Abstract
PURPOSE
Symptoms are vital outcomes for cancer clinical trials, observational research, and population-level surveillance. Patient-reported outcomes (PROs) are valuable for monitoring symptoms, yet there are many challenges to collecting PROs at scale. We sought to develop, test, and externally validate a deep learning model to extract symptoms from unstructured clinical notes in the electronic health record.
METHODS
We randomly selected 1,225 outpatient progress notes from among patients treated at the Dana-Farber Cancer Institute between January 2016 and December 2019 and used 1,125 notes as our training/validation data set and 100 notes as our test data set. We evaluated the performance of 10 deep learning models for detecting 80 symptoms included in the National Cancer Institute's Patient-Reported Outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE) framework. Model performance as compared with manual chart abstraction was assessed using standard metrics, and the highest performer was externally validated on a sample of 100 physician notes from a different clinical context.
RESULTS
In our training and test data sets, 75 of the 80 candidate symptoms were identified. The ELECTRA-small model had the highest performance for symptom identification at the token level (ie, at the individual symptom level), with an F1 of 0.87 and a processing time of 3.95 seconds per note. For the 10 most common symptoms in the test data set, the F1 score ranged from 0.98 for anxious to 0.86 for fatigue. For external validation of the same symptoms, the note-level performance ranged from F1 = 0.97 for diarrhea and dizziness to F1 = 0.73 for swelling.
CONCLUSION
Training a deep learning model to identify a wide range of electronic health record–documented symptoms relevant to cancer care is feasible. This approach could be used at the health system scale to complement to electronic PROs.
INTRODUCTION
Patients with cancer experience a multitude of distressing symptoms related to their disease and the side effects of treatment. Symptoms have a major effect on patients' quality of life,1,2 treatment tolerance, and prognosis.3,4 Symptoms are therefore critical outcomes to monitor in therapeutic clinical trials, observational research, and population-level surveillance.
CONTEXT
Key Objective
Is it feasible to rapidly capture patients' symptoms from unstructured clinical notes in the electronic health record (EHR) using deep learning?
Knowledge Generated
Deep learning models trained to detect clinically relevant symptoms exhibited high performance. Deep learning allowed for symptom detection to occur rapidly, as opposed to much longer times demanded for symptom detection during manual chart review.
Relevance
Health systems could use deep learning models to scale detection of symptoms documented in the EHR, which are relevant to cancer care; EHR-based symptom assessments could be automated for a variety of research, clinical, and regulatory purposes.
In recent years, standardized patient-reported outcome (PRO) measures have become a common method to assess patients' symptoms within clinical research and routine clinical care. Electronic systems to systematically assess PROs have demonstrated benefits of symptom control, health care utilization, and even survival.5 Despite the promise of electronic PRO tools, many patients do not complete PROs or do so only intermittently.6 This is particularly true among historically disadvantaged populations, such as the elderly, the poor, or those living in rural areas, who may lack reliable internet access or the devices required to use PRO systems.7-9 Complementary data sources and assessment methods are therefore required to monitor symptoms at scale and to avoid the biases inherent to direct patient reporting.
The electronic health record (EHR) is a rich source of data regarding symptoms owing to the fact that providers routinely assess and document systems within clinical notes. Yet, this resource remains underutilized for symptom extraction at scale given the difficult and time-consuming process of manual chart abstraction. Unlike discrete structured data points such as vital signs or laboratory data, symptoms are traditionally recorded in clinical notes as narrative free text. Extraction of symptom information from unstructured clinical notes is time-consuming, expensive, and error-prone and requires clinical expertise.10
Deep learning models are increasingly important tools for extracting oncologic end points from unstructured EHR text data.10-12 Models have been developed that can extract data on cancer progression and response and documentation of end-of-life care preferences.13-15 To date, valid, reliable models for identifying cancer-related symptoms have not been developed. The purpose of this study was to (1) develop and test a deep learning model for symptom extraction from unstructured clinical notes and (2) externally validate the method in a data set from another health care system.
METHODS
Data Source and Study Sample
Our training and test data sets were derived from the Dana-Farber Cancer Institute (DFCI) instance of the Epic EHR (Verona, WI). Clinician (medical, surgical, radiation oncologist, nurse practitioner, and physician assistant) progress notes were randomly selected from among all patients seen in breast, GI, thoracic, gynecologic, psychosocial, and palliative care clinics at DFCI between January 2016 and December 2019. Of the 1,225 selected notes, 1,125 notes were randomly selected for our model training/validation data set and 100 were used for our test data set.
