Abstract
Among several bacterial species belonging to the general Gordonia, Mycobacterium, Micromonospora, Pseudomonas, and Rhodococcus, only two mycobacterial isolates, Mycobacterium fortuitum strain NF4 and the new isolate Mycobacterium ratisbonense strain SD4, which was isolated from a sewage treatment plant, were capable of utilizing the multiply branched hydrocarbon squalane (2,6,10,15,19,23-hexamethyltetracosane) and its analogous unsaturated hydrocarbon squalene as the sole carbon source for growth. Detailed degradation studies and high-pressure liquid chromatography analysis showed a clear decrease of the concentrations of squalane and squalene during biomass increase. These results were supported by resting-cell experiments using strain SD4 and squalane or squalene as the substrate. The degradation of acyclic isoprenoids and alkanes as well as of acids derived from these compounds was also investigated. Inhibition of squalane and squalene degradation by acrylic acid indicated the possible involvement of β-oxidation in the degradation route. To our knowledge, this is the first report demonstrating the biodegradation of squalane by using defined axenic cultures.
Squalane (2,6,10,15,19,23-hexamethyltetracosane) is a multiply branched saturated hydrocarbon. It is structurally related to the highly unsaturated isoprenoid oligomeric hydrocarbon squalene but is much less susceptible to spontaneous chemical oxidation. It is a colorless, odorless, transparent, and homogeneous oil that is widely used in skin care products; it has a very high coagulation point as well as a very high melting point, making it very suitable for lubrication (16). Furthermore, squalane was also reported to be present as emulsified solvent during the degradation of polycyclic aromatic hydrocarbon (pyrene), facilitating pyrene's mass transfer without being utilized itself (7).
The degradation of alkanes, acyclic isoprenoids, and the analogous unsaturated compound squalene has been reported in detail for several microorganisms (4, 5, 21–26). However, only few microorganisms are able to utilize branched-chain hydrocarbons. Certain alkyl branched compounds, such as quaternary carbon and β-alkyl-branched (anteiso) compounds, are often recalcitrant and thus accumulate in the biosphere (1). The reason for this phenomenon may be either that the alkyl branches hinder the uptake of the hydrocarbon into the cell or that the branched-chain hydrocarbons are not susceptible to the enzymes of the β-oxidation pathway (19). On the other hand, reports of the degradation of squalane are rather scarce. Based on studies using different analytical methods, the degradation of squalane as a model for polyethylene by radiation-induced oxidation was reported (9, 10); however, the biological degradation of squalane by unspecified bacteria was mentioned by McKenna and Kallio (14) 36 years ago.
In this communication, we demonstrate that squalane is susceptible to microbial degradation and that actinomycetes, in particular those belonging to the genus Mycobacterium, are potent degraders of this multibranched saturated hydrocarbon. Furthermore, we investigated the degradation of squalane in parallel to the degradation of acyclic isoprenoids, alkanes, and acids which may be derived from this compound. Additionally, inhibition of squalane degradation by acrylic acid indicated that β-oxidation is involved in the degradation route.
MATERIALS AND METHODS
Chemicals.
All chemicals used in this study were of analytical grade and were obtained from Sigma-Aldrich Chemie (Steinheim, Germany) or Merck (Darmstadt, Germany). NR latex concentrate (Neotex Latez) was obtained from Weber & Schaer (Hamburg, Germany), and IR (SKI3) was obtained from Continental AG (Hanover, Germany).
Microorganisms.
The following microorganisms were used in this study: Gordonia sp. strains Kb1 (unpublished data) and Kb2 (DSM 44215), Gordonia sp. strain VH2 (DSM 44266), Gordonia polyisoprenivorans Kd2 (DSM 44302), Micromonospora aurantiaca w2b (DSM 44438), Mycobacterium fortuitum NF4 (DSM 44216), Pseudomonas aeruginosa AL98, Rhodococcus ruber NCIMB 40126, Rhodococcus rhodochrous (DSM 43241), Rhodococcus opacus (DSM 44193), Pseudomonas mendocina (DSM 50017), and Pseudomonas putida KT2440.
