Abstract
The chemistry of indoor surfaces and the role of microbes in shaping and responding to that chemistry are largely unexplored. We found that, over 1 month, people’s presence and activities profoundly reshaped the chemistry of a house. Molecules associated with eating/cooking, bathroom use, and personal care were found throughout the entire house, while molecules associated with medications, outdoor biocides, and microbially derived compounds were distributed in a location-dependent manner. The house and its microbial occupants, in turn, also introduced chemical transformations such as oxidation and transformations of foodborne molecules. The awareness of and the ability to observe the molecular changes introduced by people should influence future building designs.
Human presence and everyday activities such as cooking and cleaning shape the molecular profile of surfaces throughout a house.
INTRODUCTION
Modern humans spend ~70% of their time in their home environment (1) and reshape the indoor microbiome with inputs from their bodies (2, 3). To date, studies of the indoor environment have revealed that human activity inside buildings leads to potentially higher particle, pollutant, and toxin exposures than typically observed in the outdoor environment (4), but such studies often limit their measurements to one or a few molecular species. In this study, we set out to determine how humans influence the entire molecular composition throughout the home due to routine activities. This was accomplished by using an experimental test home in Austin, Texas, during summer 2018 that was sampled at two time points with 28 days apart, time point 1 (T1) and T2, to detect the distribution of molecules and microbes throughout the living spaces simulating normal activity and occupancy. After T1, the house was used for the House Observations of Microbial and Environmental Chemistry (HOMEChem) field campaign: 4 weeks of use that included cooking, cleaning, and human occupancy (5). The house experienced normal daytime human use (e.g., using one of the bathrooms, sitting on chairs, cleaning, eating and using computers on the tables, and cooking in the kitchen). Although overnight stays were not permitted, people occupied this home for 6 ± 4 hours per day for 26 days and performed scripted activities. In total, ~45 different people visited the home in those 30 days, which included an open house for the media and the local community.
RESULTS
The home was sampled to inventory the detectable molecules and microbes that were present at T1 and T2 by swabbing surfaces throughout the occupied areas of the house (fig. S1 and movie S1) (6). Extracts of the swabs were subjected to untargeted metabolomics analysis. We chose electrospray ionization (ESI) liquid chromatography–mass spectrometry (LC-MS)–based untargeted metabolomics in the positive ionization mode for this study due to its high sensitivity, potential to detect a broad range of molecules, and the large number of publicly accessible reference spectra. The molecular composition was assessed before and after human use. The ionization efficiency in ESI may change depending on the sample due to the unique chemistry of each sample, leading to potentially skewed abundances for some molecules. Thus, we emphasize the general and semiquantitative observable patterns.
Before the HOMEChem project, the test house had been cleaned thoroughly with a bleach solution, and then kitchen and table surfaces were wiped with surface cleaning wipes. At T1, despite a deep cleaning, the metabolomics analysis revealed that the house already contained traces of various molecules associated with human presence, although much less than at T2 (Fig. 1B, fig. S1C, and movie S1). Over the course of the next 4 weeks, the HOMEChem activities resulted in the introduction and changes of detectable molecules. Thousands of different molecules, observed as spectral features from tandem MS (MS/MS), were detected throughout the house (Fig. 1A), with increase in detectable chemistry at T2. Molecules are more unevenly distributed at T2, indicating possible local deposition through splatter, spills, etc. (fig. S5). We obtained spectral library matches to molecules associated with skin care products, skin (e.g., fig. S2A), drugs (e.g., antidepressants and anabolic steroids; figs. S2B and S3B), food-derived molecules (e.g., terpenes and their derivatives, flavonoids, and lignans; fig. S3, C and E), human or animal metabolites [e.g., bile acids (fig. S3D), carnitines, and long-chain fatty acids], amino acids/peptides and their various derivatives (fig. S3A), saccharides, phosphoorganic molecules, halogenated compounds (including biocides not expected to be found indoors; e.g., fig. S2C), and microbial metabolites. On the basis of the molecular profiles, the main sources of indoor surface molecules are natural products (i.e., biologically produced molecules, as opposed to synthetic compounds), food, the environment (i.e., molecules associated with outdoors), personal care products, and human-derived metabolites (many could be traced to feces; fig. S3).
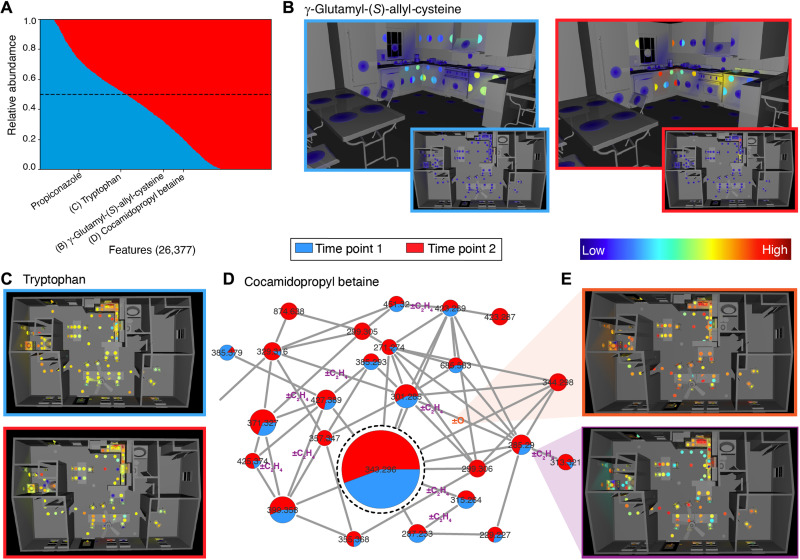
Fig. 1. Chemistry of the indoor environment and changes due to human presence.
Colored circles in 3D visualizations represent sampled surfaces. (A) Changes in chemistry of the house from T1 to T2: 26,377 spectral features obtained in the house are sorted according to their relative abundance between T1 and T2. Median value, dashed line. Examples of molecules, inferred from spectral matches level 2/3 according to the 2007 metabolomics standards initiative [Sumner et al. (12)], that decrease (propiconazole, medication) do not change [(C) tryptophan] or that increase [(B) γ-glutamyl-S-allylcysteine and (D) cocamidopropyl betaine] from T1 to T2 are marked. (B) Evidence of the previous human activity: γ-Glutamyl-S-allylcysteine, a metabolite from foods such as garlic, is already found in the kitchen at T1 (102 to 1.78 × 108). (C) Tryptophan, an amino acid and a hallmark of life, shows a comparable distribution across time points [(log10) 2 to 8]. (D) A portion of a network cluster corresponding to the family of compounds related to cocamidopropyl betaine, a common cosmetics ingredient from coconut oil (dashed node, node size = relative abundance). Multiple homologues are present as is evident from mass shifts (differences, m/z) in such as C2H4 (purple, Δm/z = ~28.03) or C4H8. These chemical shifts can be cataloged across the entire molecular network, as shown in (E). (E) 3D maps showing aggregate counts of mass shifts at T2 across the molecular network for the molecules detected within the house for the Δm/z of 15.995 Da corresponding to the O atom [orange, (log10) 1.15 to 1.87] and 28.031 corresponding to C2H4 [purple, (log10) 1.79 to 2.27]. The 3D maps are created with `ili (movie S1) (6).
