Significance
Cancer and heart disease are among the most progressive causes of mortality today. Two major regulators of these disease states are the MAP kinase and PKA signaling pathways. Ensuring proper signaling of these pathways is an essential aspect of maintaining cellular integrity and preventing disease progression. Here we identify a regulatory circuit whereby PKA phosphorylation of RKIP at S51 induces its phosphorylation by PKC, leading to up-regulation of the β-adrenergic receptor signaling and further activation of PKA. This positive feedback circuit enables rapid and robust activation of PKA in the heart and other tissues when required. This mechanism has potential applications to multiple disease states by identifying S51 as a diagnostic marker and therapeutic target.
Keywords: Raf Kinase Inhibitory Protein (RKIP), protein kinase A (PKA), phosphatidylethanolamine binding protein (PEBP), nuclear magnetic resonance (NMR)
Abstract
Raf Kinase Inhibitory Protein (RKIP) maintains cellular robustness and prevents the progression of diseases such as cancer and heart disease by regulating key kinase cascades including MAP kinase and protein kinase A (PKA). Phosphorylation of RKIP at S153 by Protein Kinase C (PKC) triggers a switch from inhibition of Raf to inhibition of the G protein coupled receptor kinase 2 (GRK2), enhancing signaling by the β-adrenergic receptor (β-AR) that activates PKA. Here we report that PKA-phosphorylated RKIP promotes β-AR–activated PKA signaling. Using biochemical, genetic, and biophysical approaches, we show that PKA phosphorylates RKIP at S51, increasing S153 phosphorylation by PKC and thereby triggering feedback activation of PKA. The S51V mutation blocks the ability of RKIP to activate PKA in prostate cancer cells and to induce contraction in primary cardiac myocytes in response to the β-AR activator isoproterenol, illustrating the functional importance of this positive feedback circuit. As previously shown for other kinases, phosphorylation of RKIP at S51 by PKA is enhanced upon RKIP destabilization by the P74L mutation. These results suggest that PKA phosphorylation at S51 may lead to allosteric changes associated with a higher-energy RKIP state that potentiates phosphorylation of RKIP at other key sites. This allosteric regulatory mechanism may have therapeutic potential for regulating PKA signaling in disease states.
Cellular responses to external and internal stimuli are mediated by protein kinase signaling cascades characterized by negative and positive feedback loops. While negative regulation resets signaling molecules back to their resting states, positive feedback loops can convert graded signals to rapid and robust outputs (1). Multiple mechanisms are utilized to achieve this task, including regulation of kinase expression by altering synthesis or degradation. Since kinase stimulation is a rapid and transient process, additional responsive and efficient mechanisms involving small protein modulators are integrated into this signaling network to affect kinase activity directly.
One ubiquitous modulator of kinase function is the Raf Kinase Inhibitory Protein (RKIP, also termed PEBP1). RKIP is a member of the highly conserved phosphatidylethanolamine binding protein (PEBP) family and regulates key kinase pathways such as the Raf/MAPK pathway and β-AR/cAMP–dependent protein kinase A (PKA) signaling cascade (2, 3). Since it inhibits kinases involved in both growth control and stress responses, RKIP loss and/or dysregulation have been linked to many diseases, including metastatic lesions, asthma, Alzheimer’s disease, systemic inflammatory response syndrome, and heart disease (4–10). Depending upon its phosphorylation state, RKIP can modulate different signaling pathways. When unphosphorylated, RKIP binds Raf, preventing its activation (11, 12). Upon phosphorylation by protein kinase C (PKC) at S153, RKIP is released from Raf and inhibits GRK2, a member of the G protein coupled receptor kinase family that modulates the function of the β-AR (10, 13). The net effect of RKIP/GRK2 interaction is to up-regulate β-AR signaling, leading to activation of its downstream effector, PKA. Therefore, RKIP is emerging as a multifunctional molecular switch that controls the MAPK kinase and PKA signaling pathways, two of the most conserved and essential cascades in mammalian cells.
Despite its importance, the cross-talk between RKIP and the β-AR signaling cascade is not well understood. Here, using a battery of biochemical, molecular, and cellular biology techniques as well as NMR spectroscopy, we show that RKIP is a substrate of the PKA catalytic subunit (PKA-C). Remarkably, we found that phosphorylation of RKIP at S51 by PKA-C increases phosphorylation of RKIP at S153 by PKC, triggering the RKIP phospho-swap of protein partners (14) that leads to further PKA activation. Thus, RKIP is a key player in a positive feedback loop to potentiate PKA activity downstream of the β-AR pathway. This allosteric regulatory mechanism highlights the multifunctional role of RKIP as a sensor, integrator, and effector of cellular kinase signaling (2, 3, 15).
Results
PKA Phosphorylates RKIP at S51 In Vitro.
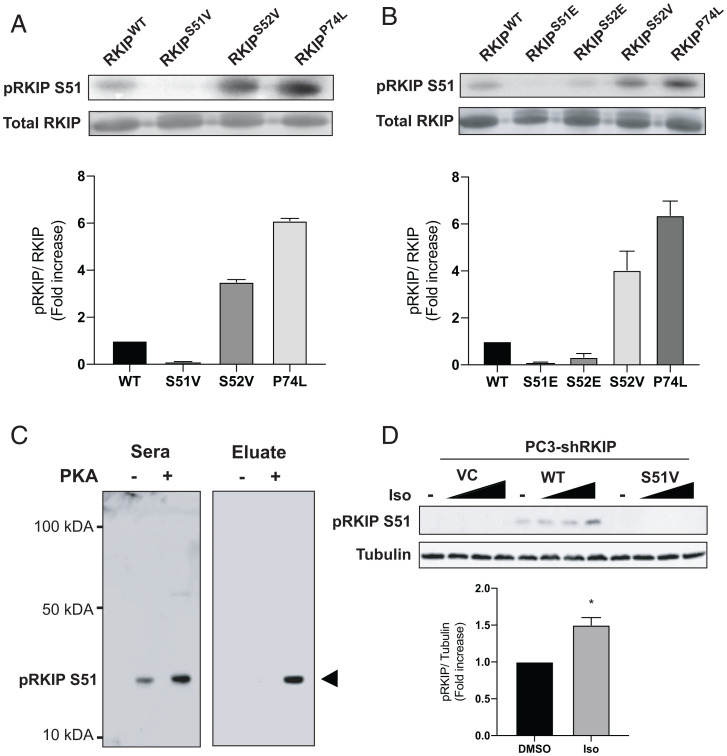
Since RKIP stimulates the β-AR and its downstream effector PKA, we hypothesized that PKA phosphorylates RKIP for feedback regulation. To test whether RKIP is a substrate for PKA, we performed kinase assays using both the recombinant catalytic subunit of PKA (PKA-C) and rat RKIP (16). Remarkably, our assays show that PKA-C consistently phosphorylates wild-type (WT) RKIP (RKIPWT; SI Appendix, Fig. S1) and that increased phosphorylation kinetics is observed for the P74L mutant of RKIP (RKIPP74L). This mutant showed similar behavior when incubated with either PKC or ERK (16). To identify the phosphorylation site, we used PhosphoSite analysis (https://www.phosphosite.org/homeAction.action?jsessionid=55A465C139104A76B09C96FC8702960F), which suggested that S51 and S52 can be both targeted by PKA-C. Indeed, these two residues are also phosphorylated under physiological conditions as detected by mass spectrometry analyses of cellular phosphoproteins (17, 18). To validate this prediction, we mutated both residues S51 and S52 to valines (RKIPS51V and RKIPS52V) and repeated the in vitro kinase assays. Although PKA-C phosphorylates RKIPWT in vitro, we could not detect any phosphorylation for RKIPS51V, confirming that S51 is the primary phosphorylation site (Fig. 1A). Interestingly, mutation of the neighboring serine to valine (S52V) significantly enhanced phosphorylation of RKIP at S51, almost to the extent of RKIPP74L (Fig. 1A), whereas substituting the phosphomimetic S52E suppressed phosphorylation at S51 (Fig. 1B). The enhanced phosphorylation at the P-1 site is consistent with previous kinetic experiments showing that substitutions with hydrophobic residues enhance PKA phosphorylation (19). By contrast, substituting a negatively charged amino acid at S52 or phosphorylation by PKA-C unfavorably affects S51 phosphorylation.
