Significance
Despite various immunomodulatory therapies for multiple sclerosis (MS), there are no curative treatments. Metabolomics, a late “omics” member, can quantitate small metabolites and discriminate between disease and a healthy state. Here, using metabolomics, we identified four perturbed metabolic pathways including structural/signaling lipids and energy in serum of patients with relapsing-remitting multiple sclerosis. Biological literature revealed that glycolysis is the common upstream feeding these altered metabolic pathways, also found to be up-regulated in the blood cells of patients with MS. Targeting glycolysis in experimental autoimmune encephalomyelitis ameliorated the disease pathology by impeding immune cell effector function. Our study outlined the metabolomics-driven application to design better therapeutic strategies for MS, which could be exploited alone or in combination with existing therapies.
Keywords: metabolomics, multiple sclerosis, EAE, glycolysis
Abstract
Metabolic aberrations impact the pathogenesis of multiple sclerosis (MS) and possibly can provide clues for new treatment strategies. Using untargeted metabolomics, we measured serum metabolites from 35 patients with relapsing-remitting multiple sclerosis (RRMS) and 14 healthy age-matched controls. Of 632 known metabolites detected, 60 were significantly altered in RRMS. Bioinformatics analysis identified an altered metabotype in patients with RRMS, represented by four changed metabolic pathways of glycerophospholipid, citrate cycle, sphingolipid, and pyruvate metabolism. Interestingly, the common upstream metabolic pathway feeding these four pathways is the glycolysis pathway. Real-time bioenergetic analysis of the patient-derived peripheral blood mononuclear cells showed enhanced glycolysis, supporting the altered metabolic state of immune cells. Experimental autoimmune encephalomyelitis mice treated with the glycolytic inhibitor 2-deoxy-D-glucose ameliorated the disease progression and inhibited the disease pathology significantly by promoting the antiinflammatory phenotype of monocytes/macrophage in the central nervous system. Our study provided a proof of principle for how a blood-based metabolomic approach using patient samples could lead to the identification of a therapeutic target for developing potential therapy.
Multiple sclerosis (MS) is a neuroinflammatory and demyelinating central nervous system (CNS) disease mainly affecting young adults. It is orchestrated by a wide continuum of immune components, which degenerate the axonal myelin sheath in the CNS (1). The most common disease course includes periods of relapse followed by episodes of partial or complete recovery (remission), known as relapsing-remitting multiple sclerosis (RRMS). Relapses involve acute autoimmune attacks on the brain and spinal cord, leading to multiple demyelinated lesions associated with the infiltration of mononuclear (MN) cells, axonal loss, and neurodegeneration (1). Despite the tremendous upsurge in the research and development of immunomodulatory drugs for MS in the past two decades, there are no satisfactory treatments that can provide long-term recovery in patients (2). The biggest challenge with most of these drugs is substantial side effects and poor tolerance (3). Thus, there is an urgent need to identify novel molecular targets and harness the immune pathways to translate into MS therapies.
Metabolomics is one of the “omics” approaches that uses sophisticated analytical and statistical methods to measure the endogenously produced end products (metabolites) of all the cellular processes in the biological matrices (4). Metabolic profiling can provide a direct window to the instantaneous physiological or pathological changes occurring in a living organism. The art of metabolomics can be implicated to differentiate between a diseased versus a nondiseased state; therefore, it has gained significant translational importance in MS studies for the possible development of MS associated surrogate biomarkers and to monitor the disease progression with or without a therapeutic intervention. A few studies have employed the untargeted metabolomics approach to identify the cardinal metabolic changes of patients with MS (5). However, most of the studies used a single platform with the ability to identify limited metabolites (5). Previously, we have also performed a comprehensive analysis of plasma and urine metabolites in the experimental autoimmune encephalomyelitis (EAE), wherein we observed alterations in several metabolic pathways and reiterated the role of metabolism in the immune response of MS (6–8). Metabolic reprogramming of the immune cells has been reported in the pathogenesis of immune-mediated diseases including systemic lupus erythematosus (9), autoimmune arthritis (10), and Guillain-Barré syndrome (11). Metabolic reprogramming within the inflammatory cells, such as activated macrophages and CD4 cells, is well documented, where a switch to glycolysis for energy generation is essentially required for their cellular functions (12). The metabolic adaptations and metabolic diversity exhibited by the immune cells provides clues to their selective regulation, by exploiting their specific metabolic requirements (13). Given that MS is an immune-mediated disease, governed by inflammation, it provides a perfect model for immune cells to undergo metabolic reprogramming.
The purpose of the present study was to characterize and identify the perturbed metabolic pathways in serum samples of patients with RRMS and target them using pharmacological approaches to examine their therapeutic potential in disease progression using classical preclinical mouse models of MS.
Results
RRMS Patients Show a Distinct Serum Metabolic Profile Compared with Healthy Subjects.
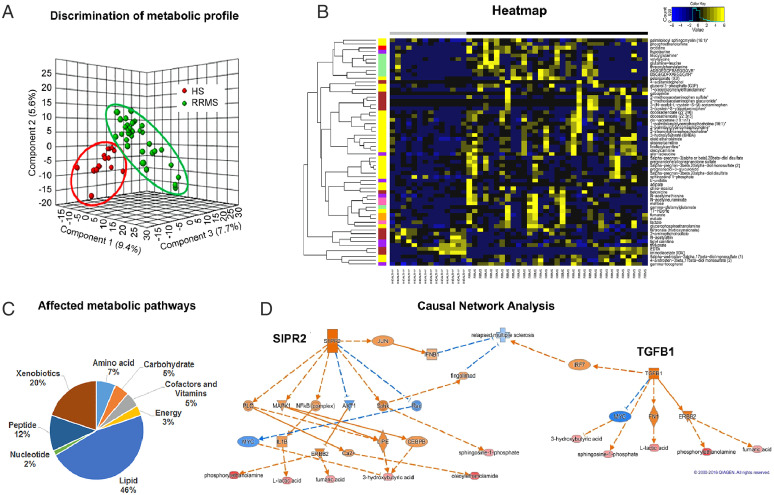
To identify the metabolic signature associated with the disease state in RRMS, we performed untargeted global metabolic profiling in serum collected from 33 RRMS-naïve patients and 14 age- and sex-matched healthy subjects (HS). Clinical characteristics of HS and untreated patients with RRMS are summarized in SI Appendix, Table S1. Of 632 known metabolites measured in RRMS, 60 metabolites mapped to various metabolic pathways were observed to be significantly altered between HS and RRMS (P < 0.05, false discover rate < 0.10), as revealed by Welch's two-sample t test. Among the altered metabolites, 53 (88.33%) metabolites were up-regulated, and only 7 (11.67%) down-regulated relative to HS. Three-dimensional partial least-squares discriminant analysis (PLS-DA) of the metabolomics data were performed to further substantiate the observed difference in serum metabolites between HS and patients with RRMS. It revealed a clear separation between the two groups based on the altered metabolites (Fig. 1A). To visualize the overall differences between the 60 altered metabolites, a heat map was drawn, which showed a clear difference of metabolite levels between the two groups (Fig. 1B). The pie chart indicates that the most altered metabolites belonged to the lipid (46%) and xenobiotics (20%) pathways, followed by peptide (12%), amino acid (7%), cofactor and vitamin (5%), carbohydrate (5%), energy (3%), and nucleotide (2%) metabolic pathways (Fig. 1C and SI Appendix, Table S2). Altogether, these data clearly show that patients with RRMS and HS have distinct serum metabolite profiles indicating unique altered metabolic pathways, which can be used to distinguish between patients with RRMS and HS.
Fig. 1.
Patients with RRMS show an altered metabolic state metabotype compared with HS. (A) Three-dimensional PLS-DA plots showing significant discrimination of RRMS and HS. (B) Pie chart depicting the classification of metabolic perturbations in the serum of patients with RRMS compared with HS. (C) Heat map representative of the hierarchal clustering of the 60 metabolites from each of the replicates of serum from RRMS and HS. Shades of yellow represent elevation of a metabolite and shades of blue represent decrease of a metabolite relative to its mean level in these samples (see color scale). (D) Ingenuity’s IPA software CNA report identified G protein-coupled S1PR2 and TGF-β1 as master regulators predicted to be activated based on the altered levels of metabolites (bottom layer). Shades of orange shows prediction of activation and shades of blue shows prediction of inhibition. Bottom layer of these CNAs are metabolites that are differential between RRMS and HS, while shades of red shows up-regulation in RRMS.
