Abstract
The avian leukosis virus (ALV) is a serious threat to sustainable and economically viable commercial poultry management world-wide. Active infections can result in more than 20% flock loss, resulting in significant economic damage. ALV detection and elimination from flocks and breeding programs is complicated by high sequence variability and the presence of endogenous virus copies which show up as false positives in assays. Previously-developed approaches to virus detection are either too labor-intensive to implement on an industrial scale or suffer from high false negative or positive rates. We developed a novel multi-locus multiplex quantitative real-time PCR system to detect viruses belonging to the J and K genetic subgroups that are particularly prevalent in our region. We used this system to eradicate ALV from our broiler breeding program comprising thousands of individuals. Our approach can be generalized to other ALV subgroups and other highly genetically diverse pathogens.
Introduction
As the global population continues to grow, agricultural systems must increase production while maintaining sustainable levels of input consumption. To achieve this goal, we must control disease outbreaks in agricultural animal production facilities. Infections cause significant economic harm and contribute to global food insecurity. An important pathogen in poultry is the avian leukosis virus (ALV), a diploid single-stranded RNA virus that belongs to the genus Alpharetrovirus of the Retroviridae family [1]. ALV leads to lymphoid and myeloid leukosis, as well as neoplasia in other tissues. Active infections result in over 20% mortality, lower productivity, and cause significant harm to industrial poultry management [1]. Even subclinical manifestations of ALV lead to decreased productivity and serious revenue loss [2, 3]. Spread of ALV presents significant problems worldwide. Several national eradication programs exist, for example in China, Australia, and Iran.
ALV types are classified into subgroups based on the GP85 envelope protein structure [4, 5]. Viruses from subgroups A through E [2], J [4], and K [5] are specific to chicken. Subgroup F and G viruses are specific to pheasants and are thus not relevant to the present study [6]. With the exception of the endogenous E group they are highly pathogenic. Endogenous virus genomes integrate into host chromosomes in the germline and are transmitted vertically via Mendelian inheritance [2]. ALV can integrate into somatic cell DNA and be induced by stress, leading to reversion of viremia [7]. Viruses belonging to the A, B, and J subgroups are common, while the C and D subgroups are encountered only rarely [2]. The K subgroup has been discovered relatively recently [5, 8–11]. Some of its variants are significantly pathogenic [12–15]. Studying the K subgroup emergence may lead to novel insights into the modes of virus spread and virulence evolution. Eradication programs typically focus on the J subgroup viruses. Russian poultry industry is severely affected by ALV belonging to this subgroup, with 70% of 223 production sites, surveyed in 46 regions, testing positive for anti-subgroup J (and 90% for all subgroups) antibodies [16]. There is evidence that susceptibility to infection has a genetic component, with a major susceptibility locus linked to the ev21 locus. ev12 and ev21 produce complete endogenous viruses and associated with significant reductions in antibody response to ALV. These genes also predispose individuals to ALV shedding [17].
Fundamentally, the virus eradication problem in a breeding program must be solved by preventing vertical transmission to subsequent generations. This has been the foundation of previous approaches [18]. However, the high horizontal transmission rates exhibited by subgroup J viruses necessitates a more intensive screening [19]. The current standard involves serological tests of cloacal and vaginal swabs at 20 weeks, followed by viremic and blood serum ELISA at 22 weeks, a serological test of the first two eggs at 23 weeks, meconium of new-born chicks at 26 weeks, and finally sero-tests of egg albumen at 40 weeks [20]. Previous studies suggest that raising poultry in small groups and identifying infected chicks before hatching prevents horizontal transmission of ALV subgroup A [21] in layers and ALV J in broilers [22]. However, there is evidence that vertical transmission can occur even when specific viral antigens are undetectable [22, 23]. Furthermore, broiler chickens appear to be particularly susceptible to ALV subgroup J horizontal transmission [2], complicating the eradication process.
ALV identification via propagation in CEFs or DF-1 cells is currently the golden standard. This method suffers from severe drawbacks: it takes seven to nine days and requires significant investment in specialized equipment and laboratory space. An ELISA assay against a group-specific ALV p27 antigen is much more widely used. However, it also has several disadvantages, chiefly a high false positive rate due to endogenous virus p27 expression [24] and low sensitivity [2, 18].