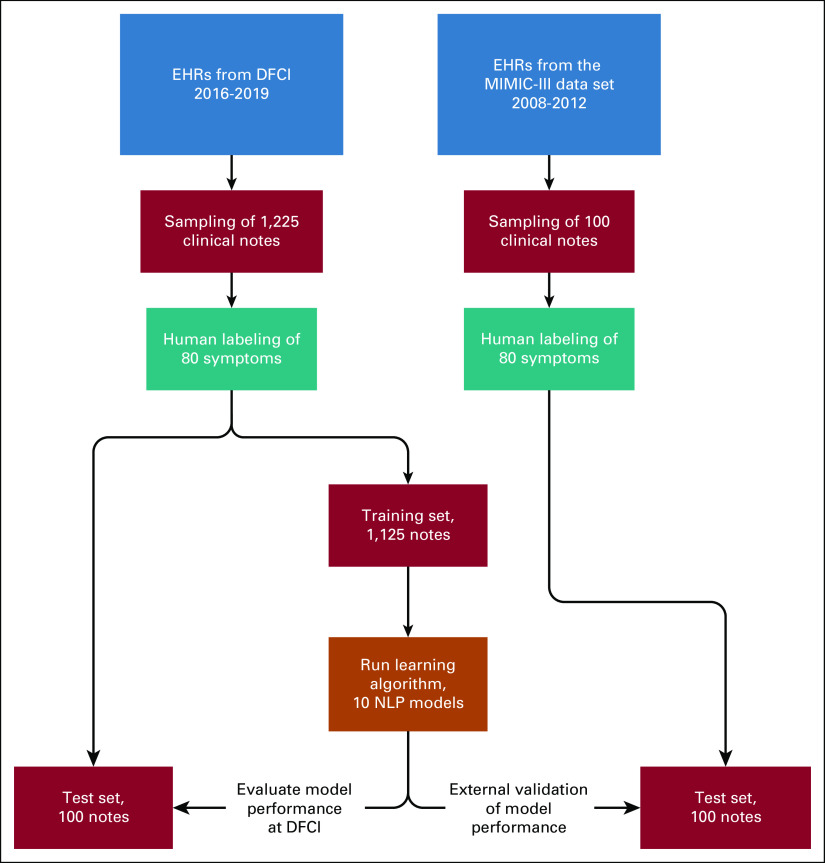
To evaluate generalizability, the model was externally validated using 100 physician notes randomly selected from a data set used in our previously published study,13 obtained from the Medical Information Mart for Intensive Care III (MIMIC-III) data set. MIMIC-III is an existing data set composed of all EHR notes for patients in intensive care units at the Beth Israel Deaconess Medical Center (Boston, MA) between January 2008 and December 2012. This data source was chosen for external validation to assess the transferability of our deep learning model for symptom detection to other care contexts (ie, symptoms documentation may differ in an ICU versus an outpatient setting). Figure 1 shows a flowchart for the derivation of the data sets and methods used. This study was approved by the DFCI Institutional Review Board (IRB 18-192); informed consent was not required.
FIG 1.
Flowchart of methods. DFCI, Dana-Farber Cancer Institute; EHR, electronic health record; MIMIC-III, Medical Information Mart for Intensive Care III; NLP, natural language processing.
Symptom Definitions and Data Annotation
We used the National Cancer Institute's Patient-Reported Outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE)16 as our framework to establish coding rules for annotating symptoms reported in EHRs. The PRO-CTCAE system was a collaborative effort between multiple stakeholders, including the US Food and Drug Administration, for creating a standardized patient-reporting tool for identification of symptoms that may be associated with adverse events.17 The PRO-CTCAE system is narrower than the full CTCAE, in that it focuses specifically on symptoms. The full PRO-CTCAE assesses a total of 80 symptoms, using between one and three survey items per symptom to assess severity, frequency, and interference. Using the PRO-CTCAE as a coding framework, a team of three internal medicine, medical oncology, and palliative care physicians independently reviewed and annotated 25 notes and labeled the text for the presence of symptoms. Discordance in annotations was discussed during in-person meeting coordinated by the project lead (C.L.). These first 25 notes were not included in the model training/validation data set. Another 50 notes were annotated by a single physician (W.M.) and then reviewed and discussed by three physicians to establish consensus labeling rules. The remaining 1,200 notes were annotated by a single physician (W.M.) for each of the 85 labels (80 symptoms and five attributes, including negation) using an open-source web-based software label studio18 (Appendix Fig A1). Label studio allows for linkage between negation and symptom(s).
Another 100 notes obtained from the MIMIC-III data set were also annotated by the physician (W.M.). The physician (W.M.) had timely access to discuss challenging labels with the project lead (C.L.).
Data Preprocessing
After data annotation and before training deep learning models, the texts were converted into the Computational Natural Language Learning-2003 (CoNLL-2003) standard format.19 First, the texts were tokenized (split up into useful semantic units for processing) using the scispaCy en_core_sci_lg tokenizer.20 Then, each token was aligned with the appropriate label by comparing the (start and end) offsets of the tokens and annotated spans.
We then extracted the History of Present Illness from each note given that this section is most likely to contain the most current information, unlike other note sections (eg, Review of Systems), which are frequently copy forwarded from previous encounters.21 The History of Present Illness in each note was identified using a set of regular expressions as described previously,21 and sentence boundary tokens were added to the token sequences. The position of each token (including words and subwords) and label tags were also recorded to process the model output.
