This review aims to provide a comprehensive and critical overview of techniques used to measure protein–protein interactions in living plants.
Keywords: BiFC, FRET, FRET-APB, FRET-FLIM, in planta, in vivo, protein–protein interaction (PPI), splitLuc
Abstract
Molecular processes depend on the concerted and dynamic interactions of proteins, either by one-on-one interactions of the same or different proteins or by the assembly of larger protein complexes consisting of many different proteins. Here, not only the protein–protein interaction (PPI) itself, but also the localization and activity of the protein of interest (POI) within the cell is essential. Therefore, in all cell biological experiments, preserving the spatio-temporal state of one POI relative to another is key to understanding the underlying complex and dynamic regulatory mechanisms in vivo. In this review, we examine some of the applicable techniques to measure PPIs in planta as well as recent combinatorial advances of PPI methods to measure the formation of higher order complexes with an emphasis on in vivo imaging techniques. We compare the different methods and discuss their benefits and potential pitfalls to facilitate the selection of appropriate techniques by providing a comprehensive overview of how to measure in vivo PPIs in plants.
Introduction
In vivo protein–protein interaction (PPI) measurements are needed to understand the dynamic and complex interactions of proteins underlying a plethora of biological processes in all living organisms. The observed PPI can also help to decipher the function of the involved proteins of interest (POIs), bringing them into a wider biological context. In this review, we focus on several different techniques that are available for PPI measurements in planta, either in heterologous systems or in stably transformed plants.
Independent of the technique used to measure PPIs, several important prerequisites must be met for PPIs of two or more POIs to occur. Here, the most important prerequisite of PPI is the spatio-temporal co-localization of POIs at a subcellular level, for example at the plasma membrane in a certain tissue at a specific time during plant development. Depending on the plant species under investigation, information about a specific POI, for example gene expression, protein localization, and putative interaction domains, might already be available and can be utilized to design subsequent PPI experiments.
To determine whether two or more proteins are co-localized, the proteins can be visualized using different techniques such as immunolocalization with specific, fluorescent dye-labelled antibodies against the POI in fixed samples, or in living cells using genetically encoded fluorescent proteins (FPs) fused to the POI. Subsequently, (co-)localization can be assessed via fluorescence microscopy. For example, the most commonly used FP, enhanced green fluorescent protein (eGFP), is a 28 kDa protein that forms a cylinder-like structure 4.2 nm long and 2.4 nm wide (Hink et al., 2000). Therefore, even when using state-of-the-art fluorescence microscopes, a mere co-localization of two POIs is no proof that they interact physically, as the lateral diffraction limit of light means the maximum resolution attainable by light microscopy is only ~250 nm. Even when super-resolution microscopy methods are applied with resolutions in the range of ~30 nm, there is still uncertainty as to whether a PPI takes place between the POIs (Won, 2009). Therefore, a number of different techniques to test for direct PPI in vitro and in vivo have been developed (Struk et al., 2019). In this review, we will focus on some of the traditional and newly emerging PPI techniques, with a focus on in vivo imaging techniques in planta.
Traditionally, techniques such as co-immunoprecipitation (co-IP) or yeast two-hybrid (Y2H) experiments were used to measure PPI. These techniques allow for the identification of numerous potential interaction candidates in a short time and are thus regarded as relatively high throughput. Although the use of these methods results in the loss of spatial and temporal information about the POIs and their interaction partners, they can still serve as a starting point to identify potential interacting POIs, which can then be verified by subsequent in vivo PPI measurements.
Traditional methods
Co-immunoprecipitation
The in vitro method co-IP is still one of the most commonly used techniques to identify PPIs (Ren et al., 2003; Phee et al., 2006). During co-IP, an immobilized antibody against the POI isolates the POI from a cell lysate, along with other proteins that directly or indirectly interact with the POI (Ransone, 1995; Ren et al., 2003; Phee et al., 2006). These potential interaction partners can then be identified by mass-spectroscopy (MS) (as reviewed in Monti et al., 2005). Furthermore, the precipitated complex can be tested for a specific target protein that has been identified by other experiments. Additionally, the putative interaction partner can be subsequently confirmed and visualized by a western blot using an antibody against the identified complex partner.
However, this is also one of the major limitations of this technique, as protein-specific antibodies are, at least in plant biology, often not available. Therefore, it is necessary to label the POI with one of the common protein tags available (e.g. His, FLAG, HA, or FPs), which can then be detected. Another concern is that co-IP is not well suited to detect weak or transient interactions as the experimental procedure includes several washing steps, often with detergents, to eliminate non-specific binding. Also, detection of proteins via western blot requires sufficient protein expression, which can be problematic if endogenous promoters are used, which are often only weakly or transiently expressed, or only expressed in a few cells (as reviewed in Tang and Takahashi, 2018). Another major drawback of this technique is that spatial information of the POI is lost through lysis of the cells. Here, proteins that are not present in the same cellular compartments are released into the lysate and PPIs may take place, even if they would not normally come into contact with each other in an intact cell. Additionally, when using co-IP, it remains unclear whether the discovered interaction is direct or indirect (as reviewed in Masters, 2004). Therefore, false-positive results must be considered, and PPI must be confirmed using other techniques that conserve spatial information. Furthermore, false-negative results are also possible due to the need for the POI to be soluble, which, for example in the case of membrane proteins, is not always the case. Nevertheless, co-IP with subsequent MS offers the possibility to identify a multitude of novel interaction partners which can subsequently be confirmed using other techniques as described below.
Yeast two-hybrid
A widely used high-throughput in vivo technique is the Y2H system, which was originally designed to identify PPIs using the yeast GAL4 transcriptional activator. GAL4 consists of two functional domains, a DNA-binding domain and an activator domain (Fields and Song, 1989). In the Y2H system, the binding of GAL4 to the upstream activation sequence triggers the transcription of an enzymatic reporter gene, for example lacZ coding for β-galactosidase. Separation of these two domains and fusion to two different POIs allows testing of their interaction with an easy read-out, such as a colour reaction triggered by the addition of a suitable substrate for β-galactosidase. In recent decades, several other variations of this technique have been developed, for example in Arabidopsis thaliana protoplasts, named protoplast two-hybrid (Ehlert et al., 2006).
A Y2H screen can give an indication of whether a PPI could take place between two POIs, and it is easy to carry out without the need for any sophisticated equipment. In addition, Y2H offers the opportunity for large-scale high-throughput approaches and allows screening of thousands of proteins for potential PPIs, which has led to the availability of Y2H interactome databases (e.g. Arabidopsis Interactome Mapping Consortium, 2011 for A. thaliana). The analysis of such large networks of interacting proteins offers the opportunity to classify POIs into larger biological contexts and enables the discovery of novel hypothetical links and putative functions of POIs. Even though these databases, if available, are a very good starting point, one major limitation of the underlying Y2H technique is the high rate of false-positive, but also false-negative results. Here, estimates suggest 70% false-positive identifications, which are caused by the overexpression of the POI and the expression in a heterologous system (as reviewed in Deane et al., 2002; Auerbach and Stagljar, 2005). Nevertheless, high-throughput Y2H screens are very useful to identify many putative POIs that could interact, even though other promising in vivo high-throughput methods have been developed recently.
Biotin-based proximity labelling
Within the last few years, an enzyme-catalysed proximity labelling technique was developed in which biomolecules, usually proteins or RNA, are labelled if they are in close proximity to the POI (Roux et al., 2012). Here, a POI is fused to a ligase that covalently labels adjacent proteins or RNA with biotin. The biotinylated proteins can then be isolated using streptavidin which has a strong affinity for biotin. The putatively interacting proteins pulled-down in this way can then be identified by MS. The bifunctional BirA isolated from Escherichia coli is one of the best studied biotin ligases (Chakravartty and Cronan, 2012). Due to its high sequence specificity, BirA was not only used for protein isolation employing streptavidin, but also to analyse binary interactions. Fusing two POIs with BirA or BAP, a short biotin acceptor peptide, respectively, leads to biotinylation in the case of interaction of the POIs (as reviewed in Kim and Roux, 2016). Since the discovery of this useful mechanism, many improvements of biotin ligases have been developed. BioID is a mutated biotin ligase derived from BirA and works independently of BAP, leading to unspecific labelling of all nearby biomolecules. Recently another modification of BirA led to the development of the biotin ligase TurboID (Branon et al., 2018). It combines advantages of other commonly used proximity labelling enzymes, such as APEX2 and BioID, by enabling non-toxic, fast labelling and an increased catalytic efficiency.
