Graphical abstract

Keywords: Spiropyrazoles, Hydrazonoyl halides, Microwaves, Antimicrobial, COVID-19
Abstract
Synthesis of a new series of spiropyrazole derivatives using microwaves irradiation with high yield in minutes was achieved through a cycloaddition reaction of nitrile imines and arylidenes of 5-bromo-indan-1-one. The structure of the new spiropyrazoles was assured based on their available spectral analyses and the comparison of the extracted data with the literature reports. Molecular docking simulations of all new synthesized spiropyrazole derivatives into leucyl-tRNA synthetase editing domain of Candida albicans (Pdb: 2WFC) indicated that about seven spiropyrazole derivatives can fit deeply in the active site via the formation of stable complexes. In addition, the docking study was utilized to tested the ability of these spiropyrazoles to inhibit COVID-19 through the interaction with COVID-19 main protease (Pdb: 6LU7). The results were surprising which revealed high docking score ranging from -7.764 to -5.9464 kcal/mol. Moreover, the nitrogen atom of pyrazole, Br atom and the C=O group of indanone are essential parts in the binding mode of almost the active derivatives. The results of the docking study are a glimmer of hope to complete the study on these compounds and examine them in the laboratory to ensure their effectiveness as antimicrobials and antiviral, especially Covid-19. Moreover, pharmacokinetics and physicochemical properties were studied.
1. Introduction
The continuation of the Corona pandemic (COVID-19) for nearly two years is a big problem, as it takes lives and affects the economic life of the whole world. The outbreak of pneumonia caused by COVID-19 is considered a pandemic according to the World Health Organization in March 2020 [1] that has resulted in millions of infections and deaths worldwide. In this scenario, the development of antiviral agents that interfere with the life cycle of the virus or in virus replication or membrane fusion is urgently needed [2]. Every day, scientists are racing to find effective heterocyclic drugs to stop this epidemic. Recently, many researches have done a theoretical search through docking of compounds with different proteins of viruses related to COVID-19 [3], [4], [5]. On the other side, overuse as well as the misuse of antibiotics has led to the expansion of multidrug resistance between various types of microorganism's strains [6]. Accordingly, new antimicrobial agents must be developed that act on new targets to overcome the increasing incidence of microbial resistance to antibiotic therapy. Scientists are also working day and night to search for new compounds with antimicrobial activity. One of the bioactive drugs is compounds containing pyrazole moiety which are known with their antimicrobial and antiviral activities [7], [8], [9], [10], [11], [12]. The class of spiropyrazoles are well known in literature with the phosphodiesterase inhibitors [13], antitumor [14,15], analgesic [16] and anti-inflammatory activities [17]. The synthesis of heterocyclic compounds with biological activity for more than one disease is a difficult goal, in addition to synthesis of these derivatives with high yield in a short reaction time is a good goal. Therefore, using microwave as an energy source increases the yield of the synthesized compounds in a short time [18,19]. Heating using microwave rays helps in the formation of organic compounds in a pure and high rate and in a time of up to a few minutes, because those irradiations can raise the temperature of the reactions to temperatures up to two to four times the boiling points of the reacting compounds in addition to maintaining the atmosphere around chemical reactions of moisture that may affect moisture-sensitive reagents and reduce the formation of unwanted by-product [20]. All the above facts with our expertise in bioactive heterocyclic synthesis [21], [22], [23], [24], we focused our effort in this context to synthesize a new series of spiropyrazoles under microwave irradiations and investigated their theoretical activity as antimicrobes and anti-COVID-19 through the docking study with peroxiredoxin 5 (Pdb: 2WFC) and COVID-19 main protease (Pdb: 6LU7); the overexpressed proteins in Candida albicans and severe acute respiratory syndrome coronavirus 2, respectively.
2. Results and discussion
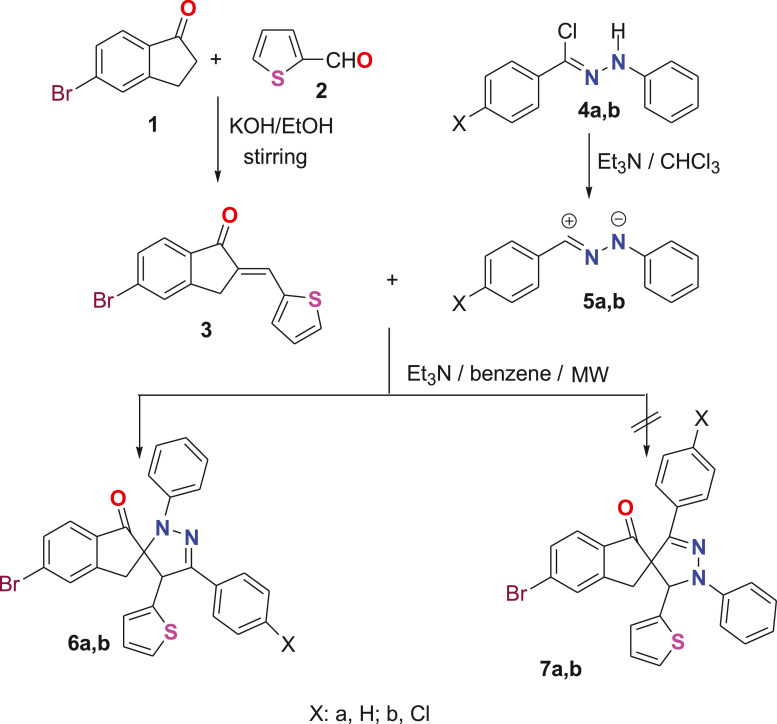
5-Bromo-2-thiophen-2-ylmethylene-indan-1-one 3 was prepared as usual way from the condensation reaction of 5-bromo-indan-1-one 1 with 2-thiophene aldehyde 2 in ethanol and in the presence of base catalyst KOH with stirring for 5 h [25]. Previously [26], it was reported that the synthesis of a series of spiropyrazoles took about 50 h under reflux. We tried herein to synthesis a related spiropyrazoles but, carrying bromide atom at the position five of indanone in short reaction time. After several trials using MW irradiations, we abled to prepare derivatives 6a,b from the reaction of hydrazonoyl chlorides 4a,b with arylidene 3 in benzene at 80 °C for about 40-50 min. Such reaction revealed only one regioisomer 6 rather than 7 (Scheme 1 ) depending on the comparison of their spectral data with the related published derivatives in literature. For instance,
Scheme 1.