Fig. 1.
Phosphorylation of RKIP by PKA. (A and B) Purified RKIP proteins were tested for in vitro kinase assays. After immunoblotting with anti-RKIP antibody, the phosphorylation levels were quantified using ImageQuant 5.2 software and normalized to RKIP protein levels to compare the phosphorylation between the various mutants. The fold induction shown in the bar graphs is an average of three experiments, and the error bars indicate SEM. (C) Immunoblotting purified WT (PKA-) and PKA-phosphorylated RKIP (PKA+) with unfractionated anti-pS51 antiserum (Sera) and purified antibody (Eluate). Rabbit serum was purified on an anti–phospho-S51 RKIP peptide column as described in Materials and Methods. (D) Immunoblotting of lysates from PC-3 prostate cancer cells treated with isoproterenol (Iso). PC-3 cells that had been stably depleted of RKIP by expression of human RKIP shRNA were transfected with a control vector (VC), RKIPWT, or the S51V RKIP mutant. Cells were lysed, and lysates were immunoblotted with anti-pS51 RKIP antibody or anti-tubulin antibody. For isoproterenol treatment, cells were serum-starved for 20 h and then either treated with DMSO or stimulated with 0.005, 0.05, or 1 μM isoproterenol (Iso) for 10 min. Cells were lysed, and lysates were immunoblotted with anti-pS51 RKIP antibody or anti-tubulin antibody. The protein bands were quantified using Licor ImageStudio, and the results were plotted for pS51 phosphorylation relative to tubulin. Mean values from four independent experiments were plotted, and P values were obtained using a Student’s t test. *P < 0.05.
Finally, we analyzed the phosphorylation kinetics of RKIPWT and RKIPS52V by protein kinase A (SI Appendix, Fig. S2, and Table 1). The Michaelis constant (Km) for RKIPWT was ∼20 μM. Although the corresponding Km value for the S52V mutant was comparable, we observed an overall increase in the phosphorylation kinetics. This discrepancy can be explained by the difference in Vmax, which was approximately fivefold higher for S52V than Vmax for RKIPWT (Table 1). These results show that RKIP phosphorylation follows the typical trends of PKA-C substrates, and S51 is the primary site for PKA-C phosphorylation.
Table 1.
Kinetic parameters for RKIPWT and mutants
| RKIP mutant | Vmax (pmol min−1 mg−1) | Km (μM) |
|---|---|---|
| WT | 1,726 ± 126 | 38 ± 7 |
| P74L | 3,620 ± 163 | 17 ± 2 |
| S52V | 9,852 ± 599 | 55 ± 7 |
| G57K | 1,020 ± 32 | 33 ± 3 |
RKIP Is Phosphorylated at S51 in Cells upon PKA Activation.
Having established that RKIP is a substrate of PKA in vitro, we asked whether PKA activation leads to enhanced phosphorylation of RKIP at S51 in cells. We first generated an antibody against an RKIP peptide containing the pS51 epitope (Materials and Methods). Purification of the antibody using a pS51 peptide column significantly increased antibody specificity toward pS51-RKIP in vitro (Fig. 1C). In PC-3 prostate cancer cells depleted of RKIP by short hairpin RNA (shRNA), the anti-pS51 antibody selectively recognized transfected RKIPWT but not the S51V mutant, which PKA-C cannot phosphorylate in vitro (Fig. 1D). Stimulation of PC-3 cells expressing RKIPWT with isoproterenol, which activates the β-AR, increased S51 phosphorylation by about twofold as shown by Western blotting with the anti-pS51 antibody (Fig. 1D). These results indicate that PKA activation in prostate cancer cells leads to RKIP phosphorylation at S51.
RKIP Phosphorylation at S51 in Cells Enhances PKC Phosphorylation at S153.
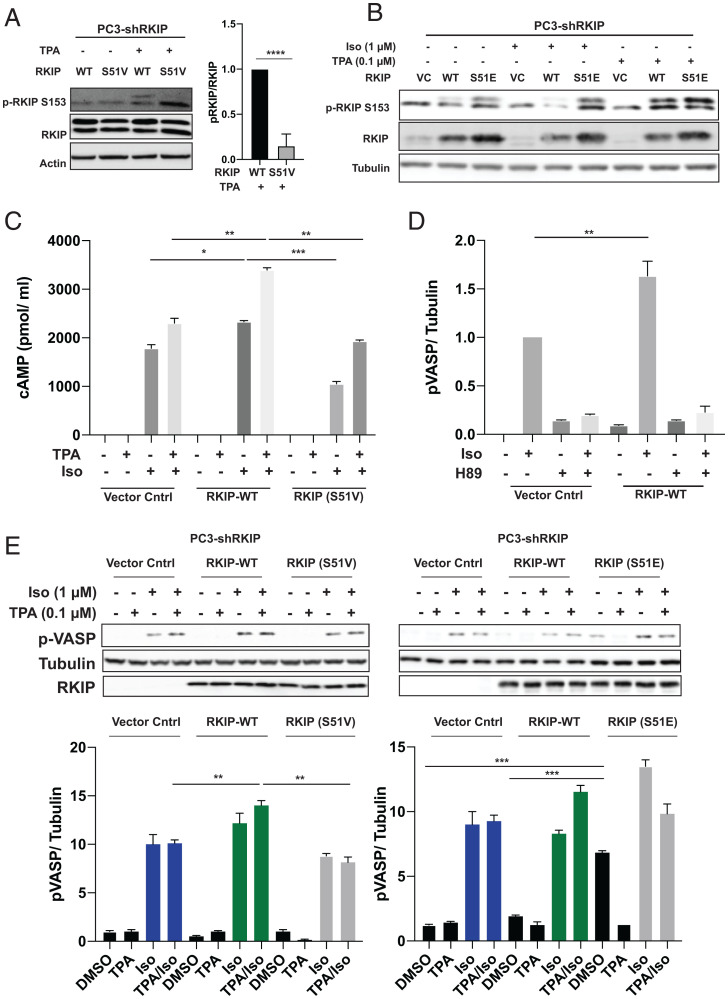
PKC up-regulates the β-AR signaling cascade and, consequently, its primary effector PKA via phosphorylation of RKIP at S153 (10). To assess the cross-talk between the S51 and S153 phosphorylation sites, we used PC-3 cells depleted of endogenous RKIP by shRNA and stably transfected with RKIPWT or RKIPS51V. These cells were serum-starved to reduce background kinase activity and then treated with tetradecanoyl phorbol acetate (TPA) for 5 min to activate PKC. We observed a dramatic decrease of RKIP phosphorylation at S153 for the RKIPS51V mutant, while the phosphorylation at this site is preserved for the WT protein (Fig. 2A).
