Abstract
Objectives
To present and validate a deep ensemble algorithm to detect diabetic retinopathy (DR) and diabetic macular oedema (DMO) using retinal fundus images.
Methods
A total of 8739 retinal fundus images were collected from a retrospective cohort of 3285 patients. For detecting DR and DMO, a multiple improved Inception-v4 ensembling approach was developed. We measured the algorithm’s performance and made a comparison with that of human experts on our primary dataset, while its generalization was assessed on the publicly available Messidor-2 dataset. Also, we investigated systematically the impact of the size and number of input images used in training on model’s performance, respectively. Further, the time budget of training/inference versus model performance was analyzed.
Results
On our primary test dataset, the model achieved an 0.992 (95% CI, 0.989–0.995) AUC corresponding to 0.925 (95% CI, 0.916-0.936) sensitivity and 0.961 (95% CI, 0.950–0.972) specificity for referable DR, while the sensitivity and specificity for ophthalmologists ranged from 0.845 to 0.936, and from 0.912 to 0.971, respectively. For referable DMO, our model generated an AUC of 0.994 (95% CI, 0.992–0.996) with a 0.930 (95% CI, 0.919–0.941) sensitivity and 0.971 (95% CI, 0.965–0.978) specificity, whereas ophthalmologists obtained sensitivities ranging between 0.852 and 0.946, and specificities ranging between 0.926 and 0.985.
Conclusion
This study showed that the deep ensemble model exhibited excellent performance in detecting DR and DMO, and had good robustness and generalization, which could potentially help support and expand DR/DMO screening programs.
Subject terms: Retinal diseases, Physical examination
Introduction
Retinal imaging had long been the most widely accepted method for screening of DR and DMO [1–4]. Nevertheless, the evaluation of the severity of retinopathy related to a person with diabetes was currently highly dependent on manual interpretation using retinal fundus images, which faced a great challenge [5–7]. As such, an automatic image grading system played a critical role in early diagnosis and evaluation of these vision-threatening diseases.
Recent studies [8–17] have demonstrated highly accurate deep learning algorithms in various medical image detection tasks, such as DMO [9], possible glaucoma [10–12], and age-related macular degeneration [13, 14]. Especially for DR, multiple works [15–17] have shown that deep learning algorithms could be leveraged to generate expert-level diagnoses for retinal fundus image grading. However, these approaches achieved good performance mostly at the expensive of increasement in time complexity. Due to the same input image size in these independent models, the robustness of their classification was relatively poor. In addition, for an automatic system to be clinically viable, it should be able to flexibly category retinal fundus images in light of clinically adopted severity scales, like international clinical diabetic retinopathy (ICDR) [18] and diabetic macular oedema disease scales [19].
To this end, we explored an ensemble approach of five classification model instances based on the improved Inception-v4 network to boost the performance and robustness in detecting DR and DMO, and evaluated its excellent performance in our primary dataset as well as the secondary dataset (publicly available Messidor-2 dataset). Further, we investigated systematically how the size and number of input images used in training affected model’s performance, respectively. In addition, we also analyzed the issue of the time budget of training/inference versus model performance.
Methods
Datasets
For algorithm development, 8739 retinal fundus images of 3285 patients in all centered on fovea and optic disc were retrospectively obtained from shanghai first people’s hospital. This study was carried out in accordance with the tenets of the Declaration of Helsinki as revised in 2013, and approved by local ethics review and institutional review board. As a result of the retrospective and anonymized nature of this study, written consent was waived by the institutional review board. In addition, another secondary publicly available Messidor-2 dataset was also used for test, which had been adopted by other groups to benchmark performance of automatic detection algorithms for DR or DMO [20, 21].