Cultivation of bacteria.
Cultivations of bacteria in liquid media were carried out in 300-ml Erlenmeyer flasks containing 30 ml of mineral salts medium (MSM) prepared as described previously (20) and supplemented with a carbon source as indicated below. Squalane and squalene were sterilized separately by filtration and added to the medium at a final concentration of 0.5% (wt/vol); levulinic and isovaleric acids were added at a final concentration of 0.1% (vol/vol); pentane, hexane, decane, pristane, hexadecane, hexanoate, and octanoate were added at a final concentration of 0.1, 0.2, or 0.3% (vol/vol). During incubation at 30°C, cultures were agitated at 120 rpm on a rotary shaker. To determine the growth kinetics of squalane- and squalene-degrading bacteria, growth was monitored with a Klett-Summerson photometer and viable-cell counts were determined by diluting cells in saline (0.9% [wt/vol] NaCl) and plating them on nutrient broth (Difco Laboratories) agar plates. Protein was determined as described previously (3). To analyze growth on acyclic isoprenoids such as R-(+)-β-citronellol, dl-citronellol, dl-geraniol, and farnesol or on citronellal, S-(−)-citronellal, R-(+)-β-citronellic acid, geranic acid, acetonyl acetone, geranylacetone, or citral, cells were exposed to a vapor of the respective compound delivered from sterile filter paper containing 50 to 100 μl of this compound and placed in the lid of the plate. Inoculated plates were incubated in an inverted position with the lid at the bottom at 30°C separately and in closed containers. All cultures were inoculated with cells obtained from a 3- to 6-day-old preculture in Luria-Bertani complex medium (18), and the cells were washed twice with sterile saline before inoculation. Cultivations of the squalane-degrading bacteria on latex agar and synthetic cis-1,4-polyisoprene (IR) was performed as described recently (12, 13).
Isolation of a potent squalane-degrading bacterium from sewage.
In another screening approach, a new bacterium was isolated from activated sewage sludge obtained from the sewage treatment plant at Münster, Germany. Approximately 5 ml of the activated sewage sludge was used to inoculate 30 ml of MSM. Sterile squalane was added as a sole carbon source at a final concentration of 0.5% (wt/vol). Cultures were incubated aerobically at 30°C with shaking. After 10 successive transfers to fresh medium using 10% (vol/vol) inocula, a squalane-degrading bacterium was isolated after a series of cultivations on MSM agar plates containing squalane as the sole carbon source. Squalane agar plates were prepared by mixing 0.5% (wt/vol) sterile squalane with the MSM agar to obtain a homogeneous suspension in which squalane was suspended in the form of fine droplets. The warm suspension was poured onto sterile petri dishes, and after being dried, the plates were used for isolation of the pure cultures by the streak plate method.
Analysis of 16S rDNA.
DNA was extracted as described previously (2). The 16S rRNA gene (rDNA) was amplified using oligonucleotide primers as described previously (17). The nucleotide sequence of the purified PCR product was determined employing a 4000L DNA sequencer (LI-COR Inc., Biotechnology Division, London, Nebr.), a Thermo Sequenase fluorescence-labeled primer cycle-sequencing kit (Amersham Life Science, Little Chalfont, United Kingdom) as described by the manufacturer, and primers described previously (17). The 16S rDNA sequence was aligned with published sequences from representative actinomycetes obtained from the EMBL database.
Analytical methods.
For quantitative determination of squalane, squalene, and the putative degradation products, culture supernatants were analyzed in a high-performance liquid chromatography (HPLC) apparatus equipped with a diode array detector (WellChrom Diodenarray-Detektor KromaSystem 2000) and a differential refractometer (Fa. Knauer, Berlin, Germany) for squalene and squalane, respectively. After each time interval, the cells were separated from the culture medium by centrifugation for 20 min at 2,772 × g and 4°C. For HPLC analysis, the cell-free culture supernatants were extracted with diethyl ether to remove soluble squalane or squalene from the aqueous phase. In separate experiments, the amounts of squalane or squalene recovered by extraction from the aqueous phase and the losses due to the adsorption of the oil substrates to the growth flasks were estimated. It was found that approximately 75% of the initial concentrations were recovered. The extracts were left to evaporate, and the remaining materials were dissolved in 1-propanol. The separation was carried out by reverse-phase chromatography on a Nucleosil-100 C18 column with 1-propanol as eluent at a flow rate of 0.5 ml/min. For quantification, squalane or squalene was used as the external standard. Squalane and squalene were identified according to their retention times and their spectra.