Fig. 2.
PKA phosphorylates RKIP at Serine 51, increasing both PKC phosphorylation of RKIP at Serine 153 and PKA activity in prostate tumor cells. For A–E, PC-3 cells stably expressing RKIP shRNA were transfected with control vector (VC), WT, or mutant RKIP (S51V, S51E) and serum-starved for 20 h before stimulation using TPA for 30 min, isoproterenol (Iso) for 10 min, or H89 for 30 min as indicated. (A) Phosphorylation of RKIP and mutant S51V RKIP at S153 in PC3 cells after TPA treatment. Cell lysates were immunoblotted with anti-pS153 RKIP antibody or anti-Actin antibody and quantified for plots. The lower band on phosphorylated RKIP and RKIP blots is nonspecific. (B) Phosphorylation of RKIP and mutant RKIP S51E at S513 after TPA and Iso treatment in PC3 cells. Cell lysates were immunoblotted with anti-pS153 RKIP antibody, anti-RKIP antibody, or anti-tubulin antibody. (C) Phosphorylation of S51 RKIP enhances cAMP levels in prostate tumor cells. Total cAMP levels were quantified as in Materials and Methods. (D) Phosphorylation of S51 RKIP enhances PKA activity in prostate tumor cells. Cell lysates were assayed for PKA activity by immunoblotting with anti-pVASP antibody. Samples were normalized to tubulin by immunoblotting with anti-tubulin antibody. (E) An S51V mutant reduces activation of PKA in TPA/Iso-stimulated cells, and S51E potentiates activation of PKA. Cell lysates were assayed for PKA activity by immunoblotting with anti-pVASP antibody. Samples were normalized to tubulin by immunoblotting with anti-tubulin antibody. Samples were quantified using Licor ImageStudio. Mean values from three independent experiments were plotted, and P values were obtained using a Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To confirm that the phosphorylation of S51 promotes PKC phosphorylation at S153, we analyzed the pS153 RKIP phosphorylation profile of the phosphomimetic mutant RKIPS51E. Our analysis showed that RKIPS51E was phosphorylated at S153 with or without TPA stimulation of PKC (Fig. 2B). Since TPA can also activate other proteins, we also pretreated cells with a TPA inhibitor, bisindolylmaleimide (BIS), and showed that the S153 phosphorylation is specific to PKC (SI Appendix, Fig. S3). Interestingly, phosphorylation of S51 does not appear to affect the phosphorylation of ERK, another kinase that is stimulated by TPA (SI Appendix, Fig. S4). As previous studies have shown that phosphorylation of RKIP by PKC results in a localized conformational change (7), these findings are consistent with the possibility that PKC phosphorylation of RKIP also requires a change in RKIP conformational state such as that induced by phosphomimetics or phosphorylation of RKIP at S51 (14).
RKIP Phosphorylation at S51 in Cells Increases Cellular cAMP Levels.
PKC phosphorylation at S153 triggers a switch in RKIP selectivity, from inhibiting Raf to inhibiting GRK2 (10). Decreased GRK2 activity suppresses the desensitization of β-AR and promotes activation of its downstream effectors. One of those effectors is adenylyl cyclase that generates the intracellular second messenger cAMP, an activator of PKA. We therefore analyzed cAMP levels in PC-3-shRKIP cells expressing either RKIPWT, RKIPS51V, or control vector. Upon stimulation with isoproterenol, the cells expressing RKIPWT show increased production of cAMP that is further enhanced by the addition of TPA (Fig. 2C). By contrast, blocking S51 phosphorylation prevents optimal stimulation of cAMP levels by isoproterenol and/or TPA (Fig. 2C). These data confirm that changes in S153 phosphorylation levels triggered by S51 phosphorylation lead to altered second messenger cAMP levels in cells.
RKIP Phosphorylation at S51 Enhances PKA Activity in Prostate Cancer Cells and Cardiomyocytes.
To determine whether S51 phosphorylation of RKIP also alters the behavior of PKA, we assayed PKA activity in vivo using prostate cancer cells. PC-3 cells were depleted of RKIP by shRNA and transfected with control vector, RKIPWT, RKIPS51V, or RKIPS51E mutants. These cells were serum-starved and then treated with isoproterenol and TPA to activate β-AR and PKC activity, respectively. PKA activity in cells was quantified by monitoring the phosphorylation of vasodilator-stimulated phosphoprotein (VASP), a substrate of PKA (20). As previously observed in other cell types (10), PKC-phosphorylated RKIP increases isoproterenol-stimulated PKA activity in PC-3 cells (Fig. 2D). As a control for specificity, we treated the cells with H89, a potent inhibitor of PKA, and found that VASP phosphorylation was dramatically suppressed (Fig. 2D). Notably, the S51V RKIP mutant, which cannot be phosphorylated on position 51, prevented potentiation of PKA activity by PKC in isoproterenol and TPA-stimulated cells (Fig. 2E). By contrast, RKIPS51E increased PKA activity in the presence of isoproterenol without added TPA stimulation (Fig. 2E). These results indicate that RKIP potentiates PKA activity in prostate cancer cells through PKA phosphorylation of RKIP at S51.
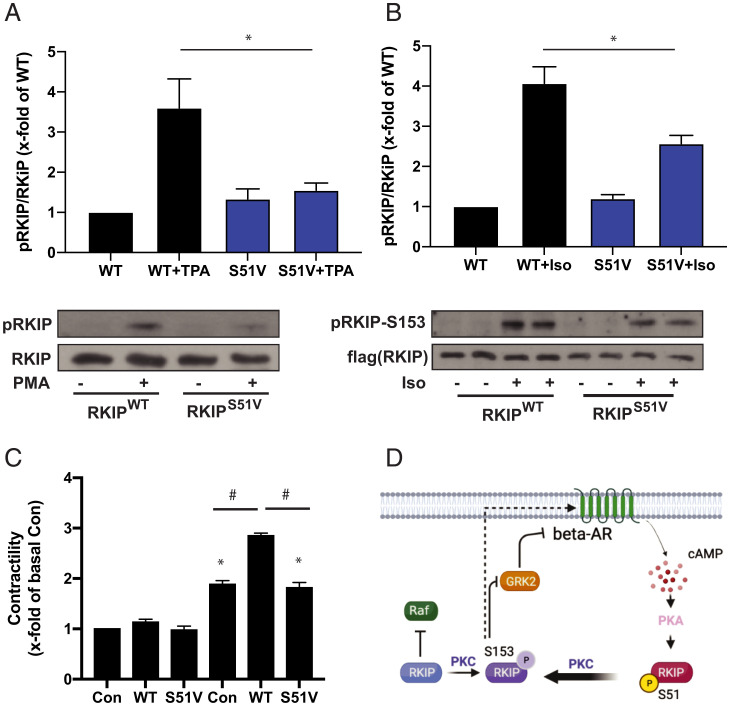
We further tested the effect of blocking PKA phosphorylation of RKIP in primary mouse cardiac myocytes. Under the conditions used for cell culture, the cardiac myocytes have higher basal levels of PKC and contract upon PKA activation (Fig. 3). As observed in PC-3 cells, the RKIP mutation S51V significantly reduced PKC phosphorylation at S153 (Fig. 3A). The S51V mutant also significantly suppressed PKC phosphorylation of S153 in response to isoproterenol treatment (Fig. 3B). Similarly, expression of RKIPS51V reduced cardiac myocyte contractile activity to control levels upon isoproterenol stimulation relative to RKIPWT (Fig. 3C). These results show that PKA phosphorylation of S51 on RKIP promotes PKC phosphorylation of RKIP at S153 and subsequent enhanced activation of PKA, leading to contraction in primary cardiac myocytes. Taken together, these results indicate that RKIP potentiates PKA activity through a positive feedback loop involving PKA phosphorylation of RKIP at S51. Phosphorylated S51 enhances PKC phosphorylation of RKIP at S153, triggering the RKIP phospho-switch. Resultant GRK2 inhibition by S153 RKIP and β-AR activation increases adenylyl cyclase activity and cAMP production and, ultimately, triggers further PKA activation (Fig. 3D).