Training Deep Learning Models
Preprocessed text was used as input for a named entity recognition (NER) task for symptom extraction. In deep learning, NER is the task of classifying short sequences of tokens or entities within a text into predefined classes. In recent years, the transformer model architecture has gained recognition for achieving state-of-the art results on several natural language processing (NLP) benchmark tasks.22,23 Since the original publication, many Transformer models have been made available that have been trained on very large general-purpose corpora, such as Wikipedia. These pretrained models can then be leveraged as a starting point for further training on new data, a generally less computationally expensive process than training a comparable model from scratch. Models selected for training included the following: BERT,24 XLNet,25 RoBERTa,26 XLM-RoBERTa,27 DistilBERT,28 ELECTRA,29 and Longformer.30 DFCI data were split into training and test sets (Fig 1). Combinations of the aforementioned batch size and learning rates were used to calculate the F1 score for every combination. The best performing combination during the cross-validation step on the validation set was selected as our final model parameters and was used for testing on both DFCI and MIMIC III data sets.
We used two NVIDIA Tesla V100 (32 GB) GPU to fine tune Electra-SMALL for NER. The maximum sequence length was fixed to 512, the minibatch size was selected from 8, 16, 32, or 64, and a learning rate of 6e-5, 3e-5, or 1e-5 was selected.
Statistical Methods
Evaluation metrics of model performance were based on the CoNLL-2003 standard,19 including precision (positive predictive value), recall (sensitivity), and F1 measure (ie, the harmonic mean between precision and recall) for token-level and note-level analysis. Negations were linked to symptoms at the sentence level. This means that if a symptom was negated (eg, no fatigue) in the gold standard, but the NLP model only picked up the symptom and not the negation (eg, fatigue), this was counted as false positive in the evaluation metrics. For note-level analysis, if there were contradictions within a document with respect to an individual symptom, such as the presence and absence of a symptom, the document was considered to be positive for that symptom. For example, if one part of a note said “denies SOB [shortness of breath],” whereas another part indicated that shortness of breath was present, this note would be considered positive for reporting the symptom.
RESULTS
Note and Patient Characteristics
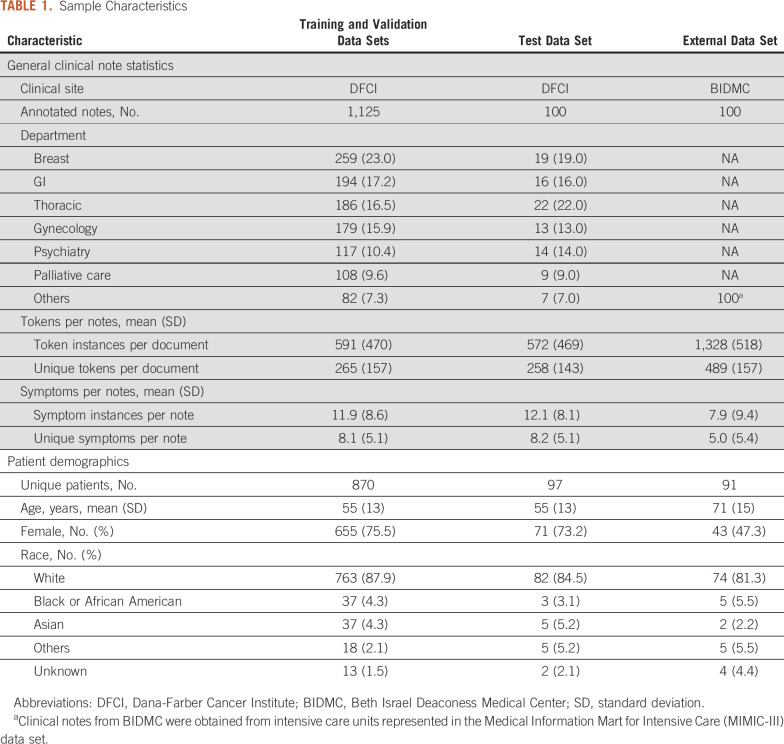
The study examined 1,125 unique notes on 870 unique patients in the DFCI training/validation data set and 100 unique notes on 97 unique patients in the DFCI test set. The MIMIC-III external test data set included 100 unique notes for 91 unique patients. Demographic information and general statistics for clinical notes are presented in Table 1. The DFCI training/validation and test data set contained a total of 2,793,511 tokens, whereas the external validation test data set contained 177,362 tokens.
TABLE 1.