Resting-cell experiment.
In the resting-cell experiment, 30 ml of a culture of Mycobacterium sp. strain SD4 cells was grown in MSM with 0.2% (wt/vol) gluconate as the sole carbon source and harvested after 3 to 4 days. Thereafter, the cells were washed twice with sterile saline (0.9% [wt/vol] NaCl) solution and used for the inoculation of 25 ml of MSM containing no ammonium chloride but either squalane (0.5% [wt/vol]) or squalene (0.25% [wt/vol] as the sole carbon source at a density of approximately 0.5 g of cells/liter. The Erlenmeyer flasks were then incubated at 30°C under aerobic conditions on a rotary shaker (120 rpm). At different time intervals, samples were withdrawn and subjected to chemical analysis as described above.
Nucleotide sequence accession number.
The 16S rRNA gene sequence data of Mycobacterium sp. strain SD4 were submitted to the EMBL nucleotide sequence database and are available under accession number AJ271863.
RESULTS
Survey for squalane-degrading bacteria.
To screen for potent squalane-degrading bacteria, several previously characterized bacterial strains capable of utilizing natural and synthetic rubber as the sole carbon source for growth were tested. The results are summarized in Table 1 and showed that only the natural- and synthetic-rubber-degrading bacterium M. fortuitum strain NF4 was capable of utilizing squalane and squalene as sole carbon sources for growth. Although G. polyisoprenivorans strain Kd2 and other strains of Gordonia (Kb1, Kb2, and VH2) are potent natural and synthetic cis-1,4-polyisoprene rubber-degrading bacteria and showed good growth on the analogous hydrocarbon squalene (11, 13), none of them was able to use squalane as a carbon source for growth. Similarly, the cis-1,4-polyisoprene-degrading bacterium Micromonospora aurantiaca strain w2b (13) exhibited no growth on either of the two substrates. Furthermore, the three Rhodococcus species (R. ruber, R. rhodochrous, and R. opacus) showed only poor growth on squalane or squalene. P. putida, P. mendocina, and P. aeruginosa AL98 were also investigated; however, none of them showed growth on squalene or squalane.
TABLE 1.
Comparison of the growth of isolate SD4 and of rubber-degrading bacteria on squalane, squalene, and natural or synthetic rubber
| Strain | Growtha on:
|
|||
|---|---|---|---|---|
| Squalane | Squalene | Latex | Synthetic IR | |
| Isolate SD4 | +++ | +++ | ++ | + |
| Gordonia sp. strain Kb1 | − | +++ | +++ | ++ |
| Gordonia sp. strain Kb2 | − | +++ | +++ | ++ |
| Gordonia sp. strain VH2 | − | +++ | +++ | +++ |
| G. polyisoprenivorans Kd2 | − | +++ | +++ | +++ |
| R. ruber | + | + | + | − |
| R. opacus | + | + | + | − |
| R. rhodochrous | + | + | + | − |
| M. fortuitum NF4 | +++ | +++ | +++ | ++ |
Growth on MSM agar plates containing the indicated carbon source was qualitatively estimated as follows: −, no growth; +, poor growth; ++, medium growth; +++, good growth.
Therefore, we enriched squalane-degrading microorganisms from environmental samples. However, we were able to isolate only one strain, which was obtained from the sewage treatment plant in Münster, that had the ability to utilize squalane or squalene as the sole carbon source for growth. This strain also grew very slowly in MSM containing natural or synthetic rubber as the sole carbon source (Table 1).
Taxonomic classification of the new isolate.
