Significance
Insects that feed on plants are thought to impose natural selection for chemical defenses in the plant tissues they eat. Here we show that seeds of the common milkweed contain a highly potent toxin, labriformin, which has evolved to higher concentrations in northern latitudes. When present, labriformin inhibits an essential cellular transport enzyme (the sodium–potassium pump) of the monarch butterfly and the large milkweed bug. The specialized seed bug biochemically modifies labriformin to less toxic compounds and sequesters these end products for its own defense. The seed bug’s highly tolerant sodium pump and biochemical modifications point to a key role for labriformin in the coevolution between milkweed and its herbivores.
Keywords: coevolution, plant–insect interactions, monarch, milkweed, chemical ecology
Abstract
Environmental clines in organismal defensive traits are usually attributed to stronger selection by enemies at lower latitudes or near the host’s range center. Nonetheless, little functional evidence has supported this hypothesis, especially for coevolving plants and herbivores. We quantified cardenolide toxins in seeds of 24 populations of common milkweed (Asclepias syriaca) across 13 degrees of latitude, revealing a pattern of increasing cardenolide concentrations toward the host's range center. The unusual nitrogen-containing cardenolide labriformin was an exception and peaked at higher latitudes. Milkweed seeds are eaten by specialist lygaeid bugs that are even more tolerant of cardenolides than the monarch butterfly, concentrating most cardenolides (but not labriformin) from seeds into their bodies. Accordingly, whether cardenolides defend seeds against these specialist bugs is unclear. We demonstrate that Oncopeltus fasciatus (Lygaeidae) metabolized two major compounds (glycosylated aspecioside and labriformin) into distinct products that were sequestered without impairing growth. We next tested several isolated cardenolides in vitro on the physiological target of cardenolides (Na+/K+-ATPase); there was little variation among compounds in inhibition of an unadapted Na+/K+-ATPase, but tremendous variation in impacts on that of monarchs and Oncopeltus. Labriformin was the most inhibitive compound tested for both insects, but Oncopeltus had the greater advantage over monarchs in tolerating labriformin compared to other compounds. Three metabolized (and stored) cardenolides were less toxic than their parent compounds found in seeds. Our results suggest that a potent plant defense is evolving by natural selection along a geographical cline and targets specialist herbivores, but is met by insect tolerance, detoxification, and sequestration.
Geographical patterns in plant defense expression have been increasingly viewed through the lens of adaptation (1–3). Be it latitudinal patterns in attack and defense (4, 5), or greater levels of damage at plant range centers where herbivores congregate (6, 7), there is ample opportunity for plant local adaptation to herbivory. Nonetheless, a key factor missing from the plethora of existing studies is a functional link between measures of plant defense traits and impacts on specific herbivores (3, 5). Given that most plants have multiple enemies with varying degrees of specialization, understanding the degree of mechanistic match between plant defense and particular herbivores is critical to gaining insight into coevolution. For example, classic work on the genetic and physiological basis of lepidopteran detoxification of plant-produced furanocoumarins has linked geographical patterns of defense with local adaptation of the herbivores (8–10).
We have been studying the multiple specialized herbivores of milkweed, each having well-characterized and genetically based physiological differences in tolerance to toxic cardenolides (11, 12). In particular, distinct genetic substitutions in the herbivores’ Na+/K+-ATPase genes provide predictable tolerance to the toxins. Nonetheless, despite characterization of overall levels of tolerance (12, 13), specific interactions between particular cardenolides and insect Na+/K+-ATPases are only beginning to emerge (14, 15) and may be a key means of understanding coevolution in a community context. In particular, because milkweed herbivores specialize on different plant parts (e.g., roots, leaves, seeds, etc.) (16), testing the functional match between defense expression and impacts on herbivore tolerance of specific cardenolides in different plant parts is a fruitful avenue to decipher coevolution between a plant and multiple attacking herbivores (13).
Here we take an integrative approach, spanning geographical patterns and whole organism feeding assays to metabolomic analyses and toxin–target site in vitro experiments to address the function of specific seed defenses of common milkweed (Asclepias syriaca). We focus on the specialized seed-feeding large milkweed bug, Oncopeltus fasciatus, the most cardenolide-tolerant insect known (13, 17, 18), but use the monarch butterfly as a key comparison group which, although less tolerant of cardenolides, shares many of the sodium pump adaptations with Oncopeltus. In particular, we addressed the following hypotheses: 1) geographical patterns in seed defense are predictable across the range of A. syriaca and phenotypic differences among populations are greater than that expected due to neutral genetic differentiation (i.e., PST > FST) (19); and 2) some cardenolide defenses will be effective against even the most tolerant seed predator specialist, but may not be sequestered unless they are converted to less toxic forms.
Results
Seed Cardenolides along the Geographical Cline.
We tested latitude as a predictor of seed cardenolide production across 24 populations of common milkweed, ranging from Quebec City, QC, Canada to Bishop, North Carolina, spanning nearly 13 degrees of latitude. High-resolution mass spectrometry revealed the relative concentrations of each of 20 cardenolides and we used high performance liquid chromatography (HPLC-UV) to quantify the total concentrations of the 13 most abundant cardenolides (Fig. 1 and SI Appendix, Fig. S1 and Tables S1–S3). For nearly all measures, including total cardenolide concentration, a quadratic model was a much better fit to the data than a linear model, with cardenolide concentrations peaking between 40 and 45° north latitude, roughly corresponding to the range center of A. syriaca from Pennsylvania to the south and New York, Vermont, and New Hampshire to the north. Two specific compounds are worth highlighting: glycosylated aspecioside (a newly described structure), the dominant cardenolide in seeds, comprising 42% of the total; and less concentrated labriformin, which is an unusual epoxy cardenolide containing nitrogen and sulfur in a thiazoline ring (20) (Fig. 1). Interestingly, in addition to 70% of the variation in labriformin being explained by latitude, this compound also had a different pattern than all others, saturating in production above 42° north latitude, but not declining further north (Fig. 1 and SI Appendix, Fig. S1). We note that two additional cardenolides, aspecioside A and glycosylated syriogenin A, decrease in relative concentration from south to north and this opposing pattern could indicate that these cardenolides are the biosynthetic precursors of labriformin (SI Appendix, Fig. S2). A correlational heat map confirms a significant negative correlation of labriformin with aspecioside A and glycosylated syriogenin A (SI Appendix, Fig. S3). The highest significant positive correlation of labriformin was found with syrioside A and B, suggesting shared biosynthesis.