Synthesis of spiropyrazoles 6a,b under microwaves irradiation.
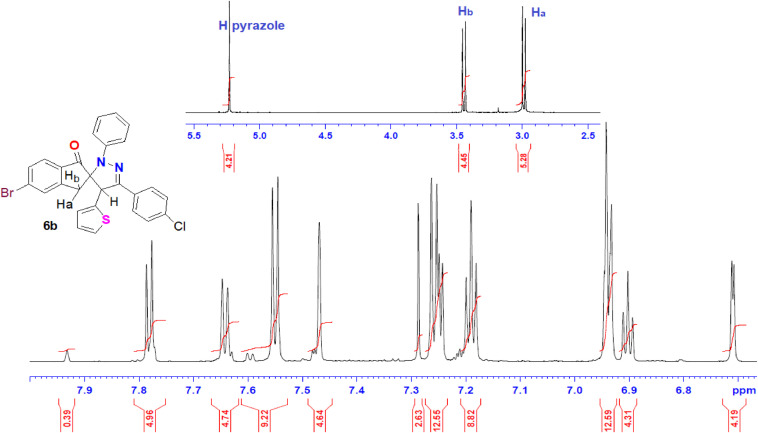
1H NMR spectrum of 6b (Fig. 1) showed all characteristic signals for the existed protons as follows: 2.98 (d, Ha), 3.44 (d, Hb), 5.23 (s, pyrazole-H) and fifteen aromatic protons at 6.71-7.78 ppm. The presence of H-4 of pyrazole at δ = 5.23 ppm [26] ruled out the other regioisomer 7.
Fig. 1.
1H NMR spectrum of spiropyrazole 6b.
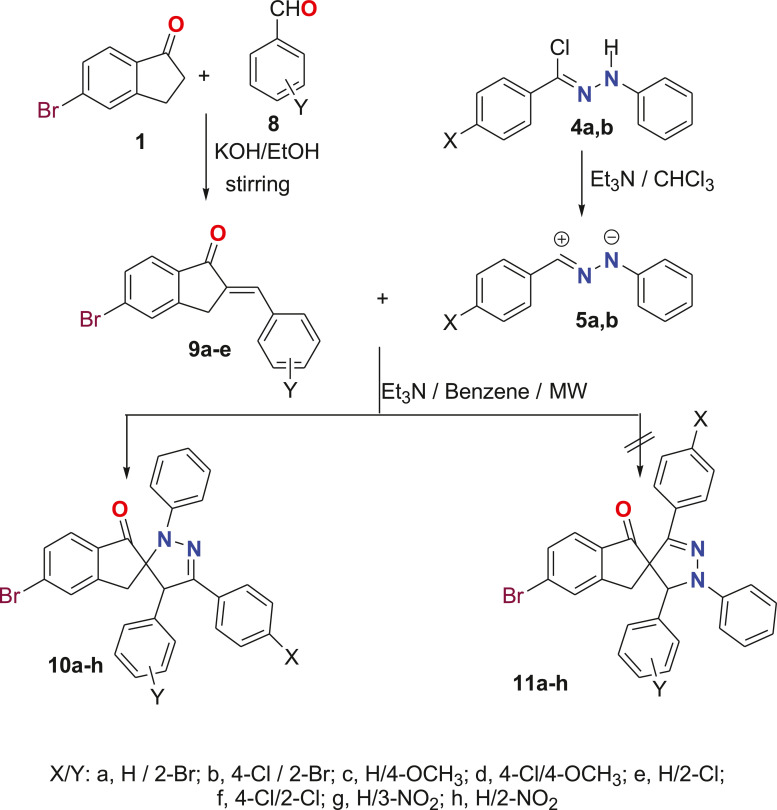
By similar way, the same reaction was repeated under the same reaction condition using arylidene derivatives 9a-h with nitrileimines 5a,b afforded the spiropyrazole derivatives 10a-h in short reaction time (40–50 min) as illustrated in Scheme 2 . The signal of pyrazole-H-4 appeared in 1H NMR of all derivatives 10a-h at δ = 4.70–5.35 ppm, these values discarded the other regioisomer 11a-h. All IR data of spiropyrazoles 10a-h have the characteristic absorption bands of C=O at ν = 1673-1755 cm−1. Another strong evidence for the formation of the spiropyrazole 10 rather than 11, the 13C NMR spectrum of compound 10d ( Fig. 2) showed two characteristic carbon signals at 61.2 (C4 pyrazole) and 78.5 (C-spiro) ppm [27,28].
Scheme 2.
Synthesis of spiropyrazoles 10a-h under microwaves irradiation.
Fig. 2.
The 13C NMR spectrum of spiropyrazole 10d.
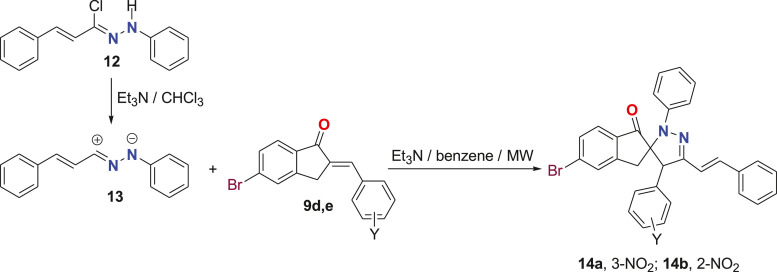
Another two new derivatives of spiropyrazoles 14a,b were prepared under the same MW irradiations in benzene in the presence of Et3N from the reaction of nitrileimine 13 and the arylidene of 5-bromo-indan-1-one 9d,e (Scheme 3 ). All the results of the spectral analyses for these two spiropyrazoles 14a,b agreed with the same results for the previous compounds 6 and 10.
Scheme 3.
Synthesis of spiropyrazoles 14a,b under microwaves irradiation.
