Abstract
Immunity-related GTPases (IRGs), also known as p47 GTPases, are a family of interferon-inducible proteins that play roles in immunity defense against intracellular pathogens. Although the molecular functions of IRGs have been well studied, the function of the family member, IRGC1, remains unclear. IRGC1 is unique among IRGs because its expression is not induced by interferon and it is expressed predominantly in the testis. Further, IRGC1 is well conserved in mammals unlike other IRGs. Here, we knocked out (KO) Irgc1 in mice using the CRISPR/Cas9 system and found that the fertility of Irgc1 KO males was severely impaired because of abnormal sperm motility. Further analyses with a transmission electron microscope revealed that the fibrous sheath (FS), an accessory structure of the sperm tail, was disorganized in Irgc1 KO mice. In addition, IRGC1 was detected in the sperm tail and fractionated with FS proteins. These results suggest that IRGC1 is a component of the FS and is involved in the correct formation of the FS.
Keywords: CRISPR/Cas9, Fibrous sheath, Flagellum, p47 GTPases, Sperm motility
Graphical abstract
Highlights
-
•
IRGC1, a testis-enriched immunity related GTPase, is important for male fertility in mice.
-
•
Irgc1 KO mice exhibited impaired sperm motility.
-
•
Fibrous sheath structures were disrupted in Irgc1 KO spermatozoa.
-
•
IRGC1 fractionates with fibrous sheath proteins, suggesting that IRGC1 is a component of the fibrous sheath.
1. Introduction
Infertility is one of the most prevailing medical problem in the world and the prevalence of infertility is one out of every six couples of reproductive age (Agarwal et al., 2019). The distribution of infertility due to male factors varies between the regions of the world and is estimated to range from 20% to 70% (Agarwal et al., 2015). Male infertility can be caused by no or low sperm production (azoospermia or oligospermia, respectively), abnormal sperm morphology (teratospermia), and/or abnormal sperm motility (asthenospermia); however, most of the causes of abnormal sperm formation and/or function remain unclear.
The flagellum is a motility apparatus of the spermatozoa and can be divided into three parts, the midpiece where mitochondria are helically arranged around the outer dense fibers (ODFs) and axoneme, the principal piece that contains the fibrous sheath (FS) surrounding the ODFs and axoneme, and the end piece without accessory structures (Eddy et al., 2003). The FS consists of the longitudinal columns (LCs) that are localized adjacent to the doublets 3 and 8 of the axoneme and the rib that connects the two LCs. The FS serves not only as a structural component that provides elastic rigidity for sperm motility but also as a scaffold for several glycolytic and signaling molecules (Krisfalusi et al., 2006). Molecular components of the FS have been studied and A-kinase anchoring protein 3 (AKAP3) and 4 (AKAP4) have been identified as primary components of the FS (Fiedler et al., 2013). Abnormal formation of the FS could result in impaired sperm motility and male infertility. This condition is called a dysplasia of the fibrous sheath, which is currently recognized as morphological abnormalities of the sperm flagella (Nsota Mbango et al., 2019).
Immunity-related GTPases (IRGs) were first identified from their expression being induced strongly by interferon γ (IFNγ) (Taylor et al., 1996; Boehm et al., 1998) due to the presence of interferon-stimulated response elements (ISRE) and/or γ-activated sequence sites (GAS) in their promoter regions (Bekpen et al., 2005). Several IRGs have been reported to have critical functions in pathogen resistance in mice (Hunn et al., 2011). For instance, Irgm1, Irgm3, and Irgd KO mice exhibit significant susceptibility to the infection of a number of pathogens and there are functional distinctions among these genes (Taylor et al., 2007). Among the IRGs, Irgc1 is a clear exception in the family in terms of lacking both ISRE and GAS elements and its expression restricted to the testis (Bekpen et al., 2005); however, its function remains to be determined.
Here, we generated Irgc1 KO mice using the CRISPR/Cas9 system to analyze its function in vivo. Irgc1 KO mice exhibit abnormal FS morphology, impaired sperm motility, and subfertility, suggesting that IRGC1 plays roles in the correct formation of the FS.
2. Results
2.1. Irgc1 is expressed predominantly in the testis
By performing Protein Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis of mouse IRGC1, other IRGs that have similar sequences to IRGC1 were identified. The phylogenetic relationship of these proteins is shown in Fig. 1A. Despite Irgc1 having close relationships with other Irgs, expression of mouse Irgc1 and its human ortholog, IRGC, is not induced by interferon (Bekpen et al., 2005). IRGs exhibit higher sequence diversity within and between species (Hunn et al., 2011), with the only exception being IRGC1, which is highly conserved between humans and mice (Fig. S1A). Further, Irgc1, but not other Irgs, is expressed predominantly in the mouse testis (Fig. 1A). We performed RT-PCR using multiple tissues of adult mice and confirmed that Irgc1 is expressed predominantly in the testis (Fig. 1B). The first cycle of spermatogenesis in mouse seminiferous tubules completes within the first 35 days after birth (Kluin et al., 1982). To investigate temporal expression of Irgc1 in mouse spermatogenesis, we conducted RT-PCR using postnatal testes of mice from different ages. This analysis reveals that Irgc1 is expressed from around postnatal three weeks, when round spermatids are observed (Fig. 1C). By analyzing the single RNA sequence database (Hermann et al., 2018), we confirmed Irgc1 expression in spermatids in both mice and humans (Figs. S1B and C). These results suggest that IRGC1 may play roles in late spermatogenesis and/or fertilization.
Fig. 1.
Irgc1 is a testis-enriched gene.