Fig. 1.
Latitudinal prediction of seed cardenolides among 24 populations of common milkweed (A. syriaca) in eastern North America. (A) Glycosylated aspecioside, the dominant compound (highly polar, 42% of total seed cardenolides). (B) Diglycosylated syriogenin, a polar cardenolide comprising 12% of the total. (C) Labriformin, the least polar seed cardenolide (8% of the total) also containing a thiazoline heterocycle with nitrogen and sulfur. (D) Total cardenolide concentration. (A–C) Quantified by high-resolution mass spectrometry and (D) quantified by HPLC-UV. A quadratic model was the best fit by far in all cases. We note that both herbivore diversity and leaf damage showed a similar humped-shaped curve in field surveys, with herbivory peaking at ∼42° north latitude (6). **P < 0.01, ***P < 0.001. Analyses of other compounds is presented in SI Appendix, Fig. S1 and the correlation among compounds is presented in SI Appendix, Fig. S3.
Next we assessed whether phenotypic differences between populations in cardenolide defenses were greater than neutral genetic differentiation using a modified PST > FST approach (following refs. 21, 22) (SI Appendix, Figs. S4 and S5 and Table S4). We assessed FST using 925 established single nucleotide polymorphisms (SNPs) (23) in each of 12 of the 24 sampled populations. Phenotypic differences between populations (PST) were substantially greater than FST for labriformin and syrioside B (but not for the other dominant cardenolides), indicating spatially divergent selection on these compounds (SI Appendix).
Sequestration and Feeding Experiment.
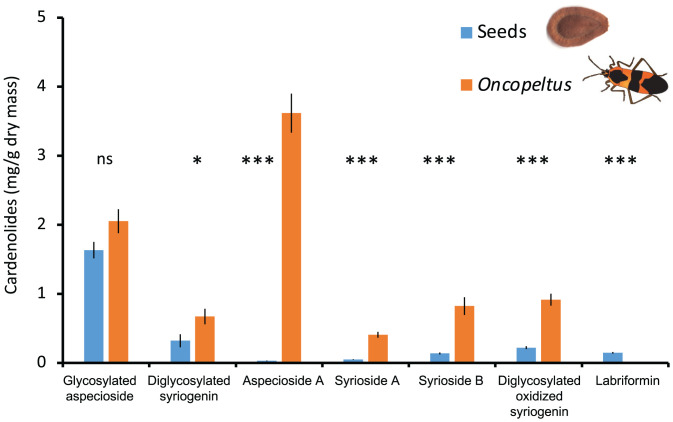
In a laboratory rearing experiment, Oncopeltus sequestered greater than four times the concentration of cardenolides in A. syriaca seeds (on a dry mass basis, mean ± SE total cardenolides in mg/g, seeds: 2.81 ± 0.61; bugs: 11.33 ± 0.61; n = 10, t = 9.21, P < 0.001). Among the six most abundant cardenolides present in milkweed seeds (accounting for 87% of the total), five were more abundant in bugs than in seeds (Fig. 2). The sole exception to this pattern was labriformin, which was not sequestered. We thus hypothesized that labriformin was highly toxic and degraded by Oncopeltus. Additionally, the dominant cardenolide in bugs was aspecioside A (33% of the total), a compound that was very low in our seeds (3% of the total in this collection from Ithaca, NY). Because of the structural similarity of the previously undescribed glycosylated aspecioside and aspecioside A (the latter simply missing one sugar compared to the former), we hypothesized that Oncopeltus accumulated aspecioside A by modification of glycosylated aspecioside (SI Appendix, Fig. S6).
Fig. 2.
Concentration of cardenolides in common milkweed (A. syriaca) seeds (collected in Ithaca, NY, 42.4440° north latitude) and sequestered by large milkweed bugs (O. fasciatus, whole adults extracted). Shown are means ± SE quantified by HPLC-UV. Although most cardenolides are concentrated in bugs compared to seeds, note that dominant (in bugs) aspecioside A is essentially absent in seeds, and labriformin is absent in bugs compared to seeds. The shown compounds comprise 87% of total seed cardenolides and are arrayed from most polar on the Left to nonpolar on the Right. *P < 0.05, ***P < 0.001, ns, not significant. Compositional differences between seeds and bugs along the labriformin degradation pathway identified by high-resolution mass spectrometry are shown in SI Appendix, Fig. S7.
Accordingly, we conducted feeding experiments, with artificial diets spiked with purified glycosylated aspecioside, labriformin, or ouabain (as a standard) compared to controls without cardenolides. Several measures of Oncopeltus growth and performance were not impacted by cardenolides (SI Appendix, Table S5), and we found evidence that glycosylated aspecioside was degraded in vivo to aspecioside A and that labriformin was converted to three products that are stored in adults (desglucosyrioside, syriobioside A, and oxidized labriformin) (SI Appendix, Fig. S7 and Table S6).
Cardenolide Toxicity Tested by Functional Na+/K+-ATPase Assays.
We next tested the inhibitory capacity of a subset of purified cardenolides on their physiological target, the Na+/K+-ATPase in vitro. Here we used ouabain as a standard, along with isolated and purified glycosylated aspecioside (polar, dominant, sequestered intact, and converted), aspecioside A (sequestered after conversion from glycosylated aspecioside), diglycosylated syriogenin (polar, subdominant, sequestered intact), and labriformin (apolar thiazoline ring–containing cardenolide, subdominant, not sequestered). All compounds were tested on the highly sensitive porcine Na+/K+-ATPase (Fig. 3, Upper Inset) and showed relatively little variation in their inhibition of this unadapted enzyme. Nonetheless, our main comparison was between the monarch enzyme compared to that of Oncopeltus, both obtained from isolated neural tissues. We focus on this comparison because Oncopeltus has the same critical amino acid substitutions in the sodium pump as the monarch (namely a combination of histidine at position 122 and a substitution of the ancestral glutamine at position 111: Threonine in bugs and valine in monarchs). In contrast to the monarch with its single gene copy, Oncopeltus expresses two versions of the sodium pump in the nervous tissue featuring additional genetic substitutions, among others at positions 786 and 797 that have been shown to further increase the enzyme’s tolerance of cardenolides (24). Thus, we are addressing the added benefit of these seed bug substitutions toward tolerance of cardenolides.
