Abstract
Eleven Acanthamoeba isolates, obtained from Acanthamoeba keratitis patients, from contact lens cases of non-Acanthamoeba keratitis patients, from asymptomatic individuals, from necrotic tissue, and from tap water and two reference strains were investigated by morphological, molecular biological, and physiological means in order to discriminate clinically relevant and nonrelevant isolates. All clinically relevant isolates showed Acanthamoeba sp. group II morphology. 18S ribosomal DNA sequencing revealed sequence type T4 to be the most prevalent group among the isolates and also the group recruiting most of the pathogenic strains. Interestingly, within T4 the strains of no clinical relevance clustered together. Moreover, physiological properties appeared to be highly consistent with initial pathogenicity and with sequence clustering. Altogether, the results of our study indicate a correlation between the phylogenetic relationship and pathogenicity.
Free-living amoebae of the genus Acanthamoeba are of medical interest as the causative agents of several infections in the immunodeficient host (8) and also of a very often seriously progressing keratitis occurring predominantly in contact lens wearers. Acanthamoeba keratitis has become increasingly important within the last 10 years correlating to the growing number of contact lens wearers. Contaminated contact lens care systems usually are the first step in an Acanthamoeba keratitis pathogenesis.
Various species have been reported to be able to cause keratitis: Acanthamoeba castellanii, A. polyphaga, A. hatchetti, A. culbertsoni, A. rhysodes, A. lugdunensis, A. quina, and A. griffini (23). However, accurate species determination remains problematic. A great advantage in the differentiation of acanthamoebae was the division of the genus into three morphological groups by Pussard and Pons (22), with a majority of keratitis causing strains belonging to group II. Recently, Stothard et al. (26) identified 12 Acanthamoeba sequence types by 18S ribosomal DNA (rDNA) sequencing and, interestingly, they found evidence that most keratitis causing strains group into sequence type T4. However, a final and generally accepted system does not yet exist. Physiological characteristics such as temperature tolerance (11), growth in defined media (4), the ability to migrate in an “under-agarose” system (28), and cytopathic effects in human cell lines (3) were found to be pathogenicity related to a certain extent. All of these methods provide some information on pathogenicity, but no single method seems to be universally valid.
The aim of our study was to correlate various qualities of Acanthamoeba strains and to evaluate any consistencies among clinically relevant isolates and among isolates of no clinical relevance. Isolates were identified by morphological and molecular biological means. Cluster analysis based on 18S rDNA sequences was performed, and the isolates were grouped according to their pathogenicity-related properties.
MATERIALS AND METHODS
Amoebae.
A total of 13 Acanthamoeba strains of various clinical relevance, including 1 strain from the brain of a mouse, 1 strain from the necrotic tissue of a basilisk, 4 strains from Acanthamoeba keratitis patients, 3 strains from non-Acanthamoeba keratitis patients, 2 strains from contact lens cases of asymptomatic individuals, 1 strain from the nasal mucosa of an asymptomatic individual, and 1 strain from tap water, were investigated and compared (Table 1).
TABLE 1.
Origin of the investigated strains
| Strain | Morphological group | Sequence type | GenBank accession no. | Identification | Origin | Source or reference |
|---|---|---|---|---|---|---|
| 72/2 | III | T5 | U94732 | A. lenticulata | Mouse brain | 6 |
| 312-2 | II | NDa | A. quina-lugdunensis | Nasal mucosa (asymptomatic individual) | 6 | |
| 40AB | II | T4 | AF114438 | A. castellanii | Necrotic tissue of a B. plumifrons | 29 |
| 4RE | II | T11 | AF251937 | A. hatchetti | Contact lens case (non-Acanthamoeba keratitis patient) | Walochnik et al., submitted |
| 9GU | II | T4 | AF251938 | A. castellanii | Contact lens case (non-Acanthamoeba keratitis patient) | Walochnik et al., submitted |
| 11DS | II | T6 | AF251939 | A. hatchetti | Keratitis patient, contact lens case | Walochnik et al., submitted |
| 36KL | III | ND | A. palestinensis | Contact lens case | 12 | |
| 7AR | II | ND | A. rhysodes | Tap water | This study | |
| 1BU | Keratitis patient, cornea | New isolate | ||||
| 2HH | Keratitis patient, cornea | New isolate | ||||
| 3ST | Keratitis patient, cornea | New isolate | ||||
| 4CL | Contact lens case | New isolate | ||||
| 5SU | Contact lens case (non-Acanthamoeba keratitis patient) | New isolate |
ND, not determined.
Non-Acanthamoeba keratitis patients are patients suffering from a keratitis not caused by Acanthamoeba. The contact lens containers of non-Acanthamoeba keratitis patients were investigated on routine basis and in all cases acanthamoebae could only be found in the contact lens containers and not in clinical specimens such as corneal swabs or corneal epithelium. Acanthamoeba keratitis patients differed from non-Acanthamoeba keratitis patients by showing typical clinical signs for Acanthamoeba keratitis, such as the presence of keratitis with severe pain and photophobia, stromal infiltrates, and radial keratoneuritis; no response to antibacterial or antiviral treatment; and detection of acanthamoebae in the corneal epithelium.
