Abstract
Commercial isolates of Saccharomyces cerevisiae differ in the production of hydrogen sulfide (H2S) during fermentation, which has been attributed to variation in the ability to incorporate reduced sulfur into organic compounds. We transformed two commercial strains (UCD522 and UCD713) with a plasmid overexpressing the MET17 gene, which encodes the bifunctional O-acetylserine/O-acetylhomoserine sulfhydrylase (OAS/OAH SHLase), to test the hypothesis that the level of activity of this enzyme limits reduced sulfur incorporation, leading to H2S release. Overexpression of MET17 resulted in a 10- to 70-fold increase in OAS/OAH SHLase activity in UCD522 but had no impact on the level of H2S produced. In contrast, OAS/OAH SHLase activity was not as highly expressed in transformants of UCD713 (0.5- to 10-fold) but resulted in greatly reduced H2S formation. Overexpression of OAS/OAH SHLase activity was greater in UCD713 when grown under low-nitrogen conditions, but the impact on reduction of H2S was greater under high-nitrogen conditions. Thus, there was not a good correlation between the level of enzyme activity and H2S production. We measured cellular levels of cysteine to determine the impact of overexpression of OAS/OAH SHLase activity on sulfur incorporation. While Met17p activity was not correlated with increased cysteine production, conditions that led to elevated cytoplasmic levels of cysteine also reduced H2S formation. Our data do not support the simple hypothesis that variation in OAS/OAH SHLase activity is correlated with H2S production and release.
Hydrogen sulfide (H2S) is an undesirable by-product of alcoholic fermentation by yeast. Very low levels of H2S can be detected due to its low aroma threshold (11), so trace amounts can have a profound effect on final product quality. Subtle changes in yeast metabolic behavior appear to have a major impact on the presence or absence of this spoilage character. It is highly desirable, therefore, to have yeast strains available for wine production that will not produce and release H2S. Lowering the amount of H2S produced could be achieved by supplementation of grape juice with one or more organic sources of sulfur: methionine, cysteine, glutathione, or the intermediate homocysteine, as transsulfuration pathways exist, thereby negating the need for sulfate reduction (7, 34). However, these sulfur-containing compounds can be metabolized to other volatile sulfur compounds by Saccharomyces and other yeasts and bacteria present in fermenting juice (8), producing characters (feces or rotten seafood) as objectionable as or more objectionable than that of H2S (rotten egg).
Several environmental factors have been implicated in the appearance of hydrogen sulfide: high residual levels of elemental sulfur (26, 27), presence of sulfur dioxide (13, 17, 31), presence of organic compounds containing sulfur (1, 19), pantothenate deficiency (35, 36), high threonine content (37), high methionine or cysteine content relative to other amino acids (1, 19, 37), and nitrogen limitation (12, 13, 37). Volatile-sulfur-compound production varies widely among commercial strains and natural isolates in response to these environmental conditions (1, 9, 12, 15, 37). This variation suggests that differences in internal levels of enzymatic activities or their regulation have a profound effect on the appearance of hydrogen sulfide. If the basis of this genetic variation can be identified, then it may be possible to construct commercial strains with reduced sulfur production.
The MET17 gene (also called MET15 or MET25) encodes a sulfhydrylase (SHLase) capable of using either O-acetylserine (OAS) or O-acetylhomoserine (OAH) as the substrate in vitro and is the last step of the sulfate reduction pathway. met17 mutants are methionine auxotrophs, and no OAS/OAH SHLase activity is detectable (22). Only O-acetylhomoserine appears to be the substrate in vivo, with O-acetylserine serving instead as an inducer of the sulfate reduction pathway (22, 23, 25), in contrast to the situation in bacteria (18). A mutational analysis indicated that the transsulfuration pathway is the only pathway by which cysteine is synthesized in Saccharomyces (7) and that the direct conversion of O-acetylserine to cysteine does not occur. The MET17 gene, therefore, encodes the only enzymatic activity responsible for incorporation of reduced sulfide in this yeast.
The levels of OAS/OAH SHLase can vary 10-fold in different strains of Saccharomyces cerevisiae (22). One hypothesis is that low levels of reduced sulfur incorporation result in “leakiness” of reduced sulfur and subsequent release of H2S (16, 19). Thus, strains that are high producers of H2S should have relatively low levels of OAS/OAH SHLase activity. In support of this hypothesis, Omura et al. (21) reported that H2S formation was reduced in a brewing yeast strain overexpressing the MET17 gene, which encodes OAS/OAH SHLase.