DNA-based detection is possible because the RNA genome of the virus is reverse-transcribed upon cell entry, with subsequent host genome integration [25]. However, identifying ALV positive birds is complicated by the background noise coming from endogenous viruses and is compounded by exogenous virus genome variability. Several of published PCR-based protocols suffer from endogenous virus driven false positives. Nevertheless, PCR-based approaches are 15 to 20% more sensitive than culture or ELISA-based protocols [26]. The major difficulty in implementing a PCR protocol is the high variability of the ALV subgroup J target sequence. For example, there is up to 5.1% divergence between gp85 DNA sequences isolated from the same individual [27]. Amino-acid identity among isolates can be as low as 86.2% [28], implying and even higher DNA sequence diversity. We describe a PCR-based system for identification of ALV subgroups A, B, J, and K. We overcame the DNA diversity problem by using multiple loci and probes. We demonstrate the effectiveness of our method by using it as a screening method to identify infected individuals and thus eradicate ALV from our Smena8 broiler cross. Our approach can be easily deployed in typical industrial settings.
Materials and methods
Ethical statement
All samples included in this study were obtained from birds with official records. Sample collection did not involve animal killing and was performed in accordance with national and European regulations. No ethical approval was necessary.
Test system development
We used BLAST (http://www.ncbi.nlm.nih.gov/BLAST) and ClustalW (http://www.genome.jp/tools-bin/clustalw) on sequences obtained from GenBank to find regions of the ALV genome that are conserved within but divergent between subgroups. GenBank entries used as reference sequences for region selection are M37980, HM452341(ALV-A); AF052428, JF826241 (ALV-B), J02342 (ALV-C), D10652 (ALV-D), EF467236, AY013303, AY013304, KC610517 (ALV-E); Z46390, JF951728, JQ935966, HM776937, JX855935, JF932002, KX058878, DQ115805, KX034517, KU997685, HM235668, HM582657(ALV-J), KF746200, KP686143, and GD14LZ (ALV K). Primers and probes for real-time PCR (Table 1) were developed based on published sequences [10, 29, 30] and synthesized by Syntol (Moscow, Russia). We took care to pick primers that did not amplify known endogenous ALV sequences. In particular, EAV-HP elements contain parts of ALV-J envelope gene. We chose primers outside of the common region, thus eliminating EAV-HP amplification.
Table 1. Primers and probes used in this study.
| Name* | Sequence (5’–3’) | Amplicon length, bp | Subgroup | Purpose |
|---|---|---|---|---|
| ALVAF | GCCACACGGTTCCTCCTTAGA | 114 | ALV A | Multiplex primer |
| ALVAR | CGCAGTACTCACTCCCCATGAA | 114 | ALV A | Multiplex primer |
| APL | (5R6G)TACGGTGG(dT-BHQ1)GACAGCGGATAG-P | ALV A | Multiplex probe | |
| ALBF1 | GGCCGAGGCCTCCCCGAAA | 77 | ALV B | Multiplex primer |
| ALVBR | GTCTCATTAATTTCCTTTGATTGA | 77 | ALV B | Multiplex primer |
| BPL1G | (Cy5)CCCATGTACC(dT-BHQ2)CCCGTGCCTTG-P | ALV B | Multiplex probe | |
| JFF1F | GCCCTGGGAAGGTGAGCAAGA | 139 | ALV J | Multiplex primer |
| JJR | GGAAATAATAACCACGCACACGA | 139 | ALV J | Multiplex primer |
| JNP | (ROX)TCCTCTCGA(dT-BHQ2)GGCAGCAAGGGTGTC-P | ALV J | Multiplex probe | |
| JJPLN | (ROX)CAGCA AGGGTG(dT-BHQ2)CTTCTCCG-P | ALV J | Multiplex probe | |
| ALVKF | CGGAGCATTGACAAGCTTTCAGA | 72 | ALV K | Multiplex primer |
| ALVKR | GTGATTGCGGCGGAGGAGGA | 72 | ALV K | Multiplex primer |
| KPL | (Cy5.5)CCACCTCGTGAG(dT-BHQ2)TGCGGCC-P | ALV K | Multiplex probe | |
| ALV-JNF [9] | TTGCAGGCATTTCTGACTGG | 214 | ALV J | Published primer |
| ALV-JNR [9] | ACACGTTTCCTGGTTGTTGC | 214 | ALV J | Published primer |