Ingenuity’s software causal network analysis (CNA; Qiagen) (14) was used to identify the underlying signaling cascade of the dysregulated metabolites in patients with RRMS and the master regulators of these signaling pathways. CNA showed G protein-coupled sphingosine-1-phosphate receptor 2 (S1PR2 or S1P2) as one of the master regulators predicted to be activated (S1PR2 activation z score = 2.449) during disease (Fig. 1D and SI Appendix, Table S3). S1PR2 emerged as the master regulator of a small causal network exhibiting two layers of six intermediate regulators, which may explain the altered six metabolites in RRMS namely phosphorylethanolamine, lactic acid, fumaric acid, 3-hydroxybutyric acid, oleoylethanolamide and sphingosine-1-phosphate shown in the bottom row of Fig. 1D. CNA also predicted that RRMS could be repressed indirectly by targeting S1PR2 with JUN, interferon-β1 (IFN-β1), and fingolimod as intermediate regulators. Since IFN-β and fingolimod are Food and Drug Administration approved drugs for patients with MS to control disease progression, this further supports the role of S1PR2 and altered metabolic pathways in MS. Another predicted master regulator identified was transforming growth factor-β1 (TGF-β1; activation z score = 2.236). TGF-β1 was connected to regulating a small causal network of three intermediate regulators, Myc, fibronectin 1, and erythroblastic oncogene B, resulting in an alteration of five metabolites in RRMS (Fig. 1D). TGF-β contributes to the MS pathogenesis in combination with another gene: interferon regulatory factor 7 (IRF-7) (Fig. 1D). To gain further insight into the signaling nodes and pathways connected with the altered metabolites, we also performed Ingenuity Pathway Analysis (IPA). To be noted, the top canonical pathway in IPA was also sphingosine and sphingosine-1-phosphate metabolism (P < 4.34E-03; SI Appendix, Fig. S1). Further, the top network (cell signaling, molecular transport, vitamin, and mineral metabolism; score 33 with 13 focus molecules; SI Appendix, Fig. S2) also revealed a signaling hub involving SIPRs. Thus, all three analyses revealed an association with the common metabolic pathway of sphingosine-1-phosphate metabolism, which is known to play a key role in regulating inflammation in MS (15). Thus, our metabolic profiling of patients with RRMS corroborates and validates that metabolite alterations are reflective of the biological processes underlying the disease.
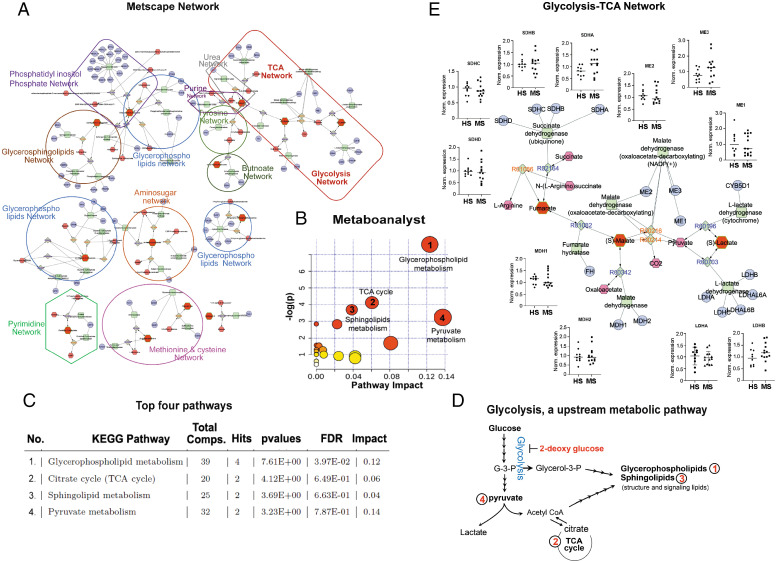
Further, to identify and visualize the enriched metabolic pathways from dysregulated metabolites in RRMS, we employed Metscape, a plug-in for Cytoscape (16). Metscape extracts and integrates metabolite database records from several public sources and provides a complete view of entire metabolic networks, including the connection between metabolites and genes, compounds networks and also provides information for reactions, enzymes, and associated pathways. A number of pathway networks were enriched in patients with RRMS compared with HS (Fig. 2A). Metabolic networks including glycolysis, tricarboxylic acid cycle (TCA), urea, purine, tyrosine, butnoate, phosphatidylinositol phosphate, glycerosphingolipids, pyrimidine, amino sugar, methionine, and cysteine were observed to be greatly impacted during MS disease and are reflected in blood (Fig. 2A). To further understand the functional role of metabolic alterations in RRMS and their link to various metabolic pathways, the Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic library was analyzed using MetaboAnalyst (Fig. 2B) as previously described (6–8). Results of each of the 80 human pathways of KEGG were simultaneously plotted to show the most significant pathways in terms of Global Test P value (vertical axis, shades of red) and impact value (horizontal axis, circle diameter). The top 4 pathways found to be linked with altered metabolites by P value or impact included 1) glycerophospholipid metabolism, 2) citrate cycle (TCA), 3) sphingolipid metabolism, and 4) pyruvate metabolism (Fig. 2 B and C). The glycerophospholipids are glycerol-based phospholipids, and their primary function is to serve as a structural component of the biological membrane. On the other hand, sphingolipids protect the cell surface against harmful environmental factors by forming a mechanical and chemically resistant outer leaflet of the plasma membrane lipid bilayer. Sphingolipid metabolites such as S1P and ceramides have been known to play an important role in various functions under normal or pathological conditions (17). Citrate cycle, also known as TCA or Krebs cycle, is an important aerobic pathway to generate energy, as cell needs to grow and divide. Pyruvate metabolism is a key player in balancing energy metabolism in the body.
Fig. 2.
Bioinformatics analysis identified glycolysis as an upstream to the altered metabolic pathways in RRMS. (A) Visualization and interpretation of metabolomic network of patients with RRMS using Metscape in the context of human metabolic networks. In Metscape, compounds are represented as hexagons, reactions are diamonds, enzymes are squares, and genes are circles. Input metabolites are represented as red hexagons with green border. (B) Sixty metabolites that displayed significantly different abundance between RRMS and HS were subjected for the pathway enrichment analysis (y axis, enrichment P values) and the pathway topology analysis (x axis, pathway impact values, and indicative of the centrality and enrichment of a pathway) in the pathway module of MetaboAnalyst 4.0. The color of a circle is indicative of the level of enrichment significance, with red being high and yellow being low. Bigger size of a circle is proportional to the higher impact value of the pathway. (C) The top four pathways that arise with low P values and with high impact are indicated in the table format. (D) Schematic indicating glycolysis is upstream to the four altered metabolic pathways identified in RRMS. (E) Schematic depicting the comparative analysis of gene expression between HS and RRMS cases for the enzymes involved in glycolytic-TCA pathway, linked with altered metabolites found in RRMS.
Based on the current biochemistry knowledge, all four identified dysregulated pathways are interconnected and point toward glycolysis as an upstream metabolic pathway (Fig. 2 D and E). Glycolysis is a sequence of 10 chemical reactions taking place in most of the cells that break down glucose, with the production of pyruvate or lactic acid and releasing energy that is captured and stored as adenosine triphosphate (ATP). Glycolysis is the major metabolic pathway that along with energy provides metabolites and mediators for various biosynthesis pathways that are essential for providing biomass for a healthy proliferating and functional cell. Pyruvate is a key player in energy metabolism and could be converted into lactate by lactate dehydrogenase (LDH) and released to extracellular space, or it could be converted to acetyl-CoA by pyruvate dehydrogenase and enter the TCA cycle, which could serve as a precursor for the synthesis of lipids (Fig. 2D). Another intermediate of glycolysis, dihydroxyacetone phosphate, gives rise to glycerol 3-phosphate, a reaction catalyzed by glycerol-3-phosphate dehydrogenase. Glycerol 3-phosphate is essential for de novo synthesis of glycerophospholipids, sphingolipids, and lysolipids (Fig. 2D). Thus, glycolysis is the upstream feeder metabolic pathway of all the four perturbed metabolic pathways identified in patients with RRMS.
Since glycolysis-TCA network emerged as the prominent altered metabolic pathway (Fig. 2E), we examined the messenger RNA expression of the key enzymes involved in the reactions of the differential altered metabolites in the peripheral blood mononuclear cells (PBMCs) of healthy and naïve patients with RRMS. Demographic detail of this cohort is listed in SI Appendix, Table S4. We did not observe any change in the expression of lactate dehydrogenase A and B (LDHA and LDHB), malate dehydrogenase-1 and -2, melic enzyme-1, -2, and -3 and succinate dehydrogenase-A, -B, -C, and -D enzymes (Fig. 2E). To further corroborate our findings, we used three datasets from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database, including GSE21942 (18), GSE26484 (19), and GSE43591 (20), and the raw data of each dataset was preprocessed with the Robust Multiarray Average (RMA) algorithm. Using Limma (or geo2R) differential expression analysis was performed on each dataset. Genes with an adjusted P value lower than 0.05 considered as significant. We analyzed the expression of genes involved in glycolysis-TCA pathway (Fig. 2E) and found that none of these genes were affected in patients with MS compared with HS, except for the LDHA and ASL genes in GSE21942 (SI Appendix, Table S5) (18). Overall, our data suggest that glycolysis-TCA pathways are being altered in the PBMCs of patients with MS without affecting their gene expression, indicating a regulation of their function or activity by the inflammatory environment of the disease.
PBMCs from Patients with RRMS Have Increased Glycolysis Compared with HS.