Fig. 3.
The neural Na+/K+-ATPase of O. fasciatus is more tolerant of inhibition by cardenolides than that of monarch butterflies. Shown is the dominant seed cardenolide (glycosylated aspecioside) and its sequestered conversion product (aspecioside A), diglycosylated syriogenin, which is a subdominant compound that is sequestered intact; and labriformin, which is not sequestered. Data are presented as the molar concentration of plant toxin necessary to cause 50% inhibition of the animal enzyme, or IC50. Higher values on the y axis indicate that the enzyme is more tolerant to the cardenolide; values within the orange bars indicate the fold increase in tolerance of the Oncopeltus enzyme over the monarch enzyme. Note that O. fasciatus is substantially more tolerant compared to monarchs for labriformin (the most potent cardenolide overall) compared to all other compounds (enlarged in Bottom Right Inset). Upper Right Inset shows the IC50 values for the same compounds on the highly sensitive porcine Na+/K+-ATPase. Each bar is a mean of 3 to 10 replicates (each based on a 6-concentration inhibition curve) ± SE.
Although the compounds had differential effects on monarch versus Oncopeltus (Fig. 3, interaction term between compounds and the two enzymes F4,30 = 18.76, P < 0.001) the latter was, on average more than sixfold more tolerant than the former. Ouabain and the highly sequestered aspecioside A were the least inhibitive, followed by glycosylated aspecioside. Strikingly, labriformin was the most potent cardenolide across the board, but Oncopeltus neural tissue was 18 times more tolerant than that of monarchs. Remarkably, the monarch enzyme was not much more tolerant of labriformin compared to the sensitive porcine enzyme (SI Appendix, Fig. S8), suggesting that the additional genetic substitutions in the two sodium pump isoforms of Oncopeltus are most beneficial against this highly toxic compound.
Oncopeltus sequestered three distinct modified compounds when fed labriformin, generated via a degradative pathway of the 3-thiazoline ring (via hydrolysis: desglucosyrioside and syriobioside A), and a nondegradative pathway (oxidation to form a thiazolidinone ring: oxidized labriformin) (Fig. 4). We were able to purify and test two of these metabolites on the Oncopeltus Na+/K+-ATPase. The sequestered compounds were, on average, eightfold less potent than labriformin, but not significantly different from the standard ouabain. Labriformin was not detected in bugs fed pure labriformin (SI Appendix, Table S6). Furthermore, the absence of labriformin and presence of oxidized labriformin in Oncopeltus extracts are the opposite pattern found in A. syriaca seeds, supporting the biotransformation of labriformin (SI Appendix, Fig. S7). Thus, the modification and breakdown of labriformin may reduce its ultimate toxicity for Oncopeltus. Monarchs are known not to sequester labriformin either, but sequester at least two breakdown products (syriobioside A and desglucosyrioside) (25, 26). Our data indicate that both syriobioside A and oxidized labriformin (unknown whether this is sequestered by monarchs) also have substantially lower toxicity to monarchs than labriformin (SI Appendix, Fig. S8).
Fig. 4.
Sequestration by O. fasciatus involves modifications of labriformin into three end products. (A) Two endogenous modification pathways exist, one with hydrolysis and reduction (loss of the thiazoline ring) and the other via oxidation (thiazoline ring maintained). Syriobioside A occurs in much lower quantities (∼1/100 concentration) compared to desglucosyrioside and oxidized labriformin. (B) The difference in inhibitory impacts of labriformin and two of the end products on O. fasciatus (shown also is the standard, ouabain). Data are presented as the molar concentration of plant toxin necessary to cause 50% inhibition of the animal enzyme, or IC50 (note log scale on the y axis). Higher values on the y axis indicate that the enzyme is more tolerant to the cardenolide. Different letters above bars represent significant (P < 0.05) pairwise comparisons based on Fisher’s least significant difference. Each bar is a mean of three to six replicates (each based on a 6-concentration inhibition curve) ± SE.
Discussion
Geographical patterns in the expression of plant defense may be driven by a variety of factors, and such patterns are widespread across latitude, elevation, and precipitation gradients (2–5, 27, 28). Yet, missing from most studies is a functional connection between measures of defense chemistry and impacts on herbivores, especially for adapted (or coevolved) species (3, 5). We previously demonstrated genetically based latitudinal clines in the expression of foliar defenses in common milkweed (A. syriaca), both in North America where it is native (6), and in Europe where it was introduced and has spread over 400 y (29). Malcolm (30) previously described a longitudinal trend in A. syriaca cardenolides, with implications for survival of monarchs. Furthermore, total cardenolides in leaves were reported to peak in midlatitudes near the range center of A. syriaca (31). In the current study, by focusing on individual cardenolide toxins in milkweed seeds, we have identified compounds that appear to be under selection, and that have tremendous potency against adapted herbivores.
Labriformin, in particular, is one of five nitrogen-containing cardenolide toxins (along with thiazolidine ring–containing uscharin, 3-thiazoline voruscharin, and two thiazolidinone ring–containing cardenolides) identified among hundreds of others (lacking nitrogen) in the genus Asclepias. In A. syriaca, 3-thiazoline ring–containing labriformin has been long known and we were able to anticipate and detect its thiazolidine derivative, reduced labriformin, which is analogous to voruscharin (SI Appendix, Fig. S9) (20, 32–34). Although labriformin was known historically to be poisonous to livestock (35) and not sequestered by monarchs (31, 36), little was known about its physiological interactions with specialist herbivores. We have shown that labriformin’s geographic pattern of expression is unique among A. syriaca’s seed cardenolides (Fig. 1 and SI Appendix, Fig. S1), it is likely subject to divergent selection among populations (SI Appendix, Fig. S5), and is among the most potent cardenolides tested against adapted sodium pumps (Fig. 3). Although we do not show whether the driver of this cline is mostly biotic, abiotic, or both, it is conceivable that evolution of more toxic defenses may be realized in more favorable locations where herbivores also tend to congregate (2). Although we previously reported greater insect diversity and herbivory on common milkweed near the range center compared to the northern and southern range edges (6), additional work specifically on the geography of seed predation and cardenolide tolerance/sequestration is needed.
