Significance
Development of effective cellular therapy strategies against acute myeloid leukemia (AML) remains a major therapeutic challenge. Memory-like natural killer (NK) cells armed with a TCR-like chimeric antigen receptor (CAR) targeting a unique neoepitope presented by HLA-A2 exhibit potent activity against nucleophosphmin-1 (NPM1)-mutated AML in vitro and in vivo. Multiomics analyses provide insight into the key pathway genes upregulated upon CAR engineering and target engagement in cytokine-induced memory-like (CIML) NK cells.
Keywords: acute myeloid leukemia, NPM1 mutation, CAR-NK cells, memory-like NK cells, TCR-like CAR
Abstract
Acute myeloid leukemia (AML) remains a therapeutic challenge, and a paucity of tumor-specific targets has significantly hampered the development of effective immune-based therapies. Recent paradigm-changing studies have shown that natural killer (NK) cells exhibit innate memory upon brief activation with IL-12 and IL-18, leading to cytokine-induced memory-like (CIML) NK cell differentiation. CIML NK cells have enhanced antitumor activity and have shown promising results in early phase clinical trials in patients with relapsed/refractory AML. Here, we show that arming CIML NK cells with a neoepitope-specific chimeric antigen receptor (CAR) significantly enhances their antitumor responses to nucleophosphmin-1 (NPM1)-mutated AML while avoiding off-target toxicity. CIML NK cells differentiated from peripheral blood NK cells were efficiently transduced to express a TCR-like CAR that specifically recognizes a neoepitope derived from the cytosolic oncogenic NPM1-mutated protein presented by HLA-A2. These CAR CIML NK cells displayed enhanced activity against NPM1-mutated AML cell lines and patient-derived leukemic blast cells. CAR CIML NK cells persisted in vivo and significantly improved AML outcomes in xenograft models. Single-cell RNA sequencing and mass cytometry analyses identified up-regulation of cell proliferation, protein folding, immune responses, and major metabolic pathways in CAR-transduced CIML NK cells, resulting in tumor-specific, CAR-dependent activation and function in response to AML target cells. Thus, efficient arming of CIML NK cells with an NPM1-mutation-specific TCR-like CAR substantially improves their innate antitumor responses against an otherwise intracellular mutant protein. These preclinical findings justify evaluating this approach in clinical trials in HLA-A2+ AML patients with NPM1c mutations.
Despite the Food and Drug Administration approval of several agents in the past several years, treatment of acute myeloid leukemia (AML) continues to be a major therapeutic challenge, with most patients succumbing to their disease. Although allogeneic hematopoietic cell transplantation (allo-HCT) can be an effective treatment, this modality has its limitations. Many patients are not suitable candidates due to age, comorbidities, or lack of an available donor. Long-term clinical success is limited by graft versus host disease (GVHD), infection, and organ toxicity. Furthermore, relapse remains the most common cause of treatment failure after allo-HCT (1). Therefore, there is a pressing need to develop less toxic and more effective targeted cellular therapies that take advantage of our evolving understanding of cancer immunotherapy. Graft versus leukemia (GVL), the underlying immune mechanism for allo-HCT, has traditionally been ascribed to T cells. However, more recent studies have found that natural killer (NK) cells also play an important role as mediators of GVL (2, 3). NK cells are innate lymphoid cells that can eliminate virus-infected and malignantly transformed cells. NK cells possess many of the key attributes critical for effective cancer therapies—“born to kill” but without an apparent risk of GVHD, cytokine release syndrome (CRS), or neurotoxicity (4). Furthermore, their intrinsic propensity to target myeloid blasts makes them particularly attractive for AML treatment (5).
The development of NK cell–based therapy remains challenging due to NK cells’ short lifespan, limited proliferative capacity, and lack of specific tumor targeting. Conventional NK (cNK) cells have shown some promise in early phase clinical trials; however, these infusions resulted in relatively short remissions in a minority of patients. Recently, paradigm-shifting studies have shown that NK cells can be induced to exhibit memory-like properties (6, 7). We and others described cytokine-induced memory-like (CIML) NK cells generated by brief (12 to 16 h) in vitro preactivation with IL-12, IL-15, and IL-18, and these cells have potent antileukemia activity (6, 8). In our first-in-human phase 1 trial, CIML NK cells induced clinical responses in >50% of patients with relapsed refractory AML, with no apparent toxicity (9). Furthermore, the transferred NK cells proliferated, expanded, and maintained enhanced antileukemia responses in patients (7, 10).
Cellular therapies that retarget T cells to recognize, respond, and kill neoplastic cells via chimeric antigen receptors (CARs) represent a promising approach for treating advanced malignancies. While CAR T cells have demonstrated promising activity in advanced B cell malignancies including lymphomas and myeloma, a paucity of tumor-specific antigens on AML has significantly limited their therapeutic efficacy in myeloid malignancies including AML. CD123 and CD33 are the most common targets that are currently being evaluated in several CAR-based trials; however, both antigens are also expressed by normal hematopoietic stem cells and/or progenitors and therefore potentially associated with significant on-target but off-tumor toxicity.
AML is a molecularly diverse malignancy. Among the most commonly occurring oncogenic mutations is a 4-base pair insertion in exon 12 of the nucleophosmin-1 (NPM1) gene, which occurs in ∼30% of all adult AML cases and in 50 to 60% of patients with an otherwise normal karyotype (1). Mutations in NPM1 result in aberrant cytoplasmic localization of the protein and are referred to as NPM1c. The NPM1c mutant protein generates a leukemia-specific neoepitope (AIQDLCLAV) that is presented by the most common HLA-A*0201 allele (∼50% of human population) (7). TCR-like CARs have shown success in T cell immunotherapies via targeting tumor neoantigens; here, we hypothesize that arming CIML NK cells with a neoepitope-specific TCR-like CAR will harness their innate cell intrinsic antitumor response and further augment it by CAR-mediated tumor targeting. To test this approach, we efficiently armed peripheral blood (PB)-derived CIML NK cells with a TCR-like CAR specific for NPM1c+ HLA-A*0201+ AML. Our results show exquisite specificity and potent antileukemia activity of NPM1c-CAR CIML NK cells in vitro and in preclinical human xenograft models. Furthermore, detailed single-cell transcriptomic and proteomic analyses provide a mechanistic understanding of the molecular basis of CAR CIML NK cells. Our proof-of-concept study demonstrates the feasibility and exciting potential for arming CIML NK cells with tumor-specific CARs and other effector molecules for cancer adoptive cell therapies.
Results
Generation of NPM1c-Specific CAR-CIML NK Cells.