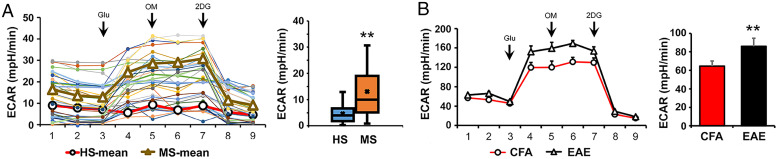
Modulation of the glycolysis pathway is reported to be directly related to the inflammatory response in immune cells (21). Our data also suggest an altered glycolysis metabolism in the PBMCs of patients with MS, which can be postulated to result in an altered immune response and neuroinflammation. We performed the glycolytic response profile, measured as extracellular acidification rate (ECAR), in HS and RRMS-naïve patient derived PBMCs using the Seahorse XFe96 analyzer (Agilent). PBMCs from both HS and RRMS responded to the various stressors and gave a typical glycolytic profile (Fig. 3A). However, the basal glycolysis was significantly (P < 0.05) higher in the RRMS PBMCs when compared with HS, indicating an inherent increase in the glycolytic activity of the RRMS PBMCs (Fig. 3A). This observation was also replicated in the preclinical mouse model of EAE, where PBMCs of the EAE group exhibited higher levels of glycolysis compared with control complete Freund's adjuvant (CFA) groups (Fig. 3B). These findings explicitly show that the PBMCs from both patients with MS and from its preclinical mouse perform glycolysis at an increased rate.
Fig. 3.
Higher glycolytic activity was observed in the PBMC of patients with RRMS and in preclinical mouse model (EAE). (A) Isolated PBMC of patients with RRMS (n = 17) and HS (n = 14) were analyzed for glycolysis (ECAR) measurement using Seahorse Bioanalyzer. (B) PBMC of EAE (n = 6) and CFA (n = 6) groups were analyzed for glycolysis measurement using Seahorse Bioanalyzer (mean + SEM). **P < 0.01; **P < 0.05 compared with HS.
Targeting Glycolysis Using 2-Deoxy-Glucose Treatment Ameliorated Disease Progression in Preclinical EAE Mouse Models.
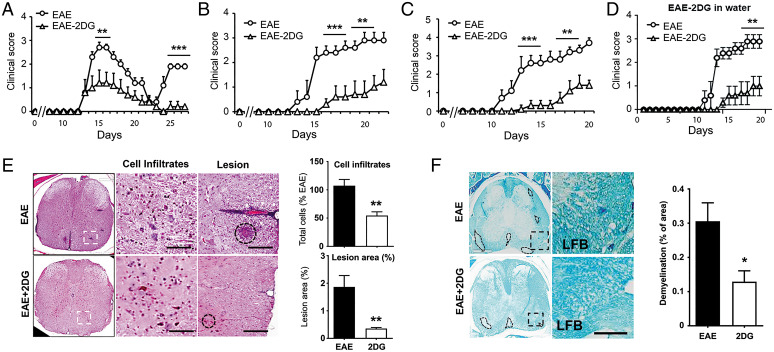
Since glycolysis is up-regulated in PBMCs and emerged as the upstream metabolic pathway regulating/connecting/feeding into the four altered metabolic pathways identified in patients with RRMS, we questioned if inhibiting glycolysis will impact MS disease progression. We employed 2-deoxy-glucose (2DG), a nonmetabolizing glucose analog and a competitive inhibitor of hexokinase, to examine the effect of glycolysis inhibition in the relapsing-remitting preclinical model of MS using SJL mice. Mice immunized for EAE with proteolipid protein (PLP) were treated daily with 2DG (50 mg/kg, intraperitoneally) from day 6 postimmunization, while the control group was injected with phosphate buffered saline (PBS). Treatment with 2DG significantly reduced the severity of the disease (2.7 ± 0.22 vs. 1.2 ± 0.54; at peak of the disease, P < 0.01) without affecting the onset of the disease (Fig. 4A). Treatment with 2DG also abrogated the relapse in the treated group compared with the PBS group (1.9 ± 0.11 vs. 0.2 ± 0.22 at day 25 postimmunization; P < 0.001, (Fig. 4A). Similar efficacy of 2DG was observed in two other mouse models of EAE, where 2DG was effective in not only in delaying the disease onset but also reduced severity (2.6 ± 0.21 vs. 0.3 ± 0.14; P < 0.001 in B6 model and 2.4 ± 0.27 vs. 0.3 ± 0.22; P < 0.001 in 2D2 T cell receptor transgenic model) (Fig. 4 B and C). We also examined the effect of 2DG when given orally in the 2D2 EAE model via drinking water at the dose of 50 mg/kg (1.25 mg per 6 mL as a mouse drink 6 to 7 mL/d) from day 6 postimmunization. The group treated with 2DG showed delayed disease onset and reduced disease severity compared with the untreated EAE group (Fig. 4D), demonstrating the positive impact of 2DG in reducing the disease progression in EAE mouse models. A decrease in clinical score of treated EAE was reflected by the reduction in the number of infiltrated MN cells and preservation of myelin content as examined by the hematoxylin and eosin and Luxol fast blue staining, respectively, in the spinal cord sections (Fig. 4 E and F). As evident from the hispothaological analyses, there is significant reduction in the infiltration of immune cells and lesion size in 2DG-treated mice as compared with vehicle treated EAE mice. Moreover, demyelination also was significantly prevented in the 2DG-treated group compared with vehicle/PBS treated EAE mice. Therefore, our findings strongly advocate that 2DG treatment following EAE reduces immune cell infiltration, prevents lesion progression, and promotes remyelination in the preclinical models of MS disease.
Fig. 4.
Targeting energy pathway ameliorates disease progression in EAE mouse models. (A–C) EAE was induced in SJL, B6, and 2D2 mice using PLP and MOG35-55, respectively. One set of the group was given daily 2DG (50 mg/kg body weight, intraperitoneally in 200 µL volume) and another set was given PBS as vehicle. Clinical score was taken until the end of the study (n = 15 per group). **P < 0.01; ***P < 0.001 compared with vehicle treated EAE group was considered as statistically significant. (D) EAE was induced in 2D2 mice with MOG and one set was given 2DG in drinking water (0.2 mg/mL; wt/vol) from day 7 postimmunization. Clinical score was taken until the end of the study (n = 6 per group). **P < 0.01 compared with vehicle-treated EAE group was considered as statistically significant. (E, F) Representative images shows histopathological changes in the spinal cord tissues in EAE mice with or without 2DG treatment. Sections were staind with hematoxylin and eosin to show cells infiltration and lesion size and Luxol fast blue to (LFB) to demonstrate changes in myelin content. Data were represented as mean ± SD (n = 6 mice/group). Scale bar, 100 μm. Statistical analyses were done with Student’s t test. Statistical significance was determined at P < 0.05.
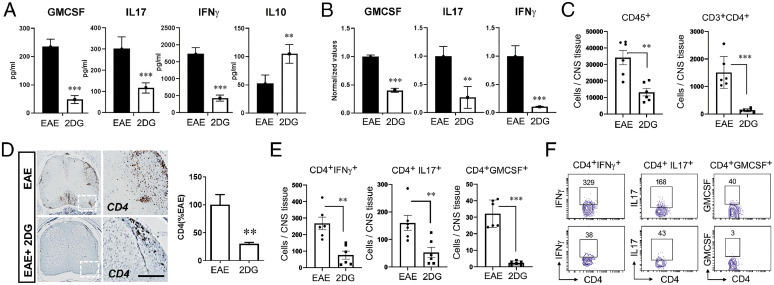
Further, to examine the effect of 2DG on myelin-specific immune response, we isolated spleen cells from 2DG treated and vehicle-EAE groups at day 20 postimmunization and stimulated them with myelin oligodendrocyte glycoprotein (MOG)35-55 (20 μg/mL) for 72 h. Cell proliferation was examined after 96 h by cell proliferation assay (Promega), and cell supernatants were used for enzyme-linked immunosorbent assay to see the status of inflammatory cytokines in both groups. 2DG treatment significantly reduced antigen-specific cell proliferation, reduced the levels of IFN-γ, interleukin (IL) 17, and granulocyte-macrophage colony-stimulating factor (GM-CSF) and induced the production of IL-10 compared with untreated EAE group (Fig. 5A). Consistent with the protein levels, qPCR data showed that the messenger RNA expression of all cytokines was reduced by 2DG treatment (Fig. 5B). Infiltration of MN cells, including myeloid cells and T cells that inflame the CNS environment, is detrimental for oligodendrocytes and neurons, leading to demyelination in MS and EAE. To examine the potential mechanism of 2DG-mediated protection in EAE, we quantified the absolute number of infiltrating MN cells in the CNS and their ability to produce proinflammatory cytokines. We observed that 2DG treatment significantly reduced the overall number of infiltrating MN cells (CD45+) and CD4 T cells (CD45+CD3+CD4+) (Fig. 5C) as examined by flow cytometry and immunohistochemical staining for CD4 (Fig. 5D). Along with the reduction of infiltration, the CD4 T cells also showed reduction in the intracellular proinflammatory cytokines being produced, including IFN-γ, IL-17, and GMCSF (Fig. 5 E and F). Thus, inhibition of glycolysis by 2DG ameliorates the EAE disease progression in various preclinical models and is able to reduce the number and capacity of the infiltrating immune cells.
Fig. 5.