Specificity in Chemical Defense–Herbivore Offense.
An emerging pattern from work on natural product inhibition of the insect sodium pump is that most cardenolides are more or less equally potent against unadapted sodium pump enzymes, while stronger differentiation is found on the more tolerant (adapted) enzymes (14, 15). Accordingly, the total concentration of cardenolides in a plant extract is typically an excellent predictor of the extent of sodium pump inhibition, irrespective of the cardenolide composition (37). However, the same compounds often show >100-fold variation in inhibition of adapted sodium pumps (14, 15) (Figs. 3 and 4 and SI Appendix, Fig. S8), and one of the approaches we have employed to understand specificity in defense is to match particular defense compounds to particular enzymes. A key result from the current work is how labriformin appears to be especially important in the milkweed–Oncopeltus interaction.
Labriformin is 1.5 to 2 times more potent than glycosylated aspecioside and the standard ouabain on the unadapted porcine sodium pump (Fig. 3). Nonetheless, when compared to the Oncopeltus enzyme, labriformin is 6 and 13 times more potent. Reciprocally, to understand the benefits of the Oncopeltus highly adapted sodium pump involving both gene duplications and substitutions (18, 38), we compare the advantage it has over the monarch’s enzyme. This comparison is especially useful because Oncopeltus has the same base-level adaptations as the monarch (functional substitutions at amino acid positions 111 and 122 of the cardenolide-binding pocket), with additional substitutions at positions 786 and 797 (24). Although Oncopeltus has an 8- and 2-fold advantage over monarchs for glycosylated aspecioside and ouabain, respectively, it has an 18-fold advantage over monarchs in terms of coping with labriformin (Fig. 3). A general agreement among pharmacological studies of enzymatic assays classifies molecules with the half maximal inhibitory concentration (IC50) >100 µM as nontoxic, 10 to 100 µM as moderately toxic, IC50 <10 µM as toxic, and IC50 <1 µM as highly toxic. With IC50 values of 5 µM, labriformin falls under the toxicity threshold for the monarch and comes close to voruscharin’s toxicity (IC50 = 2 µM) (15). Remarkably, the monarch’s enzyme, which typically shows >50-fold enhanced tolerance to cardenolides compared to nonadapted enzymes, has little advantage in the case of labriformin (SI Appendix, Fig. S8). Given labriformin’s abundance in seeds (which is substantially higher than in leaves), its high potency against adapted enzymes, and the Oncopeltus advantage in coping with this compound (IC50 = 91 µM), it appears that labriformin could be at the center of reciprocal adaptation between milkweed and seed bugs. The specific advantage of the different copies of the sodium pump and differential expression among bug tissues are worthy of further study among populations and species of the Lygaeinae.
Advances in Our Understanding of Milkweed–Herbivore Coevolution.
Nearly 50 y ago Duffey and Scudder (39) showed that Oncopeltus sequesters polar but not the most nonpolar cardenolides from A. syriaca seeds; they also showed that Oncopeltus degrades some cardenolides (e.g., the nonpolar and nonmilkweed compound, digitoxin) following ingestion, but does not degrade others (e.g., ouabain). The overall organismal tolerance of these seed bugs to cardenolides and the tremendous tolerance of their sodium pumps in vitro was also well established previously (18, 40–43). In fact, geographic variation in A. syriaca seed chemistry was briefly studied by Moore and Scudder (44) as a possible explanation for differences in sequestration of Oncopeltus reported in various studies. Nonetheless, despite being well studied, the specific cardenolides in A. syriaca seeds and those that are sequestered versus not sequestered were previously undescribed. By connecting the nonpolar labriformin, its structural attributes (e.g., the nitrogen- and sulfur-containing thiazoline ring), and its potency relative to degradation products produced by bugs, we have added key missing links in our understanding of milkweed–herbivore coevolution.
More generally, the extent to which milkweeds have independently evolved highly toxic nitrogen-containing cardenolides and different specialized lineages of herbivores have adapted to these compounds in parallel offers a test of repeated patterns in coevolutionary interactions. We previously studied interactions between nitrogen-containing voruscharin in Asclepias curassavica leaves and monarch butterflies (15). Voruscharin negatively impacts monarch growth, is not sequestered, and is transformed by monarchs to less toxic compounds (calactin and calotropin) via hydrolysis and reduction. Voruscharin has been reported from a few Asclepias spp., as has labriformin (which has a distinct genin), although their frequency in the genus and phylogenetic distribution are currently unknown (20). Oncopeltus processes labriformin in a highly parallel manner to that of monarchs consuming voruscharin, although an additional nondegradative pathway of 3-thiazoline via oxidation also exists for labriformin (Fig. 4). Mass spectrometric evidence suggests that the double oxidation occurs on the 3-thiazoline ring to form a rare S-oxothiazolidinone; this heterocyclic structure is supported by previously described thiazolidinone derivatives of cardenolides (SI Appendix, Figs. S10 and S11) (34). Here, Oncopeltus sequesters the less toxic nitrogen-containing oxidized labriformin in addition to the reduced desglucosyrioside and syriobioside A (Fig. 4). Together, these results suggest that nitrogen-containing cardenolides may be an evolved defense with apparent counter adaptations in multiple herbivores. The fact that both voruscharin and labriformin have strong inhibitory effects on adapted sodium pumps but relatively modest effects on the unadapted enzyme supports the notion that these compounds evolved in response to milkweed specialists.