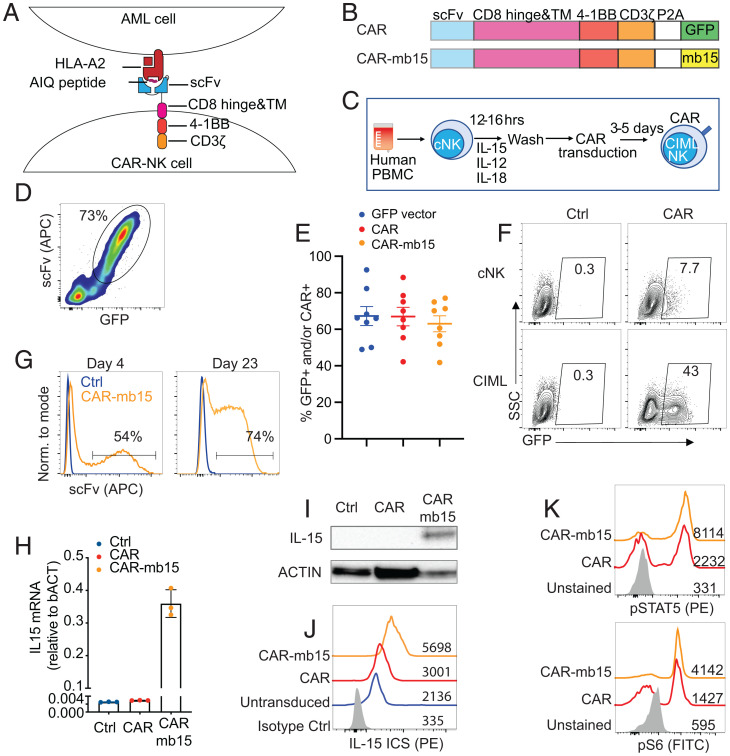
To date, TCR-like CARs have not been studied in the context of NK cells or CIML NK cells. We have recently isolated an single-chain variable fragment (scFv) that specifically recognizes the NPM1c neoepitope-HLA-A2 complex and have successfully created a CAR construct by cloning the scFv in-frame into a second-generation CAR backbone containing a CD8α hinge and transmembrane (TM) domain, a 4-1BB costimulatory domain, and a CD3ζ activation domain, followed by self-cleavage P2A and GFP (Fig. 1 A and B) (11). As both 4-1BB and CD3ζ activation domains are used naturally by NK cells for signaling and activation (12), we hypothesized that the same CAR design would likely work in CIML NK cells. We pseudotyped the NPM1c-CAR-expressing lentivirus with baboon endogenous retroviral envelope (BaEV) (13) and used them to transduce CIML NK cells derived from the PB of multiple healthy donors at a multiplicity of infection (MOI) of 10 (Fig. 1C and SI Appendix, Fig. S1). High transduction efficiency of CIML NK cells was achieved based on the coexpression of GFP and NPM1c-CAR on the cell surface (Fig. 1D). On average, ∼65% of CIML NK cells from multiple PB healthy donors were transduced to express GFP, NPM1c-CAR, or NPM1c-CAR plus mb15 (Fig. 1E). Notably, CIML NK cells were much more efficiently transduced by BaEV pseudotyped lentiviruses than their naive counterparts or cells stimulated with individual or other cytokine combinations (Fig. 1F) because ASCT2, the target of BaEV, was significantly induced following CIML NK cell differentiation (SI Appendix, Fig. S1). The transduction approach requires neither in vitro expansion of CIML NK cells nor multiple rounds of spinfection (14), while sustained CAR expression was observed in vitro for 23 d (Fig. 1G), the longest time point monitored.
Fig. 1.
Generation of NPM1c-specific CAR-CIML NK cells. (A) Schematic diagram showing the design of CAR constructs and recognition of the AIQ/HLA-A2 complex on AML cells by NPM1c-CARs in B. (C) Schematic diagram showing the differentiation of freshly purified PB cNK cells into CIML NK cells and their transduction by BaEV-pseudotyped lentivirus. PBMC, peripheral blood mononuclear cell. (D) Representative flow cytometry plot showing expression of the NPM1c-CAR and GFP on transduced CIML NK cells. (E) Summary data showing transduction efficiencies of CIML NK cells from different donors with lentiviruses expressing GFP (blue dots), NPM1c-CAR (red dots), or NPM1c-CAR-mb15 (orange dots). (F) Comparison of transduction efficiency in cNK and CIML NK cells from a representative donor. Transduced cells were analyzed for GFP expression after 72 h, using untransduced cNK or CIML NK cells as controls (Ctrl). The numbers indicate percentages of cells in the gated area. (G) Representative flow cytometry histograms showing surface CAR expression of the untransduced (Ctrl in blue) and CAR-mb15-transduced (orange) CIML NK cells at day 4 and day 23 posttransduction. (H) qPCR quantification of IL15 transcripts and (I) Western blotting of IL-15 protein in CIML NK cells without transduction (Ctrl) or transduced with CAR or CAR-mb15. (J) Intracellular staining (ICS) of IL-15 in untransduced and CAR and CAR-mb15 transduced CIML NK cells by flow cytometry. Gray, blue, red, and orange histograms show isotype controls, untransduced, and CAR- and CAR-mb15 transduced CIML NK cells, respectively. (K) Flow cytometry assays for pSTAT5 (Upper Panel) and S6 kinase (Bottom Panel) in CAR and CAR-mb15 transduced CIML NK cells. Gray, red, and orange histograms show unstained controls and CAR- and CAR-mb15 transduced CIML NK cells, respectively. Data are representative of five (D) or three (F–K) independent experiments. Data in H show representative results using CIML NK cells from one PB donor, with error bars representing mean with SD from technical triplicates. Data in E are pooled from five independent experiments, with each dot representing one different PB donor; n = 8 for each column and error bars represented mean with SEM.
IL-15 is critical for NK cell development and homeostasis and is normally transpresented by IL-15Rα-expressing dendritic cells to effector immune cells (15). Based on these observations and our prior clinical experience with an IL-15 superagonist in hematologic malignancies (16), we further engineered the CAR CIML NK cells by expressing human IL-15 from the same CAR vector (Fig. 1B). The endogenous expression of IL-15 is expected to substantially promote in vivo persistence of the transferred CAR CIML NK cells and also mitigate a need for exogenous cytokines often used in NK cell–based adoptive therapies (17). Specifically, we replaced the GFP cassette with a membrane-bound IL-15 (mb15) in the NPM1c-CAR lentiviral vector (Fig. 1B) (18). We chose a membrane-bound over the wild-type secreted IL-15 in order to minimize the toxicity reported in patients who received systemic IL-15 therapy (19, 20) while maximizing its potency within the NK cell microenvironment. Following transduction with the NPM1c-CAR-mb15 lentivirus, IL-15 expression was detected by RT-PCR for messenger RNA (mRNA) (Fig. 1H) and Western blotting and intracellular staining for protein (Fig. 1 I and J). The expression of mb15 resulted in a marked increase in the phosphorylation of STAT5 and S6 kinase in CAR transduced cells (Fig. 1K), consistent with functional signaling through the IL-15 receptor (15, 21).
In-Depth Transcriptomic and Proteomic Characterization of CAR-mb15 CIML NK Cells.
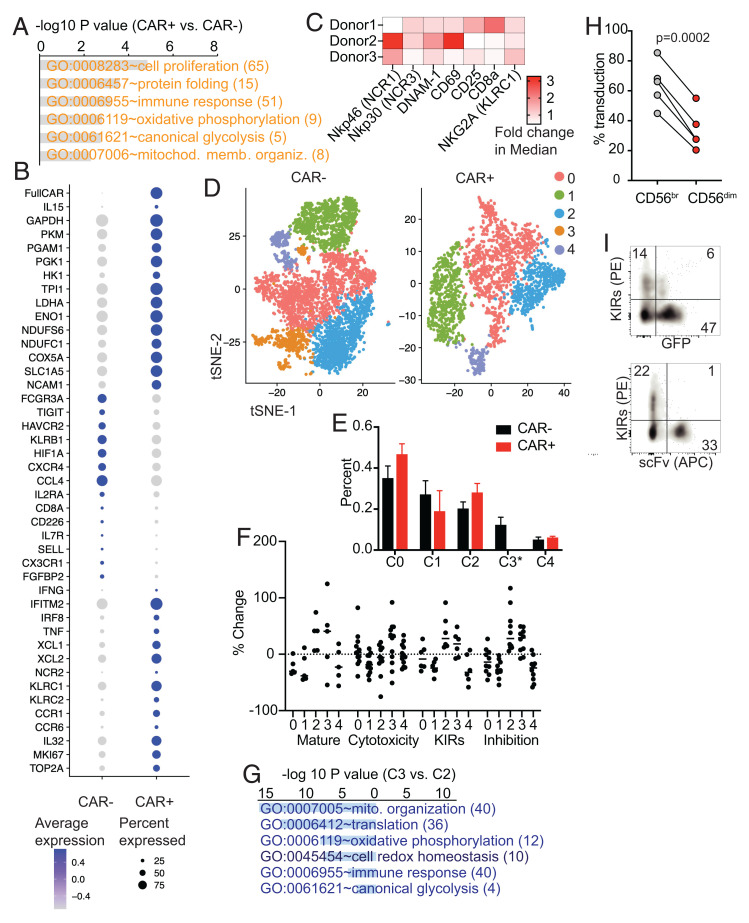
To comprehensively characterize CAR CIML NK cells, we performed single-cell transcriptome sequencing (scRNA-seq) analyses on flow-sorted NPM1c-CAR-mb15 CIML NK cells and CAR-negative CIML NK cells from the same culture generated from two different PB donors. A total of 1,864 CAR+ and 4,725 CAR− CIML NK cells were identified from a total of 7,105 sequenced cells based on the expression of the CAR construct from two different donors (cells with marginal expression of CAR were omitted, Materials and Methods). To minimize donor variation, a comparison was performed between CAR+ and CAR− CIML NK cells from the same donors. Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) identified significant up-regulation of genes in key pathways involving cell proliferation, protein folding, and immune responses as well as in major metabolic pathways involving glycolysis, OXPHOS, and mitochondrial function (Fig. 2A, with selected genes shown in Fig. 2B). This highly proliferative and metabolically active phenotype is consistent with the role of IL-15 as a master regulator of NK cell homeostatic proliferation and a major metabolic switch (15, 21) (Fig. 1K and SI Appendix, Fig. S4) and is also consistent with our previous findings that IL-15 activates the endoplasmic reticulum (ER) stress sensor IRE1/XBP1, which is critical for boosting OXPHOS in effector NK cells (22). In addition, mass cytometric analysis revealed increased levels of key surface-activating receptors and activation markers (Fig. 2C), consistent with the functionally active profile of CAR-mb15 CIML NK cells. By t-distributed stochastic neighbor embedding (tSNE) clustering analyses of scRNA-seq data (Fig. 2D), CIML NK cells generated from healthy PB donors exhibited substantial heterogeneity. Interestingly, 4 out of 5 heterogeneous clusters remained unchanged in CAR-expressing CIML NK cells after transduction, whereas only a single cluster (cluster 3 in Fig. 2 D and E) was missing that accounted for 10 to 15% of CAR-negative CIML NK cells. Functional score analyses comparing each cluster (Fig. 2F) revealed that cluster 3 and cluster 2 exhibited gene signatures associated with terminal differentiation (or “maturation”) and functional inhibition (including KIRs and exhaustion markers [23]). A further DEG analysis of cluster 3 and cluster 2 revealed profound metabolic quiescence of cluster 3 (Fig. 2G). Thus, CAR-mb15 CIML NK cells were enriched for metabolically active, highly proliferative, less mature, and less inhibitory NK cell populations, suggesting potentially favorable outcomes in vitro and in vivo against AML targets.