Strain SD4 was a gram-positive, oxidase- and catalase-negative, rod-shaped nonmotile bacterium. Its substrate utilization pattern revealed that it was able to grow on glucose, gluconate, sucrose, arabinose, urea, and phenylalanine as a sole carbon source but showed no growth on lactose, maltose, xylose, sorbitol, adonitol, mannitol, lysine, ornithine, or dulcitol.
Furthermore, taxonomic characterization of strain SD4 was carried out by analysis of the 16S rRNA gene. The PCR product obtained by the procedure described in Materials and methods comprised almost the complete gene and consisted of 1,473 nucleotides. When this sequence was compared with the 16S rDNA sequences available from the EMBL database, it revealed the greatest similarity to the corresponding sequence of Mycobacterium ratisbonense (99.5%). The next highest similarities were 99% to the 16S rDNA sequence of Mycobacterium sp. strain M0183 and 98% to those of Mycobacterium farcinogenes and Mycobacterium sp. strain HE5. SD4 is therefore referred to as Mycobacterium ratisbonense strain SD4 below.
Growth kinetics of the squalane-degrading bacteria.
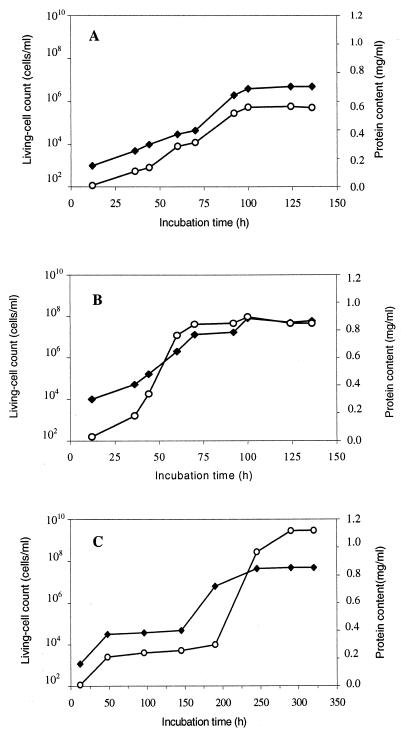
The ability of M. fortuitum NF4 and M. ratisbonense SD4 to utilize squalane and squalene as a sole source of carbon during biomass formation was investigated. The growth kinetics of both bacteria was monitored during cultivation on the two multiply branched hydrocarbons. These experiments showed that the cell biomass, which was estimated as living-cell count and as total protein content, continuously increased during growth of M. ratisbonense strain SD4 on squalane and squalene (Fig. 1A and B) and of M. fortuitum NF4 on squalane (Fig. C). While approximately 250 to 300 h was required to obtain the maximum cell density and total protein concentration with cells of strain NF4 on squalane, cells of strain SD4 entered the stationary growth phase after only 72 to 96 h. The doubling times of M. ratisbonense strain SD4 on squalane or on squalene were 4 and 7 h, respectively.
FIG. 1.
Growth of M. ratisbonense strain SD4 and M. fortuitum strain NF4 on squalane and squalene. Cells of strain SD4 (A and B) and strain NF4 (C) were grown in 30 ml of MSM in 250-ml Erlenmeyer flasks containing 0.5% (vol/vol) squalane (A and C) or 0.5% (vol/vol) squalene (B) as the sole carbon source. The cultures were inoculated from washed cells obtained from a Luria-Bertani medium preculture, and incubation was done at 30°C on a rotary shaker (120 rpm). At the indicated time intervals, samples were taken from the flasks and the indicated parameters were measured. Symbols: ⧫, living-cell count; ○, total protein content.
It was observed that cells of M. fortuitum strain NF4 tend to aggregate during growth on these hydrophobic substances (especially squalene), and it was difficult to monitor the number of living cells during growth with high accuracy. Only after consumption of the substrate did the cell aggregates disappear, and the cells became suspended in the medium. This phenomenon was not observed with cells of M. ratisbonense strain SD4.
Detection of possible degradation products.
