, 1H, Ar-H, H-2), 3.70 (q, J = 6.6 Hz, 1H, -CH-CH3), 2.21 (m, 2H, H-2′), 1.57 (d, J = 7.2 Hz, 3H, CH3-CH-), 1.42 (m, 2H, H-3′), 0.95 (t, J = 7.8 Hz, 3H, H-4′), 13C-NMR 150 MHz, MeOD δ (ppm): 177.2 (C-14), 172.4 (C-3), 161.8 (C-1′), 160.1 (C-1), 156.2 (C-7), 151.3 (C-5), 151.1 (C-10), 144.2 (C-6′), 136.8 (C-2) 131.9 (C-4), 129.9 (C-9, C-11), 129.4 (C-8, C-12), 128.7 (C-10), 45.5 (C-13), 38.1 (C-2′), 21.9 (C-3′), 19.5 (C-15), 12.2 (C-4′). LC-HRMS (ESI+): Found [M + H]+: 313.1713; C19H21FN2O.
3.4.13. (E)-2-(2-Fluoro-[1, 1′-biphenyl]-4-yl)-N′-hexylidenepropanehydrazide (4l)
Solid; yield: 76%; M.P.: 129–130 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.90 (s, 1H, -NH), 7.50 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.41 (t, J = 7.2 Hz, 2H, Ar-H, H-9, H-11), 7.34 (m, 2H, Ar-H, H-5, H-6), 7.23 (s, 1H, Ar-H, H-2), 7.11 (m, 1H, Ar-H, H-10), 4.62 (q, J = 7.2 Hz, 1H, CH-CH3), 2.23 (t, J = 7.2 Hz, 1H, -CH=N-), 1.58 (m, 4H, H-2′, H-3′), 1.29 (m, 4H, H-4′, H-5′), 0.88 (m, 3H, H-6′), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.1 (C-14), 160.2 (C-3), 158.5 (C-1′), 147.5 (C-1), 142.6 (C-7), 135.5 (C-5), 128.7 (C-9, C-11), 128.3 (C-4, C-8, C-12), 127.3 (C-10), 123.7 (C-6), 115.6 (C-2), 40.9 (C-13), 32.0 (C-3′), 31.2 (C-4′), 25.8 (C-2′), 22.4 (C-5′), 18.0 (C-15), 13.9 (C-6′). LC-HRMS (ESI+): Found [M + H]+: 341.2017; C21H25FN2O.
3.4.14. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(2-methoxybenzylidene)propanehydrazide (4m)
Solid; yield: 69%; M.P.: 131–132 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 10.45 (s, 1H, -NH), 9.05 (s, 1H, -CH=N-), 8.12 (s, 1H, Ar-H, H-2), 7.87 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.81 (d, J = 7.2 Hz, 3H, Ar-H, H-6, H-8, H-12), 7.54 (t, J = 8.4 Hz, 3H, Ar-H, H-4′, H-5′, H-10), 7.49 (d, J = 7.8 Hz, 2H, Ar-H, H-3′, H-6′), 7.39 (t, J = 7.8 Hz, 2H, Ar-H, H-9, H-11), 4.76 (q, J = 7.2 Hz, 1H, -CH-CH3), 3.82 (s, 3H, -OCH3), 1.57 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 189.7 (C-14), 175.3 (C-3), 161.6 (C-2′), 157.8 (C-1′′), 139.7 (C-1), 135.8 (C-7), 131.3 (C-4′), 130.4 (C-6′), 128.7 (C-9, C-11), 128.4 (C-4, C-8, C-12), 127.3 (C-5), 125.9 (C-10), 124.6 (C-6), 123.7 (C-5′), 121.9 (C-1′), 55.6 (C-2′′), 41.2 (C-13), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 377.1641; C23H21FN2O2.
3.4.15. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(naphthalen-1-ylmethylene)propanehydrazide (4n)
Solid; yield: 92%; M.P.: 168–170 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 10.39 (s, 1H, -NH), 8.60 (d, J = 8.4 Hz, 1H, Ar-H, H-5), 8.34 (s, 1H, -CH=N-), 8.10 (d, J = 8.4 Hz, 1H, Ar-H, H-6), 7.79 (d, J = 7.2 Hz, 1H, Ar-H, H-3′), 7.69 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 7.59 (t, J = 7.2 Hz, 3H, Ar-H, H-4′, H-5′, H-9′), 7.53 (m, 2H, Ar-H, H-9, H-11), 7.35 (m, 2H, Ar-H, H-8, H-12), 3.47 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.62 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.5 (C-14), 160.5 C-3), 158.9 (C-1′′), 143.8 (C-1), 136.6 (C-7), 135.3 (C-7′), 133.8 (C-8′), 131.4 (C-5), 130.7 (C-2′), 129.0 (C-1′), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 128.1 (C-4′, C-5′), 127.5 (C-10), 126.9 (C-10′), 125.2 (C-9′), 124.8 (C-6), 123.8 (C-3′), 115.6 (C-2), 41.6 (C-13), 18.6 (C-15). LC-HRMS (ESI+): Found [M + H]+: 397.1704; C26H21FN2O.
3.4.16. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-octylidenepropanehydrazide (4o)
Solid; yield: 62%; M.P.: 132–134 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.30 (s, 1H, -NH), 7.50 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.47 (m, 2H, Ar-H, H-5, H-6), 7.39 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 7.21 (m, 1H, Ar-H, H-2), 4.63 (q, J = 6.6 Hz, 1H, CH-CH3), 2.32 (d, J = 7.2 Hz, 3H, CH3-CH), 1.61 (m, 2H, H-3′), 1.26 (m, 4H, H-2′, H-4′), 1.21 (m, 9H, H-5′, H-6′, H-7′, H-8′), 13C-NMR 150 MHz, CDCl3 δ (ppm): 178.2 (C-14), 160.4 (C-3), 158.7 (C-1′), 148.1 (C-1), 142.7 (C-7), 135.6 (C-5), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 127.5 (C-10), 123.9 (C-6), 115.8 (C-2), 40.8 (C-13), 32.4 (C-6′), 31.7 (C-4′), 29.5 (C-5′), 28.9 (C-2′), 24.8 (C-3′), 22.6 (C-7′), 17.9 (C-15), 14.0 (C-8′). LC-HRMS (ESI+): Found [M + H]+: 369.2315; C23H29FN2O.