Fig. 3.
Positive feedback loop between RKIP and PKA leads to enhanced contractility in cardiac myocytes. (A) Cardiac myocytes were adenovirally transduced with WT or S51V RKIP, and treated with or without TPA (PMA). Cell lysates were immunoblotted for RKIP or pS153 RKIP. (B) Cardiac myocytes were adenovirally transduced with WT or S51V RKIP and then treated with or without isoproterenol. Cell lysates were immunoblotted for RKIP or pS153 RKIP. (C) Cardiac myocytes were adenovirally transduced and treated with isoproterenol as in B. For the control (Con), LacZ was adenovirally transduced. Contractility was measured as described in Materials and Methods. Mean values from five (A) or six (B) independent experiments were plotted, and P values were obtained using a one-way ANOVA and Sidak’s post hoc test (WT + PMA vs. V51 + PMA: P = 0.0178 and WT + ISO vs. V51 + ISO: P = 0.0016). (C) Mean values from 20 replicates of five independent experiments were plotted, and P values were obtained using a one-way ANOVA and Tukey’s post hoc test (*P < 0.01 vs. Con-Iso and #P < 0.0001 as indicated). (D) Scheme depicting positive feedback loop between RKIP and PKA. RKIP phosphorylated at S153 by PKC inhibits GRK2, reducing down-regulation of the β-adrenergic receptor (β-AR). β-AR activates PKA, which then phosphorylates RKIP at S51 leading to enhanced phosphorylation by PKC at S153.
Biolayer Interferometry and NMR Confirm the Binding Interaction between RKIP and PKA.
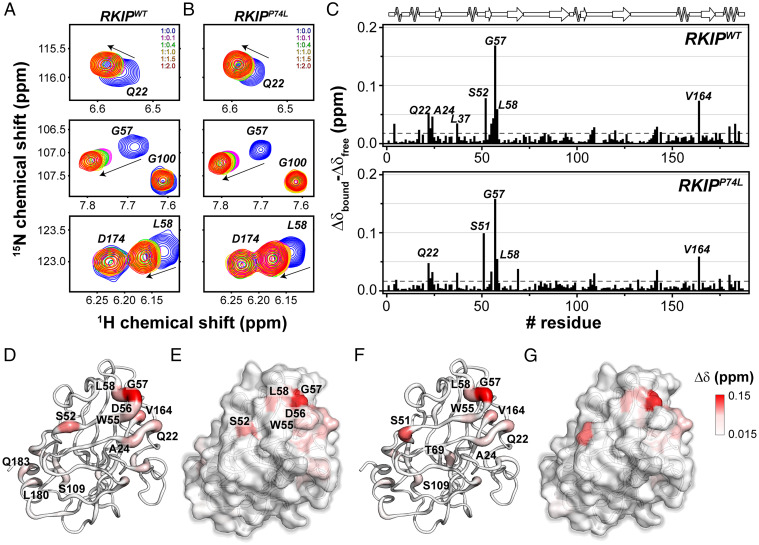
To estimate RKIP/PKA binding affinity, we expressed and purified both recombinant RKIP and His-tagged PKA catalytic subunit (PKA-C). The binding affinity was quantified using biolayer interferometry (BLI) in the presence and absence of an ATP-analog (ATPγN) (SI Appendix, Fig. S5, and Table 2). Upon fitting the binding curves, we found that RKIPWT has a dissociation constant (Kd) of ∼36 μM. Upon saturation with ATPγN, the RKIP affinity is enhanced by sixfold (∼6 μM). This effect is due to the K-type positive cooperativity between nucleotide and substrate typical of PKA-C (21, 22). The saturation of the kinase with the nucleotide shifts the ensemble of conformations of the apo state toward an intermediate state that binds the substrate with higher affinity in a 1:1 complex (21, 22). The residue-specific interactions between the kinase and RKIP were monitored using NMR spectroscopy. The unlabeled PKA-C saturated with ATPγN was titrated with uniformly 15N-labeled RKIP (U-15N-RKIP). The amide fingerprint of RKIP was analyzed using [1H-15N] heteronuclear single quantum coherence ([1H-15N]-HSQC) experiment (SI Appendix, Fig. S6). Upon binding PKA-C, several residues of RKIPWT showed marked chemical shift perturbations (CSPs) (Fig. 4A and SI Appendix, Fig. S6A). The most significant CSPs are observed for residues proximal to the putative phosphorylation site (S51) (Fig. 4C). When mapped onto the X-ray structure of RKIP, these residues populate one face of RKIP, with a few long-range (allosteric) effects observed for Q183 and L180 (Fig. 4 D and E). We repeated the NMR titration experiment with the P74L-RKIP mutant (RKIPP74L) (Fig. 4B and SI Appendix, Fig. S7). Notably, despite the marked difference in the extent of phosphorylation, the RKIPP74L mutant shows a CSP profile similar to RKIPWT (Fig. 4 B, C, F, and G). Residues like G57, L58, and Q22 show higher CSP, confirming that PKA-C engages identical residues for both proteins. Mutation of G57 to K (RKIPG57K) decreased RKIP phosphorylation by PKA relative to RKIPWT and reduced the Vmax by almost 50% (SI Appendix, Fig. S8, and Table 1), confirming the importance of the G57 residue in PKA phosphorylation of RKIP.
Table 2.
Kinetics parameters of the binding of RKIPWT to apo PKA-C and ATPγN-saturated PKA-C
| Ligand | kon [1/Ms] | koff [1/s] | Kd [μM] | Cooperativity σ | |
|---|---|---|---|---|---|
| Apo PKA-C | RKIPWT | 104 ± 3 | (3.78 ± 0.08) × 10−3 | 36.4 ± 0.7 | – |
| PKA-C/ATPγN | RKIPWT | 199 ± 4 | (1.23 ± 0.03) × 10−3 | 6.2 ± 0.7 | 6 |
Fig. 4.
NMR map of the residues of RKIPWT and P74L mutant involved in PKA-C/RKIP interaction interface. (A and B) [1H,15N]-HSQC spectra showing the backbone chemical shift changes of selected resonances of (A) RKIPWT and (B) RKIPP74L upon titration of increasing amount of PKA-C/ATPγN complex. (C) CSP of the amide fingerprint of RKIPWT and RKIPP74L mutant alone (Δδfree) and in a 1:2 complex with PKA-C/ATPγN (Δδbound) vs. residues, calculated using Eq. 1. The residues that show a CSP value two times greater than 1 SD from the average CSP (gray dashed line) are indicated in the histogram. (D–G) Cartoon (D) and surface (E) mapping of the amide CSP of RKIPWT or cartoon (F) and surface (G) of P74L mutant onto the three-dimensional structure (Protein Data Bank [PDB] ID 2IQY). The cartoons and the surfaces are colored according the CSP values ranging from 0.015 to 0.15 ppm.