Our objective in this study was to determine if low levels of OAS/OAH SHLase activity accounted for increased H2S production in two commercial yeast strains: one, UCD522, which produces high levels of H2S under commercial conditions, and another, UCD713, which is a low-to-moderate-level producer of this compound. Additional objectives of this study were (i) to overexpress the MET17 gene in two commercial strains of S. cerevisiae to further test the hypothesis that increased levels of expression would increase efficiency of incorporation of sulfur and reduce H2S formation and (ii) to study the consequences of the overproduction of OAS/OAH SHLase during alcoholic fermentation simulating wine production conditions.
MATERIALS AND METHODS
Plasmids, DNA manipulations, and transformation methods.
Restriction and modification enzymes were used according to the manufacturer's instructions. The control vector pEG25C was constructed by restriction digestion of the pEG25 (21) multicopy 2μm vector with BamHI (Gibco-BRL, Gaithersburg, Md.) in order to remove the MET17 insert, cutting the appropriate band of DNA from a 0.7% agarose gel, cleaning the DNA (QIAquick gel extraction kit; QIAGEN, Valencia, Calif.), and subsequent ligation of the DNA using T4 DNA ligase (Gibco-BRL). The resulting control vector lacked the MET17 coding region but retained the sequence corresponding to the glyceraldehyde-3-phosphate dehydrogenase gene promoter used to overexpress MET17 (21). S. cerevisiae was transformed using the lithium acetate method (28), and Escherichia coli was transformed using the method described by Inoue et al. (14). E. coli INVαF′ (Invitrogen, Carlsbad, Calif.) was used for plasmid preparations. Luria-Bertani medium with ampicillin was used for selection of transformed E. coli cells.
Yeast strains and culture conditions.
Two S. cerevisiae industrial wine yeast isolates were used from our culture collection (Department of Viticulture and Enology, University of California, Davis): UCD522 (Montrachet) and UCD713 (French White) (Universal Foods, Milwaukee, Wis.). These commonly used commercial strains are likely not true diploids and contain extra copies of some chromosomes (2).
Yeast strains were maintained and grown on yeast extract-peptone-dextrose medium with 2% glucose (YPD) (29). The same medium (YPD) with geneticin (G418, 30 ppm) was used for transformant selection and maintenance. Four transformants were constructed: UCD522 transformed with pEG25 (UCD522pMET17) and with the vector pEG25C (UCD522pVector) and UCD713 transformed with pEG25 (UCD713pMET17) and pEG25C (UCD713pVector).
Fermentation media and conditions.
In the fermentation experiments, the synthetic grape juice medium “Minimal Must Medium” (MMM) (12), modified from the original recipe described by Spiropoulos et al. (30), was used. The two nitrogen levels were generated by using 0.8 g of l-arginine/liter and 1 g of ammonium phosphate/liter for media containing 433 mg of N equivalents/liter and 0.2 g of l-arginine/liter and 0.5 g of ammonium phosphate/liter for media containing 208 mg of N equivalents/liter (30).
Fermentations were initiated at a density of 106 cells/ml by inoculation with stationary-phase cells from a culture pregrown in MMM starter medium (30). Fermentations were conducted in 500-ml Erlenmeyer flasks containing 300 ml of medium. Each flask was connected to a hydrogen sulfide trap (30). The flasks were incubated by shaking (120 rpm) on a rotary shaker at room temperature (23 to 28°C). Fermentations were monitored by using weight loss as an estimate of CO2 production (30). Completion of fermentation (absence of reducing sugars) was confirmed by using the Clinitest (Bayer, Elkhart, Ind.). Fermentations with higher nitrogen levels were performed in triplicate, while fermentations with lower nitrogen levels were performed in duplicate. All the fermentations reached dryness (less than 0.5% residual sugar), except UCD713pMET17 (2% residual sugar). Values presented represent the averages of the replicate samples. Levels of H2S in replicates run simultaneously differed by less than 5%. In replicate experiments not run at the same time, H2S values typically varied by 20% or less. This elevated variation probably was due to differences in room temperature in experiments not conducted at the same time. In the few cases where the variation was greater than 20%, the trend of peaks and valleys was the same.
Analytical methods.
H2S was measured using the cadmium trap assay as previously described (30). Protein extracts were prepared using the method of Brzywczy and Paszewski (4). OAS SHLase activity was measured using the method of Paszewski and Grabski (24). One unit of activity was defined as the amount of enzyme producing 1 nmol of cysteine per mg of protein per min. Cysteine was extracted from yeast cells by the method of Tezuka et al. (32) and was measured by the method of Gaitonde (10). The protein level was estimated by the method of Bradford with bovine serum albumin as the standard (3). Plasmid loss was monitored during the entire course of fermentation in MMM by plating samples on YPD alone and YPD plus 30 mg of G418/liter.