Targeting energy pathway moldulates CD4 cells infiltration and response in EAE mouse. (A) On day 20, splenocytes of EAE-treated and untreated with 2DG in drinking water were stimulated with MOG35-55. Post-72 h, proinflammatory (GM-CSF, IL-17 and IFN-ɣ) and antiinflammatory (IL-10) were examined in cell supernatant using their specific enzyme-linked immunosorbent assay. The data presented are the mean ± SD of two independent experiments (n = 5 per group). (B) Expression of GM-CSF, IL-17, and IFN-ɣ were examined in splenocytes of both groups using quantitative PCR and normalized with the control gene; ribosomal L27 housekeeping gene. The data presented are the mean ± SD of four values. (C) Quantitation of CD45+ and CD3+CD4+ cell number in the spinal cord and brain of EAE and 2DG treated (drinking water) groups by flow cytometry. The data presented are the mean ± SD of six animals per group. (D) Representative immunohistochemical staining and quantitative analysis for CD4+ T cell infiltration in EAE induced mice with or without 2DG treatment in drinking water. Data were shown as mean ± SD (n = 6 mice per group). Scale bar, 100 µm. Statistical analysis was done by Student’s t test. Statistical significance was determined at P < 0.05. (E, F) Quantitation of IFN-ɣ, IL-17α, and GM-CSF producing CD4+ T cell number in the spinal cord and brain of EAE- and 2DG-treated (drinking water) groups. The data presented are the mean ± SD of six animals per group. *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test, one-way analysis of variance.
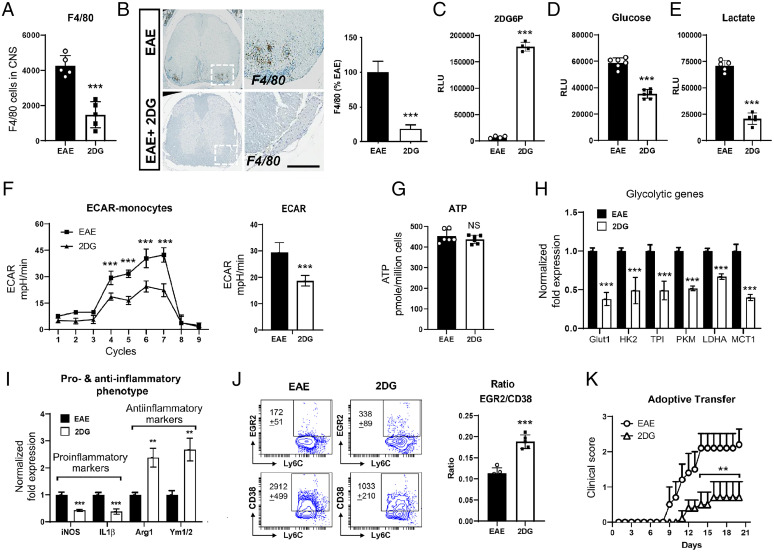
Inhibition of Glycolysis by 2DG Restored the Metabolic Programming of Monocytes/Macrophages in EAE.
Monocytes/macrophages are the major effectors of demyelination in both MS and EAE and are highly plastic. Once monocytes infiltrate into CNS during disease, depending upon the local environment, monocytes differentiate either into proinflammatory or antiinflammatory type macrophages, and their ratio determines the outcome of EAE disease. We examined the effect of 2DG on the infiltration and phenotype of monocytes/macrophages in CNS at day 20 postimmunization. We observed that 2DG treatment significantly reduced the number of macrophages (CD45+Ly6G−F4/80+) in the treated group compared with untreated EAE group (Fig. 6A). This observation was also supported by the immunohistochemical staining for F4/80 in the lumbar area of the spinal cord sections (Fig. 6B). 2DG incorporates into the cells and is phosphorylated by hexokinase, resulting in a nonmetabolizing 2 deoxy-glucose-6-phosphate (2DG6P), thereby inhibiting glycolysis. To test if activated monocytes take up 2DG during disease, we isolated monocytes from the spleens of 2DG-treated and untreated EAE groups using a monocyte isolation kit (StemCell Technologies) on day 20 postimmunization and estimated the levels of 2DG6P. The monocytes from the 2DG treated group had significantly higher levels of 2DG6P (Fig. 6C), suggesting that in treated animals 2DG is taken up by monocytes. According to our hypothesis this would mean that the monocytes isolated from the treated group would exhibit a metabolically altered phenotype represented by an altered glycolysis. Indeed, we observed that monocytes isolated from 2DG treated EAE mice demonstrated reduced glucose uptake (Fig. 6D) and decreased lactate secretion in media (Fig. 6E), indicating an inhibition of glycolysis by 2DG treatment.
Fig. 6.
2-Deoxy-glucose induced antiinflammatory phenotype of monocytes by abrogating glycolytic metabolic pathway. (A) Quantitation of CD45+F4/80+ infiltrated macrophage number in the spinal cord and brain of EAE- and 2DG-treated (drinking water) groups. The data presented are the mean ± SD of six animals per group. (B) Representative immunohistochemical staining and quantitative analysis for F4/80+ cell infiltration in EAE induced mice with or without 2DG treatment in drinking water. Data were shown as mean ± SD (n = 6 mice per group). Scale bar,100 µm. Statistical analysis was done by Student’s t test. Statistical significance was determined at P < 0.05. (C) Isolated monocytes (20,000 cells) from spleen cells from both groups were processed for the detection of 2 deoxy-glucose-6-phosphate (2DG6P) using Glucose Uptake-Glo Assay kit from Promega. The data presented are the mean ± SD of four values. (D, E) Isolated monocytes (> 92%) from spleen cells from both groups were plated (20 K per well) in 96-well plate and post 24 h, cell supernatant was processed for glucose and lactate measurement for determination of glucose consumption and lactate release in media. The data presented are the mean ± SD of six values per group. (F) Isolated monocytes from spleen cells from both groups were processed for glycolysis measurement using Seahorse Bioanalyzer (n = 8 per group). (G) ATP was measured in isolated monocytes using ATPlite luminescence assay kit from Perkin-Elmer. The data presented are the mean ± SD of six values per group. (H) Expression of glycolytic genes including glucose transporter 1 (Glut 1), hexokinase 2 (HK2), triose-phosphate isomerase (TPI), pyruvate kinase M (PKM), LDHA, and monocarboxylate transporter 1 (MCT1) were examined in isolated monocytes from both groups using quantitative PCR. Expression of glycolytic genes were normalized with the control gene; ribosomal L27 housekeeping gene. The data presented are the mean ± SD of four values per group. (I) Expression of proinflammatory (iNOS and IL-1β] and antiinflammatory (arginase 1 [Arg-1] and chitinase-like 3 [Ym1/2]) specific genes were examined in isolated monocytes from both groups using quantitative PCR and normalized with the control gene; ribosomal L27 housekeeping gene. The data presented are the mean ± SD of four values per group. (J) The ratio of EGR2 and CD38, a marker for antiinflammatory and proinflammatory were examined on Ly6C+ cells gated on CD11b+Ly6G− population by flow cytometry in the spleen of treated and untreated EAE group with 2DG on day 20 postimmunization. (K) Adoptive transfer of monocytes from EAE and 2DG treated EAE groups on day 6 in B6 mice immunized with MOG35-55. The data presented are the mean ± SEM of five animals per group. n = 4 to 6 per group; *P < 0.05, **P < 0.01, and ***P < 0.001. Student t test, one-way ANOVA.
To further determine the effects of 2DG on macrophage metabolism in vivo, on day 20 postimmunization splenic macrophages were isolated and processed for glycolysis measurement. We observed that isolated macrophages from untreated EAE exhibited an increased glycolytic rate, which was significantly inhibited upon 2DG treatment in vivo (Fig. 5F). Unchanged total cellular ATP levels suggested that enhanced glycolysis in either group's macrophages is not contributing to energy production (Fig. 6G). Increased glycolysis, as a provider of biomass, supports the de novo synthesis of fatty acids for the expansion of the endoplasmic reticulum and Golgi, which is required for the production and secretion of proteins integral to APC activation (22). Macrophages from vehicle-treated mice showed a higher metabolic phenotype, while the 2DG-treated macrophages showed a lower metabolic state, similar to that of healthy control macrophages. Decreased glycolysis in monocytes/macrophages isolated from 2DG treated EAE was associated with reduced expression of key glycolytic enzymes including glucose transporter 1, hexokinase 2, triose-phosphate isomerase, pyruvate kinase M, LDHA, and monocarboxylate transporter 1 compared with the untreated group (Fig. 6H).
As increased glycolysis is considered a characteristic of proinflammatory macrophages, we observed that splenic macrophages isolated from mice with EAE treated with 2DG displayed an M2 phenotype (higher expression of arginase 1 and Ym1/2) and reduced proinflammatory phenotype (decreased expression of inducible nitric oxide synthase [iNOS] and IL1), resulting in a higher ratio of antiinflammatory/proinflammatory monocytes/macrophages phenotype (Fig. 6I). This observation is further supported by flow cytometry where ratio of EGR2 and CD38 is significantly increased in macrophages from 2DG treated group compared with untreated EAE (Fig. 6J). Antiinflammatory macrophages are protective in EAE, and since 2DG polarized macrophages toward the antiinflammatory phenotype, we next examined if monocytes isolated from 2DG treated mice can modulate EAE disease. To test this, monocytes (1 × 106) isolated from 2DG-treated or untreated EAE groups were adoptively transferred into recipient B6 mice with active EAE on day 6 postimmunization. The adoptively transferred 2DG-treated monocytes significantly ameliorated the severity of EAE compared with the group of adoptively transferred monocytes from untreated EAE into recipient MOG immunized B6 mice (Fig. 6K). These data strongly suggest that inhibition of glycolysis by 2DG polarizes monocytes/macrophages toward an antiinflammatory phenotype, which have the capability to suppress the EAE disease course.