Retinal image grading and annotation
The six graders for the development dataset were board-certified ophthalmologists who were well educated and specialized in this field with long practicing years. They reviewed and graded each of the collected retinal images (image-level labeling) independently with respect to both DR and DMO. The training and certification process for all graders mainly included image quality examination and DR/DMO severity gradeability. For image quality, all graders were asked to examine each image on the basis of the image quality factors, which contained focus, illumination, image field definition, and artifacts. The specific image quality factors included whether the focus was good enough for grading smaller retinal lesions, whether the image was too dark/light or there existed dark or washed-out regions interfering with detailed grading, whether the primary field contained the entire optic nerve head and macular, and whether the image was sufficiently free of artifacts to ensure adequate grading. Subsequently, they were required to select the correct classification for each image quality, including gradable (like excellent, good, adequate, and insufficient for full interpretation), and ungradable. Excellent represented no problems with any image quality factors and all retinopathy lesions were gradable. Good described problems with 1-2 image quality factors, but all retinopathy lesions were gradable. Adequate denoted problems with 3-4 image quality factors, yet all retinopathy lesions were gradable. Insufficient for full interpretation was defined as the fact that one or more retinopathy lesions could not be graded while section of the image was gradable. For instance, neovascularization noted so likely proliferative diabetic retinopathy (PDR) but obscured view of the macular, thus DMO could not be graded. With respect to gradable images, graders were further asked to grade them according to ICDR [19] and diabetic macular oedema disease severity scales [20, 22], which was denoted as FDR and FDMO, respectively. If any of the image was only partially gradable, a DR/DMO grade was selected according to the gradable section. For grading quality assurance, we calculated inter-grader reliability of the panel for each grader using pairwise comparisons by taking the amount of times a grader was in agreement with another grader over the total number of pairwise comparisons, while about 12% of the development dataset were overread to determine intra-grader reliability. All graders were required to grade a test set consisting of 20 fundus images before starting formal grading so that their proficiency in reading DR/DMO fundus images could be guaranteed, while were monitored for inter-grader and intra-grader consistency. The majority decision of their labeling was served as a consensus reference standard or the ground truth.
In total, 8739 retinal fundus images from a retrospective cohort of 3285 patients were pre-selected in our study. The criterion for exclusion were (1) poor image quality, such as lack of focus, bad illumination, image field without containing the entire optic nerve head and macular, and severe artifacts; (2) existence of abnormalities other than DR and DMO. Eventually, 31 poor quality images (10 images with the lack of focus, 11 images with bad illumination, 2 images without containing the entire optic nerve head and macular, and 8 images with severe artifacts) from 12 participants and 792 abnormality images other than DR and DMO from 307 participants were excluded, while the remaining 7916 retinal images from 2966 participants were included in the current study as our primary dataset, as shown in Supplementary Fig. 1. The image data was divided into three sets at the patient level: a primary training set, a primary validation set, and a primary test set. The patients in the three sets were different and exclusive to each of the other sets. FDR and FDMO were further categorized into non-referable DR vs. Referable DR (NRDR/RDR) [15–17, 23] and non-referable DMO vs. referable DMO (NRDMO/RDMO) [19], respectively.
Image preprocessing
In order to increase heterogeneity of fundus images and prevent over-fitting, we implemented data augmentation strategies [24]. Specifically, we cropped each image to a square shape including the most tightly contained circular region of fundus to remove most of the black borders. Then, we applied the left-right, up-down flipping and rotation operations with angles of 90, 180, and 270 degrees on each image of our primary dataset (including training and validation dataset).
Development of multi-Inception-v4 ensembling approach
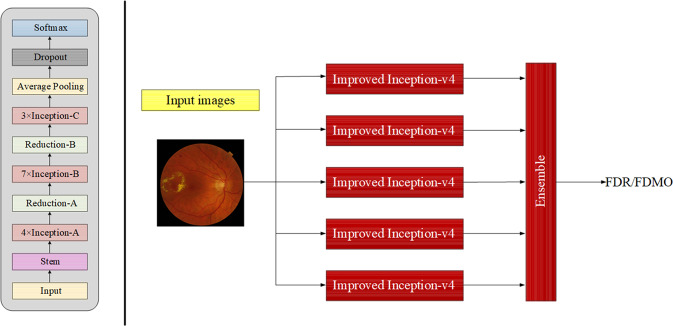
We adopted an ensemble of five classification model instances (as illustrated in the rightmost part of Fig. 1), each of which was on the basis of improved Inception-v4 architecture. The improved Inception-v4 network mainly consisted of stem, inception and reduction modules, as shown in the leftmost part of Fig. 1. In the improved Inception-v4 network, the Inception module stacking a pooling layer and convolution layers together utilized bottleneck layer with a 1 ×1 convolutions to help in reduction of computation requirements. In order to guarantee randomness in the training of each model, we fed retinal fundus images in randomized order and different random augmentations for training each of models in the ensemble model, while employed dropout regularization method. In this way, each model could learn different discriminative features even when trained with the same training data and ImageNet initialization. To accelerate the training of the single improved Inception-v4 in the ensemble model, we further fine-tuned them using pre-trained weights from ImageNet, and replaced the full connected layer of the improved Inception-v4 architecture with two consecutive full connected layers. The former one used a dropout regularization, while the latter one utilized a vanilla full connected layer with softmax nonlinearity to define DR or DMO grading probabilities. During training, we attempted to refine the network via unfreezing frozen layers and updating the corresponding pretrained weights on our primary dataset using a back propagation method. The ensemble model class probabilities were calculated by the arithmetic mean of class probabilities estimated by the constituent networks.