Molecular networking was used to illuminate the diversity of molecules across different indoor sites. Molecular networking groups molecules that fragment in a similar fashion and thus are likely to be structurally similar to each other (7). The global network of all compounds detected at each or both time points is shown in fig. S4. Networking allows exploring related compounds by noting differences in mass [Δmass/charge ratio (Δm/z)] between connected molecules (nodes) in a cluster. Cocamidopropyl betaine, an ingredient in personal care products, shampoos, and soaps made from coconut oil, highlights this linking of related structures with a diverse array of acyl chain lengths, mainly two and four carbon backbone variants connected into a cluster in the network (Fig. 1D). Both abundance and number of cocamidopropyl betaine–related molecules expectedly increased at T2 (Fig. 1A).
Co-networking of HOMEChem data with other public datasets to enable tracing of potential sources, a reference data–driven metabolomics approach (8), revealed that the molecules found in the indoor space overlap with other sample types and thus may, at least in part, originate from those sources: food (~15.7%), human-associated microbes (~1.1%), feces (~8.6%; although feces contain both food and microbial molecules), building materials and microbes that grow on them (~2.6%), and building materials in humid conditions (~4.7%).
The chemical diversity (number of observed features) increased from T1 to T2 across the house, especially on some kitchen surfaces (fig. S5); this indicates that food and its preparation was the dominant source of not only the overall observed house surface chemistry but also chemical changes. The other “hotspot” was the toilet, where the increase in molecular diversity represents a snapshot of human metabolism of various excreted endogenous and exogenous chemistries. Some kitchen surfaces were also among those whose chemical diversity decreased the most between T1 and T2, likely due to surface cleaning and sanitation associated with food preparation. It appears that, even when a subset of chemistry is removed because of the cleaning, it is only temporary and/or partial, as the sum total of cleaning and human activities overall results in an increase in accumulation of richer chemistry. Surfaces that were routinely touched by humans—tables, light switches, and knobs—also show notable changes between T1 and T2, which were measured as distances between paired samples of a Bray-Curtis dissimilarity principal coordinate analysis (PCoA), where high distances show higher dissimilarity. The change in molecular diversity was not as marked on floors, which were the other most often cleaned surfaces throughout the house during the study. The surfaces that were not in direct contact with people—windows, chairs, and doors—show the smallest changes in chemical diversity within this study.
Another way of understanding change in composition across chemical landscapes is by using the concept of “nestedness,” i.e., a measure of structure in an ecological system, used in microbial ecology, which asks whether simpler communities contain the same or different subsets of members found in more complex ones (9). Here, to explore nestedness, instead of microbial taxonomy, we considered the chemical ontology, the hierarchy of molecular families (10). This analysis revealed notable nestedness of chemistries both across the house locations and time points at all levels of chemical ontology (fig. S6, D and E). The samples at T2 were more chemically rich (greater variety of molecular classes) than T1 (fig. S6, A and C), which describes the direction of nestedness—chemistry at T1 is “nested” within T2, reflecting the idea that T2 adds molecules through the influence of human occupation rather than just replacing molecules already present. The molecules and molecular families that are introduced or formed are related to, or derived from, the chemistries that were originally present. Across the house locations, chemistries are also nested with respect to one another, with the chemistry of the kitchen being the most diverse (fig. S5, B to D), presumably reflecting the increased addition of food-derived molecules to the molecules present throughout the rest of the house. Our findings demonstrate that humans themselves and their lifestyle choices largely define the indoor surface molecular distributions (parallel to known results in microbiology) (11).
Because eating or cooking had the most significant impact on the chemistries of specific locations within the house, we used the Global FoodOmics project (8) of molecular composition of foods and a reference data–driven approach to track the overlap of molecules detected in the house with those found in various foods, as described in Materials and Methods and references therein. The test house inhabitants contributed an assortment of molecules that we could bioinformatically relate to a wide variety of possible food sources (fig. S7A), across different locations (fig. S7B), with the greatest molecular diversity contributed by plant-based foods; coffee had a particularly large trace. An evaluation of the precision and recall of the foods inferred to have been present in the test home, from detectable traces left on surfaces alone, shows reasonable recall at FoodOmics food ontology levels 1 to 4 (0.60 to 0.90) and good precision at levels 1 to 3 (0.82 to 1.0), but precision decreases substantially at levels 4 to 5 (0.29 to 0.53) (fig. S7C). The detection of molecules that are known to originate from certain foods can be used to create “molecular food maps,” which show, as an example, that molecules from herbs/spices (level 4) used during the Thanksgiving dinner experiment (5) were colocated in the kitchen and on the tables (fig. S7, D and E). The “epicenter” of food-related molecules from multiple sources was in the kitchen sink, where many foods ended up. Many food traces remained detectable on surfaces despite repeated cleanings.
Along with human activities, location within the house also corresponds to specific differences in which molecules are present and which chemical transformations occur over time during human occupation. The high-resolution MS allows for exploring changes in molecular composition, such as altering of carbon oxidation state, while molecular networking enables us to catalog the chemical shifts (differences in m/z) on the global molecular network of the detected molecules across both time points. We leveraged this information to explore possible chemical transformations related to the spatial layout within the house (Fig. 1E). For this analysis, we counted the number of times each Δm/z was found in the global molecular network of the HOMEChem data as well as in which samples (which correspond to particular locations) and time points. The observation of a certain Δm/z value could be indicative of either occurring chemical transformation or presence of chemically related molecules. Spatial distributions of these shifts may aid in understanding the reasons behind their observations. We found that these transformations occur differently at different spatial locations. The Δm/z values attributable to certain molecular differences, such as O, CO, and 2H, show very similar patterns: They occur everywhere around the house but with a universal hotspot at the kitchen sink (and, to a lesser extent, the bathroom sink) (fig. S8, A to J). The highest density of chemical shifts was consistently found at the sinks, especially the kitchen sink, and around the stove top. These correspond to locations with high amounts of organic matter and a plausible route to transformation via moisture and/or heat (fig. S8, C to G). Conversely, the Δm/z values that correspond to aliphatic homologs (CH2, C2H4, etc.; fig. S8H) are fairly evenly distributed and found at locations that humans most often interact with, such as tables, suggesting lipid contributions from skin and skincare products. These compounds reflect the presence of a mixture of homologous compounds, such as those related to cocamidopropyl betaine or other lipids, rather than chemical transformations in situ (Fig. 1, D and E). Other chemical shifts, such as glycosylation, have different patterns altogether and occur more frequently on floors (fig. S8, I and J), indicating different underlying reasons for these chemical shifts. Although such cataloging of the chemical shifts does not allow for unambiguous differentiation of introducing new molecules externally or their formation via chemical reaction(s), it does allow the understanding and visualizing of the overall changes in molecular composition that took place.
