Abstract
Background and Objectives
To develop and test the performance of the Positive Aβ Risk Score (PARS) for prediction of β-amyloid (Aβ) positivity in cognitively unimpaired individuals for use in clinical research. Detecting Aβ positivity is essential for identifying at-risk individuals who are candidates for early intervention with amyloid targeted treatments.
Methods
We used data from 4,134 cognitively normal individuals from the Anti-Amyloid Treatment in Asymptomatic Alzheimer's (A4) Study. The sample was divided into training and test sets. A modified version of AutoScore, a machine learning–based software tool, was used to develop a scoring system using the training set. Three risk scores were developed using candidate predictors in various combinations from the following categories: demographics (age, sex, education, race, family history, body mass index, marital status, and ethnicity), subjective measures (Alzheimer's Disease Cooperative Study Activities of Daily Living–Prevention Instrument, Geriatric Depression Scale, and Memory Complaint Questionnaire), objective measures (free recall, Mini-Mental State Examination, immediate recall, digit symbol substitution, and delayed logical memory scores), and APOE4 status. Performance of the risk scores was evaluated in the independent test set.
Results
PARS model 1 included age, body mass index (BMI), and family history and had an area under the curve (AUC) of 0.60 (95% CI 0.57–0.64). PARS model 2 included free recall in addition to the PARS model 1 variables and had an AUC of 0.61 (0.58–0.64). PARS model 3, which consisted of age, BMI, and APOE4 information, had an AUC of 0.73 (0.70–0.76). PARS model 3 showed the highest, but still moderate, performance metrics in comparison with other models with sensitivity of 72.0% (67.6%–76.4%), specificity of 62.1% (58.8%–65.4%), accuracy of 65.3% (62.7%–68.0%), and positive predictive value of 48.1% (44.1%–52.1%).
Discussion
PARS models are a set of simple and practical risk scores that may improve our ability to identify individuals more likely to be amyloid positive. The models can potentially be used to enrich trials and serve as a screening step in research settings. This approach can be followed by the use of additional variables for the development of improved risk scores.
Classification of Evidence
This study provides Class II evidence that in cognitively unimpaired individuals PARS models predict Aβ positivity with moderate accuracy.
Accumulation of β-amyloid (Aβ) is a defining pathologic characteristic of Alzheimer disease (AD) and may occur decades before cognitive impairment.1,2 Early diagnosis and intervention to reduce Aβ accumulation is considered a potential approach to modify disease course and has been the focus of many of the recent clinical trials involving cognitively normal (CN) participants.3,4 Most of these trials require biomarker confirmation of amyloid positivity as a criterion for enrollment, usually based on amyloid PET. Because only 30% of CN older adults are Aβ positive (Aβ+), 10 people receive a PET scan for every 3 potentially eligible participants for enrollment—a costly process. A practical, noninvasive method to identify participants likely to be Aβ+ could make the process of screening and enrolling for clinical trials more efficient.
The goal of this study was to develop a practical risk score to predict Aβ positivity of individuals prior to confirmatory PET scan. Clinical risk scores are commonly used in medical practice for diagnosis or to predict prognosis.5 Risk scores allow clinicians to quantitatively estimate the likelihood, or risk, that an individual will have a particular outcome, often based on a small number of metrics that are readily determined. Developing a practical risk score for Aβ positivity would be timely as there are several drugs in the AD research pipeline that target aggregated Aβ for treatment of AD in various stages of disease. Aducanumab, a human monoclonal antibody that selectively targets aggregated Aβ, was recently approved by the US Food and Drug Administration for the treatment of AD in mild disease stage.6 Due to the associated costs and access limitations, obtaining amyloid imaging or CSF studies from all at-risk individuals is not possible. Therefore, developing simple and reliable risk scores for determining the likelihood of Aβ positivity could serve as a support tool for decision-making before proceeding with confirmatory tests.
The Anti-Amyloid Treatment in Asymptomatic Alzheimer's (A4) Study provides an ideal setting for developing a risk score for amyloid positivity. In the A4 Study, cognitively unimpaired older adults underwent amyloid PET scanning. We used a machine learning method based on random forest (RF) algorithms to identify important predictors and determine the risk score for amyloid positivity. Predictor variables were categorized into 4 large categories of demographics, subjective measures, objective measures, and genetic information in the form of APOE4 genotypes. Risk scores were developed using different combinations of these variable categories. This is intended to provide flexibility in terms of the information needed for constructing a risk score. We hypothesized that simple risk scores developed based on machine learning predictive models can improve our ability in estimating likelihood of amyloid positivity prior to obtaining PET imaging in cognitively unimpaired older adults.
Methods
Participants
Data used for this paper come from the A4 Study. This study is an ongoing trial that is being conducted at 67 clinical sites across 4 countries with a secondary outcome being the mean composite standardized uptake value ratio (SUVR) change from baseline.7 To participate in the study, individuals must be between the ages of 65 and 85, score between 25 and 30 on the Mini-Mental State Examination (MMSE), score 0 on the Global Clinical Dementia Rating (CDR), have a Logical Memory II score at screening of 6–18 depending on educational level, have a participating study partner, and show evidence of Aβ positivity via a PET scan as defined below. Exclusion criteria included various illnesses, serious risk of suicide, or the use of particular drugs.