Fig. 2.
scRNA-seq and mass cytometry characterization of BaEV LV-transduced CAR-mb15 CIML NK cells. (A) GO functional enrichment analysis of DEGs between the CAR+ and CAR− CIML NK cells (n = 2). DEGs were computed by FindMarkers between CAR+ and CAR− populations with log2 fold-change of >0.1 and P < 0.05. (B) Dotplot of the expression of selected DEGs. (C) Heatmap of fold changes of selected NK cell activation and functional proteins between CAR+ and CAR− CIML NK cells based on mass cytometry analysis. (D) tSNE clustering analysis of scRNA-seq data from CAR+ CIML NK cells (Right) and CAR− CIML NK cells (Left). Each cluster between CAR+ and CAR− cells was annotated based on the expression similarity of 2,000 variable genes. (E) Cell proportion of each cluster in CAR+ and CAR− CIML NK cells. Note that cluster 3 was absent in CAR+ CIML NK cells. (F) The scores for maturation, cytotoxicity, KIR, and inhibition in each cluster. Each dot represents one gene in that cluster and category (SI Appendix, Materials and Methods). The score was calculated from the change of its expression to the average expression of all cells. (G) GO functional enrichment analysis of DEGs between cluster 3 and cluster 2 in CAR− CIML NK cells. (H) Flow cytometry analysis of BaEV lentiviral transduction of CD56brigh (immature) versus CD56dim (mature) NK cells. (I) Representative flow cytometry plots showing negative correlation between KIR expression and CAR transduction with BaEV pseudotyped lentivirus expressing NPM1c-CAR and GFP. n = 3 (C), 2 (E), 5 (H) PB donors. Error bars in E represented mean with SD. Data in H were analyzed by two-tailed paired t test. Data are pooled from two (C) or three (H) independent experiments, or representative of two independent experiments (I).
To investigate the differential susceptibility of various CIML NK cell populations to BaEV lentiviral transduction and how that may impact CAR-NK cell differentiation and downstream effector functions, we specifically asked whether the more mature, less proliferative subsets in CIML NK cells may be less amenable to BaEV lentiviral (LV) transduction. To account for any potential confounding effect of mb15 on transduction, we performed flow cytometry validation with the NPM1c-CAR (no mb15, as shown in Fig. 1B, Upper Panel). These data confirmed that BaEV-lentivirus-mediated CAR transduction and expression positively correlated with CD56 expression (Fig. 2H) but negatively correlated with inhibitory KIR expression (Fig. 2I) of CIML NK cells. Together, our data rule out an alternative possibility that the clustering changes could be merely attributed to mb15 expression instead of differential transduction (24) by BaEV lentiviruses.
NPM1c-CAR CIML NK Cells Exhibit Potent Anti-AML Activity In Vitro.
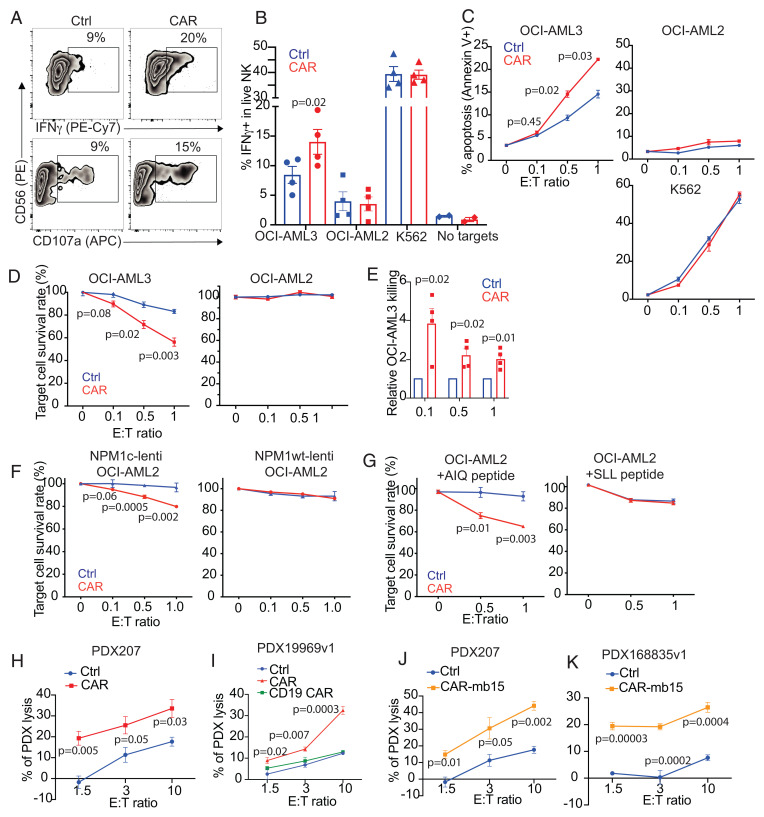
Distinct from T cells, NK cells and CIML NK cells have an intrinsic propensity to target myeloid blasts, making them particularly attractive for AML treatment (5, 25, 26). To investigate whether and to what extent CAR expression contributes to the anti-AML response, NPM1c-CAR CIML NK cells were directly compared to untransduced CIML NK cells from the same donor. When evaluated against NPM1c+ HLA-A2+ OCI-AML3 target cells, significantly higher percentages of CAR CIML NK cells expressed IFNγ and CD107a (a marker for degranulation), compared to their untransduced counterparts that exhibited basal levels of activation upon OCI-AML3 tumor stimulation (Fig. 3 A and B). Detailed phenotypic characterization of the CD107a+ IFNγ+ vs. CD107a− IFNγ− of the NPM1c-CAR-transduced CIML NK cells upon coculture with OCI-AML3 using mass cytometric analysis showed differences in several key NK cell markers, including higher levels of Np46, NKp30, CD25, and perforin expression on CD107a+ IFNγ+ CIML NK cells (SI Appendix, Fig. S2). Furthermore, CAR-dependent target cell killing was observed both in a flow cytometry–based early apoptosis detection assay (Fig. 3C) and in a luciferase killing assay (Fig. 3 D and E).
Fig. 3.