3. Docking study
3.1. Docking study for microbial activity
To investigate the ability of the new synthesized spiropyrazole derivatives 6a,b, 10a-h and 14a,b, the molecular docking study have been performed for these derivatives onto the active site of Candida albicans editing domain of cytosolic leucyl-tRNA synthetase to prove their binding affinity and orientation. We used cytosolic leucyl-tRNA synthetase editing domain co-crystallized with benzoxaborole-AMP adduct (PDB code: 2WFG) for docking using MOE 2014 program. The results of docking study were tabulated in Table 1 and the 2D pictures as well as the contact performance for selected derivatives illustrated in Fig. 3. These results revealed docking score ranging from -3.2041 to -6.4628 kcal/mol with different types of interactions as pi-H, H-donor and H-acceptor. The most widely amino-acids involved in the interaction are LYS407, LYS483, LEU317 and THR413. Focusing on the most active spiropyrazole derivatives 10c, 14b, 10g and 10h with the highest docking scores -6.4628, -6.2704, -5.6246 and -4.8742 kcal/mol, respectively, it was observed that all of them are stabilized by strong-H-bonds and pi-H interactions. Also, we noted that the presence of oxygen atoms as C=O or in nitro group and the bromide atom are essential in interaction. The presence of these strong interactions especially in derivatives 10c, 14b, 10g and 10h into the active site of Candida albicans editing domain of cytosolic leucyl-tRNA synthetase proves the effectiveness of these spiropyrazoles as antimicrobial agents. Their activity can be investigated experimentally in the future.
Table 1.
Docking results of the new synthesized derivatives 6a-b and 10a-h with the receptors of (2WFC) for microbes.
| Compd. | Ligand moiety | Receptor site | Interacting residues(Type of interaction) | Distance (oA) | E (kcal/mol) | Docking score (kcal/mol) |
|---|---|---|---|---|---|---|
| 6a | 6-ring | CD LYS 407 (A) | pi-H | 3.81 | -0.9 | - 4.5946 |
| 6b | – | – | – | – | – | - 5.7907 |
| 10a | – | – | – | – | – | -4.9668 |
| 10b | O 156-ring | NZ LYS 407 (A)CD LYS 407 (A) | H-donorpi-H | 3.062.80 | -8.6-0.9 | -3.8321 |
| 10c | O 15N 216-ring | NZ LYS 483 (A)NZ LYS 407 (A)N LEU 317 (A) | H-acceptorH-acceptorpi-H | 2.703.724.17 | -5.3-1.3-1.8 | -6.4628 |
| 10d | 6-ring6-ring | N LEU 317 (A)CD LYS 407 (A) | pi-Hpi-H | 4.644.01 | -0.6-0.8 | -3.8721 |
| 10e | – | – | – | – | – | -3.2041 |
| 10f | CL 54O 15N 216-ring | O THR 413 (A)NZ LYS 483 (A)NZ LYS 407 (A)N LEU 317 (A) | H-donorH-acceptorH-acceptorpi-H | 3.392.633.704.13 | -1.4-5.0-1.4-2.2 | -4.1968 |
| 10g | BR 16O 556-ring | OG1 THR 321 (A)NZ LYS 483 (A)CA THR 316 (A) | H-donorH-acceptorpi-H | 3.41 2.823.87 | -0.9-6.8-0.7 | -5.6246 |
| 10h | BR 16O 156-ring | OG1 THR 321 (A)NZ LYS 483 (A)CA THR 316 (A) | H-donorH-acceptorpi-H | 3.402.773.91 | -0.8-7.4-0.6 | -4.8742 |
| 14a | O 60 | N THR 409 (A) | H-acceptor | 2.80 | -1.7 | -4.1252 |
| 14b | O 59O 606-ring | CA VAL 403 (A)N LEU 404 (A)CB SER 419 (A) | H-acceptorH-acceptorpi-H | 3.253.083.86 | -0.8-1.0-0.8 | -6.2704 |
Fig. 3.
The 2D docked model and contact performance of derivatives 10c, 10h and 14b into the active site of 2WFG.
3.2. Docking study for COVID-19 proliferation
To investigate the ability of the spiropyrazoles 6a,b, 10a-h and 14a,b to interact with COVID-19 main protease (Pdb: 6LU7), the molecular docking simulation was performed using the same MOE 2014 program and the results were tabulated in Table 2 . At first glance to the results in Table 2, we noted that all spiropyrazole derivatives showed high docking score ranging from -7.764 to -5.9464 kcal/mol. The common atoms involved in the interaction in the active sile of 6lu7 are Br, oxygen atoms as well as the aromatic ring of indanone. The common amino acid residue involved in the interaction is GLY143 form pi-H with the aromatic ring of indanone. Compound 14a with the highest docking score (-7.764) is strongly fitted in the active site of 6lu7 with H-bond between Br in indanone and the oxygen atom of THR26 and pi-H between the aromatic ring of indanone and the nitrogen atom of GLY143 ( Fig. 4). The promising data for docking simulation using the major protease of COVID-19 (Pdb: 6LU7) encourage us to recommend this spiropyrazoles for in vitro investigation of these derivatives as COVD-19 inhibitors.
Table 2.
Docking results of the new synthesized derivatives 6a-b, 10a-h and 14a-b with the receptors of (6lu7) for COVID-19.
| Compd. | Ligand moiety | Receptor site | Interacting residues(Type of interaction) | Distance (oA) | E (kcal/mol) | Docking score (kcal/mol) |
|---|---|---|---|---|---|---|
| 6a | O 15 | SG CYS 145 (A) | H-donor | 3.19 | - 1.0 | - 6.5155 |
| 6b | O 15C 25N 28 | SG CYS 145 (A)OE1 GLN 189 (A)N GLU 166 (A) | H-donorH-donorH-acceptor | 3.252.863.19 | -0.8-0.8-0.9 | - 6.6631 |
| 10a | BR 16C 18N 21 | O THR 26 (A)OE1 GLN 189 (A)N GLU 166 (A) | H-donorH-donorH-acceptor | 3.363.163.27 | -1.6-1.1-2.0 | -6.5876 |
| 10b | – | – | – | – | – | -6.3476 |
| 10c | 6-ring | N GLY 143 (A) | pi-H | 4.61 | -0.6 | -6.7045 |
| 10d | 6-ring | N GLY 143 (A) | pi-H | 4.56 | -0.7 | -6.9558 |
| 10e | BR 166-ring | O THR 26 (A)N GLY 143 (A) | H-donorpi-H | 3.804.27 | -0.5-0.7 | -6.7893 |
| 10f | BR 166-ring | O THR 26 (A)N GLY 143 (A) | H-donorpi-H | 3.844.53 | -0.4-1.1 | -5.9464 |
| 10g | 6-ring | N GLY 143 (A) | pi-H | 4.18 | -0.7 | -7.0122 |
| 10h | O 15 6-ring | SG CYS 145 (A)N GLY 143 (A) | H-donorpi-H | 3.844.14 | -0.7-0.7 | -7.2934 |
| 14a | BR 166-ring | O THR 26 (A)N GLY 143 (A) | H-donorpi-H | 3.64 4.12 | -0.6-1.2 | -7.764 |
| 14b | BR 16O 60 | O THR 26 (A)NE2 HIS 163 (A) | H-donorH-acceptor | 3.603.28 | -1.4-1.3 | -7.6202 |
Fig. 4.
