Abstract
The potential of lactic acid bacteria as live vehicles for the production and delivery of therapeutic molecules is being actively investigated today. For future applications it is essential to be able to establish dose-response curves for the targeted biological effect and thus to control the production of a heterologous biopeptide by a live lactobacillus. We therefore implemented in Lactobacillus plantarum NCIMB8826 the powerful nisin-controlled expression (NICE) system based on the autoregulatory properties of the bacteriocin nisin, which is produced by Lactococcus lactis. The original two-plasmid NICE system turned out to be poorly suited to L. plantarum. In order to obtain a stable and reproducible nisin dose-dependent synthesis of a reporter protein (β-glucuronidase) or a model antigen (the C subunit of the tetanus toxin, TTFC), the lactococcal nisRK regulatory genes were integrated into the chromosome of L. plantarum NCIMB8826. Moreover, recombinant L. plantarum producing increasing amounts of TTFC was used to establish a dose-response curve after subcutaneous administration to mice. The induced serum immunoglobulin G response was correlated with the dose of antigen delivered by the live lactobacilli.
Lactic acid bacteria (LAB) are used worldwide in the preparation of fermented foods, including dairy products. They are also known for the potentially beneficial effects they may exert on the health of humans and animals (see, for example, reference 25). Their “generally recognized as safe” status (1), linked to their metabolic and technological properties, has recently led to their development as potential live-vaccine vehicles. Lactobacillus plantarum NCIMB8826 (17, 33) has been chosen for this purpose in our laboratory on the basis of its capability to persist in the mouse gastrointestinal and urogenital tracts (38). The ability to control the expression level of foreign proteins in LAB may offer certain advantages. However, while several controlled expression systems have been developed for Lactococcus lactis (9, 23), very few inducible promoters are available for lactobacilli: the xylR promoter from Lactobacillus pentosus (29), the α-amylase promoter from L. amylovorus (31), and the p-coumarate decarboxylase promoter from L. plantarum (4). One of the most promising lactococcal controlled expression systems is based on the autoregulatory properties of the L. lactis nisin gene cluster (7, 23). Nisin is an antimicrobial peptide belonging to the family of lantibiotics (19) and is used as a natural preservative in the food industry (5). Nisin induces the transcription of the genes under control of the nisA and nisF promoters, via a two-component regulatory system (34, 37) consisting of the histidine protein kinase NisK and the response regulator NisR (14, 21, 22). A transferable nisin-controlled expression (NICE) system (24) based on the combination of the nisA promoter and the nisRK regulatory genes has recently been developed (7, 20). It consists of two compatible replicons, a plasmid carrying the nisRK regulatory genes (regulatory plasmid) and an expression vector carrying the gene of interest under control of the nisA promoter. This system offers several advantages. (i) The level of expression can be controlled by the amount of nisin used for induction (6, 20, 21). (ii) Recombinant protein synthesis can reach very high levels (up to 60% of the total intracellular proteins [7]). (iii) The expression of the gene of interest is reported to be undetectable at the uninduced state, which allows the production of lethal proteins (8). (iv) This expression system has already been successfully transferred to other gram-positive bacteria (13, 20).
In this report, we describe the implementation of the NICE system in L. plantarum NCIMB8826, using the β-glucuronidase (gusA) reporter gene. This system was then used to design L. plantarum strains producing increasing amounts of TTFC (C subunit of tetanus toxin) at regulated levels, which allowed us to establish a dose effect of TTFC after subcutaneous administration to mice of recombinant lactobacilli bearing different loads of the antigen.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli MC1061 was cultured as described by Sambrook et al. (32). L. plantarum was grown at 30 or 37°C in MRS medium (Difco) without shaking. Antibiotics (Sigma) were used at the following final concentrations: for Escherichia coli, chloramphenicol (20 μg/ml), erythromycin (250 μg/ml), and ampicillin (100 μg/ml); and for L. plantarum, chloramphenicol (10 μg/ml) and erythromycin (5 μg/ml) in association with lincomycin (10 μg/ml). Double transformants of L. plantarum were grown in the presence of erythromycin (5 μg/ml) and chloramphenicol (5 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli MC1061 | araD139 Δ(ara-leu)7696 lacX74 galV galK hsr-hsm rpsL | 3 |
