Significance
A substantial component of the global nitrogen cycle is the production of biologically inaccessible dinitrogen attributed to anaerobic denitrification by prokaryotes. Recent evidence identified a eukaryote, foraminifera, as new key players in this “loss” of bioavailable nitrogen. The evolution of denitrification in eukaryotes is a rare event, and the genetic mechanisms of the denitrification pathway in foraminifera are just starting to be elucidated. We present large-scale sequencing analyses of 10 denitrifying foraminiferal species, which reveals the high conservation of the foraminiferal denitrification pathway. We further find evidence for a complementation of denitrification by the foraminiferal microbiome. Together, these findings provide insights into the early evolution of a previously overlooked component in the marine nitrogen cycle.
Keywords: foraminifera, denitrification, genomics, evolution, microbiome
Abstract
Benthic foraminifera are unicellular eukaryotes that inhabit sediments of aquatic environments. Several foraminifera of the order Rotaliida are known to store and use nitrate for denitrification, a unique energy metabolism among eukaryotes. The rotaliid Globobulimina spp. has been shown to encode an incomplete denitrification pathway of bacterial origin. However, the prevalence of denitrification genes in foraminifera remains unknown, and the missing denitrification pathway components are elusive. Analyzing transcriptomes and metagenomes of 10 foraminiferal species from the Peruvian oxygen minimum zone, we show that denitrification genes are highly conserved in foraminifera. We infer the last common ancestor of denitrifying foraminifera, which enables us to predict the ability to denitrify for additional foraminiferal species. Additionally, an examination of the foraminiferal microbiota reveals evidence for a stable interaction with Desulfobacteraceae, which harbor genes that complement the foraminiferal denitrification pathway. Our results provide evidence that foraminiferal denitrification is complemented by the foraminifera-associated microbiome. The interaction of foraminifera with their resident bacteria is at the basis of foraminiferal adaptation to anaerobic environments that manifested in ecological success in oxygen depleted habitats.
Nitrogen is an essential element for life on Earth as it forms the basis for the synthesis of nucleotides and amino acids. Nonetheless, while Earth’s atmosphere is rich in nitrogen gas (up to 78%) (1), this gas is usually inert but can be made accessible for biological processes by nitrogen fixation (2). Microbial organisms are important players in the global nitrogen cycle as they facilitate the assimilation of nitrogen into bioavailable nitrogen derivatives and the dissimilation of nitrogen derivatives into dinitrogen (N2) (2). A key dissimilatory pathway is denitrification, where nitrate (NO3−) is either partially or completely degraded and the final product, N2, is released to the atmosphere (i.e., nitrogen loss) (2). Marine organisms are considered major contributors to nitrogen loss from the environment, with benthic organisms being responsible for about two-thirds of the loss of reactive nitrogen in the ocean (3, 4). Oxygen minimum zones (OMZs) are especially worth mentioning here as they are estimated to be responsible for 20–40% of bioavailable nitrogen removal in the ocean (3, 5). The ability to perform denitrification is abundantly found in eubacteria (6), whereas it is rare among the eukaryotes. Partial or complete denitrification has been reported for only two species of fungi (7) and several foraminifera of the order Rotaliida (8–11). Denitrifying foraminifera are unicellular eukaryotes commonly found in marine sediments. Studies of foraminifera residing in the Peruvian OMZ showed that they are found in high densities of up to 600 individuals/cm2, where they are estimated to contribute 20–50% to the total benthic nitrate loss in the OMZ (10, 12).
Rotaliid foraminifera are divided into three clades based on their phylogeny (13). Only species in clades Ι and ΙΙΙ have been demonstrated to denitrify, while rotaliids classified in clade ΙΙ have been shown to lack an intracellular nitrate storage or measurable denitrification activity (9). Clade ΙΙ rotaliids typically populate environments with poor nitrate supply, such as intertidal to near-shore habitats from the tropics up to boreal bioprovinces (14), and hence it is conceivable that they lack the ability to denitrify. One example is Ammonia tepida (clade ΙΙ), which can survive episodic oxygen depletion events via dormancy (15). A study of clade ΙΙΙ species sampled from a hypoxic environment (Gulmarfjord, Sweden), Globobulimina spp., showed that that their genome encodes several genes along the denitrification pathway that are of ancient bacterial origin (11). These include the copper-containing nitrite reductase (NirK) and nitric oxide reductase (Nor) but not nitrate reductase (NapA/NarG) and nitrous oxide reductase (NosZ). In addition, the Globobulimina genome contains a diverse gene family encoding for nitrate transporters (Nrt) (11), a finding that is consistent with the accumulation of an intracellular NO3− storage in denitrifying rotaliids (8–10, 16). More recent studies have reported the presence of those genes in at least two additional species of foraminifera and provide alternative suggestions on the nitrate reductase homolog missing from the earlier model of denitrification in foraminifera (17, 18). The rotaliids’ ability to respire both oxygen (O2) and NO3− marks them as facultative anaerobes that are able to thrive in both aerobic and anaerobic conditions. Furthermore, while oxygen is generally considered to be preferred over NO3− as electron acceptor, rotaliids from the Peruvian OMZ were reported to prefer NO3− over O2 (10). These findings suggest that genes of the denitrification pathway are widespread in rotaliids; however, their distribution remains largely unknown.
Foraminifera, like other eukaryotes (19), are populated by bacterial organisms that reside outside and inside their cells (i.e., tests) (20–24). Microscopic observations showed that the bacteria are localized in food vacuoles or in the cytoplasm of the cell, which led to the suggestion that interactions of foraminifera with their microbiota may vary from prey-predator interactions or parasitism to metabolic symbiosis (22). Further studies of the microbiota in clade ΙΙ rotaliids identified sulfate-reducing and sulfur-oxidizing bacteria that were suggested to participate in sulfur cycling, thus providing carbon and other nutrients to the host (20, 21, 25). However, previous studies of foraminifera-associated bacteria used microscopic observations or sequencing of marker genes (e.g., 16S ribosomal RNA [rRNA]) that are limited in their resolution (20–25). It has been suggested that the microbiota of denitrifying foraminifera include denitrifying bacteria that use the foraminiferal NO3− storage, similarly to observations in other bacteria-protist associations (e.g., gromiids (26), allogromiids (27)). Indeed, metagenomics of the Globobulimina microbiota revealed the presence of a taxonomically diverse species community where several members encode homologs of NapA and NosZ (11). This finding gave rise to the hypothesis that the partial foraminiferal denitrification pathway may be complemented by microbiota functions.
Here we investigate the evolutionary history of genes along the denitrification pathway in foraminifera and examine the functional repertoire of the foraminiferal microbiome. For that purpose, we studied populations of 10 rotaliid species known to denitrify (10) (except Globobulimina pacifica, for which no information is available). Our study supplies insights into the evolution of rotaliids and their microbiota and lays the basis for further research of foraminiferal genome evolution.
Results
Transcriptomes of 10 Peruvian rotaliids.
For the purpose of our study, we sampled benthic foraminifera in the Peruvian OMZ. Individual foraminifers from among 10 focal species were manually identified and picked with a stereomicroscope (see sampled species in Fig. 1A). Eukaryotic transcriptomes and whole metagenomes were sequenced from the sampled foraminifera with two biological replicates per species (ca. 160 individuals per sample). Our computational analysis includes five additional publicly available transcriptomes of foraminifera (Dataset S1) and the genome data of one monothalamid species (Reticulomyxa filosa). The assessment of transcriptome completeness showed that ∼90% of the eukaryotic marker proteins are present in the data of the 10 newly sequenced species. Furthermore, we examined the purity of the focal species in each transcriptome by testing for redundancy of eukaryotic marker proteins that are expected as single-copy genes. As single-copy genes are expected to exist only once in a given species, their redundancy might indicate additional eukaryotic species present in our samples. The highest proportions of such bystander species were found in the Bolivina spissa (78%) and Bolivina costata (50%) transcriptomes (Dataset S1).