NPM1c-CAR CIML NK cells exhibit potent and specific anti-AML function in vitro. (A) Representative flow cytometry staining profiles of IFNγ or CD107a versus CD56 of untransduced (Ctrl) and NPM1c-CAR transduced (CAR) CIML NK cells. Indicated NK cells were cocultured with OCI-AML3 target cells for 6 h and CD56+ cells were analyzed. (B) Summary data showing percentages of IFNγ-expressing cells in CAR-expressing vs. untransduced (Ctrl) CIML NK cells following incubation with OCI-AML3 (NPM1c+ HLA-A2+), OCI-AML2 (NPM1c− HLA-A2+), K562 (HLA−), or no target cells. (C) Killing of OCI-AML3, OCI-AML2, and K562 target cells by untransduced (Ctrl) and CAR-transduced CIML NK cells at the indicated effector (E): target (T) ratios. CIML NK cells and target cells were incubated for 4 h, and percentages of annexin V+ tumor cells were assayed by flow cytometry. (D) Killing of OCI-AML3 and OCI-AML2 target cells by untransduced (Ctrl) and CAR-transduced CIML NK cells at the indicated E: T ratios. CIML NK cells and luciferase-expressing target cells were incubated for 24 h, and luciferase activity was quantified. (E) Summary data showing OCI-AML3 target killing by NPM1c-CAR CIML NK cells as assessed by luciferase assay. Data were normalized to the percentage of killing by the corresponding untransduced (Ctrl) CIML NK cells. (F) OCI-AML2 cells were transduced with lentivirus expressing NPM1c (Lenti-NPM1c, Left Panel) or the wild-type NPM1 control (Lenti-NPM1wt, Right Panel). Transduced cells were sorted, expanded, and used as target cells for killing assays as described in D. Killing of the transduced OCI-AML2 target cells by Ctrl and CAR-transduced CIML NK cells were measured at the indicated E:T ratios. (G) Comparison of killing of OCI-AML2 cells pulsed with 1 μM of AIQ (Left) or SLL (Right) peptides by NPM1c CAR-CIML NK cells or untransduced (Ctrl) CIML NK cells. NK cells were cocultured with peptide-pulsed OCI-AML2 target cells at the indicated E:T ratios for 24 h. Target cell killing was measured by the luciferase activity of surviving target cells. (H) Killing of low-passaged NPM1c+ HLA-A2+ PDX AML target cells by NPMc1c-CAR CIML NK cells in comparison with their untransduced CIML NK cells from the same PB donor (n = 1 PDX donor and 1 PB donor for each graph; error bars from technical triplicates). (I) Killing of NPM1c+ HLA-A2+ PDX AML target cells by untransduced (Ctrl), NPM1c CAR-CIML (CAR), and irrelevant CD19 CAR CIML NK cells. (J and K) Killing of NPM1c+ HLA-A2+ PDX target cells by NPMc1c-CAR-mb15 CIML NK cells (n = 2 PDX donors and 2 PB donors). Each dot in B and E represent one different PB donor. n = 4 in B and E. Error bars in B and E represented mean with SEM. Error bars in C, D, and F–K represented mean with SD from 3 to 5 technical replicates. Data were analyzed by two-tailed paired Student’s t test (B), two-tailed one sample t test (E), and two-tailed unpaired t test (C, D, and F–K). Data are pooled from two (B and E) independent experiments, or representative of three (C, D, F, and G) independent experiments.
To assess the specificity of NPM1c-CAR CIML NK cells for the NPM1c-derived neoepitope presented by the HLA-A2 allele, we performed IFNγ and cytotoxic assays using OCI-AML2 target cells that were HLA-A2+ but expressed the wild-type NPM1 protein. In contrast to their superior activity against NPM1c+ HLA-A2+ AML targets, NPM1c-CAR NK cells did not show any enhanced IFNγ expression and killing of NPM1 wild-type AML beyond the innate activity shown in donor-matched untransduced CIML NK cells (Fig. 3 B–D). In addition, coculturing with NK-sensitive K562 target cells (HLA−, derived from a patient with chronic myeloid leukemia) resulted in comparably robust IFNγ expression and tumor killing irrespective of CAR expression (Fig. 3 B and C), suggesting a minimal impact of CAR-mediated tonic signaling on the antitumor response.
To further demonstrate the specificity of NPM1c-CAR NK cells in targeting NPM1c+ HLA-A2+ AML cells, OCI-AML2 cells were transduced with lentivirus to stably express NPM1c. Following exogenous NPM1c expression, HLA-A2+ OCI-AML2 cells were sensitized to NPM1c CAR-NK cell-mediated killing in an effector-to-target ratio-dependent manner (Fig. 3F, Left Panel). In contrast, stable overexpression of wild-type NPM1 in OCI-AML2 did not induce CAR-dependent killing by CIML NK cells (Fig. 3F, Right Panel). We also used OCI-AML2 cells loaded with different peptides to demonstrate the antigen specificity of NPM1c CAR-NK cells (Fig. 3G). OCI-AML2 cells were pulsed with either NPM1c peptide (AIQ) or an irrelevant NY-ESO-1 peptide (SLL), and consistently, NPM1c CAR-NK cells killed AIQ-pulsed OCI-AML2 cells in an effector-to-target ratio-dependent manner but not SLL-pulsed OCI-AML2 cells (Fig. 3G). These results further support the specificity of NPM1c CAR-NK cell recognition and killing of target cells with the NPM1c-HLA-A2 complex on the cell surface. Of note, HLA-A2 can bind to the inhibitory receptor LIR1 on NK cells. To test whether CAR transduction may influence LIR1 expression, CIML NK cells isolated from multiple PB donors were assessed. A modest yet statistically significant decrease of LIR1 expression was observed in CAR+ CIML NK cells compared to the CAR− control cells (29% vs. 24% mean values, SI Appendix, Fig. S3), suggesting that slightly reduced LIR1 levels may also contribute to the enhanced cytotoxicity of anti-NPM1c CAR CIML-NK cells. Together, these data show that, while retaining their innate propensity to target AML, engineered NPM1c-CAR CIML NK cells significantly enhance responses that are antigen specific and require antigen-dependent CAR activation.
In addition, NPM1c-CAR CIML NK cells with or without mb15 exhibited significantly enhanced cytotoxicity against patient-derived-xenograft (PDX) AML cells carrying the NPM1 mutation, compared to untransduced CIML NK cells (Fig. 3 H–K). As expected, CIML NK cells without CAR expression exhibited significant cytotoxicity against OCI-AML3 and PDX AML cells as the E:T ratio increased (Fig. 3 C, D, and H–K). Consistent with these findings, anti-CD19 CAR CIML NK cells using the same CAR backbone killed NPM1c+ HLA-A2+ PDX comparably to donor-matched untransduced control CIML NK cells (Fig. 3I). Thus, human PB-derived CIML NK cells exhibited innate activity against AML blasts, and CAR expression substantially enhanced antitumor effector functions including cytotoxicity against NPM1c+ AML.
NPM1c-CAR CIML NK Cells Reduce AML Burden in NSG Mice.
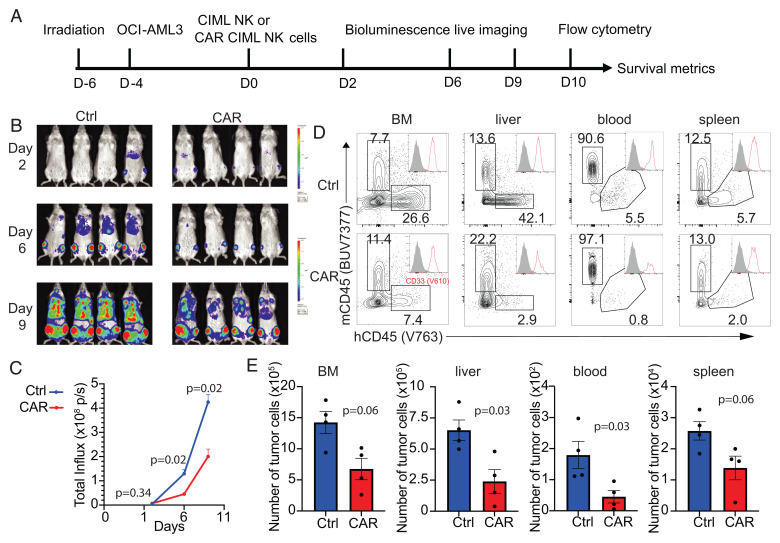
The successful generation of functional NPM1c-CAR CIML NK cells and their abundant expansion with sustained CAR expression (Figs. 1 and 2) allowed us to examine the impact of NPM1c-CAR expression on CIML NK cell functions in vivo using NOD-scid IL2Rgnull (NSG) mice engrafted with human AML xenografts (Fig. 4A). Briefly, NSG mice were irradiated on day −6, engrafted with luciferase-expressing OCI-AML3 leukemic cells on day −4, and adoptively transferred with NPM1c-CAR CIML NK cells (with 50% of the NK cells positive for CAR expression) or untransduced CIML NK cells from the same donor. NSG mice were sublethally irradiated in order to achieve stable and reproducible engraftment of human AML cells as previously described (27–29). The leukemia burden was monitored by bioluminescent imaging (BLI) on day 6, 10, and 13. Tumor burden was significantly decreased in mice transferred with NPM1c-CAR CIML NK cells compared to mice who received untransduced CIML NK cells (Fig. 4 B and C). To examine the antileukemia activity of CAR-NK cells, we collected PB, spleen, bone marrow, and liver 14 d post-NK cell transfer and analyzed them for the presence of hCD33+ hCD45+ mCD45− leukemic cells by flow cytometry. Consistent with BLI analysis, the numbers of hCD33+ leukemic cells were lower in all four tissues in mice treated with NPM1c-CAR CIML NK cells than those treated with untransduced CIML NK cells (Fig. 4 D and E). These data show that a NPM1c-CAR CIML NK cell product provides enhanced tumor control in vivo.