| JCP [9] | (FAM)CCTGGGAAGGTGAGCAAGAAGGA-BHQ1 | ALV J | Published probe | |
| H5 [10, 11] | GGATGAGGTGACTAAGAAAG | 545 | ALV J | Published primer |
| H7 [10, 11] | CGAACCAAAGGTAACACACG | 545 | ALV J | Published primer |
| Probe [10, 11] | (FAM)CTCTTTGCAGGCATTTCTGACTGGGC(TAMRA) | ALV J | Published probe | |
| SEQA-KR | CGCGATCCCCACAAATGAGGAAA | 443 | ALV A | Sequencing primer |
| SEQA-KR | CGCGATCCCCACAAATGAGGAAA | 253 | ALV B | Sequencing primer |
| SEQJF | CCCTGGGAAGGTGAGCAAGAA | 498 | ALV J | Sequencing primer |
| SEQJR | CCTTTATAGCACACCGAACCGAA | 498 | ALV J | Sequencing primer |
| SEQA-KR | CGCGATCCCCACAAATGAGGAAA | 466 | ALV K | Sequencing primer |
| JEF | CCTATTCAAGTTGCCTCTGTGGA | 72 | ALV J | LTR primer |
| JER | GCTTGCTCTATTTGGCCGTCAGA | 72 | ALV J | LTR primer |
| JEP | (Cy5)CCATCCGAGC(dT-BHQ2)GCCTCCAGTCC-P | ALV J | LTR probe |
*References provided for previously published primers
DNA isolation and quantitative real-time PCR
We isolated DNA from feathers (one feather per sample) using the M-Sorb-OOM kit from Syntol (catalog # OOM-502). Calamus fragments 0.3 to 0.5 cm long or chick cloacal smears (all birds from the Smena8 cross) were placed in 1.5 mL tubes and treated with 0.4 mL of the lysis buffer at 70°C for 15 minutes while stirring. We separated debris by a three-minute centrifugation at 13 000 rpm. We transferred the supernatant to a fresh tube and continued the isolation protocol according the manufacturer’s instructions. We used 1.5 μL of the resulting DNA solution in each PCR reaction. While screening the breeding population, individuals were re-tested up to eight times, unless determined to be positive and eliminated from the flock.
RNA isolation and reverse-transcription were performed using kits from Syntol (M-Sorb-OOM for extraction and OT-1 for reverse transcription). We used 1 feather from each bird. We confirmed quantitative PCR specificity by amplicon sequencing using primers ALVKF, SEQA-KR, SEQJF, SEQJR, ALVAF, and SEQA-KR on the Nanophore 05 genetic analyzer (Institute for Analytical Instrumentation RAS, St. Petersburg, Russia). Subsequent generations were tested for ALV-J and -K by pooling feathers from 10 birds in one sample.
We ran real-time PCR reactions on an ANK-48 machine (Institute for Analytical Instrumentation RAS, St. Petersburg, Russia). We ran 50 cycles, with the following steps: denaturation step (10 seconds, 93°C), annealing and elongation step (30 s, 55°C). We used 10 μL of the quantitative real-time PCR reaction mix from a commercial kit (M-428, Syntol, Moscow, Russia). Primer concentration was 450 nM and we used 100 nM of each probe per reaction.
ELISA
We used the IDEXX ALV-J Ab Test system (IDEXX Laboratories, Inc., USA) and the Avian leukosis virus antigen test kit from Synbiotics (USA) to perform ELISA on blood plasma samples from randomly selected birds according to the manufacturers’ instructions.
Results and discussion
Multiplex quantitative real-time PCR of common ALV subgroups
We designed our multiplex test system to identify all common ALV subgroups. We focused on the locus coding for the gp85 coat protein to target amplification of viruses of the A, B, J, and K subgroups (Table 1). Fig 1 shows primer positions and typical real-time fluorescence curves. We used synthetic DNA fragments corresponding to each subgroup, as well as field samples, as positive controls during test system development (Table 1). We performed serial ten- and subsequently two-fold dilutions of the positive control samples to estimate assay sensitivity and employed a previously described procedure [30, 31] to estimate sensitivity from our serial dilution data. Our essays can detect 25 genome-equivalents of A and B subgroup viruses, and 10 genome-equivalents of J and K.