Fig. 1.
The architecture of the improved Inception-v4 (leftmost side) and the ensemble of five classification model (rightmost side).
Statistical analysis
For binary classification tasks, we used the receiver operating characteristic (ROC) curve as well as accuracy, sensitivity, and specificity. Also, the area under the ROC curve (AUC) was calculated. In the case of multi-class classification tasks, we computed the area under macro average of ROC (macro-AUC) for each class in one-vs-all fashion, accuracy, and quadratic-weighted kappa score. With respect to each metric in the binary classification tasks, we used Clopper–Pearson method [25] to calculate the exact 95% confidence interval (CI). All statistical analyses were implemented using Python 3.6.3.
Results
Hyper-parameter selection
The hyper-parameters were standardized on each network of our ensemble network. The weights of the network were initially loaded with pre-trained weights from ImageNet except for the classification layer. We fine-tuned the network parameters using stochastic gradient descent (SGD) algorithm on different input image sizes, and employed the categorical cross entropy loss function, an initial learning rate of 0.001, a dropout rate of 0.5, a momentum of 0.9, a batch size of 64, and weight decay factor of 0.0005. The network was then trained for total of 30 epochs, updating all weights. Our framework was implemented in Python based on Keras with Tensorflow backend, using NVIDIA GTX1080 GPU.
The performance of our model on the primary test dataset
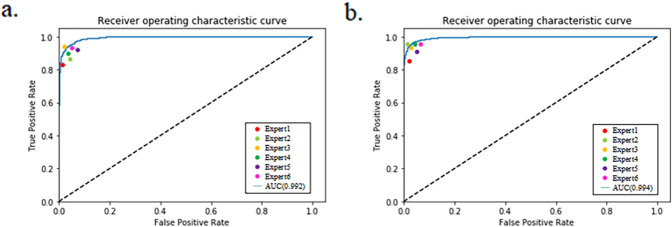
Our algorithm was optimized for the 5-point ICDR grading and the 4-stage diabetic macular oedema grading using the largest 1488 × 1488 pixels input image size, respectively. On the primary test dataset, our model achieved an 0.972 (95% CI, 0.966–0.978) accuracy and 0.992 (95% CI, 0.989–0.995) AUC for the NRDR/RDR binary classification task. This corresponded to a sensitivity of 0.925 (95% CI, 0.916–0.936) and specificity of 0.961 (95% CI, 0.950–0.972). In the NRDMO/RDMO classification, our model yielded a high accuracy of 0.974 (95% CI, 0.967–0.981) with a 0.930 (95% CI, 0.919–0.941) sensitivity and 0.971 (95% CI, 0.965–0.978) specificity, while the AUC value reached up to 0.994 (95% CI, 0.992–0.996). Further, we compared the performance in the NRDR/RDR and NRDMO/RDMO classification between our model and ophthalmologists, as shown in Fig. 2 and Supplementary Table 1. It could be observed that our model could make automated grading for DR and DMO from retinal fundus images with performance equal to or exceeding ophthalmologists.
Fig. 2. Performance of the model and ophthalmologists for classifying NRDR/RDR and NRDMO/RDMO on our primary test dataset.
a NRDR/RDR (b) NRDMO/RDMO.
Model generalizes to a secondary test dataset
We also applied the model to a secondary test dataset (publicly available Messidor-2 dataset), and performed comparison of classification results for detecting DR and DMO with reported state-of-the-art methods [26–28] to assess its generalizability, as summarized in Table 1. As we can see, the highest AUC values (0.977 (95% CI, 0.974–0.981) for DR, and 0.948 (95% CI, 0.943–0.951) for DMO) for detecting DR and DMO were achieved by our model. In the NRDR/RDR classification, the sensitivity of our model was on par with 0.922 sensitivity reported by Abbas et al. [27], and outperformed the results presented in Sahlsten et al. [26] and Gonzalez-Gonzalo et al. [28]. For the identification of DMO task, our model achieved the best results in sensitivity, and its specificity was better than the result (0.860) proposed by Gonzalez-Gonzalo et al. [28], whereas underperforming in comparison to 0.992 specificity reported by sahlsten et al. [26].
