Abstract
Biosurfactants produced by Lactobacillus fermentum RC-14, L. rhamnosus GR-1 and 36, and L. casei Shirota were found to contain proteins that bind to both collagen types III and VI, as determined by surface-enhanced laser desorption/ionization (SELDI)–time of flight mass spectrometry. Both collagen types III and VI immobilized on SELDI preactivated ProteinChip arrays detected several different sizes (2 to 48 kDa) of collagen-binding proteins. Overall, the RC-14-produced biosurfactant contained the greatest number of collagen-binding proteins (RC-14 > GR-1 > 36 > Shirota), including the mature form of a previously cloned 29-kDa collagen-binding protein (referred to in its mature 26-kDa form). Although biosurfactants isolated from L. casei Shirota and L. rhamnosus 36 and GR-1 also contain several collagen-binding proteins, they do not contain the 26-kDa collagen-binding protein. Together, these results demonstrate the utility of the SELDI system as a means of rapidly characterizing clinically important but complex biosurfactant solutions.
Biosurfactants produced by various strains of lactobacilli are complex biological mixtures that have been shown to inhibit the adhesion of urogenital pathogens to both biomaterial and uroepithelial cell surfaces (10–12). Recent biochemical analysis of Lactobacillus fermentum RC-14 biosurfactant identified a 29-kDa protein (p29) that exerts anti-adhesive effects against a well-known uropathogen, Enterococcus faecalis 1131 (2a). N- terminal amino acid sequence analysis of p29 showed 100% identity to a 29-kDa collagen binding protein previously isolated from Lactobacillus reuteri NCIB 11951 (6) and 90% identity to a 32-kDa basic surface protein from L. fermentum BR11 (9). Although very little is known about the function of either the 32-kDa basic surface protein or p29 collagen-binding protein, both are thought to regulate the adhesion of lactobacilli to host tissues. This notion is supported by studies that show that specific binding occurs between lactobacilli and components of the extracellular matrix (ECM), including collagen and fibronectin (1, 4).
The ability of microorganisms to adhere to distinct components of the ECM is a characteristic shared by many pathogenic bacteria, including Staphylococcus aureus. In fact, several genes encoding ECM-binding proteins, termed adhesins, have been cloned from S. aureus and shown to be important virulence factors that help initiate infections (2). In light of these and other observations, it has been proposed that the release of ECM-binding proteins by probiotic microorganisms may provide at least one possible mechanism for bacterial antagonism and thus help explain how lactobacilli exert their beneficial effects.
Considering the potential utility of Lactobacillus-derived ECM-binding proteins in preventing colonization by infectious pathogens, we have attempted to identify other ECM-binding proteins that might be present within Lactobacillus biosurfactants by using a novel ProteinChip system termed surface-enhanced laser desorption/ionization (SELDI)–time of flight (TOF) mass spectrometry. This recently developed ProteinChip technology (Ciphergen Biosystems, Inc., Palo Alto, Calif.) couples the sensitive analytical capabilities of mass spectrometry with novel surface chemistry (3, 7). Our objective was to determine whether the SELDI-TOF system could identify collagen-binding proteins produced by four Lactobacillus strains having potential clinical importance. In particular, two aims were to determine if the previously identified 26-kDa collagen-binding protein (p26 is used here as the designation for the mature form of p29) produced by L. fermentum RC-14 is the only collagen-binding protein present in this strain and to determine if this protein is produced by the three other isolates.
MATERIALS AND METHODS
Bacterial strains.
Four Lactobacillus strains were tested in this study. L. fermentum RC-14 was selected because it produces a biosurfactant that inhibits attachment of many urogenital pathogens to surfaces (10). Lactobacillus rhamnosus GR-1 and 36 also produce a biosurfactant, but it is less inhibitory to enterococci than the RC-14 biosurfactant (11). Lactobacillus casei Shirota is present in Yakult, a probiotic drink shown to colonize the intestine and improve recovery from rotavirus infection (8).