3.4.17. (E)-4-((2-(2-(2-Fluoro-[1,1′-biphenyl]-4-yl)propanoyl)hydrazono)methyl)benzoic acid (4p)
Solid; yield: 90%; M.P.: 176–177 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 8.20 (s, 1H, -NH), 8.09 (t, J = 7.8 Hz, 2H, Ar-H, H-9, H-11), 7.97 (s, 1H, -CH=N-), 7.89 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.81 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 7.54 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.46 (m, 4H, Ar-H, H-2′, H-3′, H-5′, H-6′), 7.38 (t, J = 7.2 Hz, 1H, Ar-H, H-10), 7.27 (s, 1H, Ar-H, H-2), 3.83 (q, J = 6.6 Hz, 1H, CH-CH3), 1.60 (d, J = 6.6 Hz, 3H, CH3-CH), 1.61-1.26 (m, 16H, -CH=N, H-1′, H-2′, H-3′, H-4′, H-5′, H-6′, H-7′), 13C-NMR 150 MHz, MeOD δ (ppm): 177.7 (C-14), 173.1 (C-2′′), 148.4 (C-3), 144.3 (C-1′′), 139.9 (C-1′), 132.0 (C-1), 131.0 (C-7), 129.9 (C-3′, C-5′), 129.5 (C-9, C-11), 128.7 (C-2′, C-6′), 127.9 (C-4′), 125.1 (C-5), 124.7 (C-6), 116.3 (C-10), 115.9 (C-2), 57.4 (C-13), 42.5 (C-15). LC-HRMS (ESI+): Found [M + H]+: 391.1461; C23H19FN2O3.
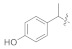
3.4.18. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-(4-hydroxyphenyl)ethylidene)propanehydrazide (5a)
Solid; yield: 83%; M.P.: 152 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 7.88 (d, J = 7.8 Hz, 2H, Ar-H, H-2′, H-6′), 7.60 (d, J = 7.8 Hz, 2H, Ar-H, H-3′, H-6′), 7.49 (d, J = 7.8 Hz, 2H, Ar-H, H-8, H-12), 7.42 (m, 3H, Ar-H, H-2, H-5, H-6), 6.86 (t, J = 7.2 Hz, 3H, Ar-H, H-9, H-10, H-11), 4.78 (q, J = 6.6 Hz, 1H, -CH-CH3), 2.53 (s, 3H, -CH3), 1.56 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.8 (C-14), 160.0 (C-4′), 156.9 (C-3), 143.5 (C-1′′), 130.9 (C-1), 130.6 (C-7), 130.4 (C-2′, C-6′), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 127.8 (C-3′, C-5′), 127.5 (C-10), 115.7 (C-5), 115.5 (C-2), 41.3 (C-13), 18.3 (C-2′′), 12.6 (C-15). LC-HRMS (ESI+): Found [M + H]+: 377.1657; C23H21FN2O2.
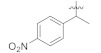
3.4.19. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-(4-nitrophenyl)ethylidene)propanehydrazide (5b)
Solid; yield: 75%; M.P.: 175 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.75 (s, 1H, -NH), 8.30 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 8.24 (d, J = 8.4 Hz, 2H, Ar-H, H-3′, H-5′), 8.10 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 7.84 (d, J = 8.4 Hz, 2H, Ar-H, H-2′, H-6′), 7.49 (d, J = 7.8 Hz, 2H, Ar-H, H-8, H-12), 7.41 (t, J = 7.8 Hz, 2H, Ar-H, H-9, H-11), 7.21 (m, 2H, Ar-H, H-2, H-10), 4.73 (q, J = 6.6 Hz, 1H, -CH-CH3), 2.22 (s, 3H, -CH3), 1.58 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.8 (C-14), 148.1 (C-3), 144.7 (C-4′), 143.7 (C-1′′), 135.4 (C-1′), 130.8 (C-1), 129.3 (C-7), 128.9 (C-9, C-11), 128.4 (C-4, C-8, C-12), 127.6 (C-10), 126.8 (C-2′, C-6′), 123.7 (C-3′, C-5′), 123.7 (C-6), 115.5 (C-2), 41.7 (C-13), 18.5 (C-2′′), 12.7 (C-15). LC-HRMS (ESI+): Found [M + H]+: 406.1557; C23H20FN3O3.
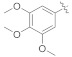
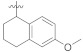
3.4.20. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(6-methoxy-3,4-dihydronaphthalen-1(2H) -ylidene)propanehydrazide (5c)
Solid; yield: 73%; M.P.: 187 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 9.10 (s, 1H, -NH), 8.00 (d, J = 9.0 Hz, 1H, Ar-H, H-5), 7.49 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.40 (t, J = 7.2 Hz, 2H, Ar-H, H-9, H-11), 7.34 (m, 2H, Ar-H, H-3′, H-4′), 7.25 (t, J = 7.8 Hz, 1H, Ar-H, H-10), 6.82 (d, J = 9.0 Hz, 1H, Ar-H, H-6), 6.62 (s, 1H, Ar-H, H-2), 3.81 (s, 3H, -OCH3), 2.72 (t, J = 6.0 Hz, 2H, H-8′), 2.49 (q, J = 6.6 Hz, 1H, -CH-CH3), 1.91 (m, 4H, H-9′, H-10′), 1.56 (d, J = 7.2 Hz, 3H, CH3-CH), 13C-NMR 150 MHz, CDCl3 δ (ppm): 176.4 (C-14), 175.3 (C-5′), 160.5 (C-3), 148.1 (C-1′), 142.9 (C-1), 142.8 (C-7), 141.7 (C-5), 130.6 (C-9, C-11), 128.9 (C-4, C-8, C-12), 127.5 (C-10), 126.4 (C-6), 125.0 (C-7′), 123.9 (C-2′), 115.6 (C-4′, C-6′), 113.2 (C-3′), 112.6 (C-2), 55.3 (C-11′), 41.0 (C-13), 29.6 (C-8′), 24.9 (C-9′), 21.5 (C-10′), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 417.1989; C26H25FN2O2.