Sample Characteristics
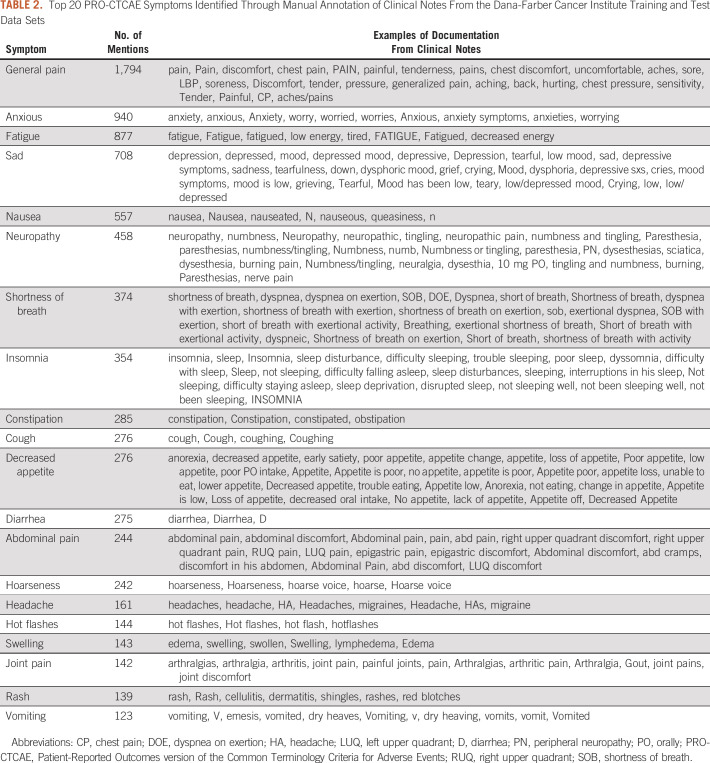
In the DFCI training/validation and test data set, we identified an average of 12 symptoms per clinical note. Of the 80 total PRO-CTCAE symptoms, only five were not identified in any notes: decreased sweating, delayed orgasm, ejaculation, no orgasm, and stretch marks. Table 2 presents a qualitative demonstration of the variety of contexts in which the 20 most frequently reported PRO-CTCAE symptoms were documented. The 20 and 10 most frequently documented symptoms represented 96% and 75% of all symptom occurrences, respectively. General pain was the most commonly documented symptom representing 20% of symptom occurrences.
TABLE 2.
Top 20 PRO-CTCAE Symptoms Identified Through Manual Annotation of Clinical Notes From the Dana-Farber Cancer Institute Training and Test Data Sets
Token-Level Model Performance
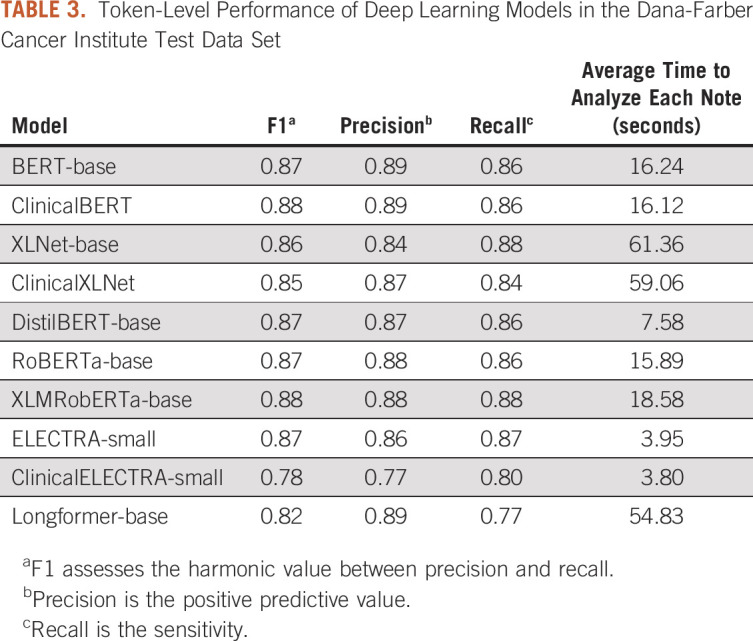
When examining the performance of the 10 deep learning models in the DFCI test data set at the token level (ie, sensitivity and specificity for distinct mentions of a symptom), BERT, ClinicalBERT, XLNet, RoBERTa, XLM-RoBERTa, DistilBERT, and ELECTRA all achieved an F1 score > 0.85. XLM-RoBERTa-base, which has the largest pretrained corpus among all the models, achieved the highest performance (F1 = 0.88). However, DistilBERT and ELECTRA-small had faster processing times (7.58 seconds and 3.95 seconds, respectively) and a similar performance (F1 = 0.87) as the XLM-RoBERTa-base model. The F1 scores, recall, precision, and processing time for each of the 10 models are presented in Table 3.
TABLE 3.
Token-Level Performance of Deep Learning Models in the Dana-Farber Cancer Institute Test Data Set
Note-Level Model Performance
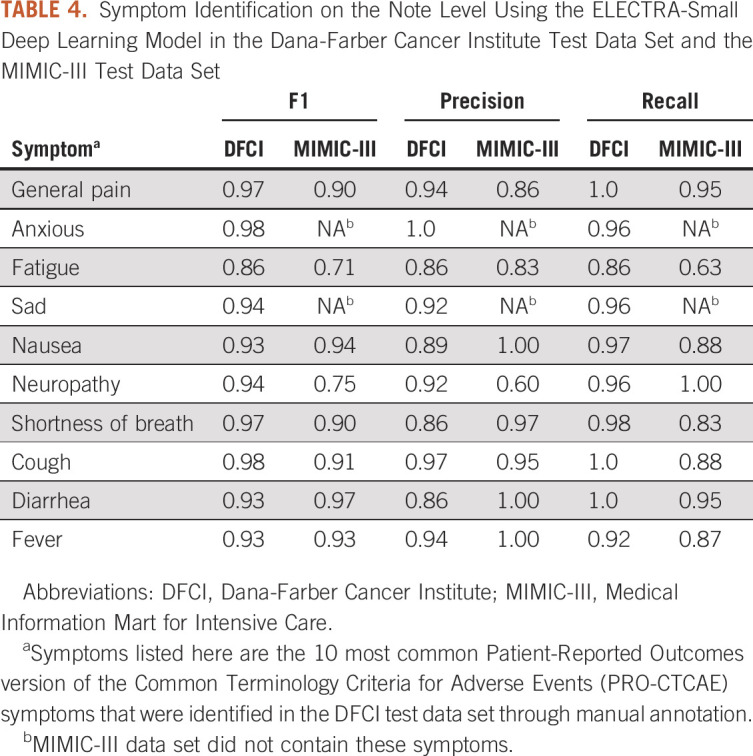
Given the combination of high performance with a fast processing time, we chose the ELECTRA-small model for validation on the note level. The F1 scores using ELECTRA-small ranged from 0.98 for identification of anxious to 0.86 for identification of fatigue. The F1 scores, precision, and recall for the 10 most common symptoms in the DFCI test data set are presented in Table 4.