(A) Phylogenetic analyses of IRGC paralogues. Z-scores of averaged gene expression values across mouse adult tissues (Li et al., 2017) are shown on the right. There are no RNA-seq data about Iigp1b. Irgc1 is expressed predominantly in the testis. Br: brain, Th: thymus, Lu: lung, He: heart, Li: liver, Sp: spleen, Ki: kidney, Te: testis, Ut: uterus, and Ov: ovary. (B) RT-PCR of Irgc1 and Iigp1b in mouse adult tissues. Actb as control. (C) RT-PCR of Irgc1 using RNAs obtained from mouse at various postnatal testes. Actb as control. (D) CRISPR/Cas9 targeting scheme. White boxes indicate untranslated regions while black boxes indicate protein coding regions. There are three variants in Irgc1. (E) Genotyping of Irgc1 mutant mice. Fw #1-Rv #1 and Fw #1-Rv #2 primers in Fig. 1D were used.
2.2. Generation of Irgc1 knockout mice
To uncover the function of Irgc1 in vivo, we generated Irgc1 KO mice using the CRISPR/Cas9 system. Using an in silico search (www.ensembl.org), we found that Irgc1 has three splice variants. Two guide RNAs (gRNAs) were designed to remove the entire open reading frame of these variants (Fig. 1D). Ribonucleoprotein complex containing the gRNA and CAS9 was then electroporated into 50 pronuclear embryos and 40 two-cell embryos were transferred into the oviducts of pseudopregnant females. Twelve of the 17 pups had large deletions, and one was caged with two wild-type (WT) females to obtain the F1 generation. Subsequent mating resulted in a KO mouse with a 1266 bp deletion (Fig. S1D), which was confirmed by PCR (Fig. 1E). No overt abnormalities were found in the KO mice.
2.3. Fertility of Irgc1 KO males is severely impaired
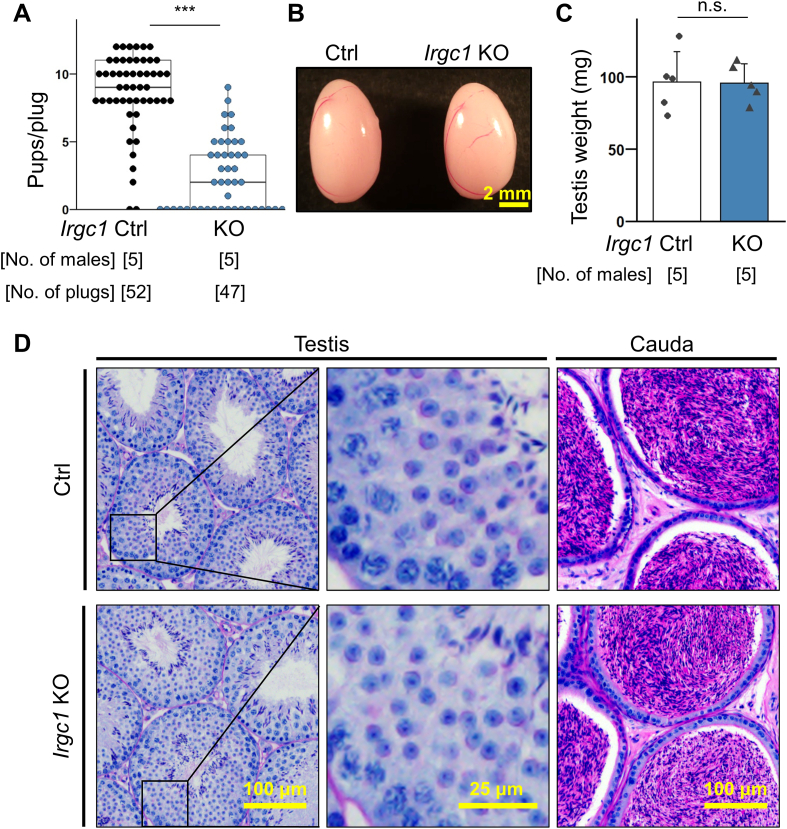
To examine whether Irgc1 is required for male fertility, individual WT or KO males were caged with WT females over a period of three months. During the course of the mating test, KO males showed severe subfertility (Fig. 2A). In order to investigate the cause of subfertility observed in Irgc1 KO mice, we performed morphological and histological analysis of the testes. First, no significant differences were found in the size and appearance of the testes between the control and Irgc1 KO mice (Fig. 2B and C). In addition, Periodic acid–Schiff (PAS) staining of testis cross-sections revealed no apparent defects in spermatogenesis (Fig. 2D). Further, PAS staining of the cauda epididymis showed that the tubules were filled with spermatozoa even in Irgc1 KO mice (Fig. 2D).
Fig. 2.
IRGC1 is important for male fertility.
(A) Number of pups born per plug detected. Three WT females were caged with each male for three months. Average pups/plug was Ctrl = 8.65 ± 2.88 pups/plug; Irgc1 KO = 2.38 ± 2.63 pups/plug. P = 1.88 × 10−19. (B) Gross morphology of control and Irgc1 KO testes. (C) Testis weight of control and Irgc1 KO mice. Average weight of testis was Ctrl = 96.30 ± 20.81 mg; Irgc1 KO = 95.54 ± 13.37 mg. P = 0.945. (D) PAS staining of testis and cauda epididymis sections.