| L. plantarum NCIMB8826 | Isolated from human saliva | NCIMBa |
| L. plantarum NCIMB8826 Int-1 | NCIMB8826 containing the nisRK genes stably integrated to the tRNASer locus; obtained by transformation with pMEC10 | This work |
| Plasmids | ||
| pNZ9520 | Eryr, nisRK cloned in pIL253, expression of nisRK driven by rep read-through, high copy number | 20 |
| pNZ9521 | Eryr, nisRK cloned in pIL253, expression of nisRK driven by nisR promoter and rep read-through, high copy number | 20 |
| pNZ9530 | Eryr, nisRK cloned in pIL252, expression of nisRK driven by rep read-through, low copy number | 20 |
| pNZ9531 | Eryr, nisRK cloned in pIL252, expression of nisRK driven by nisR promoter and rep read-through, low copy number | 20 |
| pNZ9500 | Ampr, Eryr, pUC19 derivative containing a 2.7-kb chromosomal DNA fragment from L. lactis NZ9700, which contains the 3′ end of nisP and nisRK | 20 |
| pNZ8008 | Cmr, pNZ273 carrying the gusA reporter gene transcriptionally fused to the nisA promoter | 21 |
| pNZ8032 | pNZ8008 derivative, carrying the gusA gene translationally fused to the nisA promoter | 7 |
| pNZ8037 | pNZ8008 derivative carrying a multiple-cloning site; allows translational fusion to the nisA promoter | 7 |
| pMC1 | pRC1 derivative, containing the φmv4 int-attP sequence | 12 |
| pMEC3 | pZErO -2 derivative, containing the TTFC gene cloned into the multiple cloning site | Chagnaud et al., unpublished results |
| pMEC10 | Integration plasmid, int-attP cassette of pMC1 cloned into pNZ9500, expression driven by ery read-through | This work |
| pMEC46 | Replicative plasmid derived from pNZ8037, which contains the TTFC gene from pMEC3 translationally fused to the nisA promoter | This work |
NCIMB, National Collection of Industrial and Marine Bacteria, Terry Research Station, Aberdeen, Scotland.
Transformation and DNA manipulations.
E. coli and L. plantarum were electrotransformed as described by Dower et al. (10) and Josson et al. (18), respectively. E. coli MC1061 was used as an intermediate host for cloning. Molecular biology techniques were performed essentially as described by Sambrook et al. (32). Restriction endonucleases, T4 DNA ligase, and Taq polymerase were purchased from Boehringer Mannheim and were used following the recommendations of the manufacturer. L. plantarum plasmid DNA was extracted according to the method of Posno et al. (30). Chromosomal DNA of L. plantarum was isolated according to the method of Ferain et al. (15). For Southern blotting, chromosomal DNA was extracted, digested, and transferred onto a nitrocellulose membrane after agarose gel electrophoresis. DNA was hybridized with digoxigenin-labeled pMEC10 (see below) using the DIG DNA labeling kit supplied by Boehringer Mannheim.
Plasmid constructions.
The plasmid allowing integration of nisRK in the tRNASer chromosomal locus of L. plantarum was constructed as follows. An EcoRV-ScaI fragment containing the mv4 bacteriophage int-AttP cassette from pMC1 (Table 1) was inserted into the SmaI site (130 bp downstream of the Emr marker) of pNZ9500, which carries nisRK under control of its own promoter, giving rise to pMEC10 (Table 1). The replicative plasmid carrying the TTFC coding sequence under control of the nisA promoter was obtained by cloning the NcoI-BamHI fragment from pMEC3 (P. Chagnaud et al., unpublished results) into pNZ8037 (Table 1) digested with the same enzymes. The resulting plasmid, pMEC46, harbors the TTFC gene fused to the ATG of the nisA gene.
Plasmid stability.
The integrant L. plantarum containing pNZ8008 or pNZ8032 was grown at 37°C in MRS medium without antibiotic. Serial subcultures were performed by inoculating 10 μl of the previous culture in 10 ml of fresh medium. After 10 subcultures, the number of chloramphenicol-resistant CFU was determined from the total number of CFU by plating equal numbers of bacterial dilutions on control MRS agar plates and MRS agar plates supplemented with 10 μg of chloramphenicol/ml. To determine the structural plasmid stability, six randomly chosen colonies plated from the 10th subculture were examined by plasmid DNA extraction and restriction analysis.
Nisin induction and β-glucuronidase activity.
Nisin induction of recombinant L. plantarum NCIMB8826 strains was first performed as described for L. lactis (6), except for the induction time. Briefly, bacteria were grown at 30°C to an A600 of 0.5, after a 1/20 inoculation (vol/vol) of fresh medium with an overnight culture. Nisin was added at different concentrations (0 to 100 ng/ml), and cultures were further grown for 3 h at 30°C. β-Glucuronidase assays were performed as described previously (6).