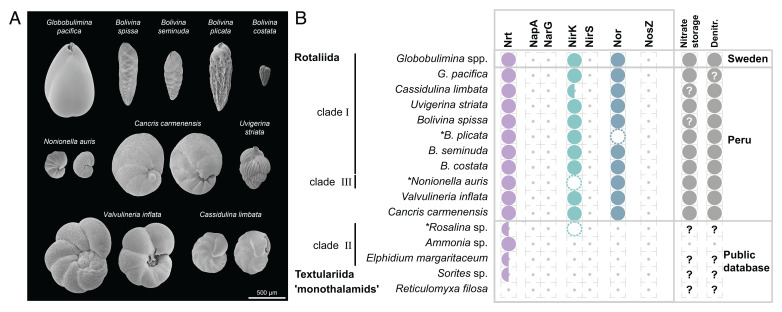
Fig. 1.
Morphological characteristics of the sampled rotaliids and denitrification gene repertoire. (A) Scanning electron micrographs of the sampled species. Species with clearly distinct lateral views are shown from two sides. For more detailed views see SI Appendix, Fig. S1. (B) Presence/absence of homologs of denitrification proteins identified for different foraminifera. Half circles indicate species where only a single, of multiple subtypes, was found. Dashed circles illustrate homologs discarded due to low coverage, and asterisks highlight corresponding species. Evidence for NO3− storage or denitrification activity is illustrated by closed circles in the last column. Question marks denote missing information. Foraminifera sampled in this study are highlighted by the location “Peru.” Protein symbols: Nrt, nitrate/nitrite transporter; NapA, periplasmic nitrate reductase; NarG, membrane-bound nitrate reductase; NirK, copper-containing nitrite reductase; NirS, cd1-containing nitrite reductase; Nor, nitric oxide reductase; NosZ, nitrous oxide reductase. Note that the identification of homologous genes is based not only on sequence similarity but also on transcript abundance in order to exclude bystander species in the data. The additional data filtration stage affected our findings for Rosalina sp., B. plicata, and N. auris (Datasets S2 and S3), where the presence of at least one the crucial homologs (i.e., NirK or Nor) remained in the B. plicata and N. auris metatranscriptomes.
The Peruvian rotaliids harbor genes of the denitrification pathway.
To explore the denitrification mechanisms in the Peruvian foraminifera, we searched for homologs to denitrification pathway genes. Our results reveal that all sampled species harbor homologous genes to Nrt, NirK, and Nor (Fig. 1B; note that genes are termed by their corresponding protein symbol). Our search for homologs in the publicly available data yielded a putative homolog of NirK in Rosalina sp. and Nrt homologs in all but R. filosa (Fig. 1B). The additional species we included here, especially Ammonia sp., Elphidium margaritaceum, and Sorites sp., are considered to reside mostly in oxygenated habitats, and hence they are not expected to be able to perform denitrification. These species lack homologs to the denitrification genes but encode nitrate transporters (Nrt, Fig. 1B). Among this group, only A. tepida was so far experimentally tested for denitrification ability and reported as a nondenitrifying species (9). The reconstructed phylogenies indicate that all the examined genes are indeed encoded in the foraminiferal genomes, as all of them group with homologs previously reported to originate from the nuclear genomes of Globobulimina spp. (11). Furthermore, the phylogenies of Nrt and NirK reveal two major subclades including most, but not all, of the species sampled (full and half circles in Fig. 1B and SI Appendix, Fig. S2 and Dataset S2). Previously we reported the presence of subclades in the Nrt and NirK phylogenies (11) but could not clarify whether those were explained by sequencing errors, gene duplication, or speciation events. The phylogenetic trees presented here show that each subclade is represented in most of the rotaliids sampled in the current study. Consequently, we conclude that ancient gene duplications in the Nrt and NirK gene families led to the evolution of two or more subclades that encode for different (paralogous) protein subtypes. Overall, our results demonstrate that the denitrification pathway is highly conserved in the Peruvian rotaliids.
Foraminiferal denitrification evolved in the Rotaliida ancestor.
The presence of NirK and Nor homologs the tested rotaliids suggests that those genes may have an ancient origin in this order. To study the origin of denitrification in foraminifera, we reconstructed the phylogenetic relations between species in our data by using 81 eukaryotic marker proteins that have homologs in all species analyzed here (SI Appendix, Fig. S3 and Dataset S4). Here, we applied a phylogenomic approach where the root position is inferred from all marker genes independent of a single species tree (28) (see Methods). Our results show that the best-supported root position (42% of the gene trees) was at the branch leading to R. filosa (SI Appendix, Fig. S3). This result is in agreement with previous studies that assumed monothalamids to be an outgroup in foraminiferal phylogenies (29). The second most frequent root position was found at the branch leading to the miliolid Sorites sp. (15% of single gene trees), which was followed closely by a root position on the branch leading to Rosalina sp. (12%). Taken together, the inferences of rooted topologies show that the Peruvian rotaliids, together with Globobulimina spp., form a monophyletic group. Thus, our results reveal a shared origin of denitrification in foraminifera and hence the evolution of denitrifying species within the order Rotaliida.
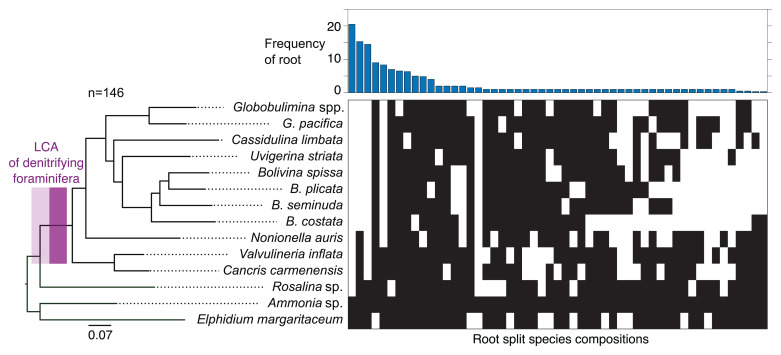
For the inference of the denitrifying rotaliids’ last common ancestor (LCA; i.e., the rotaliids’ root), we reconstructed phylogenetic trees from an extended set of 146 eukaryotic marker protein coding genes, considering only members of the Rotaliida (Fig. 2 and Dataset S4). An examination of the rooted tree topologies revealed that the branch leading to Ammonia sp. and E. margaritaceum was the most frequently supported root position (14% of single gene trees). This root inference is further supported by an alternative root position at a branch that splits Ammonia sp., E. margaritaceum, and Rosalina sp. from the remaining species (10%). Overall, the different alternative root positions support by 35% a Rotaliida LCA within clade ΙΙ foraminifera (i.e., among Ammonia sp., E. margaritaceum, and Rosalina sp.; Fig. 2). The absence of denitrification genes in clade ΙΙ members (Fig. 1) further supports the suggestion that the evolution of denitrification in rotaliids occurred by lateral gene transfer from a prokaryotic donor rather than being inherited from the rotaliid LCA (11).
Fig. 2.
Phylogenetic rooting of rotaliids. A maximum-likelihood phylogenetic reconstruction of foraminiferal species from 146 eukaryotic protein marker sequences is shown on the Left. The branch with the LCA of denitrifying foraminifera is highlighted in purple, where a lighter color includes lower bound for the origin of denitrification. Nonparametric bootstrap support is 1,000/1,000 at all the branches. A split representation of root splits determined via the MAD approach for the 146 single gene trees is shown on the Right. Each column represents a putative root branch reported as split of two groups (black and white boxes) indicating the species found on either side of the branch. The bar graph reports the single gene tree count supporting the corresponding root split.
Ancient origin of denitrification in foraminifera.
To further study the denitrifying foraminifera LCA, we examined the phylogenetic position of the Peruvian species in the context of a foraminiferal species tree. For that purpose, we extracted sequences of the 18S rRNA subunit from our transcriptome data and used it in combination with publicly available 18S rRNA sequences to reconstruct a foraminiferal species phylogeny (Fig. 3A and Dataset S5). The resulting rooted topology was mostly in agreement with the rotaliids’ phylogeny, where the focal species are found next to their closest relatives. Most species grouped well with their previously defined clades with the exception of Nonionella and Globobulimina representatives. Furthermore, several taxa were not monophyletic, as has been previously observed in foraminiferal 18S rRNA phylogenies (29, 30).