2.4. IRGC1 is required for sperm tail formation and motility
We observed mature spermatozoa collected from the cauda epididymis. Intriguingly, some Irgc1 KO spermatozoa exhibit short tails (Fig. 3A and B; Fig. S1E). Because no abnormalities were found in the midpiece (Fig. 3A), it is likely that the tips of the tails were shortened. Consistent with abnormal flagellar morphology, we found that the percentage of motile spermatozoa was significantly lower in Irgc1 KO mice than that of the control mice using computer-assisted sperm analysis (CASA) (Fig. 3C; Movies S1,S2). Further analyses showed that progressive motility and velocity parameters such as average path velocity (VAP), straight line velocity (VSL), and curvilinear velocity (VCL) were significantly impaired in Irgc1 KO mice after both 10 and 120 min incubation in a capacitating medium (Fig. 3C; Figs. S2A and B). These results suggest that abnormal sperm flagellar morphology and motility result in male subfertility.
Fig. 3.
Irgc1 KO spermatozoa exhibit abnormal tail morphology and impaired motility.
(A) Observation of spermatozoa obtained from cauda epididymis. White arrowheads indicate short tails. (B) Analysis of sperm tail length. Irgc1 KO spermatozoa exhibit short tails. Average length of sperm tail was Ctrl #1 = 119.82 ± 3.15 μm, Ctrl #2 = 118.28 ± 4.44 μm, Ctrl #3 = 120.44 ± 3.62 μm; Irgc1 KO #1 = 92.65 ± 21.46 μm, Irgc1 KO #2 = 100.40 ± 25.22 μm, Irgc1 KO #3 = 103.05 ± 22.93 μm. (C) Sperm motility was analyzed 10 min and 120 min after incubation in a capacitation medium. Average percentage of motile spermatozoa was Ctrl = 87.57 ± 6.91%, Irgc1 KO = 31.43 ± 15.41% for 10 min (P = 0.013); Ctrl = 73.33 ± 7.07%, Irgc1 KO = 30.03 ± 10.54% for 120 min (P = 0.0062). Percentage of spermatozoa with progressive motility was Ctrl = 70.90 ± 14.91%, Irgc1 KO = 15.70 ± 4.54% for 10 min (P = 0.0036); Ctrl = 59.27 ± 7.40%, Irgc1 KO = 10.10 ± 1.71% for 120 min (P = 0.00036). VAP (average path velocity) was Ctrl = 168.37 ± 20.79 μm/s, Irgc1 KO = 76.46 ± 6.99 μm/s for 10 min (P = 0.010); Ctrl = 159.79 ± 6.98 μm/s, Irgc1 KO = 72.95 ± 11.88 μm/s for 120 min (P = 0.0012). (D) IVF analyses using cumulus-intact oocytes. Ctrl = 87.34 ± 11.99%; Irgc1 KO = 22.39 ± 27.80%. P = 0.012. (E) IVF analyses using cumulus-free oocytes. Ctrl = 72.50 ± 21.89%; Irgc1 KO = 13.31 ± 19.97%. P = 0.0073. (F) IVF analyses using zona pellucida (ZP)-free oocytes. Ctrl = 68.80 ± 23.65%; Irgc1 KO = 62.54 ± 22.23%. P = 0.71.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.ydbio.2022.05.011
The following is/are the supplementary data related to this article:
When we performed in vitro fertilization (IVF) using cumulus intact oocytes, fertilization rates were lower in Irgc1 KO mice (Fig. 3D) consistent with the mating exam (Fig. 2A). Removing cumulus cells could not rescue lower fertilization rates (Fig. 3E); however, when the zona pellucida, an extracellular matrix surrounding the oocyte, was removed, Irgc1 KO spermatozoa could fertilize oocytes at comparable rates to the control spermatozoa (Fig. 3F). These results suggest that Irgc1 KO spermatozoa can undergo the acrosome reaction and fuse with eggs, but cannot penetrate the zona pellucida likely due to impaired sperm motility.
2.5. IRGC1 is important for the FS formation
Because the short tails observed in Irgc1 KO spermatozoa are similar to Akap3 or Akap4 KO spermatozoa that exhibit not only short tails but also abnormal FS morphology (Xu et al., 2020; Miki et al., 2002), we observed the ultrastructure of the Irgc1 KO spermatozoa (Fig. 4A) using transmission electron microscopy (TEM). No differences were found in the mitochondrial sheath of the midpiece between the control and Irgc1 KO spermatozoa (Fig. 4B). In contrast, the cross-sections of principal pieces revealed that Irgc1 KO spermatozoa possessed disorganized FS (Fig. 4B and C; Fig. S3A). The longitudinal columns (LCs) that are localized adjacent to the doublet microtubules 3 and 8 were not thickened in Irgc1 KO spermatozoa (Fig. 4B). Further, the ribs that circumferentially connect LCs were torn (Fig. 4B). Besides the FS structure, abnormal numbers of microtubule doublets were observed in both midpiece and principal piece of Irgc1 KO spermatozoa (Fig. 4B,D; Figs. S3B,C,D) and even some KO spermatozoa exhibit the microtubule doublets outside of the FS (Fig. 4B). These abnormalities in microtubule doublets were more frequently observed in the distal part of the principal piece rather than the midpiece and the proximal part of the principal piece (Fig. 4D; Fig. S3E).
Fig. 4.
Irgc1 KO spermatozoa exhibit abnormal FS structures.