The strain 11DS was isolated from a patient suffering from a severe Acanthamoeba keratitis, and the strains 9GU and 4RE were derived from the contact lens cases of non-Acanthamoeba keratitis patients (J. Walochnik, E.-M. Haller-Schober, H. Kölli, O. Picher, A. Obwaller, and H. Aspöck, submitted for publication). Strain 36KL was isolated from the contact lens case of an asymptomatic individual (12). The 40AB strain is a veterinary isolate and was isolated from necrotic tissue of a Basiliscus plumifrons with undefined clinical relevance (29). Strain 7AR was isolated from tap water 4 years earlier and has been kept in an axenic culture in our lab.
Five new isolates were included in this study. Three were isolated from the clinical specimens of patients who had developed a severe keratitis (1BU, 2HH, and 3ST), one derived from a contact lens case of an asymptomatic individual (4CL), and one was from the contact lens case of a non-Acanthamoeba keratitis patient (5SU).
Strain 312-2 (A. quina-lugdunensis) (6) representing morphological group II functioned as a reference strain. We also included a group III reference strain (A. lenticulata strain 72/2) for the 36KL strain and as the 11DS isolate, although exhibiting a group II morphology shows a group III-related sequence type. Strains 312-2 and 72/2 had originally been isolated from the nasal mucosa of healthy individuals (17); strain 72/2 had proven to be highly virulent to mice after intranasal instillation (6). The strain used in this study is the reisolate from the brain of the mouse.
Isolation.
The amoebae were isolated from clinical specimens by the plate culture method. Corneal epithelium and swabs from contact lens cases were inoculated onto non-nutrient agar plates seeded with a 48-h-old culture of Escherichia coli. Initial cultures were diluted in order to eliminate cocontaminants by cutting a small piece of agar with a sterile scalpel and applying it centrally onto a fresh plate. All isolates were cloned by transferring a single cyst to a fresh plate employing a micromanipulator.
Axenic cultures were obtained by harvesting cysts from the plate cultures, washing them three times in sterile saline, and incubating them in 3% HCl overnight in order to eliminate coexisting bacteria. After another washing step cysts were harvested by centrifugation at 500 × g for 7 min and then transferred into the liquid culture medium. Sterile-filtrated PYG (proteose peptone-yeast extract-glucose) (20) was used as the standard medium. Amoebae were cultured in 150-cm2 tissue culture flasks (Corning Costar, Bodenheim, Germany) at 30°C. All strains were also maintained as plate cultures.
Morphological classification.
Morphological studies included measurement of trophozoites and cysts and assessment of the size and shape of the endo- and ectocysts and of the mean number of opercula and resulted in the classification of the isolates as Acanthamoeba sp. groups I, II, or III. Species identification was achieved according to the identification key of Page (20) based mainly on cyst morphology and temperature tolerance.
Isolation of DNA.
Whole-cell DNA was isolated by a modified UNSET (14) procedure. In brief, amoebae (∼106 cells) were harvested from extensively growing axenic cultures by centrifugation at 500 × g for 7 min. From strains which would not grow in axenic culture, DNA was isolated directly from the plate culture as described previously (29). The amoeba pellet was resuspended in 500 μl of UNSET lysis buffer, overlaid with 500 μl of phenol-chloroform-isoamylalcohol (PCI), and shaken gently for 5 h. The suspension was centrifuged at 3,000 × g for 10 min, and the upper, aqueous phase was transferred to a new tube. PCI extraction was repeated two times for 10 min each time. Nucleic acids were precipitated by ethanol (15 min at −80°C), pelleted at 12,000 × g for 30 min at 4°C, washed in 70% ethanol, air dried, and resuspended in 30 μl of sterile double-distilled water. The 18S rRNA gene was amplified using the SSU1 and SSU2 primers (9), which are complementary to the 5′ and the 3′ end of the gene, respectively. We used 0.8 μl of whole-cell DNA and a standard amplification program (30 cycles of 95°C for 1 min, 50°C for 2 min, and 72°C for 3 min). Amplification of the 18S rRNA gene was visualized by ethidium bromide staining in agarose gel electrophoresis. The amplified gene was sequenced stepwise by direct sequencing from the PCR product using the Thermo Sequenase II sequencing kit (Amersham Pharmacia Biotech GmbH, Vienna, Austria) and the subsequent construction of complementary internal primers. Sequences were obtained from both strands. Sequencing was carried out in a 310 ABI PRISM automated sequencer (PE Applied Biosystems, Langen, Germany), and sequence data were processed with the GeneDoc (18) sequence editor.
Identification of sequence types and dissimilarity calculation.