Table 1.
Comparison of classification results for detecting DR and DMO with reported state-of-the-art methods.
| Author | Retinopathy | AUC (95% CI) | Sen (95% CI) | Spe (95% CI) |
|---|---|---|---|---|
| Sahlsten et al. [26] | DR | 0.967 (0.955–0.976) | 0.859 (0.826–0.888) | 0.971 (0.956–0.982) |
| DMO | 0.946 (0.932–0.958) | 0.597 (0.530–0.662) | 0.992 (0.984–0.996) | |
| Abbas et al. [27] | DR | 0.924 | 0.922 | 0.945 |
| DMO | N/A | N/A | N/A | |
| Gonza̒lez-Gonzalo et al. [28] | DR | 0.975 | 0.920 | 0.921 |
| DMO | 0.927 | 0.858 | 0.860 | |
| Ours | DR | 0.977 (0.974–0.981) | 0.923 (0.917–0.925) | 0.947 (0.937–0.954) |
| DMO | 0.948 (0.943–0.951) | 0.886 (0.881–0.892) | 0.908 (0.898–0.912) |
DR diabetic retinopathy, DMO diabetic macular oedema, AUC Area Under Curve, CI confidence interval, Sen Sensitivity, Spe Specificity.
The effect of the input image size on model performance
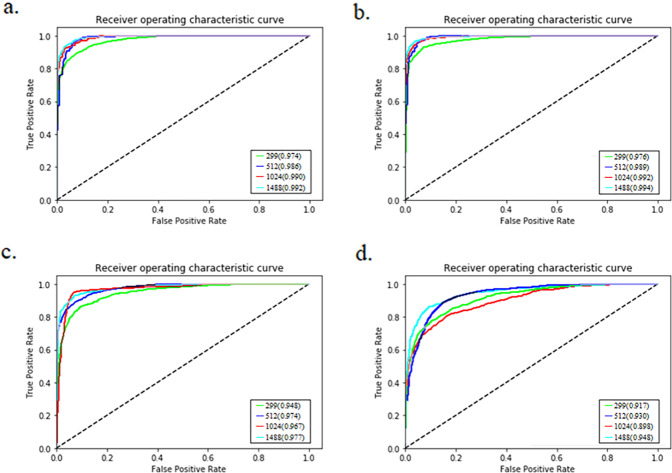
Fig. 3 displayed ROC curves for NRDR versus RDR, and NRDMO versus RDMO on our primary test dataset and the secondary publicly available Messidor-2 dataset, respectively. From these curves we could see that the dependence of the model performance on the input image size for our primary test dataset was obvious, while not clear for Messidor-2 dataset.
Fig. 3. The impact of the input image size on model performance.
a NRDR/RDR classification on our primary test set. b NRDMO/RDMO classification on our primary test set. c NRDR/RDR classification on the second publicly available Messidor-2 set. d NRDMO/RDMO classification on the second publicly available Messidor-2 set.
In the multiclass classification tasks on our primary test dataset, our model obtained the best results when the performance was measured on the basis of macro-AUC, accuracy and quadratic-weighted kappa for FDR and FDMO, using 1488 × 1488 resolution input retinal fundus images, as described in Supplementary Table 2.
More data leads to better model performance
We conducted sub-sampling experiments to measure the effect of data size on model performance on our primary dataset, in which new models were trained by means of titrated fractions of the dataset [29]. The results showed that as training dataset increased, the performance of model kept on increasing (view Supplementary Fig. 2), which meant that the accuracy of model prediction will likely continue to improve if the model was trained with more data than that in this study.
The time budget of training/inference vs. model performance
To investigate the time budget of training/inference versus model performance, we did additional experiments using the ensemble of five deep learning models with input 299 × 299 sized retinal fundus images against the single image sized model, as listed in Table 2. These experiment results demonstrated improved performance compared to a single model with the same input image size or using larger input image size under certain circumstances. Additionally, it took approximate 79 min to train the ensemble of five deep learning models with 299 × 299 sized retinal fundus images, while about 307 min for training a single model with 1488 × 1488 sized retinal fundus images.