A total of 6,763 individuals were prescreened to determine eligibility for the study. Of those, 4,486 individuals met criteria and were selected to undergo Aβ PET scans. In compiling the data set to use for this study, 352 individuals had at least one variable of missing data; 122 individuals did not have a Memory Complaint Questionnaire (MACQ) score from either visit 1 or 3 of the study; 83 individuals had unknown marital status; and for 88 individuals, race and ethnicity was unknown or missing. Therefore, as the methods required complete feature sets, a total of 4,134 individuals met criteria for inclusion in this study (Figure 1).
Figure 1. Flow Chart of the Study Design.
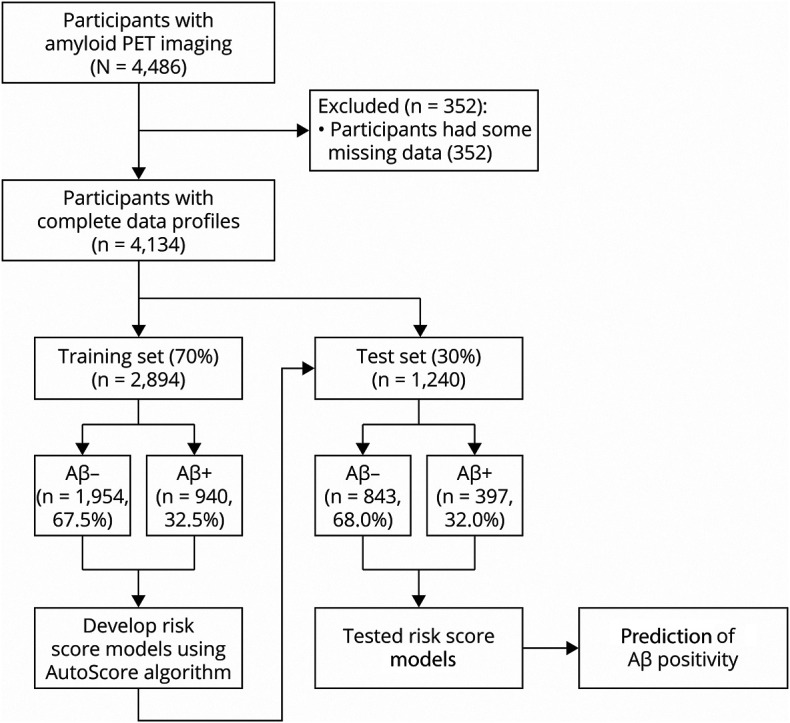
Aβ = β-amyloid.
Study Measures
Study Outcome
Amyloid PET imaging was done using 18F-florbetapir data acquired 50–70 minutes postinjection. Images were realigned and averaged and then spatially aligned to a standard space template. 18F-florbetapir, sampled in a global neocortical region for Aβ, was expressed as an SUVR with a cerebellar reference region. Aβ positivity was defined as 18F-florbetapir PET SUVR >1.10.8
Study Features (Predictors)
To develop the Positive Aβ Risk Scores (PARS), we categorized variables used as predictors into one of the following 4 broad categories to simplify systematic development of the risk score. To develop risk scores that are practical and simple, total scores for each variable are used when applicable rather than component scores of the individual measures.
Demographics
This category included age, sex, education, race, ethnicity, marital status, family history of dementia or memory impairment, and body mass index. Age and years of education were initially treated as continuous variables. Body mass index (BMI) was initially included as a continuous variable and subsequently grouped as low (0–25), intermediate (25–30), and high (30+) during the fine-tuning step of the algorithm. Marital status was a binary variable between married and not married (single, widowed, or divorced). Family history consisted of the number of parents (0, 1, or 2) with history of dementia or significant memory impairment. Race and ethnicity were categorized as white or non-White and Hispanic or non-Hispanic, respectively.
Subjective Measures
There were 3 self-report scales in this category: an adapted version of the Alzheimer's Disease Cooperative Study Activities of Daily Living–Prevention Instrument (ADCS-ADL)9; the 15-item version of the Geriatric Depression Scale (GDS)10; and the MACQ.11 The ADCS-ADL assesses 6 basic activities of daily living and 17 instrumental activities where a lower score indicates a more severe condition. GDS consists of 15 questions that assess depressive symptoms in seniors; scores range from 0 to 15, with scores above 5 indicating potential depression and above 10 likely depression. The MACQ consists of 6 questions where the first 5 are about situations often found to be troubling to those with declining memory and a final question that broadly measures one's perception of his or her own memory decline.
Objective Test Measures
This category included the following measures: the MMSE, a global measure of cognitive functioning12; the Digit Symbol Substitution, a measure of executive function13,14; free recall (FR) from the Free and Cued Selective Reminding Test (FCSRT), a measure of word recall under controlled learning conditions15,16; and immediate and delayed recall of the Logical Memory stories.17
Genetic Measures
The ε4 allele of APOE4 is the major genetic risk factor for AD. APOE4 status was entered into models as a categorical variable (0, 1, or 2 ε4 alleles).18,19
Data Analysis
Hierarchical Selection of Feature Sets
The predictors were grouped by category: demographics (D), subjective measures (S), objective measures (O), and genetic information (G). This allowed for the systematic study of the effect of each predictor group on the development of a risk score. To assess this, 5 feature sets were studied where D was a constant feature. These feature sets were D, DS, DSO, DG, and DSOG (Figure 2).
Figure 2. Feature Selection Criteria.