RESULTS
Characterization of H2S formation in commercial wine strains of Saccharomyces.
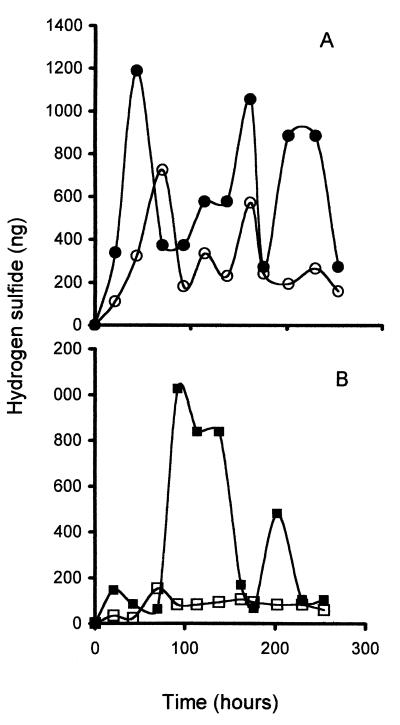
The formation of H2S by two commercial wine strains of Saccharomyces was evaluated during fermentation in synthetic grape juice media to determine if our assay conditions would yield variation in H2S production similar to that observed for these strains under commercial conditions. UCD522 produced significantly higher amounts of sulfide than did UCD713 under nutrient-sufficient conditions (Fig. 1), consistent with reports for these strains during wine production.
FIG. 1.
Pattern of production of H2S in UCD522 (A) and UCD713 (B) at two concentrations of nitrogen. The levels of H2S represent the total amount accumulated in the cadmium trap for that interval. For example, the point at 43 h represents the amount accumulated between 21 and 43 h. Open symbols, high nitrogen; solid symbols, low nitrogen.
Nitrogen limitation affects the production of H2S during alcoholic fermentation (12, 13, 37). Most strains release increased H2S when nitrogen is limited. We monitored sulfide production during an alcoholic fermentation of synthetic grape juice media but with approximately half (208 mg/liter) of the original nitrogen (433 mg/liter) (Fig. 1). Sulfide formation increased significantly for both isolates, but overall UCD522 produced almost twice as much total H2S as UCD713 (Table 1). While these two wine isolates display distinctly different behavior in the basal level of production of H2S, both react similarly to a reduction in medium nitrogen levels and increase the amount of H2S produced.
TABLE 1.
Hydrogen sulfide production under low- and high-nitrogen conditions in transformed and untransformed strains
| Nitrogen concn (mg/liter) | Strain | Maximum fermentation rate (g/h)a | Total H2S (ng) | % Maximum of H2S |
|---|---|---|---|---|
| 433b | UCD522 | 0.123 | 3,300 | 100 |
| UCD522pVector | 0.115 | 3,100 | 93 | |
| UCD522pMET17 | 0.102 | 5,000 | 150 | |
| UCD713 | 0.104 | 890 | 100 | |
| UCD713pVector | 0.127 | 850 | 96 | |
| UCD713pMET17 | 0.098 | 210 | 24 | |
| 208c | UCD522 | 0.149 | 6,800 | 100 |
| UCD522pVector | 0.144 | 6,300 | 93 | |
| UCD522pMET17 | 0.166 | 8,000 | 118 | |
| UCD713 | 0.146 | 3,900 | 100 | |
| UCD713pVector | 0.154 | 3,700 | 95 | |
| UCD713pMET17 | 0.106 | 680 | 17 |
The maximum fermentation rate was calculated from the fermentation rate data by using time points corresponding to the steepest decline in weight.
Values represent the average of independent determinations of three replicates. Replicate values varied by less than 3%.
Values represent the average of independent determinations of two replicates. Replicates varied by less than 5%.
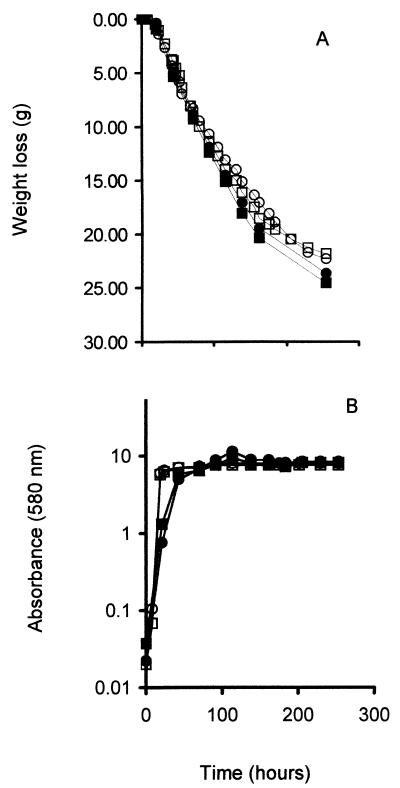
The H2S produced by yeast strains is driven from the fermentation medium into the trap by the CO2 produced from the catabolism of sugar. Differences in the fermentation rate may therefore impact the detection of H2S. The nitrogen levels used were chosen because there were no significant differences in fermentation behavior (Fig. 2A), growth rate (Fig. 2B), or maximal fermentation rate (Table 1) for these two strains under these conditions. Both displayed a slightly higher maximal fermentation rate under low-nitrogen conditions. Thus, the two commercial strains displayed differences in H2S formation not associated with differences in growth or fermentation performance.