The 2D docked model and contact performance of derivatives 6b, 14a and 14b into the active site of 6LU7.
3.3. Pharmacokinetics, and ADME activity
ADMET investigation on the selected synthesized spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b was done using SwissADME website (http://www.swi ssadme.ch/index.php). SwissADME is a web tool that computes the key pharmacokinetic, physicochemical parameters and drug-likeness for molecules [29]. The data of Physicochemical properties were collected in Tables 3a & 3b which including molecular weight (MW), molecular refractivity (MR), polar surface area (PSA), partition coefficient (log Po/w) hydrogen bond acceptors and donors counts, and the number of rotatable bonds in a molecule count of specific atom types. Drug similarity can be assumed as a correct balance between molecular parameters that influence the pharmacokinetics and pharmacodynamics of molecules that finally influence their absorption, distribution, metabolism and excretion (ADME) in the human body as a drug. From Tables 3a & 3b, the molecular weight of only derivative 6a < 500 [30] which increases the transmissibility of this spiropyrazole derivative to membranes and speed the transport and absorption. All spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b showed topological polar surface area (TPSA) between 41.90 and 78.49 Å2 which are in the correct range < 140 Å2 [31]. The Lipophilicity (Log P) value of spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b are ranged between 4.14 – 3.18 Which agrees with Lipinski's rule of five. All derivatives contain only the number of HBA (hydrogen bond acceptors) in agreement value with Lipinski's rule for drug-likeness (for HBA < 10). The above results indicated that the physicochemical properties of the tested indole derivatives within the acceptable range. In addition, tested spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b showed high GI absorption with no BBB permeant except 10c. all derivatives able to inhibit CYP2C9 in addition derivatives 10c, 10g and 10h able to inhibit CYP2C19.
Table 3a.
Physiochemical and Pharmacokinetics properties for tested spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b.
| Properties | Compounds |
||
|---|---|---|---|
| 6a | 10c | 10g | |
| Formula | C27H19BrN2OS | C30H23BrN2O2 | C29H20BrN3O3 |
| Molecular weight | 499.42 g/mol | 523.42 g/mol | 538.39 g/mol |
| Num. of heavy atoms | 32 | 35 | 36 |
| Num. of aromatic heavy atoms | 23 | 24 | 24 |
| Num. of rotatable bonds | 3 | 4 | 4 |
| Num. H-bond acceptor (HBA) | 2 | 3 | 4 |
| Num. H-bond donors (HBD) | 0 | 0 | 0 |
| Molar reactivity | 140.44 | 149.06 | 151.39 |
| Topological Polar Surface area (TPSA) | 60.91 Ų | 41.90 Ų | 78.49 Ų |
| Lipophilicity (Log P) | 3.90 | 4.14 | 3.18 |
| Water solubility (Log S) | -7.67 (InSoluble) | -7.90 (InSoluble) | -7.88 (inSoluble) |
| Pharmacokinetics (GI absorption) | High | High | High |
| Pharmacokinetics (BBB permeant) | No | Yes | No |
| Pharmacokinetics (CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 inhibitors) | No, No, Yes, No, No (respectively) | No, No, Yes, No, No (respectively) | No, Yes, Yes, No, No (respectively) |
| Druglikeness (Lipinski) | Yes; 1 violation: MLOGP>4.15 | No; 2 violations: MW>500, MLOGP>4.15 | No; 2 violations: MW>500, MLOGP>4.15 |
Table 3b.
Physiochemical and Pharmacokinetics properties for tested spiropyrazole derivatives 6a, 10c, 10g, 19h, 14a and 14b.
| Properties | Compounds |
||
|---|---|---|---|
| 10h | 14a | 14b | |
| Formula | C29H20BrN3O3 | C31H22BrN3O3 | C31H22BrN3O3 |
| Molecular weight | 538.39 g/mol | 564.43 g/mol | 564.43 g/mol |
| Num. of heavy atoms | 36 | 38 | 38 |
| Num. of aromatic heavy atoms | 24 | 24 | 24 |
| Num. of rotatable bonds | 4 | 5 | 5 |
| Num. H-bond acceptor (HBA) | 4 | 4 | 4 |
| Num. H-bond donors (HBD) | 0 | 0 | 0 |
| Molar reactivity | 151.39 | 161.32 | 161.32 |
| Topological Polar Surface area (TPSA) | 78.49 Ų | 78.49 Ų | 78.49 Ų |
| Lipophilicity (Log P) | 3.86 | 3.54 | 3.59 |
| Water solubility (Log S) | -7.88 (inSoluble) | -8.22 (inSoluble) | -8.22 (inSoluble) |
| Pharmacokinetics (GI absorption) | High | High | High |
| Pharmacokinetics (BBB permeant) | No | No | No |
| Pharmacokinetics (CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 inhibitors) | No, Yes, Yes, No, No (respectively) | No, No, Yes, No, No (respectively) | No, No, Yes, No, No (respectively) |
| Druglikeness (Lipinski) | No; 2 violations: MW>500, MLOGP>4.15 | No; 2 violations: MW>500, MLOGP>4.15 | No; 2 violations: MW>500, MLOGP>4.15 |
4. Conclusion
In this context, we synthesized a new series of spiropyrazole derivatives from cycloaddition reaction of nitrile imines and arylidenes of 5-bromo-indan-1-one under microwaves irradiation. The new spiropyrazoles were formed in excellent yield in short reaction time. The structure of the new spiropyrazoles was assured based on their available spectral analyses and the comparison of the extracted data with the literature reports. Molecular docking simulations of all new synthesized spiropyrazole derivatives into leucyl-tRNA synthetase editing domain of Candida albicans (Pdb: 2WFC) indicated that about seven spiropyrazole derivatives can fit deeply in the active site via the formation of stable complexes. In addition, the docking study was utilized to tested the ability of these spiropyrazoles to inhibit COVID-19 through the interaction with COVID-19 main protease (Pdb: 6LU7). The results were surprising which revealed high docking score ranging from -7.764 to -5.9464 kcal/mol. The results of the docking study are a glimmer of hope to complete the study on these compounds and examine them in the laboratory to ensure their effectiveness as antimicrobials and antiviral, especially Covid-19. Moreover, pharmacokinetics and physicochemical properties were studied.