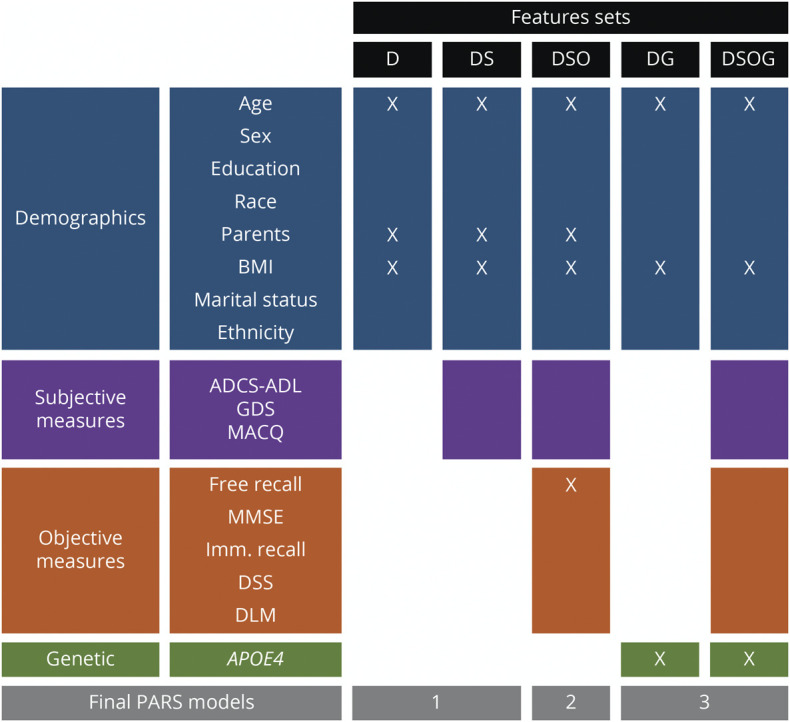
Five feature sets were considered as combinations of the 4 categories demographics (D), subjective measures (S), objective measures (O), and genetic information (G): D, DS, DSO, DG, and DSOG. Categories considered in each feature set are highlighted by color. Only the variables marked with an X were retained in the final models, resulting in 3 risk scores from the 5 feature sets considered. Parents refers to parents with dementia. ADCS-ADL = Alzheimer's Disease Cooperative Study–Activities of Daily Living; BMI = body mass index; DLM = delayed recall of the Logical Memory stories; DSS = Digit Symbol Substitution; GDS = Geriatric Depression Scale; MACQ = Memory Complaint Questionnaire; MMSE = Mini-Mental State Examination; PARS = Positive Aβ Risk Score.
Training and Test Samples
To train and test the risk scores, the data set was randomly divided into 2 independent subsets (Figure 1): training set (70%, Ntraining = 2,894) and test set (30%, Ntest = 1,240) of entire sample (N = 4,134). The training set was used to train, develop, and fine-tune the risk scores. The test set was independent from the training of the risk score and used to internally validate the performance of the final risk scores.
AutoScore Method
AutoScore is an algorithm and software package that was developed to aid clinicians and researchers in developing risk scores based on machine learning techniques.20 The AutoScore algorithm consists of 6 modules:
Module 1: Variable Ranking
RF, an ensemble machine learning algorithm, was used to identify the top-ranking predictors of subsequent score generation.21 Variables were ranked for inclusion in the risk score based on the mean decrease in impurity determined by the Gini index derived from RF outputs. An advantage of using RF to identify importance of variables for prediction over other methods such as backward stepwise regression or LASSO is that RF can rank variables on the basis of their nonlinear and heterogeneous effects. As the Gini index may exhibit bias toward continuous or high-cardinality variables, alternatives such as the permutation importance or conditional permutation importance may be used.22 In the AutoScore framework, the final list of variables is decided by the ranking, in addition to the parameter m, which is the number of final selected variables. Parameter m can be chosen case by case to accommodate clinical preference, expert or domain knowledge, or the needs of real-world applications.
Module 2: Variable Transformation
In this section, all selected variables that are continuous are preprocessed for variable transformation, that is, continuous variables are converted into categorical variables. Creating categorical variables allows for the modeling of nonlinear effects. In AutoScore, the maximum number of categories (e.g., K = 5) for each variable is predefined to ensure it is practical to use in the final risk score.
Module 3: Score Derivation
Next, the selected and transformed variables are used to create a risk score for prediction of the outcome. The method allows for the choice of maximum score depending on preference and application. The maximum score was chosen to be 100 points for this article. In this module, each category of an individual variable is weighted and given an integer point. As is commonly done with risk scores, logistic regression is used as the default setting for score weighting, with which the points can be easily interpreted.23
Module 4: Model Selection and Parameter Determination
A model is considered parsimonious when it is both sparse (small number of variables, m, possible) and has high prediction performance. To cope with the tradeoff between accuracy and complexity, different parameters m can be examined on the training set and a parsimony plot (i.e., model performance vs complexity) to which the user can refer for deciding the tradeoff in deriving the risk scores. The optimal parameter m is determined by when m continues to increase but the prediction performance is no longer improving significantly. In this study, to determine the final set of variables for risk scores, variables were incrementally added to the risk score and performance was assessed using the training set. A variable was included in the final risk score if it increased the area under the receiver operating characteristic (ROC) curve (AUC) of the predictive model by at least 1%. If adjustments were made in this step, then modules 2 and 3 are rerun to find an updated risk score.