FIG. 2.
Fermentation (A) and growth (B) of UCD522 and UCD713 in low-nitrogen (solid symbols) and high-nitrogen (open symbols) media. ○, ●, UCD522; □, ■, UCD713.
Comparison of OAS/OAH SHLase activity and H2S formation.
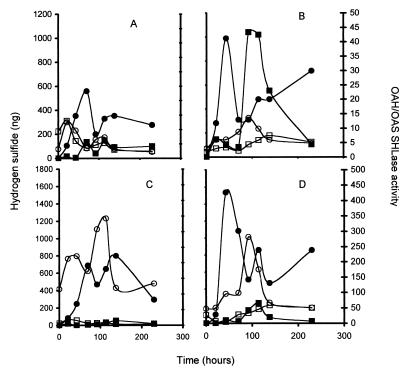
To determine if differences in the production of H2S were associated with differences in rates of reduced sulfur incorporation, we measured OAS/OAH SHLase activity in both strains, as well as in strains that were transformed with the vector encoding G418 resistance. Under high-nitrogen conditions across the time course of fermentation, activity values ranged from 2.2 to 12 U for UCD522 and from 2.5 to 13 U for UCD713. The two strains showed a similar pattern of expression of activity, with activity peaking within the first 24 to 48 h and then gradually declining to a low value late in fermentation (Fig. 3A) (data shown for strains carrying the vector only). At most time points, the activity value for UCD713 is generally 30 to 50% lower than for UCD522.
FIG. 3.
H2S and OAS/OAH SHLase activity in transformed strains of UCD522 (○, ●) and UCD713 (□, ■). Solid symbols, H2S production; open symbols, OAS/OAH SHLase activity. (A) High nitrogen, strains transformed with vector; (B) low nitrogen, strains transformed with vector; (C) high nitrogen, strains transformed with pMET17; (D) low nitrogen, strains transformed with pMET17. OAS/OAH SHLase activity is measured in arbitrary units.
Under low-nitrogen conditions, activity in UCD522 similarly ranged from 3 to 13 U, with the peak in activity occurring around 92 h into the fermentation (Fig. 3B). OAS/OAH SHLase activity was reduced in UCD713 under nitrogen-limiting conditions and ranged from 1.7 to 7.5 U. Except for very late time points, activity was two- to threefold higher in UCD522 than in UCD713.
Hydrogen sulfide production also was evaluated for the time course of fermentation (Fig. 3). Under high-nitrogen conditions, UCD522 produced higher levels of H2S throughout the time course of fermentation than did UCD713. Both strains showed the same pattern of peaks and valleys of H2S release. Thus, while enzymatic activity did not vary significantly under these conditions for these two strains, UCD522 consistently produced greater levels of reduced sulfur. Note that the enzymatic levels represent activity in a cell extract made from a sample taken at that point in time, while the H2S levels represent the amount accumulated in the trap over the time period indicated. As enzymatic activity decreased in UCD522, there was a tendency for H2S levels to rise. If the change in enzymatic activity between time points was plotted versus the level of H2S produced, an R2 value of 0.83 was obtained for UCD522, while that for UCD713 was only 0.018. Changes in activity were therefore somewhat correlated with changes in H2S production in one strain, UCD522, but not in the other.
Both strains produced higher levels of H2S under low-nitrogen conditions. The total amount of H2S formed for UCD522 doubled with a 50% decrease in nitrogen content (Table 1). Total H2S production increased over fourfold for UCD713. Thus UCD713 displays a much stronger response in increasing H2S formation under nitrogen limitation. The peak of H2S production occurred earlier in UCD522 than in UCD713 (Fig. 3B). H2S formation did not correlate well with changes in OAS/OAH SHLase activity in either UCD522 or UCD713 under low-nitrogen conditions (R2 values of 0.12 or 0.50, respectively). OAS/OAH SHLase activity differed in the two strains, as did H2S formation, but there was not a good correlation between changes in the level of activity of this enzyme and release of H2S, with the exception of UCD522 under high-nitrogen conditions.