Optimization of the protocol for L. plantarum NCIMB8826 (see Results) led to the following modifications. Bacteria were grown at 37°C by inoculating fresh medium 1/50 (vol/vol) with an overnight culture. After 1 h of growth (beginning of the exponential growth phase), nisin was added at a concentration of 25 ng/ml and cells were propagated for 5 h at 37°C before harvest and the β-glucuronidase assay.
Protein analysis.
TTFC production was examined by Western blotting (32) after induction of 10-ml cultures by nisin. Cells were harvested by 10 min of centrifugation at 3,000 × g at 4°C and were washed with 10 mM Tris-HCl (pH 7.5) and resuspended in 2 ml of 10 mM Tris-HCl (pH 7.5). Cell extracts were prepared with a French-Press (Bioritech) and immediately mixed with an antiprotease cocktail (Complete; Boehringer Mannheim). Cell debris was removed by centrifugation for 5 min at 1,300 × g at 4°C in a microcentrifuge. When relevant, the protein concentration was determined by the Bradford method (2) using the Bio-Rad protein assay kit. The soluble proteins were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (Mini Protean II; Bio-Rad) and blotted (semidried system; OWL) onto nitrocellulose membranes (Optitran BA-S85; Schleicher & Schuell). Polyclonal rabbit anti-TTFC antibodies (provided by E. Sablon, Innogenetics N.V., Ghent, Belgium) and goat anti-rabbit–alkaline phosphatase antibodies (Promega) were used as primary and secondary antibodies, at dilutions of 1/1,000 and 1/7,000, respectively. Detection was performed by addition of 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Boehringer Manneim). Semiquantification of the TTFC bands after scanning was performed by using GelCompar (Applied Maths, Kortrijk, Belgium). Intensity (pixels per band area) of the strongest band was considered 100% and used as a reference to estimate percentages of intensity in the other lanes.
Mouse immunization experiments.
Bacterial inocula were prepared as follows. Fresh medium was inoculated 1/50 (vol/vol) with an overnight culture and incubated at 37°C. Induction was achieved by adding nisin at a concentration of 0, 0.5, 2, or 25 ng/ml after 1 h of growth. Cultures were harvested by centrifugation (3,000 × g, 4°C) after 5 h of incubation at 37°C. The collected cells were washed twice with sterile phosphate-buffered saline (pH 7.2) (GibcoBRL) and resuspended in phosphate-buffered saline to a final concentration of 1010 CFU/ml. Groups (four female mice/group) of 8-week-old C57BL6 mice were immunized subcutaneously on days 0, 15, and 37 with 109 CFU (100 μl) of L. plantarum NCIMB8826 Int-1(pMEC46) that produced increasing amounts of TTFC after nisin induction. Control mice were inoculated with L. plantarum NCIMB8826 Int-1 (nonexpressor strain). Serum samples were taken before immunization (preimmune serum) and 7 days after primer and booster injections. A last sample was collected 70 days after the last boost, to evaluate the persistence of the immune response.
ELISA.
The anti-TTFC immunoglobulin G (IgG) response was analyzed by enzyme-linked immunosorbent assay (ELISA) with the end point titer method (KC4 software; Bio-Tek Instruments). The microtiter plates (Immulon 3; Dynatech) were coated with 200 ng of TTFC (Boehringer Manneim). Sequential dilutions of serum samples were distributed in the wells, and IgG detection was performed with biotinylated goat anti-mouse IgG (Southern Biotechnology Associates). Streptavidin-horseradish peroxidase complex (Amersham) was added, and plates were read with an ELX800GUV machine (Bio-Tek Instruments) at 490 nm after developing with o-phenylenediamine (Sigma) and hydrogen peroxide. The end point titer was calculated as the dilution of serum producing an optical density of three times the background level. The results were evaluated by the Mann-Whitney U test, and differences were considered significant when P values were <0.05.
RESULTS
Implementation of the NICE system in L. plantarum NCIMB8826.