Module 5: Fine-Tuning
For continuous variables, the variable transformation (module 2) is a data-driven process, in which domain knowledge is not integrated. In this module, the automatically generated cutoff values for each continuous variable can be fine-tuned by adjusting the cutoffs according to the standard clinical norm. If adjustments were made in this step, modules 2 and 3 are rerun to find an updated risk score.
Module 6: Predictive Performance Evaluation
The performance of the risk score is evaluated using ROC analysis, which includes AUC, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the final risk score on the test set.
Comparison of PARS Models With Other Machine Learning Models
Three RF algorithms were run for comparison of performance: full RF (fRF), RF with variable selection (vRF) performed using the stepwise selection procedure implemented in the VSURF R package,24 and RF regression (RFR) on the raw PET SUVR value where the predicted PET SUVR was subsequently binarized for classification using the cutoff value of >1.10 for Aβ positivity.
Standard Protocol Approvals, Registrations, and Patient Consents
All work was performed on data collected as part of the A4 Study (NCT02008357). The A4 Study was approved by the institutional review boards of all participating institutions. Informed written consent was obtained from all participants. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guidelines for cross-sectional studies.
Data Availability
Data used in this article are available for download from the Laboratory of NeuroImaging (LONI).25 All variables were extracted from files posted at LONI.
Results
Characteristics of the Study Sample
Participants had an average age of 71.28 years (SD 4.68), had an average number of years of education of 16.64 (SD 2.84), 94.4% were White, and 59.8% were women. The Aβ+ group, compared with the Aβ− group, was older, performed equally or worse on the different cognitive tests, and a larger percentage of them had at least 1 APOE4 ε4 allele. Sample characteristics are summarized in Table 1.
Table 1.
Sample Characteristics
Variable Selection and Score Derivation
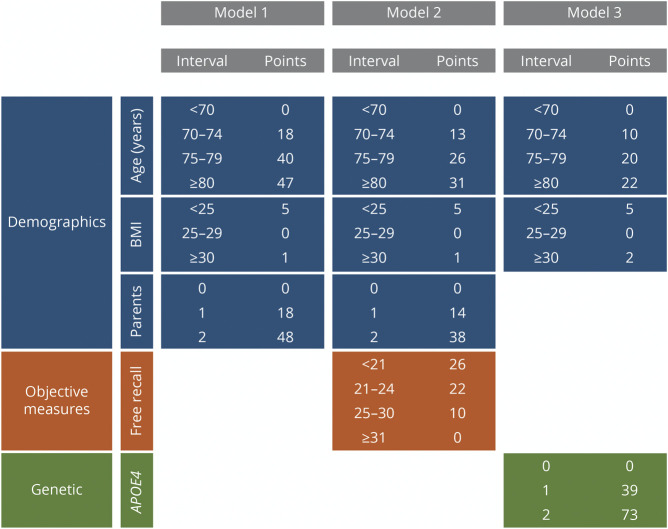
Five feature sets were considered in the development of the PARS models (Figure 2). With the goal reducing the number of variables used in order to achieve models that are simple, each variable was included in the final model only if it improved AUC by at least 1% (see Training and Test Samples for details). Therefore, 3 risk score models were derived from the initial 5 feature sets (Figure 2). The first model (PARS model 1), derived from feature set D, the final demographic model, included age, BMI, and family history. For the DS model, none of the subjective measures were retained. The DSO feature set resulted in model 2 (PARS model 2), which comprised 3 demographic features (age, BMI, and family history) and 1 objective feature (FR from the FCSRT). The last 2 feature sets both resulted in the model 3 (PARS model 3), which consisted of age, BMI, and APOE4 status. The 3 risk score models are summarized in Figures 2 and 3.
Figure 3. Chart of Point Assignments for Each of the PARS Models.
BMI = body mass index; PARS = Positive Aβ Risk Score.
Supplementary analysis on these models showed inclusion of additional variables that did not meet the inclusion criterion or improve AUC or overall performance of models. In cases where adding new variables improved one metric of model performance (e.g., specificity), it was at the cost of deterioration of another performance metric (e.g., sensitivity). The inclusion criterion worked to optimize the AUC of the model and balance the results of the other performance metrics (eFigure 1, links.lww.com/WNL/B979).
Performance of the Risk Scores
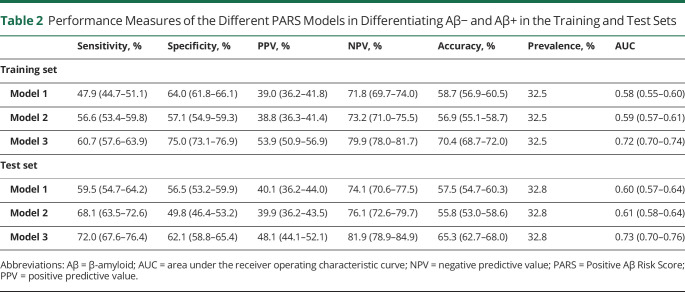
Performance of the 3 models on the training and test sets are reported in Table 2. In the test set, the ROC curves for models 1 and 2 were similar, but performance of model 3 was significantly higher (Figure 4). Models 1, 2, and 3 had AUCs of 0.60, 0.61, and 0.73, respectively (Table 2 and eFigure 2, links.lww.com/WNL/B979). Addition of FR to model 2 improved sensitivity of the model, but this was at the cost of a decrease in specificity. Model 3, the only model that included APOE4 allele as a predictor, replacing family history of dementia, performed better than all other models based on all performance metrics.