Preparation of biosurfactants.
The different strains of bacteria were grown in DeMan-Rogosa-Sharpe (MRS) broth (Merck, Darmstadt, Germany) overnight, and the biosurfactants were isolated as previously described (11). Briefly, following culture overnight at 37°C bacteria were harvested by centrifugation (10,000 × g, 20 min, 4°C), washed twice in demineralized water, and then resuspended in phosphate-buffered saline (PBS) (10 mM KH2PO4-K2HPO4 and 150 mM NaCl, pH 7.0) for 2 h at room temperature to induce release of the biosurfactant. Following release, the cells were pelleted by centrifugation (10,000 × g, 10 min, 4°C), and the supernatant was collected, filtered (22-μm-pore-size filter; Millipore), and dialyzed against demineralized water at 4°C in a Spectrapor membrane tube (molecular weight cutoff, 6,000 to 8,000; Spectrum Medical Industries Inc., Los Angeles, Calif.). The dialysate was lyophilized and stored at −20°C.
Analysis of biosurfactant with SELDI WCX-1 ProteinChip technology.
Biosurfactant isolated from each of the strains of Lactobacillus was resuspended in distilled H2O (dH2O). Aliquots (1 μl) were then added to the designated spots of a weak cation-exchange (WCX-1) ProteinChip array (Ciphergen Biosystems, Inc.) and incubated in a humidity chamber at room temperature for 1 h. The biosurfactant-bound chip arrays were then washed with dH2O containing 0.5% Triton X-100 for 20 min, spotted with 2 μl of a saturated solution of alpha-cyano-4-hydroxycinnamic acid (50% acetonitrile, 0.2% trifluoroacetic acid), and allowed to dry. The ProteinChip arrays were then analyzed by the SELDI-TOF technique (Ciphergen SELDI Protein Biology System I) and mass identification performed by averaging at least 35 laser shots (laser intensity = 25, 45, or 75) of various regions of the ProteinChip surface. Only molecular weight peaks whose intensities had signal-to-noise ratios of >5 were designated “true” protein peaks.
Analysis of biosurfactant with collagen cross-linked PS-1 ProteinChip arrays.
Collagen types III (bovine skin; Sigma) and VI (human placenta; Sigma) were resuspended in dH2O (3 mg/ml) and added (2 μl) to the spots of a preactivated (PS-1) SELDI ProteinChip array (Ciphergen Biosystems, Inc.). The collagen-modified PS-1 (Cn/PS-1) chip arrays were then incubated in a humidity chamber at room temperature for 1 h to permit covalent cross-linking of collagen to the chip surface. The arrays were spotted with 3 μl of buffer containing 0.5 M Tris (pH 7) to inactivate non-cross-linked areas on the chip surface. Whole Cn/PS-1 chip arrays were then washed with PBS containing 0.5% Triton X-100 for 20 min. Lyophilized biosurfactants were reconstituted in a buffer solution containing 20 mM Tris (pH 7), 200 mM NaCl, and 0.1% Triton X-100, added (2 μl) to spots on Cn/PS-1 ProteinChip arrays, and incubated in a humidity chamber for 1 h. Following incubation, whole Cn/PS-1 arrays were washed with PBS containing 0.1% Triton X-100 for 20 min, an aliquot (1 μl) of a saturated alpha-cyano-4-hydroxycinnamic acid solution was added to each spot on the Cn/PS-1 chip arrays, and the preparations were dried thoroughly. The samples were analyzed by the SELDI-TOF technique (Ciphergen SELDI Protein Biology System I), with mass identification of collagen-binding proteins performed by averaging at least 35 laser shots (laser intensity = 25 or 45) of various regions of a ProteinChip surface.
Amino acid analysis.
