Graphical abstract

Keywords: Epigenetics, Pathogenicity estimation, Molecular Dynamics simulation
Abbreviations: LUSC, lung squamous cell carcinoma; MD, molecular dynamics; GaMD, gaussian accelerated molecular dynamics; IDR, intrinsically disordered region; RMSD, root mean squared deviation; PCA, principal component analysis; DCCM, dynamic cross-correlation map; PMF, potential of mean force
Abstract
KDM6A is the disease causative gene of type 2 Kabuki Syndrome, a rare multisystem disease; it is also a known cancer driver gene, with multiple somatic mutations found in a few cancer types. In this study, we looked at eleven missense variants in lung squamous cell carcinoma, one of the most common lung cancer subtypes, to see how they affect the KDM6A catalytic mechanisms. We found that they influence the interaction with histone H3 and the exposure of the trimethylated Lys27, which is critical for wild-type physiological function to varying degrees, by altering the conformational transition.
1. Introduction
One of the mechanisms of gene regulation is determined by the access to DNA and the recruitment of transcription factors. These processes depend on the histones modification states. For example, tri-methylation of the 27th lysine residue on histone H3 (H3K27me3) impacts the accessibility to chromatin, with a direct consequence on gene expression. Although largely thought of as a genetic disease, alterations in these epigenetic processes were also crucial in cancer origin and progression [1].
Among the several isoforms of histone demethylases, the Ubiquitously transcribed Tetratricopeptide repeat, X chromosome (UTX), better known as KDM6A, is one of the most frequently mutated histone modifiers [2] and has been recently identified as a cancer driver gene [3]. KDM6A is an X-encoded histone demethylase that escapes X chromosome inactivation and is ubiquitously expressed. It specifically mediates the removal of di- and trimethylation markers on the histone H3 Lys27 and its loss or inactivation has been often correlated with the onset of a broad spectrum of congenital anomalies, particularly with type 2 Kabuki Syndrome. Notably, KDM6A somatic mutations can be found in multiple cancer types [4], including B-cell lymphoma, bladder urothelial carcinoma, head and neck squamous cell carcinoma, pancreatic adenocarcinoma, lung squamous cell carcinoma (LUSC), and kidney renal papillary cell carcinoma. However, their contribution to oncogenesis and tumor progression is still poorly characterized [5].
Several single nucleotide variants and deletions were found in the two primary, well-structured, functional domains, the N-terminal and the C-terminal, or the intrinsically disordered region (IDR) connecting them, and proved to affect the molecular mechanisms of the KDM6A protein and its interactions with the cellular environment. The catalytically active C-terminal, which includes the fundamental Jumonji domain, has been extensively characterized, and its alteration was mainly correlated with the reduction of the demethylase activity [6]. On the contrary, mutations occurring in the N-terminal, containing multiple tetratricopeptide repeat elements (TPR domain) and mainly described as a protein interaction motif, were poorly described due to the lack of an available structure model. In [7], we investigated the impact of seven missense mutations associated with type 2 Kabuki Syndrome using an enhanced Molecular Dynamics (MD) approach. From a structural point of view, we found that their pathogenic mechanisms could be ascribed to the disruption of the interaction between specific subdomains, with a putative impact on the recognition and demethylation of histone H3K27me3. Chi et al. [8] came to similar conclusions by characterizing the functional alteration of the demethylase function caused by multiple Kabuki mutations using a combination of genomic and structural features.
The same researchers recently evaluated 197 new somatic KDM6A mutations, reclassifying them based on their overall impact on protein structure and dynamics, as well as providing insight into their putative role in altering biophysical and biochemical mechanisms [9]. On this line, we focused on LUSC and analyzed eleven variants in greater atomic detail, by taking advantage of recent advances in structural modeling [10], which allowed us to predict and use the entire KDM6A protein, and exploiting multiple replicas of one order of magnitude longer MD simulations.
Lung cancer is one of the most common cancers and is considered the leading cause of cancer-related death worldwide [11]. Non-small-cell lung cancer represents the most frequent lung cancer type and can be subdivided into lung adenocarcinoma (LUAD) and LUSC. Clinically, both cancer types are highly heterogeneous, and the definition of the best course of treatment mainly depends on extensive subclassification and the identification of molecular targets. Interestingly, KDM6A has been recently identified as a fundamental tumor suppressor in lung cancer, representing a promising therapeutic target [12]. However, the functional consequences of its alterations are still a matter of debate [13], [14]. Hence, this work aims to comprehend whether a selected set of LUSC-associated variants may impact the demethylase activity of KDM6A in LUSC cancer and therefore provide helpful insights for diagnostic, prognostic, and therapeutic purposes.
2. Materials and methods
2.1. KDM6A missense variants in LUSC
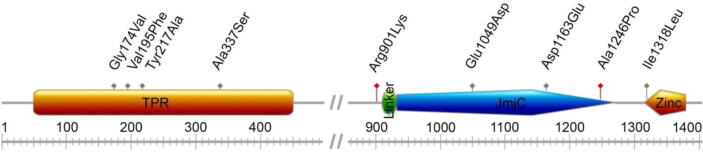
Missense variants in the KDM6A gene were retrieved from the Integrative Onco Genomics database (https://www.intogen.org, accessed on 1st February 2021), which fetches data from the TCGA/PanCanAtlas database. Variants were 7 truncating and 11 missense originated from 16 individuals included in the TCGA’s LUSC cohort. The missense variants under consideration were Gly174Val, Val195Phe, Tyr217Ala, Ala337Ser, Glu745Asp, Gly795Arg, Arg901Lys, Glu1049Asp, Asp1163Glu, Ala1246Pro, and Ile1318Leu. Allelic frequencies were retrieved from gnomAD v3.1.2 [15] and COSMIC [16] variant databases. Their pathogenicity was predicted in-silico by CADD v1.6 [17]. Potential aberrant splicing events of the pre-mRNA were predicted by four different splice site algorithms, Human Splicing Finder 3.1 [18], MaxEnt [19], Trap-score [20], and SpliceAI [21]. Gene expression profiles of KDM6A in LUSC were retrieved as FPKM-UQ expression values from TCGA using the R (https://www.r-project.org) Bioconductor’s TCGAbiolinks module [22].