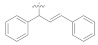
3.4.21. N′-((1Z, 2Z)-1,3-Diphenylallylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (5d)
Solid; yield: 76%; M.P.: 189 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.24 (s, 1H, -NH), 8.01 (d, J = 7.2 Hz, 1H, Ar-H, H-5), 7.81 (s, 1H, Ar-H, H-2), 7.63 (d, J = 7.2 Hz, 1H, Ar-H, H-6), 7.35 (t, J = 7.2 Hz, 6H, Ar-H, H-3′, H-4′, H-5′, H-9′, H-10′, H-11′), 7.28 (t, J = 6.6 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.19 (d, J = 7.8 Hz, 2H, Ar-H, H-2′, H-6′), 7.11 (m, 2H, Ar-H, H-8′, H-12′), 4.82 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.54 (m, 4H, -CH2-CH2-). 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.0 (C-14), 160.8 (C-3), 158.8 (C-1′′), 144.8 (C-3′′), 138.2 (C-1), 137.4 (C-7), 136.0 (C-1), 135.7 (C-7′), 132.7 (C-6′, C-10′), 130.6 (C-9′, C-11′), 129.9 (C-4′, C-4, C-8, C-12), 128.9 (C-8′, C-12′, C-9, C-11), 127.9 (C-2′, C-6′), 123.5 (C-10), 120.8 (C-5), 115.8 (C-2), 40.8 (C-13), 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 449.2042; C30H25FN2O.
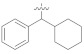
3.4.22. (E)-N′-(Cyclohexyl(phenyl)methylene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (5e)
Solid; yield: 90%; M.P.: 167 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.14 (s, 1H, -NH), 7.93 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.54 (d, J = 7.8 Hz, 2H, Ar-H, H-2′, H-6′), 7.42 (t, J = 7.2 Hz, 6H, Ar-H, H-3′, H-4′, H-5′, H-9, H-10, H-11), 7.25 (m, 3H, Ar-H, H-2, H-8, H-12), 7.05 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 4.73 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.86 (m, 6H, H-7′′, H-8′′, H-9′′), 1.50 (d, J = 7.2 Hz, 3H, CH3-CH-), 1.24 (m, 4H, H-10′′, H-11′′), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.0 (C-14), 158.8 (C-3), 158.0 (C-1′′), 135.7 (C-1), 133.2 (C-7), 132.7 (C-4′), 130.5 (C-6), 129.4 (C-4, C-8, C-12), 128.9 (C-3′, C-5′), 128.8 (C-2′, C-6′), 127.7 (C-10), 115.8 (C-2), 45.8 (C-13), 30.5 (C-8′, C-12′), 29.4 (C-10′), 26.1 (C-9′, C-11′) 18.2 (C-15). LC-HRMS (ESI+): Found [M + H]+: 429.2332; C28H29FN2O.
3.4.23. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-phenylethylidene)propanehydrazide (5f)
Solid; yield: 71%; M.P.: 181 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 8.55 (s, 1H, -NH), 7.95 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.70 (d, J = 7.2 Hz, 2H, Ar-H, H-2′, H-6′), 7.55 (d, J = 7.2 Hz, 2H, Ar-H, H-3′, H-5′), 7.50 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.45 (t, J = 7.2 Hz, 1H, Ar-H, H-10), 7.39 (m, 3H, -CH3), 4.80 (q, J = 7.2 Hz, 1H, -CH-CH3), 2.17 (s, 3H, -CH3), 1.57 (d, J = 6.6 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 175.8 (C-14), 160.5 (C-3), 158.8 (C-16), 147.2 (C-1′), 142.8 (C-1), 137.8 (C-7), 135.6 (C-4′), 133.1 (C-5), 130.6 (C-9, C-11), 129.6 (C-3′, C-5′), 128.9 (C-4, C-8, C-12), 127.8 (C-2′, C-6′), 126.1 (C-10), 126.1 (C-10), 123.9 (C-6), 115.7 (C-2), 41.3 (C-13), 18.3 (C-15), 12.7 (C-17). LC-HRMS (ESI+): Found [M + H]+: 361.1707; C23H21FN2O.
3.4.24. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-phenylpropylidene)propanehydrazide (5g)
Solid; yield: 78%; M.P.: 179 °C; 1H-NMR (600 MHz, CDCl3; ⸹, ppm): 7.95 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.70 (d, J = 7.2 Hz, 2H, Ar-H, H-2′, H-6′), 7.54 (d, J = 6.6 Hz, 2H, Ar-H, H-8, H-12), 7.44 (t, J = 7.8 Hz, 6H, Ar-H, H-3′, H-4′, H-5′, H-9, H-10, H-11), 7.24 (s, 1H, Ar-H, H-2), 7.04 (m, 1H, Ar-H, H-6), 2.99 (q, J = 7.2 Hz, 1H, -CH-CH3), 1.41 (m, 2H, -CH2-), 1.15 (t, J = 7.8 Hz, 3H, CH3-CH2-), 13C-NMR 150 MHz, CDCl3 δ (ppm): 200.9 (C-14), 136.9 (C-7), 132.8 (C-1), 129.5 (C-9, C-11), 128.9 (C-4, C-8, C-12), 128.8 (C-5), 127.9 (C-2′, C-6′), 126.1 (C-4′), 31.7 (C-17), 19.4 (C-13), 8.2 (C-18). LC-HRMS (ESI+): Found [M + H]+: 375.1858; C24H23FN2O.