Humans are not the only occupants of a home: Indoor surfaces are covered with bacteria, fungi, and other microbes (13). To test whether any detected molecules were of bacterial origin or whether the bacterial communities are altered by human occupancy, we evaluated the microbiome of the sampled surfaces alongside the metabolome (fig. S1). Repetitive surface cleaning depleted existing microbial populations and allowed different microbial taxa to be reintroduced and detected (3). We found that the bacterial portion of the indoor microbiome was reshaped after 1 month of simulated human occupation. During the month of home use, less than half of the house’s original microbiome remained at T2. The persistent microbiome fraction represents 42.6% of the observed suboperational taxonomic units (sOTUs) but encompasses 96.2% of all microbial feature counts in the study. The remaining 3.8% of the microbial feature counts exclusive to either time point are mostly rare observations with low counts. Using fast expectation-maximization for microbial source tracking (FEAST) (14), we found that the microbiome of the house at T2 had a higher proportion of sOTUs derived from human hosts, mainly commensal species on human skin or in the gut, relative to T1 (fig. S9). Correspondingly, free-living, environment-associated microbes are depleted by human activities inside the house (fig. S9).
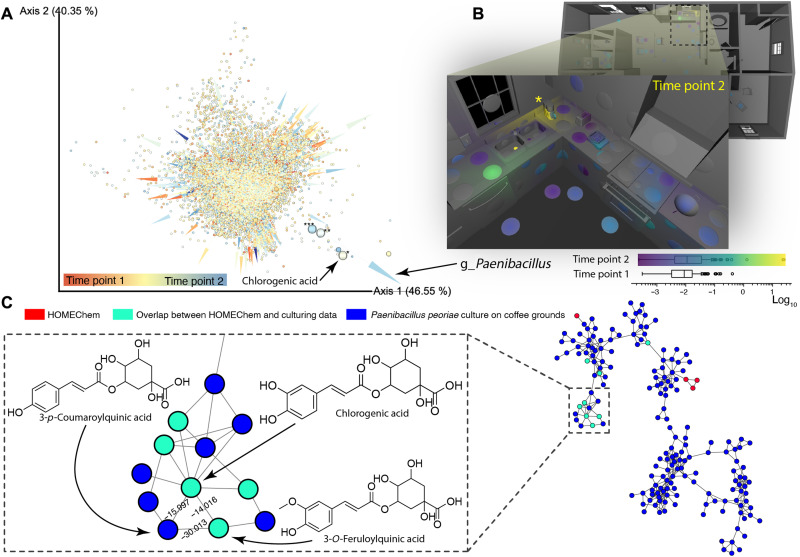
There is a human-microbiome-home relationship in the indoor environment at the molecular level. Co-occurrence analysis of microbes and metabolites using neural networks (15) reveals microbial metabolism of molecules introduced by human activity. As noted above, at least 1% of indoor molecules are directly attributable to microbes, and the actual fraction is likely much higher, as many endogenous and exogenous molecules are transformed by microbes, both in human host and the house surfaces. These molecules may have an outsized health effect (2). As an example, Paenibacillus sp. was associated with molecules from coffee, one of the dominant sources of food-derived indoor molecules (e.g., caffeine, trigonelline, and chlorogenic acid) (Fig. 2A and fig. S10). In the home, especially at T2, Paenibacillus was observed in and around the area where coffee was prepared (Fig. 2B and movie S1), and this genus has been found to grow in coffee machines (16). We observed that Paenibacillus cultures transformed coffee-derived molecules into the molecules that we detected inside the house (fig. S11); chlorogenic acid was detected in the culture of these strains when grown on spent coffee grounds (SCGs), and its metabolized versions were also found in the house (Fig. 2C), supporting this causal hypothesis about its origin.
Fig. 2. Exploration of microbial chemistry.
(A) 3D embedding using singular value decomposition of cooccurrence probabilities, which are highest for microbial genera (arrows) pointing in the same direction as metabolites (dots). The color indicates association of the metabolite or microbe with T1 or T2, as determined by multinomial regression (17). An example of three metabolites (***caffeine, **trigonelline, and *chlorogenic acid) that are some of the most positively associated with Paenibacillus are highlighted as large spheres, while medium-sized spheres are other metabolites with no annotation within the top 10 most positively associated with Paenibacillus. All of the annotated compounds can be found in coffee. (B) A spatial map of normalized read counts (log [−3.5 to 1.5]) at T2 of the sOTUs taxonomically classified to g_Paenibacillus. The main growth of this microbe appears to have occurred on the coffee machine (marked with yellow asterisk). The color bar legend shows a comparison of the distributions of normalized g_Paenibacillus read counts as boxplots for T2 (3D plotted) and T1 (not plotted). (C) A cluster of the molecular co-network of HOMEChem metabolome with the culturing experiment of P. peoriae DSM 8320 on SCGs. The joint network shows overlap of the chemistry detected in the house and chemistry of the microbe grown on coffee. The shown cluster (inlet) contains chlorogenic acid, a coffee-related compound; several of the microbially modified versions of the molecules (nodes in cyan) were detected in the house.
DISCUSSION
Humans introduce many molecules and drive alteration of the indoor microbiome; the microbiome generates its own chemistry, including transforming the molecules introduced by humans, all of which contribute to the changing chemical makeup of the house. Our indoor habitat appears to be not just a reflection of human activities but rather is in a mutualistic relationship with its inhabitants. Such household-microbial chemistry, its potential impact on health and well-being of the house inhabitants, and possible ways to control and how to optimize such chemistries to promote beneficial effects are factors that should be considered in the engineering of indoor environments; we hope that this work will stimulate interest in future studies.
MATERIALS AND METHODS
The test house and HOMEChem campaign
Details of the setup and research activities are given elsewhere (5). Briefly, the HOMEChem study was conducted in a three-bedroom, two-bathroom manufactured test house (111 m2 floor area and ~250 m3 volume), located at the University of Texas at Austin. Before experimental activities, the house was thoroughly cleaned with bleach (one-fourth cup of bleach solution per gallon of water per the manufacturer’s instructions).
Several scripted activities were conducted repeatedly in the duration of the study that included cleaning (bleach and “natural” cleaning agent) (17), people congregation, and cooking. In addition to the cleaning products, commercial surface cleaning wipes were used to wipe down the kitchen counter surfaces after cooking events on “layered days.” People were allowed to use house bathrooms and kitchen, but no overnight stay was permitted. Two types of activities were conducted: sequential, to explore semi-independent perturbations, and layered, a more realistic representation of a home where several activities were happening simultaneously and/or in quick succession. Commercial surface cleaning wipes were used to wipe down the kitchen counter surfaces after cooking events on layered days. Blanks were not collected.