2.2. System preparation
The model of the entire human KDM6A protein (UniProt ID: O15550) was retrieved from the AlphaFold Protein Structure Database [23]. Per-residue confidence scores (pLDDT) showed that both the TPR-region (residues 50–450) and the C-terminal domain (residues 880–1401) were modeled by an accuracy ranging from “well” to “very high.” Conversely, the first 50 residues, the 1058–1076 loop, and the entire intrinsically disordered region (IDR, residues 450–880) were modeled with very low accuracy (pLDDT < 50). Noteworthy, two of our selected variants, Glu745Asp and Gly795Arg, fall in the latter regions and, therefore, were excluded from any further analysis.
The interaction with the histone H3 was recreated using the X-ray structure of the KDM6A C-terminal fragment in its bound-form with histone H3K27me3 peptide (PDB ID: 3avr) as a template and extracting the atomic coordinates of the peptide (residues 17–38). Moreover, the Zn and Fe ions, together with the cofactor 2-oxoglutarate (2OG), were added to their respective binding sites. The final complex was refined using MODELLER v9.16 [24]. Finally, the wild-type structure was mutated in-silico using ChimeraX [25] to introduce the variants above, obtaining nine mutant complexes.
According to standard MD guidelines, each candidate system was subjected to multiple preparatory steps [26]. The Leap module of AmberTool21 [27] was employed to embed both wild-type and mutant complexes into a simulation box filled with TIP3P water. Na+ and Cl- counter ions were added to neutralize the overall charge of the models. The distance between the solute surface and the box was set to 12 Å. We used the Amber ff14SB force field to parametrize the amino acids, while the Zinc AMBER force field (ZAFF) [28] was employed for the Zn(II) ion. Each system was first energy-minimized using the steepest descent method, followed by the conjugate gradient method, and, thus, gradually heated and equilibrated for approximately five ns using a time-step of 1 fs. Electrostatic interactions were computed using the particle-mesh Ewald (PME) method, while a cutoff of 10 Å was used for non-bonded short-range interactions. Temperature and pressure were set at 300 K and 101.3 kPa, respectively, using the Langevin dynamics and Piston methods. Finally, additional constraints were applied to the IDR regions during both equilibration and production steps to avoid large system fluctuations during the simulations.
2.3. Gaussian accelerated molecular dynamics simulation
We implemented an enhanced MD simulation protocol similar to the one described in our previous paper [7]. GaMD [29] is an accelerated MD technique that works by adding a harmonic boost to smooth the system potential energy, reducing system energy barriers and, thus, enhancing the conformational sampling. This boost potential can be applied in a single or dual-boost scheme, so GaMD does not require any predetermined reaction coordinates or collective variables (CV). Hence, it is optimal for studying the dynamics of complex biological systems.
Here, both dihedral and total potential boost were used. Maximum, minimum, average, and standard deviation values of the system potential were obtained from an initial ∼ 10 ns simulation with no boost potential applied. Each GaMD simulation proceeded for ∼ 40 ns, in which the boost potential was updated every 1.6 ns, thus reaching equilibrium values. Finally, ∼200 ns of GaMD production was carried out by applying the dual boost scheme through the AMBER parameter “igamd = 3” and setting a time-step of 2 fs. Atomic positions were recorded every 500 steps (1.0 ps) for subsequent analysis. Each system was simulated three times using the GPU version (pmemd.cuda) of AMBER 20 on 3 NVIDIA RTX 2080Ti graphic cards [30].
2.4. Analysis of the trajectories
The obtained trajectories were analyzed from a geometric and energetic point of view, excluding the initial simulated 50 ns. In more detail, MDAnalysis [31] was used to calculate the Root Mean Squared Deviation (RMSD) profiles. We have aligned each frame of the trajectories to the starting reference structure and then investigated the relative movements of the backbone atoms of the entire protein and specific domains. We then measured their average distances to the reference structure.
We relied on Principal Component Analysis (PCA) to probe conformational changes occurring in our systems during the simulation. We inferred large-scale collective fluctuations of atoms and then predicted low-dimensional subspaces where essential protein motions were expected to take place. Hence, the covariance matrix was generated using the gmx_covar function implemented in GROMACS v2018, which captures the degree of collinearity of the atomic motions of each pair of atoms. The conformational changes of the systems caused by the variants under investigation were explored by resorting to the Dynamic Cross-Correlation Maps (DCCMs). These were plotted by a custom Python script that takes covariance matrices as input and generates the correlation matrices. DCCMs allowed us to study the long-range interactions between all pairs of atoms and highlight any correlated and anticorrelated motion.
PyReweighting, a toolkit of Python scripts, was used to reweight the GaMD simulations and detect the original free energy. The Potential of Mean Force (PMF) profiles were obtained by 2D-projecting each trajectory on eigenvectors and by setting different bin sizes until convergence. The cutoff for the number of simulation frames in one bin was 500. The DBSCAN algorithm implemented in AmberTools21 was employed to find the representative structures of the MD simulations and the clusters characterized by similar molecular density values, which, in turn, included the frames belonging to the same minimum points in the PMF profiles. The GetContacts [32] package was utilized to rapidly compute and compare the atomic interactions occurring in the frames composing each cluster. Then, the get_contact_frequencies.py script was used to calculate the frequency of each interaction. Finally, cluster-specific binding energy profiles were obtained using the MM/GBSA method, as implemented in the MMPBSA.py script from the AmberTools21 suite; it was run with default settings and sampling one frame every 100 ps of simulation. 3D figures and motions were generated using the UCSF ChimeraX software package.