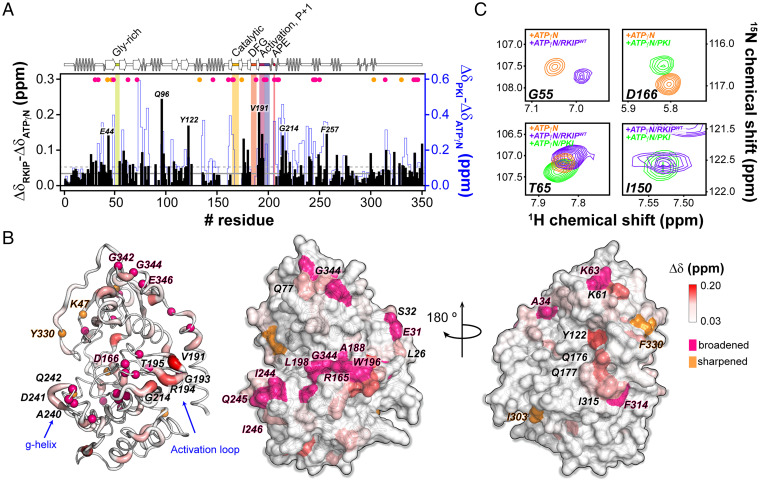
We repeated the titrations inverting the labeling scheme to detect the U-15N PKA-C fingerprint upon binding unlabeled RKIPWT (Fig. 5 and SI Appendix, Fig. S9). The amide fingerprint of PKA-C changes significantly upon nucleotide and RKIPWT binding (23–25). Several resonances belonging to the catalytically critical loops of PKA-C, such as the Gly-rich loop, activation loop, DFG loop, and the catalytic loop, broaden beyond detection, suggesting the presence of an intermediate conformational exchange on the chemical shift time scale (SI Appendix, Table S1). The residues surrounding the activation loop and the β9-sheet experience the largest CSP. Overall, the RKIP CSP profile revealed a typical trend observed for substrates and pseudosubstrates of the kinase (22, 24–26). A close comparison between the CSP profiles of RKIP and the pseudosubstrate PKI shows similar chemical shift changes for several PKA-C domains. However, the extent of the CSP for RKIP is attenuated (Fig. 5A). The CSP map on the X-ray structure of PKA-C shows that most of the chemical shift changes and the resonance broadening occur on the face of PKA-C that harbors the substrate-binding site at the interface between the small and large lobe (Fig. 5 B and C). For both BLI and NMR experiments, we utilized ATPγN to prevent hydrolysis. However, the affinity of ATPγN for the kinase is lower than ATP. Consequently, the cooperativity coefficient σ between nucleotide and substrate is significantly lower. For ATP and PKI, σ is ∼400, while for ATPγN and PKI, σ is ∼100 (21, 27). The latter may explain the discrepancy between the in vitro binding studies and the in-cell assays.
Fig. 5.
NMR mapping of the PKA-C residues involved in the PKA-C/RKIP interactions. (A) CSPs of the PKA-C/ATPγN fingerprint upon interaction with RKIPWT (black) and PKI. The shaded colors indicate the Gly-rich loop (green), catalytic loop (yellow), DFG loop (orange), activation loop (purple), P + 1 loop (blue), and APE (red). (B) Chemical shift changes mapped onto the PKA-C structure (PDB ID 4wb5). The hot pink spheres highlight residues that broaden out, while the orange spheres indicate those residues that sharpened out upon interacting with RKIPWT. (C) [1H,15N]-TROSY-HSQC spectra overlay for specific residues of PKA-C in complex with ATPγN (orange), ATPγN/RKIP (purple), and ATPγN/PKI (green).
Discussion
We discovered that RKIP is a substrate of PKA-C and functions in a positive feedback loop to potentiate PKA activity via β-AR signaling. An essential feature of this feedback loop is the phosphorylation of RKIP at S153, which inhibits GRK2 activity, limiting down-regulation of the β-AR (10). Notably, PKA phosphorylation of RKIP at S51 further increases pS153 RKIP levels, promoting the β-AR response and enhancing PKA activity. These functional effects identify components in a circuit that generates an enhanced activation of PKA function (Fig. 3D). Since RKIP inhibits Raf and the downstream target of Raf, MAP kinase, phosphorylates RKIP at T42 (16), this type of feedback control may be a general mechanism by which RKIP regulates the kinome in cells.
The relatively small size of RKIP and correspondingly high-resolution HSQC spectra afford a unique opportunity to investigate the interaction of PKA-C with a physiological substrate. Although the elegant studies of the crystal structures of PKA-C have served as models to explain the mechanism of action for the entire kinase family, the vast majority of the kinase crystals have been obtained in the presence of potent PKA inhibitors (28, 29). Only recently have PKA-C interactions been captured with the cytoplasmic domain of the physiological substrate phospholamban (24, 29). The CSP enabled us to identify the PKA-C/RKIP interaction interface. Unlike other peptide substrates, the interaction interface is larger as RKIP contacts both the small and large lobe. As observed with other peptide substrates, the main sites affected by the interactions are located near the glycine-rich loop and the catalytic cleft as well as the peptide-positioning loop. It is likewise striking that more distal interactions also seem to engage the backbone of PKA-C at A-helix. However, these interaction studies do not fully explain the mechanism by which phosphorylation of RKIP at S51 potentiates S153 phosphorylation. Previous studies on the P74L mutant of RKIP suggest the presence of long-range effects that destabilize the pocket loop, augmenting kinase binding (16). It is possible that the introduction of negative charges on pS51 may lead to disruption and increased loop flexibility, promoting PKC binding to RKIP and increased S153 phosphorylation. In addition, it is quite interesting to see that the P74L mutant does not show major structural differences with the RKIPWT. The NMR CSP cannot differentiate between PKA-C binding to this mutant and RKIPWT, but there are significant differences in the Kms suggesting a difference in the binding interactions (Table 1). This might be explained by the presence of a high-energy state associated with the P74L mutant (30). More in-depth analyses will be required to elucidate the detailed molecular mechanism.
Our results suggest that PKA phosphorylation of RKIP at S51 has direct functional consequences. In both prostate tumor cells and cardiac myocytes, the S51V mutation blocks RKIP-mediated PKA activation. Furthermore, RKIP potentiation of cardiomyocyte contractility in response to isoproterenol-activated PKA requires S51 phosphorylation. Finally, we show that the S51V mutation also inhibits S153 phosphorylation by PKC. Previous studies have demonstrated that mutation of RKIP at the PKC phosphorylation site to S153A prevents RKIP from improving cardiac contractility and increases cardiomyocyte death and collagen deposition (31). Together, these studies suggest that phosphorylation of RKIP at both S51 and S153 are essential regulators of cardiac integrity.
Since cAMP is a key second messenger in the β-AR/PKA pathway, its levels before and after stimulus need to be strictly controlled (32); therefore, PKA is subject to negative and positive feedback regulation. There are several mechanisms for ensuring negative feedback, including down-regulation of adenylyl cyclase by PKA, depletion of cAMP by phosphodiesterases, and restricted spatial localization directed by AKAPs (33). Interestingly, direct phosphorylation of GRK2 by kinases such as PKC and even PKA also promotes negative regulation of PKA by increasing GRK2 activity (34, 35). By contrast, RKIP acts to promote PKA activity. This is accomplished by at least two mechanisms: increasing PKC activity to enhance phosphorylation of S153 RKIP and positive feedback by PKA to potentiate S153 phosphorylation. Generation of an optimal, relatively noise-free output signal such as PKA activity requires a combination of fast and slow positive feedback loops (1). Whether RKIP generates such discrete PKA outputs or plays other roles in combination with negative feedback to fine-tune homeostatic control will require further analysis of these intricate regulatory circuits. Either way, the discovery of the direct interaction between PKA and its substrate RKIP reveals another regulatory mechanism for PKA activity.