Effect of overexpression of MET17.
We transformed both strains with a multicopy plasmid carrying the MET17 gene, leading to overproduction of OAS/OAH SHLase, and evaluated Met17p enzymatic activity and H2S production. In UCD522 under high-nitrogen conditions, the presence of the MET17 gene on the plasmid resulted in a 10- to 70-fold increase in Met17p activity (Fig. 3C). However, total H2S production was not reduced but instead was slightly increased (Table 1). Met17p activity displayed a similar increase under low-nitrogen conditions (Fig. 3D). H2S production did not decrease and also displayed a very slight but reproducible elevation in the strain carrying the MET17 gene, compared to the strain without plasmid or with the vector.
Transformation of UCD713 with MET17 increased OAS/OAH SHLase activity and decreased H2S formation at both nitrogen concentrations (Fig. 3C and D). The increase in OAS/OAH SHLase activity over basal level was about 10-fold under low-nitrogen conditions but was only slightly higher than the levels found in the control strain under high-nitrogen conditions. H2S production under high-nitrogen conditions was much less than under low-nitrogen conditions in spite of the much higher levels of OAS/OAH SHLase activity in the latter case. At both nitrogen levels, H2S production in UCD713pMET17 was far below that of UCD522 and of UCD713 with or without the vector.
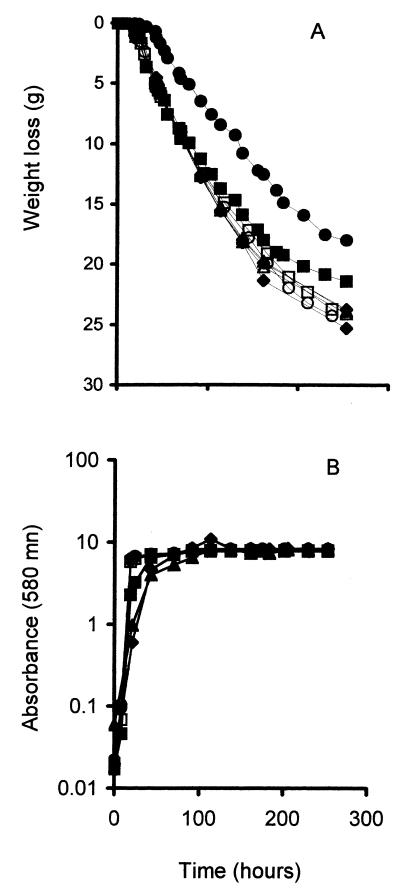
The fermentation and growth profiles for the transformed strains at both nitrogen concentrations also were evaluated (Fig. 4). Neither the control vector nor the MET17 plasmid had any dramatic impact on growth or fermentation in UCD522. The presence of the control vector did not impact UCD713. Overexpression of the MET17 gene decreased the fermentation rate and growth in UCD713 under high-nitrogen conditions. This observation suggests that high levels of expression of OAS/OAH SHLase may be deleterious in certain genetic backgrounds.
FIG. 4.
Fermentation (A) and growth (B) of pMET17-(solid symbols) and vector-transformed (open symbols) strains of UCD522 and UCD713. Testing was performed on UCD522 at high-nitrogen (○, ●) and low-nitrogen (◊, ⧫) concentrations and on UCD713 at high-nitrogen (□, ■) and low-nitrogen (▵, ▴) concentrations.
The differences in levels of enzymatic activity of the two strains and of UCD713 at different nitrogen concentrations might be explained by differences in the stability of the MET17 plasmid in these two genetic backgrounds. Plasmid loss was not detected in either transformant (data not shown). This stability suggests that differences in the level of overexpression between the two strains may reflect differences in plasmid copy number or in the transcriptional or posttranscriptional regulation of the protein.
Analysis of cysteine levels.
