Abstract
Precursor messenger RNA (pre-mRNA) splicing is catalyzed by an intricate ribonucleoprotein complex called the spliceosome. Although the spliceosome is considered to be general cell “housekeeping” machinery, mutations in core components of the spliceosome frequently correlate with cell- or tissue-specific phenotypes and diseases. In this review, we expound the links between spliceosome mutations, aberrant splicing, and human cancers. Remarkably, spliceosome-targeted therapies (STTs) have become efficient anti-cancer strategies for cancer patients with splicing defects. We also highlight the links between spliceosome and immune signaling. Recent studies have shown that some spliceosome gene mutations can result in immune dysregulation and notable phenotypes due to mis-splicing of immune-related genes. Furthermore, several core spliceosome components harbor splicing-independent immune functions within the cell, expanding the functional repertoire of these diverse proteins.
Keywords: spliceosome, splicing, cancer, innate immunity, immune dysregulation
Introduction
Pre-mRNA is transcribed from a gene’s DNA template. The pre-mRNA undergoes splicing to remove introns, forming mature messenger RNA (mRNA) that directs the synthesis of the protein during translation. Intron sequences in the pre-mRNAs contain several conserved sequences that facilitate proper splicing, including a 5′ splice site, a 3′ splice site, and the branch point typically 18 to 40 base pairs upstream of the 3′ splice site. Higher eukaryotes also have a polypyrimidine tract (PPT) following the branch point that is essential for recruiting splicing factors to the 3′ splice site (Taylor and Lee 2019). During splicing, two transesterification reactions remove introns from pre-mRNA (Moore 1993). In the first reaction, the 5′ splice site is cleaved and the 5′ end of the intron ligates to the branch adenosine of the intron. In the second reaction, the 5′ and the 3′ exons are ligated after the 3′ splice site is cleaved by the 3′ OH group of the 5′ exon.
In addition to constitutive splicing, a single pre-mRNA can be alternatively spliced, often in a tissue- or cellular condition-specific manner. In alternative splicing, exons can be skipped or extended as well as introns retained to produce different forms of the mRNA. Except for isoforms that undergo nonsense-mediated mRNA decay or are retained in the nucleus, alternatively spliced mRNAs can be translated into multiple protein products that can have unique functions (Nilsen and Graveley 2010).
The splicing of pre-mRNA is executed by the spliceosome, a multi-megadalton ribonucleoprotein complex. Although all eukaryotic cells have spliceosomes, mutations in core components of the spliceosome frequently correlate with specific phenotypes and diseases. In this review, we discuss the important roles of spliceosome components and splicing factors in cancer and immunity.
The eukaryote spliceosome
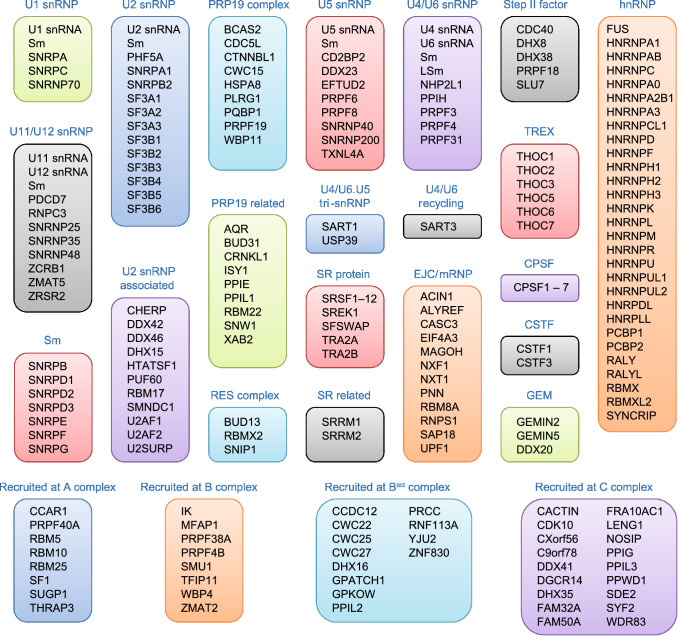
The eukaryote spliceosome consists of several small nuclear ribonucleoproteins (snRNPs). Each snRNP is composed of a uridine-rich small nuclear RNA (U snRNA), Sm proteins (i.e., SNRPB/B′, D1, D2, D3, E, F, and G), and a variable number of associated proteins (Will 2006; Fabrizio et al. 2009; Cvitkovic and Jurica 2013) (Fig. 1). At any point during splicing, over 170 proteins associate with the spliceosome (Jurica and Moore 2003; Wahl et al. 2009; Cvitkovic and Jurica 2013). Approximately 45 of the proteins are components of the snRNPs, while the other proteins are non-snRNP proteins that mediate spliceosome assembly, pre-mRNA splice site recognition, and pre-mRNA binding (Matlin and Moore 2007; Staley and Woolford 2009). Several of the spliceosome-associated proteins have redundant functions and/or are loosely associated, indicating that each spliceosome-associated protein is not required to splice every pre-mRNA substrate (Wahl et al. 2009).
Figure 1.
The human spliceosome machinery. Human spliceosome consists of several snRNPs. Each snRNP is composed of a uridine-rich small nuclear RNA (U snRNA), Sm proteins (i.e., SNRPB/B′, D1, D2, D3, E, F, and G) or LSm proteins (i.e., LSm2–8), and a variable number of particle-specific proteins. The U4/U6.U5 tri-snRNP contains two sets of Sm proteins and one set of LSm proteins. Classification is based on molecular features (e.g., SR proteins), association with stable spliceosome sub-complexes like the snRNPs or PRP19 complex, and other common designations (e.g., hnRNP, step II factors). Proteins that do not associate specifically with a given complex, such as general RNA binding factors, are not included in this figure. All protein names listed are official symbol from the National Center for Biotechnology Information (NCBI) database. The figure was illustrated based on the complex analysis results that were collected in the Spliceosome Database (Cvitkovic and Jurica 2013). Abbreviations: RES, retention and splicing; SR, serine and arginine-rich; EJC, exon junction complex; mRNP, messenger ribonucleoprotein; TREX, transcription-export; CPSF, cleavage and polyadenylation specificity factor; CSTF, cleavage stimulation factor; GEM, gemini of coiled bodies; hnRNP, heterogeneous nuclear ribonucleoprotein
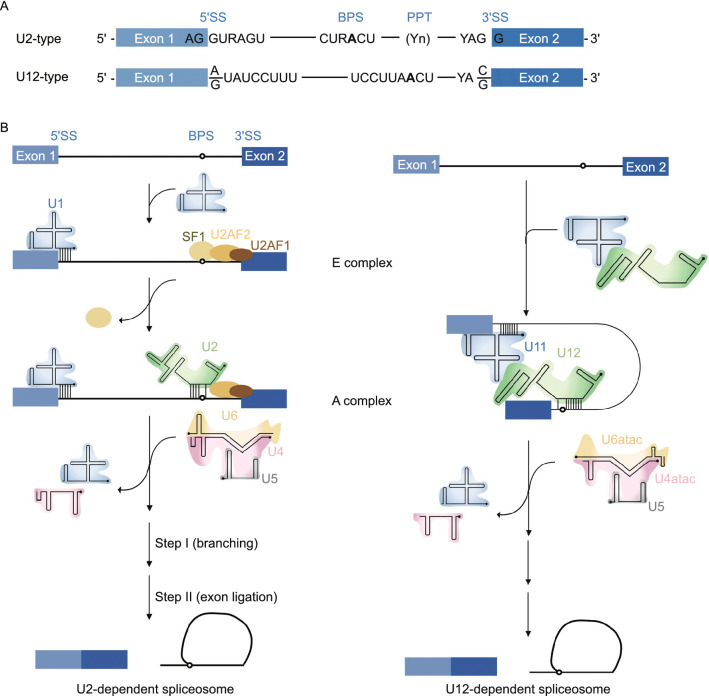
Eukaryotic cells have two types of spliceosomes: the U2-dependent (major) spliceosome and the U12-dependent (minor) spliceosome (Patel and Steitz 2003) (Fig. 2). Both spliceosomes are structurally and functionally similar in that each has five U snRNAs, but the types of snRNAs in each of the spliceosomes are unique. The U2 spliceosome contains the U1, U2, U4, U5, and U6 snRNAs, while the U12 spliceosome contains the unique U11, U12, U4atac, and U6atac snRNAs; both the U2 and U12 spliceosomes contain U5 snRNA (Patel and Steitz 2003; Turunen et al. 2013; Scotti and Swanson 2016; Shi 2017a, b). Most of the spliceosome-associated proteins are shared between the U2 and U12 spliceosomes, except those associated with the unique snRNPs (i.e., U1 and U2 in the U2 spliceosome and U11 and U12 in the U12 spliceosome) (Verma et al. 2018). The U2 spliceosome processes approximately 95.5% of all U2-type introns (Turunen et al. 2013), while the U12 spliceosome functions in the splicing of rare U12-type introns. U12-type introns occur in approximately 0.35% of all human introns in 700 to 800 genes. Most of these genes encode proteins that function in DNA replication and repair, translation, RNA processing, transcription, cytoskeletal organization, voltage-gated ion channel activity, and vesicular transport (Burge et al. 1998).
