Abstract
Functional precision medicine in AML often relies on short-term in vitro drug sensitivity screening (DSS) of primary patient cells in standard culture conditions. We designed a niche-like DSS assay combining physiologic hypoxia (O2 3%) and mesenchymal stromal cell (MSC) co-culture with multiparameter flow cytometry to enumerate lymphocytes and differentiating (CD11/CD14/CD15+) or leukemic stem cell (LSC)-enriched (GPR56+) cells within the leukemic bulk. After functional validation of GPR56 expression as a surrogate for LSC enrichment, the assay identified three patterns of response, including cytotoxicity on blasts sparing LSCs, induction of differentiation, and selective impairment of LSCs. We refined our niche-like culture by including plasma-like amino-acid and cytokine concentrations identified by targeted metabolomics and proteomics of primary AML bone marrow plasma samples. Systematic interrogation revealed distinct contributions of each niche-like component to leukemic outgrowth and drug response. Short-term niche-like culture preserved clonal architecture and transcriptional states of primary leukemic cells. In a cohort of 45 AML samples enriched for NPM1c AML, the niche-like multiparametric assay could predict morphologically (p = 0.02) and molecular (NPM1c MRD, p = 0.04) response to anthracycline-cytarabine induction chemotherapy. In this cohort, a 23-drug screen nominated ruxolitinib as a sensitizer to anthracycline-cytarabine. This finding was validated in an NPM1c PDX model.
Subject terms: Acute myeloid leukaemia, Translational research
Introduction
The outcome of patients diagnosed with acute myeloid leukemia (AML) remains unsatisfactory and new therapeutic approaches are required [1]. Drug sensitivity patterns differ across AML genetic subsets, among which NPM1-mutated (NPM1c) AML is the most frequent [2].
After the failure of conventional therapies, there is no standard salvage regimen and patients are increasingly proposing personalized therapies based on their physical status and the genetic make-up of their disease. However, genetics may not suffice to choose the optimal therapy, because some targets may have multiple available inhibitors (e.g., FLT3 mutations), and some active drugs may lack strong genetic biomarkers (e.g., venetoclax) [3]. Functional precision medicine based on ex vivo drug sensitivity screening (DSS) is gaining momentum to overcome these limitations [4–6].
DSS datasets can be leveraged at the population level, e.g., to identify drugs active in pre-defined AML subsets, or at the individual level, to tailor therapy to each patient. So far, most AML DSS platforms have tested drugs across broad concentration ranges in standard culture conditions, using the global viability of minimally fractionated primary samples to estimate drug sensitivity [4–6]. However, clinical drug exposure is bounded by bioavailability and dose-limiting hematological toxicity [1]. The leukemic niche provides resistance signals [7, 8]. Plasma-like medium improves the metabolic fidelity of in vitro assays [9, 10]. Notably, amino-acid levels differ between cancer patients and healthy subjects [11]. The tumor purity of primary AML samples is not absolute and leukemic cells can exist in different states including stem and differentiated states [12, 13]. Several surface markers have been proposed to enrich leukemic stem cells (LSCs) within the leukemic bulk [14]. Among those, expression of the G-coupled receptor GPR56 appears relatively universal across AML subsets [15, 16] and is stable upon short-term ex vivo culture [15].
Here we report the development and validation of an ex vivo drug screening platform for primary human AML cells that combines a niche-like culture system with a multiparametric flow cytometry readout. Focusing on NPM1-mutated AML, we perform clinical and genetic validation of the platform and nominate novel sensitizers to “7 + 3” daunorubicin (DNR) and cytarabine (AraC) combination chemotherapy in AML.
Methods
Detailed Methods are provided in the Supplementary Appendix (online only).
Primary AML samples
Fresh or viably frozen Bone Marrow (BM) or peripheral blood (PB) mononuclear cells (MNCs) from AML patients were collected after informed consent at the time of diagnosis or relapse by the INCa-labeled Hôpital Saint-Louis Tumor Bank. The project was approved by INSERM IRB (CEEI-15-220). Clinical and genetic annotations are provided in Supplementary Tables 1 and 2.
Short-term ex vivo culture
All experiments were conducted in 96-well plate format. hTERT-MSC-GFP immortalized human mesenchymal stromal cells (MSCs) were provided by JP Bourquin) [17]. AML MNCs were seeded at 50,000 cells per well in 90 µL of MEMα standard medium, or in plasma-like culture medium (Supplementary Table 7), both supplemented with 25% dialyzed FBS, 100 IU/mL penicillin, and 100 µg/mL streptomycin. TPO 1 ng/mL and EPO 2.5 ng/mL (Peprotech, Neuilly-sur-Seine) were added to reach “plasma-like” cytokine concentrations. Drugs were resuspended in medium and immediately added to each well, with a maximum DMSO 0.1% final concentration. Plates were incubated for 72 hours at 37 °C in 20% or 3% O2 and 5% CO2 (hypoxia, MCO-19M-PE incubator, Panasonic, Genevilliers). Details are provided in the Supplementary Appendix.
Multiparametric flow cytometry
Cells were washed and stained with Fixable Viability Stain eFluor 780 (Thermo Fisher Scientific), anti-CD45 PerCPCY5.5, anti-GPR56 PE, anti-CD11b APC, anti-CD14 APC, anti-CD15 APC, anti-CD3 BV421 and anti-CD19 BV421 (all BD Biosciences, Le Pont de Claix) and processed on an Attune Next (Thermo Fischer Scientific) flow cytometer. Cell counts were obtained after manual gating on FlowJo V10.6.2 (Beckton Dickinson, Le Pont de Claix). Details are provided in the Supplementary Appendix.
AML drug screening data analyses
Cell counts in each gate were normalized to negative controls (DMSO 0.1% vehicle wells). Drug activity was determined as the actual (trapezoidal) area over the curve (AOC) of cell counts (ie without fitting a dose-response curve) without truncation, using the PhamarcoGx R package [18]. With AOCs, higher values indicate greater drug activity. Details are provided in the Supplementary Appendix.
Statistical analyses
Statistical analyses were conducted in Prism 8.0.1 (GraphPad, San Diego, CA) or R version 3.6.0 (https://www.R-project.org/). Details are provided in the Supplementary Appendix.
Data accessibility
Bulk and single-cell RNA-Seq will be available at European Genome-phenome Archive (EGA) under accession code EGAS00001006265. Other data will be available upon reasonable request to the principal investigator.