3.4.25. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(2-methyl-1-phenylpropylidene)propanehydrazide (5h)
Solid; yield: 67%; M.P.: 177 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.55 (m, 4H, Ar-H, H-5, H-6, H-2′, H-6′), 7.49 (t, J = 6.6 Hz, 4H, Ar-H, H-4′, H-9, H-10, H-11), 7.41 (t, J = 7.2 Hz, 2H, Ar-H, H-3′, H-5′), 7.31 (m, 2H, H-8, H-12), 7.04 (m, 1H, Ar-H, H-2), 2.90 (m, 1H, H-2′′), 1.17 (m, 6H, 2 x –CH3), 13C-NMR 150 MHz, MeOD δ (ppm): 177.2 (C-14), 172.1 (C-3), 166.9 (C-1′′), 161.7 (C-1), 161.4 (C-7), 160.4 (C-4′), 136.9 (C-1′), 134.6 (C-5), 132.2 (C-10), 131.8 (C-6), 130.5 (C-9, C-11), 129.9 (C-2′, C-6′), 129.5 (C-8, C-12), 128.3 (C-3′, C-5′), 125.1 (C-2), 45.5 (C-13), 42.7 (C-2′′), 20.5 (C-3′′, C-4′′), 18.6 (C-15). LC-HRMS (ESI+): Found [M + H]+: 389.2032; C25H25FN2O.
3.4.26. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-phenylpentylidene)propanehydrazide (5i)
Solid; yield: 89%; M.P.: 165 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.82 (s, 1H, -NH), 7.75 (d, J = 6.6 Hz, 1H, Ar-H, H-5), 7.54 (d, J = 7.8 Hz, Ar-H, 2H, H-2′, 6′), 7.50 (t, J = 8.4 Hz, 2H, Ar-H, H-9, H-11), 7.46 (t, J = 8.4 Hz, 2H, Ar-H, H-3′, H-5′), 7.41 (m, 4H, Ar-H, H-4′, H-8, H-10, H-12), 7.26 (d, J = 7.8 Hz, 1H, Ar-H, H-6), 7.21 (m, 1H, Ar-H, H-2), 4.06 (q, J = 7.2 Hz, 1H, -CH-CH3), 2.87 (m, 2H, H-2′′), 2.63 (t, J = 7.2 Hz, 1H, H-1′′), 1.35 (m, 4H, H-3′′, H-4′′), 0.90 (t, J = 7.2 Hz, 3H, H-5′′), 13C-NMR 150 MHz, MeOD δ (ppm): 173.2 (C-14), 160.9 (C-7′), 154.4 (C-3), 145.0 (C-1), 144.5 (C-7), 138.8 (C-3′), 138.5 (C-5), 136.9 (C-10), 132.0 (C-6), 131.7 (C-2), 129.9 (C-9, C-11), 129.5 (C-2′, C-6′), 129.4 (C-5, C-12), 128.7 (C-4), 127.5 (C-4′), 125.1 (C-13), 11.4 (C-1′), 29.7 (C-9′), 27.9 (C-8′, C-10′), 23.6 (C-15), 14.2 (C-11′). LC-HRMS (ESI+): Found [M + H]+: 403.2188; C26H27FN2O.
3.4.27. (E)-N′-(1-Cyclopropylethylidene)-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (5j)
Solid; yield: 76%; M.P.: 160 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.53 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.45 (t, J = 7.2 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.38 (d, J = 7.2 Hz, 1H, Ar-H, H-5), 7.30 (m, 3H, H-1′, H-2′), 0.85 (m, 2H, H-2′), 13C-NMR 150 MHz, MeOD δ (ppm): 177.5 (C-14), 172.6 (C-4′), 165.2 (C-3), 161.7 (C-1), 160.1 (C-7), 156.8 (C-5), 144.6 (C-6), 136.9 (C-10), 131.8 (C-2), 129.9 (C-8, C-12), 129.4 (C-9), 128.9 (C-11), 124.7 (C-4), 49.5 (C-13), 49.1 (C-5′), 45.0 (C-15), 19.1 (C-1′), 12.3 (C-2′, C-3′). LC-HRMS (ESI+): Found [M + H]+: 325.1712; C20H21FN2O.
3.4.28. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-phenylbutylidene)propanehydrazide (5K)
Solid; yield: 85%; M.P.: 183 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.83 (d, J = 7.8 Hz, 2H, Ar-H, H-2′, H-6′), 7.75 (d, J = 7.2 Hz, 1H, Ar-H, H-5), 7.54 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.50 (t, J = 8.4 Hz, 1H, Ar-H, H-4′), 7.46 (t, J = 7.2 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.34 (m, 2H, Ar-H, H-3′, H-5′), 7.21 (m, 1H, Ar-H, H-6), 7.13 (m, 1H, Ar-H, H-2), 4.07 (q, J = 6.6 Hz, 1H, -CH-CH3), 2.76 (t, J = 7.8 Hz, 2H, H-2′′), 1.60 (m, 2H, H-3′′), 0.96 (m, 3H, H-4′′), 13C-NMR 150 MHz, MeOD δ (ppm): 178.2 (C-14), 173.3 (C-1′′), 136.9 (C-3), 131.9 (C-1), 130.7 (C-7), 130.6 (C-4′), 129.9 (C-5), 129.5 (C-9, C-11), 128.1 (C-2′, C-6′), 127.6 (C-8, C-12), 125.1 (C-6), 124.8 (C-2), 11.4 (C-3′), 49.4 (C-2′′), 45.1 (C-3′′), 29.8 (C-15),14.1 (C-4′′). LC-HRMS (ESI+): Found [M + H]+: 389.2024; C25H25FN2O.