Table 2.
Performance Measures of the Different PARS Models in Differentiating Aβ− and Aβ+ in the Training and Test Sets
Figure 4. Receiver Operating Characteristic Curves of the 3 PARS Models Performance on the Test Dataset.
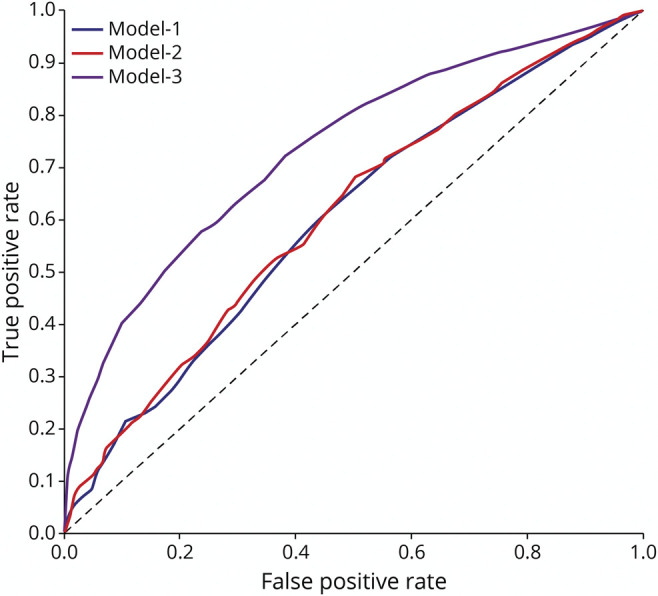
PARS = Positive Aβ Risk Score.
With a prevalence of 32.8% in the test set, model 3 showed the highest, but still moderate, performance metrics in comparison with other models: sensitivity 72.0%, specificity 62.1%, PPV 48.1%, NPV 81.9%, and accuracy 65.3%. Models 1 and 2 had PPVs approximately 22% (7.3 and 7.1 percentage points, respectively) above prevalence and model 3 had a PPV 47% (15.3 percentage points) above prevalence.
Although model 3 outperformed the other models in terms of higher risk score values corresponding to larger percentages of Aβ-positive individuals, with AUC of 0.73, the performance may be considered moderate (Figure 5). Whereas the percentages of amyloid-positive individuals in the bottom 2 quartiles of points for each model were similar, the percentages of amyloid-positive individuals with risk score points in the third quartile were 42%, 44%, and 65%, respectively, and the percentages of amyloid-positive individuals in the fourth quartile of points for risk score models 1, 2, and 3 were 71%, 60%, and 90%, respectively.
Figure 5. Percentage of Individuals for Each PARS Model Who Are Aβ Positive Within Each Quartile of Risk Score Points Calculated Using Test Dataset Where the Maximum Risk Score is 100 Points.

Aβ = β-amyloid; PARS = Positive Aβ Risk Score.
To compare the performance of the 3 PARS models, 3 RF models (fRF, vRF, and RFR) were developed on each feature set. ROC curves for the PARS and RF models were found to be comparable (eFigure 3, links.lww.com/WNL/B979). Detailed performance metrics of the PARS and RF models can be found in eTable 1. When using the feature set corresponding to all the available variables (DSOG), PARS model 3 achieved a higher sensitivity, but lower specificity, than the other models. Furthermore, model 3 performed comparably with respect to NPV, but had a slightly lower PPV.
Because the outcome only has a slight class imbalance with a ratio of 1:2, data balancing methods were not implemented. However, because the following additional metrics are considered to have advantages for imbalanced classes,26 they are reported in eTable 1 (links.lww.com/WNL/B979): F1 score, Balanced Accuracy, and the Matthews correlation coefficient (MCC). PARS models 1 and 2 performed better on all 3 metrics compared with the RF models when using the D, DS, and DSO feature sets. The PARS and RF models perform comparably on the F1 score and Balanced Accuracy on both the DG and DSOG feature sets. However, on the DG feature set, the fRF and vRF both outperform model 3 and the RFR model on the MCC metric. Lastly, on the DSOG feature set, all RF models perform moderately well and slightly outperform model 3 on the MCC metric.
Classification of Evidence
This study provides Class II evidence that in cognitively unimpaired individuals, PARS models predict Aβ positivity with moderate accuracy.
Discussion
In this study, we developed 3 PARS models for predicting Aβ positivity determined by 18F-florbetapir PET imaging. These risk scores are simple and practical to use as they only require features that are easily and inexpensively obtainable from all individuals. PARS model 3 included APOE4 genotypes as a predictor and outperformed the other models in all performance measures with moderate overall performance. In situations where APOE4 genotype information is not available, model 2, derived from the DSO feature set, could be used. In addition, although model 3 performed better than the other models, measuring FR is more inexpensive and easier than obtaining APOE4 information and may make model 2 preferable for use in some situations.