The amino acid compositions of the crude biosurfactants isolated from L. fermentum RC-14, L. casei Shirota, and L. rhamnosus GR-1 and 36 were analyzed with a Beckman 6300 amino acid analyzer. Briefly, hydrolysis was carried out by treating biosurfactant samples (3 μl) with 6 N HCl (Pierce Chemicals) for 1 h at 150°C in a Pico Tag workstation (Waters). Under these conditions asparagine and glutamine are deaminated to aspartic and glutamic acids, respectively, and tryptophan and cystine cannot be quantified accurately.
RESULTS
SELDI-TOF analysis: rapid resolution of biosurfactant proteins.
Table 1 shows the amino acid analysis results for hydrolyzed biosurfactant samples produced by L. fermentum RC-14, L. casei Shirota, and L. rhamnosus GR-1 and 36, as well as a collagen-binding protein (p26) purified from L. fermentum RC-14. Although there appears to be some free amino acid (alanine) in two of the samples (36 and Shirota), the moles percents of amino acids are quite typical of proteins.
TABLE 1.
Amino acid compositions of hydrolyzed Lactobacillus biosurfactants
| Sample | Amino acid composition (mol%)a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asxb | Thr | Ser | Glxb | Gly | Ala | Val | Met | Ile | Leu | Phe | Tyr | His | Lys | Arg | Pro | |
| 36 | 8.23 | 3.6 | 2.98 | 10.59 | 8.4 | 33.98c | 5.41 | 1.07 | 3.58 | 5.82 | 1.29 | 1.97 | 1.27 | 5.9 | 2.39 | 3.52 |
| GR-1 | 10.3 | 7.0 | 12.3 | 18.4 | 18.7 | 7.86 | —d | — | 2.02 | 10.7 | — | — | 4.1 | — | 1.05 | 6.94 |
| RC-14 | 10.4 | 4.67 | 5.81 | 12.5 | 10.1 | 8.91 | 6.19 | 1.02 | 3.24 | 9.5 | 2.67 | 3.54 | 3.64 | 7.6 | 5.76 | 4.52 |
| Shirota | 4.23 | 1.91 | 1.91 | 6.04 | 5.03 | 63.0c | 2.97 | 0.42 | 1.76 | 3.15 | 0.47 | 0.79 | 1.46 | 3.17 | 1.54 | 2.14 |
| p26/p29 | 10.2 | 4.84 | 8.26 | 11.8 | 16.7 | 17.5 | 5.47 | 0.89 | 3.93 | 6.71 | — | 0.8 | — | 8.12 | 0.56 | 4.24 |
Due to analysis conditions cystine and tryptophan could not be accurately quantified.
Sample preparation resulted in deamination of asparagine and glutamine into aspartic acid and glutamic acid, respectively.
The unlikely high values may indicate the presence of free alanine in the sample.
—, not detected.
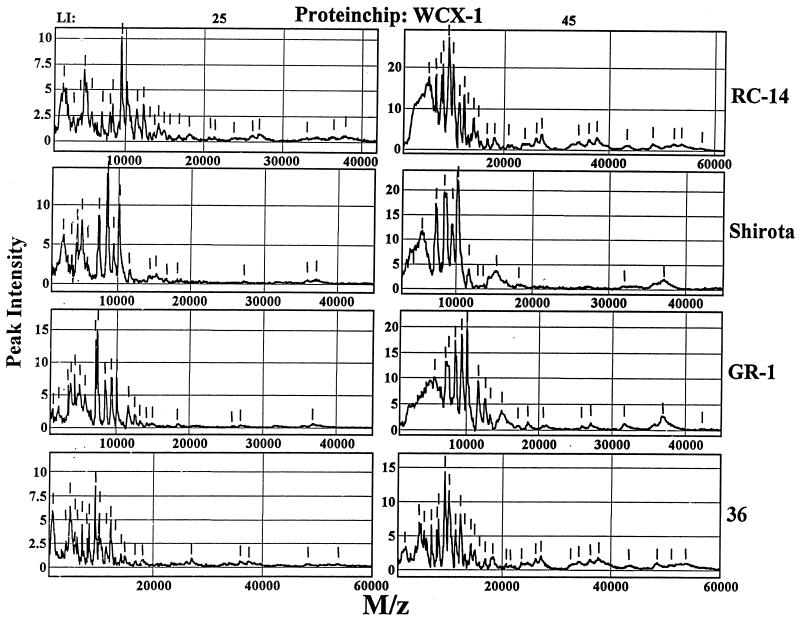