Inhibition of Glycolysis by 2DG Restricts Neuroinflammation.
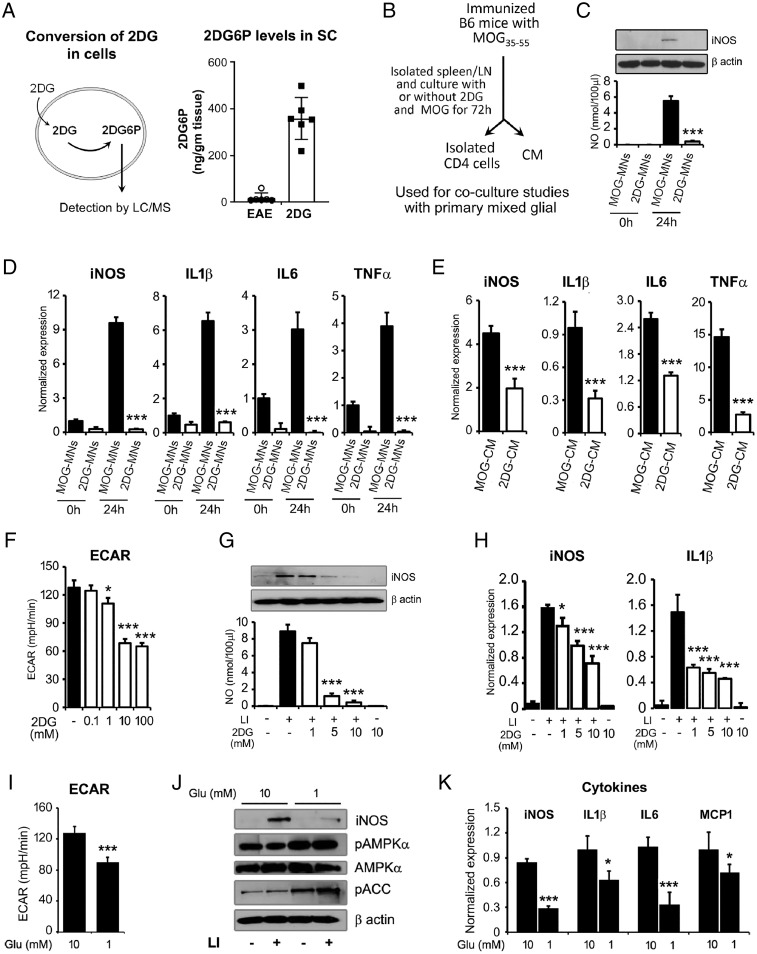
To further examine if 2DG is accumulated in CNS tissues, we examined the levels of 2DG6P in spinal colrd of EAE and 2DG-treated groups using liquid chromatography-tandem mass spectrometry (LC-MS/MS). We found that 2DG-treated EAE animals exhibited significantly higher levels of 2DG6P in spinal cord compared with EAE group (Fig. 7A) suggesting that 2DG was able to go inside CNS and may inhibit neuroinflammation directly along with modulating immune cells. To delineate the mechanism of 2DG-mediated protection, we tested if 2DG treatment impacts the MOG-primed MN cell’s ability to induce inflammation in the CNS. We used in vitro coculture model, where we treated spleen/lymph node single suspension cells with MOG35-55 in the presence or absence of 2DG (1 mM) in vitro. After 72 h, conditioned media (CM) and isolated MN cells from both treatment groups (MOG-CM and MOG-2DG-CM) were used for further studies (Fig. 7B). First, isolated MOG-primed or 2DG treated MOG-primed MNs cells were cocultured with primary brain glial cells (4:1 ratio of MNs cells:mixed glial cells) and after 24-h, cocultured cells were assessed for inflammatory markers and mediators. Mixed glial cells cocultured with MOG-MNs showed increased expression of iNOS and production of nitric oxide (NO), while, 2DG-treated MOG-primed MNs cells were unable to induce iNOS expression in the mixed glial cells (Fig. 7 B and C). Similarly, MOG-primed MNs cells induced the expression of proinflammatory cytokines including IL-1β, TNF-α, and IL-6 in mixed glial cells; however, 2DG treated MOG-primed MNs could not induce cytokines production when cocultured with mixed glial cells (Fig. 7D). Similar to MOG-primed MNs mixed glial coculture system, MOG-CM also induced the expression of inflammatory mediator (iNOS) and cytokines (IL-1β, IL6, and TNF-α) in the mixed brain glia cells while 2DG-CM inhibited the iNOS expression and cytokines production when added in culture media of mixed glia (1:1 ratio) compared with MOG-CM (Fig. 7E). These sets of data suggest that 2DG altered MN cells could not stimulate proinflammatory cytokines and iNOS expression in mixed glial cells. Overall, these data strongly show that inhibition of glycolysis by 2DG alters the MN function and phenotype resulting in protection from inflammation and demyelination.
Fig. 7.
Reduction in glycolysis inhibits activation of brain glial cells. (A) 2-deoxyglucose (2DG) is taken up by cells/tissues and phosphorylated to produce 2-deoxyglucose-6-phosphate (2DG6P), which cannot be metabolized further. Accumulated levels of 2DG6P was detected by LC/MS in the spinal cord of EAE and 2DG-treated EAE. Data were shown as mean ± SD (n = 6 mice per group). (B) Schematic flow of experimental design to generate CM and activated MNs cells in the presence or absence of 2DG (1 mM) and used for further experiments. (C, D) For in vitro coculture model, treated and untreated MOG-primed MNs cells with 2DG were cocultured with primary brain glial cells (4:1 ratios of MNs cells:mixed glial cells) for 24h and were processed for nitric oxide (NO), iNOS, and cytokines expression by immunoblot and qPCR. The data presented are the mean ± SD of three values per group. (E) MOG-CM and 2DG-CM were added in culture media of mixed glia (1:1 ratio with serum-free RPMI) and expression of inflammatory mediator (iNOS) and cytokines (IL-1β, IL-6, and TNF-α) was examined by qPCR. (F) Dose-dependent effect of 2DG (0.1 to 100 mM) on ECAR in brain glial cells using Seahorse Bioanalyzer (n = 6 per group). (G, H) Brain glial cells were treated with 2DG (1 to 10 mM) for 30 min prior the treatment of LI (0.5 µg/10 ng per milliliter). Post 18 h, cells were processed for iNOS immunoblot analysis and cell supernatant was processed for NO. Under similar experimental condition, at 6 h of LI stimulation, cells were processed for RNA isolation and qPCR for the detection of iNOS and IL-1β. The data presented are the mean ± SD of three values per group. (I) Impact of glucose on ECAR in brain glial cells using Seahorse Bioanalyzer (n = 6 per group). (J) Primary brain glial cells were cultured in normal (10 mM) and low (1 mM) glucose condition in RPMI for 24 h followed by stimulation with LI for 18 h. The levels of iNOS, pAMPKα, AMPKα, and pACC were examined by immunoblot analysis using their specific antibodies. Beta actin was used to examine the equal protein loading. Represented blots are the one of the two independent experiments. (K) Primary brain glial cells were cultured in normal (10 mM) and low (1 mM) glucose condition in RPMI for 24 h followed by stimulation with LI for 6 h. Cells were processed for RNA isolation and qPCR for the detection of iNOS, IL-1β, IL6, and MCP1. The data presented are the mean ± SD of three values per group. *P < 0.05, **P < 0.01, and ***P < 0.001. Student t test, one-way analysis of variance.
Glycolysis has been shown to be an important metabolic pathway that regulates inflammation in myeloid cells. Since we observed that 2DG-treated animals showed altered inflammatory phenotype of CD4 T cell and monocytes/macrophages, we examined the in vitro effect of glycolysis on inflammation of mixed glial cells under inflammatory conditions (LPS/IFN-γ; LI, treatment). We used various doses of 2DG ranging from 1 to 100 mM and examined the ECAR profile in mixed glial cells and found that lower dose of 2DG reduced the glycolysis in a dose dependent manner (Fig. 7F). Treatment with 2DG inhibited the production of NO and the expression of iNOS in a dose dependent manner (Fig. 7G). 2DG treatment also significantly abrogated the messenger RNA expression of iNOS, IL-1β (Fig. 7H), and MCP1, suggesting that inhibition of glycolysis by 2DG results in an antiinflammatory process. To further support the role of glycolysis, we applied the approach of modulating glucose levels to inhibit glycolysis. We cultured brain primary glial cells in low (1 mM) and normal glucose (10 mM) RPMI in the presence of normal glutamine levels (2 mM). Mixed glial cells cultured in low glucose exhibited significantly lower glycolysis compared with the cells cultured with normal glucose as expected (Fig. 7I). When cells were treated with LPS/IFN-γ (LI), the low glucose condition (1 mM) reduced the expression of iNOS by more than tenfold compared with that observed under normal glucose condition (10 mM) (Fig. 7J). Low glucose is known to activate the cellular energy sensor, AMP-activated protein kinase (AMPK), which has emerged as a metabolic regulator of energy metabolism and the ensuing functional activity of various immune cells. We observed higher phosphorylation of AMPK and its endogenous substrate, acetyl-CoA carboxylase, in the mixed glial cells cultured in lower glucose compared with glial cells in normal glucose levels (Fig. 7J). In addition, inhibition of glycolysis by glucose deprivation also resulted in lower expression of iNOS, IL-1β, IL-6, and MCP1 compared with glial cells with normal glucose upon LI stimulation (Fig. 7K). Overall, these data suggest that targeting glycolysis either by pharmacological approach (2DG) or by substrate (glucose) deprivation results in reduction in inflammatory response in brain glial cells and may be the underlying mechanism of the protective effect of 2DG in MS/EAE.