(A) Schematic drawing of the internal structures of mouse spermatozoa. Proximal and distal regions of the principal piece were defined as having 5–9 and 0–4 outer dense fibers, respectively. (B) Cross-sections of the midpiece and principal piece (proximal and distal regions). Extra microtubules are indicated by white asterisks. Disruption of LCs is indicated by white arrowheads. Torn FS is indicated by black arrows. Doublet microtubules located outside of the FS are indicated by black arrowheads. (C) Quantification of the morphological analysis of FS. The number of flagellar sections analyzed are shown above each bar. Average percentage of abnormal FS was Ctrl = 7.82 ± 5.79%, Irgc1 KO = 95.77 ± 3.70% for the proximal region (P = 8.5 × 10−5); Ctrl = 0.62 ± 1.07%, Irgc1 KO = 96.06 ± 4.40% for the distal region (P = 0.00038). (D) Quantification of axonemal structures (AX) with abnormal number of doublet microtubules (MTs). The number of flagellar sections analyzed are shown above each bar. Average percentage of AX with abnormal number of doublet MTs was Ctrl = 0.00 ± 0.00%, Irgc1 KO = 33.96 ± 4.89% for the midpiece (P = 0.0068); Ctrl = 0.00 ± 0.00%, Irgc1 KO = 43.70 ± 6.05% for the proximal region (P = 0.0063); Ctrl = 1.36 ± 2.36%, Irgc1 KO = 70.76 ± 12.02% for the distal region (P = 0.0080).
During spermatogenesis, LCs are formed before the ribs (Eddy et al., 2003). To examine whether the defects in the FS of Irgc1 KO spermatozoa are due to abnormalities in initial LC formation, we examined testicular spermatozoa using TEM. Cross-sections of step 9 Irgc1 KO spermatids revealed that LCs were formed correctly (Fig. S4), suggesting that the initial formation of the LCs was not disrupted. In contrast, thin LCs and torn ribs were observed in step 16 spermatids of Irgc1 KO mice, indicating that the FS formation was disrupted in later stages of spermatogenesis. Further, no abnormalities were found in the axoneme at step 9 or 16 (Fig. S4), suggesting that the displacement of the axoneme occurs at the very late stages of spermatogenesis or in the epididymis. Taken together, these results indicate that Irgc1 KO spermatozoa exhibit abnormal tail morphology and motility likely due to impaired FS formation.
2.6. IRGC1 is localized in the sperm tail
We performed immunoblotting analysis to confirm the deletion of IRGC1 in Irgc1 KO mice (Fig. 5A). A single band of approximately 50 kDa was detected in the control but was absent in the KO lysates, suggesting that the antibody works for immunoblotting. To analyze the localization of IRGC1 in mature spermatozoa, we tried immunofluorescence, but the antibody did not work. We then separated sperm heads and tails for immunoblotting analysis and detected the IRGC1 band in the tail fraction (Fig. 5B). Further, we performed a sperm fractionation assay as described previously (Cao et al., 2006; Castaneda et al., 2017) and found that IRGC1 is localized in the SDS-resistant fraction that contains FS and ODF proteins including AKAP4 (Fig. 5C). Considering that Irgc1 KO spermatozoa exhibit abnormal FS structures, these results suggest that IRGC1 is a component of the FS.
Fig. 5.
IRGC1 is localized in the sperm tail.
(A) IRGC1 was detected in the testis and cauda epididymal spermatozoa in the control, but not in Irgc1 KO mice. Acetylated tubulin as a loading control. Representative images of three experiments are shown. (B) Head and tail separation of mouse spermatozoa. IRGC1 is found in the tail fraction. IZUMO1 and acetylated tubulin were used as a marker for heads and tails, respectively. The experiment was done in duplicate. (C) Fractionation of the spermatozoa using different lysis buffers. IRGC1 was found in the SDS-resistant fraction that contains FS proteins. SLC2A3, acetylated tubulin, and AKAP4 were used as a marker for Triton-soluble, SDS-soluble, and SDS-resistant fraction, respectively. The experiment was done in duplicate. (D) Immunoblotting analysis of phosphorylated tyrosine residues (p-Tyr) in spermatozoa. Spermatozoa were incubated in a capacitation medium for 10 min and 120 min. Acetylated tubulin as a loading control. The experiment was done in duplicate. (E) No differences in the amounts of FS proteins were observed between control and Irgc1 KO spermatozoa. Acetylated tubulin as a loading control. (F) MS analysis of SDS-resistant fraction. IRGC1 was detected in the SDS-resistant fraction of the control mice, but not in Irgc1 KO mice. No differences were found in other FS proteins. Number of males = 3 for the control and 4 for Irgc1 KO.
During sperm incubation in a capacitation medium, several sperm proteins are tyrosine-phosphorylated (Visconti et al., 1995). Because tyrosine-phosphorylated proteins are predominantly localized in the FS (Zhao and Kan, 2019), we analyzed the tyrosine-phosphorylation status, but no differences were found between the control and Irgc1 KO spermatozoa (Fig. 5D). To determine whether the absence of IRGC1 affects the amount of other FS proteins, we performed immunoblotting analysis of AKAP3, AKAP4 and CABYR (Xu et al., 2020; Miki et al., 2002; Young et al., 2016), but no differences were observed (Fig. 5E). Further, we performed mass spectral analysis of the SDS-resistant fraction. Consistent with the immunoblotting result (Fig. 5C), we detected IRGC1 in the SDS-resistant fraction in the control, but not in the KO (Fig. 5F; Tables S1 and S2). No apparent differences were detected in other FS proteins including AkAP3, AKAP4, and CABYR. These results suggest that IRGC1 is not involved in the tyrosine-phosphorylation status or localization of major known FS components.
3. Discussion
IRGC1 was identified as a member of immunity-related GTPases, but its function was unclear especially because its expression was not induced by interferons (IFNs) (Bekpen et al., 2005). In this study, we knocked out Irgc1 in mice using the CRISPR/Cas9 system and found that Irgc1 KO males showed reduced fertility. Further analyses revealed that IRGC1 is important for normal sperm motility. Although it remains to be determined if IRGC1 plays direct roles in regulating sperm motility in mature spermatozoa, our results suggest that sperm motility is impaired due to abnormal FS formation in the testis. These results indicate that the function of IRGC1 is different from other IRGs that play roles in pathogen resistance.