Sampling
The chemical distributions in the house were mapped using high-performance LC-MS (HPLC-MS) using the established protocols (6). The general sampling procedure is described in (6). Briefly, the cotton swabs (Puritan, no. 25-806 2WC) were soaked in LC-MS (Optima)–grade ethanol before sampling to remove contaminants and then shipped to the sampling location in a sealed jar with swab tips immersed in ethanol. For sampling, a single moist swab was removed from the jar, and the surface of approximately 5 cm in diameter was vigorously swabbed for ~1 min (a closely adjacent spot was also simultaneously swabbed for microbiome analysis as described below). The swab was then placed into a well of a deep 96-well plate and the stick cutoff. While sampling, the wells with collected swabs were covered with aluminum foil to prevent cross-contamination. The last column on each plate was filled with swab blanks.
For microbiome sampling, dual cotton swabs (BD SWUBE, no. 281130) were presoaked in tris-EDTA buffer (Fisher Scientific) and then pressed and vigorously dragged against a directly adjacent but not overlapping surface to the metabolomics sampling site, with an approximately equal surface area. The dual cotton swabs were then resealed in their screw cap container and stored on dry ice while in transit to the laboratory, where they were ultimately stored at −20°C until analysis.
Sample preparation
Upon collection, the filled plate was sealed with a silicone top and stored on dry ice. All sealed plates were shipped on dry ice and stored at −80°C until analysis. Before analysis, the plates were removed from the freezer, and 500 μl of ethanol added to each well, resealed, and extracted overnight at 4°C. The swabs were then removed and discarded, and the 50-μl aliquot of each sample was transferred to an MS plate.
Data acquisition
Reverse-phase LC-MS was performed using a Thermo Vanquish ultra-HPLC (UHPLC) system coupled to a Q Exactive Orbitrap mass spectrometer. Data were acquired using data-dependent acquisition (m/z, 80 to 1200), subsequently fragmenting the five most abundant precursor ions. A 5-μl aliquot of sample was injected. The injected samples were chromatographically separated using a Vanquish UHPLC (Thermo Fisher Scientific, Waltham, MA) controlled by Thermo SII for Xcalibur software (Thermo Fisher Scientific, Waltham, MA), using a C18 Kinetex (1.7 μM, 100 × 2.1 mm, 100 Å) column (Phenomenex, Torrance, CA), kept at 40°C column temperature, with a flow rate of 0.5 ml/min. The mobile phase used was water (phase A) and acetonitrile (phase B), both containing 0.1% formic acid (Fisher Scientific, Optima LC/MS), using the following gradient: 0 to 1 min, 5% B; 1 to 8 min, linear ramp to 100% B; 8 to 10.9 min, maintain at 100% B; 10.9 to 11 min, linear decrease to 5% A; and 11 to 12 min, 5% B for all samples.
MS analysis was performed on an Orbitrap (Q Exactive, Thermo Fisher Scientific, Waltham, MA) MS equipped with heated ESI-II probe sources and controlled by Xcalibur 3.0 software. The following probe settings were used for both MS for flow aspiration and ionization: spray voltage of 3500 V, sheath gas (N2) pressure of 35 psi, auxiliary gas pressure (N2) of 10 psi, ion source temperature of 270°C, S-lens radio frequency level of 50 Hz, and auxiliary gas heater temperature at 440°C.
For Orbitrap MS, spectra were acquired in positive ion mode over a mass range of 100 to 1500 m/z. An external calibration with Pierce LTQ Velos ESI positive ion calibration solution (Thermo Fisher Scientific, Waltham, MA) was performed before data acquisition, with an error rate less than 1 parts per million (ppm). Data acquisition parameters were set as follows: minutes 0 to 0.5 were sent to waste, and minutes 0.1 to 12 were recorded with data-dependent MS/MS acquisition mode. Full scan at MS1 level was performed with resolution of 35,000 in profile mode. The 10 most intense ions with 2 m/z isolation window with m/z of 0.5 offset per MS1 scan were selected and subjected to normalized collision-induced dissociation with 30 eV. MS2 scans were performed at 17,500 resolution with maximum IT (ion trap) time of 60 ms in profile mode. MS/MS active exclusion parameter was set to 5.0 s.
LC-MS data processing
The LC-MS/MS .raw data files were converted to mzXML format, and feature detection was performed with the MZmine 2 software (18). The software settings were as follows: Mass detection was performed with a signal threshold of 1.0 × 103 for MS1 and 1.0 × 102 for MS2. For the chromatogram building, the mass tolerance was set to 10 ppm, the minimum peak time span to 0.01 s, and the minimum height to 5.0 × 103. For chromatographic deconvolution, the local minimum search algorithm was used; m/z range for MS2 scan pairing was set at 0.025 Da and retention time (RT) at a 0.1-min range. The peaks were deisotoped within 25 ppm m/z and 0.2-min RT tolerances, aligned, gap-filled using the same tolerances, and then filtered to retain only peaks that appear in at least two samples, with minimum two peaks in isotope pattern to create the feature table. Peaks present in any of the blanks were removed from the final feature table unless at least one sample contained the peak at abundance three times or above.
3D data visualization
The aligned features were then exported as .csv with the metadata and combined in RStudio(R) with the feature table to create a master table for further statistical analysis. The master table was split into individual tables for T1 and T2 for further mapping. The tables were then normalized using quantile normalization using MetaboAnalyst (19) and exported out. The three-dimensional (3D) model was created for the computer-aided design drawing of the test house. A target was placed at each location of the 3D model where the corresponding sampling in the test house was collected. The coordinates for each target in the house 3D model were then added to the normalized tables. For visualization, the 3D model of the house was dragged and dropped into `ili (https://ili.embl.de/) (6), followed by the feature table with coordinates. The input tables used for mapping are available at https://github.com/aaksenov1/HOMEChem-3D-mapping-input-files.
For visualization, the “jet” color scheme was used throughout for molecular mapping, and “viridis” was used for microbiome. When the visualized data were divergent and centered around zero (log ratios), a “blue-red” color scheme was used. The scale was set either as linear or logarithmic for visualization clarity (the scale is indicated on each figure as appropriate).
Molecular networking
A molecular network was created with classical (20) and the Feature-Based Molecular Networking (FBMN) workflow (21) on Global Natural Product Social Molecular Networking (GNPS) (https://gnps.ucsd.edu) (22). The MS data were first processed with MZmine 2 (18), and the results were exported to GNPS for FBMN analysis. The data were filtered by removing all MS/MS fragment ions within ±17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top six fragment ions in the ±50-Da window throughout the spectrum. The precursor ion mass tolerance was set to 0.02 Da, and the MS/MS fragment ion tolerance was set to 0.02 Da. A molecular network was then created, where edges were filtered to have a cosine score above 0.7 and more than six matched peaks. Furthermore, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Last, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS spectral libraries (22). The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least six matched peaks. The molecular networks were visualized using Cytoscape software (23).