3. Results
We report the analysis of the simulated trajectories of the human wild-type KDM6A protein in complex with the known interacting portion of the H3 histone. Comparatively, we assessed the conformational transitions that occurred during the simulation of a set of KDM6A mutants found in a LUSC cohort. The considered missense mutations spanned the whole protein-coding region of the gene: Gly174Val, Val195Phe, Tyr217Ala, and Ala337Ser map on the KDM6A TPR domain, while Arg901Lys, Glu1049Asp, Asp1163Glu, Ala1246Pro, and Ile1318Leu hit its catalytic domain (Fig. 1). Finally, Glu745Asp and Gly795Arg are located in the IDR region and, therefore, were excluded from this study.
Fig. 1.
Schematic representation of the KDM6A protein domains. Variants potentially affecting the pre-mRNA splicing mechanism are highlighted in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Wild-type RMSD and DCCMs
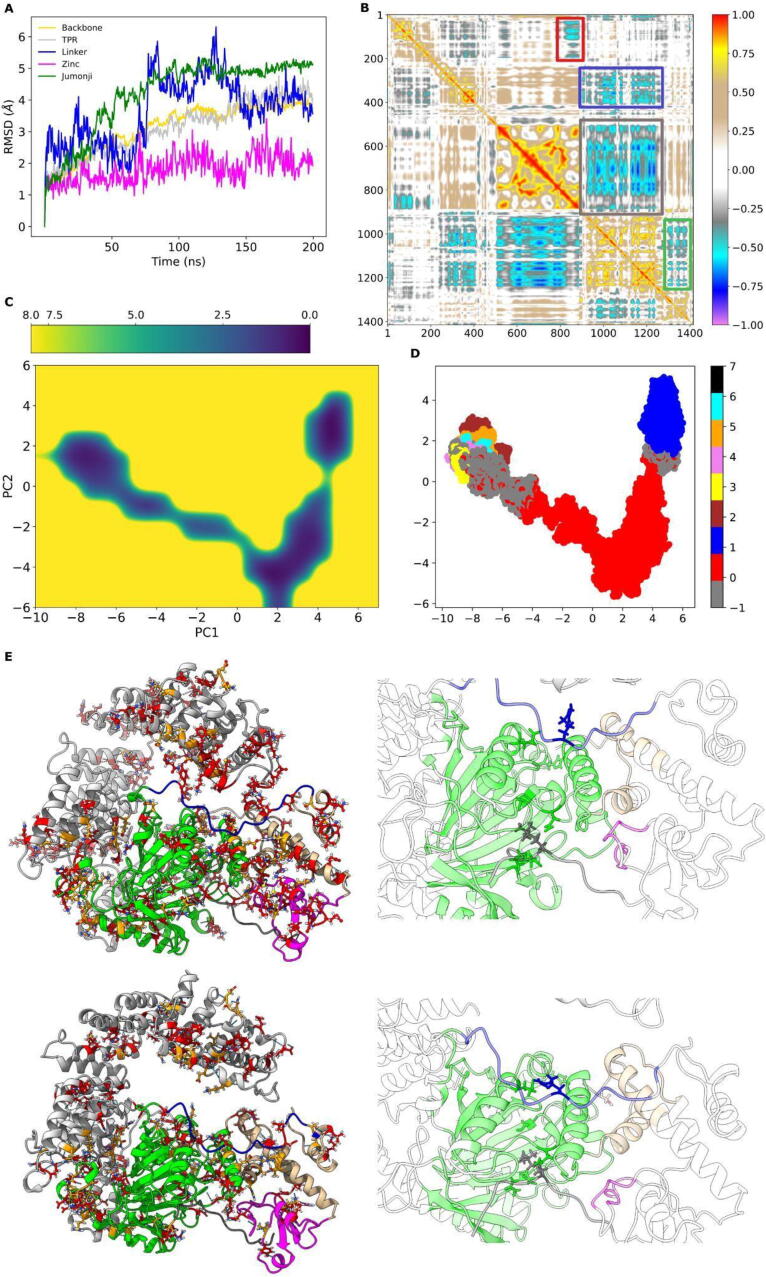
The observed structural rearrangements during simulations were assessed by computing the RMSD values of the alpha carbons (Cα) of the systems under investigation compared with those of the reference starting structure. Fig. 2A shows how the simulation of the wild-type system reached stable backbone RMSD values, ranging from 3 to 4 Å, after 100 ns. We also report the RMSD profiles subdivided for each domain. In detail, the Jumonji and Linker domains were the most flexible regions, with the former reaching a stable state after 100 ns around 5 Å and the latter at ∼ 3.5–4.5 Å after 150 ns. Finally, the Zinc domain remained stable during the entire simulation time.
Fig. 2.
MD analysis of the wild-type protein. A. RMSD profiles of the heavy atoms (yellow) and domains (Jumonji in green, Linker in blue, TPR in gray, and Zinc in magenta) of the KDM6A protein. B. DCCM: perfect correlations are highlighted in red (direct) or violet (inverse). The boxes in the DCCM highlight: red, anticorrelated movements between the first 100 residues of the TPR domain and the flaking portion of the catalytic domain (residues 810–900); blue, anticorrelated movements between the Linker and Jumonji domains (residues 920 to 1250) with part of TPR (residues 220 to 420); green, anticorrelated movements between the Linker and Jumonji domains (residues 920 to 1250) with the Zinc domain (residues 1300 to 1390); the gray box surrounds the IDR region. C. PMF. The first (PC1) and second (PC2) principal component projections are reported on the X and Y-axes, respectively. Colors represent PMF levels, from the absolute minimum (dark blue) to the maximum (yellow). D. Clusters identification in the PCA projection plane. The gray dots represent all unclustered frames. E. 3D structures of the KDM6A-H3 complex of the most representative clusters (cluster0 top-left, cluster1 bottom-left). The TPR domain is colored in gray, the Linker (910–932) in blue, Jumonji (933–1268) in green, Zinc (1315–1378) in magenta, and helical (886–902/1269–1314/1379–1395) in orange. H3 is colored in dark gray. Cluster-exclusive contacts are colored in red; contacts that change residue partners during the transition between cluster0 and cluster1 are colored in orange. The region where the interaction between the catalytic domain and the histone H3 is displayed on the right. Residues involved in fundamental interactions for the cluster0-cluster1 transition are highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A DCCM was computed starting from a pre-calculated covariance matrix with the aim of studying the role of long-range interactions between all pairs of atoms and, thus, describing the correlated and anticorrelated movements of each domain. We observed that the first 100 residues of the TPR domain moved in an anticorrelated way with the flaking portion of the catalytic domain (residues 810–900, represented by the red box in Fig. 2B). Additionally, the Linker and Jumonji domains (residues 920 to 1250) exhibited anticorrelated movements with a part of the TPR region (residues 220 to 420, blue box) and with the Zinc domain (residues 1300 to 1390, green box). It is worth noticing that, in all our analyses, we did not report any results regarding the region spanning residues 450–850 (gray box) due to constraints imposed on this region that could influence their reliability.