Our EM analyses revealed that the formation of not only LCs and ribs but also the axoneme is disrupted in Irgc1 KO mice. Because disrupted axonemes were also found in Cabyr or Eno4 KO mice that exhibit impaired FS formation (Young et al., 2016; Nakamura et al., 2013), the abnormal axonemal structures can be a secondary effect of disrupted FS. This idea is supported by the fact that IRGC1 was detected in the SDS-resistant fraction that contains FS proteins, but not in the SDS-soluble fraction that contains axonemal proteins. Because the axoneme structures were normal at step 9 and 16 of spermiogenesis (Fig. S4), the displacement of the axoneme occurs after its formation likely due to abnormal FS structures. Because it is unlikely that extra doublet microtubules are generated de novo, the axoneme may be disrupted due to abnormal microtubule sliding, which could result in the axoneme with less or more microtubules in the cross section. Microtubule abnormalities were more frequently observed in the distal part of the principal piece (Fig. 4D; Fig. S3E). Because the FS is thinner and the number of ODF is less in the distal region, the axoneme structure may be less stabilized in this region. Disruption of the axoneme after its formation has also been reported in KO mice of other genes such as Dnah17 (Zhang et al., 2020) and Cfap97d1 (Oura et al., 2020), and more frequent microtubule abnormalities in the distal region were found in Dnah17 KO mice as well.
Involvement of IRGC1 in the FS formation is also supported by its conservation among species. For example, by searching the National Center for Biotechnology Information (NCBI) database, IRGC1 was not found in zebrafish or Xenopus whose spermatozoa have an axoneme but not FS although other IRGs were found in these species. In contrast, the number of IRGs were diminished or reduced in birds and humans likely due to dispensable function of IRGs in pathogen resistance in these species; however, IRGC1 is conserved well in these species whose spermatozoa possess FS. IRGs were found even in cephalochordates and suggested to play conserved immune functions in vertebrates and cephalochordates (Li et al., 2009). IRGC1 may be evolved later as spermatozoa acquire the FS in species such as reptiles, birds, and mammals.
AKAP3, AKAP4, and CABYR have been identified as proteins that comprise the FS. Absence of these proteins in spermatozoa results in disruption of the FS. For example, Akap4 KO spermatozoa show disrupted structures of both LCs and ribs (Miki et al., 2002). Akap3 KO spermatozoa show lack of ribs but not LCs (Xu et al., 2020). Further, Cabyr KO spermatozoa show abnormalities preferentially in ribs (Young et al., 2016). Recent report suggested that Akap3 or Akap4 KO mice exhibited a global change in the sperm proteome, especially, in proteins that are related to the FS (Xu et al., 2020). For example, both Akap3 and Akap4 KO spermatozoa exhibited significant decrease of ropporin 1 (ROPN1), ropporin 1-like (ROPN1L), and sperm autoantigenic protein 17 (SPA17) (Xu et al., 2020). However, we did not find significant differences in the amount of these proteins between the control and Irgc1 KO mice (Table S1). Consistent with fewer impacts on the proteome of the FS, the disruption of the FS is milder in Irgc1 KO mice than that of Akap3 or Akap4 KO mice. These results indicate that IRGC1 may play roles in fine-tuning the FS formation. Because our fractionation assay detected IGRC1in the fraction of FS proteins (Fig. 5C), IRGC1 may be a structural component of the FS. IRGC1 has a GTP-binding motif like other IRGs and analysis of this domain may lead to further functional elucidation of IRGC1.
In summary, our results reveal that IRGC1 is important for the formation of the FS, a unique accessory structure of the spermatozoa, which may give insight into the molecular evolution of IRGC1 that is apart from other IRGs. IRGC1 is conserved well in humans (Fig. S1A) and a variation in IRGC1 that causes a frameshift (p.Leu14AlafsTer19) was found (variant ID: 19-44222636-CCT-C, allele frequency: 0.0002060) (gnomAD). Further analysis of IRGC1 may provide a better understanding of the mechanism that regulates the FS formation and may lead to better treatment for male infertility, especially a dysplasia of the fibrous sheath.
4. Materials and methods
4.1. Animals
All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University. Mice were purchased from CLEA Japan (Tokyo, Japan) or Japan SLC (Shizuoka, Japan). WT or Irgc1 heterozygous mice were used as controls.
4.2. Phylogenetic analysis
IRGC1 paralogues in Mus musculus were identified using Protein Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the proteins with more than 30% identity are listed. Phylogenetic analysis was performed using the Neighbor-Joining method with MEGA X (Kumar et al., 2018). The evolutionary distances (the units of the number of amino acid differences per site) were computed using the p-distance method (Nei and Kumar, 2000).
4.3. Isolation of RNA and RT-PCR
Mouse adult multi-tissues and mouse testes at different ages were obtained from C57BL/6N mice. RNA samples were isolated and purified using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). RNA was reverse transcribed to cDNA using SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific) using an oligo (dT) primer. PCR was performed using primers that are listed in Table S3.
4.4. In silico expression data analysis
Single-cell transcriptome data in the mouse and human testis was obtained (Hermann et al., 2018). Irgc1 expression in those cells was analyzed using Loupe Cell Browser 3.3.1 (10X Genomics, Pleasanton, CA, USA).