The transferable NICE system consists of an expression vector and a regulatory plasmid (20) which are derived from two compatible broad-host-range replicons (36). The expression plasmid (pSH71 derivative) carries the E. coli gusA reporter gene transcriptionally or translationally fused (ATG fusion) to the nisA promoter (pNZ8008 or pNZ8032, respectively) (Table 1). The regulatory nisRK genes are carried on different pAMβ1-derived replicons, leading to increasing NisRK dosage in the following order: pNZ9530, pNZ9531, pNZ9520, and pNZ9521 (Table 1). To assess the NICE system in L. plantarum NCIMB8826, the following combinations of the two plasmids were introduced into this host: pNZ8008 associated with pNZ9530, pNZ9531, pNZ9520 or pNZ9521 and pNZ8032 associated with pNZ9530. The resulting double transformants were first cultured at 37°C in the presence of erythromycin (7.5 μg/ml) and (chloramphenicol) (10 μg/ml). Under these conditions, the strains containing pNZ9520 or pNZ9521 were severely disabled, while those carrying lower nisRK dosage (pNZ9530 and pNZ9531) appeared to be viable but unstable upon propagation. However, their stability could be improved by growth at 30°C in the presence of erythromycin (5 μg/ml) and chloramphenicol (5 μg/ml). Viable double transformants (pNZ8008 or pNZ8032 combined with pNZ9530 or pNZ9531) grew approximately half as fast as the plasmid-free strain, while recombinant L. plantarum containing a single plasmid behaved like the parent strain.
The progressive induction of the nisA promoter by increasing doses of nisin was followed by measuring the β-glucuronidase activity in the different double-transformant cultures. At the maximal dose of nisin (100 ng/ml), the most stable plasmid combination was pNZ9530 associated with pNZ8008. The corresponding recombinant strain showed no basal activity in the absence of the inducer (0.002± 0.002 activity units [AU]). As in the case of the pNZ9530- and pNZ8032-containing strain, β-glucuronidase activity was found to be induced by nisin in a dose-dependent manner (data not shown). However, the maximal β-glucuronidase activity remained low and variable from one culture to another in NCIMB8826(pNZ9530) and NCIMB8826(pNZ8008) (0.028±0.018 AU). When nisRK genes were transcribed at a slightly higher level (NCIMB8826-pNZ9531 and -pNZ8008), the strains exhibited a higher β-glucuronidase activity after nisin induction (0.073 AU) but also a basal activity (0.025±0.009 AU). In view of these results, the dual-plasmid NICE system seemed poorly suited to L. plantarum NCIMB8826.
Since high expression levels of the nisin regulatory genes might be detrimental to L. plantarum, the nisRK genes were integrated with their own promoter into the genome of this host. Site-specific integration of nisRK in L. plantarum NCIMB8826 was achieved by applying the system that was based on the integration property of the mv4 temperate bacteriophage and originally developed for L. plantarum LP80 (12). Chromosomal integration of mv4 is mediated by site-specific recombination between the homologous attachment sites attP and attB, carried by the phage and the bacterial genome, respectively. attB is located at the 3′ end of the tRNASer locus, and integration into this site, catalyzed by a phage-encoded integrase (int), is nondisruptive. The promoterless mv4 int-attP cassette carried by pMC1 (12) was introduced into pNZ9500 (Table 1), which contains nisRK under the control of the nisR promoter. The int-attP sequence was inserted into pNZ9500 downstream of the erythromycin resistance marker, in order to allow read-through transcription from this gene, which lacks a functional transcription terminator. The resulting integrative plasmid, pMEC10 (Table 1), is derived from pUC19 and is thus unable to replicate in L. plantarum. Transformation of L. plantarum NCIMB8826 with pMEC10 and selection for erythromycin- and lincomycin-resistant colonies yielded putative integrants that appeared after 3 days of incubation at 37°C. Analysis of these clones by PCR and Southern blotting confirmed integration of pMEC10 at the tRNASer locus (data not shown). One integrant presenting the correct structure, L. plantarum NCIMB8826 Int-1, was kept for further studies. The plasmid-containing integrant grew on 10 μg of chloramphenicol/ml as fast as the corresponding plasmid-free strain or the wild-type NCIMB8826 strain on nonselective medium. The segregational and structural stability of pNZ8008 and pNZ8032 were then evaluated in the NCIMB8826 Int-1 background. After 100 generations in nonselective medium, 70 colonies out of 70 retained the Cmr phenotype. Analysis of the plasmid content of 6 randomly picked colonies showed no structural rearrangements.
Optimization of the nisin induction conditions in L. plantarum NCIMB8826 Int-1.