3.4.29. N′-Cyclohexylidene-2-(2-fluoro-[1,1′-biphenyl]-4-yl)propanehydrazide (5l)
Solid; yield: 80%; M.P.: 177 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.53 (d, J = 7.2 Hz, 2H, Ar-H, H-8, H-12), 7.54 (t, J = 7.8 Hz, 3H, Ar-H, H-9, H-10, H-11), 7.38 (m, 1H, Ar-H, H-5), 7.29 (m, 2H, Ar-H, H-2, H-6), 3.87 (q, J = 6.6 Hz, 1H, -CH-CH3), 2.44 (m, 2H, H-2′), 2.37 (t, J = 6.0 Hz, 2H, H-6′), 1.74 (m, 2H, H-4′), 1.68 (m, 4H, H-3′, H-5′), 1.54 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, MeOD δ (ppm): 177.7 (C-14), 172.9 (C-1′), 167.7 (C-3), 161.7 (C-1), 160.1 (C-7), 144.5 (C-5), 136.9 (C-6), 131.8 (C-10), 129.9 (C-9, C-11), 129.4 (C-8, C-12), 128.7 (C-2), 124.7 (4), 45.0 (C-13), 36.1 (C-6′), 28.7 (C-2′), 28.3 (C-3′), 27.3 (C-5′), 26.5 (C-4′), 19.1 (C-15). LC-HRMS (ESI+): Found [M + H]+: 339.1870; C21H23FN2O.
3.4.30. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-(2-methoxyphenyl)ethylidene)propanehydrazide (5m)
Solid; yield: 78%; M.P.: 198 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.55 (t, J = 7.2 Hz, 2H, Ar-H, H-9, H-11), 7.49 (m, 2H, Ar-H, H-5, H-6), 7.41 (m, 2H, Ar-H, H-8, H-12), 7.33 (d, J = 9.0 Hz, 1H, Ar-H, H-3′), 7.22 (t, J = 8.4 Hz, 1H, Ar-H, H -4′), 7.05 (t, J = 6.6 Hz, 1H, Ar-H, H-5′), 4.02 (q, J = 7.2 Hz, 1H, -CH-CH3), 3.70 (s, 3H, CH3O), 1.59 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, MeOD δ (ppm): 178.1 (C-14), 158.9 (C-1′′), 132.4 (C-2′), 132.1 (C-3), 131.9 (C-1), 131.7 (C-7), 131.4 (C-4′), 130.6 (C-6′), 129.9 (C-9, C-11), 129.4 (C-8, C-12), 128.9 (C-6), 128.6 (C-5), 125.2 (C-2), 124.8 (C-1′), 122.2 (C-3′), 55.9 (C-7′), 42.5 (C-13). LC-HRMS (ESI+): Found [M + H]+: 391.1813; C24H23FN2O2.
3.4.31. (E)-2-(2-Fluoro-[1,1′-biphenyl]-4-yl)-N′-(1-(4-methoxyphenyl)ethylidene)propanehydrazide (5n)
Solid; yield: 70%; M.P.: 192 °C; 1H-NMR (600 MHz, MeOD; ⸹, ppm): 7.92 (d, J = 7.8 Hz, 2H, Ar-H, H-2′, H-6′), 7.53 (d, J = 7.8 Hz, 1H, Ar-H, H-5), 7.43 (t, J = 7.2 Hz, 2H, Ar-H, H-9, H-11), 7.35 (d, J = 7.2 Hz, 1H, Ar-H, H-6), 7.11 (m, 1H, Ar-H, H-2), 6.92 (d, J = 8.4 Hz, 2H, Ar-H, H-3′, H-5′), 4.02 (q, J = 7.2 Hz, 1H, -CH-CH3), 3.85 (s, 3H, CH3O), 2.54 (s, 3H, -CH3), 1.65 (d, J = 7.2 Hz, 3H, CH3-CH-), 13C-NMR 150 MHz, MeOD δ (ppm): 196.8 (C-14), 163.6 (C-4′), 132.4 (C-1′′), 130.6 (C-9, C-11), 128.9 (C-3), 128.4 (C-7), 127.8 (C-1), 127.8 (C-1), 113.6 (C-8, C-12), 55.4 (C-7′), 26.3 (C-2′′), 17.9 (C-15). LC-HRMS (ESI+): Found [M + H]+: 391.1819; C24H23FN2O2.
3.5. In Vitro α-Glucosidase Inhibition Assay
α-Glucosidase (E.C.3.2.1.20) enzyme inhibition assay was developed by using 0.1 M phosphate buffer (pH 6.8) at 37 °C [39,40,41]. The enzyme (0.2 μ/mL) was incubated in phosphate-buffered saline with different concentrations of tested compounds at 37 °C for 15 min. The substrate (0.7 mM, p-nitrophenyl-α-d-glucopyranoside) was then added and the variation in absorbance at 400 nm was observed for 30 min using a spectrophotometer (xMark™ Microplate Spectrophotometer, BIO-RAD). Test compounds were substituted with DMSO-d6 (7.5% final) in the control. Acarbose was used as the standard inhibitor. The % inhibition was calculated by using the following formula:
% Inhibition = 100 − (OD test well/OD control) × 100.
3.6. Molecular Docking
The docking study of the active compounds was conducted in molecular operating environment [42] on the X-ray crystal structure of isomaltase from Saccharomyces cerevisiae in complex with alpha-D-glucopyranose (PDB code: 3A4A, resolution: 1.60 Å) [43]. In our previous studies [11,40,44], we have thoroughly checked the docking performance of applied docking protocol via re-docking of acarbose in the active site of α-glucosidase where MOE was found efficient with an RMSD value of 0.36 Å. The enzyme file was handled through QuickPrep module of MOE to add missing hydrogen atoms on each residue and to calculate partial charges using pre-defined force field (Amber10:EHT force field). The structures of active compounds were drawn by ChemDraw and converted into three-dimensional (3D)-format by MOE-WASH module after importing all the prepared structures in the MOE database where hydrogen atoms and partial charges were added to all the compounds simultaneously and the structure of each ligand was minimized until RMS gradient of 0.1 RMS kcal/mol/Å was obtained. In subsequent step, triangle matcher docking algorithm in combination with London dG scoring function of MOE was applied during docking. In the end, the best-docked conformation of each ligand was selected based on the good docking score and appropriate binding interaction.