4.5. Generation of Irgc1 KO mice using the CRISPR/Cas9 system
KO mice were generated as described previously (Abbasi et al., 2018). The gRNAs with fewer off-target sites were designed utilizing the online source CRISPRdirect (Naito et al., 2015). The gRNA target sequences were 5′- AGACTCAAAAGCAGTGCGCA -3′ and 5′- TTAGTGGAAAAGCGGAGCAC -3′. KO mice were maintained on a B6D2 background. Irgc1 mutant mice have been assigned labels (B6D2-Irgc1<em1Osb>) and deposited into the RIKEN BioResource Research Center (ID#: RBRC11221) and the Center for Animal Resources and Development (CARD), Kumamoto University (ID#: 3025).
4.6. Fertility test
For the in vivo fertility analysis, sexually mature KO male mice or control male mice were caged with three 8-week-old B6D2F1 female mice for three months and plugs were checked every morning. The number of pups was counted on the day of birth. For the in vitro fertility assay, in vitro fertilization (IVF) analysis was performed as previously described (Morohoshi et al., 2021) with some minor changes. For ZP-free oocytes, sperm insemination was performed at a final density of 2 × 104 spermatozoa/mL.
4.7. Histological analysis
PAS staining of sections were performed as previously described (Morohoshi et al., 2020). Testes or cauda epididymis were fixed at 4 °C in Bouin's solution (Polysciences, Inc., Warrington, PA, USA) and were processed for paraffin embedding. Paraffin sections were cut at a thickness of 5 μm using a HM325 microtome (Microm, Walldorf, Germany). After rehydrating the sections, they were stained with 1% periodic acid (Nacalai Tesque, Kyoto, Japan) and Schiff's reagent (FUJIFILM WakoPure Chemical, Osaka, Japan) for 20 min each at room temperature. The sections were then counterstained with Mayer hematoxylin solution (FUJIFILM WakoPure Chemical). The sections were observed with an Olympus BX-53 microscope (Tokyo, Japan).
4.8. Morphological and motility analysis of spermatozoa
Spermatozoa extracted from cauda epididymis were suspended in TYH medium (Muro et al., 2016). After 10 min incubation, spermatozoa were collected to observe morphology. Sperm motility was analyzed as previously described (Miyata et al., 2020, 2021). The motility of more than 200 spermatozoa was measured after incubation at 10 min and 120 min in TYH medium using CEROS II (software version 1.4; Hamilton Thorne Biosciences, Beverly, MA, USA). For movies, sperm motility was observed using an Olympus BX-53 microscope equipped with a high-speed camera (HAS-L1, Ditect, Tokyo, Japan).
4.9. Protein extraction from testes and spermatozoa
Testes or spermatozoa were homogenized in sample buffer containing 1 M Tris-HCL pH 6.8, 2% SDS, 10% Glycerol, and 0.005% Bromophenol Blue, and boiled for 5 min. Supernatants were obtained after centrifugation at 15,300×g for 15 min at 4 °C.
4.10. Immunoblot analysis
Immunoblot analysis was performed as described previously (Young et al., 2016). Samples were subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) under reduced conditions using 2-ME before transferred onto a polyvinylidene difluoride membrane. For phosphorylated tyrosine, anti-phosphorylated-tyrosine antibody clone 4G10 (1/1000, 05–321, Merck Millipore, Burlington, MA, USA); for IRGC1, anti-IRGC1 polyclonal antibody (1/1000, 14090-1-AP, ProteinTech, Rosemont, IL, USA); for AKAP3, anti-AKAP3 polyclonal antibody (1/1000, 13907-1-AP, ProteinTech); for AKAP4, anti-AKAP82 monoclonal antibody (1/1000, 611564, BD Transduction Laboratories, Franklin Lakes, NJ, USA); for SLC2A3, anti-SLC2A3 monoclonal antibody (1:1000, KS63-10, Fujihara et al., 2012); for IZUMO1, anti-IZUMO1 monoclonal antibody (1:1000, KS64-125, Ikawa et al., 2011); for acetylated tubulin, anti-acetylated tubulin monoclonal antibody (1:5,000, T7451, Sigma-Aldrich, St. Louis, MO, USA) were used. Immunoreactive proteins were detected using Chemi-Lumi One Super (Nacalai Tesque) or Chemi-Lumi One Ultra (Nacalai Tesque).
4.11. Sperm head-tail separation
Sperm head-tail separation was performed as previously described (Miyata et al., 2015) with some minor changes. Spermatozoa obtained from the cauda epididymis were suspended in 1 mL PBS and sonicated to separate tails from heads on ice (Sonifier SLPe, Branson Ultrasonics, Brookfield, CT, USA). The sample was centrifuged at 15,000×g for 15 min. The pellet was then resuspended with 200 μL PBS and mixed with 1.8 mL of 90% Percoll solution (GE Healthcare, Chicago, IL, USA) in PBS. After centrifugation at 15,000×g for 15 min, sperm heads were in the bottom of the tube and the tails were in the top layer of the solution. The separated sample was diluted with five-fold PBS and centrifuged at 10,000×g for 15 min. The pellets were washed twice with PBS and dissolved in sample buffer.
4.12. Fractionation of spermatozoa
Fractionation of spermatozoa obtained from the cauda epididymis was performed as described previously (Cao et al., 2006; Castaneda et al., 2017).
4.13. Mass spectral analysis of the SDS-resistant fraction
Spermatozoa from the cauda epididymis were collected and washed in PBS. In order to enrich the FS proteins, fractionation of spermatozoa was conducted as previously described (Cao et al., 2006; Castaneda et al., 2017). After the SDS-soluble fraction was removed, the pellet was dissolved in sample buffer and boiled for 5 min. LC-MS/MS was performed by the BIKEN mass spectrometry core at Osaka University, Japan. UniProt mouse database was used to match the detected proteins. Exponentially modified protein abundance index (emPAI), which enables us to estimate relative quantitation of proteins in a mixture (Ishihama et al., 2005), was calculated using Mascot (version: 2.7.0, Matrix Science, London, UK).