Results
Multiparametric flow cytometry readout for ex vivo culture of primary AML cells
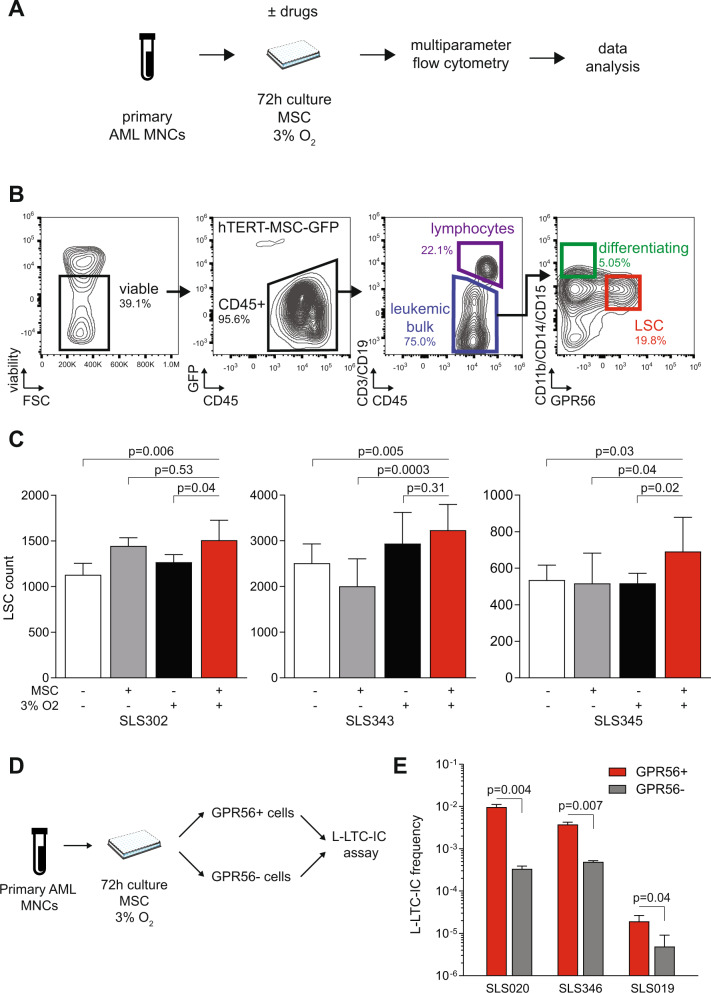
We first sought to develop a core flow cytometry panel to enumerate residual cells following a 72-hour co-culture of human primary AML MNCs with human immortalized MSC-hTERT-GFP stromal cells in 3% O2 (Fig. 1a), a culture system that has been previously shown to maintain primary human LSCs and to recapitulate micro-environment-driven drug resistance [7, 8]. The gating strategy allows exact counting of viable cells within the leukemic bulk after exclusion of GFP + MSCs and CD3/CD19+ lymphocytes. Within the leukemic bulk, cells are assigned to a GPR56+ (henceforth LSC) state, to a CD11b/CD14/CD15+ (Diff+) differentiating state, or to the basal GPR56-Diff- blast state (Fig. 1b). We chose GPR56 to define the LSC population because its gene belongs to the core 17-gene stemness gene expression signature of AMLs and because its surface expression enriches for AML initiating potential across a broad spectrum of AML subsets (including CD34- NPM1-mutated AMLs) and is stable upon short-term ex vivo culture [15, 16, 19]. We validated that combination of hypoxia (3% O2) and MSC co-culture enhanced the number of viable GPR56+ leukemic cells after 72-hour culture ex vivo over each feature alone or standard (O2 20%, no MSC) culture (Fig. 1c). In three primary AML samples, residual GPR56+ leukemic cells sorted after 72-hour hypoxic MSC co-culture were enriched ~10-fold in leukemic long-term initiating potential compared to GPR56- cells (Fig. 1d, e).
Fig. 1. Multiparametric readout after ex vivo culture of primary AML cells.
A Summary of the workflow for multiparametric flow cytometry following short-term ex vivo co-culture of primary AML MNCs with MSCs in 3% O2. B Gating strategy. C Number of GPR56+/Diff− LSCs after 72-hour culture with or without MSCs and with 20% (standard) or 3% (hypoxia) O2 in three samples; 6–10 technical replicates per condition. Mean ± SD. T tests with Welch’s correction. D, E Experiment design (D) and L-LTC-IC output (E) expressed as the ratio of colonies after 5-week culture per seeded GPR56+ or GPR56- cells sorted after a 72-hour culture with MSCs and 3% O2 in three different AML samples. Technical triplicates. Mean ± SD. Two-sided T tests with Welch’s correction. Clinical, phenotypic, and genetic annotations of primary AML samples are available in Supplementary Table 1.
Different patterns of drug activity at clinically relevant concentrations
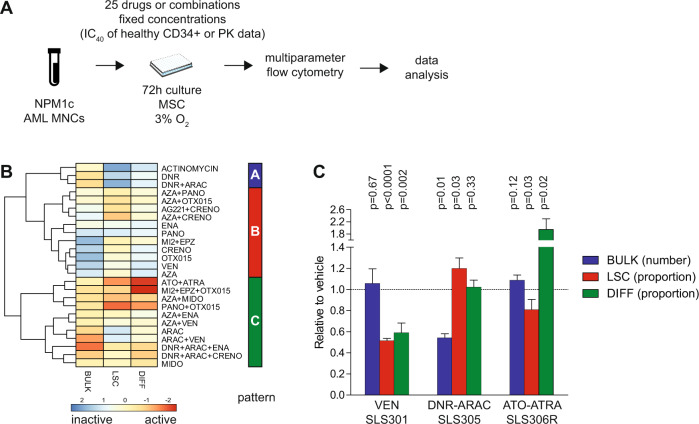
We next interrogated the patterns of activity of 25 drugs (or combinations) at fixed concentrations in the niche-like culture system. Clinically relevant maximal concentrations for ex vivo drug testing were defined as either the peak plasma drug concentration in available pharmacokinetics (PK) studies, or as the concentration inhibiting 40% (IC40) growth of CD34+ hematopoietic stem/progenitor cells from healthy donors in niche-like (MSC, 3% O2) co-culture dose-response assays, whichever was lowest (Fig. 2a, Supplementary Tables 3 and 4). In one illustrative NPM1-mutated AML (SLS305), unsupervised clustering revealed different patterns of drug activity, including one with predominant activity on the leukemic bulk with lower efficacy on LSCs and differentiation (“cytotoxic” pattern A), one with predominant activity on the proportion of LSCs (“stemness-specific” pattern B) and one with prominent differentiating activity (pattern C, Fig. 2B). Expectedly, the “cytotoxic” cluster encompassed known chemotherapeutic agents including the standard AML combination therapy DNR and cytarabine (AraC) or actinomycin D, both of which have been shown to be clinically active in NPM1-mutated AML [20, 21]. The anti-stemness cluster was enriched in epigenetic drugs or combinations targeting BET bromodomains (OTX015), menin-MLL interactions (MI-2), and DOT1L (EPZ-5676), which have demonstrated pre-clinical activity on the NPM1c leukemia-initiating program [22–24]. Finally, the combination of arsenic trioxide (ATO) and all-trans retinoic acid (ATRA) was the most prominent member of the differentiating cluster, along with other doublets or triplets of epigenetic agents. Figure 2c displays the prototypical output of drugs or combinations from each cluster across different NPM1-mutated AMLs, highlighting the reduced proportion of LSCs (and DIFF cells) without impact on the total number of leukemic cells upon single-agent exposure to the BCL-2 inhibitor venetoclax, the relative sparing of LSCs with the cytotoxic DNR-AraC combination, and the differentiating activity of ATO-ATRA, also resulting in a seemingly unchanged count of total leukemic bulk. Altogether, these results are in keeping with known pre-clinical or clinical data regarding these agents in NPM1-mutated AML [25–28], and highlight the relevance of a multiparametric readout for ex vivo drug screening of primary AML samples.
Fig. 2. Different patterns of drug activity at clinically relevant concentrations.
A Workflow of ex vivo drug screening of primary NPM1-mutated AML MNCs with 25 drugs or combinations at fixed, clinically relevant drug concentrations nominated based on published pharmacokinetics data or on dose–response assays in healthy CD34+ cells. B Heatmap of drug activity patterns for one illustrative case (SLS305, relapsed AML with trisomy 4 and NPM1 mutation) displaying the number of residual cells in the leukemic bulk (BULK), the proportion of GPR56+/Diff− LSCs and that of GPR56−/Diff+ differentiating cells (DIFF). Results of the heatmap are displayed after scaling per output so that higher arbitrary values indicate a lower number of bulk leukemic cells, a lower proportion of LSCs, and a higher proportion of DIFF cells, hence higher drug activity. C Illustrative output of single-agent venetoclax (VEN), DNR-AraC, or ATO-ATRA in the three NPM1-mutant tested samples (SLS301 relapsed AML with isolated del(15q), NPM1, DNMT3A, RAD21, CUX1, NRAS, KIT, and GATA2 mutations; SLS305 as supra; SLS306R relapsed AML with complex karyotype, NPM1, FL3-ITD, DNMT3A, and SMC3 mutations, details on patient samples and gene mutations are provided in Supplementary Tables 1, 2). Results are shown normalized to DMSO vehicle wells. Mean ± SD of technical triplicates. Unpaired two-sided t tests.