The costs of coping with highly toxic nitrogen-containing cardenolides were elucidated for monarchs as a reduction in growth associated with conversion of voruscharin and sequestration of its metabolites (15). On the contrary, there is a long history of finding no such costs of cardenolide exposure or sequestration for Oncopeltus (40, 41, 45), likely due to their remarkable morphological and physiological adaptations (18, 42). Our results from feeding Oncopeltus diets with purified glycosylated aspecioside and labriformin support these past studies. Nonetheless, another recent study fed Oncopeltus prepared diets of pure cardenolides (a mix of ouabain and digitoxin), revealing some costs (46). Although Oncopeltus showed faster nymphal growth, increased body mass, and enhanced longevity when fed diets with cardenolides, fecundity was reduced by 50% when cardenolides were continued on their diets as adults (at a dose of 6 mg/g dry mass). This high but realistic concentration of cardenolides was higher than that in the current study (we used 0.5 mg/g cardenolides). Furthermore, current work in our laboratory suggests negative effects of A. syriaca seed cardenolides on Oncopeltus, although only for male bugs. Understanding the costs of adaptation, cardenolide conversion, and sequestration for Oncopeltus thus remains an important yet unresolved question.
In summary, geographic, chemical, and functional evidence supports the notion that labriformin may be important in the evolutionary interactions between common milkweed seeds and Oncopeltus. Expression of labriformin peaks in northern latitudes and appears to be evolving by natural selection; nonetheless, Oncopeltus seed predators have a remarkable tolerance to these compounds, they chemically modify them to reduce toxicity, and sequester the end products for their own defense.
Materials and Methods
Common milkweed, A. syriaca (Apocynaceae), is a long-lived perennial plant that reproduces both sexually (through seed, largely by outcrossing) and asexually (by producing spreading underground stems). Its native geographic range spans much of eastern North America (east of Nebraska) roughly from Quebec, Canada to North Carolina. A. syriaca is attacked by a community of at least 12 specialized herbivorous insects (16), with two species of lygaeid bugs that feed on seeds. Other insect herbivores feed on roots, stems, leaves, and phloem sap. Here we employ field-collected seeds previously used in Woods et al. (6), with three to five independent seed collections from each of 24 populations (total n = 113) (SI Appendix, Table S7). We focus on latitude as a predictor of defense expression across these populations because it is strongly correlated with mean annual temperature as well as three other measures of climate (6). Mean annual precipitation had little predictive power. Population locations and basic climatic information are given in SI Appendix, Table S7.
Seed Cardenolides along the Geographical Cline.
Based on methods in Agrawal et al. (15) we employed HPLC-UV analysis on an Agilent 1100 HPLC with diode array detector and a Gemini C18 reversed-phase, 3 µm, 150 mm × 4.6 mm column. Here we used 50 mg of ground freeze-dried tissue (seeds or bugs), added 1.5 mL of 100% methanol, a 20 µg digitoxin as an internal standard, and 20 FastPrep beads, extracted by agitating twice on a FastPrep-24 homogenizer for 45 s at 6.5 m/s. We then centrifuged the extract at 20,800 × g for 12 min. Supernatants were dried down in a vacuum concentrator at 35 °C and resuspended in 200 μL methanol, filtered using 0.45-µm hydrophobic membranes in a filter plate, and 15 μL was injected into the HPLC running a constant flow of 0.7 mL/min with a gradient of acetonitrile and water as follows: 0 to 2 min at 16% acetonitrile; 2 to 25 min from 16 to 70%; 25 to 30 min from 70 to 95%; 30 to 35 min at 95%; followed by 10 min of reconditioning with 16% acetonitrile. As is standard for cardenolide analysis (15), peaks were recorded at 218 nm and absorbance spectra were recorded between 200 nm to 300 nm. Peaks showing a characteristic single absorption maximum between 214 and 222 nm correspond to the unsaturated lactone, which is diagnostic of cardenolides in our system. Concentrations were standardized by peak area to the internal standard (digitoxin).
We recorded 28 cardenolides by HPLC; however, most were rare and occurred in less than half of the populations. Six cardenolides accounted for 87% of the total across populations and were considered the dominant peaks. Compound concentrations were also assessed using mass spectrometry, here pooling three samples per population (n = 24).
Following methods in Agrawal et al. (15), here we use high-resolution mass spectrometry to identify compounds (by exact mass and pattern of fragmentation) and compare their relative concentration. We employed a reversed-phase chromatography Dionex 3000 LC coupled to an Orbitrap Q-Exactive mass spectrometer controlled by Xcalibur software (Thermo Fisher Scientific). Methanolic extracts were separated on an Agilent Zorbax Eclipse XDB-C18 column (150 mm × 2.1 mm, particle size 1.8 µm) maintained at 40 °C with a flow rate of 0.5 mL/min. Solvent A contained 0.1% formic acid (FA) in water; solvent B contained 0.1% FA in acetonitrile. The A/B gradient was started at 5% B for 2 min after injection and increased linearly to 98% B at 11 min, followed by 3 min at 98% B, then back to 5% B over 0.1 min and finally at 5% B held for an additional 2.9 min to reequilibrate the column. Mass spectrometer settings were as follows: Spray voltage (−3.0 kV, +3.5 kV), capillary temperature 380 °C, probe heater temperature 400 °C; sheath, auxiliary, and sweep gas 60, 20, and 2 AU, respectively. S-Lens radio frequency level was 50, resolution 240,000 at m/z 200, automatic gain control (AGC) target 3e6. Each sample was analyzed in positive electrospray ionization mode with m/z ranges 70 to 1,000, and each isolated cardenolide was analyzed in both positive and negative electrospray ionization modes. Parameters for data-dependent tandem mass spectrometry (MS/MS) (dd-MS2) were MS1 resolution 60,000; AGC target 1e6. MS2 resolution was 30,000, AGC target 2e5, maximum injection time 50 ms, isolation window 1.0 m/z, stepped normalized collision energy (NCE) 10, 30; dynamic exclusion 1.5 s, top five masses selected for MS/MS per scan. LC-MS data were analyzed using MZmine software (see below) and MS2 spectra were obtained via Excalibur software (Thermo Fisher Scientific).