Over the past decade, many research groups have tried to predict Aβ positivity using a wide variety of predictors including demographics, neuropsychological tests, genetic risk factors, MRI, and CSF or blood-based biomarkers.27-32 A comparison of performance between these models and PARS can be found in eTable 2 (links.lww.com/WNL/B979). With the recent approval of aducanumab as the first AD treatment that targets Aβ,33 developing tools that facilitate clinical decision-making for obtaining confirmatory tests (i.e., amyloid PET scans or CSF studies) becomes even more important. One study found RF models utilizing demographics, APOE4, and longitudinal cognitive tests could predict Aβ positivity in cognitively healthy individuals.28 A second study comparing Aβ positivity stratification models using a RF approach with minimally invasive and low-cost measures via blood-based biomarkers showed promise toward reducing the cost and difficulty of predicting Aβ positivity.34 Another study found neuropsychological and MRI measures used separately performed equally well as when used jointly on individuals with amnestic mild cognitive impairment (MCI).29 Although some of these studies developed high-performance models, it has been exceedingly difficult to draw conclusions from them not only because of the breadth of approaches used, but also because studies mix participants at different stages of the AD spectrum (normal, MCI, early dementia, or a combination).
The high-performance models for predicting Aβ positivity developed in previous work are often not practical for use by clinicians or in prospective studies. This impediment exists either because the models are too complicated to be used or the inputs used as predictor variables are too costly or burdensome to obtain. Many models require the performance of multiple neuropsychological examinations and evaluation of multiple genetic risk factors, structural MRIs, CSF, or blood-based biomarkers.35
A recent study also looked at predicting amyloid burden using data from the A4 Study with the purpose of providing solutions to challenges related to participant selection for clinical trials.36 Using the Extreme Gradient Boosting (XGBoost) algorithm, a tree-based machine learning method, researchers limited predictive variables to demographics, cognitive and functional assessments, and APOE4 genotype and developed models with AUCs between 0.60 (without APOE4 genotypes) and 0.73 (with APOE4 genotypes). The performance of these models is moderate and comparable to PARS model 1 and 2, with AUCs equal to 0.60 and 0.61, and model 3, with AUC equal to 0.73. Differences between the 2 studies are as follows: (1) although many variables are similar, different variables are considered, such as Preclinical Alzheimer Cognitive Composite (PACC) (versus individual components of PACC), Cogstate scores, and Cognitive Function Instrument scores; (2) distinction between remote and in-clinic collection of certain variables; and (3) direct use of machine learning for prediction instead of use of machine learning to develop a simplified risk score used for prediction of amyloid positivity.
For studies of presymptomatic AD, models like the PARS developed from cognitively normal individuals are required. The advantage of PARS is the ability to compute multiple risk scores that can be used situationally depending on the data available to, or easily obtainable by, clinicians and researchers. In addition, once the predictor variables are determined by the clinician, the risk scoring models require only a simple calculation to determine an individual's total risk score. To improve practicality of using PARS, we developed an online calculator, which could be accessed at Neurodiction.37 There is great potential to expand this scoring system by addition of other predictors not included in this study, such as structural MRIs, genetic information, or biofluid biomarkers.38,39
A parsimonious set of readily ascertainable risk factors like PARS would facilitate enrollment in clinical trials of very early AD and inform decisions about amyloid PET at the time of the initial evaluation. Requiring longitudinal cognitive tests will impede ascertainment and delay clinical decisions for individuals at most risk for AD progression. To address this issue and to create a model that is practical to use by clinicians, a recent study used a probabilistic approach to develop 2 algorithms with robust performance across 3 different cohorts: Alzheimer's Disease Neuroimaging Initiative (ADNI), Australian Imaging, Biomarkers, and Lifestyle Flagship Study of Ageing (AIBL), and Mayo Clinic Olmsted Study of Aging (MCSA).30 The first algorithm required only age and immediate recall test score as inputs while the second also included APOE4 genotype information. Our findings were similar to this study even though theirs was conducted in a population that included both CN individuals and patients with MCI whereas ours was conducted only in CN individuals. We also found that age and APOE4 genotype were among the most informative measures for detecting Aβ positivity. Additional predictors identified as important in our study were FR on the FCSRT, which was not available in all the study datasets used by Maserejian et al.,30 BMI, and parental history of dementia, which was not included in their final models. As cognitive status (CN vs MCI) is a predictor of amyloid positivity, models may operate differently in persons who are CN and those who have MCI.
Whereas the PARS models may require further refinement and need to be validated on independent datasets, the potential effect of the PARS models for use in clinical trials can be demonstrated by the following hypothetical thought experiment. Assume there is a goal to enroll 300 Aβ+ patients into each of 3 treatment arms (placebo, low dose, and high dose) of a potential therapy. In the first scenario, where risk scores are not utilized, 1,000 individuals need to be identified as Aβ+, due to other exclusions, where amyloid PET scans are administered on all potential participants. At the prevalence of 33%, approximately 3,300 would need to undergo costly amyloid PET scans. If instead individuals are first screened using the risk scores of PARS models 1, 2, or 3, approximately 2,500, 2,500, and 2,000 individuals need to be screened by amyloid PET scans, respectively. Savings include the reduced cost of the amyloid PET and the reduced exposure of participants to radiation, albeit at low levels. Those savings are offset by increased cost of participant identification and screening.
Some limitations should be noted. First, the models were trained, validated, and internally tested on data from the A4 Study. The A4 sample is predominantly White, is highly educated, has strict inclusion/exclusion criteria for participation in the study, and has other characteristics that might not be representative of the general population. Whereas this approach can be easily applied to other groups or populations, for these models to be considered generalizable they will need to be tested on other populations. Second, whereas performance of these risk scores is comparable to previous studies that used similar predictors, their performance is moderate. Third, only data for individuals with complete data profiles were included in this analysis. Alternatively, methods of data imputation could be used to include data from more individuals enrolled in the A4 Study. Lastly, the risk scores would likely benefit from the addition of other predictors such as blood-based biomarkers or longitudinal variables by improving prediction performance and robustness.