Fig. 4.
NPM1c-CAR CIML NK cells are effective in controlling AML in vivo in a xenograft model. (A) Scheme of the experimental setup. NSG mice were sublethally irradiated, and 2 d later, luciferase+ OCI-AML3 cells (5 × 105 cells) were injected IV. After 4 d, 1 × 106 untransduced or transduced CIML NK cells were injected IV into the tumor-bearing mice. BLI was performed on the indicated days to monitor tumor burden. Mice were euthanized at 10 d post-NK cell adoptive transfer for flow cytometry analysis. (B and C) Comparison of tumor burden in recipient mice given untransduced (Ctrl) and transduced (CAR) CIML NK cells by BLI (B) and quantification of luciferase activity (C). (D) Representative flow cytometry staining profiles of hCD45 versus mCD45 gating on live cells in the indicated tissues. Inserts show CD33 histogram of hCD45+ cells. (E) Summary data showing absolute numbers of leukemic cells in the indicated tissues of recipient mice at day 10 after adoptive transfer with untransduced CIML (blue) vs. NPM1c CAR CIML NK cells (red) (n = 4 mice per group). Error bars in C and E represented mean with SEM. Data were analyzed by two-way ANOVA with the Sidak posttest (C) or two-tailed unpaired Student’s t test (E). Data are representative of three (B–E) independent experiments.
Optimizing CAR Design by Incorporating Membrane-Bound IL-15 Enhances In Vivo Persistence and Antileukemia Activity of CAR CIML NK Cells.
NSG mice lack key homeostatic survival signals for human NK cells. Given that the expression of mb15 promoted the survival and proliferation of CAR CIML NK cells when cultured in media with limited cytokine support (Fig. 5A and SI Appendix, Fig. S4), we assessed the in vivo efficacy of NPM1c-CAR-mb15 as described in Fig. 4A except using a later endpoint in xenograft NSG mouse models. Compared to NPM1c-CAR CIML NK cells, treatment with NPM1c-CAR-mb15 CIML NK cells resulted in a substantial reduction in tumor burden in AML-bearing NSG mice as revealed by BLI (Fig. 5 B and C). Flow cytometry analyses of PB, spleen, bone marrow, and liver showed lower numbers of hCD33+ leukemic cells in the CAR-mb15 treated mice than in the CAR-treated mice (Fig. 5 D and E). Importantly, NSG mice that received CIML NK cells expressing NPM1c-CAR-mb15 displayed increased numbers of CAR-expressing NK cells in all four tissues compared to NPM1c-CAR recipients (Fig. 5 D and F). Thus, NPM1c-CAR-mb15 CIML NK cells are capable of persisting in the absence of exogenous cytokine support for homeostatic maintenance and can kill AML target cells in bone marrow and other key organs to substantially control disease progression. Together, these results show that the expression of mb15 confers improved in vivo persistence, expansion, and anti-AML activity of NPM1c-CAR CIML NK cells.
Fig. 5.
Incorporation of membrane-bound IL-15 (mb15) into the CAR design improves the efficacy of CAR CIML NK cells. (A) Representative flow cytometry plots showing cell viability (Upper Panel) and Ki-67 staining (Bottom Panel) of CAR- versus CAR-mb15-transduced CIML NK cells. CIML NK cells were cultured in cytokine-limited media for 5 d before flow cytometry assay. (B and C) Comparison of tumor burden in recipient mice given untransduced (Ctrl) or CAR- or CAR-mbIL15-transduced CIML NK cells by BLI (B) and quantification of luciferase activity (C) (n = 4 mice per group). (D–F) Comparison of AML burden and persistence of the transferred human NK cells in the indicated tissues of recipient mice treated with CIML NK cells expressing NPM1c-CAR or NPM1c-CAR-mb15 at day 19 post-NK cell adoptive transfer. (D) Representative flow cytometry staining profiles of hCD45 versus mCD45 gating on live cells (Left Panel) from liver, CD33 versus hCD45 gating on hCD45+ cells (Middle Panel), and CD56 versus anti-scFv gating on hCD45+ CD33− cells (Right Panel). (E) Quantifications of OCI-AML3 leukemic cells in the indicated tissues. (F) Quantifications of CAR-expressing human NK cell percentages in the indicated tissues. n = 4 mice per group. Error bars in C, E, and F represented mean with SEM from biological replicates. Data were analyzed by two-way ANOVA with the Sidak posttest (C) or one-way ANOVA with the Tukey posttest (E and F). Data are representative of two (A) or three (B–F) independent experiments.
We next compared CIML NK and T cells expressing the same CAR to control leukemia growth in vivo. Equal numbers of transduced CIML NK cells or T cells derived from the same PB donor and expressing the same NPM1c CAR and IL-15/IL-15Rα fusion (mb15/Ra) (18) (SI Appendix, Fig. S5 A and B) were adoptively transferred into NSG mice bearing OCI-AML3 leukemic cells. Comparable antitumor responses were observed in the two cohorts (SI Appendix, Fig. S5 C and D). Furthermore, both the adoptively transferred CAR CIML NK and T cells were detected at similar levels in the PB, spleen, bone marrow, and liver of the AML-bearing mice 19 d after adoptive transfer (SI Appendix, Fig. S5E). Thus, in this experimental setting, anti-NPM1c CAR CIML NK and CAR T cells are equally effective in controlling NPM1-mutated AML growth in vivo.
scRNA-seq Analyses Reveal CAR-Dependent Crosstalk between CAR-mb15 CIML NK Cells and AML Target Cells.