3.2. Potential of Mean force and cluster analysis
The first (PC1) and second (PC2) principal component projections were used as reaction coordinates to assess the system free-energy landscape, i.e., the PMF. Then, the frames were clustered into the most representative conformations using the density-based clustering approach provided by DBSCAN. The aim was to identify the most representative 3D structures corresponding to low-energy, long-lived conformations among all. In detail, three low energy conformational states could be identified, corresponding to the following absolute coordinates (-8, 2), (2, -4), and (4, 3) in Fig. 2C. As detailed in Fig. 2D, two of these minima correspond to highly populated clusters of frames, while the third aggregates multiple scarcely populated clusters. In detail, the red cluster represents 41% of the trajectory (henceforth cluster0), while the blue cluster represents 23% (cluster1). They are likely to represent two well-defined states of a conformational transition.
The average structures of the two main clusters were reported in Fig. 2E, where we mapped the differential interaction network. The contacts between all the amino acids in the average structures relative to these two clusters are available in Supplementary Table 2. Fig. 2E also zooms on the most flexible regions, as suggested by the RMSD and DCCM profiles and described in [7]. These regions play a fundamental role in the interaction with the H3K27me3 peptide. During the transition from cluster0 to cluster1, the Linker domain showed, in fact, an increase of the interaction frequency between Arg922 with Glu1171 (from 0.213 to 0.853) and with Glu1254 (from 0.230 to 0.977) and His957 with Ser1400 (from 0.059 to 0.615). Similarly, the histone exhibited an increase in the affinity of H3K27me3 for Asn1149 (from 0.101 to 0.765) but a decrease in its interaction with Asn1087 (from 0.532 to 0.046) and Ser1154 (from 0.505 to 0.239).
3.3. Arg901Lys
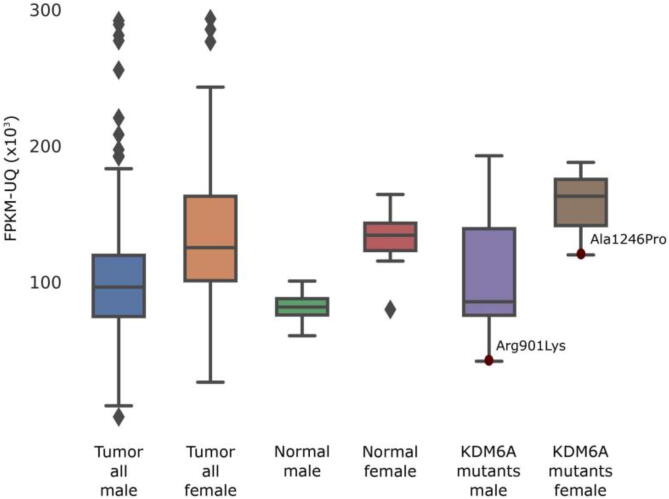
Arg901Lys was absent in gnomAD and associated with LUSC and large intestinal adenocarcinoma in COSMIC. Considering that the 901 codon overlaps an exon-exon junction, we suggest a different pathogenic mechanism. It may, in fact, affect exon splicing, as consistently confirmed by all calculated in-silico functional scores, which predicted a highly deleterious impact of the variant on the splicing mechanism (Supplementary Table 1). Gene expression analysis disclosed a significantly lower abundance of KDM6A transcript carrying this variant, thus confirming a potential effect on splicing or transcript stability. A similar hypothesis could hold for Ala1246Pro also (Fig. 3). However, we cannot ensure the two mutant proteins are expressed, especially considering that Arg901Lys and Ala1246Pro occur distantly, the latter much closer to the 3′ end of the protein than the former.
Fig. 3.
KDM6A expression profiles of TCGA samples grouped by sex and tissue type. Expression analysis of patients carrying Single Nucleotide Variants (SNVs) causing Arg901Lys and Ala1246Pro disclosed the lowest transcript abundance compared to sex-matched mutant cases.