To date, one of the major limitations of proximity labelling in plants has been that experiments had to be carried out at 37 °C due to the temperature requirements of the labelling enzyme, which could cause heat stress in plants. However, TurboID can be successfully used at room temperature in transiently and stably transformed Nicotiana benthamiana and stably transformed Arabidopsis, thereby avoiding unnecessary abiotic stress in the plants (Mair et al., 2019; Zhang et al., 2019). In contrast to co-IP approaches, proximity labelling is also suitable to detect low-affinity or transient interactions, due to the promiscuity of the biotin ligases and the strong biotin–streptavidin affinity (as reviewed in Kim and Roux, 2016). Additionally, the labelling of the putative interaction/complex partner occurs under native spatio-temporal conditions in vivo; only the subsequent identification takes place ex vivo. Nevertheless, some caveats must be considered even when TurboID or the smaller version, called miniTurbo, are used; for example, biotin might not be accessible to some organelles such as peroxisomes and vacuoles because of their acidic environments (Mair et al., 2019). Furthermore, unspecific background labelling can result in false-positive potential complex/interaction partners, and therefore appropriate negative controls must be included. In addition, as when using co-IP to monitor PPIs, isolated interaction partners must be identified by MS. Lastly, it must be taken into account that the labelling process requires a negatively charged amino acid on the surface of the protein, which could lead to false-negative results if negatively charged amino acid are not available in some POIs (as reviewed in Yang et al., 2021).
All of the three above-mentioned techniques can identify a multitude of different putatively interacting proteins. Nevertheless, these interactions must be verified individually by other PPI methods, and we will focus on these PPI methods in the following sections.
Shedding light on in vivo PPI measurements
In vivo visualization and quantification of PPIs has greatly profited during the past decades from the use of luminescent proteins and, notably, FPs that emit photons in a specific spectral range. The high spatio-temporal resolution at which FP-tagged POIs can be detected in vivo without the need for further substrates or cofactors has led to the application of a multitude of different quantitative methods involving FPs in planta (Grossmann et al., 2018). We will describe techniques that allow for in vivo PPI detection using fluorescence or bioluminescence and discuss potential benefits and pitfalls of the different applications. Next, we introduce two techniques to measure PPI by protein fragment complementation using either fluorescence or luminescence as a read-out.
Bimolecular fluorescence complementation (BiFC)
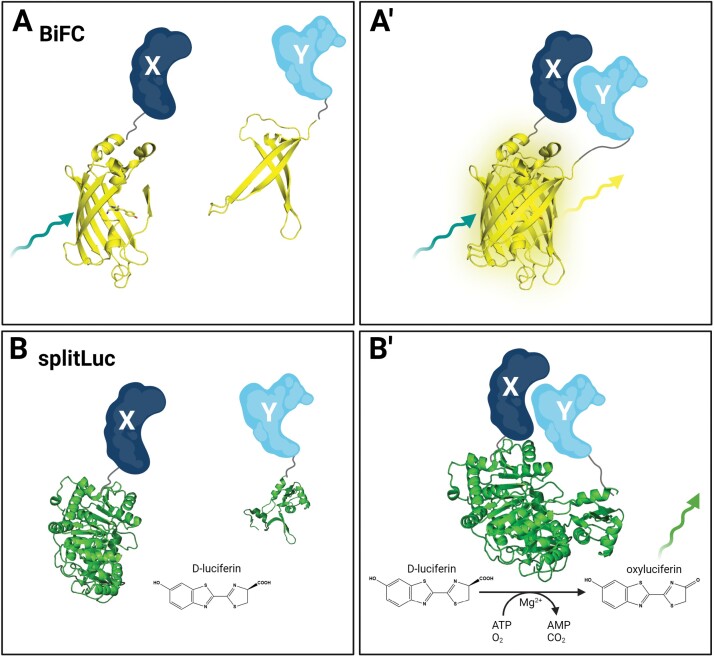
The in vivo BiFC assay is based on the complementation of an FP (Hu et al., 2002; Bracha-Drori et al., 2004). Here, the two POIs are fused to the N- or C-terminal part of the FP, respectively. If the two POIs are in close proximity to each other, the FP is reconstituted and its fluorescence restored, thus indicating interaction of the POIs (see Fig. 1B, Bʹ). This technique was first described using yellow fluorescent protein (YFP), but since then other split FPs have become available (Hu et al., 2002; Fan et al., 2008). This opens up the possibility to simultaneously visualize several PPIs within the same cell, known as multicolour BiFC (Hu and Kerppola, 2003; Waadt et al., 2008). The relatively simple principle of detecting interactions makes BiFC a widely used technique, since it does not require any specialized equipment other than a fluorescence microscope and appropriate filters for excitation and emission. Additionally, PPIs can be directly visualized in different cell compartments and, because of the cellular resolution of modern fluorescence microscopes or laser scanning microscopes, BiFC can provide information about the spatial characteristics of the investigated interaction. Because of its simplicity, many PPIs could be verified by BiFC in vivo (Table 2), for example in transient experiments in N. benthamiana and Arabidopsis leaf epidermal cells (Bracha-Drori et al., 2004), mustard seedlings, and also in protoplasts of Arabidopsis (Olejnik et al., 2011) and rice (Chen et al., 2006).
Fig. 1.
Split systems for measuring PPI in vivo. (A) In the bimolecular fluorescence complementation (BiFC) system, two POIs (X, dark blue; Y, light blue) are fused to the N- or C-terminal part of an FP (here, 3D structures of YFP in yellow), respectively. (Aʹ) If the two POIs interact, the two YFP parts are reconstituted and, after excitation (teal arrow), can emit light (yellow arrow). (B) In the split-luciferase (splitLuc) system, two POIs (X, dark blue; Y, light blue) are fused to the N- or C-terminal part of a luciferase (here, 3D protein structure of firefly luciferase in green). (Bʹ) If the two POIs interact, the luciferase fragments are reconstituted and can produce light (green arrow) in the presence of the substrate d-luciferin, in an ATP- and oxygen-dependent reaction. Figure created with BioRender.com.
Table 2.
Examples of PPI measured in planta
| Technique | Recent example and biological context | Organism | References |
|---|---|---|---|
| BiFC and FRET-FLIM | PII localizes to foci within chloroplasts where it interacts with NAGK and BCCP and thereby regulates protein degradation. | Transient expression in N. benthamiana | Krieger et al. (2021) |
| SplitLuc (firefly) | MKK2 and MPK2, both known to be involved in plant immunity, interact with ACO2 and with ACO2, CHH, and PBP1, respectively. | Transient expression in N. benthamiana | Leissing et al. (2021) |
| SplitLuc (firefly), BiFC, Y2H | OsUEV1B interaction with OsVDAC1 is suggested to be required for phosphate homeostasis in rice | Transient expression in N. benthamiana and Arabidopsis protoplasts | Liu et al. (2021) |
| FRET-APB | CRN and CLV2 interact at the plasma membrane, where they perceive CLV3 peptide and regulate stem cell number in the shoot apical meristem of A. thaliana. | Transient expression in N. benthamiana | Bleckmann et al. (2010) |
| FRET-FLIM | Cell type-specific interaction of the transcription factor network SHR–SCR–JKD regulates gene expression and thereby specifies cell fate in the A. thaliana root. | Stable expression in A. thaliana | Long et al. (2017) |
| FRET-FLIM, Y2H | OsFD7 involved in floral transition and panicle development in rice, was found to interact with OsFTL1, Hd3a, and RFT1. | Transient expression in onion peel cells | Kaur et al. (2021) |
| BRAVO and WOX5 interaction is involved in quiescent centre quiescence. Both transcription factors can interact with the BES1–TPL repressor complex. | Transient expression in N. benthamiana | Betegón-Putze et al. (2021) | |
| Triple FRET | BRI1 and BAK1 can form a trimeric complex with RLP44 which is thought to sense cell wall integrity in response to BR signalling. | Transient expression in N. benthamiana | Glöckner et al. (2020) |
| FRET-FLIM and homo-FRET | CLV1 and ACR4 form homo- and heteromeric complexes depending on their subcellular localization and thereby control distal root meristem maintenance in A. thaliana. | Transient expression in N. benthamiana | Stahl et al. (2013) |
| Cluster formation of CRN, CLV2, and CLV1 at the plasma membrane in presence of CLV3 regulates shoot meristem maintenance in A. thaliana. | Transient expression in N. benthamiana | Somssich et al. (2015) | |
| FCCS | Exogenous BR application leads to an increased co-localization of BRI1 and AtFlot1 and stimulation of the membrane microdomain-associated pathway of BRI1 internalization. Co-diffusion of BRI1 and CLC demonstrates that BRI1 internalization is clathrin dependent. | Stable expression in A. thaliana | Wang et al. (2015) |
| RICS and N&B | SHR was found to exist in a monomeric and dimeric state in the endodermis and both forms can interact with SCR, indicating a stoichiometric complex composition of 1:1 or 2:1. | Stable expression in A. thaliana | Clark et al. (2016) |
| KSP | Proof of concept as shown for the transcription factors: SACL and LHW; BES1 and BIN2 and the endocytosis protein complex: TPLATE and TML | Transient expression in N. benthamiana | Winkler et al. (2021) |
Nevertheless, one major limitation of BiFC is the high frequency of false-positive PPIs caused by the intrinsic affinity of the two parts of the FP for each other (see Table 1) (as reviewed in Horstman et al., 2014; Romei and Boxer, 2019). This is especially problematic when the expression is driven by a constitutive promoter in a heterologous system and thus the concentration of the proteins no longer reflects the endogenous expression level. In addition, expression in a heterologous system could also lead to false (co)-localization of proteins that, under native conditions, are localized in distinct compartments; for example, a protein expressed in the cytoplasm which is highly expressed in epidermal leaf cells of N. benthamiana could partially co-localize with a plasma membrane protein. Additionally, several attempts to diminish self-assembly, such as by changing the split position, have been made, but none of these approaches was generally applicable in plants (as reviewed in Horstman et al., 2014).