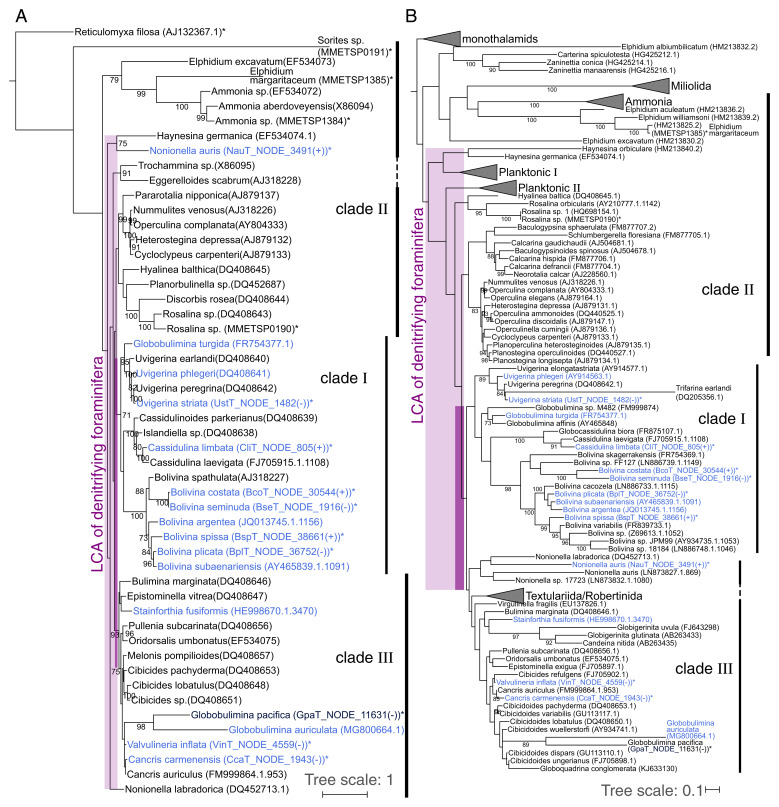
Fig. 3.
The origin of denitrification in context of the whole foraminiferal group. (A) Large-scale phylogenetic representation of Peruvian foraminifera in context of different foraminiferal taxa based on 18S sequences available in public databases. Clades Ι and ΙΙΙ of the order Rotaliida are highly supported by bootstrap values. Questionable branching of Nonionella outside clade ΙΙΙ is not characteristic of data in the current study, as exemplified for N. labradorica, which also does not group directly with clade ΙΙΙ. G. pacifica forms a clade with G. auriculata but within clade ΙΙΙ. However, we consider the branching of these two species as uncertain due to the long branches and the low bootstrap support, which we did not observe for the marker protein phylogeny (Fig. 2). Lower (light purple; including N. labradorica) and higher (dark purple) boundaries for the origin of foraminiferal denitrification are highlighted by boxes. (B) Extended phylogenetic representation of foraminifera including planktonic species. Lower (light purple) and higher (dark purple) boundaries for the origin of foraminiferal denitrification are highlighted by boxes. “Planktonic Ι” and “Planktonic ΙΙΙ” designate two to three distinct clusters comprising planktonic foraminifera. Species experimentally shown to denitrify are highlighted in blue. The only exception is Stainforthia fusiformis, where denitrification activity has been shown for an unspecified species of the same genus. Species also considered in Fig. 1 or SI Appendix, Fig. S3 are marked with asterisks. Several species labels contain the contig ID and orientation of 18S sequences in the corresponding transcriptome assemblies. Bootstrap support values (≥70) with 1,000 replicates are shown at the branches. The trees were rooted by the clade of monothalamids containing R. filosa. A detailed phylogeny is presented in SI Appendix, Fig. S4.
The inference of the denitrifying foraminiferal LCA enabled us to further reconstruct the origin of denitrification within the foraminiferal species tree. For that purpose, we examined an extended phylogeny with additional foraminiferal groups including planktonic species (Fig. 3B and SI Appendix, Fig. S4). While the general taxonomic relationships in the tree were recovered as before (i.e., Fig. 3A), many branches had low statistical support. For example, the rotaliid clades ΙΙ and ΙΙΙ were paraphyletic, in contrast to the phylogeny with less taxa (Fig. 3A). Nonetheless, an LCA of clades Ι and ΙΙΙ could be inferred; the previously unassigned genera Valvulineria and Cancris (9) grouped well with members of clade ΙΙΙ in both 18S rRNA phylogenies, and hence they can be classified as clade ΙΙΙ.
Since both clades Ι and ΙΙΙ harbor several denitrifying foraminifera, it is most parsimonious to conclude that the LCA of clades Ι and ΙΙΙ was probably a denitrifying organism. Our inference thus suggests the presence of denitrification genes (and hence denitrification capability) in all members of clades Ι and ΙΙΙ, including the genera Cibicidoides and Virgulinella, which were previously not considered as denitrifying species. Indeed, Cibicidoides wullerstoerfi has been assumed for a long time to be unable to withstand O2 depletion (31). However, recent studies observed living Cibicidoides spp. thriving in environments of <2 µmol/kg O2 (32) and fossil specimens in the paleorecord during periods of severe O2 depletion (33). These recent studies support our prediction that several Cibicidoides spp. are able to denitrify.
Composition of the foraminiferal microbiome is species-specific.
To test for bacterial contribution to foraminiferal denitrification, we examined the foraminiferal microbiome. For that purpose, we compared the composition of bacterial communities between foraminifera in our sample. In the absence of species-specific interactions, we would expect a strong impact of the environment (i.e., sampling location and water depth) on the microbiome composition. Thus, we compared the microbial community composition between foraminiferal species and their sampling depths (Fig. 4A). Our results show that the individual microbiomes are clustered by the foraminiferal species rather than the sampling location, indicating the presence of species-specific bacterial communities in foraminifera. Our results reveal a similar taxonomic composition of bacterial communities associated with Globobulimina spp. from Sweden and G. pacifica from the Peruvian OMZ (Fig. 4A). The similarity in microbiome composition among the Globobulimina species indicates the presence of genus-specific microbiomes.
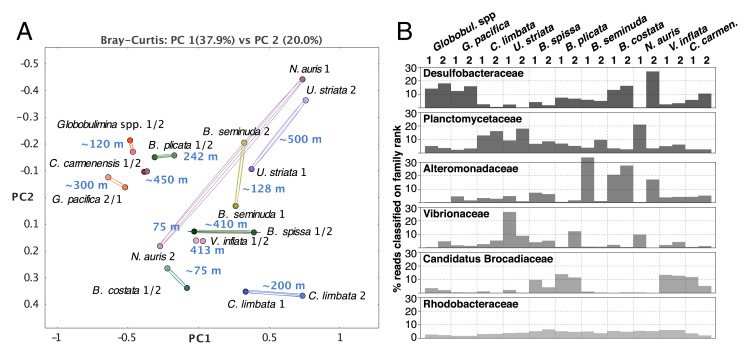
Fig. 4.
Comparison of microbial communities associated with various foraminifera. (A) Principal coordinate analysis clustering of bacterial communities. Each circle represents a bacterial community; duplicates per species are connected by lines. The text next to the circles indicates the species name, replicate number, and sampling depth. The clustering of most species-associated communities indicates species specificity. Grouping by higher taxonomy was observed only for Globobulimina spp. from Sweden and G. pacifica from the Peruvian OMZ. (B) Bar graphs of the six most abundant bacterial families found in the foraminiferal bacterial communities beginning at the Top. Each bar represents the proportion of reads assigned to the individual taxon relative to the total number of reads in a sample classified on family rank or lower. Note the prevalence of Desulfobacteraceae shared between the Globobulimina samples, while none of the other bacterial families showed high prevalence in the genus. C. carmenensis, Cancris carmenensis; C. limbata, Cassidulina limbata; U. striata, Uvigerina striata.