Refinement of niche-like culture with plasma-like medium
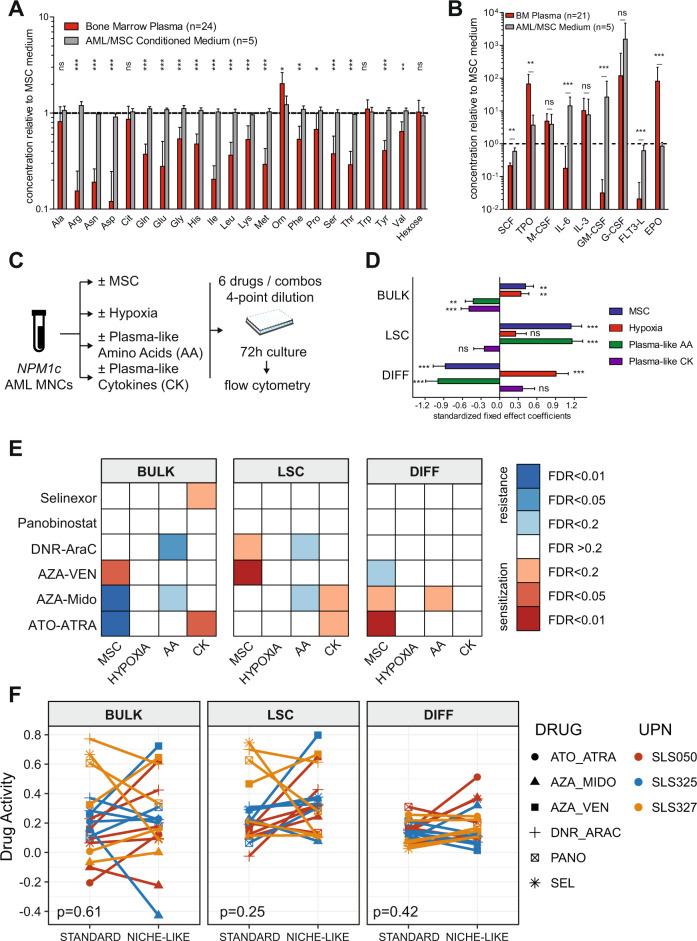
Conventional culture media poorly reflect the endogenous levels of metabolites such as amino acids and cytokines from normal human plasma [9, 10]. Furthermore, the metabolome and cytokinome of AML patients may differ from healthy subjects [29–32]. We thus undertook targeted metabolomics and multiplex cytokine dosages on the plasma of BM aspirates collected at AML diagnosis in 24 patients and compared them to conditioned media of primary AML samples co-cultured on an MSC layer in 3% O2, or to the conditioned medium of MSC alone. While amino-acid levels from conditioned media of AML co-cultures minimally differed from that of MSC alone co-culture, they significantly exceeded 1.4 to 3.6-fold those from diagnostic AML plasma for 16 of 21 tested amino acids (false discovery rate [FDR] < 0.01, Fig. 3A, Supplementary Table 5). Myeloid cytokine levels were highly variable in AML conditioned media but were systematically lower than BM plasma levels for TPO (mean 62.2 ± 61.1 versus 1128.2 ± 1012.9 pg/mL, q = 0.001) and EPO (mean 29.9 ± 7.9 versus 2852.0 ± 4506.0 pg/mL, q < 0.001, Fig. 3B, Supplementary Table 6).
Fig. 3. Plasma-like interacts with other pseudo-niche factors to dictate leukemic cell states and drug response ex vivo.
A, B Amino-acid and hexose levels (A AbsoluteIDQ® p180 Assay, Biocrates) and cytokine levels (B custom U-Plex®, MesoScale Discoveries) from 21–24 diagnostic AML BM plasma samples and five AML/MSC co-cultures conditioned media relative to conditioned medium of MSC alone. Mean+SD. FDR q values from Mann–Whitney tests for the comparison of BM plasmas versus AML/MSC conditioned media. C Workflow of a mini-screen to systematically interrogate the contribution of the four pseudo-niche components (MSC, 3% O2, plasma-like amino acids, and cytokines) on leukemic states and drug sensitivity. D Fixed effects estimates (with SD) from mixed effect models investigating the contribution of each pseudo-niche component on each readout in the vehicle (DMSO 0.1%) treated cells from three NPM1-mutated AMLs, with random effect terms for patients. P values are from the mixed effect models. *p or q < 0.05, **p or q < 0.01, ***p or q < 0.001. E Heatmap of FDR q values for the impact of each pseudo-niche component on drug activity for each readout and drug, based on mixed-effect models with a random effect for patients. F Drug activities for each readout in standard versus niche-like conditions. P values from Wilcoxon signed-rank tests. Drug activity ranking in each patient is provided in Supplementary Fig. 3.
We could thus design a custom medium based on amino-acid-free MEMa medium and dialyzed FBS supplemented with plasma-like AA levels and/or plasma-like EPO and TPO concentrations (Supplementary Table 7). We next systematically interrogated the contribution of each of the four components of our pseudo-niche culture system (addition of MSCs, lowering of O2 from 20% to 3%, substitution of the standard to plasma-like amino-acid levels, the addition of plasma-like levels of EPO and TPO cytokines) to the output of short-term ex vivo cultures of primary AML cells using multiparametric flow cytometry. Cells were treated with vehicle (DMSO) or with a mini-panel of six drugs/combinations with reported activity in NPM1-mutated AML in 4-point 10-fold dilution dose-response assays (Fig. 3c). Focusing first on vehicle-treated cells, each pseudo-niche component was found to have a distinct imprint on leukemic states ex vivo. Notably, both MSCs and hypoxia improved the viability of the leukemic bulk ex vivo while MSCs and plasma-like amino acids maintained leukemic cells in the LSC state at the expanse of differentiation (Fig. 3d). Importantly, up to 40% of the variance between the output of ex vivo culture in these different conditions could be explained by interactions between pseudo-niche factors (Supplementary Fig. 1). We next inspected the impact of each pseudo-niche component on drug activity. Overall, MSCs, plasma-like amino acids, and cytokines conveyed significant but variable sensitization or resistance to the 6 investigated drugs or combinations (Fig. 3e). Again, interactions between pseudo-niche components had a strong impact on drug responses, accounting for up to 70% of the variance (Supplementary Fig. 2). We finally compared drug activities in ‘standard’ culture conditions (no MSCs, 20% O2, standard medium without cytokine addition) to those in full “pseudo-niche” conditions (MSC layer, 3% O2, plasma-like amino acids, and cytokines). Overall, there was no systematic bias between culture conditions (Fig. 3f), and the ranking of drugs in each patient for each readout was not correlated (Supplementary Fig. 3), indicating that drug screening outputs performed in standard conditions cannot predict the results of those performed in niche-like conditions.