Reference data–based metabolomics (8) was performed to conetwork HOMEChem data with other public datasets to trace the potential source. The common features were considered an overlap. The datasets that were co-networked are food [FoodOmics (8); overlap, ~15.7%], human microbes [Human Microbiome Project (24); overlap, ~1.1%], and feces [American Gut (25); overlap, ~8.6%; feces contain both food and microbial molecules (25)]. Overlap with bacterial and fungal growth on wetted wood (26) was ~2.6% and on common building materials in humid conditions (27) was ~4.7%.
Mass shift analysis
For chemical shift analyses, unbiased by annotation of chemical shift discretization on the chemical shift space was performed by binning into 3000 bins and removing bins with no occupancy, resulting in 575 discrete count features. Chemical shift distances between locations were computed using the Bray-Curtis distance in vegan (2.5 to 6). PCoA projections were computed using ape (5.3) PCoA function.
Clear separation was observed for 13 locations because of very low feature coverage in either of the two underlying samples from that location (fig. S5). These were removed as outliers. Distances to the centroid were calculated using the betadisper function in vegan.
FoodOmics analysis: Determination of the food sources
Reference data–driven analysis using the Global FoodOmics data
A description of the methods, code, and tutorial for generation of fig. S7A can be found at https://ccms-ucsd.github.io/GNPSDocumentation/tutorials/rdd/ and is linked out to GitHub and the MassIVE Repository, as shown in Gauglitz et al. (8).
Qemistree
The chemical molecular ontology analysis using Qemistree (28) for the FoodOmics (8) and nestedness analysis (9) were performed using the GNPS workflow. The results can be viewed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=732ee57912dc4420afc7e5b83f3d8594.
Microbiome sample prep and sequencing
Both swabs of each microbiome sampling kit (BD SWUBE, no. 281130) were extracted following the standardized Earth Microbiome Project protocols (www.earthmicrobiome.org/protocols-and-standards/16s) (29). Briefly, DNA was extracted using the MagAttract PowerSoil DNA Kit (QIAGEN) on a KingFisher Flex (Thermo Fisher Scientific). The V4 region of the 16S ribosomal RNA gene was targeted for polymerase chain reaction (PCR) amplification using the 515f-806r primers with Golay error-correcting barcodes. The barcoded 16S amplicons were pooled in equal concentrations, and the pool was purified with a QIAquick PCR purification kit (QIAGEN). The purified pool was sequenced with a MiSeq V2 300 cycle kit (Illumina) with the appropriate sequencing primers.
Microbiome data analysis
Sequence data were demultiplexed, quality-filtered, and trimmed to 150 base pairs using Qiita (30). Trimmed sequences were error filtered using Deblur (31), resulting in a sOTU (31) feature table. Taxonomy was assigned on representative sOTU sequences using a prefitted Greengenes classifier in QIIME 2 (32). Upon analysis of rarefaction curves, which consisted of an evaluation of plateaus and trends in alpha diversity curves (Shannon, Faith’s phylogenetic diversity, and observed OTUs) of samples grouped at different metadata categorical levels (time point, indoor_space, object, and surface_material) with increasing rarefaction depth, a 5000 sequencing depth rarefaction was applied, resulting in the retention of 80% of the samples and ~25% of features with relatively even distribution of sample retention between T1 (n = 232) and T2 (n = 221).
Because of the compositional nature of sequencing data, feature count comparisons between time points were done as log ratios with a reference frame (33) in the denominator. The reference frame feature (“k__Bacteria;p__Cyanobacteria;c__Chloroplast;o__Streptophyta;f__;g__”) was chosen because it was observed across most samples.
Tree visualization
A phylogenetic tree visualization (34) was built on assigned taxonomy collapsed at the genera level. Changes in normalized feature counts (log ratios on genera) between T1 and T2 were averaged across all sampled surfaces. Features that were exclusive to either time point (only 3.8% of all observed feature counts) were excluded from quantitative tree visualization.
FEAST
Informed by redbiom (35), a collection of public 16S data with comparable sequencing methods was compiled through Qiita (30) for microbial source tracking meta-analysis. The source tracking meta-analysis included samples from the following environments and studies: food (8), water (36), stool (25), soil (29, 37, 38), and skin (25, 39). Because of the experimental design behind the American Gut project (25), the meta-analysis sequencing data compendium was filtered for blooms associated with room temperature shipment (40). FEAST (14) was performed in R on nonrarefied data (https://github.com/cozygene/FEAST).
Culturing of Paenibacillus on SCGs
Microbial strains
The following strains were purchased from the Leibniz Institut Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH: Paenibacillus alvei DSM 29, National Center for Biotechnology Information (NCBI): txid1206781; Paenibacillus polymyxa DSM 36, NCBI: txid1406; and Paenibacillus peoriae DSM 8320, NCBI: txid1087481. The LC-MS data were analyzed using the same approach as for the analysis of swabs. The workflow on GNPS for the analysis is found as classical molecular network, https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4c366dcc969e47aa92df68d0db6b8acc, and feature-based molecular network, https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=936f6c81421447dc9e37a07e1fceb61a.
Medium
The media used to reactivate the lyophilized strains was prepared as recommended by the provider, available on their website (www.dsmz.de/collection/catalogue/microorganisms/culture-technology/list-of-media-for-microorganisms). Medium 1 (nutrient broth) was used in this study for initial cultures of the three microorganisms.
Spent coffee ground
Commercial ground coffee (Peet’s coffee dark roast Major Dickason’s blend) was used for culturing the bacterial strains. The SCG was prepared by brewing 15 g of ground coffee into 250 ml of distilled water and sterilized for 15 min at 121°C. The brewed and sterile coffee was filtered using a Corning Filter System (ref. 430756). This SCG was added into 12-well plates, 500 mg per well, resulting in three biological replicates per bacterial strains and an experimental control (SCG with culture medium instead of bacterial inoculum). The filtration step was not exhaustive to eliminate the total content of water. Although the amount of water was not quantified, it was sufficient to support bacterial growth until the end of the experiment (day 8), which was also evidenced by the detection of microbial associated metabolites during the time-course experiment (fig. S11). The humidity of the incubator (Fisher Scientific Isotemp Laboratory Incubator) was maintained by keeping a glass beaker with water (200 ml) throughout the experiment (8 days in total).
Culture conditions
The microorganisms were initially grown in 50-ml Erlenmeyer flasks containing 25 ml of medium 1 (nutrient broth) in a rotary shaker (MaxQ 4450, Thermo Fisher Scientific) at 200 rpm with controlled temperature of 30°C for 48 hours. From each microorganism, a 500-μl microbial inoculum from a 48-hour culture was transferred into 12-well plates containing SCG or medium 1 agar (nutrient agar) and incubated at 30°C until required. Plates were used for time-course monitoring, resulting in samples for LC-MS/MS corresponding to time of 0, 2, 4, 6 and 8 days.