5. Experimental
5.1. Material and method
Reaction's progress was monitored by thin-layer chromatography (TLC) on silica gel-(G60 F254 (Merck)) of 0.5 mm thickness, visualizing with ultraviolet light (254 and 365 nm) or with iodine vapour. Melting points were determined using a Buchi B-540 capillary apparatus. NMR spectra were recorded on a Bruker Advance 850 MHz spectrometer (850 MHz for 1H NMR and 212.5 MHz for 13C NMR) respectively in CDCl3 solvent and tetramethyl silane as reference. Elemental analysis was carried out on Euro EA 3000 elemental analyser. Mass spectra were recorded on a Shimadzu GC-MS-QP-2010 mass spectrometer in EI (70 eV) model using the direct inlet probe technique. IR spectra of all spiropyrazole compounds were recorded on the IR Affinity 1S spectrometer (Shimadzu). Microwave synthesiser CEM mars machine was used for microwave irradiation.
5.1.1. Synthesis of spiropyrazole derivatives 6a,b, 10a-h and 14a,b
Equimolar quantity (0.002 mol) of arylidene derivatives of 5-bromo-indan-1-one 3 or 9 and hydrazonoyl halides 4a,b or 12 placed in Microwave-vial (HP-500) in 20 mL benzene for each reaction. All derivatives in each vial were irradiated at the same time by microwaves irradiation (400 W power) under pressure at 80 °C for 40–50 min. We tracked the reaction using TLC test. After making sure that the reactions were completed, the products were collected after the solvent was evaporated. The solid-colored products were crystalized after drying from dioxane/ethanol mixture.
5.1.2. Spiropyrazole derivative 6a
87% Yield; mp = 212-214 °C; yellow solid; IR (KBr): v 3058 (sp2 CH), 1720 (C=O), 1596 (C=N), 1498, 1376, 1208, 1088 cm−1. 1H NMR (CDCl3): δ 2.98 (d, J = 17.85 Hz, 1H, Ha), 3.43 (d, J = 17.85 Hz, 1H, Hb), 5.23 (s, 1H, pyrazole-H), 6.71 (d, J = 4.25 Hz, 1H, thiophene-H), 6.89 (t, J = 6.8 Hz, 1H, ArH), 6.93-7.29 (m, 9H, ArH and thiophene-H), 7.46 (s, 1H, Ar-H), 7.55 (d, J = 8.5 Hz, 2H, Ar-H), 7.63 (d, J= 7.7 Hz, 1H Ar-H), 7.77 (d, J = 8.5 Hz, 1H, Ar-H). MS m/z (%): 499 (M+, 16), 472 (74), 437 (46), 393 (39), 320 (19), 171 (25), 142 (54), 139 (100), 130 (50), 79 (43). Anal. Calcd for C27H19BrN2OS (499.42): C, 64.93; H, 3.83; N, 5.61%. Found C, 64.76; H, 3.79; N, 5.53%.
5.1.3. Spiropyrazole derivative 6b
90% Yield; mp = 207-210 °C; yellow solid; IR (KBr): v 3058 (sp2 CH), 2965 (sp3 CH), 1720 (C=O), 1596 (C=N), 1491, 1377, 1317, 1267, 1246, 1208, 1154, 1119, 1088, 1011 cm−1. 1H NMR (CDCl3): δ 2.98 (d, J = 18.7 Hz, 1H, Ha), 3.44 (d, J = 18.7 Hz, 1H, Hb), 5.23 (s, 1H, pyrazole-H), 6.71 (d, J = 3.4 Hz, 1H, thiophene-H), 6.89 (t, J = 7.65 Hz, 1H, ArH), 6.94 (d, J = 7.65 Hz, 3H, Ar-H), 7.17-7.26 (m, 5H, ArH and thiophene-H), 7.45 (s, 1H, Ar-H), 7.55 (d, J = 7.65 Hz, 2H, Ar-H), 7.68 (d, J= 8.5 Hz, 1H Ar-H), 7.78 (d, J = 7.65 Hz, 1H, Ar-H). MS m/z (%): 534 (M++1, 19), 533 (M+, 24), 516 (46), 475(24), 463 (22), 451 (34), 439 (25), 433 (46), 385 (44), 334 (100), 328 (36), 296 (46), 281 (62), 246 (24), 213 (29), 208 (57), 187 (72), 167 (28), 147 (42), 132 (49), 106 (42), 79 (37), 77 (25). Anal. Calcd for C27H18BrClN2OS (533.87): C, 60.74; H, 3.40; N, 5.25%. Found C, 60.65; H, 3.31; N, 5.09%.
5.1.4. Spiropyrazole derivative 10a
91% Yield; mp = 155-160 °C; yellow solid; IR (KBr): v 3060 (sp2 CH), 2934 (sp3 CH), 1755 (C=O), 1485, 14713, 1345, 1260, 1098, 1064 cm−1. 1H NMR (CDCl3): δ 2.77 (d, J = 17.5 Hz, 1H, Ha), 3.56 (d, J = 17.5 Hz, 1H, Hb), 4.70 (s, 1H, pyrazole-H), 6.35-7.84 (m, 17H Ar-H). MS m/z (%): 572 (M+, 56), 566 (56), 559 (50), 549 (100), 544 (62), 529 (53), 509 (76), 460 (45), 294 (55), 194 (64), 172 (55), 122 (69), 115 (55), 96 (70), 83 (51). Anal. Calcd for C29H20Br2N2O (572.29): C, 60.86; H, 3.52; N, 4.89%. Found C, 60.75; H, 3.48; N, 4.76%.