Short-term niche-like ex vivo culture preserves intra-tumor heterogeneity
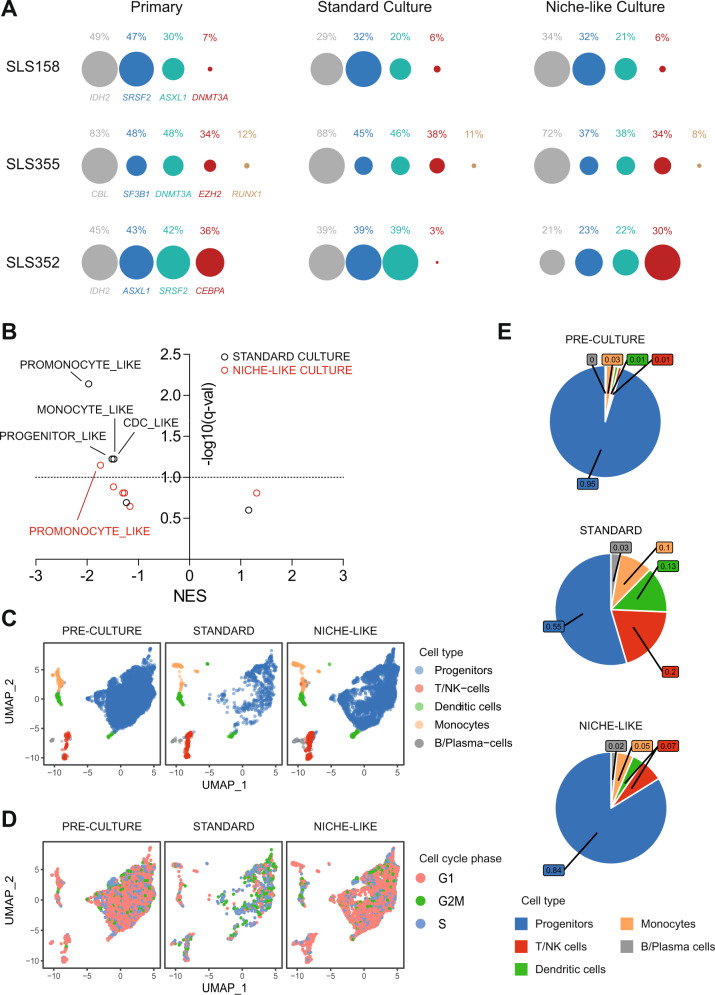
To determine whether short-term culture distorts the intra-tumor genetic heterogeneity of leukemic cells, we performed targeted sequencing of a 43 gene panel in archived BM MNCs obtained from 7 AML patients (median 4 gene mutations per patient, range 2–5, Supplementary Table 8) and performed amplicon-based sequencing of residual viable blasts after 72-hour culture in either standard or niche-like conditions. All 25 gene mutations were detectable in both culture conditions, including sub-clonal mutations with variant allele frequencies (VAF) < 20%, and limited distortion in VAF distribution compared to the primary specimen. In one sample (SLS352), niche-like culture better maintained CEBPA-mutant cells compared to standard culture (Fig. 4a and Supplementary Fig. 4). We similarly performed bulk RNA-Seq after a 6-hour culture and compared the transcriptome of cells exposed to niche-like versus standard culture conditions in 4 patients. When inspecting the gene expression signatures of leukemic cells reported at the single-cell level (Supplementary Table 9) [13], both culture conditions depleted the promonocyte-like signature but standard culture also significantly depleted three additional signatures that were preserved in niche-like conditions (FDR < 0.1, Fig. 4b). Finally, single-cell RNA-sequencing revealed that the distribution of BM mononucleated cell populations was preserved after a 72-hour culture in niche-like condition compared to the reference pre-culture specimen, while standard culture led to selective attrition of leukemic progenitors, with skewed cell cycle distribution (Fig. 4c). Of genes expressed in at least 10% of leukemic progenitor cells, 338 (103 up, 235 down) were differentially expressed ([fold change] >1.5 and FDR-adjusted q value <0.05) after standard culture versus pre-culture, compared to only 244 (up 57, down 187) after niche-like culture (Fisher exact test p = 0.001, Supplementary Tables 10–12, Supplementary Fig. 5). Collectively, these results suggest that short-term niche culture preserves the clonal heterogeneity and phenotypic diversity of primary AML specimens.
Fig. 4. Clonal architecture and transcriptomes after short-term ex vivo culture.
A Variant Allele Frequencies in primary AML MNCs and residual blasts after 72-hour culture in standard (no MSC, 20% O2, standard MEM-alpha medium) or niche-like (MSC co-culture, 3% O2, plasma-like amino acids, and cytokines) culture. Circles are proportional to VAFs, normalizing circle diameter based on the highest VAF in each sample. Additional cases are reported in Supplementary Fig. 4 and detailed mutations are provided in Supplementary Table 7. B Normalized Enrichment Scores (NES) and FDR-adjusted q values for the six leukemic state gene expression signatures from Van Galen et al. [13] (Supplementary Table 8) from RNA-Seq of four cryopreserved NPM1-mutated samples (SLS244, SLS354, SLS381, and SLS389) immediately after thawing (primary) or after 6 hours of ex vivo culture in niche-like or standard culture conditions. C, D UMAP plots of single-cell RNA-sequencing in pre-culture cells (n = 7008 cells) from SLS393 (normal cytogenetics, NPM1c and FLT3-ITD, additional details in Supplementary Tables 1, 2), after 72 hours of ex vivo culture in standard (n = 1079 cells) or niche-like (n = 4928 cells) conditions. C, D Projection of cell-type identity (C) or cell cycle phase identity (D). E Pie chart of the distribution of cell-type frequencies from C.
Validation of the niche-like multiparametric drug screening platform
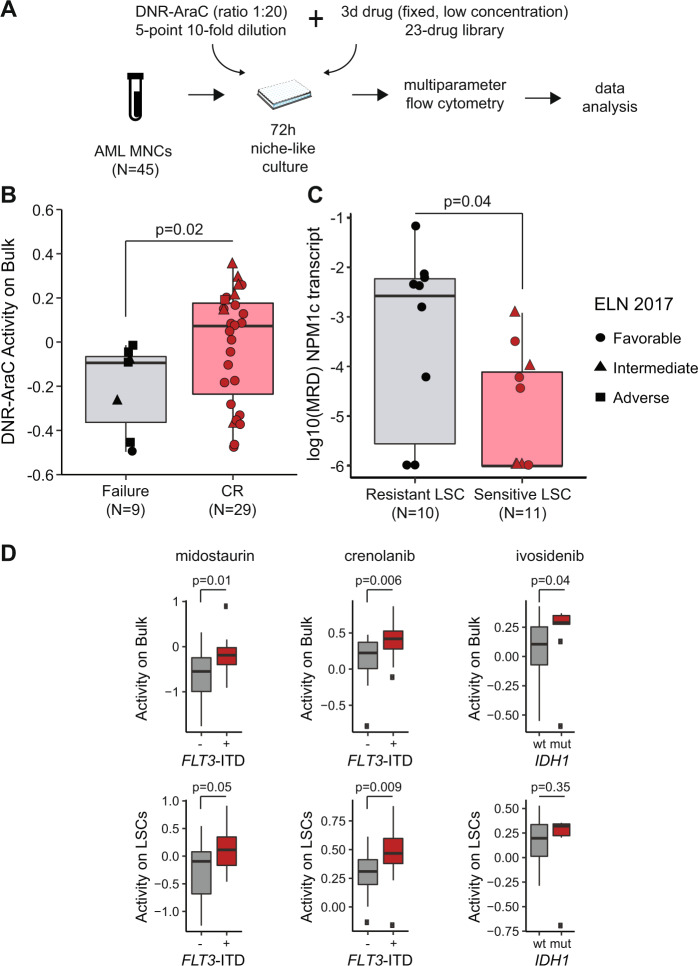
We next investigated the clinical relevance of our niche-like, multiparametric drug screening platform. We performed a screen of a 5-point, 10-fold serial dilution of the conventional chemotherapy combination of daunorubicin and cytarabine (DNR-AraC) in 45 patient samples, including 37 NPM1c AMLs (Supplementary Table 1). The DNR-AraC combination was delivered at a fixed 1:20 ratio reflective of the average ratio observed in vivo upon administration of the combination [33], either alone, or with the addition of a fixed, low concentration of a 23-drug panel (Fig. 5a, Supplementary Figs. 6–8). Low dose concentrations were chosen as the IC10 for healthy CD34+ cells (see supra) or 20% of Cmax from PK data, whichever was lower (Supplementary Tables 3, 4). Drug activity on blasts was higher in patients achieving CR after induction chemotherapy, compared to those with induction failure (p = 0.02, Fig. 5B). When adjusting the drug activity on lymphocytes as an internal reference, all nine patients whose blasts were more sensitive to DNR-AraC compared to their lymphocytes (chemosensitive blasts) achieved CR, compared to 20 of the 29 (69%) patients with lower drug activity on blasts relative to lymphocytes (chemoresistant blasts; Fisher’s test p = 0.08). In 21 NPM1-mutated AML patients with available post-induction NPM1c transcript MRD data, MRD levels did not differ between patients with chemosensitive