Discussion
MS is a disease where interplay between inflammation and immune cells dictates the disease progression. Recent advances have now established a strong link between the infiltrated immune cell function and altered cellular metabolism (15, 23). In the present study, we adapted the unique approach of interrogating the serum metabolite profile of patients with RRMS to arrive at a targetable metabolic pathway. By integrating the patient based untargeted metabolomics and functional metabolism, which were validated by extensive preclinical and ex vivo experimentation, we report that glycolysis is the key pathway regulating the metabotype and consequently the function of immune cells including CD4 and monocytes/macrophages in RRMS/EAE. Modulating the cellular metabolism by inhibition of glycolysis, can curtail the EAE disease process possibly by reverting immune cells toward an antiinflammatory phenotype of macrophage and resulting in an antiinflammatory protective environment.
Our untargeted metabolomics approach revealed 60 significantly altered metabolites in patient samples compared with HS, which strongly suggests the presence of a distinct dysregulated metabolic state in RRMS. Analyzing the biological implications of these altered metabolites by various analytical tools identified SIPR2 (intermediate regulators JUN, IFN-β1, and fingolimod) and TGF-β1 (intermediate regulator IRF7) as master regulators associated with the metabolite changes during the disease state. Studies have shown that sphingolipid metabolites such as S1P and ceramides strongly influence many cellular functions under physiological or pathological conditions (24–26). S1P is a bioactive lipid that mediates signaling and controls various physiological functions through members of the S1PR subfamily except for SIPR2 (24). According to the CNA, S1PR2, IFN-β1, and fingolimod have an inhibitory effect on MS. IFN-β1 is a cytokine in the interferon family used to treat RRMS as one of the first-line drugs. Fingolimod (also known as FTY720) is the first Food and Drug Administration approved oral disease-modifying drug for MS (27), and its phosphorylated form is considered as a functional antagonist of S1PR1 (28), by which it impacts lymphocyte trafficking and stops infiltration into CNS and mitigates the inflammatory process. The other master regulator identified, TGF-β1 cytokine plays a pleiotropic regulatory role and a synergistic role in disease activation in conjunction with transcription factor IRF-7 (29), which regulates interferon genes (30). Thus, the altered metabolites identified in our study are connected with or are a consequence of the aberrant biological processes that drive the MS disease. Metscape analysis revealed a global metabolomic derangement during MS disease resulting in perturbation of several metabolic networks that include glycolysis, TCA, urea, purine, tyrosine, butnoate, phosphatidylinositol phosphate, glycerophospholipids, glycerosphingolipids, pyrimidine, amino sugar, methionine, and cysteine. This observation is supported by some of the previous studies in patients with MS and preclinical mouse models of EAE (6, 7, 31). MetaboAnalyst analysis revealed the four major pathways encompassing the altered metabolites pertained to glycerophospholipid, citrate, sphingolipid, and pyruvate metabolism, where glycolysis is the common upstream pathway feeding and connecting to these pathways, reaffirming the fact that enhanced glycolysis is a peculiar feature of the autoimmune diseases (32–34). Apart from fulfilling the cellular energy expenses, glycolysis has a pivotal role in regulating the proper functioning of cells by generating biosynthetic intermediates and precursors required not only for cellular growth and proliferation, but also normal functioning of any cell. Recent studies have shown that enhanced glycolysis in various immune cells is a hallmark for carrying out successful immune processes like cell activation, proliferation, and effector response (35). We show that the altered glycolysis related serum metabolites are reflective of the immune cell’s energy status as the immune cells from patients with RRMS had inherent higher functional glycolysis. This suggests that PBMCs of patients with RRMS have an altered metabotype that relies mainly on glycolysis to meet their increased energy needs in the inflammatory environment. To ascertain if glycolysis is a valid target with potential to treat MS, we adopted a pharmacological approach to inhibit glycolysis and examine its effect on the inhibition of disease progression in EAE. Few studies have suggested glycolytic genes as therapeutic targets, including pyruvate kinase M2 (36–38), LDHA (34), and glucose phosphate isomerase (39). We selected a general inhibitor of glycolysis, 2-DG, which acts as a glucose analog, phosphorylated by hexokinase to 2DG6P, and cannot be further metabolized, resulting in revoking glycolysis in cells (40). We show that 2DG can offer significant protection in the clinical score, which implies a better clinical course and an overall improvement in various models of relapsing-remitting and chronic EAE. 2DG treatment blocked the infiltration of all MN cells, resulting in the protection of EAE. Previosuly, Shi et al. (41) reported that glycolytic activity in CD4 T cells could contribute to the lineage choices between Th17 and Treg cells and affect the EAE disease outcome. Here, using 2DG as a glycolytic inhibitor, we did not observe any effect on Tregs, which is in contrast to the previous study, and 2DG may play different roles in vivo versus in vitro polarization conditions (41). Here we examined the detailed mechanism of 2DG-mediated protection in various models of EAE. Our most striking observation is that 2DG polarizes monocytes/macrophages into an antiinflammatory phenotype and increases the ratio of antiinflammatory to proinflammatory macrophages in CNS. It is well established that both proinflammatory (the classically activated) and antiinflammatory (also known as alternatively activated macrophage) population is identified in MS (42–44). Their dual function is well defined in EAE as peripheral depletion of monocytes by clodronate liposomes reduced the severity of EAE (45) and the administration of antiinflammatory monocytes ameliorates EAE (46, 47). Monocytes isolated from 2DG treated EAE group could restrict the ongoing EAE disease supporting that antiinflammatory phenotype of monocytes/macrophages are protective in EAE and MS. Further, the therapeutic potential of antiinflammatory monocytes in MS has been highlighted by recent studies showing MS approved drugs, glatiramer acetate, laquinimod, and fasudil, suppress EAE by promoting the development of M2 activated monocytes (47–49). We show that the mechanism of 2DG-mediated promotion of antiinflammatory phenotype is via the inhibition of glycolysis, since monocytes isolated from 2DG treated mice maintained a lower glycolytic function compared with the untreated EAE group. Higher glycolysis in immune cells during EAE (34, 39) and other diseases (9–11, 50) clearly supports that metabolic reprograming in inflammatory cells is an attractive target for immune therapy in autoimmune disorders. The outcome of our clinical and preclinical studies strongly underlines the role of glycolysis in the regulation and functioning of immune cells during the inflammatory state within the body. This makes it entirely plausible to target the metabolic pathways for developing new therapeutics for autoimmune diseases, including MS. Apart from the glycolysis pathway, an interesting metabolite to emerge from the CAN analysis downstream of both SIPR2 and TGF-β1 is the 3-hydroxybutryric acid or β- hydroxybutyric acid (BHB), which was elevated by more than twofold in patients with RRMS. BHB is a ketone body formed in response to fatty acid oxidation and ketogenic amino acids. Its endogenous levels are elevated in response to fasting and ketogenic diets. BHB can cross the blood-brain barrier and is utilized by brain during conditions of low glucose. Ketogenic diets have been shown to provide protection against EAE and improve motor and neurological deficits along with reduced demyelination (51). Moreover, BHB treatment alleviated the clinical symptoms and pain behavior in chronic model of EAE (52). Thus, increased levels of BHB observed in our study, may be a byproduct or feedback mechanism to counter metabolic dysregulation and may be involved in ketogenic diets induced disease protective/modulatory effect. Future studies directly analyzing effect of BHB are needed to determine its therapeutic ability to improve the outcomes and quality of life in patients with MS (53). Another interesting metabolite 17-HDoHe was significantly increased in patients with RRMS. It is a downstream metabolite of omega-3 fatty acid docosahexaenoic acid (DHA) and also a precursor of D-series resolvins (54). Increased levels of 17-HDoHe in patients with RRMS either could be a protective mechanism to encounter pain sensitivity (55) or its failed metabolism into downstream resolvins during disease. We have previously demonstrated resolvin D1-mediated protection in a preclinical mouse model of MS (7) and ability of other SPMs to suppress activation and cytokine production in monocytes derived from patients with MS (56). Thus, these studies further support the therapeutic potential of resolvins in MS through modulation of omega-3 fatty acid and DHA pathway.