Table 2.
Performance comparison in AUC between the ensemble model and the single image sized model.
| Dataset | Classification | The ensemble model with 299 × 299 sized image | The single model with 1488 × 1488 sized image |
|---|---|---|---|
| Primary test dataset | NRDR/RDR | 0.974 (95% CI, 0.971–0.977) | 0.971 (95% CI, 0.968–0.974) |
| NRDMO/RDMO | 0.976 (95% CI, 0.973–0.979) | 0.972 (95% CI, 0.969–0.975) | |
| Messidor-2 dataset | NRDR/RDR | 0.948 (95% CI, 0.945–0.952) | 0.951 (95% CI, 0.947–0.954) |
| NRDMO/RDMO | 0.917 (95% CI, 0.914–0.921) | 0.915 (95% CI, 0.912–0.918) |
CI confidence interval, AUC Area Under Curve.
Discussion
In this study, we explored the use of an ensemble of five classification model instances based on the improved Inception-v4 architecture to detect DR and DMO, and evaluated its performance on our primary dataset and secondary dataset (publicly available Messidor-2 dataset). The results demonstrated that our deep learning model could identify DR and DMO with performance equal to or exceeding ophthalmologists, obtained the state-of-the-art results in the NRDR/RDR and NRDMO/RDMO classification, and had good generalization. In addition, a likely increase in performance when training contained additional data or used high resolution and quality images was also validated, respectively. In contrast to a single model with the same input image size or using larger input image size in some cases, our ensemble model had overall improved performance, with the huge savings in the time budget of training/inference. It could potentially enable automated DR or DMO screening programs using fundus images worldwide, and was both scientifically interesting and clinically impactful.
In the course of image grading, we took the majority decision from a group of six board-certified ophthalmologists who were well educated and specialized in this field with long practicing years to serve as a consensus reference standard or the ground truth. We used a small number of images (including different severity level of DR and DMO) with the consensus reference grades as a test set. Considering that many ophthalmologists had never been trained in grading images in clinical practice, over and beyond just their long work history, we also calculated the quadratic-weighted kappa score to examine the agreement between ophthalmologists without additional specific training and the reference standard, so that their ability to grade in the test set could be evaluated. This metric typically varies from 0 (random agreement between raters) to 1 (complete agreement between raters). According to the guidelines proposed by Landis and Koch [30], the kappa values ranging from 0 to 0.2, 0.21 to 0.40, 0.41 to 0.60, and 0.61 to 0.80 represented slight agreement, fair agreement, moderate agreement and substantial agreement, respectively. A value above 0.80 was considered to be almost perfect agreement. Only ophthalmologists with quadratic-weighted kappa scores over 0.80 could act as graders in our study.
Automatic DR evaluation had been previously made by other recent studies [31–34]. Despite differences in the dataset compared to these studies, our study extended this body of work by the use of multiple improved Inception-v4 ensembling to generate an algorithm with high sensitivity and specificity. Although our model exhibited slightly lower performance on the secondary dataset than that on the primary dataset, the better performance of our model was still supported in comparison with the present reported advanced methods [26–28, 34]. The best results (0.977 (95% CI, 0.974–0.981), 0.923 (95% CI, 0.917–0.925) for NRDR/RDR, and 0.948 (95% CI, 0.943–0.951), 0.886 (95% CI, 0.881–0.892) for NRDMO/RDMO) in AUC and sensitivity were still achieved when compared to the recent studies [26–28]. These results reflected that there existed inherent differences between the two datasets, but excellent performance of our model was still demonstrated on both datasets. Considering the results of the sub-sampling experiments, it was possible that the prediction accuracy of our model could continue to improve with larger dataset sizes.
As illustrated in Fig. 3, the performance of the model in AUC generally increased with the input image size on our primary dataset and the secondary dataset. This could be attributed to the fact that the number of information and features in the images increased with the image size. For our primary dataset, the performance of the model could continue to improve with the input image resolution increasing. Yet, the reliance of the model performance on the input image size was not obvious for the Messidor-2 dataset. This occurred mainly due to the fact that images on the Messidor-2 dataset had various resolutions, which were resized into the sizes depicted in Fig. 3c, d. Other attributing aspects possibly included the fact that the imaging equipment used on the Messidor-2 dataset [35] were different from those on our primary dataset.