versus chemoresistant blasts (p = 0.35). Conversely patients with chemosensitive GPR56 + LSCs (n = 11) had lower post-induction MRD than those with chemoresistant LSCs (n = 10, p = 0.04, Fig. 5C). As a continuous variable, higher relative drug activity on LSCs (i.e., adjusted to activity on lymphocytes) was associated with longer Event-Free Survival (EFS, Cox model hazard ratio [HR] = 0.25, 95% confidence interval [CI] 0.08–0.78, p = 0.02), independently of adverse European LeukemiaNet risk (HR = 4.79, 95% CI 1.90–12.10, p = 0.001). The niche-like platform also validated known genetically targeted therapies (Fig. 5d). Specifically, addition of the pan-kinase inhibitor midostaurin to DNR-AraC lead to superior combination activity on blasts (p = 0.01), and to a lesser extent on LSCs (p = 0.05) of FLT3-ITD mutated samples (n = 16), while addition of the more potent FLT3-ITD inhibitor crenolanib had superior activity on both blasts (p = 0.006) and LSCs (p = 0.009). Finally, the IDH1 inhibitor ivosidenib led to enhanced killing of blasts (p = 0.04), but not LSCs (p = 0.35) in IDH1-mutated samples (n = 9). Of note, no difference was noted for the activity of the combination with the IDH2 inhibitor enasidenib between IDH2 mutated (n = 7) and wildtype (n = 38) samples on bulk (p = 0.20) or LSC (p = 0.47) populations (not shown). Systematic inspection of chemogenomic relations further revealed that FLT3 mutations significantly sensitized blasts and LSCs to the addition of venetoclax to DNR-AraC (q < 0.05, Supplementary Fig. 9).
Fig. 5. Validation of the niche-like multiparametric drug screening platform.
A Ex vivo drug screening of primary MNCs from 45 AML patients (Supplementary Table 1, including 37 with NPM1 mutations) with a 5-point, 10-fold DNR-AraC serial dilution in fixed concentration ratio (1:20) with or without fixed, low concentrations of each of 23 drugs (Supplementary Tables 3, 4). B DNR-AraC combination activity on the leukemic bulk in patients achieving CR (N = 29) or induction failure (N = 9) after an anthracycline-cytarabine induction course. Mann–Whitney test. Circles, triangles, and squares indicate individual data of samples with favorable, intermediate, and adverse genetic risk according to ELN 2017 classification [60]. Details on patient information and gene mutations are reported in Supplementary Tables 1, 2. C Log-transformed MRD on NPM1 transcripts in 21 NPM1-mutated AML in CR, according to DNR-AraC ex vivo activity on LSC relative to lymphocytes (sensitive LSCs: activity on LSCs >activity on lymphocytes, N = 11; resistant LSCs: activity on LSCs ≤ activity on lymphocytes, N = 10). Mann–Whitney test. E Activity of the indicated drug on the bulk population (top panel) or the LSC population (bottom panel) according to the presence (n = 16) or absence of FLT3-ITD (n = 29) or the IDH1 status (wildtype [wt] n = 36 or mutated [mut] n = 9). P values from Mann–Whitney tests.
Discovery of novel drug combinations with niche-like multiparametric screening
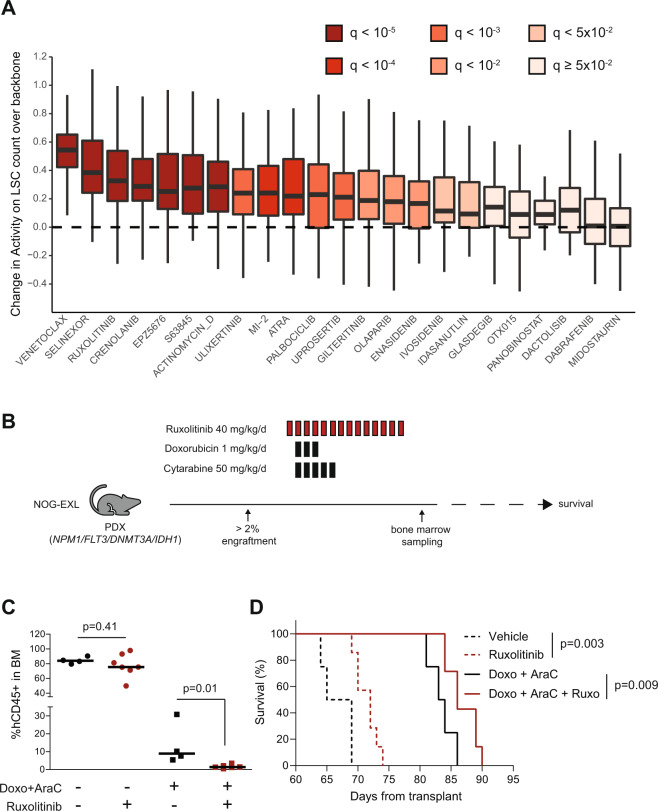
Focusing on drug activity against GPR56 + LSCs, the results of this DNR-AraC screen could be interpreted at the individual level to nominate the optimal combination in each patient. Though the DNR-AraC-venetoclax triplet scored as the top combination for 27 (60%) of 45 tested patients, “private” optimal combinations with nine different third agents were nominated for 17 (38%) patients, while only 1 (2%) had no benefit of any of the 23 third agents tested (Supplementary Fig. 10), stressing the potential role for functional assays to tailor individual therapies in AML. Inspecting the screen at the population level to identify the average benefit of each third agent over the DNR-AraC backbone alone, akin to parallel clinical trials, confirmed the benefit of adding venetoclax or selinexor to DNR-AraC, both regimens being currently tested in clinical trials [34, 35], but also revealed a significant activity of the addition of the JAK inhibitor ruxolitinib (q < 10−5, Fig. 6a), which has so far been explored in patients with AML secondary to myeloproliferative neoplasms [36], but never in the setting of NPM1 mutations. Of note, the activity of ruxolitinib in this setting was not dependent on FLT3 status (Supplementary Fig. 9). To validate prospectively this finding, we treated xenotransplanted NPM1-mutated AML cells also harboring mutations in DNMT3A, IDH1, and FLT3 with a combination of the anthracycline doxorubicin and cytarabine at maximal tolerated dose, ruxolitinib or the triplet combination (Fig. 6b). Post-treatment BM biopsies showed no reduction in leukemic burden with ruxolitinib alone compared to vehicle (p = 0.41), whereas the addition of ruxolitinib to doxorubicin-cytarabine further reduced leukemic infiltration over chemotherapy alone (p = 0.01, Fig. 6c). Ruxolitinib as a single-agent prolonged the survival of mice over vehicle (p = 0.003) and the addition of ruxolitinib to chemotherapy also significantly improved survival compared to chemotherapy alone (p = 0.009, Fig. 6d), providing in vivo confirmation of the ex vivo screen.