Figure 2.
U2-dependent (major) spliceosome versus U12-dependent (minor) spliceosome. (A) The main determinants for distinguishing U2- and U12-type introns are the 5′ splice site (5′SS) and branch point sequence (BPS). U12-type introns are subdivided into AT-AC or GT-AG minor introns based on their terminal dinucleotides. (B) U2- and U12-type introns are recognized differently by their respective spliceosomes. U2-type introns are initially recognized by the U1 snRNP binding to the 5′SS, splicing factor 1 (SF1) binding to the BPS and U2 auxiliary factor (U2AF2/1) heterodimer binding to the polypyrimidine tract (PPT) and 3′SS, respectively. Subsequently, the U2 snRNA associates with the BPS and displaces SF1, converting the E complex to A complex. In contrast to the U2-type introns, the 5′SS and BPS of U12-type introns are recognized cooperatively by the U11 and U12 snRNAs of the di-snRNP, respectively, thereby forming the U12-type A complex. The following steps in the splicing process are similar between the U2- and U12-dependent pathway, and lead to similar catalytic structures and catalytic reactions of splicing
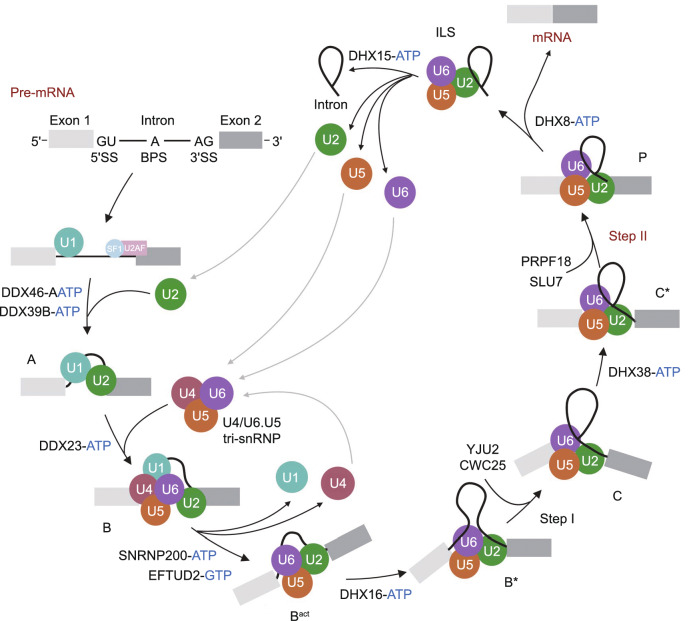
There are at least seven configurations of the spliceosome complex during splicing: the pre-catalytic complex (B), the activated complex (Bact), the catalytically activated complex (B*), the catalytic step I spliceosome (C), the step II catalytically activated complex (C*), the post-catalytic complex (P), and the intron lariat spliceosome (ILS) (Fabrizio et al. 2009; Wahl et al. 2009) (Fig. 3). In the initial spliceosome complex (designated the E complex), the U1 snRNP (U11 snRNP in the U12 spliceosome) is recruited to the 5′ splice site allowing binding of the U1 (U11) snRNA (Taylor and Lee 2019). Non-snRNP factors such as splicing factor 1 (SF1) and U2 auxiliary factor (U2AF) interact with the branch point and the 3′ splice site, respectively. Subsequently, the U2 snRNP (U12 snRNP in the U12 spliceosome) associates with the branch point and displaces SF1 via binding of the U2 (U12) snRNA to the branch point, forming the prespliceosome (alternatively, the A complex) (Wassarman and Steitz 1992). A U4/U6.U5 tri-snRNP complex (the U4atac/U6atac.U5 tri-snRNP complex in the U12 spliceosome) is recruited to the prespliceosome to subsequently form the B complex. Rearrangements in RNA-RNA and RNA-protein interactions result in destabilization of the U1 (U11) and U4 (U4atac) snRNPs. The 5′ end of the U6 (U6atac) snRNA base pairs with the 5′ splice site, the U2 (U12) snRNA forms a duplex with the branch point, and the U1 (U11) and U4 (U4atac) snRNAs are displaced, resulting in formation of the Bact complex. The Bact complex is catalytically activated by the DEAH-box ATP-dependent RNA helicase DHX16 to form the B* complex, which catalyzes the first transesterification reaction during splicing. The first splicing reaction results in spliceosome rearrangement to form the C complex. Subsequently, the DEAH-box ATPase DHX38 catalytically activates the C complex to form the C* complex, which catalyzes the second transesterification reaction. After the reaction, the P complex contains ligated exons (mRNA) and the excised lariat-intron. The DEAH-box ATPase DHX8 releases the spliced mRNA from the P complex, which forms the ILS. The ILS dissociates with assistance from the DEAH-box ATPase DHX15, and the snRNPs are able to be reused in additional splicing (Will and Luhrmann, 2011) (Fig. 3).
Figure 3.
Eukaryotic U2-dependent splicing cycle. The 5′SS, BPS and 3′SS are first recognized by the U1 snRNP, SF1 and U2AF, respectively, forming an early spliceosome (E complex). SF1 is displaced by the U2 snRNP to form the pre-spliceosome (A complex), which associates with the U4/U6.U5 tri-snRNP to assemble into the pre-catalytic spliceosome (B complex). B complex undergoes a series of rearrangements to form a catalytically active Bact complex and then B* complex, which carries out the first catalytic step of splicing, generating C complex. C complex undergoes additional rearrangements and then carries out the second catalytic step, resulting in a post-catalytic spliceosome (P complex) that contains the lariat intron and spliced exons. Release of the spliced exons from P complex generates the intron lariat spliceosome (ILS complex). Finally, the U2, U5 and U6 snRNPs are released from the mRNP particle and recycled for additional rounds of splicing. Each complex has a unique composition, and conversions between complexes are driven by highly conserved RNA-dependent ATPase/helicases (including DDX46, DDX39B, DDX23, SNRNP200, DHX16, DHX38, DHX8 and DHX15, and the GTPase EFTUD2)
Links between spliceosome mutations and diseases
Proper recognition of intron sequences by the spliceosome is essential for intron removal. Mutations in core consensus sequences (i.e., 5′ or 3′ splice sites or the branch point sequence), exon and intron splicing enhancer or silencer elements, splicing factors, spliceosome assembly factors, and spliceosome components can result in pre-mRNA processing defects, truncated or aberrant protein products, and/or increased nonsense-mediated decay of the affected mRNAs (Wang and Cooper 2007; Turunen et al. 2008; Singh and Cooper 2012; Inoue et al. 2016; Ruzickova and Stanek 2017; Verma et al. 2018).