Next, the molecular weight spectra of the proteins were determined for each of the strain-specific biosurfactants (RC-14, Shirota, GR-1, and 36) by using SELDI-TOF ProteinChip arrays with the same selectivity. The ProteinChip surfaces were chosen to highlight differences between biosurfactants and to select proteins that bind to collagen. As shown in Fig. 1, a weak cation-exchange (WCX-1) ProteinChip array was used to determine the molecular weight ranges of proteins in the biosurfactant samples. When a number of laser intensity settings (25 and 45) were used, a broad range of protein peaks was detected. The ability of the SELDI-TOF technique to readily resolve proteins of various sizes was very similar to that of sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the biosurfactants (data not shown), except that the SELDI technique was better for low-molecular-weight components (molecular weights, <10,000), which are more difficult to resolve by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
FIG. 1.
Analysis of Lactobacillus biosurfactants with WCX-1 ProteinChip arrays. Aliquots (2 μl) of biosurfactants produced by Lactobacillus strains RC-14, Shirota, GR-1, and 36 were spotted onto the surfaces of weak cation-exchange (WCX-1) ProteinChip arrays for SELDI analysis by using the laser intensity (LI) settings indicated. Individual spectra show the relative peak intensities versus mass-to-charge ratios (M/z) of proteins for each biosurfactant. The short vertical lines indicate individual protein peaks.
Identification of collagen type III- and VI-binding proteins in Lactobacillus biosurfactants by SELDI-TOF mass spectrometry.
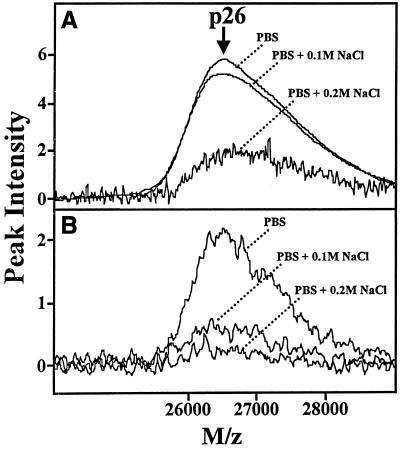
Lactobacillus-derived biosurfactants were screened for collagen-binding protein activity based on the affinity of a known collagen-binding protein, p26 (the mature form of p29) purified from L. fermentum RC-14 (2a). Figure 2 shows the binding affinity of p26 to collagen types III and VI, as determined with collagen cross-linked ProteinChip arrays (Cn-III/PS-1, Cn-VI/PS-1). By varying the salt (NaCl) concentration, the binding of p26 could be significantly inhibited.
FIG. 2.
PS-1 ProteinChip array analysis of an ∼26-kDa collagen-binding protein purified from L. fermentum RC-14. The binding affinity of a ∼26-kDa collagen-binding protein purified from L. fermentum RC-14 biosurfactant was analyzed by using preactivated surface arrays (PS-1) containing immobilized collagen types III (PS-1/Cn-III) (B) and VI (PS-1/Cn-VI) (A). The overlaid spectra show the effects of different salt (NaCl) concentrations on the chip binding affinity of p26. The relative peak intensities versus mass-to-charge ratios (M/z) of p26 are plotted.