TABLE 4.
Symptom Identification on the Note Level Using the ELECTRA-Small Deep Learning Model in the Dana-Farber Cancer Institute Test Data Set and the MIMIC-III Test Data Set
External Validation
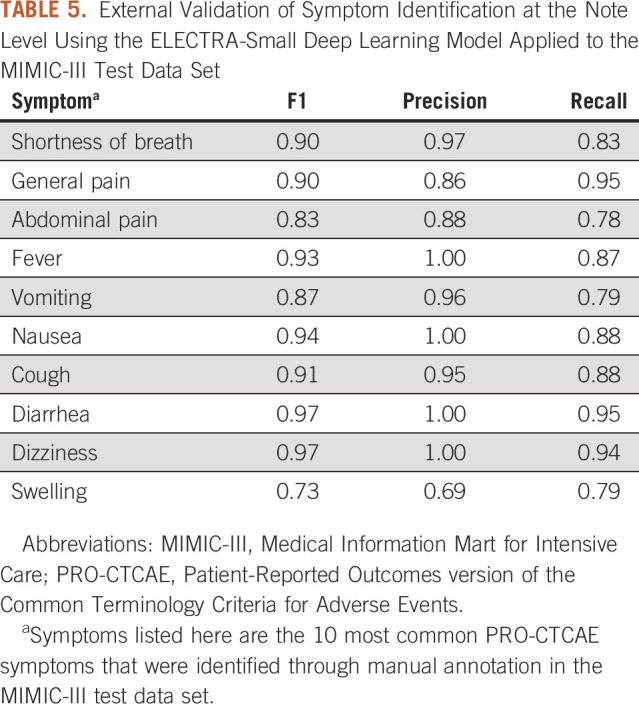
In the MIMIC-III data set used for external validation, an average of eight symptoms were identified per clinical note and 38 PRO-CTCAE symptoms were identified across the data set. Six of the 10 most frequently documented symptoms in the DFCI test data set achieved an F1 score > 0.85 in external validation (Table 4). For the top 10 most common frequently documented symptoms in the MIMIC-III data set, F1 scores ranged from 0.73 for swelling to 0.97 for diarrhea and dizziness (Table 5).
TABLE 5.
External Validation of Symptom Identification at the Note Level Using the ELECTRA-Small Deep Learning Model Applied to the MIMIC-III Test Data Set
DISCUSSION
We built, tested, and externally validated a deep learning model that extracts symptoms directly from clinical notes in EHRs. To our knowledge, this is the first creation of a deep learning model using the PRO-CTCAE framework, thus ensuring that clinically relevant patient symptoms are captured. The ELECTRA-small model achieved the highest performance for symptom identification at the token level, and at the note level, it achieved an F1 score > 0.90 for the 10 most frequently documented symptoms. This suggests that our model had high performance in identifying documentation containing relevant clinical symptoms for review. With a processing time of only 3.95 seconds per note, deep learning could greatly accelerate evaluations of patient symptoms as compared with manual chart abstraction, which typically requires over an hour of chart review per patient.31 The high performance of our model suggests that deep learning would be suitable for automated, EHR-based symptom assessments for a variety of research, clinical, and regulatory purposes. Deep learning can be used in retrospective studies of cancer symptoms, postmarketing drug surveillance programs, and for novel care delivery innovations to improve monitoring and proactive intervention in response to patient symptoms.
Electronic PROs are arguably one of the most important innovations in cancer care delivery, yet they have important limitations that could be ameliorated by deep learning. The sensitivity of PROs in the real world depends upon the frequency of patient reporting. PRO's may therefore miss important symptoms if patients may choose not to complete PROs or if they selectively report when they are feeling well. Clinically important symptoms are likely to be documented within a clinical encounter; therefore, our model could identify symptoms that might otherwise be missed by PROs. Moreover, several studies suggest that symptoms documented by clinicians may be more predictive of serious clinical events (eg, emergency department visits or mortality) than symptoms directly reported by patients.5 Therefore, symptoms identified by deep learning may complement and ultimately provide more meaningful and actionable information than PROs alone. Finally, there are well-documented sociodemographic disparities in the use of electronic PROs, with lower completion rates among patients who are racial/ethnic minorities and elderly and have limited English proficiency, cognitive disabilities, psychiatric disorders, and visual impairment.9,32 Therefore, relying solely on electronic PROs could magnify racial/ethnic disparities in the quality of cancer care. Our methods could help overcome this problem and promote equity in cancer care.