The results on our primary dataset using an ensemble of five classification model instances suggested improved performance, while the time budget of training/inference was dramatically saved. It was noticeable that identical features were not learned by deep learning models trained on a small set of retinal images. Even though we identically initialized deep neural networks to weights pretrained on the ImageNet, there still existed variation in the predictive results between them. Moreover, we could conclude from our ensemble model experiment that the robustness of the classification was improved through the ensemble approach. Apart from the NRDR/RDR classification task, we also verified multi-class classification performance of the model. The more granular 5-point grading output would be helpful in particular for DR screening programs where patient treatment varied at each level of severity, and would also be more robustness to guideline changes.
Our algorithm is designed to detect DR and DMO and have not been trained to identify other diseases that might be present. Prior to training our model, we performed image grading and excluded ungradable images, and abnormality images apart from DR and DMO so that high quality retinal images could be obtained. In the real clinical settings, the ability to automatically detect an ungradable image is also an important component using the algorithm. In the future work, we will incorporate image quality algorithm into our model to determine whether the exam quality was sufficient for our model to make a diagnostic decision. In the future clinical practice, we will try to deploy the well trained and optimized model on a server, physicians capture several fundus images including optic disc and macula centered for each eye using the camera, and feed them into our model over the network. After analyzing lesions characteristic for DR and DMO by our model, such as hemorrhages, exudates and neovascularization, several types of outputs (including DR/DMO severity, referable DR, and referral DMO) were provided. These results will be required to make a second confirmation by the attending physicians. As such, our algorithm could be regarded as a two-way confirmation system. In this case, the algorithm could provide an initial diagnosis based on retinal fundus images, which could then be confirmed by the attending physicians, drastically minimizing both human and machine error.
This study has several limitations. First, we used the majority decision of six experienced and qualified ophthalmologists as a consensus reference standard or the ground-truth during retinal image grading, which could unavoidably introduce grader biases. Future studies should validate the effect of different image grading references (e.g., a tiered grading) on the model performance. Second, human graders were required to pre-screen the collected retinal images for quality and other diseases. The resulting high quality retinal images constituted the development dataset, which was used for our model training and intra-dataset validation. The established dataset was not necessarily a good representation of data from screening programs in clinical practice. Thereby, the built dataset was not sufficient to reflect the algorithm’s performance in broader clinical use. In the future, we will use more and larger datasets from real screening programs to validate our algorithm’s capability to automatically detect DR and DMO without the need for human grader’s pre-screening for quality and other diseases.
Conclusions
This study demonstrated that multiple improved Inception-v4 ensembling could be leveraged to detect DR and DMO using retinal fundus images, at a performance level matching or exceeding those of ophthalmologists on our primary dataset. Meanwhile, we validated its excellent generalization performance on the publicly available Messidor-2 dataset. In addition, the results also indicated a likely improvement in performance when training involved additional data or used high resolution and quality images. Further, overall improved performance of our ensemble algorithm was demonstrated by comparing with a single model with the same input image size or using larger input image size under certain circumstances, yet with the huge savings in the time budget of training/inference. As such, it raised the possibility of clinical use as an automated screening tool in primary screening situations with low accessibility to specialists.
Summary
What was known before
For an automatic system to be clinically viable, it should be able to flexibly category retinal fundus images in light of clinically adopted severity scales, like international clinical diabetic retinopathy (ICDR) and diabetic macular oedema disease scales.
Although deep learning algorithms could be leveraged to generate expert-level diagnoses for retinal fundus image grading, these approaches achieved good performance mostly at the expensive of increasement in time complexity, which was more obvious specifically for larger input image size.
Due to the same input image size in these independent models, the robustness of their classification was relatively poor.
What this study adds
This research developed an ensemble approach of five classification model instances based on the improved Inception-v4 network to boost the performance and robustness in detecting DR and DME according to the 5-point ICDR grading and the 4-grade diabetic macular oedema scales using retinal fundus images.
This study analyzed the issue of the time budget of training/inference versus algorithm’s performance This research evaluated algorithm’s performance in our primary dataset as well as the secondary dataset (publicly Messidor-2 dataset), while investigated systematically how the size and number of input images used in training affected model’s performance, respectively.
Supplementary information
The whole experimental design. DR: diabetic retinopathy; DMO: diabetic macular oedema.
Effect of data size on model performance in our primary dataset.
Performance comparison in detecting DR and DMO between our model and ophthalmologists.