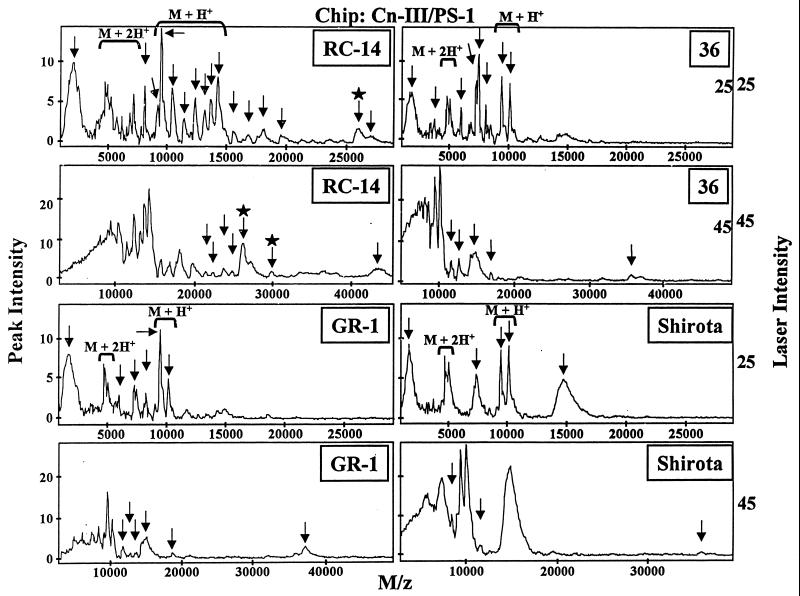
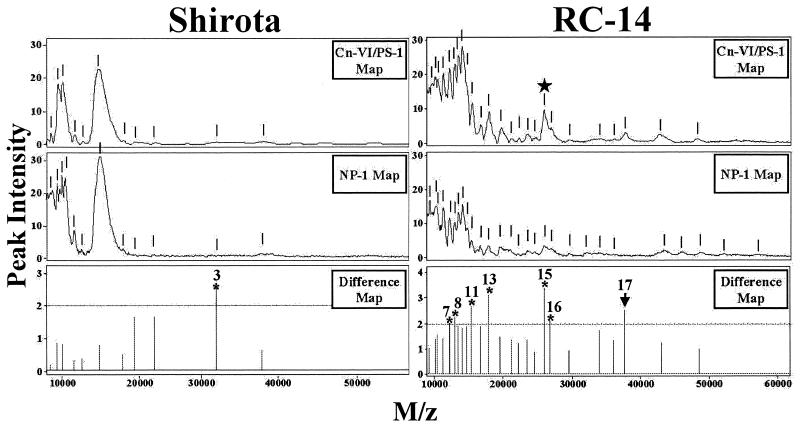
Screening was done for proteins with similar collagen binding characteristics by dissolving the lyophilized biosurfactants in a buffer solution containing PBS and 0.1 M NaCl. When Cn-III/PS-1 (Fig. 3) and Cn-VI/PS-1 (data not shown). ProteinChip arrays were used, a number of potential collagen-binding proteins were detected in each of the biosurfactants tested. When the molecular weight spectra generated by the two types of Cn/PS-1 arrays were compared, there appeared to be a remarkable degree of similarity in terms of the number of collagen-binding protein peaks and their relative abundances (peak intensities). Since the binding conditions used may not have been stringent enough to exclude binding of very abundant proteins, an additional SELDI step was performed with normal-phase (NP-1, silica) ProteinChip arrays. The NP-1 chips are good general protein-binding arrays and therefore useful for determining the relative abundances of proteins in a given biological sample. The ProteinChip software is designed to combine and compare profiles and is capable of generating difference SELDI maps (Fig. 4). By subtracting the NP-1-generated spectra from either the Cn-III/PS-1 or Cn-VI/PS-1 spectra, screening for proteins with collagen binding activity was more stringent.
FIG. 3.