3.4. KDM6A missense variants in LUSC
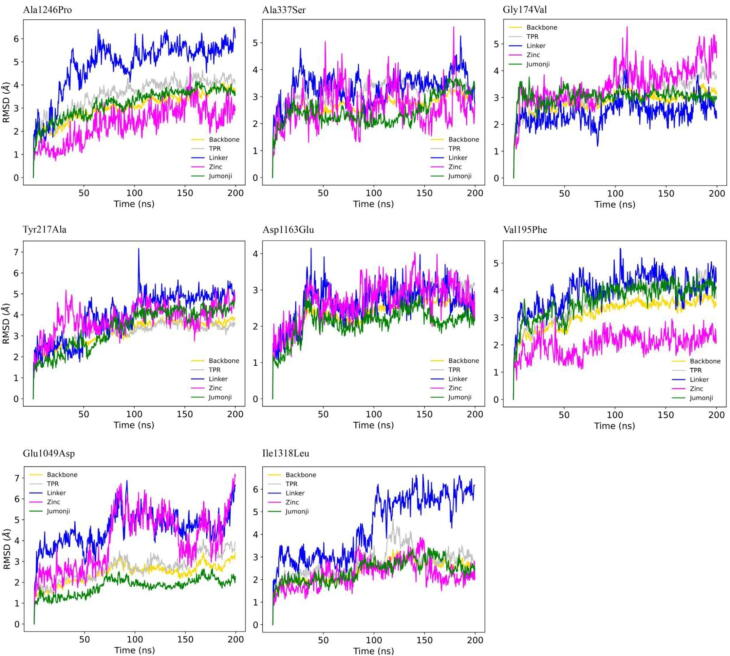
All the considered KDM6A missense variants were absent from gnomAD, most of them with highly predicted deleterious effects (CADD > 20). Instead, they were reported in COSMIC as single cases of LUSC (Supplementary Table 1). For these reasons, the same analytical protocol was applied to the mutant trajectories. In Fig. 4, we report their domain-specific RMSD profiles. In detail, Gly174Val, Glu1049Asp, Asp1163Glu, Ala1246Pro, and Ile1318Leu showed lower RMSD values for the Jumonji domain compared to the wild-type, while Ala337Ser reached ∼ 3.5 Å for the same domain only during the final 50 ns. Furthermore, Gly174Val (only in the final simulation steps), Tyr217Ala, Ala337Ser, and Glu1049Asp displayed a remarkably increased flexibility of the Zinc domain, while Asp1163Glu reached stable RMSD profiles for all the domains during the entire simulation time. Finally, Val195Phe showed an RMSD profile comparable to the wild-type for all domains.
Fig. 4.
RMSD profiles of the heavy atoms of the KDM6A protein (yellow) and its domains (Jumonji in green, Linker in blue, TPR in gray, and Zinc in magenta) for all the simulated mutants. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Mutant clustering and local interaction network analysis
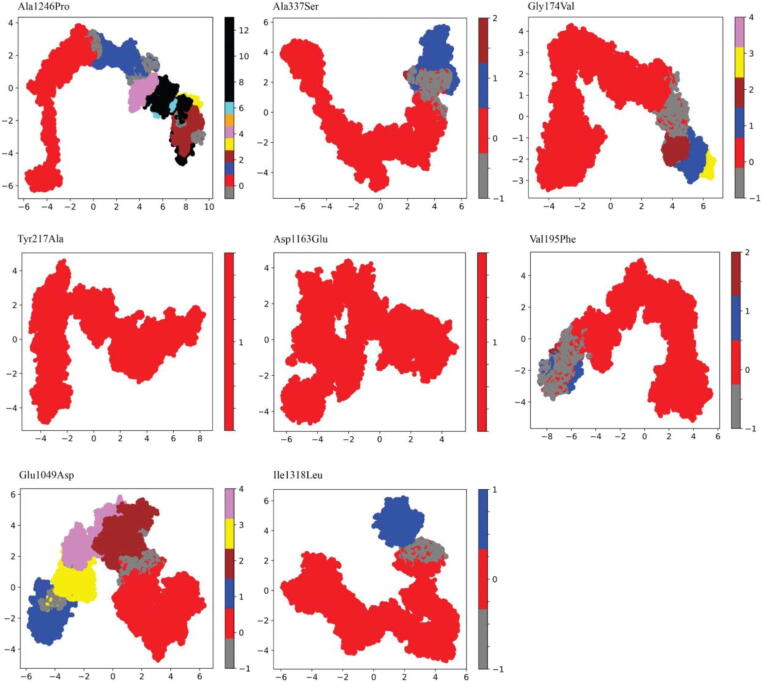
In this section, we report the results of a cluster analysis applied to the mutant trajectories and document the contacts that exhibit at least a moderate alteration in frequency, i.e., ∼0.3 to wild-type. This analysis focused on the interactions occurring in the proximity of the variants or within the H3K27me3 binding pocket. We identified only one cluster for Val195Phe, Tyr217Ala, and Asp1163Glu and two differently populated intermediate configurations for Gly174Val, Ala337Ser, Glu1049Asp, Ala1246Pro, and Ile1318Leu (Fig. 5). We considered only the clusters that included the local minimum values.
Fig. 5.
The first and second principal components projections subdivided by clusters for each mutant system.
Locally, Val195Phe mainly affected the interaction with Leu192, which caused increased stability of the mutant protein compared to the wild-type, where the interaction frequency between the two residues decreased during the transition between cluster0 and cluster1 (Table 1). Tyr217Ala lost its interaction with Lys178 causing the impairment of crucial movements needed to make the transition between the two conformations occur, as reported in Supplementary Table 2. Furthermore, Tyr217Ala decreased its interaction frequency with Tyr183 compared to both wild-type clusters. Asp1163Glu lost its interaction with Pro1214 and Gln472 residues, which were highly frequent in both wild-type clusters, and compensated by two novel interactions with the Cys1234 and Gln475 residues. Gly174Val increased its interaction frequency with Phe177 and decreased with His189. Similarly, Ala337Ser increased its interaction with Gly325 compared to the wild-type clusters. Cluster1 of the Glu1049Asp mutant and wild-type proteins displayed comparable interaction frequencies with the residues Glu1045 and Lys1053; however, a new high-frequency interaction was established with Lys1080. Moreover, the interaction with its neighbor residue, Arg1048, decreased in cluster0 and was lost in cluster1 of the mutant. The resulting local interaction network caused the loss of crucial movements that were observed in the two wild-type conformations, determining the rotation of a specific residue fragment (residues 1047–1050). Ala1246Pro caused the increase in the interaction frequency with Asp1285 compared to the wild-type. Finally, the interaction frequency rates with Ile1318Leu were similar to wild-type in cluster1 and reduced in cluster0.
Table 1.