Aberrant pre-mRNA splicing due to mutations in snRNP components and snRNP-associated proteins are linked to several human diseases (Table 1) (Novoyatleva et al. 2006; Wang and Cooper 2007; Li et al. 2016; Scotti and Swanson 2016; Verma et al. 2018; Taylor and Lee 2019). For example, mutations affecting snRNP-associated proteins PRPF3, PRPF4, PRPF6, PRPF8, PRPF31, and SNRNP200 have been identified in patients with autosomal dominant retinitis pigmentosa, a condition that results in retinal degeneration and eventual blindness (Scotti and Swanson 2016; Ruzickova and Stanek 2017). Mutations in RNU12 (encoding the U12 snRNA) that lead to reduced expression of U12 snRNA are linked to early-onset cerebellar ataxia (Elsaid et al. 2017) and amyotrophic lateral sclerosis (Ishihara et al. 2013). Mutations in RNU4ATAC (encoding the U4atac snRNA) are linked to cases of Roifman syndrome (Merico et al., 2015) and microcephalic osteodysplastic primordial dwarfism type I (Edery et al. 2011; He et al. 2011; Scotti and Swanson 2016; Verma et al. 2018). These mutations affect the formation of the U4atac/U6atac.U5 tri-snRNP (Verma et al. 2018). However, patient cells exhibit some correctly spliced mRNAs indicating that these mutations cause only a partial loss of U12 spliceosome function.
Table 1.
Human diseases associated with mutations in spliceosome genes
| Gene | OMIM prevalent mutation sites | Spliceosome complex | Disease | Inheritance | References |
|---|---|---|---|---|---|
| SNRPB | Various mutations in an alternatively spliced regulatory exon | Sm | Cerebro-costo-mandibular syndrome | AD | (Lynch et al. 2014; Bacrot et al. 2015; Tooley et al. 2016) |
| SNRPE | c.1A>G (p.M1?); c.133G>A (p.G45S) | Sm | Hypotrichosis 11 | AD | (Pasternack et al. 2013) |
| SF3B1 | Mutations clustered in exons 12 to 15; p.K700E; Other hotspots: p.E622; p.R625; p.H662; p.K666; p.I704; G742 | U2 snRNP | MDS; CMML; AML; solid tumors | (Papaemmanuil et al. 2011; Quesada et al. 2011; Wang et al. 2011; Harbour et al. 2013) | |
| SF3B4 | None prevalent; various de novo mutations occur at several sites | U2 snRNP | Nager acrofacial dysostosis | AD | (Bernier et al. 2012; Petit et al. 2014) |
| EFTUD2 | Several either de novo 17q21.31 deletions encompassing EFTUD2 or de novo heterozygous EFTUD2 mutations | U5 snRNP | Mandibulofacial dysostosis type Guion-Almeida | AD | (Wieczorek et al. 2009; Lines et al. 2012) |
| PRPF6 | c.2185C>T (p.R729W) | U5 snRNP | RP | AD | (Tanackovic et al. 2011a) |
| PRPF8 | Various sites clustered within exon 42 | U5 snRNP | RP; MDS | AD | (McKie et al. 2001; Towns et al. 2010; Maubaret et al. 2011; Kurtovic-Kozaric et al. 2015; Ruzickova and Stanek 2017) |
| SNRNP200 | c.2653C>G (p.Q885E) | U5 snRNP | RP | AD | (Zhao et al. 2006; Liu et al. 2012) |
| TXNL4A | Most patients are compound heterozygous for a 34-bp deletion in the TXNL4A promoter (chr18:77,748,581-77,748,614del, GRCh37) and a truncating point mutation (e.g., c.131delT [p. Val44AlafsTer48] or another deletion (e.g., Ex3DEL) | U5 snRNP | Burn-McKeown syndrome | AR | (Wieczorek et al. 2014) |
| PRPF3 | Various (e.g., c.1482C>T [p.T494M]; c.1478C>T [p.P493S]) | U4/U6 snRNP | RP | AD | (Chakarova et al. 2002; Tanackovic et al. 2011b) |
| PRPF4 | c.944C>T (p.P315L); c.-114_-97del | U4/U6 snRNP | RP | AD | (Chen et al. 2014) |
| PRPF31 | Various | U4/U6 snRNP | RP | AD | (Vithana et al. 2001; Deery et al. 2002; Rivolta et al. 2006) |
| RNPC3 | Compound heterozygosity (p.P474T and p.R502*) | U11/U12 snRNP | Isolated growth hormone deficiency, type V | AR | (Argente et al., 2014) |
| RNU4ATAC | Various | U4atac snRNP | Lowry-Wood syndrome; Roifman syndrome; Microcephalic osteodysplastic primordial dwarfism type I | AR | (Edery et al., 2011; He et al., 2011; Merico et al., 2015; Scotti and Swanson, 2016) |
| PUF60 | Various | U2 snRNP; A complex | Verheij syndrome | AD | (Dauber et al., 2013; Low et al., 2017) |
| U2AF1 | c.101G>A (p.S34F); c.101G>T (p.S34Y); c.470T>C (p.Q157R) | U2 snRNP; A complex | MDS | (Graubert et al., 2011) | |
| RBM10 | Various | Recruited at A complex | TARP syndrome | XLR | (Johnston et al., 2010) |
| PQBP1 | Various | PRP19 complex; B complex | Renpenning syndrome | XLR | (Kalscheuer et al., 2003) |
| CWC27 | Various | Recruited at Bact complex | RP with or without skeletal anomalies | AR | (Xu et al., 2017) |
| DHX16 | Various | Recruited at Bact complex | Neuromuscular disease and ocular or auditory anomalies with or without seizures | AD | (Paine et al., 2019) |
| PRCC | Reciprocal translocation between chromosomes X and 1, t(X;1)(p11.2;q21.2) creating a PRCC-TFE3 hybrid transcript | Recruited at Bact complex | Papillary renal cell carcinoma | (Sidhar et al., 1996) | |
| RNF113A | Various | Recruited at Bact complex | Nonphotosensitive trichothiodystrophy 5 | XLD | (Corbett et al., 2015) |
| CXORF56 | c.159_160insTA (p.D54*) | Recruited at C complex | Mental retardation, X-linked 107 | XL | (Verkerk et al., 2018) |
| DDX41 | Various | Recruited at C complex | Susceptibility to familial myeloproliferative/lymphoproliferative neoplasms (e.g., MDS and AML) | AD | (Polprasert et al., 2015) |
| EIF4A3 | Expanded 16-repeat allele (initial CACA-20-nt motif followed by 13 repeats of CGCA-20-nt, 1 CACA-20-nt, and 1 final CA-18-nt motif) in the 5′ UTR | EJC/mRNP | Robin sequence with cleft mandible and limb anomalies | AR | (Favaro et al., 2014) |
| RBM8A | 200-kb deletion on 1q21.1 plus an additional RBM8A mutation (site varies; e.g., c.-21G>A) | EJC/mRNP | Thrombocytopenia-absent radius syndrome | AR | (Albers et al., 2012) |
| FUS | Various | hnRNP | ALS | (Reber et al., 2016) | |
| HNRNPA1 | c.941A>T (p.D314V); c.940G>A (p.D314N); c.956A>G (p.N319S) | hnRNP | Inclusion body myopathy with early-onset Paget disease without frontotemporal dementia 3; ALS | AD | (Kim et al., 2013) |
| SMN1 | Various | All Sm snRNPs | Spinal muscular atrophy; myeloid neoplasms | AR | (Li et al., 2014; Yoshida and Ogawa, 2014; Malcovati et al., 2015; Verma et al., 2018) |
AD autosomal dominant, ALS amyotrophic lateral sclerosis, AML acute myeloid leukemia, AR autosomal recessive, CMML, chronic myelomonocytic leukemia, EJC exon junction complex, hnRNP heterogeneous nuclear ribonucleoprotein, MDS myelodysplastic syndromes, mRNP messenger ribonucleoprotein, OMIM Online Mendelian Inheritance in Man®, RES retention and splicing, RP retinitis pigmentosa, XLD X-linked dominant, XLR X-linked recessive
Aberrant splicing in cancer
Aberrant splicing is observed in many types of cancers, including pancreatic (Bailey et al. 2016), lung (Imielinski et al. 2012; Brooks et al. 2014), breast (Maguire et al. 2015; Nik-Zainal et al. 2016), colorectal (Adler et al. 2014), uveal melanoma (Furney et al. 2013; Harbour et al. 2013; Martin et al. 2013), acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), and myelodysplastic syndromes (MDS) (Graubert et al. 2011; Malcovati et al. 2011; Papaemmanuil et al. 2011, 2016; Quesada et al. 2011; Rossi et al. 2011; Wang et al. 2011; Yoshida et al. 2011; Bejar et al. 2012; Damm et al. 2012; Visconte et al. 2012a, 2012b; Scott and Rebel 2013; Ilagan et al. 2015; Kim et al. 2015; Lindsley et al. 2015; Madan et al. 2015; Shirai et al. 2015; Scotti and Swanson 2016; Desai et al. 2018). Splicing alterations in cancer cells often contribute to cancer progression (Rahman et al. 2020).