Table 1.
Evaluation of PPI techniques in planta involving imaging
| Method | Cellular resolution | Dynamics | False positives | False negatives | Applicability | Special features and characteristics |
|---|---|---|---|---|---|---|
| BiFC | ● | ○ | ● ● | ○ ● | ● ● | Suitable for weak/transient PPIs |
| Split-Luc | ○ | ● | ● | ● | ● a | Dynamic assembly and disassembly of PPIs can be studied |
| FRET-APB | ● | ● | ○ ● | ● | ● ● b | Fast data acquisition and analysis |
| FRET-FLIM | ● ● | ● | ○ ● | ● | ○ ● c.d | High quality of acquired data |
| BiFC and FRET | ● ● | ○ ● | ● ● | ● ● | ○ ● d,e | Analysis of trimeric complexes |
| Triple FRET | ● ● | ○ ● | ○ ● | ● ● | ○ ● c,d,e | Analysis of trimeric complexes |
| Homo-FRET | ● ● | ● ● | ○ ● | ● ● | ○ ● d | Analysis of higher order complexes Can be combined with FRET-FLIM |
| FCCS | ○ ● | ● ● | ○ | ○ ● | ○d | Low concentration of POI needed Simultaneous detection of PPI and dynamics |
| RICS and N&B | ○ ● | ● ● | ○ ● | ● | ● c | Simultaneous detection of PPI and dynamics |
| KSP | ○ ● | ○ ● | ○ ● | ○ ● | ○ ● f | Analysis of higher order complexes. Inducible visualization of PPI. Can be added to other methods as intrinsic positive control |
‘○’ = ‘no or low’, ‘○ ●’ = ‘medium’, ‘●’ = ‘high’, ‘● ●’ = ‘very high’.
a Phosphorescence of chlorophyll can mask the signal of luciferase.
b Not suitable for moving proteins.
c Data acquisition and analysis can be time consuming.
d Special technical equipment might be needed,
e Appropriate controls needed.
f Rapamycin-induced effects must be considered.
Therefore, appropriate negative controls are absolutely necessary to minimize false-positive results (Horstman et al., 2014). In addition, BiFC does not give any information about the temporal dynamics of PPI since the FP is highly unlikely to dissociate after reconstitution, as FPs have a half-life >24 h (Hu et al., 2002). On the other hand, this phenomenon can also be advantageous in detecting weak and/or transient PPIs (Morell et al., 2007). Recently, a split fluorescence reporter has been developed that also allows monitoring of assembly and disassembly of PPIs in living cells (Tebo and Gautier, 2019). To date, this reporter has only been applied in mammalian cell culture. Another in vivo protein fragment complementation assay based on a split-luciferase (splitLuc) can detect dynamic changes of PPI, and has been applied in plants; this is described next.
Split-luciferase
The in vivo splitLuc assay is based on the complementation of two fragments of a luciferase enzyme fused to two POIs that, in the case of interaction of the POIs, leads to enzyme reconstitution and subsequent substrate conversion. The turnover of the substrate, and therefore the interaction of the POIs, can be monitored by the emission of bioluminescence (Fig. 2A, Aʹ). In planta, two luciferases are most frequently used for splitLuc assays: one is obtained from the North American firefly (Photinus pyralis), referred to as firefly luciferase, and one from the sea pansy (Renilla reniformis), referred to as Renilla luciferase. The firefly luciferase uses d-luciferin as a substrate which is converted in a two-step ATP- and oxygen-dependent reaction to oxyluciferin, AMP, carbon dioxide, and light (de Wet et al., 1987). Interestingly, the emission spectrum of the firefly luciferase peaks at 560 nm (green light), but can undergo a red shift in an acidic environment or at higher temperatures (reviewed in Fraga, 2008). The Renilla luciferase converts its substrate coelenterazine in an oxygen-dependent reaction to coelenteramide, carbon dioxide, and blue light, with an emission maximum at 480 nm, and is ATP independent (Matthews et al., 1977). Compared with the firefly luciferase with an approximate mol. wt of 62 kDa, the Renilla luciferase is relatively small, measuring 37 kDa (Matthews et al., 1977; de Wet et al., 1987). The distinguishable emission spectra of these two different enzymes combined with a high substrate specificity allows for the simultaneous detection of the activity of both luciferases (McNabb et al., 2005).
Fig. 2.
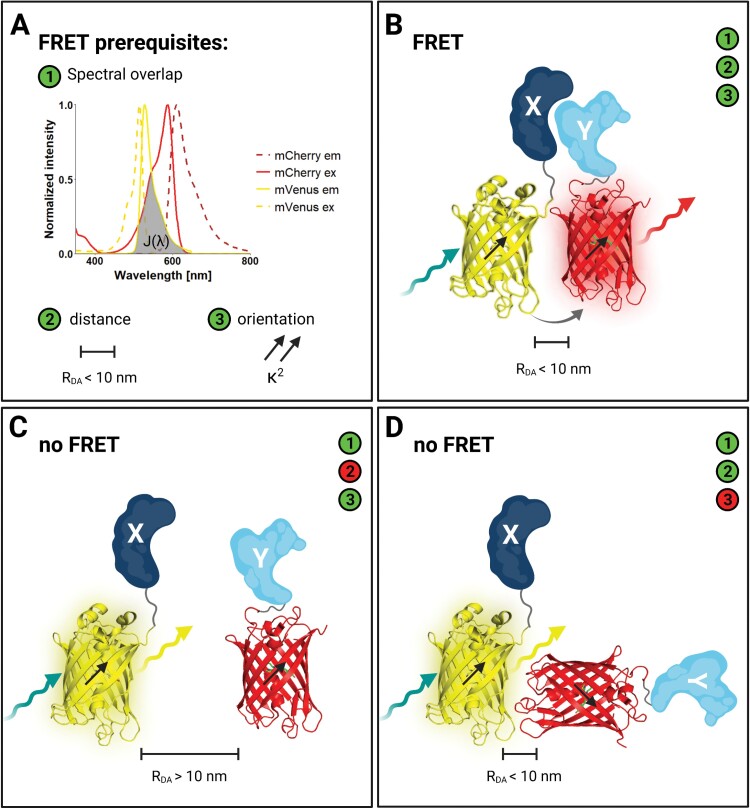
FRET prerequisites and possible scenarios when measuring PPI in vivo. (A) Prerequisites for FRET to take place between the two fluorophores or FPs of the chosen FRET pair, in this case the FPs mVenus (yellow) and mCherry (red), are: ① spectral overlap of donor emission and acceptor excitation [J(λ)]; ② distance between the donor and acceptor is <10 nm (RDA); ③ dipole orientation of the donor and acceptor are parallel (κ2). (B) PPI of two POIs can be measured by FRET in vivo if the donor FP (mVenus 3D structure in yellow) and the acceptor FP (mCherry 3D structure in red) are fused to POI X (dark blue) or Y (light blue), respectively. In the case that all three FRET prerequisites are met (①–③ green), FRET takes place by the energy migration after exciting (teal arrow) the donor FP to the acceptor FP (grey arrow) which is excited and can emit light (red arrow). (C) No FRET can be measured if the distance between donor and acceptor FPs (RDA) is >10 nm (red number ②). (D) No FRET can be measured if the dipole orientation (κ2) of donor and acceptor are not parallel (red number ③). Figure created with BioRender.com.