Overall, through our approach, we identified the distinct metabolite separation between normal and relapsing-remitting subjects and identified four significant pathways, with glycolysis being the central connector of these pathways being impacted due to disease in patients. On one hand, the observed metabolite signature in the biofluid of patients with MS can be exploited for biomarker development in the future. On the other hand, bioinformatics revealed a common upstream testable metabolic pathway, glycolysis, which we demonstrate can be successfully targeted in mouse models of MS using a glycolysis inhibitor. The 2DG-mediated inhibition of glycolysis strongly suggests the beneficial effect of this strategy on the disease progression in EAE. Moreover, an in-depth study of the altered pathway(s) could provide insight into the disease’s immunopathological mechanisms. In conclusion, our study provides additional evidence to support the existence of metabolic reprogramming in MS along with other autoimmune diseases with the observation of an altered metabotype in the patient’s immune cells, defined by a higher glycolysis, which certainly entails the potential to selectively target the glycolytic pathway for future therapeutic interventions.
Limitations.
MS is a chronic disease, and we recognize the limitation of the 2DG being translated for MS treatment due to poor drug-like characteristics including rapid metabolism and short half-life (57). To enhance its drug-like property, new analogs of 2DG are being tested (58). 2DG has been approved by the Indian Defense Research and Development Organization as adjunct therapy in patients with moderate to severe COVID-19. 2DG-treated patients with COVID-19 showed a significant improvement symptomatically and became free from oxygen supplementation by 3 d compared with untreated patients. 2DG is also being extensively tested against various cancers in clinical trials (58). We acknowledge that the therapeutic use of 2DG will have to be specifically designed around inflammatory episodes. Nevertheless, this is a proof-of-principle study to demonstrate the implication of metabolomics to identify a therapeutic target. In support, glycolytic inhibitors have been shown to be protective in EAE (34, 36–39), further supporting our metabolomics-driven interpretation to identify a therapeutic target for MS.
Materials and Methods
Patient Recruitment and Blood Sampling.
Blood samples collected from 35 unrelated patients with RRMS were obtained for the present study from the Biobank facility at the Department of Neurology, Henry Ford Hospital, Detroit, MI. Patients were diagnosed for MS, according to the 2010 revised McDonald criteria (59). The portion of the study on patient samples stands approved by the Institutional Review Board of Henry Ford Health System. All the patients/guardians were informed of the study, and informed consent was acquired from them in accordance with the ethical standards laid down by the World Health Organization and Declaration of Helsinki 1964 and its later amendments or comparable ethical standards. Inclusion criteria for selecting patients included radiological-confirmed patients with MS, while exclusion criteria included patients who did not satisfy McDonald diagnostic criteria, patients with previous or family history of other neurodegenerative or inflammatory diseases, and patients who rebutted from participating in this study.
Simultaneously, blood samples from 14 age- and sex-matched HS (to confine the confounding effect) were also used to serve as controls. Inclusion criteria for the selection of controls included healthy individuals with minor neurological problems like back pain and headache and those with no previous or family history of MS or any other autoimmune diseases. Exclusion criteria included individuals who denied participating in the study and with previous self and family history of inflammatory disease.
Serum and PBMCs.
For serum analysis, blood samples from patients with RRMS without treatment and HS were collected in red-capped vacutainer tubes. Serum was separated from blood by centrifugation at 1,500 rpm for 10 min. The clear yellow liquid supernatant part was collected from the top and stored in respective yellow capped serum tubes at −80 °C until further processing. Simultaneously, PBMCs were isolated from blood using the Ficoll-Hypaque gradient method. Blood collected in ethylenediaminetetraacetic acid-coated vacutainer tubes were mixed with an equal volume of 1 × sterile PBS (in the ratio of 1:1), and it was subsequently transferred onto the equal volume of Ficoll contained in 15-mL Falcon tube. After that, the samples were centrifuged at 3,000 rpm with zero deceleration for 30 min. Ficoll sets up the gradient, and the PBMC layer was separated between the topmost plasma and Ficoll. The collected PBMCs were again washed with 1 × PBS at 1,500 rpm for 10 min, and then cells were counted using BioRad Cell Counter. They were stored in liquid nitrogen until further use in 10% fetal bovine serum and 10% dimethylsulfoxide. The details of peptides and reagents are given in Star Methods.
Animals, EAE induction and recall response, histopathology and luxol fast blue staining and flow cytometry are described before (6, 7, 60) and details are provided in SI Appendix.
qPCR and NCBI GEO Database Analysis.
Total RNA was isolated from PBMCs of patients with RRMS without treatment and HS (SI Appendix, Table S4) using Direct-zol RNA kit (Zymo Research). The complementary DNA was prepared using iScript cDNA synthesis kit (Bio-Rad) as per the manufacturer’s protocol. The qPCR was performed for the genes of glycolytic-TCA network using their specific primers and SYBER Green PCR master mix (Bio-Rad) and a Bio-Rad iCycler iQ PCR (Bio-Rad). We selected three gene expression profile datasets about MS, GSE21942 (18), GSE26484 (19), and GSE43591 (20). All three datasets are based on Affymetrix. Raw data of each dataset is downloaded from NCBI GEO database with GEOquery in R and is preprocessed with RMA algorithm. We performed differential expression analysis with Limma between the MS group and the HS on each dataset separately. Isolation of total RNA and preparation of cDNA was performed as described above from isolated monocytes of treated and untreated groups with 2DG. The qPCR was performed using specific primers against specific genes including iNOS, IL-1β, arginase 1, chitinase-like 3 (Ym1/2) and glycolytic genes. Expression of these genes were normalized with the control gene, ribosomal L27 housekeeping gene. The sequence of primers used here is listed in SI Appendix.
2DG6P, Glucose, Lactate, and ATP Measurements.
The levels of 2DG6P in treated and untreated EAE with 2DG were measured in the spinal cord by ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) (Waters) as described in SI Appendix. The glucose and lactate levels in media were measured using Glucose Glo and Lactate Glo assay kits (Promega) as per the manufacturer's instructions. In brief, isolated monocytes using EasySep mouse monocyte isolation kit (Stemcell Technologies) from the spleen of treated and untreated EAE with 2DG were plated (25K/well) in a 96-well plate in RPMI media containing 10 mM glucose, 2 mM glutamine and 5% dialyzed fetal bovine serum. After 24 h, culture media was processed for the measurement of glucose and lactate using their respective assay kits from Promega. Total cellular levels of ATP were measured using ATPlite luminescence assay kit from Perkin-Elmer. Ten millimolar of the ATP solution was used to set up an ATP standard curve, and all samples were within the linear range of the assay.
ECAR.
Seahorse Bioscience XFe96 Extracellular Flux Analyzer (Agilent) was used to measure the glycolytic activity of intact PBMCs by monitoring the ECAR as described before (61). In brief, the XFe96 sensor cartridge was soaked in Calibrant overnight. PBMCs from HS and patients with RRMS (0.1 × 106 cells per well) or CFA and EAE groups (0.25 × 106 cells per well) were seeded in poly-lysine coated XFe 96-well seahorse culture microplate in 100 µL of Agilent Seahorse XF DMEM, pH 7.4 containing 2 mM glutamax (Thermofisher). The plate was centrifuged for 60 s at 400 RPMI. Seventy-five microliters of Dulbecco modified Eagle medium (DMEM) media was added in seahorse culture microplate and kept at a CO2-free incubator at 37 °C for degassing. Injection of glucose as fuel via first port induced the ECAR levels in cells. The basal glycolytic activity was calculated as the difference between ECAR reading following the preinjection and the injection of glucose. Blanks without cells were automatically subtracted during analysis by Agilent seahorse XF technology. ECAR values were normalized to cell number.
Metabolomic Analysis.
Metabolomic profiling analysis was performed by Metabolon Inc., as previously described (6–8). Detail of sample accessioning and preparation, analysis by UPLC-MS/MS and gas chromatography-mass spectroscopy, quality assurance/quality control, data extraction and compound identification, curation, metabolite quantification and data normalization are provided in SI Appendix.
Statistical Analysis.
Metabolite intensities (scaled imputed) obtained from Metabolon. Statistical analyses are performed with R software (cran.r-project.org/) and using MetaboloAnalyst (https://www.metaboanalyst.ca/) (62). The first two components of a principal components analysis were plotted to identify whether samples or batch effects have contributed disproportionally to the variance in the data. PLS-DA was used for the assessment of the separability of the samples by supervised clustering of samples based on metabolites intensities. Per metabolite, comparisons were made using two-sample t tests on the study cohort. Significant differences were determined at P < 0.05. False discovery rates were calculated by the Q value method from the Bioconductor R package and are provided for reference. Heatmaps are drawn for significantly differential metabolites using blue (low) to yellow (high) coloring to depict standardized intensity differences from the metabolite-level mean. Hierarchical clustering on Pearson's correlation coefficient is used to generate all dendrograms. Metabolites were mapped into the IPA software using HMDB IDs and KEGG knowledgebase of compound IDs, PubChem, CAS IDs, and networks of altered metabolites were constructed using Fisher exact test including CNA (14). The goal of IPA's CNA is to prioritize regulatory networks that explain observed biological changes based on altered metabolites. Its algorithm constructs a multiedge path from a regulator molecule to regulated metabolites. To construct a gene-enzyme-compound network, Metscape-based on the KEGG database was performed to build a metabolic network on significantly altered metabolites. In Metscape, the construction of the metabolic Network pathway is based on HUMDB, KEGG, and EHMN databases (63).