The sequence types for the 312-2, 36KL, 7AR, 1BU, 2HH, 3ST, 4CL, and 5SU strains were determined by searching for the published sequence with the greatest nucleotide identity employing the BLAST application (1) and by subsequently calculating the percentage of nucleotide dissimilarity using all sites of the gene. Stothard et al. (26) have established their sequence types with <4.9% nucleotide dissimilarity within the sequence types and >5% nucleotide dissimilarity between different sequence types. Multiple sequence alignment was performed by subsequent pairwise alignment using the CLUSTAL X application (27) and showed homologous positions. A dissimilarity matrix was calculated using all sites of the gene.
Cluster analysis.
Cluster analysis was performed for the T4 isolates by constructing a cladogram using the PHYLIP (7) package. The tree was rooted by sequence types T3 and T11 using the T3 strains S-7 (A. griffini; GenBank accession no. U07412), Hay & Seal (A. griffini; S81337), Panola Mt. (A. polyphaga; AF019052), and Sawyer (A. pearcei; AF019053) and the T11 strains RB:F:1 (A. stevensoni; AF019069), BH-2 (A. hatchetti; AF019068), and our T11 isolate 4RE (A. hatchetti; AF251937) as an outgroup.
Primer sites, unique gaps, and introns were excluded from the analysis. The nucleotide sequence alignment was bootstrapped using the SEQBOOT application with a generation of 1,000 replicates. Distance matrices were calculated using DNADIST. Data were also analyzed with maximum parsimony using DNAPARS. Bootstrapped matrices were analyzed with the Neighbor-joining application and a Kimura two-parameter correction. A consensus tree was generated from the resulting trees using CONSENSE and prepared as a figure with the TREEVIEW (21) application.
Growth rates.
Since several strains did not grow in axenic culture, growth rates were evaluated on a lawn of living E. coli in the plate culture. Amoeba cysts were harvested from plate cultures, counted in a hemacytometer, and brought to a concentration of 105 cells/ml. Then, 1 μl was inoculated into the center of an agar plate seeded with 100 μl of a 48-h-old culture of E. coli. The cultures were incubated at a range of temperatures (30, 34, 37, 40, and 42°C) and observed daily using phase-contrast microscopy. In the presence of bacteria the amoebae excyst and, feeding on the bacteria, constantly proliferate by binary fission. Replacing the bacteria, the amoeba front moves toward the edges of the plate. Plates were marked with three concentric circles spaced at 15-mm intervals on the bottom side. Growth rates were recorded after 48 h and assessed according to the number of circles already traversed by the amoeba front. The ability of the amoebae to migrate into the agar was recorded after 72 h, and the temperature tolerances were recorded after 2 weeks.
Cytopathic effects.
HEp-2 cells were cultured in a 1:1 mixture of PC-1 (BioWhittaker, Walkersville, Md.) and CO2-independent medium (Life Technologies, Ltd., Paisley, Scotland) supplemented with l-glutamine (2 mM) in 75-cm2 tissue culture flasks (Corning/Costar, Bodenheim, Germany) at 37°C under sterile conditions. Then, 1 ml of a 105-cell/ml axenized suspension of each isolate was inoculated onto a monolayer of HEp-2 cells. Cocultures of amoebae and tissue cells were incubated at 30, 37, and 42°C. Pathogenicity was defined as complete lysis of the monolayer within 48 h at 30°C. All experiments were carried out in triplicate and were repeated after 6 weeks.
Sequence data.
Sequence data were deposited in GenBank and are available under the following accession numbers: AF260718 (strain 312-2), AF260719 (strain 36KL), AF260720 (strain 7AR), AF260721 (strain 1BU), AF260722 (strain 2HH), AF260723 (strain 3ST), AF260724 (strain 4CL), and AF260725 (strain 5SU).
RESULTS
Identification of strains.
The five new isolates (1BU, 2HH, 3ST, 4CL, and 5SU) all exhibited a typical Acanthamoeba group II morphology. 2HH and 3ST were assigned to A. hatchetti. The 1BU and the 4CL strains were designated to A. castellanii, and the 5SU strain was identified as A. polyphaga. Altogether, the species spectrum in our study comprised four strains of A. hatchetti, four strains of A. castellanii, and one strain each of A. lenticulata, A. palestinensis, A. polyphaga, A. quina-lugdunensis, and A. rhysodes.
Sequence typing in all cases revealed >97% identity to published strains, and all isolates could be designated as belonging to one of the published 12 sequence types. Strains 312-2 (AF260718), 36KL (AF260719), 7AR (AF260720), 1BU (AF260721), 2HH (AF260722), 3ST (AF260723), 4CL (AF260724), and the 5SU (AF260725) all exhibited sequence type T4. Including the isolates with known sequence types, T4 was represented by 10 isolates; exceptions were isolate 72/2 with sequence type T5, isolate 11DS with sequence type T6, and isolate 4RE with sequence type T11. In several cases the sequence typing was inconsistent with the morphological species identification. This was most apparent in the four isolates which were identified as A. hatchetti, two of which exhibited sequence type T4, one of which exhibited sequence type T11, and one of which exhibited sequence type T6. In the 11DS and the 36KL strains the molecular biological typing even interferes with the gross morphological group. The 11DS isolate shows a typical A. hatchetti and thus Acanthamoeba group II morphology but also shows a T6 sequence type, which represents morphological group III. Strain 36KL shows a group III morphology but exhibits sequence type T4, associated with Acanthamoeba group II. Pairwise nucleotide dissimilarity calculation revealed sequence dissimilarities of 0.62 to 3.84% among the T4 isolates (Table 2). Interestingly, the 11DS strain identified as type T6 showed more sequence similarity to the T4 strains, representing morphological group II, than to the 72/2 strain (T5), even though T6 and T5 both represent Acanthamoeba group III, and the 4RE strain (T11) showed <5% sequence dissimilarity to the T4 strains 5SU, 1BU, and 3ST, which conflicts with the perception of >5% dissimilarity between different sequence types.