To identify common key players in foraminiferal microbiota, we examined the relative abundance of all bacterial families in their microbiome (Fig. 4B and Dataset S6). Our results revealed that the most prevalent bacterial families are Desulfobacteraceae, followed by Planctomycetaceae. The relative abundance of these two families varied between species and samples. Furthermore, communities with a high relative abundance of Desulfobacteraceae are characterized by a low abundance of Planctomycetaceae and vice versa (rs = –0.77, P < 0.01, according to Spearman correlation coefficient and t test). Desulfobacteraceae comprise sulfate-reducing bacteria and several representatives that are able to grow chemoautotrophically (34). Members of this group were previously observed in association with other rotaliids like Virgulinella fragilis and members of clade ΙΙ (20, 25). Planctomycetes are often found in association with macroalgae (35); they are characterized by a diversified metabolism, allowing them to colonize a wide range of habitats (35). The next most abundant families were Alteromonadaceae, Vibrionaceae, and ‘Candidatus Brocadiaceae’ that were more diverse in their relative abundances across species and replicates. Most reported ‘Candidatus Brocadiaceae’ members are autotrophic, obligately anaerobic bacteria performing anammox, which is a dissimilatory pathway where ammonium and nitrite (NO2−) are metabolized into N2 (36). The presence of nitrate storage in foraminifera and NirK suggests that nitrite is readily available inside the foraminiferal test. Vibrionaceae are a diverse group including multiple species that colonize marine organisms either as symbionts (37, 38) or pathogens (39). Alteromonadaceae were isolated from diverse marine environments including eukaryotic microbiota (40). They are considered to be aerobes or facultative anaerobic bacteria that generally lack denitrification capabilities (41). Finally, another abundant family, Rhodobacteraceae, is worth mentioning, as it is always found in low but similar abundance in all samples. Marine Rhodobacteraceae species are considered ecological generalists (42–44) and have been reported to colonize marine animals (e.g., fish larvae or sponges) (45, 46). The uniform distribution of Rhodobacteraceae in the foraminifera microbiome suggests that members of this group are permanent residents in the foraminiferal microbiota.
Our data thus show that rotaliids are habitat to bacterial communities whose composition is akin to the microbiota of other marine eukaryotes.
An ancient interaction between Desulfobacteraceae and Globobulimina hints at metabolic dependency.
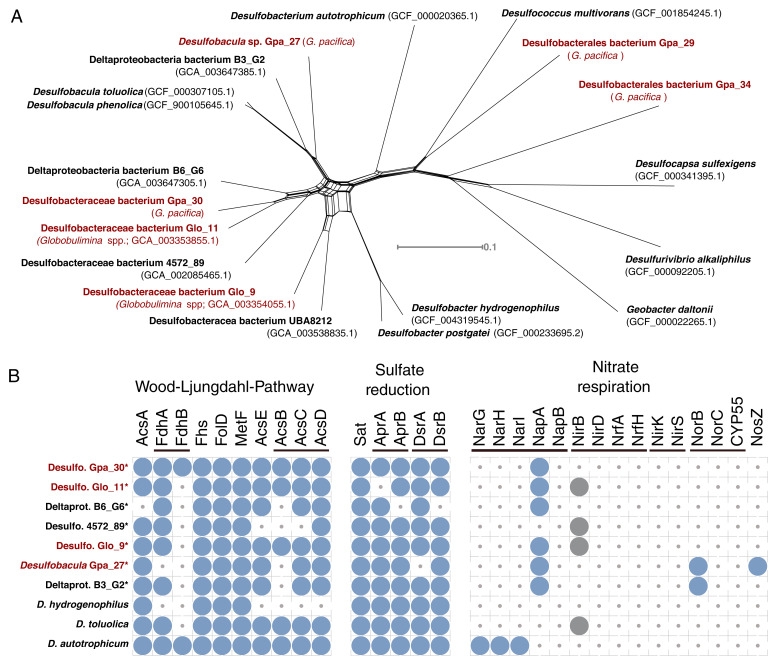
The metagenomic analysis and classification into bacterial metagenome-assembled genomes (MAGs) resulted in a total of 263 high-quality MAGs (i.e., draft bacterial genomes) of foraminifera-associated bacteria (Dataset S1). The strongest signals for a stable core microbiome in our data were observed in the comparison between the Globobulimina species, whose microbiome is characterized by a high frequency of Desulfobacteraceae (Fig. 4B). To further explore the association between Globobulimina and Desulfobacteraceae we examined the Desulfobacteraceae MAGs in our data. A total of 40 high-quality MAGs were obtained from the G. pacifica metagenome, including four high-quality draft MAGs classified as Desulfobacteraceae. These were compared with the 26 previously published MAGs for Globobulimina from Sweden, which include two Desulfobacteraceae MAGs (11) (Dataset S7). A phylogenetic network of the Desulfobacteraceae MAGs reveals two MAGs from the Globobulimina spp. and G. pacifica metagenomes (Glo_11 and Gpa_30) that appear as sister taxa (Fig. 5A). The average nucleotide identity (ANI) between both MAGs was 84%, which is within the range expected for interspecies sequence similarity (e.g., within genera) (47). The common ancestry of these two MAGs suggests that the association between Desulfobacteraceae and Globobulimina has an ancient origin. Our results thus indicate that the interaction fidelity between Globobulimina and Desulfobacteraceae is high, similarly to observations in other eukaryote-bacterial symbioses (e.g., oligochaete worms and sulfur bacteria (48) or marine sponges with bacteria of the Poribacteria phylum (49)).
Fig. 5.
Characteristics of Desulfobacteraceae bacteria associated with Globobulimina. (A) Neighbor-net network for Desulfobacteraceae associated with the genus Globobulimina from Sweden and Peru (Globobulimina spp. and G. pacifica, respectively) and additional Deltaproteobacteria. The network is based on a concatenated alignment of 33 single copy orthologous protein sequences found in all isolates shown. Red fonts highlight isolates found in association with members of the genus Globobulimina. The monophyly of the MAG Gpa_30 and MAG Glo_11 suggests a common ancestry. Note that the closest relatives to that pair are two uncultivated MAGs (Deltaproteobacteria bacterium B6_G6 and Desulfobacteraceae bacterium 4572_89) sampled from sediments in deep-sea hydrothermal vent sites at the Gulf of California. (B) Presence of predicted proteins encoding metabolic pathways in representative members of Desulfobacteraceae. Three pathways are shown: Wood-Ljungdahl pathway, (dissimilatory) sulfate reduction and nitrate respiration. Asterisks highlight genomes in draft state. Circles indicate for the presence of homologs identified by the KEGG KAAS server. The thin bars link enzymes that participate in the same reaction step either by being an alternative enzyme or being part of one protein complex. The gray circles indicate for cases that were discarded following a manual inspection.
A central foundation in the evolution of symbiotic interaction is an exchange of currencies (i.e., resources) between the partners (50). The versatile metabolic capabilities of Desulfobacteraceae and the availability of intracellular NO3− storage in foraminifera may suggest that the interaction between Desulfobacteraceae and Globobulimina is based on nutritional currencies. To further examine possible symbiotic interaction between Desulfobacteraceae and Globobulimina, we searched for metabolic properties of the Desulfobacteraceae MAGs that could serve as nutritional currencies in the symbiosis. For that purpose, we surveyed the metabolic pathways of representative Desulfobacteraceae MAGs with respect to carbon, sulfur, and nitrogen metabolism (Fig. 5B and Dataset S8). Our results reveal that all MAGs sampled encode the genetic repertoire needed in order to perform dissimilatory sulfate reduction as well as the Wood-Ljungdahl pathway. Anaerobic respiration via the dissimilatory sulfate reduction to sulfide is widespread among Desulfobacteraceae. Members of this family that encode the Wood-Ljungdahl pathway are able to oxidize organic compounds to carbon dioxide (51). Alternatively, the Wood-Ljungdahl pathway may function in CO2 carbon fixation during autotrophic growth.
Genes along the denitrification pathway (or NO3− respiration) are mostly absent from all Desulfobacteraceae MAGs, except for periplasmic NO3− reductase (NapA) homologs that were found in most foraminifera-associated MAGs (for a review in NapA function see ref. (52)). All MAGs lack the typical NapB protein forming a complex with NapA. However, nap operons without NapB genes have been reported for other members of Deltaproteobacteria, such as Desulfovibrio desulfuricans (53). Notably, we found that the genome of Desulfobulbus propionicus (accession: GCA_000186885.1), a member of the order Desulfobacterales that has been demonstrated to grow based on NO3− (54), is lacking a gene for NapB, and hence NO3− respiration in Desulfobacterales in the absence of NapB is possible. We found that the NapA sequences of all MAGs (and D. desulfuricans) include a signal peptide of the twin-arginine translocation pathway (reviewed in ref. (55)), indicating that those proteins could be translocated across cellular membranes. It is tenable to hypothesize that NapA is secreted by the bacteria to Globobulimina intracellular environment. An alternative scenario is nitrate uptake by the bacterium followed by the reduction to nitrite within the bacterial cellular compartments that are surrounded by membranes. The nitrite is then released from the bacterial cell and can be used by the foraminiferal host. The latter scenario may be more beneficial for the bacterium. Overall, our results suggest that the Globobulimina-associated Desulfobacteraceae are able to reduce NO3− to NO2− and thus contribute to the foraminiferal denitrification by performing the first reaction in the denitrification pathway for which we found no evidence in the foraminiferal transcriptomes.