5.1.5. Spiropyrazole derivative 10b
90% Yield; mp = 125-126 °C; yellow solid; IR (KBr): v 3058 (sp2 CH), 1755 (C=O), 1489, 1471, 1316, 1262, 1091, 1024, 1014 cm−1. 1H NMR (CDCl3): δ 2.92 (d, J = 17.5 Hz, 1H, Ha), 3.42 (d, J = 17.5 Hz, 1H, Hb), 5.12 (s, 1H, pyrazole-H), 6.75-7.81 (m, 16H Ar-H). MS m/z (%): 606 (M+, 40), 598 (24), 556 (66), 476 (30), 439 (46), 308 (43), 289 (34), 223 (53), 217 (54), 208 (49), 199 (40), 177 (43), 175 (100), 156 (65), 145 (34). Anal. Calcd for C29H19Br2ClN2O (606.73): C, 57.41; H, 3.16; N, 4.62%. Found C, 57.27; H, 3.04; N, 4.58%.
5.1.6. Spiropyrazole derivative 10c
89% Yield; mp = 142-144 °C; yellow solid; IR (KBr): v 3058 (sp2 CH), 2996, 1947 (sp3 CH), 1724 (C=O), 1593 (C=N), 1494, 1380, 1269, 1246, 1028 cm−1. 1H NMR (CDCl3): δ 2.80 (d, J = 18.0 Hz, 1H, Ha), 3.38 (d, J = 18.0 Hz, 1H, Hb), 3.77 (s, 3H, OCH3), 4.91 (s, 1H, pyrazole-H), 6.74 (d, J = 8.5 Hz, 1H, Ar-H), 6.82 – 6.85 (m, 2H, ArH), 6.91 (d, J = 7.65 Hz, 1H, Ar-H), 7.06-7.75 (m, 13H Ar-H). MS m/z (%): 523 (M+, 27), 445 (54), 417 (36), 369 (25), 263 (20), 207 (32), 193 (15) 152 (35), 106 (67), 77 (100). Anal. Calcd for C30H23BrN2O2 (523.42): C, 68.84; H, 4.43; N, 5.35%. Found C, 68.67; H, 4.32; N, 5.19%.
5.1.7. Spiropyrazole derivative 10d
92% Yield; mp = 162-164 °C; yellow solid; IR (KBr): v 3060 (sp2 CH), 2836 (sp3 CH), 1727 (C=O), 1593 (C=N), 1309, 1247, 1154 cm−1. 1H NMR (CDCl3): δ 2.79 (d, J = 18.7 Hz, 1H, Ha), 3.36 (d, J = 18.7 Hz, 1H, Hb), 3.76 (s, 3H, OCH3), 4.87 (s, 1H, pyrazole-H), 6.75-6.86 (m, 5H, Ar-H), 6.89 (d, J = 8.5 Hz, 2H, Ar-H), 7.10 (t, J = 9.35 Hz, 2H, Ar-H), 7.18 (d, J = 8.5 Hz, 2H, Ar-H), 7.36 (s, 1H, Ar-H), 7.46 (d, J = 9.35 Hz, 2H, Ar-H), 7.59 (d, J = 7.65 Hz, 1H Ar-H), 7.74 (d, J = 8.5 Hz, 1H, Ar-H). 13C NMR (CDCl3): δ 55.2 (OCH3), 61.2 (C4 pyrazole), 78.5 (C-spiro), 114.4, 114.8, 115.8, 120.9, 126.0, 126.7, 127.7, 128.3, 128.4, 129.1, 129.9, 130.4, 131.6, 131.8, 133.9, 134.2, 142.4, 146.5, 152.3, 159.3, 203.3 (C=O). MS m/z (%): 557 (M+, 24), 522 (20), 348 (41), 338 (72), 329 (30), 229 (23), 206 (32), 111 (32), 107 (25), 105 (33), 77 (100). Anal. Calcd for C30H22BrClN2O2 (557.86): C, 64.59; H, 3.97; N, 5.02%. Found C, 64.41; H, 3.84; N, 4.95%.
5.1.8. Spiropyrazole derivative 10e
88% Yield; mp = 183-184 °C; yellow solid; IR (KBr): v 3063 (sp2 CH), 2928 (sp3 CH), 1713 (C=O), 1593 (C=N), 1475, 1413, 1313, 1260, 1210, 1107, 1037 cm−1. 1H NMR (CDCl3): δ 2.73(d, J = 18 Hz, 1H, Ha), 3.27 (d, J = 18 Hz, 1H, Hb), 4.70 (s, 1H, pyrazole-H), 7.03 (s, 1H, Ar-H), 7.05-7.18 (m, 10H, Ar-H), 7.22 (d, J = 7.65 Hz, 2H, Ar-H), 7.32 (t, J = 8.5 Hz, 1H, ArH), 7.40 (d, J = 8.5 Hz, 1H, Ar-H), 7.54 (d, J= 7.65 Hz, 1H, Ar-H), 7.62 (d, J = 8.5Hz, 1H, Ar-H). MS m/z (%): 527 (M+, 12), 525 (6), 519 (22), 505 (20), 465 (28), 441 (36), 405 (42), 399 (52), 367 (48), 362 (85), 346 (53), 316 (56), 294 (88), 275 (100), 244 (35), 138 (20), 121 (41), 96 (68), 78 (19). Anal. Calcd for C29H20BrClN2O (527.84): C, 65.99; H, 3.82; N, 5.31%. Found C, 65.78; H, 3.73; N, 5.20%.
5.1.9. Spiropyrazole derivative 10f
93% Yield; mp = 190-192 °C; yellow solid; IR (KBr): v 3063 (sp2 CH), 2927 (sp3 CH), 1716 (C=O), 1592 (C=N), 1438, 1413, 1313, 1260, 1210, 1107, 1037 cm−1. 1H NMR (CDCl3): δ 2.73(d, J = 17.85 Hz, 1H, Ha), 3.30 (d, J = 17.85 Hz, 1H, Hb), 5.35 (s, 1H, pyrazole-H), 7.03 (s, 1H, Ar-H), 7.09-7.80 (m, 15H, Ar-H). MS m/z (%): 562 (M+, 51), 537 (68), 498 (84), 432 (72), 390 (100), 305 (80), 201 (83), 191 (91), 149 (82), 142 (74), 102 (59). Anal. Calcd for C29H19BrCl2N2O (562.28): C, 61.95; H, 3.41; N, 4.98%. Found C, 61.78; H, 3.34; N, 4.86%.