TABLE 2.
Percentages of sequence dissimilarities of investigated strains
| Strain | % Sequence dissimilarity in:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 312-2 | 7AR | 36KL | 5SU | 9GU | 4CL | 1BU | 3ST | 40AB | 2HH | 4RE | 11DS | 72/2 | |
| 312-2 | 0 | 0.98 | 1.66 | 1.52 | 1.75 | 1.25 | 2.01 | 1.88 | 2.10 | 3.09 | 5.28 | 8.90 | 11.19 |
| 7AR | 0.98 | 0 | 2.05 | 1.87 | 2.14 | 1.83 | 2.68 | 2.54 | 2.90 | 3.84 | 5.75 | 9.28 | 11.69 |
| 36KL | 1.66 | 2.05 | 0 | 0.62 | 1.43 | 1.56 | 2.76 | 2.54 | 2.72 | 3.12 | 5.08 | 9.27 | 11.68 |
| 5SU | 1.52 | 1.87 | 0.62 | 0 | 1.25 | 1.34 | 2.63 | 2.41 | 2.58 | 2.90 | 4.99 | 9.22 | 11.49 |
| 9GU | 1.75 | 2.14 | 1.43 | 1.25 | 0 | 1.69 | 2.90 | 2.67 | 2.76 | 3.65 | 5.57 | 9.40 | 11.85 |
| 4CL | 1.25 | 1.83 | 1.56 | 1.34 | 1.69 | 0 | 2.28 | 2.14 | 2.19 | 2.64 | 5.05 | 9.02 | 11.17 |
| 1BU | 2.01 | 2.68 | 2.76 | 2.63 | 2.90 | 2.28 | 0 | 0.90 | 2.65 | 3.36 | 4.89 | 8.47 | 11.25 |
| 3ST | 1.88 | 2.54 | 2.54 | 2.41 | 2.67 | 2.14 | 0.90 | 0 | 2.29 | 2.92 | 4.53 | 8.53 | 11.00 |
| 40AB | 2.10 | 2.90 | 2.72 | 2.58 | 2.76 | 2.19 | 2.65 | 2.29 | 0 | 3.42 | 6.12 | 9.58 | 12.02 |
| 2HH | 3.09 | 3.84 | 3.12 | 2.90 | 3.65 | 2.64 | 3.36 | 2.92 | 3.42 | 0 | 5.42 | 9.01 | 11.46 |
| 4RE | 5.28 | 5.75 | 5.08 | 4.99 | 5.57 | 5.05 | 4.89 | 4.53 | 6.12 | 5.42 | 0 | 9.68 | 11.82 |
| 11DS | 8.90 | 9.28 | 9.27 | 9.22 | 9.40 | 9.02 | 8.47 | 8.53 | 9.58 | 9.01 | 9.68 | 0 | 12.30 |
| 72/2 | 11.19 | 11.69 | 11.68 | 11.49 | 11.85 | 11.17 | 11.25 | 11.00 | 12.02 | 11.46 | 11.82 | 12.30 | 0 |
T4 isolates are indicated in boldface.
The four A. hatchetti isolates (4RE, 11DS, 2HH, and 3ST) showed sequence dissimilarities ranging from 2.92 to 9.68% and the four A. castellanii isolates (1BU, 4CL, 9GU, and 40AB) differed by 1.69 to 2.90%.
Altogether, 10 isolates, including four clinically relevant and six not clinically relevant isolates, displayed sequence type T4. The closest related isolates were the 36KL and the 5SU strains (0.62%), both of which were isolated from contact lens cases, the 36KL strain from an asymptomatic contact lens wearer and the 5SU strain from a non-Acanthamoeba keratitis patient. Among the clinical strains, the 1BU and 3ST strains showed the lowest nucleotide dissimilarity (0.9%). It is remarkable that the nonclinical isolates (36KL, 5SU, 9GU, 4CL, 7AR, and 312-2) among type T4 showed lower dissimilarities to one another than to any of the clinical isolates.
Cluster analysis.