Discussion
Our results demonstrate that foraminifera are habitat to bacterial communities that may play a role in their ability to thrive in oxygen-depleted habitats. A recent study demonstrates the endosymbiotic contribution to denitrification within a ciliate host (56). However, previous studies of denitrification in foraminifera argued against the possibility of bacterial contribution to foraminiferal denitrification (8, 11). Notably, most species in the foraminiferal microbiota are considered strict anaerobes; hence, when exposed to oxygen the foraminifera may lose their associated microbiota. Metagenomic sequencing of samples frozen directly after sampling is thus an important source of information on the composition and function of the foraminiferal microbiome.
Our results indicate that Desulfobacteraceae members of the foraminiferal microbiota can use the NO3− storage accumulated by their host. Whether the NO3− reduction by bacteria is beneficial to the foraminifera remains an open question, however. The bacteria may use the NO3− for their own respiratory processes or to build up organic compounds via the assimilation of NO3−. For example, it has been suggested that a proportion of NO3− taken up by the foraminiferal species Ammonia beccari was used for amino acid synthesis, probably by resident bacteria (21). Previously we speculated about a foraminiferal sulfite reductase homolog performing the conversion of NO3− (11). Yet, considering the absence of foraminiferal homologs to NO3− reductase and the presence of NapA in all sampled Desulfobacteraceae MAGs, it is tenable to hypothesize that the reduction of NO3− to NO2− in foraminifera is performed by resident bacteria.
An association between foraminifera and Desulfobacteraceae has been previously reported. For example, the presence of a putative Deltaproteobacterium (Desulfobacteraceae) was previously described for Virgulinella fragilis (20), a foraminiferal species that we here predict to be a denitrifying species (Fig. 3B). Furthermore, foraminifera from the genera Ammonia, Elphidium, and Haynesina that are not expected to denitrify were found to harbor members of Desulfobacterales bacteria (25). Hence, the interaction between foraminifera and Desulfobacteraceae may involve alternative nutritional currencies. Previous studies refer to the role of Desulfobacteraceae in sulfur cycling and carbon/nutrient acquisition (20, 21, 25). The carbon fixation capabilities via Wood-Ljungdahl pathway in particular would be highly beneficial for the heterotrophic lifestyle of foraminifera, similarly to other symbioses that involve fixed carbon as a nutritional currency (e.g., as in deep-sea mussels (57) or sponges (49)).
The radiation of early foraminiferal species has been estimated to occur between 690 and 1,150 million years ago (mya), and the first primitive rotaliids appeared in the late Permian around 260 mya (58, 59). Most rotaliid superfamilies diverged in the mid to late Triassic between ∼240 and 200 mya (29, 60, 61), and the majority of extant species diverged during the Miocene (23.8–5.5 mya) (62). Our phylogenetic analyses suggest that clades Ι and ΙΙΙ diversified from within clade ΙΙ, and the LCA of clade Ι probably originated from within clade ΙΙΙ (Figs. 2 and 3). Although most species reported to denitrify were reported from the Peruvian OMZ and Sweden, representatives of corresponding genera are found all over the world (Dataset S5). Therefore, most extant rotaliids probably diversified from a denitrifying LCA far back in time. The origin of foraminiferal denitrification within or after diversification from members of clade ΙΙ may coincide with the rapid increase of fossil records at the onset of the late Cretaceous ∼100 mya (60, 61). Our results therefore indicate that eukaryotic denitrification by rotaliids emerged late in foraminiferal evolution, possibly during the worldwide Cretaceous Ocean Anoxic Events (63, 64).
Considering the high conservation of nitrate transporters (Fig. 1) and the observation of nitrate storage in many divergent species (9), it is tenable to speculate that foraminifera had the mechanisms for nitrate import and storage long before they evolved the ability to denitrify. NO3− transporters are ancient eukaryotic enzymes that play a role in the NO3− assimilation machinery, and they are encoded in genomes of photoautotrophs such as plants and saprotrophs like fungi (65, 66). In contrast to bacteria, foraminifera (as most heterotrophs) are unable to perform NO3− assimilation on their own. Thus, NO3− accumulated by the foraminiferal hosts could have fueled bacterial NO3− metabolism of associated bacteria in exchange for organic compounds. We propose that denitrification by symbiotic bacteria was indeed the ancestral state of denitrification in foraminifera, similarly to gromiids (26), which share a common evolutionary origin with foraminifera (67). The finding of bacterial-like denitrification genes in rotaliids furthermore suggests that foraminifera may have acquired those genes from bacteria (11). Thus, a rare gene acquisition from a foraminifera-associated, denitrifying bacterium could have been at the origin of foraminiferal (eukaryotic) denitrification. Notably, the rotaliids’ denitrification gene set is incomplete, and it varies between the species sampled here (i.e., Nonionella auris, Bolivina plicata, or even Rosalina sp.). Species reported to release N2O instead of N2 (9) further support our observation. Thus, the presence of a partial denitrification pathway in rotaliids as well as in their resident Desulfobacteraceae may suggest that the acquisition of denitrification ability in foraminifera occurred in multiple stages and is not yet complete.
Methods
Foraminifera sampling.
Samples were collected off the Peruvian coast in the 2017 Austral winter (R/V Meteor M137) as described by Glock et al., 2019 (10). Briefly, sediment samples were taken with a video-guided sediment multiple corer (MUC) containing six liners along a depth transect at 12°S. The top 1–3 cm of the sediment cores were sampled and immediately wet sieved with surface water through staked sieves with mesh size of 2,000 μm to 63 μm to retrieve benthic foraminifera. The foraminifera were rinsed in sterile seawater obtained by filtering core-overlying seawater with a sterile bottle top filtration system (Durapore filter, 0.2 µm) and a vacuum pump within 40 min of MUC arrival on deck. Focal species were manually picked with a stereomicroscope. These were characterized morphologically according to the literature (68–71). Up to 160 individuals (classified into focal species) were pooled in one cryo vial (2 mL, RNase free) and flash frozen in liquid nitrogen.
Microscopy.
Individual foraminifers were dehydrated in a graded ethanol series (70%, 80%, 90%, 96%, and two times 100%; 15 min each), air-dried at 20 °C for 12 h in a desiccator and mounted on aluminum stubs (PLANO GmbH) with conductive and adhesive carbon pads (PLANO GmbH). Subsequently, the preparations were sputter-coated with a 10-nm-thick gold-palladium (80/20) layer with a high-vacuum sputter coater (Leica EM SCD500, Leica Microsystems GmbH) and visualized with a Hitachi S-4800 field emission scanning electron microscope (Hitachi High-Technologies Corporation) at an acceleration voltage of 3 kV and an emission current of 10 mA applying a combination of the upper detector and the lower detector. The B. costata specimen was taken from previous samplings by the General Direction of Research in Oceanography and Climate Change IMARPE (71). The corresponding sample was air-dried at 35–38 °C, sputter-coated with gold, and visualized with an FEI Inspect S50 scanning electron microscope (Thermo Fisher Scientific Inc.) at an acceleration voltage of 3 kV.
Nucleic acid extraction and sequencing.