Fig. 6. Niche-like multiparametric drug screening platform identifies chemosensitization by ruxolitinib.
A Boxplot of the difference between drug activity on the count of GPR56+ /Diff− LSCs for each of the 23 triplets tested in the DNR-AraC screen (Fig. 5) and that of DNR-AraC backbone. FDR q values from t tests. B Experimental plan of the in vivo assessment of vehicle (n = 4), ruxolitinib (n = 8, 40 mg/kg/d orally, d1–14), chemotherapy (n = 4, doxorubicin 1 mg/kg/d IV d2-4 and cytarabine 50 mg/kg/d IP d2-6), or chemotherapy + ruxolitinib (n = 8) treatment of a PDX model (NPM1c, FLT3ITD, DNMT3AR88H, and IDH1R132H mutations) after engraftment into sub-lethally irradiated NOG-EXL recipient mice. C Proportion of hCD45+ leukemic cells in bone marrow aspirates performed at day 16 (hence 2 days after the last ruxolitinib or vehicle administration). P values from Mann–Whitney tests. D Overall survival of the four mice groups since the first day of treatments. P values from log-rank tests.
Discussion
With the ongoing expansion of therapeutic options in AML, ex vivo DSS of primary AML cells has gained renewed interest to tailor personalized treatment decisions [4, 37–39], reposition existing therapies [40, 41], discover novel agents or combinations [42–44], or perform chemo-genetic correlations [6, 43]. Ex vivo DSS is often limited by the lack of niche mimicry and the limited information obtained by global viability assessment. Little is known of the impact of short-term ex vivo culture on the intra-tumor genetic and transcriptional heterogeneity of AML [13, 45, 46].
We report the development of a niche-like multiparametric platform for ex vivo drug screening of primary AML samples. Combining an immortalized MSC stromal layer with low oxygen concentrations recapitulating BM oxygen tension [7, 47], we could validate the choice of GPR56 as a surface marker to enrich residual LSC activity after short-term culture. Our simple 6-color flow panel was sufficient to capture distinct drug-induced phenotypes, including cytotoxicity sparing LSCs, inhibition of stemness potential, and differentiating activity, recapitulating known features of selected approved AML drugs or combinations [25–28].
A systematic investigation of our niche-like culture system on leukemia growth and drug response revealed the crucial role of interactions between MSCs, low oxygen, plasma-like amino acids, and cytokines on the number and phenotype of leukemic cells, and on their response to selected drugs or combinations. MSC and low oxygen-limited the attrition of primary AML cells upon ex vivo culture, with stroma and plasma-like amino acids maintaining the GPR56 + LSC-enriched phenotype, while hypoxia-induced phenotypic differentiation. Response to five of six tested drugs or combinations was significantly affected by the addition of stroma, plasma-like amino acids, or cytokines. Though oxygen level had no impact on sensitivity to any of the 6 drugs tested, we cannot exclude that it may modulate drug response on larger screens. Further work is thus needed to determine that hypoxia is a critical component of niche-like culture for primary AML cells. Importantly, in none of the three tested primary AML samples could results of the drug screen in niche-like culture be predicted by conducting the screen in standard conditions. In seven AML samples, we could show that short-term ex vivo culture does not distort clonal representation, minor sub-clones (variant allelic frequency <20% in the primary specimen) still being detectable in all tested patients, although the full resolution of clonal architecture would require whole-exome sequencing. Our bulk RNA-Seq results suggest a benefit of niche-like culture over standard culture in preserving leukemic transcriptional states, though a significant depletion of the promonocyte-like state was also noticeable upon niche-like culture, while single-cell RNA-sequencing revealed a skewed cell cycle distribution in residual cells after standard, but not niche-like culture.
Akin to several DSS studies [38, 48–51], we could show that ex vivo response of the leukemic bulk to a DNR-AraC combination was correlated to the achievement of remission after anthracycline-cytarabine “7 + 3” therapy. Focusing on NPM1-mutated AMLs, most of whom achieve remission after 7 + 3 chemotherapy [20], and using non-leukemic cells (lymphocytes) as an internal reference as previously proposed [38], we could show that the activity of DNR-AraC on GPR56+ LSC-enriched cells but not on the leukemic bulk, could predict the depth of remission as assessed by NPM1-transcript MRD, but also event-free survival. Of note, CD34 expression was not accounted for in this study, given its variable expression in NPM1-mutated LSCs [52]. This clinical validation was completed by a genetic one, whereby the presence of FLT3 or IDH1 mutations predicted the response to combinations including FLT3 or IDH1 inhibitors, respectively. Of note, no such correlation was noted with the IDH2 inhibitor enasidenib. Though this could simply reflect the limited size of the studied cohort (n = 45, including seven with an IDH2 mutation), this could reflect a potential IDH2-independent activity of enasidenib [53].
The standard randomized clinical trial approach, even when selecting patients based on a genetic biomarker, has so far yielded limited benefit, owing to the variable benefit of adding a third agent to the standard 7 + 3 backbone. Indeed, by screening the addition of 23 drugs at concentrations deemed clinically relevant based on available PK data, or on drug testing of healthy CD34+ cells, we could show that in 39% of patients, the optimal triplet therapy is a private one. This result praises the development of robust DSS assays and innovative clinical trial design to foster functional precision medicine in AML [54]. Analyzing the average benefit of adding each of those 23 drugs to erode the GPR56 + LSC-enriched pool in a cohort of AMLs (many with NPM1 mutation) confirmed the benefit of adding venetoclax or selinexor to intensive chemotherapy [34, 35]. More surprisingly, the addition of the JAK1/2 inhibitor ruxolitinib also markedly improved anti-LSC activity over DNR-AraC alone ex vivo. This benefit was not confined to the subset of patients harboring FLT3 or signaling mutations and is reminiscent of the sensitization to BCL-2 inhibition by this kinase inhibitor recently reported by an ex vivo DSS study [55]. A combination of ruxolitinib with intensive chemotherapy in patients selected based on a DSS assay is currently explored by the BEAT-AML master trial (NCT03013998) [54], but to date, no clinical trial has reported the activity of this combination in de novo AML. The benefit of ruxolitinib addition could be validated in vivo in an NPM1-mutated PDX model, where engrafted mice received a ruxolitinib regimen in the lower range of reported in vivo experiments [56–58], and/or a combination of the anthracycline doxorubicin with cytarabine at maximally tolerated doses. Further work will be required to determine the mechanism, which could be non-cell-autonomous, through which ruxolitinib chemosensitizer NPM1-mutated AML cells [55].
Prospective analytical and clinical validation of our proposed niche-like ex vivo drug testing assay and benchmarking to existing platforms are important future steps [54, 59]. Further improvements to the assay will rely on miniaturization to improve throughput, incorporation of patient-derived MSCs, and culture medium refinement by accounting for plasma concentrations of polar metabolites [9]. More sophisticated flow cytometry panels will need to include additional LSC and differentiation markers and may also allow studying T cell activation. Collectively, our work contributes to the growing interest in functional assays to complement genomics-based precision oncology in leukemias.
Supplementary information
Supplementary Methods and Figures (Clean)
Acknowledgements
We are indebted to Karine Bailly and Céline Bertholle from the Cytomorphology and Immunology (CYBIO) core facility of Institut Cochin for technical support with electrochemiluminescence assays, Véronique Parietti, Sophie Duchez, and Niclas Setterblad from the Saint-Louis Research Institute Animal Models and Flow Cytometry Core Facilities, and Lionel Faivre from the Cell Therapy Department of Hopital Saint-Louis for CD34+ cells from healthy donors. This work was supported by Gilead’s Research Scholars Program in Hematology/Oncology, Association Laurette Fugain (ALF2017-15 and ALF2020-01), French National Cancer Institute (INCa), and Ministry of Health (DGOS) PRT-K15-125, Association Princesse Margot and Fondation Leucémie Espoir.