Extraction of metabolites
Following a time-course experiment (time of 0, 2, 4, 6 and 8 days), plates containing microbial cultures were submitted to three freeze-thaw cycles of 10 min each. After that, an aliquot of 30 to 50 mg from SCG and agar were transferred to a 96-well plate. These samples were extracted with methanol, followed by sonication for 15 min (Branson 5510, Marshall Scientific, Hampton, NH, USA), centrifugation for 15 min at 2000 rpm (865g) using a Sorvall Legend RT centrifuge (Marshall Scientific, Hampton, NH, USA). The obtained supernatant was transferred to a clean 96-well plate and dried out in a Centrifugal Vacuum Concentrator, CentriVap (Labconco, Kansas City, MO, USA). Samples were resuspended with 200 μl of 80% methanol-water containing internal standard (1 μM sulfamethazine) for LC-MS/MS acquisition.
Acknowledgments
We thank the Alfred P. Sloan Foundation Microbiology of the Built Environment Program and Chemistry of Indoor Environments Program (G-2017-9944) for support of this study, and also for its support of previous development of the 3D molecular cartography methodology used in this work. We are grateful to Dr. Atila Novoselac and the HOMEChem science team for their work on the model house. J. Gilbert has provided data for the building materials metabolome analysis.
Funding: A.M.C.-R. and P.C.D. were supported by the Gordon and Betty Moore Foundation through grant GBMF7622; the U.S. National Institutes of Health for the Center (P41 GM103484 and R01 GM107550); and Department of Energy, Federal subaward DE-SC0021340.
Author contributions: P.C.D. and R.K. formulated the study. A.V.M., A.A.A., R.A.S., and C.B. collected the metabolomics and microbiome samples. A.V.M. and A.A.A. performed the LC-MS analysis. R.A.S. and C.B. performed the microbiome sequencing. A.V.M. and A.A.A. analyzed the LC-MS data. R.A.S. analyzed the sequencing data. A.B. and A.A.A. performed the chemical shifts analysis. F.L. and A.A.A. performed the mmvec analysis. F.L. and A.A.A. performed the nestedness analysis. J.M.G. performed the FoodOmics analysis. A.M.C.-R. performed the Paenibacillus culturing experiment and LC-MS acquisition. M.E.V. and D.K.F. coordinated the HOMEChem campaign. A.A.A., R.A.S., R.K., and P.C.D. wrote the manuscript.
Competing interests: P.C.D. is a scientific advisor for Sirenas, Cybele, and Galileo and a scientific advisor and founder of Enveda and OMETA with approval by University of California San Diego. A.A.A. is a founder of Arome Science Inc. The authors declare that they have no other competing interests.
Data and materials availability: All data generated in this study are publicly available. The raw data are available on MassIVE Repository (massive.ucsd.edu) under the following dataset accession numbers MSV000083320 and MSV000087141. The annotation and molecular networking have been conducted using GNPS. The links to analyses are provided at classical molecular network: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=9355dfd0f2174a2f883b363bc3a8ca26; feature-based molecular network: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=ba54bdcb526344bdb5d25fa4f77f4a15; classical molecular network for culturing of Paenibacillus on SCGs: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4c366dcc969e47aa92df68d0db6b8acc; and feature-based molecular network for culturing of Paenibacillus on SCGs: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=936f6c81421447dc9e37a07e1fceb61a. The microbiome data are available on Qiita, study ID 11865: https://qiita.ucsd.edu/study/description/11865. GitHub repository to recreate main figures: https://github.com/RodolfoSalido/HOMEChem_main_figures.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Klepeis N. E., Nelson W. C., Ott W. R., Robinson J. P., Tsang A. M., Switzer P., Behar J. V., Hern S. C., Engelmann W. H., The National Human Activity Pattern Survey (NHAPS): A resource for assessing exposure to environmental pollutants. J. Expo. Anal. Environ. Epidemiol. 11, 231–252 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Reynolds L. A., Brett Finlay B., Early life factors that affect allergy development. Nat. Rev. Immunol. 17, 518–528 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lax S., Sangwan N., Smith D., Larsen P., Handley K. M., Richardson M., Guyton K., Krezalek M., Shogan B. D., Defazio J., Flemming I., Shakhsheer B., Weber S., Landon E., Garcia-Houchins S., Siegel J., Alverdy J., Knight R., Stephens B., Gilbert J. A., Bacterial colonization and succession in a newly opened hospital. Sci. Transl. Med. 9, eaah6500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Martín J., Kraakman N. J. R., Pérez C., Lebrero R., Muñoz R., A state–of–the-art review on indoor air pollution and strategies for indoor air pollution control. Chemosphere 262, 128376 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Farmer D. K., Vance M. E., Abbatt J. P. D., Abeleira A., Alves M. R., Arata C., Boedicker E., Bourne S., Cardoso-Saldaña F., Corsi R., DeCarlo P. F., Goldstein A. H., Grassian V. H., Ruiz L. H., Jimenez J. L., Kahan T. F., Katz E. F., Mattila J. M., Nazaroff W. W., Novoselac A., O’Brien R. E., Or V. W., Patel S., Sankhyan S., Stevens P. S., Tian Y., Wade M., Wang C., Zhou S., Zhou Y., Overview of HOMEChem: House observations of microbial and environmental chemistry. Environ Sci Process Impacts 21, 1280–1300 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Protsyuk I., Melnik A. V., Nothias L.-F., Rappez L., Phapale P., Aksenov A. A., Bouslimani A., Ryazanov S., Dorrestein P. C., Alexandrov T., 3D molecular cartography using LC–MS facilitated by Optimus and `ili software. Nat. Protoc. 13, 134–154 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Watrous J., Roach P., Alexandrov T., Heath B. S., Yang J. Y., Kersten R. D., van der Voort M., Pogliano K., Gross H., Raaijmakers J. M., Moore B. S., Laskin J., Bandeira N., Dorrestein P. C., Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U.S.A. 109, E1743–E1752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauglitz J. M., Bittremieux W., Williams C. L., Weldon K. C., Panitchpakdi M., Di Ottavio F., Aceves C. M., Brown E., Sikora N. C., Jarmusch A. K., Martino C., Tripathi A., Sayyari E., Shaffer J. P., Coras R., Vargas F., Goldasich L. D., Schwartz T., Bryant M., Humphrey G., Johnson A. J., Spengler K., Belda-Ferre P., Diaz E., McDonald D., Zhu Q., Nguyen D. S., Elijah E. O., Wang M., Marotz C., Sprecher K. E., Robles D. V., Withrow D., Ackermann G., Herrera L., Bradford B. J., Marques L. M. M., Amaral J. G., Silva R. M., Veras F. P., Cunha T. M., Oliveira R. D. R., Louzada-Junior P., Mills R. H., Galasko D., Dulai P. S., Wittenberg C., Gonzalez D. J., Terkeltaub R., Doty M. M., Kim J. H., Rhee K. E., Beauchamp-Walters J., Wright K. P., Dominguez-Bello M. G., Manary M., Oliveira M. F., Boland B. S., Lopes N. P., Guma M., Swafford A. D., Dutton R. J., Knight R., Dorrestein P. C., Reference data based insights expand understanding of human metabolomes. bioRxiv , 2020.07.08.194159 (2020). [Google Scholar]