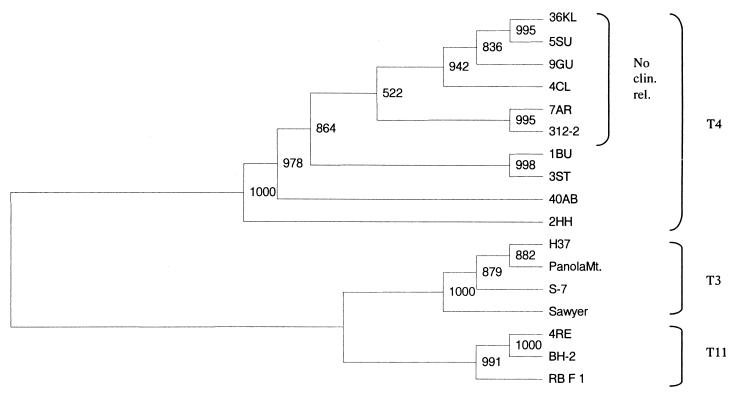
In order to evaluate whether this grouping is also valid in a phylogenetic context, the T4 isolates were grouped according to their 18S rDNA sequences by constructing a cladogram (Fig. 1). T3 and T11 strains were used as an outgroup since these are the closest related sequence types to type T4 (26). Our analysis revealed high bootstrap values and also, if maximum parsimony or other outgroups were used, the clustering of strains remained in the same order. Cluster analysis revealed that all T4 isolates formed a monophyletic group. Interestingly, within type T4 the strains of no clinical relevance (36KL, 5SU, 9GU, 4CL, 7AR, and 312-2) clustered together (Fig. 1).
FIG. 1.
18S rDNA neighbor-joining tree of the 10 sequence type T4 isolates. The tree was rooted by T3 and T11 using the T3 strains S-7, Hay & Seal, Panola Mt., and Sawyer and the T11 strains RB:F:1, BH-2 and the T11 isolate 4RE as an outgroup. Bootstrap values are based on 1,000 replicates. The numbers at the nodes represent bootstrap values based on 1,000 replicates.
Physiological characterization.
Generally, isolates that clustered together in the sequence analysis were rather similar in their physiological properties, and moreover, physiological characteristics of the isolates corresponded very well to their initial clinical relevance values (Table 3). While virulent strains exhibited high growth rates, high temperature tolerances, and high cytopathic effects, the strains isolated from contact lens cases or sanitary facilities grew only moderately, preferentially at lower temperatures, and showed low cytopathic effects. All strains except the 36KL isolate showed growth at 37°C, and eight strains showed the ability to grow at 40°C. However, all five strains growing at 42°C were among the clinical isolates. The physiological characteristics of the 40AB strain with previously unknown clinical relevance suggest that this strain is virulent.
TABLE 3.
Physiological characters of the isolates, listed according to decreasing pathogenicity of related charactersa
| Strain | Origin | Morphological group | Sequence type | Growth rate at:
|
Under-agarose migration | Axenic culture | Cytopathic effect | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 30°C | 34°C | 37°C | 40°C | 42°C | |||||||
| 72/2 | Mouse brain | III | T5 | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ |
| 11DS | Keratitis | II | T6 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 1BU | Keratitis | II | T4 | +++ | +++ | +++ | +++ | ++ | ++ | +++ | ++ |
| 2HH | Keratitis | II | T4 | +++ | +++ | +++ | +++ | ++ | ++ | +++ | +++ |
| 3ST | Keratitis | II | T4 | +++ | +++ | +++ | + | − | ++ | +++ | ++ |
| 40AB | Necrosis | II | T4 | +++ | +++ | +++ | ++ | ++ | ++ | − | ++ |
| 5SU | Lens case | II | T4 | ++ | ++ | ++ | + | − | − | ++ | + |
| 4CL | Lens case | II | T4 | +++ | +++ | ++ | − | − | + | ++ | + |
| 9GU | Lens case | II | T4 | +++ | +++ | ++ | − | − | + | ++ | ++ |
| 312-2 | Nasal mucosa | II | T4 | +++ | ++ | ++ | − | − | − | ++ | + |
| 36KL | Lens case | III | T4 | ++ | ++ | − | − | − | ++ | − | − |
| 7AR | Tap water | II | T4 | ++ | + | + | + | − | − | − | − |
| 4RE | Lens case | II | T11 | + | + | + | − | − | − | + | − |
Response: +++, very strong; ++, strong; +, moderate; −, none.
Under-agarose migration was shown by nine strains, including all of the clinical isolates. All isolates except 4RE, 36KL, 7AR, and the 40AB strain showed good growth in an axenic culture. In contrast to 40AB, isolates 4RE, 36KL, and 7AR showed poor growth even on living bacteria.
Cytopathic effects were restricted to 10 strains and were proven to be independent of incubation temperature, except in strain 40AB, which only showed cytopathic effects at 30°C. Temperature tolerances and cytopathic effects appeared to be constant characteristics and did not alter after multiple subcultures. The reference strain 72/2 isolated from the brain of a mouse and strains 11DS and 2HH strain deriving from patients with a seriously progressing keratitis showed the best growth, were highly temperature tolerant and, moreover, were the only three strains resulting in complete lysis of a HEp-2 monolayer within 24 h.
DISCUSSION
Identification.