Splicing alterations can result from several mechanisms, including differential regulation during transcription, alternative splicing, nonsense-mediated decay, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and aberrant post-translational modifications due to cancer-related changes in intracellular signaling (Urbanski et al. 2018). Splicing alterations can also be due to mutations in spliceosome components or splicing factors. Any of the above alterations can subsequently result in altered function and/or nuclear-cytoplasmic localization of the splicing factors, splicing changes to downstream targets of the splicing factors, changes in cellular signaling in tumor cells, and changes in tumor initiation and progression (Fig. 4) (Zhang and Manley 2013; Dvinge et al. 2016).
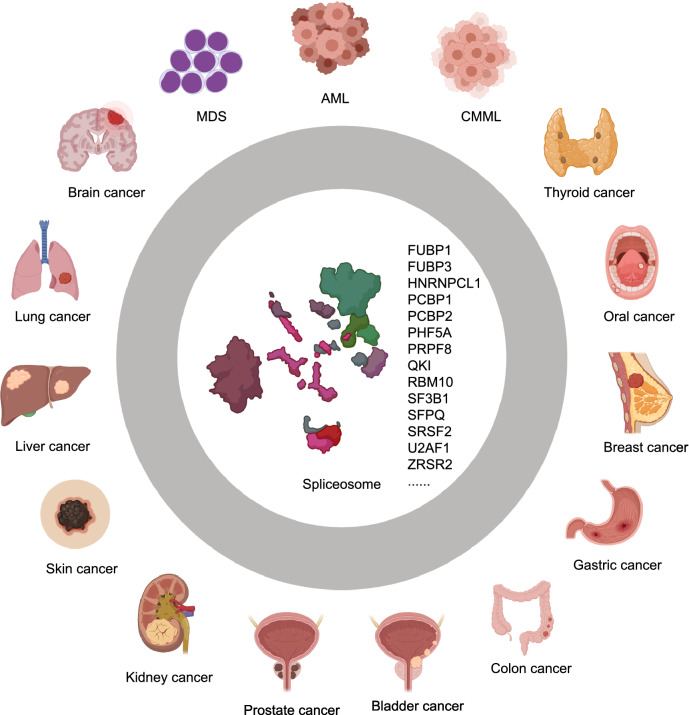
Figure 4.
Schematic representation of human cancers linked to mutations or abnormal expressions of splicing factors and spliceosome genes. Mutated or aberrantly expressed splicing factors and spliceosome genes including FUBP1, FUBP3, HNRNPCL1, PCBP1, PCBP2, PHF5A, QKI, RBM10, SF3B1, and SFPQ, are linked to a variety of solid tumors. While alterations in some other spliceosome genes, e.g., PRPF8, SF3B1, SRSF2, U2AF1, and ZRSR2, are associated with hematologic disorders and malignancies, such as myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), and chronic myelomonocytic leukemia (CMML)
Transcription
Transcription of splicing factors and spliceosome proteins can be differentially regulated by several factors in cancer cells. For example, the transcription factor and oncogene MYC regulates the transcription of splicing factors (e.g., HNRNPA1, PTBP1) (Zhang et al. 2016a) and the core snRNP particle assembly genes (e.g., PRMT5) in cancer cells (Koh et al. 2015), consequently leading to enhanced cell survival. The Wnt signaling pathway also controls the transcriptional activation of splicing factors in some cancers (Goncalves et al. 2008; Corbo et al. 2012).
Alternative splicing
RNA-binding protein expression can be regulated through the splicing of their pre-mRNAs. Members of the SR protein family (e.g., SRSF1, SRSF3, etc.) are able to regulate the inclusion of premature stop codon-containing exons within their own mRNAs. These aberrant transcripts subsequently undergo nonsense-mediated decay, reducing the expression levels of SR protein (Jumaa and Nielsen 1997; Sureau et al. 2001). The splicing of some RNA-binding factors can also be regulated by other splicing factors such as RBFOX2 in cancer tissues, resulting in altered function or transcript degradation (Jumaa and Nielsen 1997; Rossbach et al. 2009; Venables et al. 2013).
MicroRNAs
MiRNAs are small single-stranded noncoding RNAs that can bind target mRNAs, promoting the target mRNA’s degradation. Splicing factors are among the many targets of miRNAs. SRSF7 expression is suppressed by the miRNAs miR-30a-5p and miR-181a-5p in renal tumors, subsequently leading to altered splicing of apoptosis regulators, oncogenes, and tumor suppressors (Boguslawska et al. 2016). In tumors, the expression of splicing factor SRSF1 is putatively increased due to lymphoma/leukemia-related factor-associated repression of miR-28 and miR-505 (Verduci et al. 2010). Also, retinoic acid-induced miR-10a and miR-10b upregulation in neuroblastoma cells, and subsequent repression of SRSF1, putatively results in terminal differentiation of neuroblastoma cells (Meseguer et al. 2011).
Long non-coding RNAs
LncRNAs facilitate splicing factors’ binding to exonic or intronic splicing silencer elements to regulate alternative splicing (Urbanski et al. 2018). The lncRNAs PCGEM1 and BC200 interact with splicing factors HNRNPA1, U2AF65 or HNRNPA2B1 to regulate the alternative splicing of AR (Zhang et al. 2016b) and BCL-x (Singh et al. 2016). Some lncRNAs (e.g., MALAT1) also regulate alternative splicing by regulating nuclear localization of SR proteins (Tripathi et al. 2010). The lncRNA LINC01133 promotes nuclear sequestration of SRSF6, preventing SRSF6-associated epithelial-mesenchymal transition and metastasis in colorectal cancer mouse models (Kong et al. 2016).
Post-translational modifications
Post-translational modifications affect the localization and function of many proteins, including splicing factors. For example, SR protein phosphorylation by SR-specific protein kinases (SRPKs) regulates SR activation and SR-mediated splicing. Aberrant SR protein phosphorylation (either hypo- or hyper-phosphorylation) inhibits splicing (Zhong et al. 2009). SRPK localization is also regulated by its phosphorylation, which promotes nuclear import through interaction with the nuclear import receptor transportin-SR2 (Koizumi et al. 1999; Lai et al. 2000). The CDC-like kinase CLK2 alters splicing, putatively by altering the function of SR proteins through phosphorylation (Yoshida et al. 2015). AKT phosphorylates SRSF1, SRSF7, and SRSF5 as well as SRPKs (Blaustein et al. 2005). The AKT-SRPK-SR axis promotes epidermal growth factor signaling and regulation of alternative splicing (Zhou et al. 2012).
Mutations in spliceosome components and splicing factors
Splicing alterations in cancers can also be caused by mutations affecting splicing-regulatory elements or splicing factors. The most common spliceosome-associated mutations in cancers are in SF3B1, SRSF2, U2AF1, and ZRSR2. Mutations in these genes result in changes in the RNA recognition preferences of the encoded proteins (Rahman et al. 2020); more information about these proteins in cancer can be found in the next section. Recurrent mutations in cancer have been identified in several RNA splicing factors (e.g., PRPF8, RBM10, SFPQ, PHF5A, HNRNPCL1, PCBP1, PCBP2, FUBP1, FUBP3, and QKI), but these mutations have not been fully functionally characterized (Rahman et al. 2020).