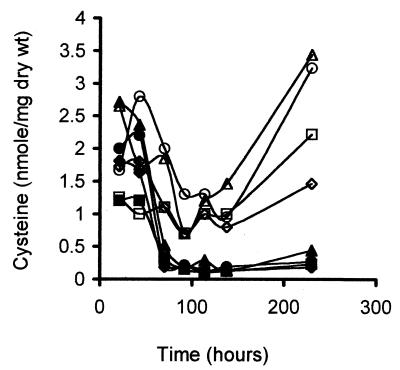
To determine if increased Met17p activity leads to increased incorporation of reduced sulfate, we measured the cellular pool levels of cysteine. Cytoplasmic cysteine levels were higher in strains grown under high-nitrogen conditions than in strains grown under low-nitrogen conditions (Fig. 5). All strains showed a similar pattern for internal cysteine levels. Levels were high early in the time course, peaking by 48 h. Pool levels then decreased but remained at a higher level under high-nitrogen conditions than under low-nitrogen conditions. Cysteine levels tended to increase late in fermentation in the case of high-nitrogen conditions but remained low in the nitrogen-limiting state. Overexpression of OAS/OAH SHLase activity in UCD522 had little or no effect on cysteine levels and, in some cases, led to an apparent decrease in the internal concentration of the amino acid. Overexpression of OAS/OAH SHLase resulted in slightly higher internal levels of cysteine in UCD713, especially under high-nitrogen conditions, where the decrease in H2S formation was greatest. Thus, there is a relationship between high pool levels of cysteine and low levels of H2S release. High cysteine levels may indicate more efficient incorporation of reduced sulfur. Alternately, high cysteine levels may alter the level of activity or expression of the enzymes involved in sulfate reduction, as has been proposed (22).
FIG. 5.
Cysteine levels in transformed strains of UCD522 and UCD713 under high-nitrogen (open symbols) or low-nitrogen (solid symbols) conditions. Testing was performed on vector-transformed UCD522 (○, ●) and pMET17-transformed UCD522 (◊, ⧫) and on vector-transformed UCD713 (□, ■) and pMET17-transformed UCD713 (▵, ▴).
DISCUSSION
The discovery of variation in the levels of OAS/OAH SHLase activity led us to the hypothesis that differences in the activity of this enzyme might explain the differences in hydrogen sulfide formation observed with different yeast genetic backgrounds. Low enzymatic activity was predicted to be correlated with reduced efficiency of incorporation of reduced sulfur and increased release of H2S. Our analysis of commercial strains known to produce high and low levels of H2S does not support this hypothesis. OAS/OAH SHLase activity was generally higher in UCD522, the strain that produced more H2S, than in UCD713 under all conditions. Overexpression of the MET17 gene did not inhibit H2S formation in UCD522 but did reduce volatile sulfur production in UCD713. Thus, enzyme activity appears to be limiting only in the strain that produced low levels of H2S and cannot explain the difference between the two strains.
The lack of an effect of manipulation of OAS/OAH SHLase activity on H2S production in UCD522 suggests that if decreased incorporation of reduced sulfur is responsible for H2S release, then it is due to other factors such as the limiting substrate, OAH. If OAH is limiting, then increasing enzymatic activity will have no impact on incorporation. The fact that H2S production in UCD522 is high at both nitrogen concentrations compared to that in UCD713 may reflect generally lower pool levels of the substrate rather than of enzymatic activity. This situation is also consistent with there being little to no change in the cytoplasmic levels of cysteine in this strain. Cysteine is synthesized via the transsulfuration pathway, involving conversion of homocysteine to cystathionine and then to cysteine (7). Low levels of OAH would prevent formation of cysteine. We predict that H2S formation would be greater under low-nitrogen conditions, when pool levels of OAH would be even further reduced.
The presence of the MET17 gene on a multicopy plasmid led to a slight increase in H2S formation in UCD522 that was reproduced in all experiments under both nitrogen conditions. This result suggests that increased activity of OAS/OAH SHLase decreases efficiency of incorporation. Increasing activity, while not affecting cysteine concentration, may lead to an increase in the level of methionine and S-adenosylmethionine, both of which are thought to repress the expression of genes encoding enzymes upstream in the sulfate reduction pathway (5, 6, 20, 33). However, if the pathway were repressed, we would expect a reduction in the production of H2S, not an increase. Methionine also represses expression of the MET2 gene, encoding homoserine-O-transacetylase, which would lead to reduced levels of OAH (5, 6, 20, 33). If the reduction in OAH levels were greater than the decrease in activity of the sulfate reduction pathway, greater H2S release would occur. This hypothesis also is consistent with reduced levels of OAH being primarily responsible for increased release of H2S in UCD522 in general. It is also possible that our knowledge of the regulation of sulfate reduction in Saccharomyces is incomplete and that other regulatory mechanisms also impact reduced sulfur incorporation. For example, if the sulfate reduction sequence existed as a multienzyme complex, then overexpression of MET17 could interfere with formation of the complex by titering other components, thus reducing the efficiency of incorporation. Other speculative models are equally plausible.