Genomic DNA and total RNA from different biological samples were extracted with either the DNeasy Plant Mini kit (QIAGEN) for genomics (DNA) or simultaneously purified with an InnuPREP DNA/RNA Mini Kit (Analytik Jena) for transcriptomics (RNA). Foraminiferal cells in each sample (ca. 160 individuals) were disrupted by pestle-crushing on ice after immersion of the containing cryo-vial in liquid nitrogen. Samples for genomics were treated with lysozyme (200 μL of 10 mg/mL TE) and proteinase K (1 mg/100 μL). Samples for transcriptomic samples were treated only with lysozyme (6 μL of 20 mg/mL TE) before additional crushing, and 2-min incubation was performed. After disruption and initial lysis, manufacturer protocols for the respective nucleic acid extraction were followed by modifications to the elution volumes (2 × 50 μL for DNA and 2 × 25 μL for RNA). Libraries for genomics were produced after DNA fragmentation (Covaris target 400, intensity 5, duty cycle 5% cycles per burst 200, 55 s treatment time) with NEBNext Ultra II DNA Library Prep Kit for Illumina, whereas transcriptomic libraries were produced with a NEBNext Ultra RNA Library Prep Kit for Illumina with mRNA isolation performed with poly-A mRNA beads. All libraries were produced in duplicate from two different sets of pooled individuals for each species and were produced without protocol interruption. Before sequencing, each library was quantified with a Qubit fluorometer (Invitrogen by Life Technologies) and qualified with a TapeStation (Agilent Technologies). The libraries were sequenced paired-end (2 × 150 bp) on an Illumina HiSeq 4000 platform.
Sequencing datasets of foraminifera.
Sequencing resulted in 4.6 billion paired-end reads covering 1.4 terabases in total. This includes transcriptome and metagenome datasets (BioProject accessions PRJNA494828 and PRJNA503328). Reads were quality-checked by FastQC ver. 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc; August 2016). Filtering and trimming of reads was performed in Trimmomatic (72) ver. 0.36 (Parameters: ILLUMINACLIP:primers.fa:2:30:10 LEADING:5 TRAILING:5 SLIDINGWINDOW:4:5 MINLEN:21; the file ‘primers.fa’ contained adaptor and contaminant sequences provided by Trimmomatic and FastQC). Processed reads from transcriptomes of the two samples per species were assembled into transcript contigs in SPAdes (73) ver. 3.11.1 (“–rna” option). Protein sequences were translated from transcripts as open reading frames in TransDecoder (74) ver. 5.0.2 (LongOrfs; “-m 30” option). Final protein names consist of the contig IDs followed by the sequence positions covered by the coding sequences and an indicator for the forward (+) or reverse (–) strand. Transcript abundance of individual transcriptome datasets are referring to transcripts per million determined by the Trinity pipeline (74) 2.4.0 (Trinity script ‘align_and_estimate_abundance.pl’) via RSEM (75) ver. 1.2.30 and Bowtie (76) ver. 2.1.0 with paired-end reads. Additional raw sequencing reads were obtained from the Marine Microbial Transcriptome Project (Sequence Read Archive [SRA] accessions: Rosalina sp., SRR1296887; Sorites sp., SRR1296734; Ammonia sp., SRR1300434; Elphidium margaritaceum, SRR1300475) and processed as described above. We found that the assembly obtained for Sorites sp. contained a high proportion of sequences probably originating from an algal species (probably Symbiodinium sp.), which was removed via a binning approach considering only contigs with GC-content ≤38%. Finally, we included data for Globobulimina spp. sampled in Sweden from our previous study (transcriptome data: GloT15) (11) and proteins annotated on the genome assembly of R. filosa (National Center for Biotechnology Information [NCBI] accession: GCA_000512085.1).
Species phylogenies and rooting.
Transcriptome completeness and heterogeneity were determined by assessing genome completeness via Benchmarking Universal Single-Copy Orthologs (77) (BUSCO ver. 3; lineage ‘eukaryota’) method. Orthologous proteins determined as Complete (or Duplicated) by the BUSCO analysis were merged into protein clusters to study phylogenetic relationships between the species. In case of duplicated BUSCOs in metatranscriptomes, one representative was inferred based on sequence similarity. Therefore, all orthologs retrieved for the same metatranscriptome were compared to all members of the BUSCO protein cluster by global pairwise alignments with needle (EMBOSS tools ver. 6.6.0) (78). The ortholog with the highest median sequence similarity over all comparisons was picked as the representative sequence. Multiple sequence alignments used in the current study were obtained in MAFFT (79) (ver. 7; parameter: ‘linsi’), and the phylogenetic trees were reconstructed in IQTREE (80) (ver. 1.5.5 or 1.6.9; default parameters; note that ModelFinder is enabled by default). Phylogenies were reconstructed for the individual BUSCO clusters. Due to their morphological differences from other foraminiferal groups, monothalamids like R. filosa have been previously used as an outgroup for phylogenetic studies of foraminifera (29). Since the choice of a distantly related outgroup may lead to erroneous rooted topology due to long branch attraction (81), we further tested the robustness of the root position independent from outgroup species. Root positions were determined in MAD (28) (parameters: ‘-bsnn’). The support in each root split is calculated as the proportion of gene trees where the root position was inferred as such (28). For the overall species trees, multigene phylogenies were reconstructed in IQTREE (parameter: ‘-spp’) considering all protein cluster alignments of either foraminifera or Rotaliida.
Reference 18S rRNA sequences used for the large-scale phylogenies (Dataset S5) were obtained from NCBI, Marine Microbial Transcriptome Project, SILVA (82), the foram barcoding project (83), and the Planktonic Foraminifera Ribosomal Reference database (84). These sequences were used as database sequences to identify 18S sequences from Peruvian species transcriptomes based on BLAST (85) (ver. 2.2.28+; options: ‘-task blastn -evalue 1e−10’) searches. Among the hits retrieved we searched for a single representative transcript contig per transcriptome assembly. The transcripts with the highest product of sequence length and sequencing depth (i.e., coverage given by SPAdes) were determined as representative sequences for most of the focal species. However, for B. costata, Valvulineria inflata, G. pacifica, and N. auris different representative sequences were chosen by giving sequence information (i.e., longer sequences) a higher weight than sequencing depth. To obtain 18S phylogenies, first the reference sequences were aligned with MAFFT, and subsequently representative 18S rRNA sequences of the Peruvian transcriptomes were added (MAFFT options: ‘–addfragments –adjustdirection’) to the alignment, followed by the tree reconstruction in IQTREE.
Identification of foraminiferal denitrification proteins.
To identify homologs to enzymes in the denitrification pathway, we used a similar approach as previously established by Woehle et al., 2018 (11), using the corresponding protein database expanded by the protein sequences identified for Globobulimina (11). The search for denitrification enzyme homologs in the transcriptome assemblies was performed with BLASTP (parameter: ‘-max_target_seqs 1000000 e-value 1e-5’). Protein sequences of hits with query coverage ≥40% and sequence identity ≥20% were extracted to obtain a first set of homologs. We further applied a cutoff to discard lowly represented transcripts with transcripts per million <2 in at least one of the two replicates sequenced. With the resulting protein set, we reiterated searches in the nonredundant NCBI protein (NR; version May 2018) and the RefSeq 88 database (86) by using diamond version 0.9.22 and applying the ‘–more-sensitive’ option. First best hit sequences per query were obtained and clustered with CD-HIT 4.6 (87) (option: “-c 0.98”) to reduce sequence redundancy. All obtained protein sequences of a given enzyme were aligned with MAFFT, and phylogenetic trees were reconstructed with IQTREE (parameters: ‘-bb 1000 -alrt 1000’). The trees were rooted with an outgroup, if available, or MAD.
Metagenomic processing.
For the visualization of metagenomic composition, trimmed paired-end reads from Peruvian species samples and the Globobulimina ‘Ambient’ samples (11) were subsampled via BBMap (ver. 36.84; https://sourceforge.net/projects/bbmap/; ‘reformat.sh’ script; parameters: ‘samplereadstarget = 10000000 addslash = t’). The resulting reads were mapped against the NR database with ac-diamond (88) and classified in MEGAN6 (89) (ver. 6.15.0; options ‘-a2t prot_acc2tax-Nov2018 × 1.abin -a2eggnog acc2eggnog-Oct2016 × .abin -a2seed acc2seed-May2015XX.abin’). MEGAN6 was used for assessment of metagenomic communities and visual representation (see Fig. 4) that were classified up to the order rank level with different taxon sets (e.g., all nodes or only the bacterial subtree). Metagenome assemblies were obtained by combining all trimmed reads per focal species in MEGAHIT (90) (ver. 1.1.3; default settings). Individual bacterial genomes were obtained via the binning approach implemented with MaxBin2 (91) (ver. 2.2.4; parameter: ‘-min_contig_length 500’) with coverage information of the two samples per species of foraminifera. Binning statistics were assessed in CheckM (92) (ver. 1.0.11); a threshold for completeness of at least 80% and contamination of maximum 20% was applied to classify bacterial bins as draft genomes. The protein sequences obtained via checkM were used for further classifications. The ANI was calculated with a perl script obtained from https://github.com/chjp.