For both luciferases, a split variant is available for the detection of PPI (Ozawa et al., 2001; Paulmurugan and Gambhir, 2003; Fujikawa and Kato, 2007; Chen et al., 2008). These have been successfully utilized in different experimental approaches in plants, such as in transient expression in N. benthamiana (Liu et al., 2021) or in Arabidopsis protoplasts (Li et al., 2011) (see Table 2). Which luciferase is more suitable for the detection of PPI in vivo strongly depends on the characteristics of the putative interaction partners. For example, the enzymatic reaction catalysed by Renilla luciferase does not require ATP, which might be limiting in some cellular compartments. On the other hand, the substrate coelenterazine is unstable and can undergo spontaneous oxidation (Zhao et al., 2004), whereas d-luciferin is more stable.
Compared with other protein fragment complementation assays, luciferase fragments do not spontaneously reconstitute, thus avoiding false-positive interactions often encountered in BiFC. Furthermore, splitLuc assays enable the investigation of dynamic changes of PPIs since the reconstitution of the firefly luciferase is reversible, as shown in large-scale approaches in Arabidopsis mesophyll protoplasts (Table 1) (Li et al., 2011). The high turnover rate of reconstituted luciferase and its short half-life time allow the visualization of formation and dissociation of protein complexes (Luker et al., 2004; Xing et al., 2016). Although the splitLuc assays require the addition of a substrate, it can be easily applied exogenously, either by incubation or by watering (Chen et al., 2008), or even through infiltration of the diluted substrate (Schatlowski et al., 2010).
Despite its advantages, splitLuc assays are less commonly used than other protein fragment complementation assays. For one thing, luciferase activity can only be measured in the dark. Therefore, light-dependent processes are not suitable for this technique. Furthermore, for splitLuc assays in green tissue, for example tobacco leaves or Arabidopsis leaf protoplasts, a special filter is needed that excludes the phosphorescence of Chl a and b (reviewed in Krasnovsky and Kovalev, 2014) as this could interfere with the detection of the emitted light from the luciferase. Although splitLuc experiments can be used in quantitative high-throughput assays, such as on floating leaf discs or on protoplasts using plate readers (Chen et al., 2006; Gehl et al., 2011), these experiments do not provide information on the subcellular localization of the PPI and are not suitable for low-affinity interactions (reviewed in Xing et al., 2016). Another concern is that compared with commonly used fluorescent proteins, such as GFP or mCherry, the firefly luciferase is particularly large, which could cause problems when monitoring interactions of small proteins as this could sterically hinder PPIs (Matthews et al., 1977; de Wet et al., 1987; Cormack et al., 1996; Shaner et al., 2004). Recently, a smaller so-called Nano luciferase (Nluc) with a total molecular mass of 19.1 kDa was established to address this concern (Wang et al., 2020).
Nonetheless, neither BiFC nor splitLuc assays can provide information on whether the interaction of the proteins is direct or indirect or whether an interaction could be considered strong or weak, thereby making the quantification of PPI of different interaction partners impossible. Additionally, either spatial or temporal resolution of PPI is lost in splitLuc or BiFC experiments, respectively. Therefore, other quantitative techniques to measure in vivo PPI that preserve the spatio-temporal resolution are additionally required and will be discussed in the next sections.
Measurement of PPI by energy transfer
Förster resonance energy transfer (FRET)
FRET, first described by Theodor Förster (Förster, 1948), is a physical phenomenon in which the energy of an excited donor fluorophore is transferred by a radiation-free process to an acceptor chromophore, for example chlorophyll in the light-harvesting complexes necessary for plant photosynthesis. This process of energy transfer is strongly dependent on the distance between donor and acceptor fluorophores (RDA) (see Fig. 2A, C), the overlap integral of donor emission and acceptor absorption spectra [J(λ)] (see Fig. 2A), as well as the parallel orientation of the donor and acceptor dipoles (κ2) (see Fig. 2A, D). FRET efficiency (EFRET) is directly dependent on the distance between the donor and acceptor fluorophores, as it is inversely proportional to the sixth power of the distance between donor and acceptor, and can be described by the following equation:
Here, RDA represents the actual distance between the two fluorophores, and R0 the so-called Förster distance between the two fluorophores, a characteristic distance between a pair of fluorophores at which the FRET efficiency (EFRET) is 50%, which is usually well below 10 nm. For PPI measurements, two POIs are fused to suitable donor or acceptor fluorophores or FPs, also called FRET pairs. Widely used FP FRET pairs are: eCFP–eYFP, eGFP–mRFP, eGFP–mCherry, or mVenus–mCherry. The Förster radius (R0) of the FRET pair eCFP–eYFP is 4.9 nm (Bajar et al., 2016). Because of its strong distance dependency, FRET can be utilized to quantitatively determine PPI in vivo. To figure out which FRET pair is most suitable to detect PPI in a specific in planta experiment can be quite challenging, as it depends on several aspects, such as spectral properties, photostability, folding, localization, and activity of the labelled POI within the respective cellular context (Denay et al., 2019). The development of new FPs or new variants of already established FPs starts with analysing important characteristics in vitro, such as photostability, pH stability, and maturation time (reviewed in Day and Davidson, 2009). For some FPs, these attributes have been at least partially characterized in vivo, albeit mostly in bacteria (Megerle et al., 2008; Hebisch et al., 2013; Balleza et al., 2018) and also in yeast or mammalian cell culture (Zhong et al., 2019; Lee et al., 2020). Therefore, when starting with a recently developed FP, its applicability in plants first needs to be tested, as its properties can vary quite significantly in comparison with published results from other organisms (Denay et al., 2019).
Furthermore, when choosing a suitable FRET pair for PPI experiments, the maturation time of the individual fluorophores should also be considered. The specific maturation time of an FP can vary between different species or even within different strains of the same species, as shown for E. coli (Hebisch et al., 2013). As higher amounts of acceptor increase the possibility for FRET to occur, an equal or shorter maturation time for the acceptor fluorophore compared with the donor fluorophore is preferred (Bajar et al., 2016; Denay et al., 2019).
One important property of FRET is that it also affects the mean fluorescence intensity and lifetime of the donor fluorophore, as the fraction of excited donor fluorophores is depopulated faster in the presence of a suitable acceptor fluorophore in close proximity (Bücherl et al., 2014; Weidtkamp-Peters and Stahl, 2017). The resulting decrease of donor fluorescence intensity and lifetime, also called quenching, and the consequent increase of acceptor fluorescence can be measured by different approaches.
Donor and acceptor fluorescence intensities can be simultaneously measured by either acquiring a complete spectrum covering the donor and acceptor emission or by using appropriate filter sets for acceptor and donor fluorescence, known as sensitized emission. These techniques are often applied in genetically encoded FRET-based biosensors, detecting changes of intramolecular FRET in response to specific biological stimuli, such as calcium, reactive oxygen species, pH, phytohormones, and nutrients (Walia et al., 2018).
The two techniques most widely used for quantitative measurements of PPI by FRET are acceptor photobleaching (APB) and fluorescence lifetime imaging microscopy (FLIM), which we describe in more detail below.
Acceptor photobleaching (APB)
One of the most accessible FRET-based techniques to monitor PPI in vivo is acceptor APB. APB makes use of the differences in fluorescence intensity of the donor molecule in the presence or absence of the acceptor. Here, if FRET takes place, the energy transfer from the donor to the acceptor fluorophore is inhibited by bleaching the acceptor with a strong laser pulse (Bastiaens and Jovin, 1996; Bastiaens et al., 1996; Wouters et al., 1998; Kenworthy and Edidin, 1999; Kenworthy, 2001; Karpova et al., 2003; Albertazzi et al., 2009). Therefore, if the two POIs interact, bleaching of the acceptor leads to an increase of the donor fluorescence intensity because the energy is no longer transferred to the acceptor. This technique has successfully been applied in planta, for example in transiently expressing N. benthamiana leaf epidermal cells expressing different receptor proteins (Bleckmann et al., 2010). An apparent FRET efficiency (EFRET, as a percentage) can be calculated if the intensity of the donor fluorescence (ID) is recorded before (ID before) and after (ID after) bleaching of the acceptor and is described by the following equation:
This method does not require expensive or complicated equipment, and does not need time-consuming training or extensive experience (see Table 1). Additionally, data acquisition is relatively fast compared with other FRET-based methods. Drawbacks of FRET-APB are the need for high laser powers during bleaching of the acceptor fluorophore, potentially leading to phototoxic effects, as well as low spatial resolution due to the necessary high acquisition speed and the analysis of only a small region of interest. Furthermore, filter sets and/or bandwidths should be carefully chosen to avoid possible crosstalk of donor and acceptor emission.