5.1.10. Spiropyrazole derivative 10g
92% Yield; mp = 176-180 °C; yellow solid; IR (KBr): v 3065 (sp2 CH), 1717 (C=O), 1593 (C=N), 1529, 1491, 1350, 1317, 1208, 1058 cm−1. 1H NMR (CDCl3): δ 2.92 (d, J = 18 Hz, 1H, Ha), 3.43 (d, J = 18 Hz, 1H, Hb), 5.05 (s, 1H, pyrazole-H), 6.93 (d, J = 8.5 Hz, 1H, Ar-H), 7.15-7.83 (m, 14H, ArH), 8.01 (d, J = 7.65 Hz, 1H, Ar-H), 8.09 (d, J = 8.5 Hz, 1H, Ar-H). MS m/z (%): 540 (M++2, 2), 538 (M+, 36), 514 (11), 480 (29), 383 (13), 316 (15), 259 (34), 72 (51), 64 (100). Anal. Calcd for C29H20BrN3O3 (538.39): C, 64.69; H, 3.74; N, 7.80%. Found C, 64.56; H, 3.66; N, 7.73%.
5.1.11. Spiropyrazole derivative 10h
89% Yield; mp = 222-224 °C; yellow solid; IR (KBr): v 3060 (sp2 CH), 1674 (C=O), 1596 (C=N), 1491, 1368, 1297, 1195, 1090 cm−1. 1H NMR (CDCl3): δ 3.09 (d, J = 17.85 Hz, 1H, Ha), 3.45 (d, J = 17.85 Hz, 1H, Hb), 4.98 (s, 1H, pyrazole-H), 6.67 – 8.42 (m, 17H, ArH). MS m/z (%): 572 (M+, 20), 599 (84), 476 (33), 416 (34), 339 (65), 294 (100), 249 (72), 187 (44), 94 (45), 65 (86). Anal. Calcd for C29H20BrN3O3 (572.84): C, 60.80; H, 3.34; N, 7.34%. Found C, 60.72; H, 3.18; N, 7.20%.
5.1.12. Spiropyrazole derivative 14a
91% Yield; mp = 200-203 °C; yellow solid; IR (KBr): v 3060-3030 (sp2 CH), 1717 (C=O), 1595 (C=N), 1498, 1348, 1265, 1122, 1058 cm−1. 1H NMR (CDCl3): δ 2.98 (d, J = 17.5 Hz, 1H, Ha), 3.71 (d, J = 17.5 Hz, 1H, Hb), 4.85 (s, 1H, pyrazole-H), 6.0 (d, J = 12 Hz, 1H, CH=CH), 6.87 – 8.27 (m, 18H, Ar-H and CH=CH). MS m/z (%): 564 (M+, 25), 516 (30), 460 (19), 441 (32), 355 (42), 344 (25), 220 (25), 209 (26), 122 (52), 105 (36), 103 (12), 77 (52). Anal. Calcd for C31H22BrN3O3 (564.43): C, 65.97; H, 3.93; N, 7.44%. Found C, 65.78; H, 3.90; N, 7.36%.
5.1.13. Spiropyrazole derivative 14b
90% Yield; mp = 205-207 °C; yellow solid; IR (KBr): v 3058- 3026 (sp2 CH), 1663 (C=O), 1596 (C=N), 1496, 1339, 1195, 1071cm−1. 1H NMR (CDCl3): δ 3.08 (d, J = 17.85 Hz, 1H, Ha), 3.70 (d, J = 17.85 Hz, 1H, Hb), 4.75 (s, 1H, pyrazole-H), 6.20 (d, J = 12 Hz, 1H, CH=CH), 7.03 – 8.70 (m, 18H, ArH and CH=CH). MS m/z (%): 564 (M+, 26), 561 (48), 538 (47), 501 (52), 468 (64), 400 (28), 371 (37), 348 (69), 306 (33), 284 (48), 253 (100), 241 (60), 192 (66), 106 (52), 77 (26) Anal. Calcd for C31H22BrN3O3 (564.43): C, 65.97; H, 3.93; N, 7.44%. Found C, 65.85; H, 3.79; N, 7.32%.
Declaration of Competing Interest
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.133581.
Appendix. Supplementary materials
References
- 1.https://www.who.int/news-room/detail/27-04-2020-who-timeline—covid-19
- 2.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domínguez-Villa F.X., Durán-Iturbide N.A, Ávila-Zárraga J.G. Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl)indol-4-ones: potential inhibitors of SARS CoV-2 main protease. Bioorg. Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asha R.N., Nayagam B.R.D., Bhuvanesh N. Synthesis, molecular docking, and in silico ADMET studies of 4-benzyl-1-(2,4,6-trimethyl-benzyl)-piperidine: potential inhibitor of SARS-CoV2. Bioorg. Chem. 2021;112 doi: 10.1016/j.bioorg.2021.104967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoud H.K., Asghar B.H., Harras M.F., Farghaly T.A. Nano-sized formazan analogues: synthesis, structure elucidation, antimicrobial activity and docking study for COVID-19. Bioorg. Chem. 2020;105 doi: 10.1016/j.bioorg.2020.104354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Report on Surveillance of Antibiotic Consumption (2018). https://www.who.int/medicines/areas/rational_use/oms-amr-amc-report-2016-2018/en/.