Recent studies have examined aberrant spliceosome component function in different types of cancers. Mutations in the U1 snRNA have been reported in medulloblastoma, chronic lymphocytic leukemia, hepatocellular carcinoma, B cell non-Hodgkin lymphoma, and pancreatic adenocarcinoma (Suzuki et al. 2019). Mutations at the fifth nucleotide (r.5A>G) of the U11 snRNA gene (RNU11) have been identified in medulloblastoma (Suzuki et al. 2019). Tumors expressing mutant snRNAs exhibit aberrant splicing, notably excess cryptic 5′ splice site events (Rahman et al. 2020).
Compared to hematological cancers that often exhibit point mutations in core spliceosome genes (Lee and Abdel-Wahab 2016), few recurrent mutations in spliceosome genes have been detected in prostate cancer (Sebestyen et al. 2016). Instead, splicing changes, namely intron retention, in prostate cancer cells are attributed to copy number variation of splicing-regulatory genes (Zhang et al. 2020). Prostate cancer cells exhibit variations in the genomic copy number of almost 70% of genes encoding spliceosome core subunits and auxiliary splicing regulatory proteins during prostate cancer development and progression (Zhang et al. 2020). These splicing alterations correlate with disease progression, prostate cancer stemness, and tumor aggressiveness (Zhang et al. 2020).
The spliceosome component RNF113A is overexpressed in pulmonary adenocarcinomas (Shostak et al. 2020). RNF113A promotes cell survival in lung cancer cells treated with the chemotherapeutic drug Cisplatin by promoting the splicing of prosurvival candidate genes SAT1 and NUPR1. RNF113A also stabilizes the prosurvival protein MCL1 by an unknown mechanism, putatively through a spliceosome-independent manner. Loss of RNF113A expression presumably enhances cell death through MCL1 downregulation.
Spliceosome Mutations are frequently observed in myelodysplastic syndromes
MDS are a group of serious cancers in which the bone marrow can not produce enough healthy and mature blood cells (Heaney and Golde 1999; Pellagatti and Boultwood 2015). Early on, there are typically no symptoms. As the disease progresses, symptoms may include feeling tired, shortness of breath, easy bleeding, or frequent infections. Patients with MDS show increased numbers of bone marrow myeloblasts (i.e., immature cells) over time and are at risk of developing AML (Heaney and Golde 1999; Pellagatti and Boultwood 2015; Bersanelli et al. 2021).
Approximately 50% of MDS patients have a mutation in SF3B1, U2AF1, SRSF2, or ZRSR2 (Yoshida et al. 2011; Haferlach et al. 2014; Nguyen et al. 2018). Mutations in SRSF2 and U2AF1 are the most frequent splicing-associated mutations found in the more aggressive subtypes of MDS, including refractory anemia with excess blasts I (RAEB I) and RAEB II (Inoue et al. 2016). The MDS-associated spliceosome mutations identified thus far are heterozygous mutations as opposed to nonsense mutations, indicating that these mutations are neomorphic or dominant-negative (Inoue et al. 2016).
SF3B1 encodes a protein that stabilizes U2 snRNP binding to the branch point sequence (Yip et al. 2016). Greater than 90% of refractory anemia with ringed sideroblasts (RARS) MDS patients and approximately 70% of non-RARS forms of MDS with ringed sideroblasts (i.e., refractory cytopenia with multilineage dysplasias; ringed sideroblasts and RARS associated with marked thrombocytosis) have mutations in SF3B1 (Inoue et al. 2016). MDS-causing SF3B1 mutations result in aberrant 3′ splice site usage, leading to coding of premature stop codons and subsequent nonsense-mediated decay of the affected transcripts (Darman et al. 2015).
U2AF1-encoded protein recognizes the AG dinucleotide at 3′ splice sites (Wu et al. 1999; Yip et al. 2016). Mutations in U2AF1 have been identified not only in patients with MDS (Graubert et al. 2011; Yoshida et al. 2011), but also in patients with CMML, secondary acute AML (Graubert et al. 2011; Yoshida et al. 2011; Papaemmanuil et al. 2013), hairy cell leukemia (Waterfall et al. 2014), pancreatic ductal adenocarcinomas (Bailey et al. 2016), and non-small cell lung adenocarcinomas (Imielinski et al. 2012). The cancer-causing mutations in U2AF1 typically affect either S34 or Q157 within the zinc finger domains of the U2AF1 protein, resulting in changes in U2AF1 3′ splice site recognition. Furthermore, a U2AF1 mutation (p.S34F) causes accumulation of a transcription intermediate containing RNA:DNA hybrids and single-stranded DNA (i.e., R loops) (Nguyen et al. 2018). The R loops elicit a DNA damage response induced by ataxia telangiectasia mutated- and Rad3-related kinase (ATR). S34F mutant U2AF1-expressing cells undergo ATR inhibitor-induced cell death, which also promotes DNA damage. The U2AF1 mutations cause mis-splicing of hundreds of transcripts by exon skipping, exon inclusion, or alternative 3′ splice site usage (Ilagan et al. 2015; Okeyo-Owuor et al. 2015; Shirai et al. 2015).
SRSF2 encodes a splicing factor that binds mRNA exon splicing enhancer motifs (Yip et al. 2016). SRSF2 mutations are linked to poor prognosis and a higher risk of transformation in AML (Damm et al. 2012; Meggendorfer et al. 2012; Zhang et al. 2012; Papaemmanuil et al. 2013). SRSF2 mutations that affect P95 near the RNA recognition motif in the SRSF2 protein result in changes to SRSF2’s RNA-binding characteristics, altered splicing patterns, and impaired hematopoietic cell differentiation (Kim et al. 2015; Komeno et al. 2015; Zhang et al. 2015).
ZRSR2 encodes a spliceosome component required for the recognition of the 3′ splice site for both U2- and U12-type introns. ZRSR2 is an X-linked gene; therefore, mutations in ZRSR2 are predominantly linked to MDS in male patients (Yoshida et al. 2011). The ZRSR2 mutations are sporadic across the entire coding region. These loss-of-function mutations lead to increased retention of U12 introns, but not U2 intron splicing. Notably, loss-of-function mutations in ZRSR2 cause widespread minor intron retention and thus enhance hematopoietic stem cell self-renewal as well as drive diverse cancer predisposition, which putatively results from LZTR1 minor intron retention (Inoue et al. 2021).
Anti-cancer strategies targeting the spliceosome machinery
Mutations affecting the spliceosome or splicing factors result in perturbation in downstream splicing targets, many of which are in signaling pathways involved with cancer. Alternatively, the copy number or expression level of a splicing factor can be changed in tumors without the presence of a mutation (Anczukow and Krainer 2016). Therefore, identifying anti-cancer strategies towards problematic factors is imperative.
Antisense oligonucleotides (ASOs) are synthetic, single-stranded oligodeoxynucleotides that inhibit gene expression by binding cellular RNA. ASOs can also be developed to target non-coding RNAs, including lncRNAs and miRNAs. ASOs can be used to alter splicing, namely the selective removal or inclusion of a particular exon (Spitali and Aartsma-Rus 2012; Havens and Hastings 2016). These ASOs alter splicing by changing the spliceosome recognition sites on the target RNAs. ASOs can potentially restore splice defects in mutated genes, allowing a normal protein to be expressed (Dominski and Kole 1993). Numerous ASOs (e.g., Danvatirsen, Trabederen, Custirsen) designed for different oncogenic targets (e.g., STAT3, TGFB2, clusterin, respectively) are being investigated in clinical trials or are in development (Quemener et al. 2020).