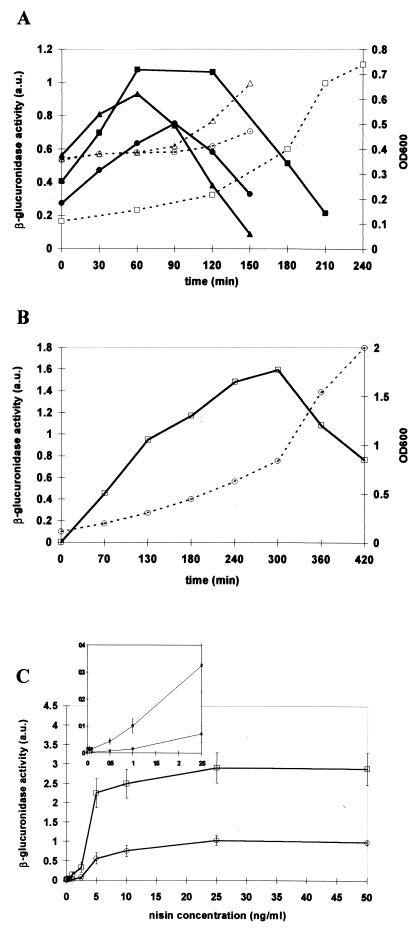
The optimal growth phase for nisin induction was the first parameter studied. β-Glucuronidase activity was determined in NCIMB8826 Int-1(pNZ8008) cultures incubated at 30 or 37°C after inoculation at 1/20 (vol/vol) or 1/50 (vol/vol) with an overnight culture. Nisin was added at 50 ng/ml every 30 min during 3 h after the start of the culture, and β-glucuronidase activity was measured 2 h 30 min after induction. As shown in Fig. 1A, maximal levels were reached when the inducer was added early in the exponential phase, i.e., 1 h or 1 h 30 min after bacterial inoculum, depending on the inoculation conditions (1/50 inoculum, 37°C; 1/20 inoculum, 37°C; or 1/20 inoculum, 30°C). The higher β-glucuronidase activity was obtained with cultures grown at 37°C with 1/50 inoculum. These conditions were kept for further experiments, and nisin was added after 1 h of growth.
FIG. 1.
β-Glucuronidase activity (AU and optical density at 600 nm [OD600]) of L. plantarum NCIMB8826 Int-1(pNZ8008) cell extracts. (A) β-Glucuronidase activity (solid lines) determined during growth (dotted lines) in different conditions (squares, 1/50 inoculum and incubation at 37°C; triangles, 1/20 inoculum and incubation at 30°C; and circles, 1/20 inoculum and incubation at 37°C) after induction with 50 ng of nisin/ml at different times after inoculation. β-Glucuronidase activity was measured after 2 h 30 min of contact with nisin. (B) β-Glucuronidase activity (solid line) after different periods of contact with 50 ng of nisin/ml. Optical density (dotted line) was measured at 600 nm (OD600). (C) Dose-response curves of gusA expression in L. plantarum NCIMB8826 Int-1 harboring pNZ8008 (circles) or pNZ8032 (squares) induced with increasing concentrations of nisin (0 to 50 ng/ml). Error bars represent standard deviations. The inset shows β-glucuronidase activity for nisin concentrations ranging from 0 to 2.5 ng/ml.
Next, we determined the optimal production levels as a function of induction time. β-Glucuronidase activity was measured at several time points after the addition of nisin and was found to quickly rise after induction, the maximum level being reached 4 h 30 min to 5 h after induction (Fig. 1B).
These conditions (addition of nisin 1 h after inoculation at 1/50 [vol/vol] with an overnight culture and incubation at 37°C for 5 h) were finally applied to characterize the induction of the nisA promoter by different nisin concentrations. Both transcriptional (pNZ8008) and translational (pNZ8032) fusions of PnisA-gusA were analyzed. Repeated β-glucuronidase assays were performed with six individual clones of each recombinant strain to evaluate the reproducibility of induction and stability of the expression system. Cultures were induced with nisin concentrations ranging from 0 to 50 ng/ml. With both plasmids, β-glucuronidase activity was found to be nisin dose dependent, essentially in the range of 0 to 5 ng of nisin/ml, and remained very low in uninduced cultures (Fig. 1C). The maximum level was reached with 25 ng of nisin/ml and was three times higher for strains containing pNZ8032 (2.9±0.4 AU) than for those containing pNZ8008 (1.0±0.1 AU), which was comparable to the results obtained in L. lactis (6). All clones exhibited reproducible enzymatic activities (Fig. 1C).
Controlled production of TTFC by the integrant NCIMB8826 Int-1.