The acquired LC-MS data files were converted to mzXML files using the ProteoWizard MSconvert tool. LC-MS data were then preprocessed with the open-source MZmine 2 software (47) and consisted of peak detection, removal of isotopes, alignment, filtering, and peak filling. Peak detection was performed in three steps: 1) mass detection with noise value = 15,000; 2) automated data analysis pipeline (ADAP) chromatogram builder with minimum group size in number of scan = 5, group intensity threshold = 25,000, minimum height = 30,000, and m/z tolerance = 10 ppm; 3) wavelet ADAP deconvolution with S/N = 3, minimum feature height = 1,000, coefficient area threshold = 5, peak duration range = 0.01 to 3 min, and retention time wavelet range = 0.01 to 0.04 min. Isotopes were removed using the isotopic peak grouper with m/z tolerance = 10 ppm, retention time tolerance = 0.5 min, and maximum charge = 3. Chromatograms were aligned using the join aligner with m/z tolerance = 10 ppm, weight for m/z = 75, retention time tolerance = 0.5 min, and weight for retention time = 25. Filtering minimum peak in a row = 4, minimum peak in an isotopic pattern = 2, and keep-only peaks with MS2 scan. Gap filling was applied using the method peak finder with retention time correction with intensity tolerance = 10%, m/z tolerance = 10 ppm, and retention time tolerance = 0.5 min. Quality control (QC) metabolites with a coefficient of variation (CV) greater than 30% were removed from the whole data matrix. Correlation heat map, and t tests were performed with MetaboAnalyst 5.0 (48).
Glycosylated aspecioside, diglycosylated syriogenin, aspecioside A, glycosylated syriobioside, syrioside A and B, diglycosylated oxidized syriogenin, diglycosylated digitotoxin, and labriformin were isolated and characterized by NMR spectrometry. The chemical structures of glycosylated syriobioside, syrioside A and B, diglycosylated oxidized syriogenin, and diglycosylated digitoxigenin will be reported in a separate manuscript. The high-resolution mass spectrometry (HRMS) and MS/MS data of isolated cardenolides were used to retrieve the cardenolide ion adducts in A. syriaca seed samples. We mined the HRMS data for additional cardenolides based on reported structures in A. syriaca and anticipated cardenolides structures. After normalization using a standard, the relative concentration based on ion counts was reported.
Cardenolide concentrations (28 peaks) were assessed with latitude as a predictor. Because most plots clearly showed a nonlinear quadratic relationship, we included latitude squared in the model. Statistical analyses were conducted using JMP Pro-14. To test whether phenotypic differences between populations in cardenolide defenses were greater than neutral genetic differentiation, we used a modified PST > FST approach detailed in SI Appendix.
Sequestration and Feeding Experiment.
We quantified cardenolides in an A. syriaca seed pool (collected in Tompkins Co.) used to rear Oncopeltus, as well as whole Oncopeltus adults reared on these seeds, to address which compounds were sequestered (n = 5, 50-mg samples of each), using HPLC and HRMS as described above. Oncopeltus were collected in Ithaca, NY and reared in the laboratory on A. syriaca seed for fewer than five generations.
To address the specific sequestration of purified compounds, we conducted a feeding assay starting with freshly molted third instar Oncopeltus nymphs (from a laboratory colony obtained in 2015 from the University of Hamburg, Germany, and previously raised on sunflower seeds) on four artificial diets: Control (sunflower seed agar-based diet [SSABD]), SSABD spiked with ouabain octahydrate (0.59 mg/g dry diet), SSABD spiked with glycosylated aspecioside (0.58 mg/g dry diet), and SSABD spiked with labriformin (0.5 mg/g dry diet). The three doses were equimolar. Details on the diet ingredients and its preparation are given in Pokharel et al. (46). Purified compounds were isolated from A. syriaca seeds using a preparative HPLC fractionation method as in Agrawal et al. (15) and determined to be at least 95% pure based on NMR spectrometry. We dispersed the purified toxins with water and added them to the diet. Nymphs were placed in groups of three in a 90-mm Petri dish with vents (n = 10 Petri dishes per treatment) lined with filter paper and supplied with the corresponding artificial diet in a pellet and a separate source of water. Environmental conditions were set at 27 °C, 60% relative humidity at a light:dark cycle of 16 h:8 h (Binder KBWF 240), and all Petri dishes were spatially randomized. Diets were replaced once after 2 wk.
We measured the following parameters to assess effects of ingesting cardenolides: Mass (average in milligrams of the three individuals in each Petri dish after 3 wk), time until adulthood (day when all insects from the Petri dish reached adulthood), adult length (length in millimeters from the ventral tip of the head to the penultimate external abdominal segment), total eggs produced per adult female (virgin females mated to males from the same treatment/treatment, n = 8 except control n = 7), and total hatchlings per female (n = 8 except control n = 7, as above). A total of 29 adult bugs were also assessed for cardenolide sequestration (using HPLC for quantification and mass spectrometry for compound identification), equally spread across the four treatments. Analyses were carried out using the statistical software R (version 4.0.3). We conducted one-way ANOVAs followed by Tukey’s honestly significant difference post hoc analyses. For time until adulthood we used a Kruskal–Wallis test followed by a post hoc analysis Dunn test with the Benjamini–Hochberg P value adjustment method.
Functional Na+/K+-ATPase Assays.
We quantified the inhibitory potential of isolated cardenolides using Na+/K+-ATPase from the porcine cerebral cortex (Sigma-Aldrich), monarch butterflies, and Oncopeltus following methods of Petschenka et al. (14). Compounds were purified from A. syriaca seeds or Oncopeltus adults. Each compound was dissolved fully in methanol assayed by HPLC to determine concentration, and then dried and resuspended in 20% dimethyl sulfoxide (DMSO)/H2O to 5 mM. Due to solubility issues, labriformin was instead dissolved in acetonitrile for HPLC analysis, and then dried and resuspended in 20% DMSO/H2O to 1 mM. We then prepared 1/10 serial dilutions to produce a six-point inhibition curve for each compound, incubated with each of the three enzyme preparations.