For the taxonomic assignment of genome bins, we used the diamond tool to find first best hits for each protein of a genome bin against the NR database (option: ‘-k 10’; e-value ≤ 1e-10) and retrieved the corresponding taxonomic assignments (as we previously described in ref. (11)). For each bin, identical taxonomic hierarchies were counted with the genus being the lowest rank considered, sorted accordingly and stepwise searched for the lowest taxonomic rank supporting >50% of protein bin hits starting with the most abundant taxonomy. ‘Environmental samples’ and ‘Cellular organisms’ were not considered. For the phylogenetic reconstruction of Desulfobacteraceae we determined protein families as protein clusters by using sequences of Desulfobacteraceae bins associated with Globobulimina and additional Desulfobacteraceae genomes downloaded from NCBI. First, we determined reciprocal best BLAST hit pairs (rBBH; parameter: ‘-evalue 1e-5’) between all the Desulfobacteraceae protein-coding sequences (93). Then rBBH pairs were globally aligned with the needle tool, and pairs with ≥30% identical amino acids were sorted into clusters with the Markov clustering algorithm (94) (ver. 12-135). The 33 resulting protein clusters that contained a single copy for each of the Desulfobacteraceae strains (i.e., universal single-copy clusters) were aligned with MAFFT. The resulting alignments were concatenated, and a splits network was reconstructed with SplitsTree (95) (ver. 4.15.1). Prediction of protein function for bacterial genomes were obtained with the KEGG Automatic Annotation Server (96) (BBH method via BLAST and the following species set: hsa, dme, ath, sce, pfa, eco, sty, hin, pae, nme, hpy, rpr, mlo, bsu, sau, lla, spn, cac, mge, mtu, ctr, bbu, syn, aae, mja, afu, pho, ape, geo, dvu, dat, dpr, dol, dal, dak, dps, drt, dba, dao, dbr). Global pairwise identities for NapA protein sequences were inferred with needle. Signal peptides were predicted via the SignalP-5.0 webservice for Gram-negative bacteria (97).
Supplementary Material
Acknowledgments
We thank Devani Romero Picazo and Anne Kupczok for critical comments on the manuscript. We gratefully acknowledge the scientific party and crew of the R/V Meteor cruise M137 as well as Asmus Petersen and Matthias Türk for their support at sea. Samples from Peru were obtained according to Peruvian access and benefit sharing regulations. We thank the Nagoya officer of Kiel University, Dr. Scarlett Sett, for her support of our research. We also thank Natalia Bernabe Lopez for her assistance in collecting metadata for the 18S sequences. The micrograph of B. costata was possible due to the General Direction of Research in Oceanography and Climate Change. Genome sequencing was performed in the Centre for Genome Analysis Kiel, funded by the German Research Foundation. The study was supported by the German Research Foundation via the SFB 754 on Climate–Biogeochemistry Interactions in the Tropical Ocean and the Collaborative Research Centre 1182 on the Origin and Function of Metaorganisms, the cluster of excellence The Future Ocean, and the European Research Council (grant no. 281357 awarded to T.D.). C.W. was supported by the Kiel Life Science Young Scientist Programme and would like to thank the Max Planck-Genome-Centre Cologne for their support in data analysis.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200198119/-/DCSupplemental.
Data Availability
Sequencing reads are deposited in the single read archive accessions SRR8144071 to SRR8144090 and SRR7971179 to SRR7971198. The assemblies are available in the transcriptome sequencing archive (see Dataset S1 for accessions) and as whole genome shotgun projects (see Dataset S7 for accessions). All other information on accessing data analyzed in this study is included in the manuscript or in the SI Appendix.
Change History
June 22, 2022: Due to a production error, the SI Appendix appeared incorrectly. The SI Appendix has been updated.
References
- 1.Cox A. N., Ed., Allen’s Astrophysical Quantities (Springer, New York, NY, 2015). [Google Scholar]
- 2.Thamdrup B., New pathways and processes in the global nitrogen cycle. Annu. Rev. Ecol. Evol. Syst. 43, 407–428 (2012). [Google Scholar]
- 3.Gruber N., The Dynamics of the Marine Nitrogen Cycle and Its Influence on Atmospheric CO2 Variations. The Ocean Carbon Cycle and Climate (Springer, Dordrecht, Dordrecht, ed. 2, 2004), pp. 97–148. [Google Scholar]
- 4.Gruber N., Galloway J. N., An Earth-system perspective of the global nitrogen cycle. Nature 451, 293–296 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Lam P., et al. , Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U.S.A. 106, 4752–4757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philippot L., Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577, 355–376 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Shoun H., Kim D. H., Uchiyama H., Sugiyama J., Denitrification by fungi. FEMS Microbiol. Lett. 73, 277–281 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Risgaard-Petersen N., et al. , Evidence for complete denitrification in a benthic foraminifer. Nature 443, 93–96 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Piña-Ochoa E., et al. , Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc. Natl. Acad. Sci. U.S.A. 107, 1148–1153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glock N., et al. , Metabolic preference of nitrate over oxygen as an electron acceptor in foraminifera from the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U.S.A. 116, 2860–2865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woehle C., et al. , A novel eukaryotic denitrification pathway in foraminifera. Curr. Biol. 28, 2536–2543.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glock N., et al. , The role of benthic foraminifera in the benthic nitrogen cycle of the Peruvian oxygen minimum zone. Biogeosciences 10, 4767–4783 (2013). [Google Scholar]
- 13.Schweizer M., Pawlowski J., Kouwenhoven T. J., Guiard J., van der Zwaan B., Molecular phylogeny of Rotaliida (Foraminifera) based on complete small subunit rDNA sequences. Mar. Micropaleontol. 66, 233–246 (2008). [Google Scholar]
- 14.Murray J. W., Ecology and Applications of Benthic Foraminifera (Cambridge University Press, 2006). [Google Scholar]
- 15.LeKieffre C., et al. , Surviving anoxia in marine sediments: The metabolic response of ubiquitous benthic foraminifera (Ammonia tepida). PLoS One 12, e0177604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernhard J. M., et al. , Potential importance of physiologically diverse benthic foraminifera in sedimentary nitrate storage and respiration. J. Geophys. Res. 117, G03002 (2012). [Google Scholar]
- 17.Gomaa F., et al. , Multiple integrated metabolic strategies allow foraminiferan protists to thrive in anoxic marine sediments. Sci. Adv. 7, eabf1586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orsi W. D., et al. , Anaerobic metabolism of Foraminifera thriving below the seafloor. ISME J. 14, 2580–2594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zilber-Rosenberg I., Rosenberg E., Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Tsuchiya M., et al. , Cytologic and genetic characteristics of endobiotic bacteria and kleptoplasts of Virgulinella fragilis (Foraminifera). J. Eukaryot. Microbiol. 62, 454–469 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Nomaki H., et al. , Nitrate uptake by foraminifera and use in conjunction with endobionts under anoxic conditions. Limnol. Oceanogr. 59, 1879–1888 (2014). [Google Scholar]
- 22.Bernhard J. M., Martin J. B., Rathburn A. E., Combined carbonate carbon isotopic and cellular ultrastructural studies of individual benthic foraminifera: 2. Toward an understanding of apparent disequilibrium in hydrocarbon seeps. Paleoceanography 25, PA4206 (2010). [Google Scholar]
- 23.Bernhard J. M., Habura A., Bowser S. S., An endobiont‐bearing allogromiid from the Santa Barbara Basin: Implications for the early diversification of foraminifera. J. Geophys. Res. 111, 399 (2006). [Google Scholar]
- 24.Bernhard J. M., Potential symbionts in bathyal foraminifera. Science 299, 861 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Salonen I. S., Chronopoulou P. M., Bird C., Reichart G. J., Koho K. A., Enrichment of intracellular sulphur cycle -associated bacteria in intertidal benthic foraminifera revealed by 16S and aprA gene analysis. Sci. Rep. 9, 11692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Høgslund S., Cedhagen T., Bowser S. S., Risgaard-Petersen N., Sinks and sources of intracellular nitrate in gromiids. Front. Microbiol. 8, 617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernhard J. M., Edgcomb V. P., Casciotti K. L., McIlvin M. R., Beaudoin D. J., Denitrification likely catalyzed by endobionts in an allogromiid foraminifer. ISME J. 6, 951–960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tria F. D. K., Landan G., Dagan T., Phylogenetic rooting using minimal ancestor deviation. Nat. Ecol. Evol. 1, 193 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Pawlowski J., et al. , The evolution of early Foraminifera. Proc. Natl. Acad. Sci. U.S.A. 100, 11494–11498 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ertan K. T., Hemleben V., Hemleben C., Molecular evolution of some selected benthic foraminifera as inferred from sequences of the small subunit ribosomal DNA. Mar. Micropaleontol. 53, 367–388 (2004). [Google Scholar]