4. Conclusions
Novel acyl hydrazones of flurbiprofen (4a-4p) and (5a-5n) were synthesized and evaluated for in vitro inhibition against the α-glucosidase enzyme. Among the synthesized compounds, 5k, 4h, 5h, 4d, 4b, and 5i showed excellent inhibitory potential. The structure–activity relationship (SAR) advocated that the deviation in the activity of the synthesized compounds may be due to the attachment and nature of substituents. Likewise, in silico studies have shown that the synthesized hydrazones (ligands) have widespread binding interactions within the active site of the enzyme, and due to their different substituent, their conformation is different in the active site of the enzyme. This study recognized numerous lead candidates derived from flurbiprofen and further exploration of these molecules for coming research to obtain novel α-glucosidase inhibitors.
Acknowledgments
We are thankful to the Higher Education Commission of Pakistan for financial support under the NRPU research program (no. 10646). The authors are also grateful to The Oman Research Council (TRC) for the funded project (BFP/RGP/CBS/21/002). Many thanks to the Convell Pharmaceutical Laboratory Saidu Sharif, Swat for providing the active raw material of flurbiprofen.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph15060672/s1, Figures S1–S16: 1H, 13C NMR and HRMS (ESI+) of compounds 4a–4p, Figures S17–S30: 1H, 13C NMR and HRMS (ESI+) of compounds 5a–5n.
Author Contributions
A.A., Z. and O.U. synthesized all these compounds. N.U.R. and performed structural elucidation and wrote original draft of the manuscript. S.U. and A.K. conducted α-glucosidase inhibition of the compounds. S.A.H., M.A. (Mumtaz Ali) and A.L. performed molecular docking studies. M.A. (Manzoor Ahmad), S.A. and A.A.-H. assisted in reviewing and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
All authors declare that they have no conflicts of interest concerning this publication.
Funding Statement
The project was supported by a grant from The Oman Research Council (TRC) through the funded project (BFP/RGP/CBS/21/002).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khan K.M., Taha M., Rahim F., Fakhri M.I., Jamil W., Khan M., Rasheed S., Karim A., Perveen S., IQBAL M. Acylhydrazide Schiff bases: Synthesis and antiglycation activity. J. Chem. Soc. Pak. 2013;34:930–938. [Google Scholar]
- 2.Taha M., Ismail N.H., Jamil W., Yousuf S., Jaafar F.M., Ali M.I., Kashif S.M., Hussain E. Synthesis, evaluation of antioxidant activity and crystal structure of 2, 4-dimethylbenzoylhydrazones. Molecules. 2013;18:10912–10929. doi: 10.3390/molecules180910912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohammed Khan K., Shah Z., Uddin Ahmad V., Khan M., Taha M., Rahim F., Ali S., Ambreen N., Perveen S., Iqbal Choudhary M. 2,4,6-Trichlorophenylhydrazine Schiff bases as DPPH radical and super oxide anion scavengers. Med. Chem. 2012;8:452–461. doi: 10.2174/1573406411208030452. [DOI] [PubMed] [Google Scholar]
- 4.Anouar E.H., Raweh S., Bayach I., Taha M., Baharudin M.S., Di Meo F., Hasan M.H., Adam A., Ismail N.H., Weber J.-F.F. Antioxidant properties of phenolic Schiff bases: Structure–activity relationship and mechanism of action. J. Comput.-Aided Mol. Des. 2013;27:951–964. doi: 10.1007/s10822-013-9692-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng K., Zheng Q.-Z., Qian Y., Shi L., Zhao J., Zhu H.-L. Synthesis, antibacterial activities and molecular docking studies of peptide and Schiff bases as targeted antibiotics. Bioorg. Med. Chem. 2009;17:7861–7871. doi: 10.1016/j.bmc.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Avupati V.R., Yejella R.P., Parala V.R., Killari K.N., Papasani V.M.R., Cheepurupalli P., Gavalapu V.R., Boddeda B. Synthesis, characterization and in vitro biological evaluation of some novel 1,3,5-triazine–Schiff base conjugates as potential antimycobacterial agents. Bioorg. Med. Chem. Lett. 2013;23:5968–5970. doi: 10.1016/j.bmcl.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 7.Jarrahpour A., Khalili D., De Clercq E., Salmi C., Brunel J.M. Synthesis, antibacterial, antifungal and antiviral activity evaluation of some new bis-Schiff bases of isatin and their derivatives. Molecules. 2007;12:1720–1730. doi: 10.3390/12081720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popp F.D., Kirsch W. Synthesis of potential anticancer agents. V. Schiff bases and related Compounds1-2. J. Org. Chem. 1961;26:3858–3860. doi: 10.1021/jo01068a056. [DOI] [Google Scholar]
- 9.Sinha D., Tiwari A.K., Singh S., Shukla G., Mishra P., Chandra H., Mishra A.K. Synthesis, characterization and biological activity of Schiff base analogues of indole-3-carboxaldehyde. Eur. J. Med. Chem. 2008;43:160–165. doi: 10.1016/j.ejmech.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 10.You Z.-L., Shi D.-H., Xu C., Zhang Q., Zhu H.-L. Schiff base transition metal complexes as novel inhibitors of xanthine oxidase. Eur. J. Med. Chem. 2008;43:862–871. doi: 10.1016/j.ejmech.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Rafiq K., Khan M., Muhammed N., Khan A., Rehman N.U., Al-Yahyaei B.E.M., Khiat M., Halim S.A., Shah Z., Csuk R. New amino acid clubbed Schiff bases inhibit carbonic anhydrase II, α-glucosidase, and urease enzymes: In silico and in vitro. Med. Chem. Res. 2021;30:712–728. doi: 10.1007/s00044-020-02696-0. [DOI] [Google Scholar]