Detection of collagen-binding proteins from Lactobacillus biosurfactants with collagen type III immobilized on PS-1 ProteinChip arrays. Aliquots (2 μl) of biosurfactants prepared from Lactobacillus strains RC-14, Shirota, GR-1, and 36 were spotted onto the surfaces of PS-1 ProteinChip arrays containing immobilized collagen type III (Cn-III/PS-1) and analyzed by the SELDI-TOF technique by using the laser intensity settings indicated. The plotted spectra depict the relative peak intensities versus mass-to-charge ratios (M/z) of proteins for each biosurfactant. The arrows indicate distinct collagen-binding peaks. The brackets indicate dual-charge peaks (M + 2H+) and their parental single-charge peaks (M + H+). The arrows with stars identify p29 collagen-binding protein and its mature processed form (p26).
FIG. 4.
Differential protein spectral mapping of collagen-binding proteins from Lactobacillus biosurfactants by using normal-phase (NP-1) and collagen type VI immobilized preactivated surface (PS-1) ProteinChip arrays. Biosurfactant samples (2 μl) prepared from Lactobacillus strain RC-14 and Shirota were spotted onto the surfaces of either PS-1 arrays containing immobilized collagen type VI (Cn-VI/PS-1) or normal-phase (NP-1) ProteinChip arrays. SELDI-TOF analysis was carried out by using a laser intensity setting of 50. The spectra show relative peak intensities versus mass-to-charge ratios (M/z) of proteins for each of the biosurfactants indicated. The short vertical lines indicate protein peaks. The difference maps show the ratios of the common peak intensities (Cn-VI/PS-1:NP-1) for each biosurfactant. Collagen-binding proteins with peak intensity ratios of >2 (above the dotted line) are indicated by asterisks and numbered. The star identifies the 26-kDa collagen-binding protein.
p26 was successfully identified as an endogenous collagen-binding protein produced by L. fermentum RC-14 biosurfactant (2a). p26 showed more than threefold-greater binding affinity for Cn-VI/PS-1 arrays than for NP-1 arrays. The identity of the 26.5-kDa peak as the p26 collagen-binding protein was confirmed by on-chip protease digestion of the partially purified RC-14 biosurfactant, followed by mass spectrometry-mass spectrometry sequencing of the peptide fragments by the SELDI-Qq-TOF technique (data not shown). Based on this selection criterion, the RC-14 biosurfactant contained the greatest number of putative collagen-binding proteins (a total of 17 proteins), including a ∼37.8-kDa peak that is unique to the Cn/PS-1 chip array. By comparison, the SELDI analysis of the biosurfactants produced by strains GR-1, Shirota, and 36 detected eight, two, and two collagen-binding proteins, respectively.
DISCUSSION
The SELDI ProteinChip technology used in this study is capable of detecting proteins at picomolar to femtomolar levels in small native biological mixtures with little or no preparation. It was found here to identify a number of collagen-binding proteins in crude biosurfactant preparations from four probiotic Lactobacillus strains. While the p26 collagen-binding protein present in L. fermentum RC-14 exerts anti-adhesive activity against uropathogenic bacteria (2a), the roles of the other putative collagen-binding proteins identified in this study have yet to be determined and are the subject of ongoing investigations.
The ability of Lactobacillus biosurfactants to prevent uropathogens from adhering to surfaces (10–12) and subsequently infecting healthy human tissue is believed to account, in part, for the beneficial clinical effects of these probiotics (5). Based on this notion, the extent of collagen binding activity present in Lactobacillus-produced biosurfactants was determined by the SELDI-TOF technique. Collagen is a major ECM component of the dermis that facilitates attachment of invading pathogenic bacteria (e.g., S. aureus). Thus, the ability of lactobacilli to produce collagen-binding proteins could explain one aspect of the organisms' protective ability on surfaces and in the urogenital tract.
The SELDI-TOF ProteinChip array system is an excellent system for screening strains for the presence of collagen-binding proteins and for identifying other factors which may be important in potential clinical application of the bacteria. Here, L. fermentum RC-14 not only produced more collagen-binding proteins, but it alone produced p26. The fact that L. casei Shirota, a strain used in the commercial probiotic Yakult, did not produce the p26 protein illustrates that not all probiotic lactobacilli are identical, and without verification of their activity in the human urogenital tract, these organisms should not be universally given to humans to treat or prevent urogenital disease. The differences in the levels of expression of collagen-binding proteins may be of clinical significance for another reason; namely, some may better inhibit pathogenic microorganisms (such as S. aureus) binding to ECM proteins (2).