The results for FDR and FDMO classification.
Acknowledgements
The authors acknowledge Shanghai First People’s Hospital and Shanghai Ninth People’s Hospital for our help and support.
Author contributions
FL, LD and HJ have made contributions to the writing. YGW and LY implemented the deep learning algorithm. HDZ, TYX and MSJ acquired the data. XDZ and ZZW analyzed and interpreted the data. All authors were involved in the study design, and approval of the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (61905144), the National Key Research and Development Program of China (2016YFF0101400) and the National Natural Science Foundation of China (51675321).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Feng Li, Yuguang Wang, Tianyi Xu
Contributor Information
Minshan Jiang, Email: jiangmsc@gmail.com.
Xuedian Zhang, Email: xdzhang@usst.edu.cn.
Hong Jiang, Email: jianghongjiuyuan@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41433-021-01552-8.
References
- 1.Li X, Hu X, Yu L, Zhu L, Fu CW, Heng PA. CANet: cross-disease attention network for joint diabetic retinopathy and diabetic macular edema grading. IEEE Trans Med Imaging. 2020;39:1483–93. doi: 10.1109/TMI.2019.2951844. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Liu Z, Chen H, Jiang M, Zhang X, Wu Z. Automatic detection of diabetic retinopathy in retinal fundus photographs based on deep learning algorithm. Transl Vis Sci Technol. 2019;8:1–13.. doi: 10.1167/tvst.8.6.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gadekallu TR, Khare N, Bhattacharya S, Singh S, Maddikunta PKR, Srivastava G. Deep neural networks to predict diabetic retinopathy. J Amb Intel Hum Comp. 2020;13:1–14.
- 4.Rajalakshmi R, Subashini R, Anjana RM, Mohan V. Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence. Eye. 2018;32:1138–44. doi: 10.1038/s41433-018-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulshan V, Rajan RP, Widner K, Wu D, Wubbels P, Rhodes T, et al. Performance of a deep-learning algorithm vs manual grading for detecting diabetic retinopathy in India. JAMA Ophthalmol. 2019;137:987–93. doi: 10.1001/jamaophthalmol.2019.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Torre J, Valls A, Puig D. A deep learning interpretable classifier for diabetic retinopathy disease grading. Neurocomputing. 2019;396:465–76. doi: 10.1016/j.neucom.2018.07.102. [DOI] [Google Scholar]
- 7.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–31. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Keenan TD, Dharssi S, Peng Y, Chen Q, Agron E, Wong WT, et al. A deep learning approach for automated detection of geographic atrophy from color fundus photographs. Ophthalmology. 2019;126:1533–40. doi: 10.1016/j.ophtha.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keel S, Lee PY, Scheetz J, Li Z, Kotowicz MA, MacIsaac RJ, et al. Feasibility and patient acceptability of a novel artificial intelligence-based screening model for diabetic retinopathy at endocrinology outpatient services: a pilot study. Sci Rep. 2018;8:1–6. doi: 10.1038/s41598-018-22612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Chen H, Liu Z, Zhang X, Wu Z. Fully automated detection of retinal disorders by image-based deep learning. Graef Arch Clin Exp. 2019;257:495–505. doi: 10.1007/s00417-018-04224-8. [DOI] [PubMed] [Google Scholar]
- 11.Li F, Yan L, Wang Y, Shi J, Chen H, Zhang X, et al. Deep learning-based automated detection of glaucomatous optic neuropathy on color fundus photographs. Graef Arch Clin Exp. 2020;258:851–67. doi: 10.1007/s00417-020-04609-8. [DOI] [PubMed] [Google Scholar]
- 12.Son J, Shin JY, Kim HD, Jung KH, Park KH, Park SJ. Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images. Ophthalmology. 2020;127:85–94. doi: 10.1016/j.ophtha.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Burlina PM, Joshi N, Pacheco KD, Liu TYA, Bressler NM. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. JAMA Ophthalmol. 2019;137:258–64. doi: 10.1001/jamaophthalmol.2018.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Y, Dharssi S, Chen Q, Keenan TD, Agron E, Wong WT, et al. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126:565–75. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayres R, Taly A, Rahimy E, Blumer K, Coz D, Hammel N, et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology. 2019;126:552–64. doi: 10.1016/j.ophtha.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Araujo T, Aresta G, Mendonca L, Penas S, Maia C, Carneiro A, et al. DR|GRADUATE: uncertainty-aware deep learning-based diabetic retinopathy grading in eye fundus images. Med Image Anal. 2020;63:1–17. doi: 10.1016/j.media.2020.101715. [DOI] [PubMed] [Google Scholar]