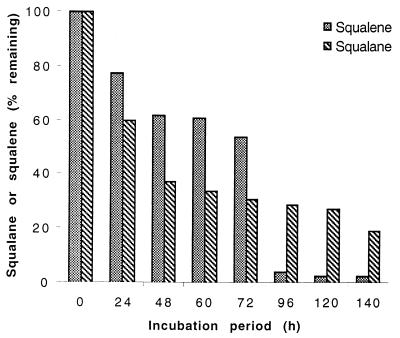
The amounts of squalane or squalene utilized during biomass formation by both strains were estimated by HPLC analysis. The results shown in Fig. 2 demonstrate that approximately 97 or 73% of the squalane or squalene present, respectively, was utilized by M. ratisbonense strain SD4 cells after 140 h of cultivation.
FIG. 2.
Degradation of squalane and squalene by M. ratisbonense strain SD4. At the indicated time intervals, the culture supernatant was extracted and treated as described in Materials and Methods. The disappearance of squalane or squalene was estimated by HPLC analysis. The data recorded represent the mean value from three different cultivation experiments.
Although the growth of M. fortuitum strain NF4 on these substrates was slower than that of M. ratisbonense strain SD4, approximately 44 and 20% of squalane or squalene, respectively, was utilized during 168 h. After 336 h, only approximately 36 and 10% of squalane or squalene remained, respectively. Due to the hydrophobic surface of the cells of M. fortuitum strain NF4, squalane or squalene droplets remained adsorbed by cells, and it was not possible to completely extract these compounds with the organic solvent and quantitatively estimate the exact amount of substrate that remained during degradation. Therefore, the disappearance of squalene or squalane in M. fortuitum strain NF4 cultures may not represent the actual utilization of these substances (data not shown).
Oxidation of squalane and squalene by resting cells of M. ratisbonense strain SD4.
To further characterize squalane and squalene degradation by M. ratisbonense strain SD4, resting-cell experiments were performed. Nitrogen-free MSM containing squalane or squalene as a sole source of carbon was inoculated with cells of this strain as described in Materials and Methods. The results shown in Table 2 represent the time courses of squalane and squalene oxidation during incubation. The decrease in the squalane concentration was approximately 80% after 100 h of incubation, and approximately 60% of the squalene was oxidized after 110 h.
TABLE 2.
Oxidation of squalane or squalene by resting cells of M. ratisbonense strain SD4a
| Time (h) | Squalane
|
Squalene
|
||
|---|---|---|---|---|
| Concn (mg/ml) | Decrease in concn (%) | Concn (mg/ml) | Decrease in concn (%) | |
| 0 | 4.0 | 0 | 2.0 | 0 |
| 22 | 2.5 | 37.5 | 1.9 | 5 |
| 44 | 2.4 | 40 | ND | ND |
| 50 | ND | ND | 1.6 | 20 |
| 55 | 1.0 | 75 | ND | ND |
| 70 | 0.9 | 77.5 | ND | ND |
| 80 | ND | ND | 0.9 | 55 |
| 100 | 0 | 100 | ND | ND |
| 110 | ND | ND | 0.8 | 60 |
Resting cells of Mycobacterium sp. strain SD4 (approximately 0.5 g of cells/liter) were used to inoculate nitrogen-free MSM supplemented with 0.5% (wt/vol) squalane or 0.25% (wt/vol) squalene as the sole carbon source as described in Materials and Methods. Results recorded in this table represent the mean values obtained from two independent experiments.