- 7.Nidhar M., Khanam S., Sonker P., Gupta P., Mahapatra A., Patil S., Yadav B.K., Singh R.K., Tewari A.K. Click inspired novel pyrazole-triazole-persulfonimide & pyrazole-triazole-aryl derivatives; design, synthesis, DPP-4 inhibitor with potential anti-diabetic agents. Bioorg. Chem. 2022;120 doi: 10.1016/j.bioorg.2021.105586. [DOI] [PubMed] [Google Scholar]
- 8.Yao H., Guo Q., Rui M.W., Xu W.Z. Discovery of pyrazole N-aryl sulfonate: a novel and highly potent cyclooxygenase-2 (COX-2) selective inhibitors. Bioorg. Med. Chem. 2021;46 doi: 10.1016/j.bmc.2021.116344. [DOI] [PubMed] [Google Scholar]
- 9.Huo H., Jiang W., Sun F., Li J., Shi B. Synthesis and biological evaluation of novel steroidal pyrazole amides as highly potent anticancer agents. Steroids. 2021;176 doi: 10.1016/j.steroids.2021.108931. [DOI] [PubMed] [Google Scholar]
- 10.Patel B., Zunk M., Grant G., Rudrawar S. Design, synthesis and bioactivity evaluation of novel pyrazole linked phenylthiazole derivatives in context of antibacterial activity. Bioorg. Med. Chem. Lett. 2021;39 doi: 10.1016/j.bmcl.2021.127853. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z., Yang W., Hou S., Xie D., Yang J., Liu L., Yang S. In vivo antiviral activity and disassembly mechanism of novel 1-phenyl-5-amine-4-pyrazole thioether derivatives against Tobacco mosaic virus. Pestici. Biochem. Physiol. 2021;173 doi: 10.1016/j.pestbp.2021.104771. [DOI] [PubMed] [Google Scholar]
- 12.Rui R., Tang C., Zhang C., Pan W., Gan K., Luo R., Wei Z., Jing F., Huang S., Yang L., Li Y., Wang Y., Xiao W., Zhang H., Zheng Y., He Y. C6-structural optimizations of 2-aryl-1H-pyrazole-S-DABOs: From anti-HIV to anti-DENV activity. Bioorg. Chem. 2022;119 doi: 10.1016/j.bioorg.2021.105494. [DOI] [PubMed] [Google Scholar]
- 13.Amata E., Bland N.D., Campbell R.K., Pollastri M.P. Evaluation of pyrrolidine and pyrazolone derivatives as inhibitors of trypanosomal phosphodiesterase B1 (TbrPDEB1) Tetrahedron Lett. 2015;56:2832. doi: 10.1016/j.tetlet.2015.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S., Li Y., Xu G., Chen S., Zhang Y., Liu N., Dong G., Miao C., Su H., Zhang W., Sheng C. Novel spiropyrazolone antitumor scaffold with potent activity: design, synthesis and structure activity relationship. Eur. J. Med. Chem. 2016;115:141–147. doi: 10.1016/j.ejmech.2016.03.039. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro Â., Gonçalves L.M., Santos M.M.M. Synthesis of novel spiropyrazoline oxindoles and evaluation of cytotoxicity in cancer cell lines. Eur. J. Med. Chem. 2014;79:266–272. doi: 10.1016/j.ejmech.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 16.P.S. Silaychev, V.O. Filimonov, A.N. Maslivets, R.R. Makhmudov, Russ. Patent (2014) RU2577528.
- 17.Tada M., Motoki N., Takahashi T.M. Synthesis and Structure-activity relationships of miticidal 4,5-dihydropyrazole-5-thiones. Pestic. Sci. 1996;48:165. [Google Scholar]
- 18.Y. Seekaew, O. Arayawut, K. Timsorn, C. Wongchoosuk. Chapter nine – synthesis, characterization, and applications of graphene and derivatives. carbon-based nanofillers and their rubber nanocomposites carbon nano-objects (2019) 259-283.
- 19.Padwa A., Flick A.C. Chapter one – intramolecular diels–alder cycloaddition of furans (IMDAF) for natural product synthesis. Adv. Heterocycl. Chem. 2013;110:1–41. [Google Scholar]
- 20.Y. Seekaew, O. Arayawut, K. Timsorn, C. Wongchoosuk. Chapter nine - synthesis, characterization, and applications of graphene and derivatives. carbon-based nanofillers and their rubber nanocomposites carbon nano-objects (2019) 259-283.
- 21.Masaret G.S. A new approach for the synthesis and biological activities of novel thiazolyl-pyrazole derivatives. Chem. Sel. 2012;6:1–10. [Google Scholar]
- 22.Omar M.A., Masaret G.S., Abbas E.M.H., Abdel-Aziz M.M., Harras M.F., Farghaly T.A. Novel anti-tubercular and antibacterial based benzosuberone-thiazole moieties: Synthesis, molecular docking analysis, DNA gyrase supercoiling and ATPase activity. Bioorg. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104316. [DOI] [PubMed] [Google Scholar]
- 23.Farghaly T.A., Masaret G.S., Muhammad Z.A., Harras M.F. Discovery of thiazole-based-chalcones and 4-hetarylthiazoles as potent anticancer agents: synthesis, docking study and anticancer activity. Bioorg. Chem. 2020;98 doi: 10.1016/j.bioorg.2020.103761. [DOI] [PubMed] [Google Scholar]
- 24.Masaret G.S. Convenient synthesis and anticancer evaluation of novel pyrazolyl-thiophene, thieno[3,2-b]pyridine, pyrazolo[3,4-d] thieno[3,2-b]pyridine and pyrano[2,3-d]thieno[3,2-b] pyridine derivatives. J. Heterocycl. Chem. 2021:1–15. [Google Scholar]
- 25.Ishimaru T., Shibata N., Horikawa T., Yasuda N., Nakamura S., Toru T., Shiro M. Angew. Chem. Int. Ed. 2008;47:4157–4161. doi: 10.1002/anie.200800717. Angew. Chem. 120 (2008) 4225–4229. [DOI] [PubMed] [Google Scholar]
- 26.Girgisa A.S., Ibrahimb Y.A., Ahmed Faraga I.S., Mishriky N. Simple regioselective synthesis of spiro[2H-indene-2,3’-[3H]pyrazoles] J. Chem. Research (S), 2000, 508–509. J. Chem. Research (M) 2000:1272–1284. [Google Scholar]
- 27.Fathi T., Ciamala K., Dinh An N., Vebrel J. Comportement particulier d’arylidènebenzothiazinonedioxydes dans les cycloadditions [3 + 2]. Compétition entre sites dipolarophiliques. Évolution des cycloadduits. Can. J. Chem. 1994;72:1424. [Google Scholar]
- 28.Mishriky N., Girgis A.S., Ibrahim Y.A. Regioselective Synthesis of Spiro[Naphthalene-2(1H),3′-[3H]Pyrazol]-1-Ones Utilizing 1,3-Dipolar Cycloaddition of Nitrilimines. J. Chem. Res. 2000:2. (S) [Google Scholar]
- 29.Daina A., Michielin O., Zoete V. A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1):1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waring M.J. Defining optimum lipophilicity and molecular weight ranges for drug candidates-Molecular weight dependent lower logD limits based on permeability. Bioorg. Med. Chem. Lett. 2009;19(10):2844–2851. doi: 10.1016/j.bmcl.2009.03.109. [DOI] [PubMed] [Google Scholar]
- 31.Ertl P., Rohde B., Selzer P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000;43(20):3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.