The development of the PARS models in this study demonstrated an approach to creating risk scores that is simple and results in models with few variables that perform comparably to more complicated algorithms such as RF models with or without variable selection. As such, the models can be refined further using the same method on datasets that have additional potentially informative variables, particularly volumetric MRI data or blood-based biomarkers.
Acknowledgment
The A4 Study is a secondary prevention trial in preclinical Alzheimer disease aiming to slow cognitive decline associated with brain amyloid accumulation in clinically normal older individuals. The A4 Study is funded by a public–private–philanthropic partnership, including funding from the NIH–National Institute on Aging, Eli Lilly and Company, Alzheimer's Association, Accelerating Medicines Partnership, GHR Foundation, an anonymous foundation, and additional private donors, with in-kind support from Avid and Cogstate. The companion observational Longitudinal Evaluation of Amyloid Risk and Neurodegeneration (LEARN) Study is funded by the Alzheimer's Association and GHR Foundation. The A4 and LEARN Studies are led by Dr. Reisa Sperling at Brigham and Women's Hospital, Harvard Medical School, and Dr. Paul Aisen at the Alzheimer's Therapeutic Research Institute (ATRI), University of Southern California. The A4 and LEARN Studies are coordinated by ATRI at the University of Southern California and the data are made available through the Laboratory for NeuroImaging at the University of Southern California. The participants screening for the A4 Study provided permission to share their de-identified data in order to advance the quest to find a successful treatment for Alzheimer disease. The authors thank the participants, the site personnel, and the partnership team members of the A4 and LEARN Studies. The complete A4 Study Team list is available at a4study.org/a4-study-team.
Glossary
- A4
Anti-Amyloid Treatment in Asymptomatic Alzheimer's
- Aβ
β-amyloid
- AD
Alzheimer disease
- ADCS-ADL
Alzheimer's Disease Cooperative Study–Activities of Daily Living
- AUC
area under the receiver operating characteristic curve
- BMI
body mass index
- CN
cognitively normal
- FCSRT
Free and Cued Selective Reminding Test
- FR
free recall
- fRF
full random forest
- GDS
Geriatric Depression Scale
- LONI
Laboratory of NeuroImaging
- MACQ
Memory Complaint Questionnaire
- MCC
Matthews correlation coefficient
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
- NPV
negative predictive value
- PACC
Preclinical Alzheimer Cognitive Composite
- PARS
Positive Aβ Risk Score
- PPV
positive predictive value
- RF
random forest
- RFR
random forest regression
- ROC
receiver operating characteristic
- SUVR
standardized uptake value ratio
- vRF
random forest with variable selection
Appendix. Authors

Footnotes
Class of Evidence: NPub.org/coe
Editorial, page 999
Study Funding
This work was supported by grants from the NIH (NIA K23 AG063993, Dr. Ezzati; NIA AG03949, Dr. Lipton), the Alzheimer's Association (2019-AACSF-641329, Dr. Ezzati), the Cure Alzheimer's Fund (Drs. Ezzati and Lipton), and the Leonard and Sylvia Marx Foundation (Dr. Lipton).
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Jack CR Jr, Bennett DA, Blennow K, et al. . NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14(4):535-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen WJ, Ossenkoppele R, Knol DL, et al. . Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA. 2015;313(19):1924-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doody RS, Thomas RG, Farlow M, et al. . Phase 3 trials of solanezumab for mild-to-moderate Alzheimer's disease. N Engl J Med. 2014;370(4):311-321. [DOI] [PubMed] [Google Scholar]
- 4.Honig LS, Vellas B, Woodward M, et al. . Trial of solanezumab for mild dementia due to Alzheimer's disease. N Engl J Med. 2018;378(4):321-330. [DOI] [PubMed] [Google Scholar]
- 5.Foote C, Woodward M, Jardine MJ. Scoring risk scores: considerations before incorporating clinical risk prediction tools into your practice. Am J Kidney Dis. 2017;69(5):555-557. [DOI] [PubMed] [Google Scholar]
- 6.Schneider L. A resurrection of aducanumab for Alzheimer's disease. Lancet Neurol. 2020;19(2):111-112. [DOI] [PubMed] [Google Scholar]
- 7.Clinical Trial of Solanezumab for Older Individuals Who May be at Risk for Memory Loss (A4). Accessed May 31, 2021. clinicaltrials.gov/ct2/show/NCT02008357 [Google Scholar]
- 8.Joshi AD, Pontecorvo MJ, Clark CM, et al. . Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer's disease and cognitively normal subjects. J Nucl Med. 2012;53(3):378-384. [DOI] [PubMed] [Google Scholar]
- 9.Galasko D, Bennett DA, Sano M, Marson D, Kaye J, Edland SD. ADCS Prevention Instrument Project: assessment of instrumental activities of daily living for community-dwelling elderly individuals in dementia prevention clinical trials. Alzheimer Dis Assoc Disord. 2006;20(4 suppl 3):S152-S169. [DOI] [PubMed] [Google Scholar]
- 10.Sheikh JI, Yesavage JA. Geriatric Depression Scale (GDS): recent evidence and development of a shorter version. Clin Gerontol. 1986;5(1-2):165-173. [Google Scholar]
- 11.Crook TH III, Feher EP, Larrabee GJ. Assessment of memory complaint in age-associated memory impairment: the MAC-Q. Int Psychogeriatr. 1992;4(2):165-176. [DOI] [PubMed] [Google Scholar]