4.14. Observation of sperm flagellum ultrastructure using TEM
The sections of testes and cauda epididymis were prepared as previously described (Shimada et al., 2019). The prepared sections were observed using a JEM-1400 plus electron microscope (JEOL, Tokyo, Japan) at 80 kV with a CCD Veleta 2 K × 2 K camera (Olympus).
4.15. Statistical analysis
Statistical analyses were carried out using the two-tailed Welch's t-test using Microsoft Office Excel 2016 (Microsoft Corporation, Redmond, WA, USA). Differences were considered significant at P < 0.05 (∗), P < 0.01 (∗∗), P < 0.001 (∗∗∗). Error bars are standard deviation.
Funding
This research was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP21K19569, JP22H03214 to H.M., JP20K16107 to K.S., and JP19H05750, JP21H04753, JP21H05033 to M.I.); Takeda Science Foundation grant to H.M., K.S., and M.I.; the Japan Agency for Medical Research and Development (AMED) grant (JP21gm5010001 to M.I.); JST FOREST (JPMJFR211F to H.M.); the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01HD087157 and R01HD088412 to M.I.); and the Bill & Melinda Gates Foundation (Grand Challenges Explorations grant INV-001902 to M.I.).
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgements
The authors would like to thank Dr. Julio M. Castaneda for critical reading of the manuscript and Ms. Eri Hosoyamada and Ms. Natsuki Furuta for technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2022.05.011.
Contributor Information
Yuki Kaneda, Email: kaneda-y@biken.osaka-u.ac.jp.
Haruhiko Miyata, Email: hmiya003@biken.osaka-u.ac.jp.
Keisuke Shimada, Email: shimada-k@biken.osaka-u.ac.jp.
Yuki Oyama, Email: oyama-y@biken.osaka-u.ac.jp.
Rie Iida-Norita, Email: iida-r@biken.osaka-u.ac.jp.
Masahito Ikawa, Email: ikawa@biken.osaka-u.ac.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abbasi F., Miyata H., Shimada K., Morohoshi A., Nozawa K., Matsumura T., Xu Z., Pratiwi P., Ikawa M. RSPH6A is required for sperm flagellum formation and male fertility in mice. J. Cell Sci. 2018;131 doi: 10.1242/jcs.221648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Majzoub A., Parekh N., Henkel R. A schematic overview of the current status of male infertility practice. World J. Mens. Health. 2019;37:308–322. doi: 10.5534/WJMH.190068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Mulgund A., Hamada A., Chyatte M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015;13:1–9. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekpen C., Hunn J.P., Rohde C., Parvanova I., Guethlein L., Dunn D.M., Glowalla E., Leptin M., Howard J.C. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U., Guethlein L., Klamp T., Ozbek K., Schaub A., Fütterer A., Pfeffer K., Howard J.C. Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 1998;161:6715–6723. [PubMed] [Google Scholar]
- Cao W., Gerton G.L., Moss S.B. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics. 2006;5:801–810. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- Castaneda J.M., Hua R., Miyata H., Oji A., Guo Y., Cheng Y., Zhou T., Guo X., Cui Y., Shen B., Wang Z., Hu Z., Zhou Z., Sha J., Prunskaite-Hyyrylainen R., Yu Z., Ramirez-Solis R., Ikawa M., Matzuk M.M., Liu M. TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E5370–E5378. doi: 10.1073/pnas.1621279114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy E.M., Toshimori K., O'Brien D.A. Fibrous sheath of mammalian spermatozoa. Microsc. Res. Tech. 2003;61:103–115. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- Fiedler S.E., Dudiki T., Vijayaraghavan S., Carr D.W. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol. Reprod. 2013;88:1–10. doi: 10.1095/biolreprod.112.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara Y., Satouh Y., Inoue N., Isotani A., Ikawa M., Okabe M. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development. 2012;139:3583–3589. doi: 10.1242/dev.081778. [DOI] [PubMed] [Google Scholar]
- Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernströer B., Law N.C., Oatley M.J., Velte E.K., Niedenberger B.A., Fritze D., Silber S., Geyer C.B., Oatley J.M., McCarrey J.R. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667. doi: 10.1016/j.celrep.2018.10.026. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunn J.P., Feng C.G., Sher A., Howard J.C. The immunity-related GTPases in mammals: a fast-evolving cellautonomous resistance system against intracellular pathogens. Mamm. Genome. 2011;22:43–54. doi: 10.1007/s00335-010-9293-3. doi.org/10.1007/s00335-010-9293-3.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa M., Tokuhiro K., Yamaguchi R., Benham A.M., Tamura T., Wada I., Satouh Y., Inoue N., Okabe M. Calsperin is a testis-specific chaperone required for sperm fertility. J. Biol. Chem. 2011;286:5639–5646. doi: 10.1074/jbc.M110.140152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Kluin P.M., Kramer M.F., de Rooij D.G. Spermatogenesis in the immature mouse proceeds faster than in the adult. Int. J. Androl. 1982;5:282–294. doi: 10.1111/j.1365-2605.1982.tb00257.x. [DOI] [PubMed] [Google Scholar]
- Krisfalusi M., Miki K., Magyar P.L., O’Brien D.A. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Qing T., Zhu J., Wen Z., Yu Y., Fukumura R., Zheng Y., Gondo Y., Shi L. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhang J., Sun Y., Wang H., Wang Y. The evolutionarily dynamic IFN-inducible GTPase proteins play conserved immune functions in vertebrates and cephalochordates. Mol. Biol. Evol. 2009;26:1619–1630. doi: 10.1093/molbev/msp074. [DOI] [PubMed] [Google Scholar]