Small molecules that target components of the spliceosome have exhibited antitumor effects in cancer cells with spliceosome component mutations (Bonnal et al. 2020). For example, prostate cancer cells and the prostate cancer cell line PC3 show sensitivity to the spliceosome inhibitor E7107 (Zhang et al. 2020). E7107 targets SF3B complex, preventing tight binding of the U2 snRNP to pre-mRNA (Kotake et al. 2007; Folco et al. 2011). Treatment with E7107 suppresses prostate cancer cell migration and invasion as well as causes cell death by promoting cell cycle arrest at the G2/M phase. E7107 putatively inhibits the splicing of pancreatic cancer-promoting genes, consequently resulting in a less aggressive phenotype. Yet more preclinical studies are necessary to determine which cancer patients would most benefit from these therapies, as well as to determine the combination strategies with other therapies (Eymin 2020).
Although small-molecule splicing modulators have been explored as anti-cancer therapeutics for a lone time (Kaida et al. 2007; Kotake et al. 2007), the mechanisms underlying the selective antitumor activity remain largely unknown. Recent studies have shown that spliceosome-targeted therapies (STTs) can cause widespread cytoplasmic accumulation of mis-spliced (e.g., intron-retained) mRNAs, many of which form double-stranded RNAs (dsRNAs). Consequently, these dsRNAs can be recognized by intracellular immune sensors, initiating antiviral signaling (viral mimicry) and extrinsic apoptosis in breast cancer (Bowling et al. 2021; Ishak et al. 2021).
Cancer-associated spliceosome mutations result in aberrant immune signaling
MDS patients often exhibit inflammation due to the overproduction of inflammatory cytokines by myeloid cells. Subsequently, the excess cytokines could contribute to impaired hematopoietic stem cell niche function and the suppression of normal hematopoiesis (Baldridge et al. 2011). Altered splicing of inflammatory and immune genes (e.g., IRAK4, MAP3K7, and CASP8) due to spliceosome mutations may contribute to MDS pathogenesis by leading to inflammation, changes in immune cell function, and increased risk of infection (Starczynowski and Karsan 2010b, 2010a; Pagano and Caira 2012; Darman et al. 2015; Ilagan et al. 2015; Kim et al. 2015; Zhang et al. 2015; Alsafadi et al. 2016; Grignano et al. 2018; Pollyea et al. 2019; Smith et al. 2019). For example, in some MDS/AML patients with U2AF1 mutations, IRAK4 is alternatively spliced to retain exon 4. This alternatively spliced IRAK4 subsequently encodes a protein (termed IRAK4-L) that assembles with the myddosome, resulting in maximal activation of NF-κB. This process is essential for leukemic cell function, and inhibition of IRAK4-L blocks leukemic growth.
Inhibition of SF3B1, SF3A1, SF3A2, SF3A3, U2AF1, SRSF2, or EFTUD2 in mouse or human macrophages results in diminished toll-like receptor (TLR)-induced inflammatory cytokine production after stimulation with the TLR4 agonist lipopolysaccharide (De Arras and Alper 2013; De Arras et al. 2014; O'Connor et al. 2015; Lv et al. 2019; Pollyea et al. 2019). Notably, knockdown of any of the genes does not affect cell viability or phagocytosis (De Arras and Alper 2013; De Arras et al. 2014; O'Connor et al. 2015). Conversely, expression of SF3B1, U2AF1, and SRSF2 MDS-associated mutations in macrophages, MDS patient-derived cell lines, and mouse and human myeloid cells results in enhanced NF-ĸB activation and subsequent lipopolysaccharide-induced inflammatory cytokine production (Pollyea et al. 2019). These inflammatory changes point to a mechanism that contributes to MDS pathogenesis.
MyD88 is an adaptor protein that functions in most TLR signaling pathways. Knockdown of Sf3a1, Sf3b1, Srsf2 or U2af1 in RAW264.7 mouse macrophage cells results in increased expression of the anti-inflammatory short form of MyD88 (MyD88s) and subsequently reduced inflammatory cytokine production; the expression level of the long isoform of MyD88 (MyD88L) is unchanged (O'Connor et al. 2015; Pollyea et al. 2019). Interestingly, MYD88 splicing is not affected in the myeloid leukemia cell line K562 upon splicing factor knockdown, indicating that the splicing of other TLR signaling genes is promoting the increased inflammatory cytokine production in this cell type.
TAK1 (alternatively, MAP3K7) functions as a regulator of NF-ĸB, JNK, and MAPK signaling pathways. A mutation in SF3B1 (p.K700E) can cause aberrant splicing of MAP3K7 (encoding TAK1) (Lee et al. 2018). Expression of mutant SF3B1 in K562 cells leads to increased production of the alternative TAK1 isoform and enhanced innate immune signaling; however, the expression of canonical TAK1 is unchanged (Lee et al. 2018; Pollyea et al. 2019).
Conditional deletion of Srsf1 results in severe defects at late stage thymocyte development, thus a notable reduction in periphery T cell pool. SRSF1 can directly bind and regulate Irf7 and Il27ra expression via alternative splicing in response to type I interferon signaling (Qi et al. 2021). SRSF1 restrains IFN-γ production and Th1 differentiation through the control of RhoH (Katsuyama et al. 2021). SRSF1 modulates the expression of several genes involved in immune system functions through multiple mechanisms (Paz et al. 2021).
Moreover, cells expressing a mutant SRSF2 (p.P95H) can express an alternative caspase 8 isoform that is truncated due to exon 6 skipping. Expression of the alternative caspase 8 isoform results in enhanced NF-ĸB activity (Lee et al. 2018; Pollyea et al. 2019).
The expanding role of core spliceosome components in immunity
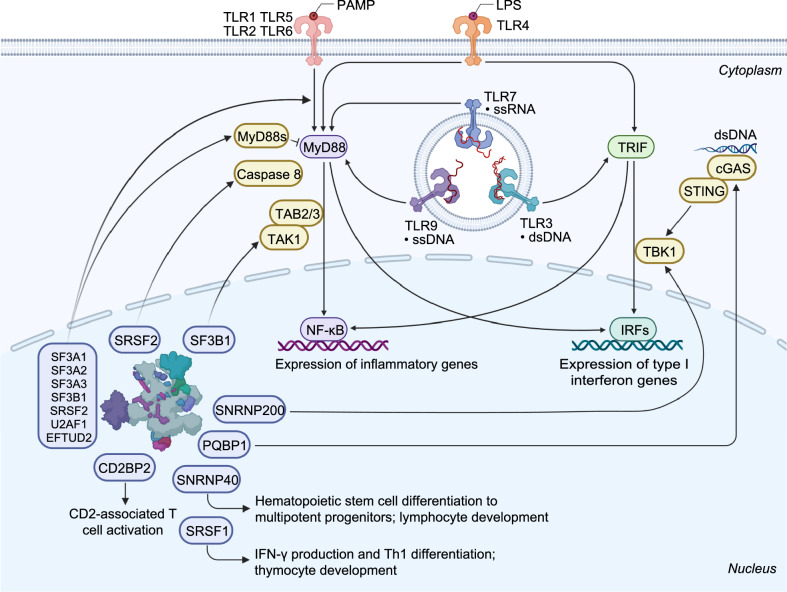
Several snRNP complex components are differentially expressed in a cell- or tissue-specific manner, and this often correlates with tissue-specific phenotypes that arise upon their inactivation. In addition, core spliceosome components such as SNRNP40, SNRNP200, CD2BP2, and PQBP1, harbor splicing-independent functions specifically related to immune signaling (Fig. 5).
Figure 5.