- 12.Rahim F., Zaman K., Taha M., Ullah H., Ghufran M., Wadood A., Rehman W., Uddin N., Shah S.A.A., Sajid M. Synthesis, in vitro alpha-glucosidase inhibitory potential of benzimidazole bearing bis-Schiff bases and their molecular docking study. Bioorg. Chem. 2020;94:103394. doi: 10.1016/j.bioorg.2019.103394. [DOI] [PubMed] [Google Scholar]
- 13.Mishra P., Gupta P., Shakya A.K., Shukla R., Srimal R. Anti-inflammatory and diuretic activity of a new class of compounds--Schiff bases of 3-amino-2-methylquinazolin 4 (3H)-ones. Indian J. Physiol. Pharmacol. 1995;39:169–172. [PubMed] [Google Scholar]
- 14.Jain J.S., Srivastava R.S., Aggarwal N., Sinha R. Synthesis and evaluation of Schiff bases for anticonvulsant and behavioral depressant properties. Cent. Nerv. Syst. Agents Med. Chem. 2007;7:200–204. doi: 10.2174/187152407781669143. [DOI] [Google Scholar]
- 15.Khan M., Alam A., Khan K.M., Salar U., Chigurupati S., Wadood A., Ali F., Mohammad J.I., Riaz M., Perveen S. Flurbiprofen derivatives as novel α-amylase inhibitors: Biology-oriented drug synthesis (BIODS), in vitro, and in silico evaluation. Bioorg. Chem. 2018;81:157–167. doi: 10.1016/j.bioorg.2018.07.038. [DOI] [PubMed] [Google Scholar]
- 16.Rao C.V., Reddy B.S. NSAIDs and chemoprevention. Curr. Cancer Drug Targets. 2004;4:29–42. doi: 10.2174/1568009043481632. [DOI] [PubMed] [Google Scholar]
- 17.Tian C., Qiang X., Song Q., Cao Z., Ye C., He Y., Deng Y., Zhang L. Flurbiprofen-chalcone hybrid Mannich base derivatives as balanced multifunctional agents against Alzheimer’s disease: Design, synthesis and biological evaluation. Bioorg. Chem. 2020;94:103477. doi: 10.1016/j.bioorg.2019.103477. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Yuan L., Zeng S. Characterizing the effect of UDP-glucuronosyltransferase (UGT) 2B7 and UGT1A9 genetic polymorphisms on enantioselective glucuronidation of flurbiprofen. Biochem. Pharmacol. 2011;82:1757–1763. doi: 10.1016/j.bcp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Sagdinc S., Pir H. Spectroscopic and DFT studies of flurbiprofen as dimer and its Cu (II) and Hg (II) complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009;73:181–194. doi: 10.1016/j.saa.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Musa K.A., Eriksson L.A. Photochemical and photophysical properties, and photodegradation mechanism, of the non-steroid anti-inflammatory drug Flurbiprofen. J. Photochem. Photobiol. A Chem. 2009;202:48–56. doi: 10.1016/j.jphotochem.2008.11.010. [DOI] [Google Scholar]
- 21.Tarafder M., Kasbollah A., Saravanan N., Crouse K.A., Ali A.M. S-methyldithiocarbazate and its Schiff bases: Evaluation of bondings and biological properties. J. Biochem. Mol. Biol. Biophys. JBMBB Off. J. Fed. Asian Ocean. Biochem. Mol. Biol. (FAOBMB) 2002;6:85–91. doi: 10.1080/10258140290027207. [DOI] [PubMed] [Google Scholar]
- 22.Mohammed Khan K., Rahim F., Ambreen N., Taha M., Khan M., Jahan H., Shaikh A., Iqbal S., Perveen S., Iqbal Choudhary M. Synthesis of benzophenonehydrazone Schiff bases and their in vitro antiglycating activities. Med. Chem. 2013;9:588–595. doi: 10.2174/1573406411309040013. [DOI] [PubMed] [Google Scholar]
- 23.Rahim F., Malik F., Ullah H., Wadood A., Khan F., Javid M.T., Taha M., Rehman W., Rehman A.U., Khan K.M. Isatin based Schiff bases as inhibitors of α-glucosidase: Synthesis, characterization, in vitro evaluation and molecular docking studies. Bioorg. Chem. 2015;60:42–48. doi: 10.1016/j.bioorg.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Taha M., Naz H., Rasheed S., Ismail N.H., Rahman A.A., Yousuf S., Choudhary M.I. Synthesis of 4-methoxybenzoylhydrazones and evaluation of their antiglycation activity. Molecules. 2014;19:1286–1301. doi: 10.3390/molecules19011286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Küçükgüzel S.G., Mazi A., Sahin F., Öztürk S., Stables J. Synthesis and biological activities of diflunisal hydrazide–hydrazones. Eur. J. Med. Chem. 2003;38:1005–1013. doi: 10.1016/j.ejmech.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Küçükgüzel G., Kocatepe A., De Clercq E., Şahin F., Güllüce M. Synthesis and biological activity of 4-thiazolidinones, thiosemicarbazides derived from diflunisal hydrazide. Eur. J. Med. Chem. 2006;41:353–359. doi: 10.1016/j.ejmech.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Nair S.S., Kavrekar V., Mishra A. In vitro studies on alpha amylase and alpha glucosidase inhibitory activities of selected plant extracts. Eur. J. Exp. Biol. 2013;3:128–132. [Google Scholar]