Apart from the reference strain 72/2 and the 36KL strain, all investigated isolates displayed an Acanthamoeba group II morphology. Also, in a previous study of 18 cases of Acanthamoeba-associated keratitis in Austria, all Acanthamoeba isolates exhibited a group II morphology (Walochnik et al., submitted). In fact, group II seems to be the most abundant group not only in the environment but also in clinical specimens (26).
All isolates revealed high sequence identities to published strains from other parts of the world. In Naegleria, the other important free-living amoeba of potential pathogenicity, a geographically defined whole-cell DNA restriction fragment length polymorphism (RFLP) was determined (15), and it was shown that in Naegleria fowleri interstrain variation correlates with geographic origin. Acanthamoeba species, however, seem to be geographically widespread (10). This may also be due to the enormously robust cysts being resistant to various opposing environmental conditions, including desiccation, and thus allowing a wide distribution.
Sequencing data revealed most obviously that T4 is the group of acanthamoebae isolated most frequently; 10 of 13 investigated strains showed sequence type T4. This was already reported by Gast et al. (9) and by Stothard et al. (25, 26). Strain 4RE (T11) was more closely related to the T4 strains than to the strain 72/2 or strain 11DS, a finding which supports T11 being besides T3 the closest relative of T4 (26). Interestingly, it showed <5% sequence dissimilarity to the T4 strains 5SU, 1BU, and 3ST, a result which interferes with the perception of >5% dissimilarity between different sequence types. However, the branching pattern in the T3, T4, and T11 regions is reported to be ambiguous (26); we therefore still hold to the described system.
In several cases sequence typing interfered with morphological identification. This was most evident in the A. hatchetti isolates. In fact, A. hatchetti, as well as A. castellanii, A. culberstoni, A. palestinensis, and A. polyphaga, has been described to be polyphyletic (26). Stothard et al. (26) generally propose to reclassify all sequence type T4 isolates as A. castellanii, since T4 includes the type strain for this species. This seems reliable to us as long as we try to use the traditional binary nomenclature also in free-living amoebae. Our examinations demonstrate once more how difficult it may be in the future to use the binary nomenclature in genetically different but morphological identical organisms with asexual reproduction. In any case, it has become apparent that a thorough molecular analysis of various strains of “species” will be a prerequisite for a new and stable nomenclature in the genus Acanthamoeba.
Pathogenicity.
Stothard et al. (26) reported on 29 of 30 analyzed Acanthamoeba keratitis causing strains belonging to sequence type T4. They anticipate that the ability to cause keratitis might have only evolved once, since the only exception in their study is a T3 strain and this sequence type is, other than type T11, the sequence type most closely related to sequence type T4.
In our study, four of the five clinical isolates showed sequence type T4; only strain 11DS diverged from the other clinical strains in displaying sequence type T6. Nevertheless, it could not be morphologically discriminated from the T4 keratitis causing strains 2HH and 3ST of A. hatchetti. Types T6 and T2 are, next to types T3 and T11, the closest relatives to T4 (26). This correlation between virulence and distinct genotypes is supported by the recent identification of two genetic markers that distinguish pathogenic and nonpathogenic strains of Acanthamoeba (13). Also, in Naegleria (5, 15) and Entamoeba (2) spp., pathogenic and nonpathogenic isolates can be discriminated by molecular biological methods.
However, it remains unclear whether these correlations are due to T4 strains being more abundant in the environment or to their increased virulence for the cornea. In fact, most environmental isolates also group into type T4, though this need not necessarily interfere with the idea of a single evolution of pathogenicity in acanthamoebae, since virulence could subsequently have been lost by several strains. It is interesting that, in our study, within type T4 all strains with no clinical relevance revealed higher nucleotide dissimilarities to all of the clinical strains than to any of the other nonclinical strains.
Cluster analysis.
This grouping was also confirmed by cluster analysis, within type T4, isolates with no clinical relevance clustered together. It has been reported that restriction endonuclease digestions of Acanthamoeba whole-cell DNA reveal a distinct RFLP type being most frequently associated with keratitis (16), and comparison of mitochondrial DNA digests also indicate the presence of coherent cladistic populations among the clinical Acanthamoeba isolates (30). In a recent study on mitochondrial DNA digestion patterns, it was shown that 80% of the human isolates studied group into 7 of 22 genotypes, whereas the other 20% were distributed among the 15 other genotypes; the authors assume that virulence may be associated with specific clusters of cladistic groups within Acanthamoeba (31). Also, in Acanthamoeba group III, a grouping according to pathogenicity has been observed with A. griffini branching between pathogenic and nonpathogenic isolates (32). The recent finding of an eye and a lung isolate being most closely related with regard to their 18S rDNA sequences confirms this assumption. The authors concluded that any pathogenic isolate may be capable of infecting more than one tissue (9). It seems at least conceivable to us that any isolate infecting the brain can also infect the eye, an immunologically underprivileged site.