We next investigated whether the nisin-inducible system would allow us to produce increasing amounts of a model antigen (TTFC). A plasmid carrying the TTFC gene translationally fused to the nisA promoter (pMEC46) (Table 1) was constructed and introduced into NCIMB8826 Int-1. Cultures were induced with increasing concentrations of nisin in the conditions defined for β-glucuronidase production, and the corresponding cell extracts (soluble proteins) were analyzed by Western blotting. As shown in Fig. 2, TTFC was produced in a nisin dose-dependent manner. However, a low production level was detected in uninduced cultures (lane 3), probably due to the high sensitivity of the anti-TTFC antibodies used, which allowed detection even of a few nanograms of the antigen. By scanning, it was estimated that TTFC constituted about 10% of the total soluble proteins in NCIMB8826 Int-1(pMEC46), which is notably higher than the results obtained with the strong constitutive promoters that we have studied so far (Chagnaud et al., unpublished results).
FIG. 2.
Dose-dependent nisin induction of TTFC expression in L. plantarum NCIMB8826 Int-1(pMEC46). Western blotting was performed on 2 μg of soluble proteins (lanes 2 to 9) from total cell extracts. Lane 1, purified recombinant TTFC; lane 2, L. plantarum NCIMB8826 Int-1 (negative control); lanes 3 to 9, NCIMB8826 Int-1(pMEC46) uninduced and induced with 0.5, 1, 1.5, 2, 5, or 20 ng of nisin/ml, respectively. Sizes as deduced from molecular mass markers are shown at the left. Relative band intensities in lanes 3 to 9 are shown at the top.
Immunization of mice by the subcutaneous route.
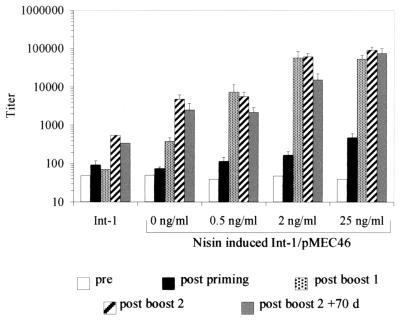
NCIMB8826 Int-1(pMEC46) was grown in the presence of 0, 0.5, 2, or 25 ng of nisin/ml as described above, and 109 CFU of each culture was administered by the subcutaneous route to C57BL6 mice. The serum anti-TTFC IgG response of the mice was analyzed by ELISA at different time points. NCIMB8826 Int-1 containing no plasmid was used as the negative control. As shown in Fig. 3, only the mice that received recombinant lactobacilli induced by 25 ng of nisin/ml showed a statistically significant (P < 0.05) immune response after priming. In contrast, all mice, including those immunized with the noninduced culture of NCIMB8826 Int-1(pMEC46), produced anti-TTFC serum IgG (P < 0.05) after the first and second boosts. Notably, the titers reached after the first boost reflected the amount of TTFC produced by the bacterial vector (Fig. 2 and 3). A 1- to 2-log increase in the anti-TTFC IgG titers was observed for mice immunized with recombinant L. plantarum strains grown in the presence of high doses of the inducer (2 and 25 ng/ml; mean titer, 55,100) compared to those which were given strains induced with low amounts of nisin (0.5 ng/ml; mean titer, 7,278). Uninduced cultures led to a mean antibody titer of 378. No difference was seen after the first boost between the two groups of mice immunized with the two highest doses of antigen. The induced immune response persisted for up to 70 days after the last booster injection, regardless of the dose of antigen delivered at priming.
FIG. 3.
Anti-TTFC IgG titers of individual sera of mice inoculated subcutaneously with L. plantarum NCIMB8826 Int-1 (negative control) (Int-1) or NCIMB8826 Int-1(pMEC46) induced by increasing nisin concentrations. Data represent the ELISA mean end-point titers for each group in the preimmune sera (pre), the sera obtained 7 days after each inoculation (postpriming, post-boost 1, and post-boost 2), and the sera obtained 70 days after the second boost (post boost 2 + 70 d). Error bars represent standard errors of the means.
DISCUSSION
In this paper, we describe the implementation of the NICE system in L. plantarum NCIMB8826, which necessitated integrating the nisRK regulatory genes into the chromosome of this strain. In contrast with the use of the dual-plasmid NICE system, the nisRK integration has led to genetic stabilization of the recombinant strain and to reproducible and dose-dependent nisin induction of the nisA promoter.