- 9.Almeida-Neto M., Guimarães P., Guimarães P. R. Jr., Loyola R. D., Ulrich W., A consistent metric for nestedness analysis in ecological systems: Reconciling concept and measurement. Oikos 117, 1227–1239 (2008). [Google Scholar]
- 10.Djoumbou Feunang Y., Eisner R., Knox C., Chepelev L., Hastings J., Owen G., Fahy E., Steinbeck C., Subramanian S., Bolton E., Greiner R., Wishart D. S., ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Chem. 8, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lax S., Smith D. P., Hampton-Marcell J., Owens S. M., Handley K. M., Scott N. M., Gibbons S. M., Larsen P., Shogan B. D., Weiss S., Metcalf J. L., Ursell L. K., Vázquez-Baeza Y., Van Treuren W., Hasan N. A., Gibson M. K., Colwell R., Dantas G., Knight R., Gilbert J. A., Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W.-M., Fiehn O., Goodacre R., Griffin J. L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A. N., Lindon J. C., Marriott P., Nicholls A. W., Reily M. D., Thaden J. J., Viant M. R., Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaprakash B., Adams R. I., Kirjavainen P., Karvonen A., Vepsäläinen A., Valkonen M., Järvi K., Sulyok M., Pekkanen J., Hyvärinen A., Täubel M., Indoor microbiota in severely moisture damaged homes and the impact of interventions. Microbiome 5, 138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shenhav L., Thompson M., Joseph T. A., Briscoe L., Furman O., Bogumil D., Mizrahi I., Pe’er I., Halperin E., FEAST: Fast expectation-maximization for microbial source tracking. Nat. Methods 16, 627–632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morton J. T., Aksenov A. A., Nothias L. F., Foulds J. R., Quinn R. A., Badri M. H., Swenson T. L., Van Goethem M. W., Northen T. R., Vazquez-Baeza Y., Wang M., Bokulich N. A., Watters A., Song S. J., Bonneau R., Dorrestein P. C., Knight R., Learning representations of microbe-metabolite interactions. Nat. Methods 16, 1306–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilanova C., Iglesias A., Porcar M., The coffee-machine bacteriome: Biodiversity and colonisation of the wasted coffee tray leach. Sci. Rep. 5, 17163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattila J. M., Lakey P. S. J., Shiraiwa M., Wang C., Abbatt J. P. D., Arata C., Goldstein A. H., Ampollini L., Katz E. F., DeCarlo P. F., Zhou S., Kahan T. F., Cardoso-Saldaña F. J., Ruiz L. H., Abeleira A., Boedicker E. K., Vance M. E., Farmer D. K., Multiphase chemistry controls inorganic chlorinated and nitrogenated compounds in indoor air during bleach cleaning. Environ. Sci. Technol. 54, 1730–1739 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Pluskal T., Castillo S., Villar-Briones A., Oresic M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong J., Xia J., Using MetaboAnalyst 4.0 for metabolomics data analysis, interpretation, and integration with other omics data. Methods Mol. Biol. 2104, 337–360 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Aron A. T., Gentry E. C., McPhail K. L., Nothias L.-F., Nothias-Esposito M., Bouslimani A., Petras D., Gauglitz J. M., Sikora N., Vargas F., van der Hooft J. J. J., Ernst M., Kang K. B., Aceves C. M., Caraballo-Rodríguez A. M., Koester I., Weldon K. C., Bertrand S., Roullier C., Sun K., Tehan R. M., Boya C. A., Christian P. M. H., Gutiérrez M., Ulloa A. M., Mora J. A. T., Mojica-Flores R., Lakey-Beitia J., Vásquez-Chaves V., Zhang Y., Calderón A. I., Tayler N., Keyzers R. A., Tugizimana F., Ndlovu N., Aksenov A. A., Jarmusch A. K., Schmid R., Truman A. W., Bandeira N., Wang M., Dorrestein P. C., Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 15, 1954–1991 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Nothias L. F., Petras D., Schmid R., Dührkop K., Rainer J., Sarvepalli A., Protsyuk I., Ernst M., Tsugawa H., Fleischauer M., Aicheler F., Aksenov A., Alka O., Allard P.-M., Barsch A., Cachet X., Caraballo M., Da Silva R. R., Dang T., Garg N., Gauglitz J. M., Gurevich A., Isaac G., Jarmusch A. K., Kameník Z., Kang K. B., Kessler N., Koester I., Korf A., Le Gouellec A., Ludwig M., Christian M. H., McCall L.-I., McSayles J., Meyer S. W., Mohimani H., Morsy M., Moyne O., Neumann S., Neuweger H., Nguyen N. H., Nothias-Esposito M., Paolini J., Phelan V. V., Pluskal T., Quinn R. A., Rogers S., Shrestha B., Tripathi A., van der Hooft J. J. J., Vargas F., Weldon K. C., Witting M., Yang H., Zhang Z., Zubeil F., Kohlbacher O., Böcker S., Alexandrov T., Bandeira N., Wang M., Dorrestein P. C., Feature-based molecular networking in the GNPS analysis environment. bioRxiv , 812404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., Nguyen D. D., Watrous J., Kapono C. A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A. V., Meehan M. J., Liu W.-T., Crüsemann M., Boudreau P. D., Esquenazi E., Sandoval-Calderón M., Kersten R. D., Pace L. A., Quinn R. A., Duncan K. R., Hsu C.-C., Floros D. J., Gavilan R. G., Kleigrewe K., Northen T., Dutton R. J., Parrot D., Carlson E. E., Aigle B., Michelsen C. F., Jelsbak L., Sohlenkamp C., Pevzner P., Edlund A., McLean J., Piel J., Murphy B. T., Gerwick L., Liaw C.-C., Yang Y.-L., Humpf H.-U., Maansson M., Keyzers R. A., Sims A. C., Johnson A. R., Sidebottom A. M., Sedio B. E., Klitgaard A., Larson C. B., Boya C. A., Torres-Mendoza D., Gonzalez D. J., Silva D. B., Marques L. M., Demarque D. P., Pociute E., O’Neill E. C., Briand E., Helfrich E. J. N., Granatosky E. A., Glukhov E., Ryffel F., Houson H., Mohimani H., Kharbush J. J., Zeng Y., Vorholt J. A., Kurita K. L., Charusanti P., McPhail K. L., Nielsen K. F., Vuong L., Elfeki M., Traxler M. F., Engene N., Koyama N., Vining O. B., Baric R., Silva R. R., Mascuch S. J., Tomasi S., Jenkins S., Macherla V., Hoffman T., Agarwal V., Williams P. G., Dai J., Neupane R., Gurr J., Rodríguez A. M. C., Lamsa A., Zhang C., Dorrestein K., Duggan B. M., Almaliti J., Allard P.-M., Phapale P., Nothias L.-F., Alexandrov T., Litaudon M., Wolfender J.-L., Kyle J. E., Metz T. O., Peryea T., Nguyen D.