It is most important that, in our study, strains were determined by clinical relevance and not by the site of isolation. The site of isolation does not necessarily correlate with pathogenicity. It has already been shown that nonclinical isolates exhibit virulence (13). Strains 1BU, 2HH, 3ST, and 11DS were the causative agents of seriously progressing cases of Acanthamoeba keratitis, while strains 4RE, 5SU, and 9GU, although they were isolated from contact lens cases of keratitis patients and thus had come into contact with an affected cornea, did not cause any disease.
Physiological properties.
Remarkably, sequence similarity clustering was strongly supported by the pathogenicity-associated physiological properties of these isolates. Altogether, clinically relevant strains grew significantly better than strains isolated from non-Acanthamoeba keratitis patients. Yagita et al. assume that the development of an eye infection may depend on the size of the inoculum, the frequency of contact with the cornea, and the virulence of the amoeba (31). In this respect the growth rate is of prime importance in order to provide a large inoculum. There was a true correlation of growth rate and cytopathic effect in our study. Moreover, the 42°C growing strains were all among the keratitis-causing isolates, although temperature tolerance is probably less important for infections of the eye since the human eye has an average temperature of only 34°C. We also observed differences in the ability of isolates to migrate into the agar. It is assumed that the amoeboidal locomotion enables the amoebae to penetrate host tissue. In Naegleria spp., migration patterns in an under-agarose system correlate with pathogenicity (28).
All Acanthamoeba keratitis isolates exhibited strong cytopathic effects to a monolayer of HEp-2 cells. Cursons and Brown (3) demonstrated that cell culture pathogenicity is correlated with mouse pathogenicity and also is dependent upon the growth rate. It has been reported that passaging amoebae through cell culture increases the virulence of the amoebae and that the virulence of Acanthamoeba attenuates during axenic culture (24). However, in our study tissue culture pathogenicity was not lost even after long-term axenic culture and was not gained either. Also, reference strains exhibited the same temperature tolerances as initially described elsewhere (6). The cytopathic effect is not yet wholly understood. Intimate contact with the cell membrane involving the amoebal pseudopodia is probably necessary. Moreover, cytolytic proteins play an important role in the lytic process (8). It has been demonstrated that A. castellanii exercises rigid host specificity. It neither produces cytopathic effects nor does it even bind to the corneal epithelium of mice, rats, horses, cows, chicken, dogs, and rabbits, whereas it causes severe damage to human, pig, or Chinese hamster cornea during a 24-h period (19).
Altogether, the results presented here indicate a correlation between clinical relevance and pathogenicity. Moreover, strains of no clinical relevance grouped together in T4 cluster analysis. There seems to be some evidence that virulence is a distinct characteristic in Acanthamoeba strains and that this might also stand in a phylogenetic context.
ACKNOWLEDGMENT
We thank Rolf Michel from the Ernst-Rodenwaldt-Institute, Koblenz, Germany, for providing reference strains 312-2 and 72/2.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Clark C G, Diamond L S. Ribosomal RNA genes of “pathogenic” and “nonpathogenic” Entamoeba histolytica are distinct. Mol Biochem Parasitol. 1991;49:297–303. doi: 10.1016/0166-6851(91)90073-f. [DOI] [PubMed] [Google Scholar]
- 3.Cursons R T M, Brown T J. Use of cell cultures as an indicator of pathogenicity of free-living amoebae. J Clin Pathol. 1978;31:1–11. doi: 10.1136/jcp.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Jonckheere J F. Use of an axenic medium for differentiation between pathogenic and non-pathogenic Naegleria fowleri isolates. Appl Environ Microbiol. 1977;33:751–757. doi: 10.1128/aem.33.4.751-757.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Jonckheere J F. Riboprinting of Naegleria spp.: small-subunit versus large-subunit rDNA. Parasitol Res. 1994;80:230–234. doi: 10.1007/BF00932679. [DOI] [PubMed] [Google Scholar]
- 6.De Jonckheere J F, Michel R. Species identification and virulence of Acanthamoeba strains from human nasal mucosa. Parasitol Res. 1988;74:314–316. doi: 10.1007/BF00539451. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP-phylogeny inference package, vers. 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- 8.Ferrante A. Free-living amoebae: pathogenicity and immunity. Parasite Immunol. 1991;13:31–47. doi: 10.1111/j.1365-3024.1991.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 9.Gast R J, Ledee D R, Fuerst P A, Byers T. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J Eukaryot Microbiol. 1996;43:498–504. doi: 10.1111/j.1550-7408.1996.tb04510.x. [DOI] [PubMed] [Google Scholar]