Another point to consider is that FRET-APB utilizes the intensity of the donor molecule fluorescence to calculate FRET efficiency and therefore is strongly affected by protein concentration. As a rule, a low donor/acceptor ratio will lead to increased FRET efficiencies whereas a high donor/acceptor ratio decreases FRET efficiency as the acceptor may be limiting. On the other hand, high expression levels of both proteins increase the possibility of the donor and acceptor fluorophore meeting by chance and could thereby artificially increase FRET efficiency. This phenomenon is known as bystander-FRET and should be taken into consideration for all FRET-based techniques. To avoid strongly differing POI concentrations in transient expression systems, both POIs can be expressed from a single T-DNA so that the preferable 1:1 ratio of donor and acceptor is achieved (Mehlhorn et al., 2018; Denay et al., 2019). Another point is, that depending on the cellular compartment and the mobility of the protein, the fluorescence of the acceptor could recover after the bleaching, even before an increase of the donor intensity can be detected. This effect might be enhanced by the inevitable delay between pre-bleach and post-bleach image acquisition. Therefore, highly mobile POIs and/or transient PPIs would be difficult to measure using APB. Another quantitative method to measure FRET overcoming some of the shortcomings of FRET-APB is FLIM, which is described next.
Fluorescence lifetime imaging microscopy (FLIM)
Fluorescence lifetime (τ) is defined as the average time, usually in the nanosecond range, that a fluorophore remains in the excited state after excitation before returning to the ground state by emitting a photon. If two proteins interact, the fluorescence lifetime of the donor is decreased, and its fluorescence intensity is quenched. The fluorescence lifetime is an intrinsic characteristic for each fluorophore and therefore strongly differs between different fluorophores. FRET-FLIM is a non-intensity-based imaging method in contrast to the above-described FRET-APB, largely independent of protein concentration, making it particularly suitable for the quantitative analysis of PPIs in living cells, where fluorescence intensity can vary significantly. In order to measure the fluorescence lifetime of an FP in the so-called time domain, a pulsed laser source and special equipment for time-correlated single photon counting (TCSPC) is required: single photon-sensitive detectors and photon counting electronics (Becker et al., 2004). The time between the laser pulse and emission of a single photon is measured for every individual photon and plotted as a histogram, which shows an exponential decay. From fitting this decay, the average fluorescence lifetime can be deduced (Biskup et al., 2007; Weidtkamp-Peters and Stahl, 2017). The resulting FRET efficiency can be determined by recording the fluorescence lifetime τ of the POI labelled with the donor fluorophore in the absence (donor only sample) or presence of the putative interactor labelled with the acceptor fluorophore (FRET sample). The following equation describes the resulting FRET efficiency (EFRET, as a percentage) depending on the donor fluorescence lifetime in the presence (τDA) or absence (τD) of the acceptor:
One major advantage of FRET-FLIM is the quality of the acquired data. In contrast to other methods to investigate PPIs, FLIM data can also be used to determine quantitative data that can potentially show differences in the interaction strength or binding of different POIs at a high spatial resolution (see Table 1) (Weidtkamp-Peters and Stahl, 2017). Nevertheless, this requires some expert knowledge and advanced data analyses, as well as careful control experiments. Additionally, FLIM is largely independent of protein concentration. Therefore, FRET-FLIM measurements are widely considered as more reliable than the detection of FRET by APB and spectral imaging, and have been successfully applied in planta (Stahl et al., 2013; Bücherl et al., 2014; Somssich et al., 2015; Long et al., 2017; Weidtkamp-Peters and Stahl, 2017; Betegón-Putze et al., 2021; Kaur et al., 2021).
On the other hand, time domain FLIM data acquisition and processing are time-consuming and require a significant amount of training and experience. Furthermore, the necessary equipment, such as pulsed laser sources, TCSPC electronics, etc. are a quite expensive additions to a widefield or confocal microscope setup. An alternative that does not require cost-intensive TCSPC electronics and fitting of the data is the so-called frequency domain FLIM. Here, a continuous, modulated light source and a synchronized modulated detector are required to determine the phase-shifted fluorescent lifetimes of the donor fluorophore. Frequency domain FLIM measurements can be carried out at high speed, which is advantageous for monitoring dynamic processes. Nevertheless, time domain FLIM measurements show a higher precision even at low signal to noise ratios and can be used for more complex donors (Datta et al., 2020).
A more general limitation of FRET-based methods, as in FRET-APB and FRET-FLIM, is the number of false-negative results resulting from inadequate photoselection of fluorophores (Bajar et al., 2016). As mentioned, the dipole orientation (κ2) of the two fluorophores used for FRET should be (close to) parallel (Fig. 2). The more precisely the dipole orientations of the fluorophores are aligned in parallel, the more efficient the energy transfer from donor to acceptor and therefore the higher the FRET efficiency (reviewed in Weidtkamp-Peters et al., 2022).
Another potentially problematic factor, apart from choosing the best possible FRET pair for the PPI experiment, is the position of the FP or enzyme at the N- or C-terminus of the POI. The protein class, the (subcellular) localization, and known functional domains can help to choose the optimal position of the reporter protein (Long et al., 2018, 2020). In addition, a linker between the POI and the fluorophore could be used to improve FRET efficiency if this leads to an increased rotation of the fluorophore without increasing the distance between the two fluorophores, which could diminish FRET (Denay et al., 2019).
Nevertheless, FRET-FLIM measurements are highly suited to validate PPIs in a quantitative way preserving spatio-temporal information of the POI. In addition, FRET-FLIM can also be used if PPI measurements of more than two POIs is necessary, as described below.
PPI measurements of higher order complexes
Combined BiFC and FRET
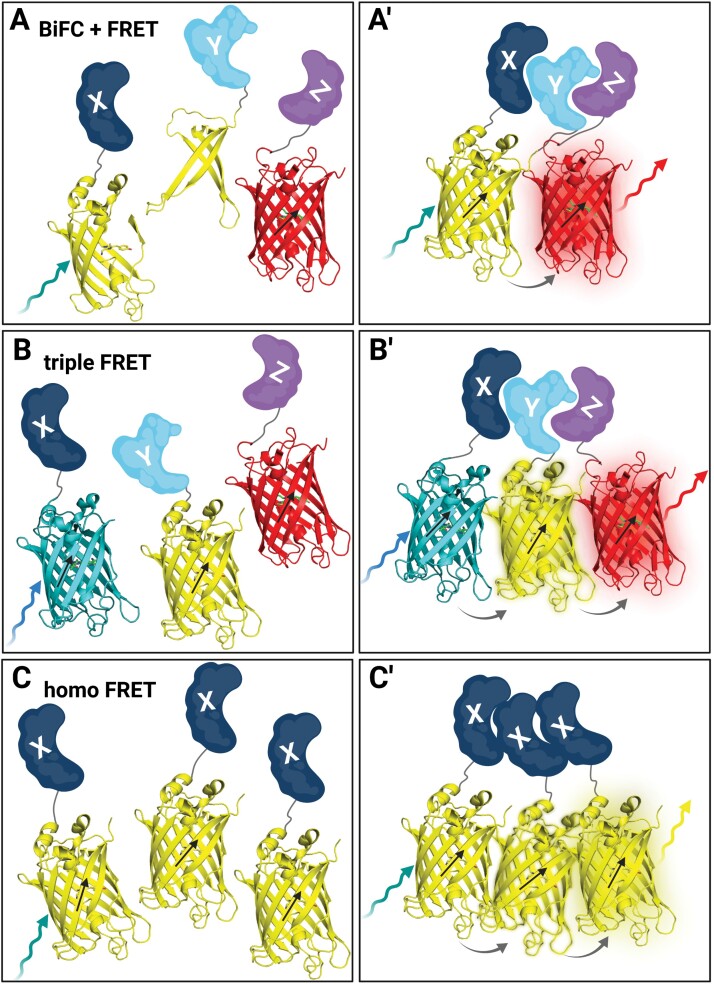
To identify and analyse putative homo- or heteromeric complex formation of more than two proteins, combinations of established methods to study PPIs are used. The combination of BiFC and FRET-FLIM offers the possibility to simultaneously test the interaction of three POIs while preserving high spatial resolution (Kwaaitaal et al., 2010). Here, two POIs are fused to the N- or C-terminal part of the donor fluorophore, respectively. The third POI is fused to the acceptor fluorophore. Only in the case of complex formation of all three POIs can a reduction of the donor fluorescence lifetime or intensity be observed, for example by FRET-FLIM or FRET-APB measurements, respectively (Fig. 3B, Bʹ). An alternative approach without BiFC is described next.
Fig. 3.