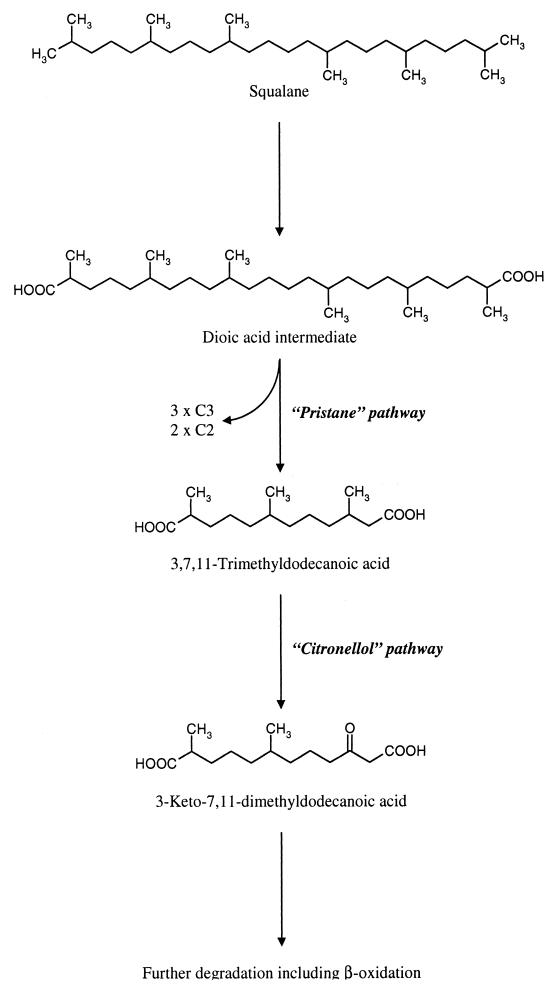
Elucidation of the degradation pathways.
To elucidate the putative pathways for the degradation of squalane by the two Mycobacterium species being studied (see Fig. 3), the utilization of various alkanes, acyclic isoprenoids, and acids derived from these compounds was investigated. The results shown in Table 3 indicated that hexadecane, octanoate, and the acidic form of the acyclic isoprenoids such as geranic acid and citronellic acid were easily utilized by both of the squalane-degrading bacteria. Similarly, both strains showed good growth on the keto and carboxylic forms of the putative degradation products of acyclic isoprenoids such as acetonylacetone, geranylacetone, levulinic acid, and isovaleric acid. On the other hand, most of the hydroxylic forms of acyclic isoprenoids such as citronellol, dl-citronellol, and geraniol (for strain NF4) were not utilized; only farnesol was utilized by both isolates. Citronellal, pentane, hexane, and heptane were not utilized at all. Furthermore, it was clearly demonstrated that acrylic acid at 0.1 or 0.3 mg/ml inhibited the growth of M. fortuitum strain NF4 and M. ratisbonense strain SD4 on squalane as well as squalene, respectively. In contrast, at these concentrations, acrylic acid showed no inhibitory effect on both bacteria during growth on MSM containing gluconate as the sole source of carbon (data not shown).
FIG. 3.
Putative pathway for the degradation of squalane in Mycobacterium spp. The initial dioic acid intermediate is oxidized via β-oxidation reactions analogous to those postulated for the pristane degradation pathway. For further degradation, the β-methyl group is replaced by a carbonyl group, as found in the citronellol pathway.
TABLE 3.
Growth of M. ratisbonense strain SD4 and M. fortuitum strain NF4 on isoprenoids, alkanes, and related compounds
| Compound tested | Growtha on carbon source of:
|
|
|---|---|---|
| M. ratisbonense SD4 | M. fortuitum NF4 | |
| Isoprenoids and related compounds | ||
| Citronellol | − | − |
| dl-Citronellol | − | − |
| Geraniol | ++ | − |
| Farnesol | ++ | +++ |
| Citronellal | − | − |
| S-(−)-Citronellal | − | − |
| Citronellal | − | − |
| Acetonylacetone | +++ | +++ |
| Geranylacetone | +++ | +++ |
| R-(+)-Citronellic acid | +++ | +++ |
| Geranic acid | +++ | +++ |
| Citral | − | − |
| Alkanes and alkanoic acids | ||
| Pentane | − | − |
| Hexane | − | − |
| Heptane | − | − |
| Decane | +++ | ++ |
| Hexadecane | +++ | ++ |
| Pristane | +++ | ++ |
| Hexanoate | +++ | ++ |
| Octanoate | +++ | ++ |
| Levulinic acid | + | + |
| Isovaleric acid | ++ | ++ |
Growth was qualitatively estimated as follows: −, no growth; +, poor growth; ++, moderate growth; +++, good growth. The strains were cultivated in MSM agar plates and were exposed to the vapor of the tested isoprenoid compound (50 to 100 μl) as described in Materials and Methods. For testing of alkanes and alkanoic acids, compounds were used at concentrations ranging from 0.1 to 0.3% (vol/vol).