- 10.Gautom R K, Lory S, Seyedirashti S, Bergeron D L, Fritsche T R. Mitochondrial DNA fingerprinting of Acanthamoeba spp. isolated from clinical and environmental sources. J Clin Microbiol. 1994;32:1020–1073. doi: 10.1128/jcm.32.4.1070-1073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin J L. Temperature tolerance of pathogenic and nonpathogenic free-living amoebae. Science. 1972;178:869–870. doi: 10.1126/science.178.4063.869. [DOI] [PubMed] [Google Scholar]
- 12.Hiti K, Faschinger C, Haller-Schober E M, Hiti H, Walochnik J, Aspöck H. Acanthamoeba in asymptomatic contact lens wearers? Examination of storage-boxes. Spektrum Augenheilkd. 2000;14:163–166. [Google Scholar]
- 13.Howe D K, Vodkin M H, Novak R J, Visvesvara G, McLaughlin G L. Identification of two genetic markers that distinguish pathogenic and non-pathogenic strains of Acanthamoeba spp. Parasitol Res. 1997;83:345–348. doi: 10.1007/s004360050259. [DOI] [PubMed] [Google Scholar]
- 14.Hugo E R, Stewart V J, Gast R J, Byers T J. Purification of amoeba mtDNA using the UNSET procedure. In: Soldo A T, Lee J J, editors. Protocols in protozoology. Lawrence, Kans: Allen Press; 1992. pp. D7.1–D7.2. [Google Scholar]
- 15.Kilvington S, Beeching J. Identification and epidemiological typing of Naegleria fowleri with DNA probes. Appl Environ Microbiol. 1995;61:2071–2078. doi: 10.1128/aem.61.6.2071-2078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilvington S, Beeching J R, White D G. Differentiation of Acanthamoeba strains from infected corneas and the environment by using restriction endonuclease digestion of whole-cell DNA. J Clin Microbiol. 1991;29:310–314. doi: 10.1128/jcm.29.2.310-314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel R, Röhl R, Schneider H. Isolation of free-living amoebae from nasal mucosa of healthy individuals. Zentbl Bakteriol Hyg. 1982;176:155–159. [PubMed] [Google Scholar]
- 18.Nicholas K B, Nicholas H B, Jr, Deerfield D W., II GeneDoc: analysis and visualization of genetic variation. Embnew News. 1997;4:14. [Google Scholar]
- 19.Niederkorn J Y, Ubelaker J E, McCulley J P, Stewart G L, Meyer D R, Mellon J A, Silvany R E, He Y G, Pidherney M, Martin J H, Alizadeh H. Susceptibility of corneas from various species to in vitro binding and invasion by Acanthamoeba castellanii. Investig Ophthalmol Vis Sci. 1992;33:104–112. [PubMed] [Google Scholar]
- 20.Page F C. Nackte Rhizopoda. In: Matthes D, editor. Protozoenfauna, Band 2. G. Stuttgart, Germany: Fischer; 1991. pp. 3–145. [Google Scholar]
- 21.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 22.Pussard M, Pons R. Morphologie de la paroi kystique et taxonomie du genre Acanthamoeba (Protozoa, Amoebida) Protistologica. 1977;8:557–598. [Google Scholar]
- 23.Schaumberg D A, Snow K K, Dana M R. The epidemic of Acanthamoeba keratitis: where do we stand? Cornea. 1998;17:3–10. doi: 10.1097/00003226-199801000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Stevens A R, O'Dell W. In vitro growth and virulence of Acanthamoeba. J Parasitol. 1974;60:884–885. [PubMed] [Google Scholar]
- 25.Stothard D R, Hay J, Schroeder-Dietrich J M, Seal D V, Byers T J. Fluorescence oligonucleotide probes for clinical and environmental detection of Acanthamoeba and the T4 18S rRNA gene sequence type. J Clin Microbiol. 1999;37:2687–2693. doi: 10.1128/jcm.37.8.2687-2693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stothard D R, Schroeder-Dietrich J M, Awwad M H, Gast R J, Ledee D R, Rodriguez-Zaragoza S, Dean C L, Fuerst P A, Byers T. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rDNA gene sequence types. J Eukaryot Microbiol. 1998;45:45–54. doi: 10.1111/j.1550-7408.1998.tb05068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thong Y, Ferrante A. Migration patterns or pathogenic and non-pathogenic Naegleria spp. Infect Immun. 1986;51:177–180. doi: 10.1128/iai.51.1.177-180.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walochnik J, Hassl A, Simon K, Benyr G, Aspöck H. Isolation and identification by partial sequencing of the 18S ribosomal gene of free-living amoebae from necrotic tissue of Basiliscus plumifrons (Sauria: Iguanidae) Parasitol Res. 1999;85:601–603. doi: 10.1007/s004360050602. [DOI] [PubMed] [Google Scholar]
- 30.Yagita K, Endo T. Restriction enzyme analysis of mitochondrial DNA of Acanthamoeba strains in Japan. J Protozool. 1990;36:570–575. doi: 10.1111/j.1550-7408.1990.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 31.Yagita K, Endo T, De Jonckheere J F. Clustering of Acanthamoeba isolated from human eye infections by means of mitochondrial DNA digestion patterns. Parasitol Res. 1999;85:284–289. doi: 10.1007/s004360050549. [DOI] [PubMed] [Google Scholar]
- 32.Yu H S, Hwang M Y, Kim T O, Yun H C, Kom T H, Kong H H, Chung D I. Phylogenetic relationships among Acanthamoeba spp. based on PCR-RFLP analyses of mitochondrial small subunit rRNA gene. Korean J Parasitol. 1999;37:181–188. doi: 10.3347/kjp.1999.37.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]