Techniques to measure PPI of more than two POIs. (A) In a combined BiFC–FRET experiment, two POIs (X, dark blue; Y, light blue) are fused to the N- or C-terminal part of a split FP (here, split 3D YFP structures in yellow), and a third POI (Z, purple) is fused to an acceptor FP (here, 3D mCherry structure in red). (Aʹ) In the case of interaction of all three POIs, the YFP parts are reconstituted and, after excitation (teal arrow), can transfer energy by FRET (grey arrow) to the acceptor FP (mCherry) which can emit light (red arrow). (B) In a triple FRET experiment, three POIs (X, dark blue; Y, light blue; Z, purple) are fused to three different FPs (here, 3D structures of CFP, cyan; YFP, yellow; and mCherry, red). (Bʹ) In the case of interaction of all three POIs, the three FPs come close enough to allow, upon excitation of CFP by blue light (blue arrow), energy transfer (grey arrow) by FRET to YFP as acceptor which then serves also as a donor and transfers energy via FRET (grey arrow) to mCherry which can then emit light (red arrow). (C) In a homo-FRET experiment, two or more of the same POI (X, dark blue) are labelled with an FP (here, 3D structure of YFP in yellow). (Cʹ) If the POI can form higher order complexes, energy can be transferred from one excited (teal arrow) FP to another, thereby depolarizing the emission of the FP (yellow arrow). Figure created with BioRender.com.
Triple FRET
Triple FRET, also referred to as three-colour FRET or two-step FRET, is another FRET-based method to simultaneously study interactions of more than two proteins, for example in higher order complexes consisting of different proteins. Here, the excited donor fluorophore (D) transfers energy to the first acceptor (A1), which at the same time serves as a donor for a second acceptor fluorophore (A2) (Horsey et al., 2000). Thereby, energy can be transferred sequentially from D via A1 to A2, but also directly from D to A1 and from D to A2 (Fig. 3C, Cʹ). For the required energy transfer, the same prerequisites must be met that were described for conventional FRET above, but in this case for three FPs. Most importantly, the emission spectrum of D must overlap with the excitation spectrum of A1, and the emission spectrum of A1 must overlap with the excitation spectrum of A2. Before triple FRET-FLIM measurements can be carried out, a series of control experiments must be performed to ensure the measured FRET effect is caused by interaction and not by an artefact, for example quantification of fluorescence emission intensity of A1 and A2 with the excitation wavelength of D, to estimate spectral bleed through and crosstalk (Table 1).
One of the first attempts of triple FRET was used in vitro to investigate structural changes in DNA using sensitized emission as well as donor fluorescence and lifetime quenching as a read-out for FRET (Watrob et al., 2003). The first experiments using triple FRET in planta in Arabidopsis mesophyll protoplasts also analysed sensitized acceptor emission and showed that 2-Cys peroxiredoxin forms decamers (Seidel et al., 2010). Recently, the first attempts to establish three-colour FRET-FLIM in planta were performed (see Table 2) (Glöckner et al., 2020, Preprint). Another technique to measure PPI of higher order complexes of the same POI is described next.
Homo-FRET detection by fluorescence anisotropy measurements
FRET can also take place between members of the same kind of fluorophores because they also fulfil the requirements of FRET. This phenomenon of energy migration between identical fluorophores is called homo-FRET (Vogel et al., 2009). The detection of homo-FRET can also be utilized to measure PPI between the same POIs, for example when they form dimeric or multimeric homomeric complexes. While homo-FRET does not impact fluorescence intensity or lifetime, it does affect the direction of the fluorescence emission of the examined fluorophores. This effect can be measured as fluorescence anisotropy r of a fluorophore (reviewed in Weidtkamp-Peters et al., 2022). Here, a microscopic setup with a polarized light source (e.g. a laser) excites the fluorophores with a parallel dipole orientation, a process called photoselection. The steady-state fluorescence anisotropy r of the excited fluorophores can be measured by two detectors detecting the same emission bandwidth of the excited fluorophores but divided by a polarized beam splitter for detecting the intensity of the emitted light parallel (I||) and perpendicular (I⊥) to the polarized excitation light. Steady-state fluorescence anisotropy r can be described by the following equation:
The fluorescence anisotropy r decreases in the case of PPI because of energy transfer to a fluorophore with the same properties in the vicinity of the next POI with a slightly different dipole orientation, thereby depolarizing the emitted fluorescence (Fig. 3C, Cʹ) (Weidtkamp-Peters et al., 2022). Fluorescence anisotropy measurements can be combined with FRET-FLIM measurements to detect dynamic hetero- and homomeric complexes and their spatial distribution within a cell at the same time, which was shown in planta (Table 2) (Stahl et al., 2013; Somssich et al., 2015). Other spectroscopic methods described below can be used to detect PPI by correlating the fluctuations of the fluorescence of POIs as they move together.
Fluorescence fluctuation microscopy to study PPI
Fluorescence correlation spectroscopy (FCS) is an advanced fluorescence technique that measures fluctuations in fluorescence of single molecules over time to quantify the concentration and diffusion coefficients of these molecules in a very small defined volume, such as in a confocal volume (Laursen et al., 2016). This is achieved by focusing excitation light on to a sample (e.g. in a confocal or two-photon microscope), and the resulting fluctuation of fluorescence due to the movement or Brownian motion of the fluorescent molecules is statistically analysed. For FCS measurements, the number of fluorescent molecules must be relatively low (in the pico- to micromolar range) (Eigen and Rigler, 1994; Schwille et al., 1999). Two POIs, if labelled with two spectrally distinct FPs, can be observed in two separate channels by fluorescence cross-correlation spectroscopy (FCCS). If the two proteins indeed interact, they will move together, as demonstrated with interacting receptor kinases in Arabidopsis roots (Table 2) (Wang et al., 2015). For FCS measurements, the same specialized microscopic equipment is needed as described above for FRET-FLIM measurements, because here too a very high temporal resolution of single photons is needed. These single molecule measurements are highly dependent on a very good signal to noise ratio which can be achieved by eliminating out-of-focus fluorescence as much as possible, such as by internal reflection microscopy, in particular variable angle total internal reflection microscopy (VA-TIRF) (Wang et al., 2015).
By using another FCS-based method called raster image correlation spectroscopy (RICS) which extracts fluorescence correlation from confocal image stacks combined with number and brightness analyses (N&B), two POIs labelled with spectrally different FPs can be analysed for their mobility, oligomeric state, and stoichiometry (including PPI) preserving spatio-temporal information, which has recently been successfully applied in Arabidopsis roots employing a conventional confocal microscopic setup (Table 2) (Clark et al., 2016). The advantage of using RICS instead of FCCS is that no specific setup other than a conventional confocal microscope is needed (Table 1). Even though the analyses of the FCCS and RICS data is not as easy as, for example, in FRET-APB experiments, these techniques provide very valuable information on the mobility at the same time as on PPI.
Knocksideways in plants (KSP)
Recently, an exciting new technique to measure and visualize PPI in transiently expressing N. benthamiana leaf epidermis was described which can be compared with an intracellular co-IP (Winkler et al., 2021). This technique combines the—in the animal field—well-established conditional ability of rapamycin to alter the localization of a bait protein and its interactors via the heterodimerization of FKBP and FRB domains. In KSP, this conditional heterodimerization is combined with rerouting interacting proteins to mitochondria upon rapamycin induction. The PPI of more than two POIs can thereby be directly visualized and quantified by FP-tagged POIs in conventional fluorescence microscopy (Table 2) (Winkler et al., 2021). So far, KSP has been used to improve the quality of interaction data acquired with split-ubiquitin, BiFC, and FRET approaches, as the addition of FKBP and FRB domains to these well-established methods can serve as an intrinsic positive control (Andersen et al., 2016). It will be very interesting to see how applicable this new technique is in stable transgenic lines in the future, as KSP also offers, aside from PPI detection, a conditional compartmentalization and thereby a protein knockout tool.
One general consideration concerning all of the above-described PPI techniques that involve imaging is that the POI must be tagged or expressed in fusion with an FP or other protein (domain), and therefore the function of the POI might be impaired. Therefore, control experiments, such as rescue experiments using stable transformants in the respective loss-of-function mutants, should be carried out to test if the labelled POI is still functioning.
Nevertheless, the outstanding benefit of all described imaging-based PPI techniques is the preservation of spatio-temporal information of the involved POI and even quantitative data on the observed PPI (Table 1).
Conclusions
In summary, many different techniques, most of them relying on the use of FPs, have been successfully applied in living plant cells, either in stably transformed plants or transiently in heterologous plant expression systems. Here, dynamic PPIs and complex formations can be investigated in a minimally invasive manner, whilst in most cases preserving the spatio-temporal characteristics of the POI. While we have summarized numerous pros and cons for each of the techniques to study PPI in this review, there is no one technique that fits all requirements. Which technique is best for a given research question depends on, for example, the expression system, POI abundance, and PPI strength and dynamics (Table 1). In the future, emerging techniques in the in vivo or correlative super-resolution microscopy field and/or in novel advances of data analyses could add more depth to the detection and/or quantification of PPIs. Here, novel, high-throughput techniques for improved visualization of PPI and the determination of dynamic in vivo binding affinities would help to decipher complex regulatory networks in plants. Additionally, the more insights into structural information on plant proteins become available, the more in silico predictions of PPI and even of PPI sites, such as in PlaPPISite, will become available (Ding and Kihara, 2019; Yang et al., 2020), which can guide in vivo and in planta experiments. Due to the numbers of PPIs already predicted and/or verified in different plant species, computational networks of PPIs will become even more necessary to understand the complex and dynamic PPIs in a wider biological context.