Materials and Methods
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed as described in the QuikChange II Site-Directed Mutagenesis kit instruction manual (Agilent Technologies). Briefly, the oligonucleotides used in these polymerase chain reactions (PCRs) were designed using the QuikChange Primer Design program available on the website of Agilent Technologies Inc. PCR was carried out in the presence of 15 ng of rat RKIP cDNA previously cloned into pGEX2T vector, 125 ng of each oligonucleotide primer, dNTP mix, and 2.5U of PfuUltra High Fidelity DNA polymerase in a total volume of 50 μL. The amplified DNA products were then used to transform XL1-Blue cells by subjecting the cells to heat shock treatment at 42 °C for 45 s. Plasmid DNA isolated from bacterial colonies grown and selected on LB agar plates containing ampicillin (50 μg/mL) was then sequenced to confirm the identity of the RKIP mutations.
Characterization of RKIP Binding Proteins.
HeLa cells stably transfected with GFP-RKIP were grown on 50 dishes (100 mm). Cells were lysed, and GFP–RKIP complexes were isolated using anti-GFP antibody bound to beads as previously described (36).
Expression and Isolation of Recombinant RKIP Mutants for In Vitro Kinase Assays.
BL21–Gold (DE3) pLysS cells transformed with pGEX-2T containing rat RKIP cDNA were grown at 37 °C in LB medium in the presence of ampicillin. Recombinant RKIP expression was induced with 0.4 mM IPTG at 30 °C for 4 h. The cell pellet collected was resuspended in PBS buffer and sonicated. This was followed by the addition of Triton-X-100 to a final concentration of 1% (vol/vol). The cell suspension was centrifuged at 17,200 × g for 20 min at 4 °C, and the supernatant was subjected to affinity chromatography on GSH Sepharose 4B resin (GE Healthcare) to allow the binding of GST-RKIP fusion proteins. In order to cleave the RKIP portion of the fusion protein from the GST portion bound to the resin, the resin was treated with biotinylated thrombin (EMD Millipore) at 4 °C for 16 h. Following thrombin-mediated digestion, eluate and washes were collected from the GSH Sepharose 4B resin and were subsequently mixed with a suspension of streptavidin agarose resin (EMD Millipore) to separate the biotinylated thrombin from RKIP. The RKIP-containing eluate and washes from the streptavidin agarose resin was dialyzed into 50 mM Tris⋅HCl, 100 mM NaCl, pH 7.40 solution at 4 °C.
Expression and Purification of RKIPWT and RKIPP74L for the NMR and BLI Studies.
Gene coding for WT rat RKIP (Rattus norvegicus, PEBP1) was subcloned into a pET SUMO vector (Thermo Fisher) and was used as a template to generate the P74L using the QuikChange Lightning mutagenesis kit (Agilent genomics). Escherichia coli BL21(DE3) pLysS cells (Invitrogen), transformed with RKIPWT or RKIPP74L-pET SUMO vector, were cultivated at 30 °C to an optical density (OD600) of 1.1, then protein expression was induced with 0.4 mM IPTG. For the BLI experiments, the cells were cultured in standard LB, while for the NMR experiment, the cells were grown in M9 minimal media containing 15NH4Cl (Cambridge Isotope Laboratories Inc.). The cells were harvested, suspended in 50 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, 20% sucrose, 0.15 mg/mL lysozyme, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM b-mercaptoethanol, and lysed using French press at 1,000 psi. The lysate was then centrifuged at 60,000 × g for 30 min, and the supernatant was batch-bound with Ni2+-NTA agarose affinity resin (Thermo Fisher), previously equilibrated with lysis buffer. The fusion protein was eluted using 250 mM of imidazole. The SUMO-tag was then cleaved out using a stoichiometric amount of recombinant UPL1 protease in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, 0.5 mM PMSF. The cleavage reaction was performed overnight at 4 °C, and all the contaminants were eliminated by performing a second purification using Ni2+-NTA agarose resin. The flow-through, containing the untagged-RKIP, was collected, and an additional size exclusion (SEC) purification step was performed with a 16/60 HiLoad Superdex 200 resin (GE Healthcare) using 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 2 mM dithiothreitol (DTT) as a mobile phase. The purified protein was concentrated and stored at 4 °C in the SEC buffer supplied with protease inhibitors. The purity was assessed using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the final purity was >98%.
Expression and Purification of Human PKA-C.
Recombinant catalytic subunit Cα (human gene) of PKA (PKA-C) was expressed and purified as reported (27). Briefly, heterologous PKA-C overexpression was induced with 0.4 mM IPTG and carried out overnight at 20 °C in E. coli BL21 (DE3) pLysS cells (Invitrogen), cultured in M9 minimal media supplemented with 15NH4Cl (Cambridge Isotope Laboratories Inc.) or in LB media. The cell pellet was collected, lysed in 50 mM Tris⋅HCl, pH 8.0, 30 mM KH2PO4, 50 mM NaCl, 200 μM ATP, 5 mM b-mercaptoethanol, 0.15 mg/mL lysozyme, 100 U/mL Dnase I (Roche Applied Science), and 1 tablet of cOmplete ULTRA EDTA-FREE protease inhibitors (Roche Applied Science) using a French press. The first purification step was carried out using Ni2+ affinity chromatography. The fraction containing His(6x)-PKA-C was collected, dialyzed against 20 mM KH2PO4, pH 6.5, 25 mM KCl, 0.1 mM PMSF, and 5 mM β-mercaptoethanol and cleaved using stoichiometric quantities of recombinant TEV (except when noted otherwise). An additional purification step was performed using a HiTrap SP HP cation exchange column (GE Healthcare Biosciences Corp.) to separate the three isoforms of PKA-C that differ in their phosphorylation profiles (37). The purified protein was then stored in phosphate buffer containing 10 mM DTT, 10 mM MgCl2, and 1.0 mM NaN3, at 4 °C. The protein activity was assessed using the PepTag Non-Radioactive Protein Kinase assay (Promega) and quantified using A280 = 52,060 M−1 cm−1.
Purification of Antiphosphoserine51 RKIP.
Rabbit anti-phosphoserine51 RKIP sera were obtained by immunizing the animals with phosphopeptide, TQVMNRPpSSIS, corresponding to residues 44 to 54 of rat RKIP. A total of 750 μL of rabbit sera was mixed with 50 μL of Sepharose beads conjugated to the phosphopeptide and preequilibrated with 10 mM Tris⋅HCl, pH 7.5, in a column under gentle agitation at 4 °C overnight. The following day, the flow-through and wash fractions were collected, and the antibody was eluted with four column volumes of 100 mM glycine, pH 2.50, into a tube that contained 40 μL of 1 M Tris⋅HCl, pH 8.0, with 6 mg/mL BSA.
In Vitro Kinase Assays.