Schematic representation of immune functions linked to splicing factors and spliceosome components. The spliceosome proteins (e.g., SF3A1, SF3A2, SF3A3, SF3B1, SRSF2, U2AF1, and EFTUD2) can regulate TLR-induced NF-ĸB activation and inflammatory cytokine production. Knockdown of Sf3a1, Sf3b1, Srsf2, or U2af1 in mouse macrophage cells results in increased expression of the anti-inflammatory short form of MyD88 (MyD88s) and subsequently reduced inflammatory cytokine production. SF3B1 can also regulate the production of an alternative TAK1 isoform, subsequently affecting innate immune signaling. SRSF1 restrains IFN-γ production and Th1 differentiation; it regulates the late stage of thymocyte development and the expression of several immune-related genes through multiple mechanisms. SRSF2 regulates the expression of an alternative caspase 8 isoform and subsequent NF-ĸB activity. SNRNP40 plays a pivotal role in hematopoietic stem cell differentiation to multipotent progenitors and further to common lymphoid progenitors. SNRNP200 activates IRF3-mediated antiviral innate immune responses through an interaction with TBK1. CD2BP2 is involved in CD2-associated T cell activation via binding to the cytoplasmic tail of CD2 molecule. PQBP1 can interact with cGAS to trigger type I interferon production in response to cytosolic nucleic acids or DNA damage
SNRNP40
Our recent studies have shown that a viable hypomorphic mutation in mouse spliceosome gene Snrnp40 causes remarkable immune phenotypes (Zhang et al. 2019). SNRNP40 is part of the U5 snRNP; however, its exact function is unknown. In the adult mouse, the SNRNP40 protein is predominantly expressed in lymphoid tissue. The Snrnp40 mutant mice show impaired hematopoietic stem cell differentiation to multipotent progenitors, and aberrant multipotent progenitor differentiation to common lymphoid progenitors, T cells, B cells and natural killer cells. A few hundred splicing errors, mostly intron retention, can be observed in the Snrnp40-deficient mouse hematopoietic stem and progenitor cells or T cells. These cumulative splicing errors lead to reduced expression of immune-related proteins and subsequent immune phenotypes (Zhang et al. 2019). Furthermore, the SNRNP40 protein also shows localization within the cytoplasm (unpublished data), implying its splicing-independent functions, most likely in immune signaling due to the severe immune-specific phenotypes observed in the Snrnp40 mutant mice.
SNRNP200
TBK1 is a downstream kinase activated by a number of pattern recognition receptor (PRR)–adaptor protein pairs, including cGAS-STING, RIG-I–MAVS, and TLR3/4-TRIF. TBK1 functions as an adaptor to subsequently promote the expression of proinflammatory and antiviral cytokines and chemokines (Liu et al. 2015). The spliceosome protein SNRNP200 which catalyzes the ATP-dependent unwinding of U4/U6 RNA duplices is able to bind RNA and interact with TBK1. Upon viral infection, SNRNP200 relocalizes into TBK1-containing cytoplasmic structures, and subsequently promotes the activation of IRF3-mediated antiviral innate immune response. Loss of SNRNP200 expression results in a decrease in antiviral innate immunity (Tremblay et al. 2016).
CD2BP2
The adaptor protein CD2BP2 is a constituent of the U5 snRNP (Kofler et al. 2004; Laggerbauer et al. 2005). Yeast two-hybrid analysis determines that CD2BP2 binds the cytoplasmic tail of CD2, a surface antigen that promotes the adhesion and activation of T cells upon engagement with its ligand (Nishizawa et al. 1998; Heinze et al. 2007). The CD2BP2 and CD2 interaction regulates CD2-associated T cell activation (Freund et al. 2002; Heinze et al. 2007), and transient transfection of a CD2BP2 fragment in Jurkat cells induces CD2-stimulated IL-2 production (Nishizawa et al. 1998). Conversely, knockdown of CD2BP2 expression does not impact cytokine expression in primary T cells in response to CD2 stimulation, indicating that CD2 signaling is not completely dependent on CD2BP2 binding (Heinze et al. 2007).
PQBP1
Polyglutamine binding protein 1 (PQBP1) is a component of the B complex in spliceosome. PQBP1 can also associate with, and inhibit, the DNA sensors cyclic GMP-AMP synthase (cGAS) and interferon-γ-inducible protein 16 (IFI16) in response to cytosolic DNA. Reduced expression of PQBP1 in THP-1 cells leads to increased type I interferon production in response to transfected cytosolic nucleic acids or DNA damage (Shannon et al. 2018). Furthermore, PQBP1 directly binds to reverse-transcribed HIV-1 DNA and interacts with cGAS to trigger an IRF3-dependent innate immune response. Primary human monocyte-derived DCs from Renpenning syndrome patients who harbor PQBP1 mutations exhibit a significantly reduced innate response to HIV-1 challenge, emphasizing the role of PQBP1 as a proximal innate sensor of HIV-1 infection (Yoh et al. 2015).
Perspectives
The immune-related findings of SNRNP200 and PQBP1 highlight that some spliceosome components can have tissue-specific functions within the cytoplasm (Tremblay et al. 2016; Shannon et al. 2018). The observation that the SNRNP40 protein also localizes outside of the nucleus suggests that additional spliceosome-independent functions of SNRNP40 are worth studying. Other spliceosome proteins with documented splicing-independent functions include survival of motor neuron 1 (SMN1) and splicing factor 3b subunit 4 (SF3B4). In the spliceosome, SMN1 functions in the early stages of cytoplasmic snRNP assembly by promoting the addition of the Sm proteins around the snRNA (Tisdale et al. 2013; Li et al. 2014). SMN1 also functions in the trafficking of mature mRNA within the cytoplasm of axons and neurites (Fallini et al. 2016). SF3B4, a subunit of the U2 snRNP, putatively functions in transcription (Devotta et al. 2016; Marques et al. 2016), translation (Ueno et al. 2019), and cell signaling (Nishanian and Waldman 2004). In Arabidopsis, SF3B4 mediates the transcription of certain genes by recruiting the RNA polymerase II complex to the RNA transcript (Xiong et al. 2019); however, the role of SF3B4 in human transcription regulation remains unclear. While during translation, SF3B4 interacts with the cofactor p180, promoting the association of mRNAs with the endoplasmic reticulum membrane and assembly of polyribosomes (Ueno et al. 2019). SF3B4 regulates the receptor kinase BMPR1A-associated signaling and specifically inhibits osteochondral cell differentiation. SF3B4 interacts with, and downregulates expression of BMPR1A at the cell surface (possibly by regulating receptor internalization), subsequently regulating craniofacial development in embryos (Nishanian and Waldman 2004; Watanabe et al. 2007).
Future studies will undoubtedly show that other spliceosome components display splicing-independent functions within the nucleus and the cytoplasm, including the functions that may be related to innate immunity, cell cycle, cell growth, cell death, or transformation. Moreover, future studies will uncover new correlations between spliceosome mutations, aberrant splicing, immune dysregulation, and human cancers.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82071780), Science and Technology Commission of Shanghai Municipality (No. 20ZR1407600), Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), and the Zhangjiang Lab. The authors thank Anne R. Murray, PhD, MWC® for writing and editorial assistance during the preparation of this manuscript. Illustrations were mainly created with BioRender.com.