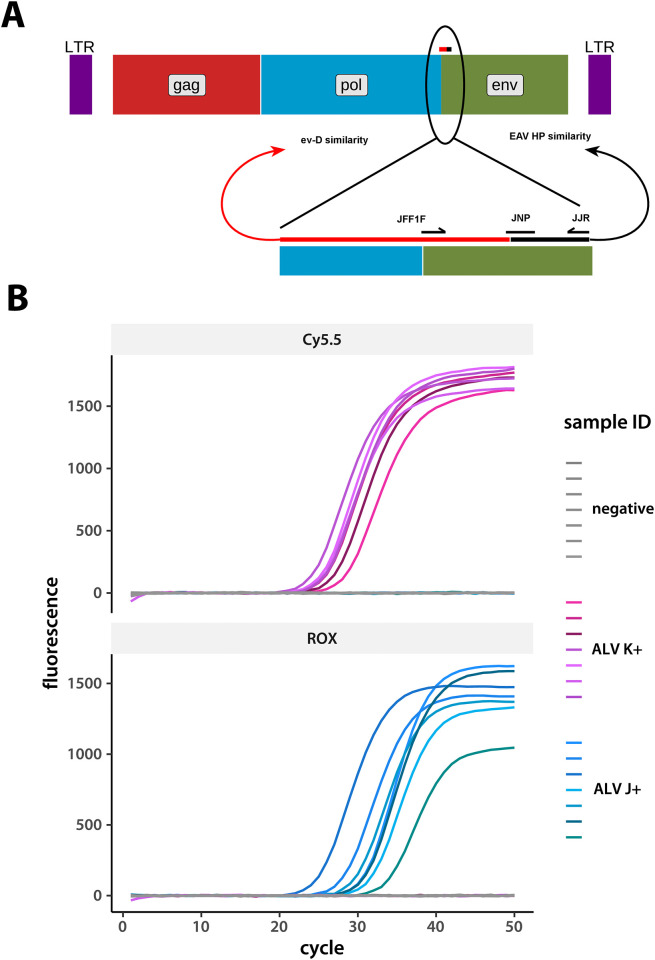
Fig 1. Specificity of the developed real-time PCR system for detecting ALV J and K.
A. Primer positioning around the ev-D/EAV-HP (GenBank accession numbers DQ500016 and AC270426) similarity breakpoint in the ALV J gene that codes for the gp85 coat protein. A diagram of the full ALV genome is on top, with the region surrounding primer placement shown in detail below. B. A representative set of detection curves from the Cy5.5 and ROX channels (the KPL and JJPLN probes, respectively, in Table 1). Each curve is a sample from a separate bird. Eight ALV J positive, ALV K positive, and negative samples each were measured on the same 96-well plate and fluorescence values recorded simultaneously. Data for this graph are included as the S1 File.
To verify the performance of our system, we compared quantitative PCR threshold cycles obtained with the set of primers and probes specific to the J subgroup to previously published results (Table 1) [26, 29, 32]. We only used PCR methods for comparison since other approaches are unacceptably invasive for our application. Field isolates from poultry production facilities in the Moscow region were used for the latter estimates. We successfully verified our results using the Qin L system [26]. This system can detect fewer than 10 virus genome copies per reaction. We made the published primers ourselves (Table 1) and used them to replicate the previously described essays [26]. In our hands, the widely used primers H5 and H7 that detect the J subgroup genomes [29, 32] exhibit 100-fold lower sensitivity, consistent with previous reports [26]. Negative controls, either empty PCR buffer or specific pathogen free chicken DNA, gave no detectable signal after 50 cycles. We tested our identification system on a sample of 1200 individuals randomly picked from DNA isolates of four base lines used to initiate the Smena8 broiler cross. We found that up to 52% of all individuals in our flock were positive for the subgroup J ALV, while up to 10% carried subgroup K (some birds were double-infected). We did not detect any subgroup A or B cases. Previous studies [1] reported around 20% mortality in flocks affected by ALV. Given that not all infected birds die, our prevalence estimates appear to be in line with these previous observations.
We then moved on to analyze DNA from several tissues (feathers, liver, spleen, and blood) from the following regions in Russia: Moscow, Orenburg, Chelyabinsk, Kemerovo, Tyumen, Kaliningrad, Leningrad, Sverdlovsk, Novgorod, and Krasnodar. Samples collected in the Kaliningrad, Leningrad, Sverdlovsk, and Novgorod regions tested positive for K subgroup ALV. We found subgroup A infected individuals in the Leningrad region, while subgroup J positive samples were identified in areas surrounding Moscow, Sverdlovsk, and Leningrad. We did not detect any subgroup B positive samples, suggesting that this ALV variant was absent from Russia during the testing period. We confirmed our test system specificity by sequencing amplicons obtained from positive samples using primers described in Table 1. We did not see any subgroup misidentification.