- 12.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189-198. [DOI] [PubMed] [Google Scholar]
- 13.Boake C. From the Binet-Simon to the Wechsler-Bellevue: tracing the history of intelligence testing. J Clin Exp Neuropsychol. 2002;24(3):383-405. [DOI] [PubMed] [Google Scholar]
- 14.Wechsler D. The Measurement of Adult Intelligence, 3rd ed. Williams & Wilkins; 1944:vii, 258-vii, 258. [Google Scholar]
- 15.Grober E, Buschke H. Genuine memory deficits in dementia. Dev Neuropsychol. 1987;3(1):13-36. [Google Scholar]
- 16.Grober E, Hall CB, Lipton RB, Zonderman AB, Resnick SM, Kawas C. Memory impairment, executive dysfunction, and intellectual decline in preclinical Alzheimer's disease. J Int Neuropsychol Soc. 2008;14(2):266-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wechsler D. WMS-R: Wechsler Memory Scale-Revised. Psychological Corporation; 1987. [Google Scholar]
- 18.Apostolova LG, Risacher SL, Duran T, et al. . Associations of the top 20 Alzheimer disease risk variants with brain amyloidosis. JAMA Neurol. 2018;75(3):328-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Battista AM, Heinsinger NM, Rebeck GW. Alzheimer's disease genetic risk factor APOE-ε4 also affects normal brain function. Curr Alzheimer Res. 2016;13(11):1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie F, Chakraborty B, Ong MEH, Goldstein BA, Liu N. AutoScore: a machine learning–based automatic clinical score generator and its application to mortality prediction using electronic health records. JMIR Med Inform. 2020;8(10):e21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TK. Random Decision Forests; 1995:278-282. [Google Scholar]
- 22.Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340-1347. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan LM, Massaro JM, D'Agostino RB Sr. Presentation of multivariate data for clinical use: the Framingham Study risk score functions. Stat Med. 2004;23(10):1631-1660. [DOI] [PubMed] [Google Scholar]
- 24.Genuer R, Poggi JM, Tuleau-Malot C. VSURF: an R package for variable selection using random forests. R J. 2015;7(2):19-33. [Google Scholar]
- 25.Laboratory of NeuroImaging website. loni.usc.edu
- 26.Zhou PY, Wong AKC. Explanation and prediction of clinical data with imbalanced class distribution based on pattern discovery and disentanglement. BMC Med Inform Decis Making. 2021;21(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandel BM, Avants BB, Gee JC, Arnold SE, Wolk DA. Neuropsychological testing predicts cerebrospinal fluid amyloid-β in mild cognitive impairment. J Alzheimers Dis. 2015;46(4):901-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Insel PS, Palmqvist S, Mackin RS, et al. . Assessing risk for preclinical β-amyloid pathology with APOE, cognitive, and demographic information. Alzheimers Dement. 2016;4:76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ezzati A, Harvey DJ, Habeck C, et al. . Predicting amyloid-β levels in amnestic mild cognitive impairment using machine learning techniques. J Alzheimers Dis. 2020;73(3):1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maserejian N, Bian S, Wang W, et al. . Practical algorithms for amyloid β probability in subjective or mild cognitive impairment. Alzheimers Dement. 2019;11:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnham SC, Faux NG, Wilson W, et al. . A blood-based predictor for neocortical Aβ burden in Alzheimer's disease: results from the AIBL study. Mol Psychiatry. 2014;19(4):519-526. [DOI] [PubMed] [Google Scholar]
- 32.Palmqvist S, Insel PS, Zetterberg H, et al. . Accurate risk estimation of β-amyloid positivity to identify prodromal Alzheimer's disease: cross-validation study of practical algorithms. Alzheimers Dement. 2019;15(2):194-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.FDA's Decision to Approve New Treatment for Alzheimer's Disease. Accessed June 7, 2021. fda.gov/drugs/news-events-human-drugs/fdas-decision-approve-new-treatment-alzheimers-disease [Google Scholar]
- 34.Tosun D, Veitch D, Aisen P, et al. . Detection of β-amyloid positivity in Alzheimer's Disease Neuroimaging Initiative participants with demographics, cognition, MRI and plasma biomarkers. Brain Commun. 2021;3(2):fcab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashford MT, Veitch DP, Neuhaus J, Nosheny RL, Tosun D, Weiner MW. The search for a convenient procedure to detect one of the earliest signs of Alzheimer's disease: a systematic review of the prediction of brain amyloid status. Alzheimers Dement. 2021;17(5):866-887. [DOI] [PubMed] [Google Scholar]
- 36.Langford O, Raman R, Sperling RA, et al. . Predicting amyloid burden to accelerate recruitment of secondary prevention clinical trials. J Prev Alzheimers Dis. 2020;7(4):213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neurodiction LLC website. neurodiction.com/pars-amyloid-positivity
- 38.Desikan RS, Fan CC, Wang Y, et al. . Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med. 2017;14(3):e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer BE, Digma LA, Jennings RG, Brewer JB. Gene‐ and age‐informed screening for preclinical Alzheimer's disease trials. Alzheimers Dement. 2021;17(3):457-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used in this article are available for download from the Laboratory of NeuroImaging (LONI).25 All variables were extracted from files posted at LONI.