Lactobacilli constitute a very broad genus including over 60 species that may be quite distant from each other from a phylogenetic point of view (35). As a consequence of this diversity, the efficacy of transcription, translation, or secretion signals may vary greatly from one Lactobacillus species to another (17, 27, 30). This is well illustrated with L. plantarum, in which the dual-plasmid NICE system appeared unstable and poorly regulated, even when it could be successfully transferred to other lactobacilli (20). However, some common features could be observed between L. plantarum and other LAB. Firstly, the expression levels of the nisRK regulatory elements in the final expression host are very important (this work and references 13 and 20). In NCIMB8826, these expression levels have to remain very low in order to retain both the inducibility to nisin and no basal activity. This is the case with the double transformant carrying pNZ9530 and with NCIMB8826 Int-1, which contains a single chromosomal copy of nisRK, thus mimicking the optimal L. lactis system (pepN::nisRK integrants [6]). Secondly, cultures and induction conditions have to be optimized for each host. For NCIMB8826 Int-1, it is necessary to add higher doses of the inducer (20 to 25 ng/ml) (this work) than those used for L. lactis (1 to 5 ng/ml) (6). However, these concentrations remain notably lower than those described for other gram-positive bacteria (13). Also, L. plantarum is more receptive to nisin when its addition occurs very early in the exponential growth phase (this work), while the best induction time for L. lactis is located in the middle of the exponential growth (7). Moreover, the optimal contact time with the inducer, leading to maximal protein production, can vary greatly depending on the host (this work and reference 20).
Adaptation of the NICE system in L. plantarum also allowed it to reach very high β-glucuronidase activity levels, especially in strains harboring pNZ8032 (2.9 AU versus 0.02 AU with the dual-plasmid system), in which initiation of translation is optimized. Induction factors, as defined by the ratio of the β-glucuronidase activity (observed after maximal induction) to the basal activity (no inducer), are equal to 500 or 200 with NCIMB8826 Int-1 containing pNZ8008 or pNZ8032, respectively. This is only two- to threefold lower than in L. lactis (7). When fully induced, the nisA promoter appears to be more efficient than strong endogenous L. plantarum promoters (unpublished observations), leading to an estimated intracellular production of about 10% in the case of TTFC. These levels are close to those obtained with the best recombinant L. lactis strains producing TTFC (39). Contrary to L. lactis, very few inducible expression systems are available for lactobacilli. Thus, the adaptation of the NICE system in L. plantarum provides a powerful, well regulatable, and stable expression tool for this bacterium.
Because of the potential health applications of certain Lactobacillus strains, it is of utmost importance to develop controlled gene expression systems which would allow dose-response curves to be established for the biological effect under study. In the second part of this work, we used live recombinant L. plantarum to deliver increasing levels of the model antigen TTFC by subcutaneous administration to mice and we showed a correlation between the amount of antigen delivered and the specific serum IgG response induced after priming and the first boost. This is one of the first successful attempts to establish dose-response curves with bacterial live vectors, based on the delivery of incremental amounts of protein by equivalent numbers of live cells. It will allow us to evaluate the contribution of protein and bacterial doses in the induction of a biological effect. This point may be particularly relevant in the case of LAB, as some Lactobacillus strains are reported to exhibit adjuvant properties (26, 31). Thus, we have taken advantage of the main property of the NICE system, i.e., controlled expression level by nisin concentration, while it is usually used at the fully induced stage for overproduction of heterologous proteins. The optimized NICE system was successfully applied to tag L. plantarum with green fluorescent protein and to follow the fate of these fluorescent lactobacilli in vivo or in cell cultures (16), providing a powerful tool to study the health-promoting effects of lactobacilli in humans or animals. In vivo induction of the NICE system by adding nisin in food, as shown by Drouault et al. (11), could be considered.
ACKNOWLEDGMENTS
This work was supported by the EU BIO4-CT96-0542 grant, FEDER funds, and Institut Pasteur de Lille funding.
We are grateful to H. Müller-Alouf and D. Goudercourt for their skillful help with animal experiments and helpful discussions. We thank C. Locht for critical reading of the manuscript. Rabbit anti-TTFC antibodies were kindly supplied by E. Sablon, Innogenetics N.V., Ghent, Belgium.