DISCUSSION
Screening procedures for the isolation of squalane-degrading bacteria led to the identification of two Mycobacterium strains, M. fortuitum strain NF4 and the new isolate from sewage, which was characterized by 16S rRNA analysis as a strain of M. ratisbonense SD4. To our knowledge, this is the first report describing the biodegradation of squalane. It was found that both isolates were also able to degrade the analogous unsaturated isoprenoid oligomer squalene as well as polyisoprenoid substrates such as latex and IR. Therefore, axenic cultures of these two bacteria are suitable for studying the degradation of multiply branched alkanes and alkenes. Growth experiments revealed that the growth of strain SD4 on these substrates was better than that of strain NF4. The cells of strain NF4 had a tendency to aggregate and to bind to the hydrophobic substrates; this probably induced feature of mycobacteria could be caused by the higher lipid content at the cell surface (15). It has been found that lipids provide a more hydrophobic region through which hydrophobic substrates may penetrate more easily (6, 8).
To elucidate the pathway for the degradation of squalane by these bacteria, the growth of both strains on alkanes, acyclic isoprenoids, and acids derived from these compounds was studied. According to the results obtained for the degradation of squalane, the putative pathway demonstrated in Fig. 3 is proposed. It seems likely that after the conversion of squalane to a dioic acid as one of the first intermediates, three propionyl coenzyme A and acetyl coenzyme A molecules are oxidatively removed by the β-oxidation route to form the 3,7,11-trimethyldodecandioic acid intermediate by a pathway analogous to that for degradation of the multiply branched alkane pristane (2,3,10,14-tetramethylpentadecane) (4). For further degradation, the β-methyl group of this intermediate could be converted into a carbonyl oxygen, thus generating a suitable substrate for β-oxidation and further degradation. Such a pathway was revealed for the degradation of citronellol in various pseudomonads and is referred to as the citronellol degradation pathway (5, 22). These results were supported by the ability of both strains to utilize acetate, propionate, and pristane as well as several acyclic isoprenoids such as farnesol, citronellic acid, geranic acid, and geranylacetone as a carbon source for growth. Indeed, inhibition of squalane and squalene degradation by acrylic acid also indicated that β-oxidation is involved. These investigations could be important to develop a new approach to solve specific bioaccumulation problems associated with alkyl-branched compounds and which confer molecular recalcitrance.
ACKNOWLEDGMENT
This work was supported in part by a grant from the Egyptian Government Fellowship Program.
REFERENCES
- 1.Alexander M. Nonbiodegradable and other recalcitrant molecules. Biotechnol Bioeng. 1973;IV:611–647. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Bühler M, Schindler J. Aliphatic hydrocarbons. In: Rehm H-J, Reed G, editors. Biotechnology. 6a. Weinheim, Germany: Verlag Chemie; 1984. pp. 329–385. [Google Scholar]
- 5.Fall R R, Susan G C, Edward P L, David S W. Biodegradation of acyclic isoprenoids by Pseudomonas species. J Bacteriol. 1978;135:324–333. doi: 10.1128/jb.135.2.324-333.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hug H, Blanch H W, Fiechter A. The functional role of lipids in hydrocarbon assimilation. Biotechnol Bioeng. 1974;16:965–985. doi: 10.1002/bit.260160709. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez I Y, Bartha R. Solvent-augmented mineralization of pyrene by a Mycobacterium sp. Appl Environ Microbiol. 1996;62:2311–2316. doi: 10.1128/aem.62.7.2311-2316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Käppeli O, Finnerty W R. Partition of alkane by an extracellular vesicle derived from hexadecane-grown Acinetobacter. J Bacteriol. 1979;140:707–712. doi: 10.1128/jb.140.2.707-712.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsumura Y, Soebianto Y S, Ishigure K, Kubo J, Hamakawa S, Kudoh H, Seguchi T. Radiation induced oxidation of liquid alkanes as a polymer model. Radiat Phys Chem. 1996;48:449–456. [Google Scholar]