Author contributions
RI designed the research, performed analyses, and drafted the manuscript. RI and AP supervised the project and obtained research funding. RDB, JP, RJ, MD, BP, PA, GDF, GS, CC, LV, LC, FL, KP, CV, JB, CB, and NF performed experiments. RK, CP, and EC provided genetic annotations of primary AML samples. NB, TB, CP, HD, ER, and LA provided primary AML samples. All authors reviewed the manuscript and approved its final version.
Funding
This study was funded by Agence Nationale de la Recherche (THEMA project, IHU-B-2018), Gilead Research Scholarship, Institut National du Cancer and French Health Ministry (DGOS, PRT-K15-125 CAMELIA), Association Laurette Fugain (ALF2017-15 and ALF2020-01) and Association Princesse Margot,
Competing interests
Ri has received research funding from Oncoethix (now Merck) and Novartis, and honoraria from Abbvie, Astellas, BMS, and Karyopharm. Other authors have no relevant conflict of interest to disclose.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-022-00689-3.
References
- 1.Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10:506–25. doi: 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiNardo CD, Tiong IS, Quaglieri A, MacRaild S, Loghavi S, Brown FC, et al. Molecular patterns of response and treatment failure after frontline venetoclax combinations in older patients with AML. Blood. 2020;135:791–803. doi: 10.1182/blood.2019003988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3:1416–29. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 5.Lavallee VP, Baccelli I, Krosl J, Wilhelm B, Barabe F, Gendron P, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–7. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 6.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griessinger E, Anjos-Afonso F, Pizzitola I, Rouault-Pierre K, Vargaftig J, Taussig D, et al. A niche-like culture system allowing the maintenance of primary human acute myeloid leukemia-initiating cells: a new tool to decipher their chemoresistance and self-renewal mechanisms. Stem Cells Transl Med. 2014;3:520–9. doi: 10.5966/sctm.2013-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moschoi R, Imbert V, Nebout M, Chiche J, Mary D, Prebet T, et al. Protective mitochondrial transfer from bone marrow stromal cells to acute myeloid leukemic cells during chemotherapy. Blood. 2016;128:253–64. doi: 10.1182/blood-2015-07-655860. [DOI] [PubMed] [Google Scholar]
- 9.Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A, Jr., et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell. 2017;169:258–72.e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vande Voorde J, Ackermann T, Pfetzer N, Sumpton D, Mackay G, Kalna G, et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv. 2019;5:eaau7314. doi: 10.1126/sciadv.aau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siminska E, Koba M. Amino acid profiling as a method of discovering biomarkers for early diagnosis of cancer. Amino Acids. 2016;48:1339–45. doi: 10.1007/s00726-016-2215-2. [DOI] [PubMed] [Google Scholar]
- 12.Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547:104–8. doi: 10.1038/nature22993. [DOI] [PubMed] [Google Scholar]
- 13.van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia S, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–81.e24. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnone M, Konantz M, Hanns P, Paczulla Stanger AM, Bertels S, Godavarthy PS, et al. Acute myeloid leukemia stem cells: the challenges of phenotypic heterogeneity. Cancers (Basel). 2020;12:3742. [DOI] [PMC free article] [PubMed]
- 15.Pabst C, Bergeron A, Lavallee VP, Yeh J, Gendron P, Norddahl GL, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127:2018–27. doi: 10.1182/blood-2015-11-683649. [DOI] [PubMed] [Google Scholar]
- 16.Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–7. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 17.Frismantas V, Dobay MP, Rinaldi A, Tchinda J, Dunn SH, Kunz J, et al. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood. 2017;129:e26–e37. doi: 10.1182/blood-2016-09-738070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smirnov P, Safikhani Z, El-Hachem N, Wang D, She A, Olsen C, et al. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016;32:1244–6. doi: 10.1093/bioinformatics/btv723. [DOI] [PubMed] [Google Scholar]
- 19.Garg S, Reyes-Palomares A, He L, Bergeron A, Lavallee VP, Lemieux S, et al. Hepatic leukemia factor is a novel leukemic stem cell regulator in DNMT3A, NPM1, and FLT3-ITD triple-mutated AML. Blood. 2019;134:263–76.. doi: 10.1182/blood.2018862383. [DOI] [PubMed] [Google Scholar]
- 20.Balsat M, Renneville A, Thomas X, de Botton S, Caillot D, Marceau A, et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with npm1 mutation: a study by the acute leukemia french association group. J Clin Oncol. 2017;35:185–93. doi: 10.1200/JCO.2016.67.1875. [DOI] [PubMed] [Google Scholar]
- 21.Falini B, Brunetti L, Martelli MP. Dactinomycin in NPM1-mutated acute myeloid leukemia. N. Engl J Med. 2015;373:1180–2. doi: 10.1056/NEJMc1509584. [DOI] [PubMed] [Google Scholar]
- 22.Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia. 2014;28:311–20. doi: 10.1038/leu.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masse A, Roulin L, Pasanisi J, Penneroux J, Gachet S, Delord M, et al. BET inhibitors impair leukemic stem cell function only in defined oncogenic subgroups of acute myeloid leukaemias. Leuk Res. 2019;87:106269. doi: 10.1016/j.leukres.2019.106269. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn MW, Song E, Feng Z, Sinha A, Chen CW, Deshpande AJ, et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov. 2016;6:1166–81. doi: 10.1158/2159-8290.CD-16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov. 2016;6:1106–17. doi: 10.1158/2159-8290.CD-16-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–41. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito Y, Kitamura H, Hijikata A, Tomizawa-Murasawa M, Tanaka S, Takagi S, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2:17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Hajj H, Dassouki Z, Berthier C, Raffoux E, Ades L, Legrand O, et al. Retinoic acid and arsenic trioxide trigger degradation of mutated NPM1, resulting in apoptosis of AML cells. Blood. 2015;125:3447–54. doi: 10.1182/blood-2014-11-612416. [DOI] [PubMed] [Google Scholar]
- 29.Muscaritoli M, Conversano L, Petti MC, Torelli GF, Cascino A, Mecarocci S, et al. Plasma amino acid concentrations in patients with acute myelogenous leukemia. Nutrition. 1999;15:195–9. doi: 10.1016/S0899-9007(98)00179-8. [DOI] [PubMed] [Google Scholar]
- 30.Islam M, Mohamed EH, Esa E, Kamaluddin NR, Zain SM, Yusoff YM, et al. Circulating cytokines and small molecules follow distinct expression patterns in acute myeloid leukaemia. Br J Cancer. 2017;117:1551–6. doi: 10.1038/bjc.2017.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Correa B, Bergua JM, Campos C, Gayoso I, Arcos MJ, Banas H, et al. Cytokine profiles in acute myeloid leukemia patients at diagnosis: survival is inversely correlated with IL-6 and directly correlated with IL-10 levels. Cytokine. 2013;61:885–91. doi: 10.1016/j.cyto.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Stevens AM, Miller JM, Munoz JO, Gaikwad AS, Redell MS. Interleukin-6 levels predict event-free survival in pediatric AML and suggest a mechanism of chemotherapy resistance. Blood Adv. 2017;1:1387–97.. doi: 10.1182/bloodadvances.2017007856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tardi P, Johnstone S, Harasym N, Xie S, Harasym T, Zisman N, et al. In vivo maintenance of synergistic cytarabine:daunorubicin ratios greatly enhances therapeutic efficacy. Leuk Res. 2009;33:129–39. doi: 10.1016/j.leukres.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 34.Chua CC, Roberts AW, Reynolds J, Fong CY, Ting SB, Salmon JM, et al. Chemotherapy and venetoclax in elderly acute myeloid leukemia trial (CAVEAT): a phase Ib dose-escalation study of venetoclax combined with modified intensive chemotherapy. J Clin Oncol. 2020:JCO2000572. [DOI] [PubMed]