A recent systematic review of automated methods to extract symptoms from EHRs found that most previous efforts have focused on very narrow sets of symptoms relevant to diseases of interest (eg, heart failure,33 multiple sclerosis,34 and acute respiratory distress syndrome)35,36 or to specific adverse drug events (eg, rash and arrhythmia).37-39 By contrast, few studies have focused on the extraction of broad sets of symptoms that are necessary within oncology or for research or care interventions focused primarily on symptom management.40,41 Efforts to perform automated extraction of symptoms from EHRs for the oncologic population have been limited, with only 11% of such previous studies featuring oncology as the clinical specialty of interest.36 These few studies are limited by the absence of a clear guiding framework to identify clinically meaningful symptoms for adverse event monitoring. By contrast, we followed the PRO-CTCAE framework, which has been rigorously developed with guidance from the National Cancer Institute, the US Food and Drug Administration, and patient advocates and is widely used in cancer care delivery research and in therapeutic clinical trials.42 Our automated method to identify PRO-CTCAE symptoms is therefore a significant advance that could accelerate research and clinical innovations to improve symptom management in the oncologic population.
Our study has several limitations. Although deep learning models were trained to identify 80 PRO-CTCAE symptoms considered to be most clinically relevant to the care of oncology patients, this framework does not encompass all the possible symptoms that a patient may experience and does not extract descriptive aspects of symptoms such as quality, severity, and frequency. The utility of this model for abstracting symptoms for patient populations outside of the oncology context may therefore be limited. Nevertheless, our study included external validation of our model in another health care system with a distinctly different validation cohort consisting of ICU patients. EHRs of ICU patients may be more likely to describe a different set of symptoms (eg, shortness of breath) compared with longitudinal progress notes. Although the high performance of the model in this patient population may suggest its transferability, further efforts for external validation in disparate patient populations may be warranted. Although our study presents an important method for supplementing ongoing efforts to identify patient symptoms using electronic PROs, ensuring that all relevant patient symptoms are captured requires innovative efforts that can use machine learning for identifying symptoms from clinician-patient conversations. An important limitation of this work is its basis in EHR data; although clinical notes comprise an important unstructured data source, they may not be fully reflective of patient-experienced symptoms, as clinicians tend to under-report symptoms compared with what patients would self-report.43 Therefore, additional efforts to extract verbally discussed symptoms would identify critical information that a clinician may not document or a patient may not report in a questionnaire. A combination of symptom abstraction from EHRs, electronic PROs, and audio recordings would represent the most comprehensive approach to date for adverse event monitoring and effective symptom management. Finally, a single physician annotated all clinical notes used for model training and testing. Discordance is common when multiple clinicians annotate symptoms in clinical notes.31,43,44 Interestingly, studies have shown that NLP models can outperform humans in identifying text-based data.13,45
In conclusion, we demonstrated that NLP methods can be applied to EHRs for extraction of symptoms at scale. The use of the PRO-CTCAE framework to guide training of deep learning ensures that the model captures a variety of symptoms considered to be most clinically meaningful in the oncology context. Implementation of this automated surveillance method in conjunction with electronic PROs can enable real-time adverse event monitoring and ongoing quality improvement efforts.
APPENDIX
FIG A1.

Example of the annotation interface, label studio, for symptom identification in clinical notations. Ground Truth represents manual annotations, and Prediction represents symptoms identified by a deep learning algorithm.
Charlotta Lindvall
Patents, Royalties, Other Intellectual Property: System and Method of Using Machine Learning for Extraction of Symptoms From Electronic Health Records patient application 2705.001 (Inst)
Chih-Ying Deng
Employment: Verily, Google Health
Travel, Accommodations, Expenses: Google Health
Renato Umeton
Patents, Royalties, Other Intellectual Property: Patent: Portable medical device and method for quantitative retinal image analysis through a smartphone, Patent: Epstein Barr virus genotipic variants and uses thereof as risk predictors, biomarkers and therapeutic targets of multiple sclerosis
Kenneth L. Kehl
Employment: Change Healthcare
Honoraria: Roche, IBM (Inst)
Andrea C. Enzinger
Consulting or Advisory Role: Five Prime Therapeutics, Merck, Astellas Pharma, Lilly, Loxo, Taiho Pharmaceutical, Daiichi Sankyo, AstraZeneca, Zymeworks, Takeda, Istari, Ono Pharmaceutical, Xencor, Novartis
Research Funding: Medtronic
No other potential conflicts of interest were reported.
SUPPORT
Supported by the Poorvu Jaffe Family Foundation and Dana-Farber Cancer Institute.
DATA SHARING STATEMENT
Code is publicly available on GitHub (https://github.com/lindvalllab/MLSym). MIMIC-III notes used in the study can be accessed after approval at https://mimic.mit.edu/. DFCI clinical notes cannot be shared due to identifiable protected health information. For questions about the code or data sets, please contact the corresponding author.