Supplementary Material
Acknowledgments
This work is in part supported by research grants from the National Multiple Sclerosis Society (US) (RG-1807-31964, RG-1508-05912), the NIH (NS112727, AI144004), and Henry Ford Hospital Internal support (A10270, A30967) to S.G. A.K. is supported by grant R01EY026964. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123265119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Compston A., Coles A., Multiple sclerosis. Lancet 372, 1502–1517 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Torkildsen Ø., Myhr K. M., Bø L., Disease-modifying treatments for multiple sclerosis—A review of approved medications. Eur. J. Neurol. 23 (suppl. 1), 18–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saidha S., Eckstein C., Calabresi P. A., New and emerging disease modifying therapies for multiple sclerosis. Ann. N. Y. Acad. Sci. 1247, 117–137 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Zhang A. H., Sun H., Wang X. J., Recent advances in metabolomics in neurological disease, and future perspectives. Anal. Bioanal. Chem. 405, 8143–8150 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Zahoor I., Rui B., Khan J., Datta I., Giri S., An emerging potential of metabolomics in multiple sclerosis: A comprehensive overview. Cell. Mol. Life Sci. 78, 3181–3203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangalam A., et al. , Profile of circulatory metabolites in a relapsing-remitting animal model of multiple sclerosis using global metabolomics. J. Clin. Cell. Immunol. 4, 1000150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poisson L. M., et al. , Untargeted plasma metabolomics identifies endogenous metabolite with drug-like properties in chronic animal model of multiple sclerosis. J. Biol. Chem. 290, 30697–30712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh J., et al. , Urinary and plasma metabolomics identify the distinct metabolic profile of disease state in chronic mouse model of multiple sclerosis. J. Neuroimmune Pharmacol. 14, 241–250 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Yin Y., et al. , Glucose oxidation is critical for CD4+ T cell activation in a mouse model of systemic lupus erythematosus. J. Immunol. 196, 80–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weyand C. M., Zeisbrich M., Goronzy J. J., Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Curr. Opin. Immunol. 46, 112–120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R. T., et al. , Enhanced glycolysis contributes to the pathogenesis of experimental autoimmune neuritis. J. Neuroinflammation 15, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalek R. D., et al. , Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettencourt I. A., Powell J. D., Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J. Immunol. 198, 999–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krämer A., Green J., Pollard J. Jr., Tugendreich S., Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohan S., Lucassen E., Smoot K., Brink J., Chen C., Sphingosine-1-phosphate: Its pharmacological regulation and the treatment of multiple sclerosis: A review article. Biomedicines 8, 227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J., et al. , Metscape: A Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 26, 971–973 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maceyka M., Spiegel S., Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemppinen A. K., Kaprio J., Palotie A., Saarela J., Systematic review of genome-wide expression studies in multiple sclerosis. BMJ Open 1, e000053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakatsuji Y., et al. , Elevation of Sema4A implicates Th cell skewing and the efficacy of IFN-β therapy in multiple sclerosis. J. Immunol. 188, 4858–4865 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Jernås M., et al. , MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol. 14, 32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill L. A., Pearce E. J., Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everts B., et al. , TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15, 323–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makowski L., Chaib M., Rathmell J. C., Immunometabolism: From basic mechanisms to translation. Immunol. Rev. 295, 5–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strub G. M., Maceyka M., Hait N. C., Milstien S., Spiegel S., Extracellular and intracellular actions of sphingosine-1-phosphate. Adv. Exp. Med. Biol. 688, 141–155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., et al. , Sphingosine-1-phosphate, a novel lipid, involved in cellular proliferation. J. Cell Biol. 114, 155–167 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W., et al. , Ceramides and other bioactive sphingolipid backbones in health and disease: Lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta 1758, 1864–1884 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Brinkmann V., et al. , Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9, 883–897 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Billich A., et al. , Phosphorylation of the immunomodulatory drug FTY720 by sphingosine kinases. J. Biol. Chem. 278, 47408–47415 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Ning S., Pagano J. S., Barber G. N., IRF7: Activation, regulation, modification and function. Genes Immun. 12, 399–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirshafiey A., Mohsenzadegan M., TGF-beta as a promising option in the treatment of multiple sclerosis. Neuropharmacology 56, 929–936 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Stoessel D., et al. , Metabolomic profiles for primary progressive multiple sclerosis stratification and disease course monitoring. Front. Hum. Neurosci. 12, 226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D., et al. , High glucose intake exacerbates autoimmunity through reactive-oxygen-species-mediated TGF-β cytokine activation. Immunity 51, 671–681.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radu C. G., Shu C. J., Shelly S. M., Phelps M. E., Witte O. N., Positron emission tomography with computed tomography imaging of neuroinflammation in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 104, 1937–1942 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaushik D. K., et al. , Enhanced glycolytic metabolism supports transmigration of brain-infiltrating macrophages in multiple sclerosis. J. Clin. Invest. 129, 3277–3292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locasale J. W., Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kono M., et al. , Pyruvate kinase M2 is requisite for Th1 and Th17 differentiation. JCI Insight 4, e127395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damasceno L. E. A., et al. , PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J. Exp. Med. 217, e20190613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angiari S., et al. , Pharmacological activation of pyruvate kinase M2 inhibits CD4+ T cell pathogenicity and suppresses autoimmunity. Cell Metab. 31, 391–405.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L., et al. , Niche-selective inhibition of pathogenic Th17 cells by targeting metabolic redundancy. Cell 182, 641–654.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wick A. N., Drury D. R., Nakada H. I., Wolfe J. B., Localization of the primary metabolic block produced by 2-deoxyglucose. J. Biol. Chem. 224, 963–969 (1957). [PubMed] [Google Scholar]
- 41.Shi L. Z., et al. , HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boven L. A., et al. , Myelin-laden macrophages are anti-inflammatory, consistent with foam cells in multiple sclerosis. Brain 129, 517–526 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z., et al. , Parenchymal accumulation of CD163+ macrophages/microglia in multiple sclerosis brains. J. Neuroimmunol. 237, 73–79 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Raine C. S., Multiple sclerosis: The resolving lesion revealed. J. Neuroimmunol. 304, 2–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huitinga I., van Rooijen N., de Groot C. J., Uitdehaag B. M., Dijkstra C. D., Suppression of experimental allergic encephalomyelitis in Lewis rats after elimination of macrophages. J. Exp. Med. 172, 1025–1033 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikita J., et al. , Altered M1/M2 activation patterns of monocytes in severe relapsing experimental rat model of multiple sclerosis. Amelioration of clinical status by M2 activated monocyte administration. Mult. Scler. 17, 2–15 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Weber M. S., et al. , Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat. Med. 13, 935–943 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Schulze-Topphoff U., et al. , Laquinimod, a quinoline-3-carboxamide, induces type II myeloid cells that modulate central nervous system autoimmunity. PLoS One 7, e33797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C., et al. , Targeting the shift from M1 to M2 macrophages in experimental autoimmune encephalomyelitis mice treated with fasudil. PLoS One 8, e54841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okano T., et al. , 3-bromopyruvate ameliorate autoimmune arthritis by modulating Th17/Treg cell differentiation and suppressing dendritic cell activation. Sci. Rep. 7, 42412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D. Y., et al. , Inflammation-mediated memory dysfunction and effects of a ketogenic diet in a murine model of multiple sclerosis. PLoS One 7, e35476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirzaeia V., Eidia A., Manahejib H., Oryana S., Zaringhalamb J., β-Hydroxybutyrate attenuates clinical symptoms and pain behaviors in MOG-induced encephalomyelitis. Neurochem. J. 15, 181–186 (2021). [Google Scholar]
- 53.Brenton J. N., et al. , Pilot study of a ketogenic diet in relapsing-remitting MS. Neurol. Neuroimmunol. Neuroinflamm. 6, e565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serhan C. N., Pro-resolving lipid mediators are leads for resolution physiology. Nature 510, 92–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valdes A. M., et al. , Association of the resolvin precursor 17-HDHA, but not D- or E- series resolvins, with heat pain sensitivity and osteoarthritis pain in humans. Sci. Rep. 7, 10748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kooij G., et al. , Specialized pro-resolving lipid mediators are differentially altered in peripheral blood of patients with multiple sclerosis and attenuate monocyte and blood-brain barrier dysfunction. Haematologica 105, 2056–2070 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen I. L., Levy M. M., Kerr D. S., The 2-deoxyglucose test as a supplement to fasting for detection of childhood hypoglycemia. Pediatr. Res. 18, 359–364 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Pajak B., et al. , 2-deoxy-d-glucose and its analogs: From diagnostic to therapeutic agents. Int. J. Mol. Sci. 21, 234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polman C. H., et al. , Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 69, 292–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mangalam A. K., et al. , AMP-activated protein kinase suppresses autoimmune central nervous system disease by regulating M1-type macrophage-Th17 axis. J. Immunol. 197, 747–760 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Dar S., et al. , Bioenergetic adaptations in chemoresistant ovarian cancer cells. Sci. Rep. 7, 8760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chong J., Wishart D. S., Xia J., Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics 68, e86 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Karnovsky A., et al. , Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 28, 373–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.