Multi-locus multiplex quantitative PCR and its use in ALV-J and K eradication
Having established a robust assay, we set out to test its effectiveness in eradicating the subgroup J and K infections we uncovered in our Smena8 broiler cross. To increase assay reliability, we implemented a multiplex approach, adding an extra probe specific to the gp85 gene (JJPLN) and another that targets the long terminal repeat (LTR) sequence. Both probes are specific to the J subgroup viruses (Table 1). We succeeded in eradicating the virus infection by the 77th generation of the selection process.
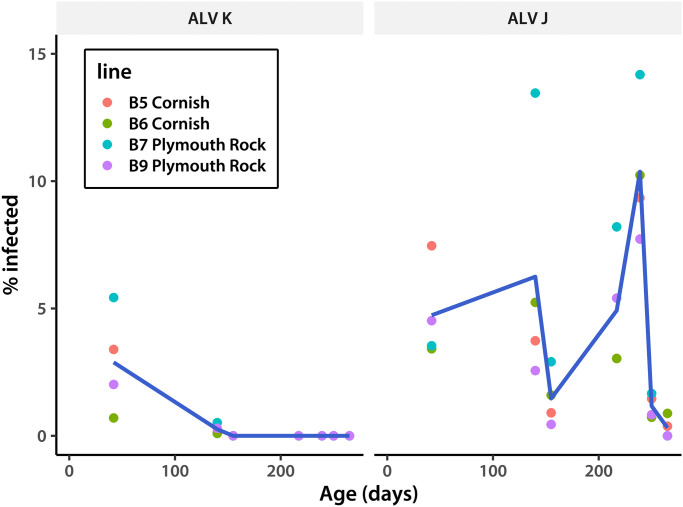
We started by employing our J-subgroup specific PCR system for four generations of the Smena8 broiler cross (Table 1). High variability of the J genomes led us to implement the multi-locus multiplex system mentioned above, containing an additional probe (JJPLN, Table 1). This resulted in an average 2.5% increase in virus identification rates. We also introduced an additional amplification of a subgroup J-specific LTR fragment (Table 1), further increasing identification rates. The latter step allows us to see additional J subgroup variants. Managing the time interval between successive tests proved crucial. Increasing the between-test duration to 62 days between cycle four and five resulted in a two-fold jump in infection rates. Therefore, from that point on testing was done no less frequently than once every two weeks. As a result, we no longer detected subgroup J viruses after cycle eight of the program in two Smena8 broiler cross lines (B7 and B9), while infection rates in two others were minimal (Table 2 and Fig 2). This in contrast to about 52% ALV subgroup J positive rate at the start of the program. Subgroup K viruses disappeared by cycle four, suggesting that they are easier to eradicate. Chickens from the next generation of the Smena8 broiler cross showed several ELISA-positive tests at day 267 after hatching. However, none of these were PCR-positive. We re-tested individuals from the generation after that at day 360 and found no positives by either PCR or ELISA. We saw no clinical manifestations of leukosis in the three years since. A handful of suspected cases proved to be PCR-negative.
Table 2. ALV eradication in the 77th generation of Smena8 broiler cross pure lines.