AUTHOR CONTRIBUTIONS
Conception and design: Charlotta Lindvall, Chih-Ying Deng, Renato Umeton, James A. Tulsky, Andrea C. Enzinger
Financial support: Charlotta Lindvall, James A. Tulsky
Administrative support: Charlotta Lindvall
Provision of study materials or patients: Charlotta Lindvall, James A. Tulsky
Collection and assembly of data: Charlotta Lindvall, Chih-Ying Deng, Warren Mackie-Jenkins, James A. Tulsky
Data analysis and interpretation: Charlotta Lindvall, Chih-Ying Deng, Nicole D. Agaronnik, Anne Kwok, Soujanya Samineni, Renato Umeton, Kenneth L. Kehl, James A. Tulsky, Andrea C. Enzinger
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Charlotta Lindvall
Patents, Royalties, Other Intellectual Property: System and Method of Using Machine Learning for Extraction of Symptoms From Electronic Health Records patient application 2705.001 (Inst)
Chih-Ying Deng
Employment: Verily, Google Health
Travel, Accommodations, Expenses: Google Health
Renato Umeton
Patents, Royalties, Other Intellectual Property: Patent: Portable medical device and method for quantitative retinal image analysis through a smartphone, Patent: Epstein Barr virus genotipic variants and uses thereof as risk predictors, biomarkers and therapeutic targets of multiple sclerosis
Kenneth L. Kehl
Employment: Change Healthcare
Honoraria: Roche, IBM (Inst)
Andrea C. Enzinger
Consulting or Advisory Role: Five Prime Therapeutics, Merck, Astellas Pharma, Lilly, Loxo, Taiho Pharmaceutical, Daiichi Sankyo, AstraZeneca, Zymeworks, Takeda, Istari, Ono Pharmaceutical, Xencor, Novartis
Research Funding: Medtronic
No other potential conflicts of interest were reported.
REFERENCES
- 1.Cleeland CS.Symptom burden: Multiple symptoms and their impact as patient-reported outcomes J Natl Cancer Inst Monogr 3716–212007 [DOI] [PubMed] [Google Scholar]
- 2.Bubis LD, Davis L, Mahar A, et al. Symptom burden in the first year after cancer diagnosis: An analysis of patient-reported outcomes J Clin Oncol 361103–11112018 [DOI] [PubMed] [Google Scholar]
- 3.Basch E, Deal AM, Dueck AC, et al. Overall survival results of a trial assessing patient-reported outcomes for symptom monitoring during routine cancer treatment JAMA 318197–1982017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basch E, Deal AM, Kris MG, et al. Symptom monitoring with patient-reported outcomes during routine cancer treatment: A randomized controlled trial J Clin Oncol 34557–5652016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basch E, Jia X, Heller G, et al. Adverse symptom event reporting by patients vs clinicians: Relationships with clinical outcomes J Natl Cancer Inst 1011624–16322009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamper EM, Nerich V, Sztankay M, et al. Evaluation of noncompletion bias and long-term adherence in a 10-year patient-reported outcome monitoring program in clinical routine Value Health 20610–6172017 [DOI] [PubMed] [Google Scholar]
- 7. Yocavitch L, Binder A, Leader A, et al. Challenges in implementing a mobile-based patient-reported outcome (PRO) tool for cancer patients. J Clin Oncol. 2019;37 suppl 27; abstr 206. [Google Scholar]
- 8. Biber J, Ose D, Reese J, et al. Patient reported outcomes—Experiences with implementation in a University Health Care setting. J Patient Rep Outcomes. 2017;2:34. doi: 10.1186/s41687-018-0059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gayet-Ageron A, Agoritsas T, Schiesari L, et al. Barriers to participation in a patient satisfaction survey: Who are we missing? PLoS One. 2011;6:e26852. doi: 10.1371/journal.pone.0026852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology: A review JAMA Oncol 2797–8042016 [DOI] [PubMed] [Google Scholar]
- 11.Savova GK, Danciu I, Alamudun F, et al. Use of natural language processing to extract clinical cancer phenotypes from electronic medical records Cancer Res 795463–54702019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehl KL, Xu W, Lepisto E, et al. Natural language processing to ascertain cancer outcomes from medical oncologist notes JCO Clin Cancer Inform 4680–6902020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan A, Chien I, Moseley E, et al. Deep learning algorithms to identify documentation of serious illness conversations during intensive care unit admissions Palliat Med 33187–1962019 [DOI] [PubMed] [Google Scholar]
- 14.Udelsman BV, Moseley ET, Sudore RL, et al. Deep natural language processing identifies variation in care preference documentation J Pain Symptom Manage 591186–1194.e32020 [DOI] [PubMed] [Google Scholar]
- 15.Kehl KL, Elmarakeby H, Nishino M, et al. Assessment of deep natural language processing in ascertaining oncologic outcomes from radiology reports JAMA Oncol 51421–14292019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Cancer Institute Patient-Reported Outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAETM) https://healthcaredelivery.cancer.gov/pro-ctcae/ [DOI] [PMC free article] [PubMed]
- 17.Kluetz PG, Chingos DT, Basch EM, et al. Patient-reported outcomes in cancer clinical trials: Measuring symptomatic adverse events with the National Cancer Institute's Patient-Reported Outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE) Am Soc Clin Oncol Ed Book 3567–732016 [DOI] [PubMed] [Google Scholar]