- Miki K., Willis W.D., Brown P.R., Goulding E.H., Fulcher K.D., Eddy E.M. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev. Biol. 2002;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- Miyata H., Abbasi F., Visconti P.E., Ikawa M. CRISPR/CAS9-mediated amino acid substitution reveals phosphorylation residues of RSPH6A are not essential for male fertility in mice. Biol. Reprod. 2020;103:912–914. doi: 10.1093/biolre/ioaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H., Oura S., Morohoshi A., Shimada K., Mashiko D., Oyama Y., Kaneda Y., Matsumura T., Abbasi F., Ikawa M. SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice. Proc. Natl. Acad. Sci. U.S.A. 2021;118:1–9. doi: 10.1073/pnas.2106673118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H., Satouh Y., Mashiko D., Muto M., Nozawa K., Shiba K., Fujihara Y., Isotani A., Inaba K., Ikawa M. Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science. 2015;350:442–445. doi: 10.1126/science.aad0836. 80. [DOI] [PubMed] [Google Scholar]
- Morohoshi A., Miyata H., Oyama Y., Oura S., Noda T., Ikawa M. FAM71F1 binds to RAB2A and RAB2B and is essential for acrosome formation and male fertility in mice. Development. 2021;148:1–12. doi: 10.1242/dev.199644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi A., Miyata H., Shimada K., Nozawa K., Matsumura T., Yanase R., Shiba K., Inaba K., Ikawa M. Nexin-Dynein regulatory complex component DRC7 but not FBXL13 is required for sperm flagellum formation and male fertility in mice. PLoS Genet. 2020;16:1–21. doi: 10.1371/journal.pgen.1008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y., Hasuwa H., Isotani A., Miyata H., Yamagata K., Ikawa M., Yanagimachi R., Okabe M. Behavior of mouse spermatozoa in the female reproductive tract from soon after mating to the beginning of fertilization. Biol. Reprod. 2016;94:1–7. doi: 10.1095/biolreprod.115.135368. [DOI] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Dai Q., Williams J., Goulding E.H., Willis W.D., Brown P.R., Eddy E.M. Disruption of a spermatogenic cell-specific mouse enolase 4 (Eno4) gene causes sperm structural defects and male infertility. Biol. Reprod. 2013;88:1–12. doi: 10.1095/biolreprod.112.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- Nsota Mbango J.F., Coutton C., Arnoult C., Ray P.F., Touré A. Genetic causes of male infertility: snapshot on morphological abnormalities of the sperm flagellum. Basic Clin. Androl. 2019;29:1–8. doi: 10.1186/s12610-019-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura S., Kazi S., Savolainen A., Nozawa K., Castañeda J., Yu Z., Miyata H., Matzuk R.M., Hansen J.N., Wachten D., Matzuk M.M., Prunskaite-Hyyryläinen R. Cfap97d1 is important for flagellar axoneme maintenance and male mouse fertility. PLoS Genet. 2020;16:1–22. doi: 10.1371/JOURNAL.PGEN.1008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Kato H., Miyata H., Ikawa M. Glycerol kinase 2 is essential for proper arrangement of crescent-like mitochondria to form the mitochondrial sheath during mouse spermatogenesis. J. Reprod. Dev. 2019;65:155–162. doi: 10.1262/jrd.2018-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G.A., Feng C.G., Sher A. Control of IFN-γ-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases) Microb. Infect. 2007;9:1644–1651. doi: 10.1016/j.micinf.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Taylor G.A., Jeffers M., Largaespada D.A., Jenkins N.A., Copeland N.G., Vande Woude G.F. Identification of a novel GTPase, the inducibly expressed GTPase, that accumulates in response to interferon γ. J. Biol. Chem. 1996;271:20399–20405. doi: 10.1074/jbc.271.34.20399. [DOI] [PubMed] [Google Scholar]
- Visconti P.E., Bailey J.L., Moore G.D., Pan D., Olds-Clarke P., Kopf G.S. Capacitation of mouse spermatozoa. 1. Correlation between the capacitation state and protein tyrosine phosphorylation. Development. 1995;121:1129–1137. doi: 10.1242/dev.121.4.1129. [DOI] [PubMed] [Google Scholar]
- Xu K., Yang L., Zhang L., Qi H. Lack of AKAP3 disrupts integrity of the subcellular structure and proteome of mouse sperm and causes male sterility. Development. 2020;147 doi: 10.1242/dev.181057. [DOI] [PubMed] [Google Scholar]
- Young S.A.M., Miyata H., Satouh Y., Aitken R.J., Baker M.A., Ikawa M. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J. Cell Sci. 2016;129:4379–4387. doi: 10.1242/jcs.193151. [DOI] [PubMed] [Google Scholar]
- Zhang B., Ma H., Khan T., Ma A., Li T., Zhang H., Gao J., Zhou J., Li Y., Yu C., Bao J., Ali A., Murtaza G., Yin H., Gao Q., Jiang X., Zhang F., Liu C., Khan I., Zubair M., Hussain H.M.J., Khan R., Yousaf A., Yuan L., Lu Y., Xu X., Wang Y., Tao Q., Hao Q., Fang H., Cheng H., Zhang Y., Shi Q. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J. Exp. Med. 2020;217 doi: 10.1084/jem.20182365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kan F.W.K. Human OVGP1 enhances tyrosine phosphorylation of proteins in the fibrous sheath involving AKAP3 and increases sperm-zona binding. J. Assist. Reprod. Genet. 2019;36:1363–1377. doi: 10.1007/s10815-019-01502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.