Because the porcine enzyme is less tolerant than those of the milkweed herbivores, the set of dilutions used for its six-point curve was shifted by a factor of 1/10. Compound solutions were diluted 1:5 with a buffered reaction mix containing Tris-buffered ATP, NaCl, KCl, MgCl2, and porcine Na+/K+-ATPase, and incubated on a BioShake iQ microplate shaker (Quantifoil Instruments) at 200 rpm and 37 °C for 20 min. Milkweed cardenolides were run alongside equivalent molar solutions of ouabain. Reactions were terminated with 10% sodium dodecyl sulfate, then inorganic phosphate was stained with Taussky–Shorr reagent and absorbance measured spectrophotometrically at 700 nm. Absorbance values of reactions were corrected by their respective backgrounds (containing 10 mM ouabain, ATP, NaCl, KCl, MgCl2, and appropriate enzyme but lacking KCL), and dose–response curves were fitted using a nonlinear mixed effects model with a four-parameter logistic function in the statistical software R (function nlme with SSfpl in package nlme v3.1-137) based on ref 49. We focus analyses on cardenolide concentration at which the enzyme is inhibited by 50% (IC50) compared to a control without toxins added. IC50 values were compared with one-way ANOVAs.
Data Availability
All study data are included in the article and/or SI Appendix.
Supplementary Material
Acknowledgments
We thank Katalin Böröczky and Ivan Keresztes for the initial identification of labriformin, Prayan Pokharel and Sarah Rissmann for help with the feeding assays, Nathaniel Carlson and Max Goldman for help with the bug colonies, and Bennett Fox and Frank Schroeder for unfailing assistance to access the high-resolution mass spectrometer at the Boyce Thompson Institute. Comments from Micah Freedman, Stephen Malcolm, and Xoaquín Moreira improved the manuscript. This research was supported by grants from the NSF (to A.A.A.) (IOS-1907491 and IOS-2209762) and the German Research Foundation Grant PE 2059/3-1 (to G.P.).
Footnotes
Reviewers: S.M., Western Michigan University; and X.M.T., Mision Biologica de Galicia.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205073119/-/DCSupplemental.
References
- 1.López-Goldar X., Agrawal A. A., Ecological interactions, environmental gradients, and gene flow in local adaptation. Trends Plant Sci. 26, 796–809 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Hahn P. G., Agrawal A. A., Sussman K. I., Maron J. L., Population variation, environmental gradients, and the evolutionary ecology of plant defense against herbivory. Am. Nat. 193, 20–34 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Anstett D. N., Nunes K. A., Baskett C., Kotanen P. M., Sources of controversy surrounding latitudinal patterns in herbivory and defense. Trends Ecol. Evol. 31, 789–802 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Anstett D. N., et al. , Can genetically based clines in plant defence explain greater herbivory at higher latitudes? Ecol. Lett. 18, 1376–1386 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Moreira X., et al. , Latitudinal variation in plant chemical defences drives latitudinal patterns of leaf herbivory. Ecography 41, 1124–1134 (2018). [Google Scholar]
- 6.Woods E. C., Hastings A. P., Turley N. E., Heard S. B., Agrawal A. A., Adaptive geographical clines in the growth and defense of a native plant. Ecol. Monogr. 82, 149–168 (2012). [Google Scholar]
- 7.Crutsinger G. M., Gonzalez A. L., Crawford K. M., Sanders N. J., Local and latitudinal variation in abundance: The mechanisms shaping the distribution of an ecosystem engineer. PeerJ 1, e100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zangerl A. R., Berenbaum M. R., Phenotype matching in wild parsnip and parsnip webworms: Causes and consequences. Evolution 57, 806–815 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Li W., Schuler M. A., Berenbaum M. R., Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: Specificity and substrate encounter rate. Proc. Natl. Acad. Sci. U.S.A. 100 (suppl. 2), 14593–14598 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calla B., Wu W. Y., Dean C. A. E., Schuler M. A., Berenbaum M. R., Substrate-specificity of cytochrome P450-mediated detoxification as an evolutionary strategy for specialization on furanocoumarin-containing hostplants: CYP6AE89 in parsnip webworms. Insect Mol. Biol. 29, 112–123 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Dobler S., Dalla S., Wagschal V., Agrawal A. A., Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc. Natl. Acad. Sci. U.S.A. 109, 13040–13045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karageorgi M., et al. , Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature 574, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Goldar X., Hastings A., Züst T., Agrawal A., Evidence for tissue-specific defence-offence interactions between milkweed and its community of specialized herbivores. Mol. Ecol., 10.1111/mec.16450 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Petschenka G., et al. , Relative selectivity of plant cardenolides for Na+/K+-ATPases from the monarch butterfly and non-resistant insects. Front. Plant Sci. 9, 1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agrawal A. A., et al. , Cardenolides, toxicity, and the costs of sequestration in the coevolutionary interaction between monarchs and milkweeds. Proc. Natl. Acad. Sci. U.S.A. 118, e2024463118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal A. A., Monarchs and Milkweed: A Migrating Butterfly, a Poisonous Plant, and Their Remarkable Story of Coevolution (Princeton University Press, Princeton, NJ, 2017), pp. 296. [Google Scholar]
- 17.Herbertz M., et al. , Different combinations of insect Na, K-ATPase α-and β-subunit paralogs enable fine tuning of toxin resistance and enzyme kinetics. bioRxiv [Preprint] (2021). 10.1101/2020.08.28.272054 (Accessed 3 June 2022). [DOI]
- 18.Lohr J. N., Meinzer F., Dalla S., Romey-Glüsing R., Dobler S., The function and evolutionary significance of a triplicated Na,K-ATPase gene in a toxin-specialized insect. BMC Evol. Biol. 17, 256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leinonen T., Cano J. M., Mäkinen H., Merilä J., Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. J. Evol. Biol. 19, 1803–1812 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Seiber J. N., Lee S. M., Benson J., “Cardiac glycosides (cardenolides) in species of Asclepias (Asclepiadaceae)” in Handbook of Natural Toxins, R. F. Keeler, A. T. Tu, Eds. (Plant and Fungal Toxins, Marcel Dekker, Amsterdam, 1983), vol. 1, pp. 43–83. [Google Scholar]