- 31.Mackensen A., Schmiedl G., Harloff J., Giese M., Deep-sea foraminifera in the South Atlantic Ocean: Ecology and assemblage generation. Micropaleontology 41, 342 (1995). [Google Scholar]
- 32.Rathburn A. E., Willingham J., Ziebis W., Burkett A. M., Corliss B. H., A new biological proxy for deep-sea paleo-oxygen: Pores of epifaunal benthic foraminifera. Sci. Rep. 8, 9456–9458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoogakker B. A. A., et al. , Glacial expansion of oxygen-depleted seawater in the eastern tropical Pacific. Nature 562, 410–413 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Widdel F., New types of acetate-oxidizing, sulfate-reducing Desulfobacter species, D. hydrogenophilus sp. nov., D. latus sp. nov., and D. curvatus sp. nov. Arch. Microbiol. 148, 286–291 (1987). [Google Scholar]
- 35.Lage O. M., Bondoso J., Planctomycetes and macroalgae, a striking association. Front. Microbiol. 5, 267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshiki M., Satoh H., Okabe S., Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ. Microbiol. 18, 2784–2796 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Noguchi T., et al. , Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu vermicularis. Mar. Biol. 94, 625–630 (1987). [Google Scholar]
- 38.McFall-Ngai M., Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 12, e1001783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Haim Y., Zicherman-Keren M., Rosenberg E., Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López-Pérez M., Rodriguez-Valera F., “The family Alteromonadaceae” in The Prokaryotes: Gammaproteobacteria, Rosenberg E., Delong E. F., Lory S., Stackebrandt E., Thompson F., Eds. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2014), pp. 69–92. [Google Scholar]
- 41.Ivanova E. P., Flavier S., Christen R., Phylogenetic relationships among marine Alteromonas-like proteobacteria: Emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int. J. Syst. Evol. Microbiol. 54, 1773–1788 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Simon M., et al. , Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton R. J., et al. , Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Buchan A., González J. M., Moran M. A., Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hjelm M., et al. , Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27, 360–371 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Webster N. S., Negri A. P., Munro M. M. H. G., Battershill C. N., Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6, 288–300 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114–5118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubilier N., et al. , Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Kamke J., et al. , Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J. 7, 2287–2300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wein T., et al. , Currency, exchange, and inheritance in the evolution of symbiosis. Trends Microbiol. 27, 836–849 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Strittmatter A. W., et al. , Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ. Microbiol. 11, 1038–1055 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.González P. J., Correia C., Moura I., Brondino C. D., Moura J. J. G., Bacterial nitrate reductases: Molecular and biological aspects of nitrate reduction. J. Inorg. Biochem. 100, 1015–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Dias J. M., et al. , Crystal structure of the first dissimilatory nitrate reductase at 1.9 A solved by MAD methods. Structure 7, 65–79 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Widdel F., Pfennig N., Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 131, 360–365 (1982). [DOI] [PubMed] [Google Scholar]
- 55.Berks B. C., Ferguson S. J., Moir J. W., Richardson D. J., Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232, 97–173 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Graf J. S., et al. , Anaerobic endosymbiont generates energy for ciliate host by denitrification. Nature 591, 445–450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duperron S., et al. , A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ. Microbiol. 8, 1441–1447 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Maklay M., Novye Rannekamennougolnye Arkhedistsidy. Min. Geol. Okhrana Nedr USSR 1, 158–159 (1964). [Google Scholar]
- 59.Crespin I., Permian foraminifera of Australia. Bur. Miner. Resour. Aust. Bull. 48, 1–207 (1958). [Google Scholar]
- 60.Tappan H., Loeblich A. R., Foraminiferal evolution, diversification, and extinction. J. Paleontol. 62, 695–714 (1988). [Google Scholar]
- 61.Loeblich A. R., Tappan H., Treatise on Invertebrate Paleontology. Part C. Protista 2. Sarcodina Chiefly “Thecamoebians” and Foraminiferida (Geological Society of America New York/University of Kansas Lawrence, New York, Lawrence, 1964). [Google Scholar]
- 62.Kucera M., Schönfeld J., The Origin of Modern Oceanic Foraminiferal Faunas and Neogene Climate Change (The Micropalaeontology Society Special Publications, 2007), pp. 409–425. [Google Scholar]
- 63.Schlanger S. O., Jenkyns H. C., Cretaceous oceanic anoxic events: Causes and consequences. Geologie en Mijnbouw 55, 179–184 (1976). [Google Scholar]
- 64.Jenkyns H. C., Cretaceous anoxic events: From continents to oceans. J. Geol. Soc. London 137, 171–188 (1980). [Google Scholar]
- 65.Forde B. G., Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 1465, 219–235 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Pao S. S., Paulsen I. T., Saier M. H. Jr., Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sierra R., et al. , Deep relationships of Rhizaria revealed by phylogenomics: A farewell to Haeckel’s Radiolaria. Mol. Phylogenet. Evol. 67, 53–59 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Figueroa S., Marchant M., Giglio S., Ramírez M., Foraminíferos bentónicos rotalínidos del centro sur de Chile (36°S - 44°S). Gayana (Concepc.) 69, 329–363 (2005). [Google Scholar]
- 69.Erdem Z., “Reconstruction of past bottom water conditions of the Peruvian Oxygen Minimum Zone (OMZ) for the last 22,000 years and the benthic foraminiferal response to (de)oxygenation,” PhD thesis, Kiel University (2016).
- 70.Mallon J., “Benthic foraminifera of the Peruvian and Ecuadorian continental margin,” PhD thesis, Kiel University (2012).
- 71.Cardich J., et al. , Calcareous benthic foraminifera from the upper central Peruvian margin: Control of the assemblage by pore water redox and sedimentary organic matter. Mar. Ecol. Prog. Ser. 535, 63–87 (2015). [Google Scholar]
- 72.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nurk S., et al. , Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Rice P., Longden I., Bleasby A., EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Felsenstein J., Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (1978). [Google Scholar]
- 82.Yilmaz P., et al. , The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pawlowski J., Holzmann M., A plea for DNA barcoding of foraminifera. J. Foraminiferal Res. 44, 62–67 (2014). [Google Scholar]
- 84.Morard R., et al. , PFR2: A curated database of planktonic foraminifera 18S ribosomal DNA as a resource for studies of plankton ecology, biogeography and evolution. Mol. Ecol. Resour. 15, 1472–1485 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R., NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Mai H., et al. , AC-DIAMOND v1: Accelerating large-scale DNA-protein alignment. Bioinformatics 34, 3744–3746 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Huson D. H., et al. , MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput. Biol. 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Wu Y.-W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tatusov R. L., Koonin E. V., Lipman D. J., A genomic perspective on protein families. Science 278, 631–637 (1997). [DOI] [PubMed] [Google Scholar]
- 94.Enright A. J., Van Dongen S., Ouzounis C. A., An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M., KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nielsen H., Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads are deposited in the single read archive accessions SRR8144071 to SRR8144090 and SRR7971179 to SRR7971198. The assemblies are available in the transcriptome sequencing archive (see Dataset S1 for accessions) and as whole genome shotgun projects (see Dataset S7 for accessions). All other information on accessing data analyzed in this study is included in the manuscript or in the SI Appendix.