Perturbations in local interaction networks caused by KDM6A variants. Missing clusters are indicated with “//”.
| Residue 1 | Residue 2 | Wild-type cluster0 |
Wild-type cluster1 |
Mutant cluster0 |
Mutant cluster1 |
|
|---|---|---|---|---|---|---|
| Ala1246Pro | 1246 | Asp1285 | 0.229 | 0.230 | 0.626 | 0.711 |
| Gly174Val | 174 | Phe177 | 0.517 | 0.598 | 0.925 | 0.910 |
| 174 | His189 | 0.904 | 0.806 | 0.579 | 0.320 | |
| Ala337Ser | 337 | Gly325 | 0.675 | 0.690 | 0.998 | 0.998 |
| Thr217Ala | 217 | Tyr183 | 0.821 | 0.901 | 0.497 | // |
| 217 | Lys178 | 0.249 | 0.356 | 0 | // | |
| Glu1049Asp | 1049 | Glu1045 | 0.464 | 0.839 | 0.912 | 0.859 |
| 1049 | Lys1053 | 0.600 | 0.944 | 0.948 | 0.979 | |
| 1049 | Arg1048 | 0.414 | 0.297 | 0.146 | 0 | |
| 1049 | Lys1080 | 0 | 0 | 0.964 | 0.855 | |
| Asp1163Glu | 1163 | Pro1214 | 0.418 | 0.561 | 0.118 | // |
| 1163 | Cys1234 | 0.101 | 0 | 0.468 | // | |
| 1163 | Gln472 | 0.887 | 0.694 | 0 | // | |
| 1163 | Gln475 | 0 | 0 | 0.657 | // | |
| Val195Phe | 195 | Leu192 | 0.819 | 0.585 | 0.864 | // |
| Ile1318Leu | 1318 | His1320 | 0.508 | 0.480 | 0.266 | 0.412 |
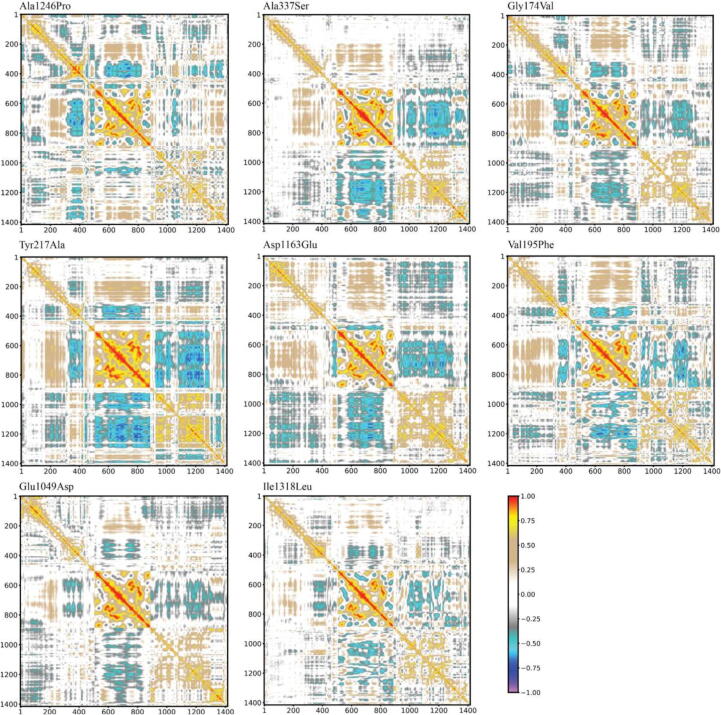
3.6. Mutants' DCCMs
In all mutant DCCMs (Fig. 6), we observed a loss of anticorrelated motions featuring the wild-type simulation between the Zinc and Jumonji domains (green box, Fig. 2B), with the only exception of Tyr217Ala, where it was partially preserved. Similarly, in Ala337Ser, Glu1049Asp, and Ile1318Leu, the correlation between TPR and Jumonji domains (blue box, Fig. 2B) was entirely lost. In Gly174Val, Ala1246Pro, Asp1163Glu and Val195Phe, the Jumonji domain moved in an anticorrelated way toward the first part of the TPR (residues 1 to 200). Furthermore, in Tyr217Ala, the same anticorrelated motions observed in the wild-type were partly replaced by a correlated movement of part of the TPR domain (residues 350 to 450). Finally, Ala337Ser, Tyr217Ala, Val195Phe, and Ile1318Leu entirely lost the anticorrelated motions between the flanking regions of the catalytic domain and part of the TPR domain (red box, Fig. 2B). Such anticorrelated dynamics were only partially preserved in Gly174Val, Glu1049Asp, Asp1246Pro, and Asp1163Glu.
Fig. 6.
DCCMs of mutant KDM6A proteins; perfect correlations are highlighted in red (direct) or violet (inverse). Altered motions with respect to the wild-type protein can be recognized by direct comparison with colored boxes in Fig. 2B. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. The roles of H3 and H3K27me3
The binding energy of the KDM6A protein with the ligand H3 was calculated for the average structures of the considered clusters using the MM/GBSA method. In the wild-type complex, cluster1 showed a reduced binding affinity for H3 than cluster0 (-82 to -103 kcal/mol). Moreover, in Ala1246Pro and Ile1318Leu, we observed two different binding configurations, showing higher binding energy values than the wild-type (-102 to -106 kcal/mol and -106 to -127 kcal/mol, respectively). Similarly, in Ala337Ser and Gly174Val, we obtained two different conformations featured by lower binding energy values (-49 to -68 kcal/mol and -86 to -95 kcal/mol, respectively). In Glu1049Asp and Val195Phe, we identified two conformations exhibiting proximal binding energy values (-111 kcal/mol and -96 kcal/mol, respectively). Finally, for Tyr217Ala and Asp1163Glu, we obtained only one cluster, with a binding energy of -74 kcal/mol and -102 kcal/mol, respectively.