| Pure line | Age (days) | Livestock | Number infected | ALV K/J ratio | % infected |
|---|---|---|---|---|---|
| Line B5 Cornish | 1 | 1886 | 208 | N/A* | 11.0 |
| 42 | 1032 | 112 | 35/77 | 10.9 | |
| 140 | 804 | 31 | 1/30 | 3.9 | |
| 155 | 775 | 7 | 0/7 | 0.9 | |
| 217 | 757 | 23 | 0/23 | 3.0 | |
| 239 | 610 | 57 | 0/57 | 9.3 | |
| 250 | 548 | 8 | 0/8 | 1.5 | |
| 265 | 531 | 2 | 0/2 | 0.4 | |
| Line B6 Cornish | 1 | 2358 | 118 | N/A* | 5.0 |
| 42 | 1142 | 47 | 8/39 | 4.1 | |
| 140 | 1088 | 58 | 1/57 | 5.3 | |
| 155 | 939 | 15 | 0/15 | 1.6 | |
| 217 | 923 | 28 | 0/28 | 3.0 | |
| 239 | 772 | 79 | 0/79 | 10.2 | |
| 250 | 687 | 5 | 0/5 | 0.7 | |
| 265 | 682 | 6 | 0/6 | 1.0 | |
| Line B7 Plymouth Rock | 1 | 2319 | 134 | N/A* | 5.8 |
| 42 | 1216 | 109 | 66/43 | 9.0 | |
| 140 | 966 | 135 | 5/130 | 14.0 | |
| 155 | 860 | 25 | 0/25 | 2.9 | |
| 217 | 841 | 69 | 0/69 | 8.2 | |
| 239 | 684 | 97 | 0/97 | 14.2 | |
| 250 | 602 | 10 | 0/10 | 1.7 | |
| 265 | 585 | 0 | 0/0 | 0.0 | |
| Line B9 Plymouth Rock | 1 | 2466 | 67 | N/A* | 2.7 |
| 42 | 1438 | 94 | 29/65 | 6.5 | |
| 140 | 1367 | 39 | 4/35 | 2.9 | |
| 155 | 1118 | 5 | 0/5 | 0.4 | |
| 217 | 1073 | 58 | 0/58 | 5.4 | |
| 239 | 919 | 71 | 0/71 | 7.7 | |
| 250 | 845 | 7 | 0/7 | 0.8 | |
| 265 | 823 | 0 | 0/0 | 0.0 |
*N/A—data are not available
Fig 2. Prevalence of infected individuals over time.
Data for ALV K and J infections are shown on separate panels. Lines trace among-line means, calculated for each day separately. The data for this plot are in Table 2.
Conclusions
The avian leukosis virus is a serious threat to sustainable and economically viable commercial poultry management world-wide. As an RNA virus, it is highly variable, making reliable detection difficult. We developed a multi-locus multiplex quantitative real-time PCR system to identify individuals infected with subgroup J and K viruses with high sensitivity and specificity. We demonstrate the effectiveness of our approach by quickly eliminating both ALV J and K subgroups from our breeding program.
Our innovation is to couple quantitative PCR of multiple genes and the use of several probes per locus to increase sensitivity in the face of high genetic variation. In addition, using pulped feathers as the DNA source makes the assay non-invasive, cheap, and easy to implement [33–35]. Preliminary data indicate that we can get an almost two order of magnitude sensitivity increase when we assay ALV J cDNA, suggesting that future assay development can improve on the current approach, albeit perhaps at the cost of increased labor. Our experience in successfully eliminating ALV from our population comprising thousands of individuals demonstrates that the proposed system has adequate sensitivity and specificity. The procedure may be further simplified by screening only parents selected for establishment of the next generation. This multilocus multiplex approach with multiple probes can also be used to detect retro- and coronaviruses, as well as any other highly variable microorganisms.
Supporting information
(CSV)
Acknowledgments
The authors are grateful to the two anonymous reviewers for comments that helped improve the manuscript.
Data Availability
All relevant data are within the manuscript.