REFERENCES
- 1.Adams M R, Marteau P. On the safety of lactic acid bacteria. Int J Food Microbiol. 1995;27:263–264. doi: 10.1016/0168-1605(95)00067-t. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen J. Analysis of gene control signals by DNA fusion cloning in E. coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Cavin J-F, Barthelmebs L, Diviès C. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification, and characterization. Appl Environ Microbiol. 1997;63:1939–1944. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delves-Broughton J, Blackburn P, Evans R J, Hugenholtz J. Applications of the bacteriocin, nisin. Antonie Leeuwenhoek. 1996;69:193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 6.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ruyter P G G A, Kuipers O P, Meijer W C, de Vos W M. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat Biotechnol. 1997;15:976–979. doi: 10.1038/nbt1097-976. [DOI] [PubMed] [Google Scholar]
- 9.de Vos W M. Gene expression systems for lactic acid bacteria. Curr Opin Microbiol. 1999;2:289–295. doi: 10.1016/S1369-5274(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 10.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouault S. Lactococcus lactis, vecteur de lipase dans le tractus digestif: application au traitement de la stéatorrhée. Ph.D. thesis. Orsay, France: University of Paris XI; 1999. [Google Scholar]
- 12.Dupont L, Boizet-Bonhoure B, Coddeville M, Auvray F, Ritzenthaler P. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J Bacteriol. 1995;177:586–595. doi: 10.1128/jb.177.3.586-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichenbaum Z, Federle M J, Marra D, de Vos W M, Kuipers O, Kleerebezem M, Scott J R. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998;64:2763–2769. doi: 10.1128/aem.64.8.2763-2769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian K D. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol. 1994;60:814–825. doi: 10.1128/aem.60.3.814-825.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferain T, Hobbs J N, Jr, Richardson J, Bernard N, Garmyn D, Hols P, Allen N E, Delcour J. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol. 1996;178:5431–5437. doi: 10.1128/jb.178.18.5431-5437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geoffroy M-C, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol. 2000;66:383–391. doi: 10.1128/aem.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hols P, Slos P, Dutot P, Reymund J, Chabot P, Delplace B, Delcour J, Mercenier A. Efficient secretion of the model antigen M6-gp41E in Lactobacillus plantarum NCIMB8826. Microbiology. 1997;143:2733–2741. doi: 10.1099/00221287-143-8-2733. [DOI] [PubMed] [Google Scholar]
- 18.Josson K, Scheirlink T, Michiels F, Platteeuw C, Stanssens P, Joos H, Dhaese P, Zabeau M, Mahillon J. Characterization of a gram-positive broad-host range plasmid isolated from Lactobacillus hilgardii. Plasmid. 1989;21:9–20. doi: 10.1016/0147-619x(89)90082-6. [DOI] [PubMed] [Google Scholar]
- 19.Klaenhammer T D. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 20.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 22.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis, requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers O P, de Ruyter P G G A, Kleerbezem M, de Vos W M. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 1997;15:135–140. doi: 10.1016/S0167-7799(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 25.Marteau P, Rambaud J C. Potential of using lactic acid bacteria for therapy and immunomodulation in man. FEMS Microbiol Rev. 1993;12:207–222. doi: 10.1111/j.1574-6976.1993.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 26.Miettinen M, Vuopio-Varkila J, Varkila K. Production of human tumor necrosis factor Alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun. 1996;64:5403–5405. doi: 10.1128/iai.64.12.5403-5405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platteeuw C, Simons G, de Vos W M. Use of Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posno M, Leer R J, van Luijk N, van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pouwels P, Leer R, Posno M. Genetic modification of Lactobacillus. In: Novel G, Le Querier J-F, editors. Les bactéries lactiques—lactic acid bacteria, Caen. Caen, France: Centre de Publications de l'Université de Caen; 1991. pp. 134–148. [Google Scholar]
- 30.Pouwels P H, Leer R J. Genetics of lactobacilli. Plasmids and gene expression. Antonie Leeuwenhoek. 1993;64:85–107. doi: 10.1007/BF00873020. [DOI] [PubMed] [Google Scholar]
- 31.Pouwels P H, Leer R J, Boersma W J A. The potential of Lactobacillus as a carrier for oral immunization: development and preliminary characteristics of vector systems for targeted delivery of antigens. J Biotechnol. 1996;44:183–192. doi: 10.1016/0168-1656(95)00140-9. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J E, Fritsch F, Maniatis J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Slos P, Dutot P, Reymund J, Kleinpeter P, Prozzi D, Kieni M P, Delcour J, Mercenier A, Hols P. Production of cholera toxin B subunit in Lactobacillus. FEMS Microbiol Lett. 1998;169:29–36. doi: 10.1111/j.1574-6968.1998.tb13295.x. [DOI] [PubMed] [Google Scholar]
- 34.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 35.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanner B L. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J Bacteriol. 1992;174:2053–2058. doi: 10.1128/jb.174.7.2053-2058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells J M, Robinson K, Chamberlain L M, Schofield K M, Le Page R W F. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]
- 39.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: a high level of expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]