- 28.Kawde A.-N., Taha M., Alansari R.S., Almandil N.B., Uddin N., Rahim F., Chigurupati S., Nawaz M., Hayat S., Ibrahim M. Exploring efficacy of indole-based dual inhibitors for α-glucosidase and α-amylase enzymes: In silico, biochemical and kinetic studies. Int. J. Biol. Macromol. 2020;154:217–232. doi: 10.1016/j.ijbiomac.2020.03.090. [DOI] [PubMed] [Google Scholar]
- 29.Taha M., Noreen T., Imran S., Nawaz F., Chigurupati S., Selvaraj M., Rahim F., Ismail N.H., Kumar A., Mosaddik A. Synthesis, α-amylase inhibition and molecular docking study of bisindolylmethane sulfonamide derivatives. Med. Chem. Res. 2019;28:2010–2022. doi: 10.1007/s00044-019-02431-4. [DOI] [Google Scholar]
- 30.Chigurupati S., Yiik E.W.K., Vijayabalan S., Selvarajan K.K., Alhowail A., Nanda S.S., Das S. Antioxidant and antidiabetic properties of Tamarindus indica leaf ethanolic extract from Malaysia. Southeast Asian J. Trop. Med. Public Health. 2020;51:559–569. [Google Scholar]
- 31.Wu B., Song H.-P., Zhou X., Liu X.-G., Gao W., Dong X., Li H.-J., Li P., Yang H. Screening of minor bioactive compounds from herbal medicines by in silico docking and the trace peak exposure methods. J. Chromatogr. A. 2016;1436:91–99. doi: 10.1016/j.chroma.2016.01.062. [DOI] [PubMed] [Google Scholar]
- 32.Tundis R., Loizzo M., Menichini F. Natural products as α-amylase and α-glucosidase inhibitors and their hypoglycaemic potential in the treatment of diabetes: An update. Mini Rev. Med. Chem. 2010;10:315–331. doi: 10.2174/138955710791331007. [DOI] [PubMed] [Google Scholar]
- 33.Chougale A.D., Ghadyale V.A., Panaskar S.N., Arvindekar A.U. Alpha glucosidase inhibition by stem extract of Tinospora cordifolia. J. Enzym. Inhib. Med. Chem. 2009;24:998–1001. doi: 10.1080/14756360802565346. [DOI] [PubMed] [Google Scholar]
- 34.Raju B.C., Tiwari A.K., Kumar J.A., Ali A.Z., Agawane S.B., Saidachary G., Madhusudana K. α-Glucosidase inhibitory antihyperglycemic activity of substituted chromenone derivatives. Bioorg. Med. Chem. 2010;18:358–365. doi: 10.1016/j.bmc.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 35.Rahim F., Ullah H., Javid M.T., Wadood A., Taha M., Ashraf M., Shaukat A., Junaid M., Hussain S., Rehman W. Synthesis, in vitro evaluation and molecular docking studies of thiazole derivatives as new inhibitors of α-glucosidase. Bioorg. Chem. 2015;62:15–21. doi: 10.1016/j.bioorg.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Ding Y., Mao L., Xu D., Xie H., Yang L., Xu H., Geng W., Gao Y., Xia C., Zhang X. C-Aryl glucoside SGLT2 inhibitors containing a biphenyl motif as potential anti-diabetic agents. Bioorg. Med. Chem. Lett. 2015;25:2744–2748. doi: 10.1016/j.bmcl.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 37.Taha M., Ismail N.H., Imran S., Wadood A., Rahim F., Saad S.M., Khan K.M., Nasir A. Synthesis, molecular docking and α-glucosidase inhibition of 5-aryl-2-(6′-nitrobenzofuran-2′-yl)-1,3,4-oxadiazoles. Bioorg. Chem. 2016;66:117–123. doi: 10.1016/j.bioorg.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Taha M., Ismail N.H., Imran S., Wadood A., Ali M., Rahim F., Khan A.A., Riaz M. Novel thiosemicarbazide–oxadiazole hybrids as unprecedented inhibitors of yeast α-glucosidase and in silico binding analysis. RSC Adv. 2016;6:33733–33742. doi: 10.1039/C5RA28012E. [DOI] [Google Scholar]
- 39.Rehman N.U., Khan A., Al-Harrasi A., Hussain H., Wadood A., Riaz M., Al-Abri Z. New α-glucosidase inhibitors from the resins of Boswellia species with structure–glucosidase activity and molecular docking studies. Bioorg. Chem. 2018;79:27–33. doi: 10.1016/j.bioorg.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Ur Rehman N., Rafiq K., Khan A., Ahsan Halim S., Ali L., Al-Saady N., Hilal Al-Balushi A., Al-Busaidi H.K., Al-Harrasi A. α-Glucosidase inhibition and molecular docking studies of natural brominated metabolites from marine macro brown alga Dictyopteris hoytii. Mar. Drugs. 2019;17:666. doi: 10.3390/md17120666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ur Rehman N., Halim S.A., Al-Azri M., Khan M., Khan A., Rafiq K., Al-Rawahi A., Csuk R., Al-Harrasi A. Triterpenic acids as non-competitive α-glucosidase inhibitors from Boswellia elongata with structure-activity relationship: In vitro and in silico studies. Biomolecules. 2020;10:751. doi: 10.3390/biom10050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molecular Operating Environment (MOE) Chemical Computing Group ULC; Montreal, QC, Canada: 2022. 2020.09. [Google Scholar]
- 43.Yamamoto K., Miyake H., Kusunoki M., Osaki S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. FEBS J. 2010;277:4205–4214. doi: 10.1111/j.1742-4658.2010.07810.x. [DOI] [PubMed] [Google Scholar]
- 44.Halim S.A., Jabeen S., Khan A., Al-Harrasi A. Rational Design of Novel Inhibitors of α-Glucosidase: An Application of Quantitative Structure Activity Relationship and Structure-Based Virtual Screening. Pharmaceuticals. 2021;14:482. doi: 10.3390/ph14050482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary Materials.