Such alterations of the binding affinity also influenced the orientation of H3K27me3 within its binding pocket. For this reason, we evaluated the H3K27me3 interaction network for each protein system (Supplementary Table 4) and reported whether fundamental interactions in the wild-type protein were partially or severely altered in the mutants. We found that Ala337Ser lacks the three bonds with H3K27me3 characterizing the wild-type protein and described above, i.e., with Asn1149, Asn1087, and Ser1154; this causes the complete disorganization of the binding site interaction network. Similarly, Gly174Val, Val195Phe, Tyr217Ala, Ala1246Pro, and Ile1318Leu lost their bonds with Ser1154. Ala1246Pro and Ile1318Leu replaced this bond with two high-frequency bonds with Asn1087 and Asn1149, which were partially conserved in Val195Phe and Asp1163Glu (both characterized by only one cluster); Tyr217Ala also lost many other essential interactions. Furthermore, Glu1049Asp has partially lost the interaction with Ser1154 and increased that with Glu1148 in one of the two sampled conformations. On the contrary, the interaction with Glu1148 was never established in Gly174Val. Finally, Ala1246Pro showed a novel bond between H3K27me3 and Leu1127 (Supplementary Table 3).
4. Discussion
MD simulation represents one of the most potent computational strategies to study the dynamical features of macromolecules, such as, for example, the conformational transitions in atomistic detail. Using an enhanced MD simulation technique, we were able to identify a key conformational transition occurring in the wild-type KDM6A protein. The RMSD and DCCM profiles, which describe the motility of protein domains during an MD simulation, remarked the great flexibility of the Linker and Jumonji domains and the clear anticorrelated motions of the Jumonji and part of the TPR and the Zinc domains. Such high motility can be mainly ascribed to the differential interaction network resulting from the transition between the two main clusters, cluster0 and cluster1. Here, the torsion of the Linker domain, consequent to the anticorrelated motion between the other catalytic subdomains, resulted in increased interaction between the Arg922 residue and the Jumonji domain and a decreased affinity for the H3 in cluster1, as pointed out by the analysis of the binding energy. Notably, this latter conformation is characterized by a significant rewiring of the H3K27me3 interaction network compared to cluster0, signaling the orientation change of the trimethylated lysine into its binding pocket.
This extensive characterization allowed us to finely describe alternative conformations corresponding to well-defined and highly populated low energy states in nine KDM6A, LUSC-related mutants, i.e., Gly174Val, Val195Phe, Tyr217Ala, Ala337Ser, Arg901Lys, Glu1049Asp, Asp1163Glu, Ala1246Pro, and Ile1318Leu, and hypothesize their mechanistic roles. Interestingly, all of them but Arg901Lys were found to have a structural impact, affecting the overall protein dynamics at different levels. As the SNV causing Arg901Lys substitution localizes on the last nucleotide of the coding exon 17 (ENST00000377967.9), a plausible explanation is that a functional effect on splicing or transcript stability could be envisaged as the pathogenic mechanism. In-silico predictions and gene expression analysis confirmed this hypothesis. This could also apply to Ala1246Pro as the residue 1246 overlaps an exon-exon junction. In-silico predictions confirmed a potential functional effect on the splicing mechanism; considering the gene expression values, a functional effect of this variant on both transcript splicing/stability and protein activity could not, therefore, be excluded.
The RMSD profiles showed how the majority of the alterations to the protein dynamics regarded the Jumonji domain, with Gly174Val, Glu1049Asp, Asp1163Glu, Ala1246Pro, and Ile1318Leu displaying a decrease in the motility of this catalytic domain. In contrast, Gly174Val, Tyr217Ala, Ala337Ser, and Glu1049Asp dynamics were characterized by a disordered movement of the Zinc domain.
Analyzing the perturbations caused by the mutants locally, we were able to identify several interactions whose alterations resulted in the loss of fundamental motions needed for the correct conformational transition of the protein. Val195Phe determined an increase of the local stability, which, in turn, caused an altered motion correlation between the TPR and the Jumonji domains. Similarly, in Ala337Ser, we verified enhanced rigidity in the variant site's neighborhood, with the consequential loss of fundamental anticorrelated motions present in the wild-type protein. Notably, this resulted in the interaction impairment between H3 and the catalytic domain. On the contrary, Tyr217Ala and Glu1049Asp altered several interactions described as fundamental for the native conformational transition, with a lack of correlation and anticorrelation motions between different domains, especially for Glu1049Asp. Alterations of the interaction frequencies were found in the surrounding Gly174Val and Asp1163Glu regions and the establishment of novel interactions. Globally, this caused anticorrelated movements of the Jumonji domain towards part of the TPR domain and a loss of correlation with the Zinc domain. Finally, Ala1246Pro and Ile1318Leu showed slight but evident differences in the interaction frequency compared to the wild-type, with the complete lack of motion correlation between Jumonji and the Zinc and TPR domains.
The analysis of the binding energy between H3 and KDM6A and the H3K27me3-specific interaction network further supported these conclusions. An evident decrease in the H3-KDM6A affinity was observed in Tyr217Ala and Ala337Ser. At the same time, Val195Phe, Glu1049Asp, and Ala1246Pro displayed similar binding energy values for different conformations, a sign of the overall protein rigidity. Finally, Gly174Val and Ile1318Leu did not show dramatic differences in binding energy but an alteration of the H3K27me3 orientation caused by the Ser1154 binding loss and the increased affinity for Asn1087 (both) and Asn1149 (Ile1318Leu only).
To summarize, this research suggests that LUSC-associated missense mutations in KDM6A affect the physiological transition between two wild-type alternative conformations, which in turn affects the H3 interaction. Compared with our previous description of Kabuki-associated mutations, this finding expanded our understanding of the demethylase mechanism, emphasizing the importance of the correct orientation of H3K27me3. This fundamental aspect, perceivable but not observable due to the overall rigidity of the Kabuki mutant KDM6A-H3 complexes, was clearly described by the interaction rewiring occurring in the simulated mutants. As a result, while the lack of quantitative data on the mutant systems' putatively altered demethylase activity limits this study, it can serve as a starting point for a therapeutic approach aimed at restoring the physiological activity in patients with KDM6A mutations.