Overexpression of MET17 dramatically reduced H2S production in UCD713 under both low- and high-nitrogen conditions. This result suggests that the activity of this enzyme may indeed be limiting for sulfide incorporation in this strain. The increase in enzymatic activity was greater under low-nitrogen conditions, but the production of H2S was greater in these fermentations. It is likely that under nitrogen-limiting conditions, OAH levels also are affected, reducing the level of the substrate, which would negate the effect of elevated activity and account for an increase in H2S production. However, the increase in H2S production does not approach the values observed for the control strains not carrying multiple copies of the MET17 gene, suggesting that in this strain, enzymatic activity may be truly limiting for incorporation.
Our work suggests that the factors resulting in inefficient reduced sulfur incorporation in these two strains are fundamentally different. In the case of UCD522, OAS/OAH SHLase activity is not limiting and the data are consistent with pool levels of OAH influencing H2S production and release. In contrast, in UCD713, enzymatic activity does appear to be limiting for reduced sulfur incorporation. This study underscores the importance of a thorough understanding of the nuances of variation in pathway regulation in different strains, as a prelude to the development of strategies for the generation of commercial strains with improved physiological characteristics.
ACKNOWLEDGMENTS
This research was supported by grants from the American Vineyard Foundation and the California Competitive Grant Program for Research in Viticulture and Enology. A. Spiropoulos was supported by scholarships from the American Society of Enology and Viticulture, the Wine Spectator, and the Jastro Shields Graduate Research Awards.
We thank K. Spiropoulou (Department of Food Science and Technology, Agricultural University of Athens, Athens, Greece) for suggestions and technical assistance during this study and F. Omura for generously providing plasmid pEG25. We also thank the reviewers of the manuscript for numerous excellent suggestions for improving the clarity of the presentation.
REFERENCES
- 1.Acree T E, Sonoff E P, Splittstoesser D F. Effect of yeast strain and type of sulfur compound on hydrogen sulfide production. Am J Enol Vitic. 1972;23:6–9. [Google Scholar]
- 2.Balakinsky A T, Snow R. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast. 1990;6:367–382. doi: 10.1002/yea.320060503. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brzywczy J, Paszewski A. Role of O-acetylhomoserine sulfhydrylase in sulfur amino acid synthesis in various yeasts. Yeast. 1993;9:1335–1342. doi: 10.1002/yea.320091207. [DOI] [PubMed] [Google Scholar]
- 5.Cherest H, Surdin-Kerjan Y, Antoniewski J, de Robichon-Szulmajster H. S-Adenosyl methionine-mediated repression of methionine biosynthetic enzymes in Saccharomyces cerevisiae. J Bacteriol. 1973;114:928–933. doi: 10.1128/jb.114.3.928-933.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherest H, Surdin-Kerjan Y, de Robichon-Szulmajster H. Methionine-mediated repression in Saccharomyces cerevisiae: a pleiotropic regulatory system involving methionyl transfer ribonucleic acid and the product of gene eth2. J Bacteriol. 1971;106:758–772. doi: 10.1128/jb.106.3.758-772.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherest H, Surdin-Kerjan Y. Genetic analysis of a new mutation confering cysteine auxotrophy in Saccharomyces cerevisiae: updating the sulfur metabolism pathway. Genetics. 1992;130:51–58. doi: 10.1093/genetics/130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mora S J, Eschenburch R, Knouels S J, Spedding D J. The formation of dimethyl sulfide during fermentation using a wine yeast. Food Microbiol. 1986;3:27–32. [Google Scholar]
- 9.Eschenbruch R, Bonish P, Fisher B M. The production of H2S by pure culture wine yeasts. Vitis. 1978;17:67–74. [Google Scholar]
- 10.Gaitonde M K. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goniak O J, Noble A C. Sensory study of selected volatile sulfur compounds in white wine. Am J Enol Vitic. 1987;38:223–227. [Google Scholar]
- 12.Guidici P, Kunkee R E. The effect of nitrogen deficiency and sulfur-containing amino acids on the reduction of sulfate to hydrogen sulfide by wine yeasts. Am J Enol Vitic. 1994;45:107–112. [Google Scholar]
- 13.Henschke P, Jiravek V. Proceedings of the International Nitrogen Symposium, Seattle, Wash. Davis, Calif: American Society of Enology and Viticulture; 1991. Hydrogen sulfide formation during fermentation: effect of nitrogen composition in model grape musts; pp. 172–184. [Google Scholar]