In conclusion, SELDI ProteinChip arrays provide researchers with a powerful means of quickly characterizing proteins and specific protein-protein interactions. The tremendous versatility of this ProteinChip technology makes it particularly useful for more extensive biochemical and microbiological applications, including the development of specific protein expression databases. For example, one could envision the development of biofilm- or probiotic-specific proteome databases. Taken together, these studies may lead to the discovery of novel, clinically important antimicrobial factors.
ACKNOWLEDGMENT
This study was funded by the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Aleljung P, Paulsson M, Emody L, Andersson M, Naidu A S, Wadstrom T. Collagen binding by lactobacilli. Curr Microbiol. 1991;23:33–38. [Google Scholar]
- 2.Flock J-I. Extracellular-matrix-binding proteins as targets for the prevention of Staphylococcus aureus infections. Mol Med Today. 1999;5:532–537. doi: 10.1016/s1357-4310(99)01597-x. [DOI] [PubMed] [Google Scholar]
- 2a.Heinemann C, van Hylckama Vlieg J E T, Janssen D B, Busscher H J, van der Mei H C, Reid G. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol Lett. 2000;190:177–180. doi: 10.1111/j.1574-6968.2000.tb09282.x. [DOI] [PubMed] [Google Scholar]
- 3.Hutchens T W, Yip T T. New desorption strategies for the mass spectrometric analysis of macromolecules. Rapid Commun Mass Spectrom. 1993;7:576–580. [Google Scholar]
- 4.Nagy E, Froman G, Mardh P-A. Fibronectin binding of Lactobacillus species isolated from women with and without bacterial vaginosis. J Med Microbiol. 1992;37:38–42. doi: 10.1099/00222615-37-1-38. [DOI] [PubMed] [Google Scholar]
- 5.Reid G. The scientific basis for probiotic strains of Lactobacillus. Appl Environ Microbiol. 1999;65:3763–3766. doi: 10.1128/aem.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roos S, Aleljung P, Robert N, Lee B, Wadstrom T, Lindberg M, Jonsson H. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol Lett. 1996;144:33–38. doi: 10.1111/j.1574-6968.1996.tb08505.x. [DOI] [PubMed] [Google Scholar]
- 7.Strauss E. New ways to probe the molecules of life. Science. 1998;282:1406–1407. doi: 10.1126/science.282.5393.1406. [DOI] [PubMed] [Google Scholar]
- 8.Sugita T, Togawa M. Efficacy of Lactobacillus preparation Biolactis powder in children with rotavirus enteritis. Jpn J Pediatr. 1994;47:2755–2762. [Google Scholar]
- 9.Turner M S, Timms P, Hafner L M, Giffard P M. Identification and characterization of a basic cell surface-located protein from Lactobacillus fermentum BR11. J Bacteriol. 1997;179:3310–3316. doi: 10.1128/jb.179.10.3310-3316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velraeds M C, van der Belt B, van der Mei H C, Reid G, Busscher H J. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J Med Microbiol. 1998;49:790–794. doi: 10.1099/00222615-47-12-1081. [DOI] [PubMed] [Google Scholar]
- 11.Velraeds M C, van der Mei H C, Reid G, Busscher H J. Inhibition of initial adhesion of uropathogenic Enterococcus faecalis by biosurfactants from Lactobacillus isolates. Appl Environ Microbiol. 1996;62:1958–1963. doi: 10.1128/aem.62.6.1958-1963.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velraeds M C, van der Mei H C, Reid G, Busscher H J. Physicochemical and biochemical characterization of biosurfactants released from Lactobacillus strains. Colloids Surf B Biointerfaces. 1996;8:51–61. [Google Scholar]