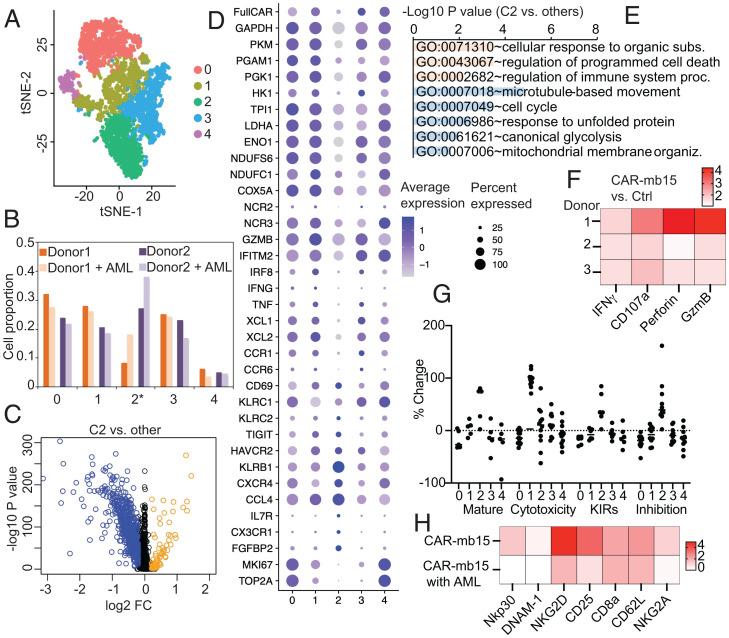
We hypothesized that the antitumor potency of CAR engagement with AML target cells might arise from specific CAR CIML NK subsets characterized by unique transcriptomic signatures. To address this question, CIML NK cells generated from two PB donors were transduced with NPM1c-CAR-mb15 lentivirus, and purified viable NK cells were cocultured with or without OCI-AML3 targets for 24 h, followed by scRNA-seq. Five clusters were identified by tSNE in CAR-expressing CIML NK cells (Fig. 6A). Interestingly, cluster 2 was substantially and consistently expanded in response to AML targets (Fig. 6B). DEG analysis revealed a distinct transcriptomic profile of cluster 2 (Fig. 6C, with selected genes shown in Fig. 6D). GO enrichment analyses showed significant up-regulation in “cellular responses to organic substances” and in “regulation of immune system process” (Fig. 6E), confirming that the strong antitumor responses in CAR CIML NK cells were particularly enriched in this cluster. In contrast, no significant clustering changes were found in AML-cocultured CAR-negative control CIML NK cells from the same culture (SI Appendix, Fig. S6 A and B), indicating that the specific responses depend on CAR engagement by tumor cells. Consistently, mass cytometric analyses revealed that CAR-mb15 CIML NK cells derived from multiple PB donors expressed significantly higher levels of IFNγ, CD107a, granzyme B, and perforin than untransduced CIML NK cells in response to OCI-AML3 target stimulation in vitro (Fig. 6F).
Fig. 6.
scRNA-seq analyses reveal crosstalk between CAR-mb15 CIML NK cells and AML target cells. (A) tSNE clustering analysis of CAR+ NK cells cocultured with or without AML target cells. CIML NK cells generated from two PB donors were transduced with NPM1c-CAR-mb15 lentivirus, and purified viable NK cells were cocultured with OCI-AML3 cells for 24 h followed by scRNA-seq. (B) Cell proportion of identified clusters in CAR+ NK cells cocultured with or without AML target cells; cluster 2 was consistently increased in CAR+ NK cells cocultured with AML targets. (C) Volcano plot of gene expression between cluster 2 and other clusters. Blue dots: down-regulated; and orange dots: up-regulated DEGs. (D) Dot plots of the expression of selected genes. (E) GO functional enrichment analysis of DEGs in C. (F) Fold-change of IFNγ, CD107a, perforin, and granzyme B in NPM1c-CAR-mb15 CIML NK cells over untransduced (Ctrl) CIML NK cells. CIML NK cells from three different donors were cocultured with OCI-AML3 target cells for 5 h followed by mass cytometry analysis. (G) The scores for maturation, cytotoxicity, KIR, and inhibition in each cluster. (H) Comparison of selected marker expression by NPM1c-CAR-mb15 CIML NK cells and the untransduced CIML NK cells from the same donor. CAR+ and CAR− CIML NK cells were cocultured with AML target cells for 5 h and analyzed by mass cytometry. The heatmap shows median values in protein expression levels of the indicated activation markers and receptors. n = 2 (A–E and G) and 3 (F) PB donors. Data are representative of two (H) independent experiments or pooled from two (F) independent experiments.
Interestingly, the GO analysis of DEGs (Fig. 6E) also highlighted an up-regulation of genes encoding programmed cell death pathways as well as dramatic down-regulation in the cell cycle, cellular metabolism, and ER stress responses. Moreover, cluster 2 exhibited high levels of KIRs and inhibitory receptors as well as terminal maturation signatures (Fig. 6G). Consistently, mass cytometric analyses revealed lower expression levels of activating receptors and markers, such as NKp30 and NKG2D, by CAR-mb15 CIML NK cells following coculture with OCI-AML3 target cells in vitro (Fig. 6H and SI Appendix, Fig. S6C). These results suggest that CAR engagement and killing of AML target cells might eventually lead to dampening of CAR-NK cell effector function.
Discussion
AML remains a major therapeutic challenge in part because of the lack of tumor-specific cell surface targets. In the current study, we aimed to take advantage of the enhanced antileukemia activity displayed by CIML NK cells by arming them with a TCR-like CAR that selectively targets AML blasts, thus avoiding toxicity against normal hematopoietic stem cells. Our CAR-CIML NK cells showed potent in vitro and in vivo activity against NPM1-mutated AML cells by specifically targeting a unique neoepitope generated from cytosolic mutant NPM1c presented by HLA-A2 on AML target cells. We selected PB-derived CIML NK cells for CAR engineering as these cells have enhanced antitumor responses and prolonged in vivo persistence in humans. We achieved high transduction efficiency of CIML NK cells by using BaEV-pseudotyped lentivirus, taking advantage of the elevated expression of ASCT2 on CIML NK cells, the amino acid transporter utilized by this lentivirus. We also integrated into our CAR membrane-bound IL-15, a cytokine critical for the survival and expansion of NK cells. Our GO enrichment analysis identified up-regulation of several key pathways consistent with a highly proliferative and metabolically active phenotype of CAR CIML NK cells, especially when mb15 is included. Furthermore, we identify several unique transcriptomic profiles induced upon CAR engagement with AML target cells. This approach may further identify key proteins that activate effector–tumor cell interactions particularly in the context of CAR-expressing CIML NK cells. Overall, our results demonstrate that innovative CAR-CIML NK cells can be developed as an efficient cellular immunotherapy for treating NPM1c+ HLA-A2+ AML with reduced on-target/off-tumor toxicity and tumor resistance.
A recent study using allogeneic cord blood–derived NK cells as a platform for expressing a CD19-CAR showed promising clinical responses in patients with relapsed B cell malignancies (6). Notably, clinical responses were achieved with minimal toxicity, and no cases of severe CRS or neurotoxicity were reported. These clinical observations as well as preclinical studies with engineered induced pluripotent stem cell–derived NK cells (30) suggest that NK cells represent an attractive cellular platform for genetically modified adoptive cell therapy. However, CAR-NK cell–based therapies are still at an early stage with existing major barriers, including lack of an efficient and safe gene transfer method in human primary NK cells (31). Canonical vesicular stomatitis virus glycoprotein (VSVG)-pseudotyped lentiviruses often lead to <5% CAR transduction even when used at an MOI as high as 100 in activated NK cells. This can be improved to 5 to 25% (14) by performing a second sequential spinfection on previously expanded and/or activated NK cells with trade-offs on cell viability that double the need for lentivirus manufacturing. We found a very high expression of ASCT2 in primary PB-derived CIML NK cells, allowing the use of BaEV-pseudotyped lentiviruses to achieve efficient CAR transduction and expression. CIML NK cells provide an attractive platform for genetically engineering NK cells, including CARs, based on their favorable safety profile, increased proliferation, prolonged persistence, and enhanced antileukemia function in preclinical animal models and in patients treated with unmodified CIML NK cells (9, 26). Furthermore, NK cells’ intrinsic propensity to target myeloid blasts makes them particularly attractive for AML where CAR T cells have shown only modest to no benefit primarily due to a lack of suitable surface target antigens. We are optimistic that high-efficiency transduction of CIML NK cells with BaEV lentiviruses will significantly accelerate efforts aimed at using NK cells as a platform for developing the next generation of NK cell immunotherapeutic products.
Using single-cell resolution transcriptomic analyses, our data reveal that BaEV lentiviruses are capable of transducing heterogeneous NK cell subsets with a significant preference for highly proliferative, highly functional and less mature NK cell populations. This differs from the unbiased transduction pattern seen in recent studies using VSVG lentivirus– and BaEV lentivirus–mediated transduction of cNK cells (13, 14). Our findings are consistent with the recent report by Bari et al. (24) that the BaEV lentivirus preferentially transduced a highly proliferative NK cell subset. We found that IL-12, which along with IL-18 is required for the CIML differentiation of cNK cells, was also essential for the optimal induction of ASCT2 expression (SI Appendix, Fig. S1). Furthermore, up-regulation of ASCT2 expression required only a brief overnight (12 to 16 h) stimulation of cNK cells in order to induce CIML differentiation. This is in contrast to Bari et al. (24) where a prolonged cytokine stimulation under non-CIML-inducing conditions (no IL-12) was used.