Acknowledgements
We would like to thank Dr Vicky Howe for critical reading of the manuscript. We would like to apologize to all colleagues whose relevant work could not be discussed due to space restrictions.
Contributor Information
Vivien I Strotmann, Institute for Developmental Genetics, Heinrich-Heine University, Universitätsstr. 1, D-40225 Düsseldorf, Germany.
Yvonne Stahl, Institute for Developmental Genetics, Heinrich-Heine University, Universitätsstr. 1, D-40225 Düsseldorf, Germany.
Aleksandra Skirycz, MPI for Molecular Plant Physiology, Germany.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) via grant STA 1212/4-1 to YS.
References
- Albertazzi L, Arosio D, Marchetti L, Ricci F, Beltram F.. 2009. Quantitative FRET analysis with the EGFP–mCherry fluorescent protein pair. Photochemistry and Photobiology 85, 287–297. [DOI] [PubMed] [Google Scholar]
- Andersen TG, Nintemann SJ, Marek M, Halkier BA, Schulz A, Burow M.. 2016. Improving analytical methods for protein–protein interaction through implementation of chemically inducible dimerization. Scientific Reports 6, 27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach D, Stagljar I.. 2005. Yeast two-hybrid protein–protein interaction networks. In: Waksman G, ed. Proteomics and protein–protein interactions. Boston, MA: Springer US, 19–31. [Google Scholar]
- Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J.. 2016. A guide to fluorescent protein FRET pairs. Sensors 16, 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Kim JM, Cluzel P.. 2018. Systematic characterization of maturation time of fluorescent proteins in living cells. Nature Methods 15, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens PI, Jovin TM.. 1996. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proceedings of the National Academy of Sciences, USA 93, 8407–8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens PI, Majoul IV, Verveer PJ, Söling HD, Jovin TM.. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. The EMBO Journal 15, 4246–4253. [PMC free article] [PubMed] [Google Scholar]
- Becker W, Bergmann A, Hink MA, König K, Benndorf K, Biskup C.. 2004. Fluorescence lifetime imaging by time-correlated single-photon counting. Microscopy Research and Technique 63, 58–66. [DOI] [PubMed] [Google Scholar]
- Betegón-Putze I, Mercadal J, Bosch N, et al. 2021. Precise transcriptional control of cellular quiescence by BRAVO/WOX5 complex in Arabidopsis roots. Molecular Systems Biology 17, e9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup C, Zimmer T, Kelbauskas L, et al. 2007. Multi-dimensional fluorescence lifetime and FRET measurements. Microscopy Research and Technique 70, 442–451. [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CAM, Simon R.. 2010. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiology 152, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, et al. 2004. Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. The Plant Journal 40, 419–427. [DOI] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, et al. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nature Biotechnology 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Bader A, Westphal AH, Laptenok SP, Borst JW.. 2014. FRET-FLIM applications in plant systems. Protoplasma 251, 383–394. [DOI] [PubMed] [Google Scholar]
- Chakravartty V, Cronan JE.. 2012. Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. Journal of Bacteriology 194, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang G-L.. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein–protein interactions in rice. Molecular Plant Pathology 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, et al. 2008. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, et al. 2016. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5, e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S.. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Datta R, Heaster TM, Sharick JT, Gillette AA, Skala MC.. 2020. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. Journal of Biomedical Optics 25, 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, Davidson MW.. 2009. The fluorescent protein palette: tools for cellular imaging. Chemical Society Reviews 38, 2887–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane CM, Salwiński L, Xenarios I, Eisenberg D.. 2002. Protein interactions: two methods for assessment of the reliability of high throughput observations. Molecular & Cellular Proteomics 1, 349–356. [DOI] [PubMed] [Google Scholar]
- Denay G, Schultz P, Hänsch S, Weidtkamp-Peters S, Simon R.. 2019. Over the rainbow: a practical guide for fluorescent protein selection in plant FRET experiments. Plant Direct 3, e00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S.. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Molecular and Cellular Biology 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Kihara D.. 2019. Computational identification of protein–protein interactions in model plant proteomes. Scientific Reports 9, 8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A, Weltmeier F, Wang X, et al. 2006. Two-hybrid protein–protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. The Plant Journal 46, 890–900. [DOI] [PubMed] [Google Scholar]
- Eigen M, Rigler R.. 1994. Sorting single molecules: application to diagnostics and evolutionary biotechnology. Proceedings of the National Academy of Sciences, USA 91, 5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J-Y, Cui Z-Q, Wei H-P, et al. 2008. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein–protein interactions in living cells. Biochemical and Biophysical Research Communications 367, 47–53. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O.. 1989. A novel genetic system to detect protein–protein interactions. Nature 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Förster T. 1948. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik 437, 55–75. [Google Scholar]
- Fraga H. 2008. Firefly luminescence: a historical perspective and recent developments. Photochemical & Photobiological Sciences 7, 146–158. [DOI] [PubMed] [Google Scholar]
- Fujikawa Y, Kato N.. 2007. Split luciferase complementation assay to study protein–protein interactions in Arabidopsis protoplasts. The Plant Journal 52, 185–195. [DOI] [PubMed] [Google Scholar]
- Gehl C, Kaufholdt D, Hamisch D, et al. 2011. Quantitative analysis of dynamic protein–protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. The Plant Journal 67, 542–553. [DOI] [PubMed] [Google Scholar]
- Glöckner N, zur Oven-Krockhaus S, Rohr L, et al. 2020. Three-fluorophore FRET enables the analysis of ternary protein association in living plant cells. BioRxiv doi: 10.1101/722124. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G, Krebs M, Maizel A, Stahl Y, Vermeer JEM, Ott T.. 2018. Green light for quantitative live-cell imaging in plants. Journal of Cell Science 131, jcs209270. [DOI] [PubMed] [Google Scholar]
- Hebisch E, Knebel J, Landsberg J, Frey E, Leisner M.. 2013. High variation of fluorescence protein maturation times in closely related Escherichia coli strains. PLoS One 8, e75991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Griep RA, Borst JW, et al. 2000. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. Journal of Biological Chemistry 275, 17556–17560. [DOI] [PubMed] [Google Scholar]
- Horsey I, Furey WS, Harrison JG, Osborne MA, Balasubramanian S.. 2000. Double fluorescence resonance energy transfer to explore multicomponent binding interactions: a case study of DNA mismatches. Chemical Communications 2000, 1043–1044. [Google Scholar]
- Horstman A, Tonaco IAN, Boutilier K, Immink RGH.. 2014. A cautionary note on the use of split-YFP/BiFC in plant protein–protein interaction studies. International Journal of Molecular Sciences 15, 9628–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C-D, Chinenov Y, Kerppola TK.. 2002. Visualization of interactions among bZIP and rel family proteins in living cells using bimolecular fluorescence complementation. Molecular Cell 9, 789–798. [DOI] [PubMed] [Google Scholar]
- Hu C-D, Kerppola TK.. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotechnology 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Baumann CT, He L, et al. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. Journal of Microscopy 209, 56–70. [DOI] [PubMed] [Google Scholar]
- Kaur A, Nijhawan A, Yadav M, Khurana JP.. 2021. OsbZIP62/OsFD7, a functional ortholog of FLOWERING LOCUS D, regulates floral transition and panicle development in rice. Journal of Experimental Botany 72, 7826–7845. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK. 2001. Imaging protein–protein interactions using fluorescence resonance energy transfer microscopy. Methods 24, 289–296. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK, Edidin M.. 1999. Imaging fluorescence resonance energy transfer as probe of membrane organization and molecular associations of GPI-anchored proteins. Methods in Molecular Biology 116, 37–49. [DOI] [PubMed] [Google Scholar]
- Kim DI, Roux KJ.. 2016. Filling the void: proximity-based labeling of proteins in living cells. Trends in Cell Biology 26, 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnovsky AA, Kovalev YV.. 2014. Spectral and kinetic parameters of phosphorescence of triplet chlorophyll a in the photosynthetic apparatus of plants. Biochemistry 79, 349–361. [DOI] [PubMed] [Google Scholar]