- 17.He J, Cao T, Xu F, Wang S, Tao H, Wu T, et al. Artificial intelligence-based screening for diabetic retinopathy at community hospital. Eye. 2020;34:572–6. doi: 10.1038/s41433-019-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellemo V, Lim ZW, Lim G, Nguyen QD, Xie Y, Yip MYT, et al. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa. Lancet. 2019;1:E35–E44. doi: 10.1016/S2589-7500(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson CP, Ferris FL, Klein RE, Lee PP, Agardh CD, Davis M, et al. Proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology. 2003;110:1677–82. doi: 10.1016/S0161-6420(03)00475-5. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Gao Y, Wang K, Guo S, Liu H, Kang H. Diagnostic assessment of deep learning algorithms for diabetic retinopathy screening. Inf Sci. 2019;501:511–22. doi: 10.1016/j.ins.2019.06.011. [DOI] [Google Scholar]
- 21.Raman R, Srinivasan S, Virmani S, Sivaprasad S, Rao C, Rajalakshmi R. Fundus photograph-based deep learning algorithms in detecting diabetic retinopathy. Eye. 2018;33:97–109. doi: 10.1038/s41433-018-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bria A, Marrocco C, Tortorella F. Addressing class imbalance in deep learning for small lesion detection on medical images. Comput Biol Med. 2020;120:1–14. doi: 10.1016/j.compbiomed.2020.103735. [DOI] [PubMed] [Google Scholar]
- 23.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124:962–9. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Kim TN, Aaberg MT, Li P, Davila JR, Bhaskaranand M, Bhat S, et al. Comparison of automated and expert human grading of diabetic retinopathy using smartphone-based retinal photography. Eye. 2021;35:334–42. [DOI] [PMC free article] [PubMed]
- 25.Zago GT, Andreao RV, Dorizzi B, Teatini, Salles EO. Diabetic retinopathy detection using red lesion localization and convolutional neural networks. Comput Biol Med. 2020;116:1–12. doi: 10.1016/j.compbiomed.2019.103537. [DOI] [PubMed] [Google Scholar]
- 26.Sahlsten J, Jaskari J, Kivinen J, Turunen L, Jaanio E, Hietala K, et al. Deep learning fundus image analysis for diabetic retinopathy and macular edema grading. Sci Rep. 2019;9:1–11. doi: 10.1038/s41598-019-47181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbas Q, Fondon I, Sarmiento A, Jimenez S, Alemany P. Automatic recognition of severity level for diagnosis of diabetic retinopathy using deep visual features. Med Biol Eng Comput. 2017;55:1959–74. doi: 10.1007/s11517-017-1638-6. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Gonzalo C, Sanchez-Gutierrez V, Hernandez-Martinez P, Contreras I, Lechanteur YT, Domanian A, et al. Evaluation of a deep learning system for the joint automated detection of diabetic retinopathy and age-related macular degeneration. Acta Ophthalmol. 2019;98:368–77. doi: 10.1111/aos.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avinash VV, Pinal B, Paisan R, Peranut C, Subhashini V, Arunachalam N, et al. Predicting optical coherence tomography-derived diabetic macular edema grades from fundus photographs using deep learning. Nat Commun. 2020;11:1–8. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 31.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 32.Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–23. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan MY, Gulshan V, Dai AM, Hinton GE. Who said what: modeling individual labelers improves classification. In: 32nd AAAI Conference on Artificial Intelligence. New Orleans, LA, USA: AAAI; 2018; 3109-18.
- 34.Krause J, Gulshan V, Rahimy E, Karth P, Widner K, Corrado GS, et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125:1264–72. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 35.Decenciere E, Zhang XW, Cazuguel G, Lay B, Cochener B, Trone C, et al. Feedback on a publicly distributed image database: the Messidor database. Image Anal Stereol. 2014;33:231–4. doi: 10.5566/ias.1155. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The whole experimental design. DR: diabetic retinopathy; DMO: diabetic macular oedema.
Effect of data size on model performance in our primary dataset.
Performance comparison in detecting DR and DMO between our model and ophthalmologists.
The results for FDR and FDMO classification.