Funding Statement
ZhVE, AMB, SVS: Ministry of Science and Higher Education of the Russian Federation, the state assignment for Breeding and Genetic Center Smena No. 075-01297-20-00. https://minobrnauki.gov.ru/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gao L, Qin L, Pan W, Wang Y, Lee QX, Gao H, et al. Avian leukosis virus subgroup J in layer chickens, China. Emerg Infect Dis. 2010;16(10):1637–8. doi: 10.3201/eid1610.100780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Payne L. Retrovirus-induced disease in poultry. Poult Sci. 1998;77(8):1204–1212. doi: 10.1093/ps/77.8.1204 [DOI] [PubMed] [Google Scholar]
- 3. Bacon LD, Fulton JE, Kulkarni GB. Methods for evaluating and developing commercial chicken strains free of endogenous subgroup E avian leukosis virus. Avian Pathol. 2004;33(2):233–243. doi: 10.1080/0307943042000195731 [DOI] [PubMed] [Google Scholar]
- 4. Payne L, Brown S, Bumstead N, Howes K, Frazier J, Thouless M. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801 [DOI] [PubMed] [Google Scholar]
- 5. Li X, Lin W, Chang S, Zhao P, Zhang X, Liu Y, et al. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in South China. Arch Virol. 2016;161(10):2717–25. doi: 10.1007/s00705-016-2965-x [DOI] [PubMed] [Google Scholar]
- 6. Fujita DJ, Chen YC, Friis RR, Vogt PK. RNA tumor viruses of pheasants: Characterization of avian leukosis subgroups F and G. Virology. 1974;60(2):558–571. doi: 10.1016/0042-6822(74)90350-X [DOI] [PubMed] [Google Scholar]
- 7. Pandiri A, Gimeno I, Mays J, Reed W, Fadly A. Reversion to subgroup J avian leukosis virus viremia in seroconverted adult meat-type chickens exposed to chronic stress by adrenocorticotrophin treatment. Avian Dis. 2012;56(3):578–82. doi: 10.1637/9949-092611-ResNote.1 [DOI] [PubMed] [Google Scholar]
- 8. Nakamura S, Ochiai K, Hatai H, Ochi A, Sunden U, Umemura T. Pathogenicity of avian leukosis viruses related to fowl glioma-inducing virus. Aian Pathol. 2011;40(5):499–505. [DOI] [PubMed] [Google Scholar]
- 9. Cui N, Su S, Chen Z, Zhao X, Cui Z. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J Gen Virol. 2014;95(11):2512–22. doi: 10.1099/vir.0.067264-0 [DOI] [PubMed] [Google Scholar]
- 10. Shao H, Wang L, Sang J, Li T, Liu Y, Wan Z, et al. Novel avian leukosis viruses from domestic chicken breeds in mainland China. Arch Virol. 2017;. doi: 10.1007/s00705-017-3344-y [DOI] [PubMed] [Google Scholar]
- 11. Chang S, Hsu M, Wang C. Gene detection, virus isolation, and sequence analysis of avian leukosis viruses in Taiwan country chickens. Avian Dis. 2013;57(2):172–177. doi: 10.1637/10387-092612-Reg.1 [DOI] [PubMed] [Google Scholar]
- 12. Lv L, Li T, Hu M, Deng J, Liu Y, Xie Q, et al. A recombination efficiently increases the pathogenesis of the novel K subgroup of avian leukosis virus. Vet Microbiol. 2019;231:214–217. doi: 10.1016/j.vetmic.2019.03.021 [DOI] [PubMed] [Google Scholar]
- 13. Su Q, Li Y, Cui Z, Chang S, Zhao P. The emerging novel avian leukosis virus with mutations in the pol gene shows competitive replication advantages both in vivo and in vitro. Emerg Microbes Infect. 2018;7(1):117. doi: 10.1038/s41426-018-0111-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang X, Gu Y, Chen X, Li T, Gao Y, Wang X, et al. Identification and characterization of a novel natural recombinant avian leucosis virus from Chinese indigenous chicken flock. Virus Genes. 2019;55(5):726–733. doi: 10.1007/s11262-019-01695-7 [DOI] [PubMed] [Google Scholar]
- 15. Borodin AM, Alekseev YI, Konovalova NV, Terentyeva EV, Efimov DN, Emanuilova ZV, et al. Detection of avian leukosis virus subgroup K in Russia and its molecular genetic analysis. Agricultural Biology (in Russian). 2018;53(4):842–850. [Google Scholar]
- 16. Plotnikov V, Grebennikova T, Dudnikova EK, Shulpin MI, Lazareva SP, Nikonova ZB, et al. About spreading the avian leukosis viruses in poultry farms in Russian Federation. Agricultural Biology (in Russian). 2013;6:36–42. [Google Scholar]
- 17. Gavora J, Spencer J, Benkel B, Gagnon C, A Emsley A, Kulenkamp A. Endogenous viral genes influence infection with avian leukosis virus. Avian Pathol. 1995;24(4):653–664. doi: 10.1080/03079459508419105 [DOI] [PubMed] [Google Scholar]