- Krieger N, Pastryk K-F, Forchhammer K, Kolukisaoglu U.. 2021. Arabidopsis PII proteins form characteristic foci in chloroplasts indicating novel properties in protein interaction and degradation. International Journal of Molecular Sciences 22, 12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R.. 2010. Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiology 152, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen T, Borch J, Knudsen C, et al. 2016. Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum. Science 354, 890–893. [DOI] [PubMed] [Google Scholar]
- Lee J, Liu Z, Suzuki P, et al. 2020. Versatile phenotype-activated cell sorting. Science Advances 6, eabb7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leissing F, Misch NV, Wang X, et al. 2021. Purification of MAP-kinase protein complexes and identification of candidate components by XL-TAP-MS. Plant Physiology 187, 2381–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-F, Bush J, Xiong Y, Li L, McCormack M.. 2011. Large-scale protein–protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS One 6, e27364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liao W, Nie B, Zhang J, Xu W.. 2021. OsUEV1B, an Ubc enzyme variant protein, is required for phosphate homeostasis in rice. The Plant Journal 106, 706–719. [DOI] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, et al. 2017. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548, 97–102. [DOI] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, et al. 2018. Optimizing FRET-FLIM labeling conditions to detect nuclear protein interactions at native expression levels in living arabidopsis roots. Frontiers in Plant Science 9, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, Blilou I.. 2020. Visualizing protein associations in living arabidopsis embryo. Methods in Molecular Biology 2122, 167–188. [DOI] [PubMed] [Google Scholar]
- Luker KE, Smith MCP, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D.. 2004. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences, USA 101, 12288–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair A, Xu S-L, Branon TC, Ting AY, Bergmann DC.. 2019. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8, e47864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters SC. 2004. Co-immunoprecipitation from transfected cells. Methods in Molecular Biology 261, 337–350. [DOI] [PubMed] [Google Scholar]
- Matthews JC, Hori K, Cormier MJ.. 1977. Purification and properties of Renilla reniformis luciferase. Biochemistry 16, 85–91. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Reed R, Marciniak RA.. 2005. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryotic Cell 4, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megerle JA, Fritz G, Gerland U, Jung K, Rädler JO.. 2008. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophysical Journal 95, 2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhorn DG, Wallmeroth N, Berendzen KW, Grefen C.. 2018. 2in1 vectors improve in planta BiFC and FRET analyses. Methods in Molecular Biology 1691, 139–158. [DOI] [PubMed] [Google Scholar]
- Monti M, Orrù S, Pagnozzi D, Pucci P.. 2005. Interaction proteomics. Bioscience Reports 25, 45–56. [DOI] [PubMed] [Google Scholar]
- Morell M, Espargaró A, Avilés FX, Ventura S.. 2007. Detection of transient protein–protein interactions by bimolecular fluorescence complementation: the Abl–SH3 case. Proteomics 7, 1023–1036. [DOI] [PubMed] [Google Scholar]
- Olejnik K, Bucholc M, Anielska-Mazur A, et al. 2011. Arabidopsis thaliana Nudix hydrolase AtNUDT7 forms complexes with the regulatory RACK1A protein and Ggamma subunits of the signal transducing heterotrimeric G protein. Acta Biochimica Polonica 58, 609–616. [PubMed] [Google Scholar]
- Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y.. 2001. Split luciferase as an optical probe for detecting protein–protein interactions in mammalian cells based on protein splicing. Analytical Chemistry 73, 2516–2521. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS.. 2003. Monitoring protein–protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Analytical Chemistry 75, 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phee B-K, Shin DH, Cho J-H, et al. 2006. Identification of phytochrome-interacting protein candidates in Arabidopsis thaliana by co-immunoprecipitation coupled with MALDI-TOF MS. Proteomics 6, 3671–3680. [DOI] [PubMed] [Google Scholar]
- Ransone LJ. 1995. Detection of protein–protein interactions by coimmunoprecipitation and dimerization. Methods in Enzymology 254, 491–497. [DOI] [PubMed] [Google Scholar]
- Ren L, Emery D, Kaboord B, Chang E, Qoronfleh MW.. 2003. Improved immunomatrix methods to detect protein:protein interactions. Journal of Biochemical and Biophysical Methods 57, 143–157. [DOI] [PubMed] [Google Scholar]
- Romei MG, Boxer SG.. 2019. Split green fluorescent proteins: scope, limitations, and outlook. Annual Review of Biophysics 48, 19–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, in Kim D, Raida M, Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. Journal of Cell Biology 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatlowski N, Stahl Y, Hohenstatt ML, Goodrich J, Schubert D.. 2010. The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. The Plant Cell 22, 2291–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P, Haupts U, Maiti S, Webb WW.. 1999. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophysical Journal 77, 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel T, Seefeldt B, Sauer M, Dietz K-J.. 2010. In vivo analysis of the 2-Cys peroxiredoxin oligomeric state by two-step FRET. Journal of Biotechnology 149, 272–279. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY.. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Somssich M, Ma Q, Weidtkamp-Peters S, et al. 2015. Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Science Signaling 8, ra76. [DOI] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, et al. 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Current Biology 23, 362–371. [DOI] [PubMed] [Google Scholar]
- Struk S, Jacobs A, Sánchez Martín-Fontecha E, Gevaert K, Cubas P, Goormachtig S.. 2019. Exploring the protein–protein interaction landscape in plants. Plant, Cell & Environment 42, 387–409. [DOI] [PubMed] [Google Scholar]
- Tang Z, Takahashi Y.. 2018. Analysis of protein–protein interaction by co-IP in human cells. Methods in Molecular Biology 1794, 289–296. [DOI] [PubMed] [Google Scholar]
- Tebo AG, Gautier A.. 2019. A split fluorescent reporter with rapid and reversible complementation. Nature Communications 10, 2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SS, Thaler C, Blank PS, Koushik SV.. 2009. Time-resolved fluorescence anisotropy. In: Periasamy A, Clegg RM, eds. FLIM microscopy in biology and medicine. Chapman and Hall/CRC, 245–290. [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J.. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. The Plant Journal 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM.. 2018. Genetically encoded biosensors in plants: pathways to discovery. Annual Review of Plant Biology 69, 497–524. [DOI] [PubMed] [Google Scholar]
- Wang F-Z, Zhang N, Guo Y-J, Gong B-Q, Li J-F.. 2020. Split Nano luciferase complementation for probing protein–protein interactions in plant cells. Journal of Integrative Plant Biology 62, 1065–1079. [DOI] [PubMed] [Google Scholar]
- Wang L, Li H, Lv X, et al. 2015. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Molecular Plant 8, 1334–1349. [DOI] [PubMed] [Google Scholar]
- Watrob HM, Pan C-P, Barkley MD.. 2003. Two-step FRET as a structural tool. Journal of the American Chemical Society 125, 7336–7343. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Rehwald S, Stahl Y.. 2022. Homo-FRET imaging to study protein–protein interaction and complex formation in plants. Methods in Molecular Biology 2379, 197–208. [DOI] [PubMed] [Google Scholar]
- Weidtkamp-Peters S, Stahl Y.. 2017. The use of FRET/FLIM to study proteins interacting with plant receptor kinases. Methods in Molecular Biology 1621, 163–175. [DOI] [PubMed] [Google Scholar]
- Winkler J, Mylle E, de Meyer A, et al. 2021. Visualizing protein–protein interactions in plants by rapamycin-dependent delocalization. The Plant Cell 33, 1101–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won R. 2009. Eyes on super-resolution. Nature Photonics 3, 368–369. [Google Scholar]
- Wouters FS, Bastiaens PI, Wirtz KW, Jovin TM.. 1998. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. The EMBO Journal 17, 7179–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Wallmeroth N, Berendzen KW, Grefen C.. 2016. Techniques for the analysis of protein–protein interactions in vivo. Plant Physiology 171, 727–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wen Z, Zhang D, et al. 2021. Proximity labeling: an emerging tool for probing in planta molecular interactions. Plant Communications 2, 100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang S, Qi H, Wang T, Li H, Zhang Z.. 2020. PlaPPISite: a comprehensive resource for plant protein–protein interaction sites. BMC Plant Biology 20, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Song G, Lal NK, et al. 2019. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nature Communications 10, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Doyle TC, Wong R, et al. 2004. Characterization of coelenterazine analogs for measurements of Renilla luciferase activity in live cells and living animals. Molecular Imaging 3, 43–54. [DOI] [PubMed] [Google Scholar]
- Zhong S, Rivera-Molina F, Rivetta A, Toomre D, Santos-Sacchi J, Navaratnam D.. 2019. Seeing the long tail: a novel green fluorescent protein, SiriusGFP, for ultra long timelapse imaging. Journal of Neuroscience Methods 313, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]