CRediT authorship contribution statement
Tommaso Biagini: Methodology, Writing – original draft. Francesco Petrizzelli: Methodology, Writing – original draft. Salvatore Daniele Bianco: Methodology. Niccolò Liorni: Investigation. Alessandro Napoli: Investigation. Stefano Castellana: Investigation. Angelo Luigi Vescovi: Funding acquisition. Massimo Carella: Funding acquisition. Viviana Caputo: Methodology, Writing – review & editing. Tommaso Mazza: Writing – review & editing, Conceptualization, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
Italian Ministry of Health (Ricerca Corrente 2022–2025); ‘5 x 1000’ voluntary contribution.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.041.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Das P., Taube J.H. Regulating Methylation at H3K27: A Trick or Treat for Cancer Cell Plasticity. Cancers. 2020;12:2792. doi: 10.3390/cancers12102792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Shilatifard A. UTX Mutations in Human Cancer. Cancer Cell. 2019;35:168–176. doi: 10.1016/j.ccell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S, Yim S, Park H. The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp Mol Med 2019;51. https://doi.org/10.1038/s12276-019-0230-6. [DOI] [PMC free article] [PubMed]
- 4.Epigenetic regulation of epithelial-mesenchymal transition by KDM6A histone demethylase in lung cancer cells. Biochem Biophys Res Commun 2017;490:1407–13. [DOI] [PubMed]
- 5.Andricovich J., Perkail S., Kai Y., Casasanta N., Peng W., Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch J., Lang A., Whongsiri P., Schulz W.A., Hoffmann M.J., Greife A. KDM6A mutations promote acute cytoplasmic DNA release, DNA damage response and mitosis defects. BMC Molecular and Cell Biology. 2021;22:1–18. doi: 10.1186/s12860-021-00394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrizzelli F., Biagini T., Barbieri A., Parca L., Panzironi N., Castellana S., et al. Mechanisms of pathogenesis of missense mutations on the KDM6A-H3 interaction in type 2 Kabuki Syndrome. Comput Struct Biotechnol J. 2020;18:2033–2042. doi: 10.1016/j.csbj.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi Y.-I., Stodola T.J., De Assuncao T.M., Leverence E.N., Tripathi S., Dsouza N.R., et al. Molecular mechanics and dynamic simulations of well-known Kabuki syndrome-associated KDM6A variants reveal putative mechanisms of dysfunction. Orphanet J Rare Dis. 2021;16:1–15. doi: 10.1186/s13023-021-01692-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi Y.-I., Stodola T.J., De Assuncao T.M., Leverence E.N., Smith B.C., Volkman B.F., et al. Structural bioinformatics enhances the interpretation of somatic mutations in KDM6A found in human cancers. Comput Struct Biotechnol J. 2022;20:2200–2211. doi: 10.1016/j.csbj.2022.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zengin T., Önal-Süzek T. Comprehensive Profiling of Genomic and Transcriptomic Differences between Risk Groups of Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Journal of Personalized Medicine. 2021;11:154. doi: 10.3390/jpm11020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Methylation L.C.T.T.H. Opportunities and Challenges. Comput Struct Biotechnol J. 2018;16:211–223. doi: 10.1016/j.csbj.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran N., Broun A., Ge K. Lysine Demethylase KDM6A in Differentiation, Development, and Cancer. Mol Cell Biol. 2020;40 doi: 10.1128/MCB.00341-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz W.A., Lang A., Koch J., Greife A. The histone demethylase UTX/KDM6A in cancer: Progress and puzzles. Int J Cancer. 2019;145:614–620. doi: 10.1002/ijc.32116. [DOI] [PubMed] [Google Scholar]
- 15.Gudmundsson S., Singer-Berk M., Watts N.A., Phu W., Goodrich J.K., Solomonson M., et al. Variant interpretation using population databases: Lessons from gnomAD. Hum Mutat. 2021 doi: 10.1002/humu.24309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmet F.-O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 20.Gelfman S., Wang Q., McSweeney K.M., Ren Z., La Carpia F., Halvorsen M., et al. Annotating pathogenic non-coding variants in genic regions. Nat Commun. 2017;8:236. doi: 10.1038/s41467-017-00141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176 doi: 10.1016/j.cell.2018.12.015. 535–48.e24. [DOI] [PubMed] [Google Scholar]
- 22.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 2016;54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biagini T., Chillemi G., Mazzoccoli G., Grottesi A., Fusilli C., Capocefalo D., et al. Molecular dynamics recipes for genome research. Brief Bioinform. 2017;19:853–862. doi: 10.1093/bib/bbx006. [DOI] [PubMed] [Google Scholar]
- 27.Case D.A., Metin Aktulga H., Belfon K., Ben-Shalom I., Brozell S.R., Cerutti D.S., et al. University of California; San Francisco: 2021. Amber 2021. [Google Scholar]
- 28.Peters M.B., Yang Y., Wang B., Füsti-Molnár L., Weaver M.N., Merz K.M., Jr. Structural Survey of Zinc Containing Proteins and the Development of the Zinc AMBER Force Field (ZAFF) J Chem Theory Comput. 2010;6:2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao Y., Feher V.A., McCammon J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J Chem Theory Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biagini T., Petrizzelli F., Truglio M., Cespa R., Barbieri A., Capocefalo D., et al. Are Gaming-Enabled Graphic Processing Unit Cards Convenient for Molecular Dynamics Simulation? Evol Bioinform Online. 2019;15 doi: 10.1177/1176934319850144. 1176934319850144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gowers R., Linke M., Barnoud J., Reddy T., Melo M., Seyler S., et al. Proceedings of the 15th Python in Science Conference. 2016. MDAnalysis: A python package for the rapid analysis of molecular dynamics simulations. [Google Scholar]
- 32.Venkatakrishnan AJ, Fonseca R, Ma AK, Hollingsworth SA, Chemparathy A, Hilger D, et al. Uncovering patterns of atomic interactions in static and dynamic structures of proteins. bioRxiv 2019:840694. https://doi.org/10.1101/840694.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.