A recent study from our laboratory (11) indicated that engineered CAR-T cells bearing an isolated scFv exhibit potent and specific cytotoxicity both in vitro and in vivo against NPM1c+ HLA-A2+ AML. This indicates that targeting the oncogenic NPM1c neoepitope is an efficient approach for developing cancer-specific immunotherapies to treat AML with reduced on-target/off-tumor toxicity and tumor resistance. In line with this, Greiner et al. (32) reported that NPM1c+ AML patients with specific CD8 T cell responses to the neoepitope had a dramatically better overall survival than NPM1c+ AML patients without CD8 T cell responses to the same neoepitope. In the current study, we proposed to utilize TCR-like CAR CIML NK cells that recognize the NPM1c neoepitope in complex with human HLA-A*0201 as a targeted immunotherapy for NPM1c+ AML. Compared to CAR T cells, CAR NK cells armed with this TCR-like CAR may offer some significant advantages, including the following: 1) better safety, 2) multiple tumor-recognition mechanisms for activating cytotoxic activity (e.g., dual targeting of myeloid blasts by the CAR protein, NK activation through MICA/MICAB engagement, and loss of or down-regulation of HLA class 1 triggering NK cell activation through the “missing self” mechanism), and 3) high feasibility for allogeneic “off-the-shelf” product manufacturing using some of the recently described expansion methods (33, 34). In particular, the CAR-dependent and CAR-independent killing of target tumor cells by NK cells may help to minimize the emergence of tumor resistance.
IL-15 is critical for NK cell survival and homeostasis, and several recent studies have included exogenous IL-15 to support the expansion and activation of adoptively transferred NK cells in clinical trials (19) (and NCT01385423, NCT01875601, and NCT04290546, for example). We added membrane-bound IL-15 as part of our CAR construct design to provide a survival and expansion advantage for these transduced NK cells. Our results demonstrate that the combination of an NPM1c-CAR and membrane-bound IL-15 enhances the antitumor efficacy of NK cells targeting AML in xenograft models. These data indicate that CAR and cytokine signaling dual-armed CIML NK cells exhibited optimal specificity and sustainability against tumor targets. The incorporation of mb15 may also help promote NK cell–mediated tumor surveillance in vivo by modulating the immunosuppressive tumor microenvironment, a key process in controlling metastatic progression and disease relapse in AML and solid tumors, such as breast cancer (35).
IL-15 is also crucial for NK cell development, as well as survival and proliferation. Our scRNA-seq data showed that NK cells expressing CAR and mb15 up-regulate the expression of genes associated with pathways of proliferation and elevated metabolic activities, suggesting mb15 as an essential factor for persistence of CAR+ NK in vivo. Interestingly, we identified one NK subpopulation with reduced metabolic activity that could become functionally suppressive by expressing several NK inhibitory receptors, such as KLRB1, TIGIT, and TIM3. Hence, the tumor–NK cell interaction may suppress NK cell function through their respective ligands, MHC-1, or secreted factors by AML. Combinatory treatment of CAR-NK with other therapeutic compounds, such as anti-KLRB1 (36) to activate NK cells, or anti-MICA to inhibit tumor suppressive function (37), could further augment the efficacy of CAR-NK cell therapy.
In summary, we demonstrate that combining ASCT2 up-regulation with an anti-NPM1c TCR-like CAR allows efficient arming of CIML NK cells and substantially improves the antitumor response against an otherwise intracellular mutant protein. Our findings also provide translational and mechanistic approaches for utilizing CIML NK cells as an attractive platform for CAR engineering.
MATERIALS AND METHODS
Key information is summarized below, and a comprehensive description of methods can be found in SI Appendix.
Construction of CAR Lentiviral Vectors.
The sequence of CAR, consisting of the completely human anti-NPM1c scFv (clone: YG1) (11), the CD8a signal peptide sequence, extracellular hinge domain and TM domain, the 4-1BB costimulatory domain, and the CD3ζ activation domain, was custom synthesized by Integrated DNA Technologies. The second fragment was synthesized by the same way consisting of self-cleavage P2A followed by either EGFP (P2A-GFP) or membrane-bound IL-15 (P2A-mb15). The pHIV vector was doubly digested with the enzymes XbaI and ClaI. After gel purification of the vector backbone, the pHIV backbone, CAR fragment, and the corresponding P2A-linked fragment were assembled basing on their overlap region at 5′ and 3′ termini using HiFi DNA Assembly Master Mix. The resulting plasmids were sequenced and the plasmid with the correct sequence was named pHIV-CAR-GFP and pHIV-CAR-mb15, respectively.
Cytotoxicity of CIML CAR-NK Cells In Vitro.
To assess the ability of CAR-NK cells to kill target cells, CAR-NK cells and untransduced control cells from the same donors were incubated with luciferase-expressing OCI-AML3 or OCI-AML2 cells at indicated effector: target ratios. At 20 h after coculture, luciferase assays (Promega) were performed according to the manufacturer’s protocol. The reduction in target cell numbers due to NK cell–mediated killing was quantified by the endpoint luciferase signals normalized to the corresponding control wells where the culture only contained target cells. To quantify the early apoptosis of target cells by flow cytometry, we prelabeled target cells with CellTrace dye before setting up similar cocultures with NK cells. Cells were harvested after 4 h, and routine viability and surface marker staining were performed as described above. Cell samples were then incubated with anti-Annexin V in Annexin V Binding Buffer (eBioscience) before analysis by a CytoFLEX flow cytometer.
In Vivo Testing of CIML CAR-NK Cells in AML Xenograft Models.
All experiments with mice were approved by the Animal Care and Use Committee at Dana–Farber Cancer Institute. Both female and male mice were evaluated, and each experimental cohort consisted of age- and sex-matched mice. Briefly, 8- to 12-wk-old NSG mice were purchased from the Jackson Laboratories and housed in the specific-pathogen-free vivarium at Dana–Farber Cancer Institute. NSG mice were irradiated with sublethal dose (250 cGy), and 2 d later, luciferase-expressing OCI-AML3 cells (5 × 105) in 200 μL phosphate-buffered saline were injected intravenously (IV). After 4 d, 1 × 106 untransduced or transduced CIML NK cells (containing about 500,000 CAR-expressing cells) were injected IV into the tumor-bearing mice. Mice were randomly allocated to different treatment groups. BLI was performed in a blinded manner at the indicated time points using a Xenogen IVIS-200 Spectrum camera.
To analyze tumor cells and NK cells by flow cytometry, mice were euthanized 2 to 3 wk post-NK cell adoptive transfer, and blood, spleen, bone marrow, and liver were harvested. Single-cell suspensions were prepared and lysed of red blood cells. Cells were washed with fluorescence-activated cell sorting buffer and counted with trypan blue staining. For flow cytometry analysis, about 1 million cells were aliquoted and mixed with 20 μL human serum to block the Fc receptor for 5 min. The cells were stained with Live/Dead Fixable Dead Cell Stain (Thermo Fisher) and anti-Human IgG(H+L) on ice for 1 h, followed by the routine surface marker staining.
Single-Cell RNA-Sequencing Analyses.
CIML NK cells were transduced with CAR-mb15 at an MOI of 5 and then cultured for 7 d. One day before the analyses, CIML NK cells (mixture of CAR+ and CAR−) were flow sorted to remove dead cells/debris. CIML NK cells were then cultured alone or cocultured with OCI-AML3 target cells in a 1:2 ratio for 24 h before harvesting for library preparation and sequencing. Cells were loaded onto 10X chromium machine (10X Genomics) and run through library preparation using the Chromium Next GEM Single Cell 3′ kits (10X Genomics). The single-cell complementary DNA libraries were sequenced by the NovaSeq S4 flowcell (Illumina). Raw sequences were demultiplexed, aligned, and filtered, and barcode counting and unique molecular identifier (UMI) counting were performed with Cell Ranger software v3.1 (10X Genomics) to digitalize the expression of each gene for each cell. The analysis was performed using the Seurat 3.0 package (38). We first processed each individual data set separately. Subsequently, samples were combined based on the identified anchors for the following integrated analysis. We ran a principal component analysis and used the first 15 principal components to perform tSNE clustering. To compare CAR+ and CAR− CIML NK cells, CAR+ cells were defined with an expression of the full CAR sequence of ≥0.1 while CAR− cells were defined with an expression of ≤0. To demonstrate the difference of subpopulations, CAR+ and CAR− cells were clustered separately, and clusters were matched based on the correlation similarities of global gene expression. To investigate the CAR+ NK response to tumor cells, CAR+ cells were isolated from the cocultured samples based on the expression of the full CAR sequence of ≥0. Differential expression analyses were performed to identify the genes significantly up-regulated in each cluster compared with all other cells by setting the log2 fold-change to ≥0.2 and P value to <0.05. For gene sets representing specific cellular functions or pathways, we performed functional enrichment analysis with the biological process of GO by the online tool DAVID (39).