- 18. Lloyd Spencer J. Progress towards eradication of lymphoid leukosis viruses—A review. Avian Pathol. 1984;13(4):599–619. doi: 10.1080/03079458408418560 [DOI] [PubMed] [Google Scholar]
- 19. Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res Vet Sci. 1999;67(2):113–119. doi: 10.1053/rvsc.1998.0283 [DOI] [PubMed] [Google Scholar]
- 20. Payne LN. HPRS-103: A retrovirus strikes back. The emergence of subgroup J avian leukosis virus. Avian Pathol. 1998;27(S1):S36–S45. doi: 10.1080/03079459808419291 [DOI] [Google Scholar]
- 21. Fadly AM, Okazaki W, Witter RL. Hatchery-related contact transmission and short-term small-group-rearing as related to lymphoid leukosis virus eradication programs. Avian Dis. 1981;25:667–677. doi: 10.2307/1589997 [DOI] [PubMed] [Google Scholar]
- 22. Witter RL, M FA. Reduction of horizontal transmission of avian leukosis virus subgroup J in broiler breeder chickens hatched and reared in small groups. Avian Pathol. 2001;30:641–654. doi: 10.1080/03079450120092134 [DOI] [PubMed] [Google Scholar]
- 23. Ignjatovic J. Congenital transmission of avian leukosis virus in the absence of detectable shedding of group specific antigen. Aust Vet J. 1990;67:299–301. doi: 10.1111/j.1751-0813.1990.tb07801.x [DOI] [PubMed] [Google Scholar]
- 24. Peng H, Qin L, Bi Y, Wang P, Zou G, Li J, et al. Rapid detection of the common avian leukosis virus subgroups by real-time loop-mediated isothermal amplification. Virol J. 2015;12:195. doi: 10.1186/s12985-015-0430-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Whitcomb J, Hughes S. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423 [DOI] [PubMed] [Google Scholar]
- 26. Qin L, Gao Y, Ni W, Sun M, Wang Y, Yin C, et al. Development and Application of Real-Time PCR for Detection of Subgroup J Avian Leukosis Virus. J Clin Microbiol. 2013;51(1):149–54. doi: 10.1128/JCM.02030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng F, Li X, Fang J, Gao Y, Zhu L, Xing G, et al. Genomic diversity of the Avian leukosis virus subgroup J gp85 gene in different organs of an infected chicken. J Vet Sci. 2016;17(4):497–503. doi: 10.4142/jvs.2016.17.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang P, Lin L, Li H, Yang Y, Huang T, Wei P. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989–2016: Coexistence of five extremely different clusters. Arch Virol. 2017;163(2):377–389. doi: 10.1007/s00705-017-3601-0 [DOI] [PubMed] [Google Scholar]
- 29. Smith L, Brown S, Howes K, McLeod S, Arshad S, Barron G, et al. Development and application of polymerase chain reaction (PCR) tests for the detection of subgroup J avian leukosis virus. Virus Res. 1998;54(1):87–98. doi: 10.1016/S0168-1702(98)00022-7 [DOI] [PubMed] [Google Scholar]
- 30. Yang S, Lin S, Kelen G, Quinn T, Dick J, Gaydos C, et al. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol. 2002;40(9):3449–54. doi: 10.1128/JCM.40.9.3449-3454.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamien C, Lelenta M, Goger W, Silber R, Tuppurainen E, Matijevic M, et al. Real time PCR method for simultaneous detection, quantitation and differentiation of capripoxviruses. J Virol Methods. 2011;171(1):134–40. doi: 10.1016/j.jviromet.2010.10.014 [DOI] [PubMed] [Google Scholar]
- 32. Kim Y, Gharaibeh S, Stedman N, Brown T. Comparison and verification of quantitative competitive reverse transcription polymerase chain reaction (QC-RT-PCR) and real time RT-PCR for avian leukosis virus subgroup J. J Virol Methods. 2002;102(1–2):1–8. doi: 10.1016/S0166-0934(01)00372-X [DOI] [PubMed] [Google Scholar]
- 33. Sung H, Reddy S, Fadly A. High virus titer in feather pulp of chickens infected with subgroup J avian leukosis virus. Avian Dis. 2002;46(2):281–286. doi: 10.1637/0005-2086(2002)046[0281:HVTIFP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 34. Zavala G, Jackwood M, Hilt D. Polymerase chain reaction for detection of avian leukosis virus subgroup J in feather pulp. Avian Dis. 2002;46(4):971–8. doi: 10.1637/0005-2086(2002)046[0971:PCRFDO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 35. Davidson I, Borenshtain R. The feather tips of commercial chickens are a favorable source of DNA for the amplification of Marek’s disease virus and avian leukosis virus, subgroup J. Avian Pathol. 2002;31(3):237–40. doi: 10.1080/03079450220136549 [DOI] [PubMed] [Google Scholar]