- 14.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 15.Jiranek V, Langridge P, Henschke P A. Regulation of hydrogen sulfide liberation in wine-producing Saccharomyces cerevisiae strains by assimilable nitrogen. Appl Environ Microbiol. 1995;61:461–467. doi: 10.1128/aem.61.2.461-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiranek V, Langridge P, Henschke P A. Determination of sulphite reductase activity and its response to assimilable nitrogen status in a commercial Saccharomyces cerevisiae wine yeast. J Appl Bacteriol. 1996;81:329–336. doi: 10.1111/j.1365-2672.1996.tb04335.x. [DOI] [PubMed] [Google Scholar]
- 17.Jiravek V, Langridge P, Henschke P. Validation of bismuth-containing indicator media for predicting H2S-producing potential of Saccharomyces cerevisiae wine yeasts under enological conditions. Am J Enol Vitic. 1995;46:269–273. [Google Scholar]
- 18.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Ingraham J L, Magasanik B, Low K B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 419–428. [Google Scholar]
- 19.Lawrence W C, Cole E R. Yeast sulfur metabolism and the formation of hydrogen sulfide in brewery fermentations. Wallerstein Commun. 1968;21:95–113. [Google Scholar]
- 20.Mountain H A, Bystrom A S, Larsen J T, Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991;7:781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- 21.Omura F, Shibano Y. Reduction of hydrogen sulfide production in brewing yeast by constitutive expression of MET25 gene. J Am Soc Brew Chem. 1995;53:58–62. [Google Scholar]
- 22.Ono B-I, Hazu T, Yoshida S, Kawato T, Shinoda S, Brzywczy J, Paszewski A. Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeast. 1999;15:1365–1375. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1365::AID-YEA468>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Ono B-I, Kijima K, Ishii N, Kawato T, Matsuda A, Paszewski A, Shinoda S. Regulation of sulphate assimilation in Saccharomyces cerevisiae. Yeast. 1996;12:1153–1162. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1153::AID-YEA16%3E3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Paszewski A, Grabski J. Studies on 2-cystathionase and O-acetylhomoserine sulfhydrylase pathways in Aspergillus nidulans. Acta Biochim Pol. 1973;20:159–168. [PubMed] [Google Scholar]
- 25.Paszewski A, Ono B I. Biosynthesis of sulfur amino acids in Saccharomyces cerevisiae: regulatory roles of methionine and S-adenosylmethionine reassessed. Curr Genet. 1992;22:273–275. doi: 10.1007/BF00317920. [DOI] [PubMed] [Google Scholar]
- 26.Rankine B C. Nature, origin and prevention of hydrogen sulfide aroma in wines. J Sci Food Agric. 1963;14:79–91. [Google Scholar]
- 27.Rauhut D, Kurbel H. The production of H2S from elemental sulfur residues during fermentation and its influence on the formation of sulfur metabolites causing off-flavors in wines. Vitic Enol Sci. 1994;49:27–36. [Google Scholar]
- 28.Schiestl R H, Gietz R D. High efficiency transformation of intact cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink G R, Lawrence C W. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 30.Spiropoulos, A., J. Tanaka, I. Flerianos, and L. F. Bisson. Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am. J. Enol. Vitic., in press.
- 31.Statford M, Rose A H. Hydrogen sulfide production from sulfite by Saccharomyces cerevisiae. J Gen Microbiol. 1985;131:1417–1424. [Google Scholar]
- 32.Tezuka H, Mori T, Okumura Y, Kitabatake K, Tsumura Y. Cloning of a gene suppressing hydrogen sulfide production by Saccharomyces cerevisiae and its expression in a brewing yeast. J Am Soc Brew Chem. 1992;50:130–133. [Google Scholar]
- 33.Thomas D, Cherest H, Sudin-Kerjan Y. Elements involved in S-adenosylmethionine-mediated regulation of the Saccharomyces cerevisiae MET25 gene. Mol Cell Biol. 1989;9:3292–3298. doi: 10.1128/mcb.9.8.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas D, Barley R, Henry D, Surdin-Kerjan Y. Physiological analysis of mutants of Saccharomyces cerevisiae impaired in sulphate assimilation. J Gen Microbiol. 1992;138:2021–2028. doi: 10.1099/00221287-138-10-2021. [DOI] [PubMed] [Google Scholar]
- 35.Tokuyama T, Kuraishi H, Aida K, Uemura T. Hydrogen sulfide evolution due to pantothenic acid deficiency in the yeast requiring this vitamin, with special reference to the effect of adenosine triphosphate on yeast cysteine desulfhydrase. J Gen Appl Microbiol. 1973;19:439–466. [Google Scholar]
- 36.Wainwright T. Hydrogen sulfide production under conditions of methionine, pantothenate or vitamin B6 deficiency. J Gen Microbiol. 1970;61:107–119. doi: 10.1099/00221287-61-1-107. [DOI] [PubMed] [Google Scholar]
- 37.Wainwright T. Production of H2S by yeasts: role of nutrients. J Appl Bacteriol. 1971;34:161–171. doi: 10.1111/j.1365-2672.1971.tb02275.x. [DOI] [PubMed] [Google Scholar]