- 35.Abboud R, Chendamarai E, Rettig MP, Trinkaus KM, Riedell PA, Abboud CN, et al. Selinexor combined with cladribine, cytarabine, and filgrastim in relapsed or refractory acute myeloid leukemia. Haematologica. 2020;105:e404–e7. doi: 10.3324/haematol.2019.236810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devillier R, Raffoux E, Rey J, Lengline E, Ronchetti AM, Sebert M, et al. Combination therapy with ruxolitinib plus intensive treatment strategy is feasible in patients with blast-phase myeloproliferative neoplasms. Br J Haematol. 2016;172:628–30. doi: 10.1111/bjh.13516. [DOI] [PubMed] [Google Scholar]
- 37.Tyner JW, Yang WF, Bankhead A, 3rd, Fan G, Fletcher LB, Bryant J, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–96. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder B, Vladimer GI, Krall N, Miura K, Schmolke AS, Kornauth C, et al. Image-based ex-vivo drug screening for patients with aggressive haematological malignancies: interim results from a single-arm, open-label, pilot study. Lancet Haematol. 2017;4:e595–e606. doi: 10.1016/S2352-3026(17)30208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swords RT, Azzam D, Al-Ali H, Lohse I, Volmar CH, Watts JM, et al. Ex-vivo sensitivity profiling to guide clinical decision making in acute myeloid leukemia: a pilot study. Leuk Res. 2018;64:34–41. doi: 10.1016/j.leukres.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baccelli I, Gareau Y, Lehnertz B, Gingras S, Spinella JF, Corneau S, et al. Mubritinib targets the electron transport chain complex i and reveals the landscape of OXPHOS dependency in acute myeloid leukemia. Cancer Cell. 2019;36:84–99 e8. doi: 10.1016/j.ccell.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Tavor S, Shalit T, Chapal Ilani N, Moskovitz Y, Livnat N, Groner Y, et al. Dasatinib response in acute myeloid leukemia is correlated with FLT3/ITD, PTPN11 mutations and a unique gene expression signature. Haematologica. 2020;105:2795–804.. doi: 10.3324/haematol.2019.240705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kivioja JL, Thanasopoulou A, Kumar A, Kontro M, Yadav B, Majumder MM, et al. Dasatinib and navitoclax act synergistically to target NUP98-NSD1(+)/FLT3-ITD(+) acute myeloid leukemia. Leukemia. 2019;33:1360–72. doi: 10.1038/s41375-018-0327-2. [DOI] [PubMed] [Google Scholar]
- 43.Lavallee VP, Lemieux S, Boucher G, Gendron P, Boivin I, Armstrong RN, et al. RNA-sequencing analysis of core binding factor AML identifies recurrent ZBTB7A mutations and defines RUNX1-CBFA2T3 fusion signature. Blood. 2016;127:2498–501. doi: 10.1182/blood-2016-03-703868. [DOI] [PubMed] [Google Scholar]
- 44.Hashimoto M, Saito Y, Nakagawa R, Ogahara I, Takagi S, Takata S, et al. Combined inhibition of XIAP and BCL2 drives maximal therapeutic efficacy in genetically diverse aggressive acute myeloid leukemia. Nat Cancer. 2021;2:340–56.. doi: 10.1038/s43018-021-00177-w. [DOI] [PubMed] [Google Scholar]
- 45.Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587:477–82.. doi: 10.1038/s41586-020-2864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O’Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014;25:379–92. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269–73. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Cuadron D, Gil C, Serrano J, Rodriguez G, Perez-Oteyza J, Garcia-Boyero R, et al. A precision medicine test predicts clinical response after idarubicin and cytarabine induction therapy in AML patients. Leuk Res. 2019;76:1–10. doi: 10.1016/j.leukres.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Lin L, Tong Y, Straube J, Zhao J, Gao Y, Bai P, et al. Ex-vivo drug testing predicts chemosensitivity in acute myeloid leukemia. J Leukoc Biol. 2020;107:859–70.. doi: 10.1002/JLB.5A0220-676RR. [DOI] [PubMed] [Google Scholar]
- 50.Onecha E, Ruiz-Heredia Y, Martinez-Cuadron D, Barragan E, Martinez-Sanchez P, Linares M, et al. Improving the prediction of acute myeloid leukaemia outcomes by complementing mutational profiling with ex vivo chemosensitivity. Br J Haematol. 2020;189:672–83. doi: 10.1111/bjh.16432. [DOI] [PubMed] [Google Scholar]
- 51.Dhami SPS, Tirincsi A, Baev D, Krawczyk J, Quinn J, Cahill MR, et al. Theranostic drug test incorporating the bone-marrow microenvironment can predict the clinical response of acute myeloid leukaemia to chemotherapy. Br J Haematol. 2020;189:e254–e8. doi: 10.1111/bjh.16684. [DOI] [PubMed] [Google Scholar]
- 52.Quek L, Otto GW, Garnett C, Lhermitte L, Karamitros D, Stoilova B, et al. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213:1513–35. [DOI] [PMC free article] [PubMed]
- 53.Dutta R, Zhang TY, Kohnke T, Thomas D, Linde M, Gars E, et al. Enasidenib drives human erythroid differentiation independently of isocitrate dehydrogenase 2. J Clin Invest. 2020;130:1843–9. doi: 10.1172/JCI133344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burd A, Levine RL, Ruppert AS, Mims AS, Borate U, Stein EM, et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: feasibility and preliminary efficacy of the Beat AML Master Trial. Nat Med. 2020;26:1852–8. doi: 10.1038/s41591-020-1089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karjalainen R, Pemovska T, Popa M, Liu M, Javarappa KK, Majumder MM, et al. JAK1/2 and BCL2 inhibitors synergize to counteract bone marrow stromal cell-induced protection of AML. Blood. 2017;130:789–802. doi: 10.1182/blood-2016-02-699363. [DOI] [PubMed] [Google Scholar]
- 56.Verbeke D, Gielen O, Jacobs K, Boeckx N, De Keersmaecker K, Maertens J, et al. Ruxolitinib synergizes with dexamethasone for the treatment of t-cell acute lymphoblastic leukemia. Hemasphere. 2019;3:e310. doi: 10.1097/HS9.0000000000000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lysenko V, Wildner-Verhey van Wijk N, Zimmermann K, Weller MC, Buhler M, Wildschut MHE, et al. Enhanced engraftment of human myelofibrosis stem and progenitor cells in MISTRG mice. Blood Adv. 2020;4:2477–88. doi: 10.1182/bloodadvances.2019001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Degryse S, de Bock CE, Demeyer S, Govaerts I, Bornschein S, Verbeke D, et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia. 2018;32:788–800. doi: 10.1038/leu.2017.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collignon A, Hospital MA, Montersino C, Courtier F, Charbonnier A, Saillard C, et al. A chemogenomic approach to identify personalized therapy for patients with relapse or refractory acute myeloid leukemia: results of a prospective feasibility study. Blood Cancer J. 2020;10:64. doi: 10.1038/s41408-020-0330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods and Figures (Clean)
Data Availability Statement
Bulk and single-cell RNA-Seq will be available at European Genome-phenome Archive (EGA) under accession code EGAS00001006265. Other data will be available upon reasonable request to the principal investigator.