The kinase assay was initiated by adding recombinant PKA-C to a final concentration of 100 nM in a reaction containing 50 mM Tris⋅HCl (pH 7.50), 10 mM MgCl2, RKIP protein, 150 μM ATP containing 10 μCi of [γ-32P] ATP (Perkin-Elmer). The total reaction volume was 35 μL. Incubation was carried out at 30 °C for 30 min, and the reaction was terminated by adding 12 μL 4× SDS-PAGE gel loading buffer followed by heating at 95 °C for 5 min. The phosphorylated protein products were resolved on a 12.5% reducing polyacrylamide gel, and the proteins were transferred onto a nitrocellulose membrane (GE Healthcare). The labeled proteins were visualized using a phospho-imager (Storm 860, GE Healthcare) and quantified using IMageQuant 5.2 (GE Healthcare). Normalization for protein loading was determined using Western blotting probed with a rabbit anti-RKIP antibody generated in the laboratory and detected with a secondary infrared labeled goat anti-rabbit antibody (LI-COR Biosciences). Protein loading was quantified using Image Studio ver2.1 software (LI-COR Biosciences).
Generation of PC3 Cells Stably Expressing RKIPWT and RKIP Mutants.
PC3 cells were first depleted of endogenous RKIP expression through transduction of lentiviral vectors carrying shRKIP as previously described (12). These cells were subsequently transduced according to the manufacturer’s instructions with HA-RKIPWT or HA-RKIPS51V cloned into the pCDH1-CMV-MCS1-EF1 lentiviral vector (Systems Bioscience). PC3 cells transduced with empty vectors served as controls.
Cell Treatment and Immunoblotting.
PC3 cells stably expressing human shRKIP were then stably transduced with lentivirus to overexpress either rat RKIPWT, RKIPS51V, or the pCDH vector. To investigate PKA activation, the cells were serum starved for about 20 h followed by stimulation with 5 nM isoproterenol for 10 min. To suppress PKA activity, cells were pretreated with 10 μM H89 for 30 min prior to exposure to isoproterenol. The cells were washed with PBS twice and lysed with RIPA buffer containing phosphatase and protease inhibitors (Mixture Set III from Calbiochem, cat 539134). Cell lysates were subjected to electrophoresis on a 12.5% SDS-PAGE gel and immunoblotted with primary antibody followed by secondary antibody conjugated to IRDye fluorophore in PBS/LI-COR blocking reagent/0.1% Tween 20/0.01%SDS for 1 h at room temperature. The membrane was analyzed on a LI-COR Odyssey CLx Imager or Odyssey Fc Imager using Image Studio ver2.1 software. The following primary antibodies were used: mouse anti-alpha tubulin (1:10,000, Santa Cruz), rabbit anti-phosphoserine157 VASP (1:1,000, Thermo Fisher), rabbit anti-phosphoserine51 RKIP (1: 5, prepared as described in Purification of Antiphosphoserine51 RKIP in Materials and Methods), rabbit anti-phosphoserine153 RKIP (1:4,000, prepared as described in ref. 13, 1:3,000 Abcam no. ab75971), anti-RKIP (1:1,000 Abcam no. ab76582), anti-ERK (1:1,000 Cell Signaling Technology no. 4695), anti-phosphorylated Thr202/Tyr204 ERK (1:1,000 Cell Signaling Technology no. 4370), and anti-Actin (1:3,000 Sigma Aldrich no. SAB5600204). Secondary antibodies used were goat anti-mouse IgM IRDye 800CW (1:10,000), goat anti-rabbit IRDye 680RD (from 1:8,000 to 1:10,000), mouse anti-rabbit IgG-HRP (1:5,000 Cell Signaling Technology no. 31464). Blots were imaged using Odyssey XF Imaging system or iBright FL1500 Imaging System (Thermo Fisher).
Cyclic AMP Assays.
Lysates of PC-3 cells expressing RKIPWT, RKIPS51V, or control vector were collected for cAMP quantification using a colorimetric ELISA method according to the manufacturer’s instruction (cAMP ELISA Kit ab133501, Promega). Diluted cell lysates (1:20, 1:100, and 1:200) were added into a 96-well plate coated with a goat anti-rabbit IgG antibody that reacts with an anti-cAMP antibody. Free cAMP in samples bound to the cAMP antibody on the plate after competition with alkaline phosphatase (AP)-labeled cAMP was washed for absorbance at 405 nm. Relative levels of cAMP were calculated based on standard curve using free cAMP.
Neonatal Cardiomyocytes and Assessment of Cardiomyocyte Contractility.
Neonatal rat cardiomyocytes were isolated as described previously (31). Cardiomyocytes were transduced adenovirally (31). To monitor the contractile activity of cardiomyocytes, live cells were examined on a Zeiss Axiovert 135 microscope. Cells were held thermostatically at 33 °C in MEM, and the number of beats per minute was determined before and 2 min after stimulation with 50 nM isoprenaline by an observer who was blind to the experimental protocol studied (10).
NMR Experiments.
NMR measurements were performed on a Bruker Avance spectrometer operating at a 1H Larmor frequency of 700 MHz equipped with a cryogenic probe or on a Bruker Avance III 850 MHz spectrometer equipped with a TCI cryoprobe. All data were acquired at 300 K. The typical NMR sample contained 20 mM KH2PO4 (pH 6.5), 90 mM KCl, 10 mM MgCl2, 10 mM DTT, 1 mM NaN3, and 10% D2O. The concentrations of U-15N labeled RKIPWT and P74L mutant were 100 μM. For the NMR titration, a standard [1H-15N] HSQC pulse sequence (38) was used, and five titration points (1:0.1, 0:0.4, 1:1, 1:1.5, and 1:2 RKIP:PKA-C/ATPγN molar ratio) were collected. The total number of scans was 48, with 2,048 and 128 complex points for 1H and 15N, respectively. All data were processed using NMRPipe (39) and visualized using NMRFAM-Sparky (40, 41). CSPs were measured using amide proton and nitrogen resonances for each residue using Eq. 1:
| [1] |
in which Δδ is the compounded CSP; ΔδΗ and ΔδN are the differences of 1H and 15N chemical shifts, respectively, between the first and last point of the titration; and 0.154 is the scaling factor for nitrogen (42). Standard [1H, 15N]-TROSY-HSQC experiment (43) was used to record the amide fingerprint spectra of PKA-C/ATPγN/RKIPWT ternary complex. The total number of scans was 96, with 2,048 and 128 complex points for 1H and 15N, respectively. The sample consisted of 100 μM 15N-PKA-C, 0.12 mM unlabeled-RKIPWT (1:1.2 molar ratio), and 120 times excess of ATPγN (12 mM).
BLI.
BLI experiments were performed using a BLItz biosensor system (ForteBio, Pall) at 20 °C. Uncleaved His(6x)-PKA-C (1 μM) was resuspended in 20 mM MOPS (pH 6.5), 90 mM KCl, 10 mM MgCl2, 10 mM DTT, and 1 mM NaN3 and immobilized on the Ni-NTA (NTA) Biosensors (ForteBio, Pall). RKIPWT was dialyzed in the same buffer, diluted to a final concentration of 280 μM, and applied in a dose-dependent manner (1:35, 1:70, 1:140, and 1:280 molar ratio) to the biosensor containing the immobilized PKA-C. For the binding of RKIPWT to nucleotide-saturated/PKA-C, the buffer was supplied with 2 mM of ATPγN (2,000 times excess), to prevent the phosphorylation of RKIPWT. Triton-X (0.05%) was also added to the buffer to avoid nonspecific protein interaction. Parallel experiments were performed for reference where the sensor was titrated with buffer to the immobilized PKA-C (with or without ATPγN), and these resulting signals were subtracted during data analysis. The association and dissociation periods were set to 300 and 450 s, respectively. The data were analyzed using the software provided by the instrument and plotted using GraphPad Prism 9 software package (GraphPad Software, Inc.). All the experiments were performed in triplicate to determine the experimental error.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM087630 and GM121735 to M.R.R., NIH grant GM100310 to G.V., and the Deutsche Forschungsgemeinschaft ( grants SFB1116/A09 and SFB/TR296/P10) to K.L. NMR experiments were carried out at the Minnesota NMR center. We thank Ya Chen and Arnold Satterthwait from the Sanford–Burnham Medical Research Institute, La Jolla, CA, for generously providing pS51-RKIP peptide for antibody isolation. We thank John Skinner and Jonggul Kim for helpful discussions and Long Nguyen for assistance with the figures.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121867119/-/DCSupplemental.