- 10.Katsumura Y. Radiation induced degradation of polymers—an approach by using liquid paraffins as a model system. Angew Makromol Chem. 1997;252:89–101. [Google Scholar]
- 11.Linos A, Steinbüchel A, Spröer C, Kroppenstedt R M. Gordonia polyisoprenivorans sp. nov., a rubber degrading actinomycete isolated from automobile tire. Int J Syst Bacteriol. 1999;49:1785–1791. doi: 10.1099/00207713-49-4-1785. [DOI] [PubMed] [Google Scholar]
- 12.Linos A, Reichelt R, Keller U, Steinbüchel A. A Gram-negative bacterium, identified as Pseudomonas aeruginosa AL98, is a potent degrader of natural rubber and synthetic cis-1,4-polyisoprene. FEMS Microbiol Lett. 2000;182:155–161. doi: 10.1111/j.1574-6968.2000.tb08890.x. [DOI] [PubMed] [Google Scholar]
- 13.Linos A, Berekaa M M, Reichelt R, Keller U, Schmitt J, Flemming H-C, Kroppenstedt R M, Steinbüchel A. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: microbial strategies and detailed surface analysis. Appl Environ Microbiol. 2000;66:1639–1645. doi: 10.1128/aem.66.4.1639-1645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna E G, Kallio R E. Hydrocarbon structure: its effect on bacteria utilization of alkanes. In: Heukelkian H, Dondero N C, editors. Principles and applications in aquatic microbiology. New York, N.Y: John Wiley & Sons, Inc.; 1964. pp. 1–4. [Google Scholar]
- 15.Minnikin D E, O'Donnell A G. Actinomycete envelope lipid and peptidoglycan composition. In: Goodfellow M, Mordarski M, Williams S T, editors. The biology of the actinomycetes. London, United Kingdom: Academic Press, Ltd.; 1983. pp. 337–388. [Google Scholar]
- 16.Neumüller O A. Römpps chemie-lexikon. 8th ed. Stuttgart, Germany: Franckh; 1987. [Google Scholar]
- 17.Rainey F A, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Schaeffer T L, Cantwell S G, Brown J L, Watt D S, Fall R R. Microbial growth on hydrocarbons: terminal branching inhibits biodegradation. Appl Environ Microbiol. 1979;38:742–746. doi: 10.1128/aem.38.4.742-746.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: wachstumphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 21.Schröder E, Rehm H J. Degradation of long chain n-alkanes by chlorococcales. Eur J Appl Microbiol Biotechnol. 1981;12:36–38. [Google Scholar]
- 22.Seubert W. Degradation of isoprenoid compounds by microorganisms. J Bacteriol. 1960;79:426–434. doi: 10.1128/jb.79.3.426-434.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams P A. Genetics of biodegradation. In: Leisinger T, Cook A M, Hütter R, Nüesch J, editors. Microbial degradation of xenobiotics and recalcitrant compounds. London, United Kingdom: Academic Press Ltd., for the Swiss Academy for Sciences and the Swiss Society of Microbiology; 1981. pp. 97–107. [Google Scholar]
- 24.Yamada Y, Moto H, Kinoshita S, Takeda N, Okada H. Oxidative degradation of squalene by Arthrobacter species. Appl Microbiol. 1975;29:400–404. doi: 10.1128/am.29.3.400-404.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada Y, Naoki I, Takuya N, Atsuko K. Oxidative pathway from squalene to geranylacetone in Arthrobacter sp. strain Y-11. Appl Environ Microbiol. 1988;54:381–385. doi: 10.1128/aem.54.2.381-385.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuhiro Y, Chull W S, Hirosuke O. Oxidation of acyclic terpenoids by Corynebacterium sp. Appl Environ Microbiol. 1985;49:960–963. doi: 10.1128/aem.49.4.960-963.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]