- 18.Label studio. https://labelstud.io/
- 19.Tjong Kim Sang EF, De Meulder F.Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition Proceedings of the Seventh Conference on Natural Language Learning at HLT-NAACL 2003 Volume 4CONLL 20003. USAAssociation for Computational Linguistics; 2003, pp 142–147 [Google Scholar]
- 20.Scispacy. https://allenai.github.io/scispacy/ [Google Scholar]
- 21. Pomares-Quimbaya A, Kreuzthaler M, Schulz S. Current approaches to identify sections within clinical narratives from electronic health records: A systematic review. BMC Med Res Methodol. 2019;19:155. doi: 10.1186/s12874-019-0792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin J, Chang M-W, Lee K, et al. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. Google AI Lang; 2018. https://arxiv.org/abs/1810.04805 [Google Scholar]
- 23.Vaswani A, Shazeer N, Parmar N, et al. 31st Conference on Neural Information Processing Systems (NIPS 2017) Long Beach, CA: 2017. Attention is all you need. [Google Scholar]
- 24.Bidirectional Encoder Representation from Transformers (BERT) https://arxiv.org/abs/1810.04805 [Google Scholar]
- 25.XLNet. https://arxiv.org/abs/1906.08237 [Google Scholar]
- 26.RoBERTa. https://arxiv.org/abs/1907.11692 [Google Scholar]
- 27.Conneau A, Khandelwal K, Goyal N, et al. Unsupervised Cross-Lingual Representation Learning at Scale. https://arxiv.org/abs/1911.02116
- 28.DistilBERT. https://arxiv.org/abs/1910.01108 [Google Scholar]
- 29.ELECTRA. https://arxiv.org/abs/2003.10555 [Google Scholar]
- 30.Beltagy I, Peters ME, Cohan A. Longformer. https://arxiv.org/abs/2004.05150 [Google Scholar]
- 31.Atkinson TM, Li Y, Coffey CW, et al. Reliability of adverse symptom event reporting by clinicians Qual Life Res 211159–11642012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Rooij BH, Ezendam NPM, Mols F, et al. Cancer survivors not participating in observational patient-reported outcome studies have a lower survival compared to participants: The population-based PROFILES registry Qual Life Res 273313–33242018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byrd RJ, Steinhubl SR, Sun J, et al. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records Int J Med Inform 83983–9922014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chase HS, Mitrani LR, Lu GG, et al. Early recognition of multiple sclerosis using natural language processing of the electronic health record. BMC Med Inform Decis Mak. 2017;17:24. doi: 10.1186/s12911-017-0418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissman GE, Harhay MO, Lugo RM, et al. Natural language processing to assess documentation of features of critical illness in discharge documents of Acute Respiratory Distress Syndrome survivors Ann Am Thorac Soc 131538–15452016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koleck TA, Dreisbach C, Bourne PE, et al. Natural language processing of symptoms documented in free-text narratives of electronic health records: A systematic review J Am Med Inform Assoc 26364–3792019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Hripcsak G, Markatou M, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: A feasibility study J Am Med Inform Assoc 16328–3372009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iqbal E, Mallah R, Rhodes D, et al. ADEPt, a semantically-enriched pipeline for extracting adverse drug events from free-text electronic health records. PLoS One. 2017;12:e0187121. doi: 10.1371/journal.pone.0187121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang H, Solti I, Kirkendall E, et al. Leveraging Food and Drug Administration Adverse Event Reports for the automated monitoring of electronic health records in a pediatric hospital. Biomed Inform Insights. 2017;9:1178222617713018. doi: 10.1177/1178222617713018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong JC, Fairchild AT, Tanksley JP, et al. Natural Language processing for abstraction of cancer treatment toxicities: Accuracy versus human experts JAMIA Open 3513–5172020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamang S, Patel MI, Blayney DW, et al. Detecting unplanned care from clinician notes in electronic health records JCO Oncol Pract 11e313–e3192015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruner DW, Hanisch LJ, Reeve BB, et al. Stakeholder perspectives on implementing the National Cancer Institute's patient-reported outcomes version of the Common Terminology Criteria for Adverse Events (PRO-CTCAE) Transl Behav Med 1110–1222011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeung AR, Pugh SL, Klopp AH, et al. Improvement in patient-reported outcomes with intensity-modulated radiotherapy (RT) compared with standard RT: A report from the NRG Oncology RTOG 1203 study J Clin Oncol 381685–16922020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fairchild AT, Tanksley JP, Tenenbaum JD, et al. Interrater reliability in toxicity identification: Limitations of current standards Int J Radiat Oncol Biol Phys 107996–10002020 [DOI] [PubMed] [Google Scholar]
- 45.Lindvall C, Deng CY, Moseley E, et al. Natural language processing to identify advance care planning documentation in a multisite pragmatic clinical trial J Pain Symptom Manage 63e29–e362022 [DOI] [PMC free article] [PubMed] [Google Scholar]