Data Availability
Raw data from BLI experiments and corresponding analysis, along with proton and amide chemical shift list files of RKIPWT and RKIPP74L free and in complex with PKA-C, have been deposited in the publicly accessible Data Repository for the University of Minnesota (DRUM) and can be accessed at the following link: https://conservancy.umn.edu/handle/11299/225840. All data needed to evalute the conclusions in the paper are present in the paper and/or SI Appendix.
References
- 1.Brandman O., Ferrell J. E. Jr., Li R., Meyer T., Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310, 496–498 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenz K., Rosner M. R., Brand T., Schmitt J. P., Raf kinase inhibitor protein: Lessons of a better way for β-adrenergic receptor activation in the heart. J. Physiol. 595, 4073–4087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yesilkanal A. E., Rosner M. R., Targeting Raf kinase inhibitory protein regulation and function. Cancers (Basel) 10, E306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dangi-Garimella S., et al. , Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 28, 347–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun J., et al. , Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 30, 4500–4514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Z., et al. , Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate 66, 248–256 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Zhao J., et al. , 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14246–14251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okita K., et al. , Analysis of DNA variations in promoter region of HCNP gene with Alzheimer’s disease. Biochem. Biophys. Res. Commun. 379, 272–276 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Wright K. T., Vella A. T., RKIP contributes to IFN-γ synthesis by CD8+ T cells after serial TCR triggering in systemic inflammatory response syndrome. J. Immunol. 191, 708–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz K., Lohse M. J., Quitterer U., Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature 426, 574–579 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Yeung K., et al. , Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Trakul N., Menard R. E., Schade G. R., Qian Z., Rosner M. R., Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J. Biol. Chem. 280, 24931–24940 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Corbit K. C., et al. , Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J. Biol. Chem. 278, 13061–13068 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Skinner J. J., et al. , Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome. Proc. Natl. Acad. Sci. U.S.A. 114, 13453–13458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yesilkanal A. E., et al. , Limited inhibition of multiple nodes in a driver network blocks metastasis. eLife 10, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granovsky A. E., et al. , Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol. Cell. Biol. 29, 1306–1320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffert J. D., Pisitkun T., Wang G., Shen R. F., Knepper M. A., Quantitative phosphoproteomics of vasopressin-sensitive renal cells: Regulation of aquaporin-2 phosphorylation at two sites. Proc. Natl. Acad. Sci. U.S.A. 103, 7159–7164 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demirkan G., Yu K., Boylan J. M., Salomon A. R., Gruppuso P. A., Phosphoproteomic profiling of in vivo signaling in liver by the mammalian target of rapamycin complex 1 (mTORC1). PLoS One 6, e21729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams J. A., Taylor S. S., Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry 31, 8516–8522 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Harbeck B., Hüttelmaier S., Schluter K., Jockusch B. M., Illenberger S., Phosphorylation of the vasodilator-stimulated phosphoprotein regulates its interaction with actin. J. Biol. Chem. 275, 30817–30825 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Kim J., Li G., Walters M. A., Taylor S. S., Veglia G., Uncoupling catalytic and binding functions in the cyclic AMP-dependent protein kinase A. Structure 24, 353–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., et al. , Globally correlated conformational entropy underlies positive and negative cooperativity in a kinase’s enzymatic cycle. Nat. Commun. 10, 799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masterson L. R., Mascioni A., Traaseth N. J., Taylor S. S., Veglia G., Allosteric cooperativity in protein kinase A. Proc. Natl. Acad. Sci. U.S.A. 105, 506–511 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masterson L. R., et al. , Dynamics connect substrate recognition to catalysis in protein kinase A. Nat. Chem. Biol. 6, 821–828 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivieri C., et al. , Multi-state recognition pathway of the intrinsically disordered protein kinase inhibitor by protein kinase A. eLife 9, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivieri C., et al. , Defective internal allosteric network imparts dysfunctional ATP/substrate-binding cooperativity in oncogenic chimera of protein kinase A. Commun. Biol. 4, 321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker C., et al. , Cushing’s syndrome driver mutation disrupts protein kinase A allosteric network, altering both regulation and substrate specificity. Sci. Adv. 5, eaaw9298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor S. S., Zhang P., Steichen J. M., Keshwani M. M., Kornev A. P., PKA: Lessons learned after twenty years. Biochim. Biophys. Acta 1834, 1271–1278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masterson L. R., et al. , cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J. Mol. Biol. 412, 155–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skinner J. J., Rosner M. R., RKIP structure drives its function: A three-state model for regulation of RKIP. Crit. Rev. Oncog. 19, 483–488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid E., et al. , Cardiac RKIP induces a beneficial β-adrenoceptor-dependent positive inotropy. Nat. Med. 21, 1298–1306 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Vandamme J., Castermans D., Thevelein J. M., Molecular mechanisms of feedback inhibition of protein kinase A on intracellular cAMP accumulation. Cell. Signal. 24, 1610–1618 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Pidoux G., Taskén K., Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J. Mol. Endocrinol. 44, 271–284 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Krasel C., et al. , Phosphorylation of GRK2 by protein kinase C abolishes its inhibition by calmodulin. J. Biol. Chem. 276, 1911–1915 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Murthy K. S., Mahavadi S., Huang J., Zhou H., Sriwai W., Phosphorylation of GRK2 by PKA augments GRK2-mediated phosphorylation, internalization, and desensitization of VPAC2 receptors in smooth muscle. Am. J. Physiol. Cell Physiol. 294, C477–C487 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Cristea I. M., Williams R., Chait B. T., Rout M. P., Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Yonemoto W., Garrod S. M., Bell S. M., Taylor S. S., Identification of phosphorylation sites in the recombinant catalytic subunit of cAMP-dependent protein kinase. J. Biol. Chem. 268, 18626–18632 (1993). [PubMed] [Google Scholar]
- 38.Bodenhausen G., Ruben D. J., Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 (1980). [Google Scholar]
- 39.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Goddard D., Kneller D. G., Sparky 3 (University of California, San Francisco, 2004). [Google Scholar]
- 41.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williamson M. P., Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Nietlispach D., Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J. Biomol. NMR 31, 161–166 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data from BLI experiments and corresponding analysis, along with proton and amide chemical shift list files of RKIPWT and RKIPP74L free and in complex with PKA-C, have been deposited in the publicly accessible Data Repository for the University of Minnesota (DRUM) and can be accessed at the following link: https://conservancy.umn.edu/handle/11299/225840. All data needed to evalute the conclusions in the paper are present in the paper and/or SI Appendix.