Mass Cytometry and Data Analyses.
CAR CIML NK cells generated from three healthy peripheral blood mononuclear cell donors were assessed by mass cytometry using a custom NK cell function panel consisting of 38 markers (SI Appendix, Table S2). A comprehensive description of sample preparation, data acquisition, and analyses can be found in SI Appendix, Materials and Methods.
Statistical Analysis.
For graphs, data are shown as mean ± SEM or SD as indicated, and unless otherwise indicated, statistical differences were evaluated using a two-tailed unpaired Student’s t test, assuming equal sample variance. P < 0.05 was considered significant. Graphs were produced and statistical analyses were performed using GraphPad Prism 7.0d for Mac OS X (GraphPad Software, www.graphpad.com) or R. P values were two sided, and a P value <0.05 was considered statistically significant after any adjustment; *=P < 0.05, **=P < 0.01, ***=P < 0.001, ****=P < 0.0001. Sample size was not specifically predetermined, but the number of mice used was consistent with prior experience with similar experiments.
Supplementary Material
Acknowledgments
R.R. was supported by Claudia Adams Barr Foundation, Doug Bell Fund, and Pasquarello Family Funds. J.C. was supported by Claudia Adams Barr Foundation, NIH Grant CA197605, and the Koch Institute Support (core) Grant P30-CA14051. L.H.G. was supported by a Dana–Farber Cancer Institute Internal Funding. H.D. was supported by Claudia Adams Barr Foundation, Elsa U. Pardee Foundation, an American Society of Hematology Scholar Award, a Helen Gurley Brown Fellowship by the Pussycat Foundation, and National Cancer Institute (NCI) DF/HCC SPORE in Myeloid Malignancies Career Enhancement Program. J.C. and J.D.H. were also supported by a Frontier award from Koch Institute for Integrative Cancer Research at Massachusetts Institute of Technology (MIT).
Footnotes
Reviewers: L.L., University of California San Francisco Medical Center at Parnassus; and J.S., Stanford University.
Competing interest statement: R.R. has a sponsored research agreement with Crispr Therapeutics, Skyline Therapeutics and serves on the scientific advisory board of Glycostem Therapeutics. L.H.G. is a former Director of Bristol-Myers Squibb and the Waters Corporation and currently serves on the Board of Directors of GlaxoSmithKline Pharmaceuticals and Analog Devices, Inc. She also serves on the scientific advisory boards of Repare Therapeutics, Abpro Therapeutics and Kaleido Therapeutics. Other authors declare that they have no competing interests.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122379119/-/DCSupplemental.
Data Availability
scRNA-seq data have been deposited in Gene Expression Omnibus (GSE186115). All study data are included in the article and/or supporting information.
References
- 1.Falini B., Tiacci E., Martelli M. P., Ascani S., Pileri S. A., New classification of acute myeloid leukemia and precursor-related neoplasms: Changes and unsolved issues. Discov. Med. 10, 281–292 (2010). [PubMed] [Google Scholar]
- 2.Venstrom J. M., et al. , HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N. Engl. J. Med. 367, 805–816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooley S., et al. , Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116, 2411–2419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bald T., Krummel M. F., Smyth M. J., Barry K. C., The NK cell-cancer cycle: Advances and new challenges in NK cell-based immunotherapies. Nat. Immunol. 21, 835–847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivori S., et al. , NK cells and ILCs in tumor immunotherapy. Mol. Aspects Med. 80, 100870 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Sun J. C., Lanier L. L., Is there natural killer cell memory and can it be harnessed by vaccination? NK cell memory and immunization strategies against infectious diseases and cancer. Cold Spring Harb. Perspect. Biol. 10, a029538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerwenka A., Lanier L. L., Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 16, 112–123 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Ni J., Miller M., Stojanovic A., Garbi N., Cerwenka A., Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J. Exp. Med. 209, 2351–2365 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romee R., et al. , Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci. Transl. Med. 8, 357ra123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berrien-Elliott M. M., et al. , Multidimensional analyses of donor memory-like NK cells reveal new associations with response after adoptive immunotherapy for leukemia. Cancer Discov. 10, 1854–1871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie G., et al. , CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat. Biomed. Eng. 5, 399–413 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanier L. L., Yu G., Phillips J. H., Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature 342, 803–805 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Colamartino A. B. L., et al. , Efficient and robust NK-cell transduction with baboon envelope pseudotyped lentivector. Front. Immunol. 10, 2873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gang M., et al. , CAR-modified memory-like NK cells exhibit potent responses to NK-resistant lymphomas. Blood 136, 2308–2318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marçais A., et al. , The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15, 749–757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romee R., et al. , First-in-human phase 1 clinical study of the IL-15 superagonist complex ALT-803 to treat relapse after transplantation. Blood 131, 2515–2527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu E., et al. , Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurton L. V., et al. , Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc. Natl. Acad. Sci. U.S.A. 113, E7788–E7797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooley S., et al. , First-in-human trial of rhIL-15 and haploidentical natural killer cell therapy for advanced acute myeloid leukemia. Blood Adv. 3, 1970–1980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldmann T. A., Dubois S., Miljkovic M. D., Conlon K. C., IL-15 in the combination immunotherapy of cancer. Front. Immunol. 11, 868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Y., et al. , IL-15 activates mTOR and primes stress-activated gene expression leading to prolonged antitumor capacity of NK cells. Blood 128, 1475–1489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H., et al. , The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 20, 865–878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni J., et al. , Single-cell RNA sequencing of tumor-infiltrating NK cells reveals that inhibition of transcription factor HIF-1α unleashes NK cell activity. Immunity 52, 1075–1087.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Bari R., et al. , A distinct subset of highly proliferative and lentiviral vector (LV)-transducible NK cells define a readily engineered subset for adoptive cellular therapy. Front. Immunol. 10, 2001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romee R., Leong J. W., Fehniger T. A., Utilizing cytokines to function-enable human NK cells for the immunotherapy of cancer. Scientifica (Cairo) 2014, 205796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romee R., et al. , Cytokine activation induces human memory-like NK cells. Blood 120, 4751–4760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malaise M., et al. , Stable and reproducible engraftment of primary adult and pediatric acute myeloid leukemia in NSG mice. Leukemia 25, 1635–1639 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Paczulla A. M., et al. , Long-term observation reveals high-frequency engraftment of human acute myeloid leukemia in immunodeficient mice. Haematologica 102, 854–864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho B. S., et al. , Antileukemia activity of the novel peptidic CXCR4 antagonist LY2510924 as monotherapy and in combination with chemotherapy. Blood 126, 222–232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Hermanson D. L., Moriarity B. S., Kaufman D. S., Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell 23, 181–192.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie G., et al. , CAR-NK cells: A promising cellular immunotherapy for cancer. EBioMedicine 59, 102975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greiner J., et al. , Immune responses against the mutated region of cytoplasmatic NPM1 might contribute to the favorable clinical outcome of AML patients with NPM1 mutations (NPM1mut). Blood 122, 1087–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Thangaraj J. L., et al. , Expansion of cytotoxic natural killer cells in multiple myeloma patients using K562 cells expressing OX40 ligand and membrane-bound IL-18 and IL-21. Cancer Immunol. Immunother. 71, 613–625 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang R. S., Lai M. C., Shih H. A., Lin S., A robust platform for expansion and genome editing of primary human natural killer cells. J. Exp. Med. 218, e20201529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correia A. L., et al. , Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature 594, 566–571 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Mathewson N. D., et al. , Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184, 1281–1298.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrari de Andrade L., et al. , Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science 359, 1537–1542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang D. W., et al. , The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data have been deposited in Gene Expression Omnibus (GSE186115). All study data are included in the article and/or supporting information.