- 21.Whitlock M. C., Guillaume F., Testing for spatially divergent selection: Comparing QST to FST. Genetics 183, 1055–1063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brommer J. E., Whither Pst? The approximation of Qst by Pst in evolutionary and conservation biology. J. Evol. Biol. 24, 1160–1168 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Boyle J. H., et al. , Temporal matches and mismatches between monarch butterfly and milkweed population changes over the past 12,000 years. bioRxiv [Preprint] (2022). 10.1101/2022.02.25.481796 (Accessed 3 June 2022). [DOI]
- 24.Dalla S., Dobler S., Gene duplications circumvent trade-offs in enzyme function: Insect adaptation to toxic host plants. Evolution 70, 2767–2777 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Seiber J. N., et al. , Cardenolide connection between overwintering monarch butterflies from Mexico and their larval food plant, Asclepias syriaca. J. Chem. Ecol. 12, 1157–1170 (1986). [DOI] [PubMed] [Google Scholar]
- 26.Brown P., von Euw J., Reichstein T., Stöckel K., Watson T. R., Cardenolides of Asclepias syriaca L., Probable structure of syrioside and syriobioside. Helv. Chim. Acta 62, 412–441 (1979). [Google Scholar]
- 27.Moreira X., Petry W. K., Mooney K. A., Rasmann S., Abdala-Roberts L., Elevational gradients in plant defences and insect herbivory: Recent advances in the field and prospects for future research. Ecography 41, 1485–1496 (2018). [Google Scholar]
- 28.Kooyers N. J., Blackman B. K., Holeski L. M., Optimal defense theory explains deviations from latitudinal herbivory defense hypothesis. Ecology 98, 1036–1048 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Agrawal A. A., et al. , Evolution of plant growth and defense in a continental introduction. Am. Nat. 186, E1–E15 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Malcolm S. B., Milkweeds, monarch butterflies and the ecological significance of cardenolides. Chemoecology 5/6, 101–117 (1995). [Google Scholar]
- 31.Malcolm S. B., Cockrell B. J., Brower L. P., Cardenolide fingerprint of monarch butterflies reared on common milkweed, Asclepias syriaca L. J. Chem. Ecol. 15, 819–853 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Agrawal A. A., Petschenka G., Bingham R. A., Weber M. G., Rasmann S., Toxic cardenolides: Chemical ecology and coevolution of specialized plant-herbivore interactions. New Phytol. 194, 28–45 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Seiber J. N., Roeske C. N., Benson J. M., Three new cardenolides from the milkweeds Asclepias eriocarpa and A. labriformis. Phytochemistry 17, 967–970 (1978). [Google Scholar]
- 34.Abe F., Yamauchi T., An androstane bioside and 3′-thiazolidinone derivatives of doubly-linked cardenolide glycosides from the roots of Asclepias tuberosa. Chem. Pharm. Bull. (Tokyo) 48, 991–993 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Benson J. M., et al. , Effects on sheep of the milkweeds Asclepias eriocarpa and A. labriformis and of cardiac glycoside-containing derivative material. Toxicon 17, 155–165 (1979). [DOI] [PubMed] [Google Scholar]
- 36.Brower L. P., Seiber J. N., Nelson C. J., Lynch S. P., Tuskes P. M., Plant-determined variation in the cardenolide content, thin-layer chromatography profiles, and emetic potency of monarch butterflies, Danaus plexippus reared on the milkweed, Asclepias eriocarpa in California. J. Chem. Ecol. 8, 579–633 (1982). [DOI] [PubMed] [Google Scholar]
- 37.Züst T., Petschenka G., Hastings A. P., Agrawal A. A., Toxicity of milkweed leaves and latex: Chromatographic quantification versus biological activity of cardenolides in 16 Asclepias species. J. Chem. Ecol. 45, 50–60 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Taverner A. M., et al. , Adaptive substitutions underlying cardiac glycoside insensitivity in insects exhibit epistasis in vivo. eLife 8, e48224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duffey S., Scudder G., Cardiac glycosides in Oncopeltus fasciatus (Dallas) (Hemiptera: Lygaeidae). I. The uptake and distribution of natural cardenolides in the body. Can. J. Zool. 52, 283–290 (1974). [Google Scholar]
- 40.Vaughan F. A., Effect of gross cardiac glycoside content of seeds of common milkweed, Asclepias syriaca, on cardiac glycoside uptake by the milkweed bug Oncopeltus fasciatus. J. Chem. Ecol. 5, 89–100 (1979). [Google Scholar]
- 41.Isman M. B., Dietary influence of cardenolides on larval growth and development of milkweed bug Oncopeltus fasciatus. J. Insect Physiol. 23, 1183–1187 (1977). [Google Scholar]
- 42.Scudder G. G. E., Moore L. V., Isman M. B., Sequestration of cardenolides in Oncopeltus fasciatus: Morphological and physiological adaptations. J. Chem. Ecol. 12, 1171–1187 (1986). [DOI] [PubMed] [Google Scholar]
- 43.Bramer C., Dobler S., Deckert J., Stemmer M., Petschenka G., Na+/K+-ATPase resistance and cardenolide sequestration: Basal adaptations to host plant toxins in the milkweed bugs (Hemiptera: Lygaeidae: Lygaeinae). Proc. Biol. Sci. 282, 20142346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore L. V., Scudder G. G., Selective sequestration of milkweed (Asclepias sp.) cardenolides in Oncopeltus fasciatus (Dallas) (Hemiptera: Lygaeidae). J. Chem. Ecol. 11, 667–687 (1985). [DOI] [PubMed] [Google Scholar]
- 45.Chaplin S. J., Chaplin S. B., Growth dynamics of a specialized milkweed seed feeder (Oncopeltus fasciatus) on seeds of familiar and unfamiliar milkweeds (Asclepias spp.). Entomol. Exp. Appl. 29, 345–355 (1981). [Google Scholar]
- 46.Pokharel P., Steppuhn A., Petschenka G., Dietary cardenolides enhance growth and change the direction of the fecundity-longevity trade-off in milkweed bugs (Heteroptera: Lygaeinae). Ecol. Evol. 11, 18042–18054 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pluskal T., Castillo S., Villar-Briones A., Orešič M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang Z., et al. , MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Züst T., et al. , Independent evolution of ancestral and novel defenses in a genus of toxic plants (Erysimum, Brassicaceae). eLife 9, e51712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.