-T., VanLeer D., Shinn P., Jadhav A., Müller R., Waters K. M., Shi W., Liu X., Zhang L., Knight R., Jensen P. R., Palsson B. O., Pogliano K., Linington R. G., Gutiérrez M., Lopes N. P., Gerwick W. H., Moore B. S., Dorrestein P. C., Bandeira N., Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Integrative HMP (iHMP) Research Network Consortium , The Integrative Human Microbiome Project. Nature 569, 641–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald D., Hyde E., Debelius J. W., Morton J. T., Gonzalez A., Ackermann G., Aksenov A. A., Behsaz B., Brennan C., Chen Y., Goldasich L. D. R., Dorrestein P. C., Dunn R. R., Fahimipour A. K., Gaffney J., Gilbert J. A., Gogul G., Green J. L., Hugenholtz P., Humphrey G., Huttenhower C., Jackson M. A., Janssen S., Jeste D. V., Jiang L., Kelley S. T., Knights D., Kosciolek T., Ladau J., Leach J., Marotz C., Meleshko D., Melnik A. V., Metcalf J. L., Mohimani H., Montassier E., Navas-Molina J., Nguyen T. T., Peddada S., Pevzner P., Pollard K. S., Rahnavard G., Robbins-Pianka A., Sangwan N., Shorenstein J., Smarr L., Song S. J., Spector T., Swafford A. D., Thackray V. G., Thompson L. R., Tripathi A., Vázquez-Baeza Y., Vrbanac A., Wischmeyer P., Wolfe E., Zhu Q.; American Gut Consortium, Knight R., American Gut: An open platform for citizen science microbiome research. mSystems 3, e00031-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao D., Cardona C., Gottel N., Winton V. J., Thomas P. M., Raba D. A., Kelley S. T., Henry C., Gilbert J. A., Stephens B., Chemical composition of material extractives influences microbial growth and dynamics on wetted wood materials. Sci. Rep. 10, 14500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lax S., Cardona C., Zhao D., Winton V. J., Goodney G., Gao P., Gottel N., Hartmann E. M., Henry C., Thomas P. M., Kelley S. T., Stephens B., Gilbert J. A., Microbial and metabolic succession on common building materials under high humidity conditions. Nat. Commun. 10, 1767 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathi A., Vázquez-Baeza Y., Gauglitz J. M., Wang M., Dührkop K., Nothias-Esposito M., Acharya D. D., Ernst M., van der Hooft J. J. J., Zhu Q., McDonald D., Gonzalez A., Handelsman J., Fleischauer M., Ludwig M., Böcker S., Nothias L.-F., Knight R., Dorrestein P. C., Chemically-informed analyses of metabolomics mass spectrometry data with Qemistree. bioRxiv, 2020.05.04.077636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., Prill R. J., Tripathi A., Gibbons S. M., Ackermann G., Navas-Molina J. A., Janssen S., Kopylova E., Vázquez-Baeza Y., González A., Morton J. T., Mirarab S., Xu Z. Z., Jiang L., Haroon M. F., Kanbar J., Zhu Q., Song S. J., Kosciolek T., Bokulich N. A., Lefler J., Brislawn C. J., Humphrey G., Owens S. M., Hampton-Marcell J., Berg-Lyons D., McKenzie V., Fierer N., Fuhrman J. A., Clauset A., Stevens R. L., Shade A., Pollard K. S., Goodwin K. D., Jansson J. K., Gilbert J. A., Knight R.; Earth Microbiome Project Consortium , A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez A., Navas-Molina J. A., Kosciolek T., McDonald D., Vázquez-Baeza Y., Ackermann G., DeReus J., Janssen S., Swafford A. D., Orchanian S. B., Sanders J. G., Shorenstein J., Holste H., Petrus S., Robbins-Pianka A., Brislawn C. J., Wang M., Rideout J. R., Bolyen E., Dillon M., Caporaso J. G., Dorrestein P. C., Knight R., Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amir A., McDonald D., Navas-Molina J. A., Kopylova E., Morton J. T., Zech Xu Z., Kightley E. P., Thompson L. R., Hyde E. R., Gonzalez A., Knight R., Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton J. T., Marotz C., Washburne A., Silverman J., Zaramela L. S., Edlund A., Zengler K., Knight R., Establishing microbial composition measurement standards with reference frames. Nat. Commun. 10, 2719 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald D., Kaehler B., Gonzalez A., DeReus J., Ackermann G., Marotz C., Huttley G., Knight R., redbiom: A rapid sample discovery and feature characterization system. mSystems 4, e00215-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji P., Rhoads W. J., Edwards M. A., Pruden A., Impact of water heater temperature setting and water use frequency on the building plumbing microbiome. ISME J. 11, 1318–1330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Brien S. L., Gibbons S. M., Owens S. M., Hampton-Marcell J., Johnston E. R., Jastrow J. D., Gilbert J. A., Meyer F., Antonopoulos D. A., Spatial scale drives patterns in soil bacterial diversity. Environ. Microbiol. 18, 2039–2051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reese A. T., Savage A., Youngsteadt E., McGuire K. L., Koling A., Watkins O., Frank S. D., Dunn R. R., Urban stress is associated with variation in microbial species composition-but not richness-in Manhattan. ISME J. 10, 751–760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouslimani A., da Silva R., Kosciolek T., Janssen S., Callewaert C., Amir A., Dorrestein K., Melnik A. V., Zaramela L. S., Kim J.-N., Humphrey G., Schwartz T., Sanders K., Brennan C., Luzzatto-Knaan T., Ackermann G., McDonald D., Zengler K., Knight R., Dorrestein P. C., The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 17, 47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amir A., McDonald D., Navas-Molina J. A., Debelius J., Morton J. T., Hyde E., Robbins-Pianka A., Knight R., Correcting for microbial blooms in fecal samples during room-temperature shipping. mSystems 2, e00199-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson E. L., Heaver S. L., Walters W. A., Ley R. E., Microbiome and metabolic disease: Revisiting the bacterial phylum Bacteroidetes. J. Mol. Med. 95, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dührkop K., Fleischauer M., Ludwig M., Aksenov A. A., Melnik A. V., Meusel M., Dorrestein P. C., Rousu J., Böcker S., SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Sohlenkamp C., Geiger O., Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Wang M., Jarmusch A. K., Vargas F., Aksenov A. A., Gauglitz J. M., Weldon K., Petras D., da Silva R., Quinn R., Melnik A. V., van der Hooft J. J. J., Caraballo-Rodríguez A. M., Nothias L. F., Aceves C. M., Panitchpakdi M., Brown E., Di Ottavio F., Sikora N., Elijah E. O., Labarta-Bajo L., Gentry E. C., Shalapour S., Kyle K. E., Puckett S. P., Watrous J. D., Carpenter C. S., Bouslimani A., Ernst M., Swafford A. D., Zúñiga E. I., Balunas M. J., Klassen J. L., Loomba R., Knight R., Bandeira N., Dorrestein P. C., Mass spectrometry searches using MASST. Nat. Biotechnol. 38, 23–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
References
Movie S1