Abbreviations
- AML
acute myeloid leukemia
- AR
androgen receptor
- ASOs
antisense oligonucleotides
- ATR
ataxia telangiectasia-mutated and RAD3-related kinase
- BMPR1A
bone morphogenetic protein receptor type 1A
- CD2BP2
CD2 cytoplasmic tail binding protein 2
- cGAS
cyclic GMP-AMP synthase
- CLK2
CDC-like kinase 2
- CMML
chronic myelomonocytic leukemia
- DDX23
DEAD-box helicase 23
- DDX46
DEAD-box helicase 46
- DHX8
DEAH-box helicase 8
- DHX15
DEAH-box helicase 15
- DHX16
DEAH-box helicase 16
- DHX38
DEAH-box helicase 38
- EFTUD2
elongation factor Tu GTP binding domain containing 2
- FUBP1
far upstream element binding protein 1
- FUBP3
far upstream element binding protein 3
- HIV-1
human immunodeficiency virus 1
- HNRNP
heterogeneous nuclear ribonucleoprotein
- IFI16
interferon-γ-inducible protein 16
- IRAK4
interleukin 1 receptor associated kinase 4
- JNK
JUN N-terminal kinase
- LZTR1
leucine zipper like transcription regulator 1
- MAPK
mitogen-activated protein kinase
- MAP3K7
mitogen-activated protein kinase kinase kinase 7
- MAVS
mitochondrial antiviral signaling protein
- MCL1
MCL1 apoptosis regulator, BCL2 family member
- MDA5/IFIH1
interferon induced with helicase C domain 1
- MDS
myelodysplastic syndromes
- MyD88
myeloid differentiation primary response 88
- NF-κB
nuclear factor kappa B
- NUPR1
nuclear protein 1, transcriptional regulator
- PCBP1
poly(rC) binding protein 1
- PCBP2
poly(rC) binding protein 2
- PHF5A
PHD finger protein 5A
- PQBP1
polyglutamine binding protein 1
- PRMT5
protein arginine methyltransferase 5
- PRPF
pre-mRNA processing factor
- PTBP1
polypyrimidine tract binding protein 1
- QKI
quaking homolog, KH domain RNA binding
- RAEB
refractory anemia with excess blasts
- RARS
refractory anemia with ringed sideroblasts
- RBFOX2
RNA binding fox-1 homolog 2
- RBM10
RNA binding motif protein 10
- RhoH
Ras homolog family member H
- RIG-I/DDX58
DExD/H-box helicase 58
- RNF113A
ring finger protein 113A
- RP
retinitis pigmentosa
- SAT1
spermidine/spermine N1-acetyltransferase 1
- SF1
splicing factor 1
- SF3B1
splicing factor 3b subunit 1
- SF3B4
splicing factor 3b subunit 4
- SFPQ
splicing factor proline and glutamine rich
- SMN1
survival of motor neuron 1
- snRNP
small nuclear ribonucleoprotein
- SNRNP40
small nuclear ribonucleoprotein U5 subunit 40
- SNRNP200
small nuclear ribonucleoprotein U5 subunit 200
- SR
serine/arginine-rich
- SRPK
SRSF protein kinase
- SRSF1
serine/arginine-rich splicing factor 1
- SRSF2
serine/arginine-rich splicing factor 2
- STAT3
signal transducer and activator of transcription 3
- STING
stimulator of interferon response cGAMP interactor 1
- TAK1
tGF-beta activated kinase 1
- TBK1
tANK-binding kinase 1
- TGFB2
transforming growth factor beta 2
- TLR
toll-like receptor
- TRIF/TICAM1
toll-like receptor adaptor molecule 1
- U snRNA
uridine-rich small nuclear RNA
- U2AF1
U2 auxiliary factor 1
- U2AF2
U2 auxiliary factor 2
- ZRSR2
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2
Declarations
Conflict of interest
Hui Yang, Bruce Beutler and Duanwu Zhang declare that they have no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by the any of the authors.
Contributor Information
Bruce Beutler, Email: bruce.beutler@utsouthwestern.edu.
Duanwu Zhang, Email: duanwu@fudan.edu.cn.
References
- Adler AS, McCleland ML, Yee S, Yaylaoglu M, Hussain S, Cosino E, Quinones G, Modrusan Z, Seshagiri S, Torres E, et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 2014;28:1068–1084. doi: 10.1101/gad.237206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA, Jolley JD, Cvejic A, Kostadima M, Bertone P, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet. 2012;44(4):435–439. doi: 10.1038/ng.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczukow O, Krainer AR. Splicing-factor alterations in cancers. RNA. 2016;22:1285–1301. doi: 10.1261/rna.057919.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argente J, Flores R, Gutierrez-Arumi A, Verma B, Martos-Moreno GA, Cusco I, Oghabian A, Chowen JA, Frilander MJ, Perez-Jurado LA. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol Med. 2014;6:299–306. doi: 10.1002/emmm.201303573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacrot S, Doyard M, Huber C, Alibeu O, Feldhahn N, Lehalle D, Lacombe D, Marlin S, Nitschke P, Petit F, et al. Mutations in SNRPB, encoding components of the core splicing machinery, cause cerebro-costo-mandibular syndrome. Hum Mutat. 2015;36:187–190. doi: 10.1002/humu.22729. [DOI] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- Baldridge MT, King KY, Goodell MA. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 2011;32:57–65. doi: 10.1016/j.it.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, Raza A, Kantarjian H, Levine RL, Neuberg D, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S, Schwartzentruber J, Buckingham KJ, Innes AM, Jabs EW, Innis JW, Schuette JL, Gorski JL, et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersanelli M, Travaglino E, Meggendorfer M, Matteuzzi T, Sala C, Mosca E, Chiereghin C, Di Nanni N, Gnocchi M, Zampini M, et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J Clin Oncol. 2021;39:1223–1233. doi: 10.1200/JCO.20.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Boguslawska J, Sokol E, Rybicka B, Czubaty A, Rodzik K, Piekielko-Witkowska A. microRNAs target SRSF7 splicing factor to modulate the expression of osteopontin splice variants in renal cancer cells. Gene. 2016;595:142–149. doi: 10.1016/j.gene.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Bonnal SC, Lopez-Oreja I, Valcarcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. 2020;17:457–474. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- Bowling EA, Wang JH, Gong F, Wu W, Neill NJ, Kim IS, Tyagi S, Orellana M, Kurley SJ, Dominguez-Vidana R, et al. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 2021;184:384–403. doi: 10.1016/j.cell.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE. 2014;9:e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–785. doi: 10.1016/S1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Chakarova CF, Hims MM, Bolz H, Abu-Safieh L, Patel RJ, Papaioannou MG, Inglehearn CF, Keen TJ, Willis C, Moore AT, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu Y, Sheng X, Tam PO, Zhao K, Chen X, Rong W, Liu Y, Liu X, Pan X, et al. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2014;23:2926–2939. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- Corbett MA, Dudding-Byth T, Crock PA, Botta E, Christie LM, Nardo T, Caligiuri G, Hobson L, Boyle J, Mansour A, et al. A novel X-linked trichothiodystrophy associated with a nonsense mutation in RNF113A. J Med Genet. 2015;52:269–274. doi: 10.1136/jmedgenet-2014-102418. [DOI] [PubMed] [Google Scholar]
- Corbo C, Orru S, Gemei M, Noto RD, Mirabelli P, Imperlini E, Ruoppolo M, Vecchio LD, Salvatore F. Protein cross-talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumorigenicity. Proteomics. 2012;12:2045–2059. doi: 10.1002/pmic.201100370. [DOI] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2013;41:D132–141. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, Della Valle V, Couronne L, Scourzic L, Chesnais V, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. Cancer-associated SF3B1 hotspot mutations induce Cryptic 3' splice site selection through use of a different branch point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Dauber A, Golzio C, Guenot C, Jodelka FM, Kibaek M, Kjaergaard S, Leheup B, Martinet D, Nowaczyk MJ, Rosenfeld JA, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am J Hum Genet. 2013;93:798–811. doi: 10.1016/j.ajhg.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arras L, Alper S. Limiting of the innate immune response by SF3A-dependent control of MyD88 alternative mRNA splicing. PLoS Genet. 2013;9:e1003855. doi: 10.1371/journal.pgen.1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arras L, Laws R, Leach SM, Pontis K, Freedman JH, Schwartz DA, Alper S. Comparative genomics RNAi screen identifies Eftud2 as a novel regulator of innate immunity. Genetics. 2014;197:485–496. doi: 10.1534/genetics.113.160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deery EC, Vithana EN, Newbold RJ, Gallon VA, Bhattacharya SS, Warren MJ, Hunt DM, Wilkie SE. Disease mechanism for retinitis pigmentosa (RP11) caused by mutations in the splicing factor gene PRPF31. Hum Mol Genet. 2002;11:3209–3219. doi: 10.1093/hmg/11.25.3209. [DOI] [PubMed] [Google Scholar]
- Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, Samuel M, Ritchie EK, Guzman ML, Ballman KV, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24:1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devotta A, Juraver-Geslin H, Gonzalez JA, Hong CS, Saint-Jeannet JP. Sf3b4-depleted Xenopus embryos: A model to study the pathogenesis of craniofacial defects in Nager syndrome. Dev Biol. 2016;415:371–382. doi: 10.1016/j.ydbio.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- Elsaid MF, Chalhoub N, Ben-Omran T, Kumar P, Kamel H, Ibrahim K, Mohamoud Y, Al-Dous E, Al-Azwani I, Malek JA, et al. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann Neurol. 2017;81:68–78. doi: 10.1002/ana.24826. [DOI] [PubMed] [Google Scholar]
- Eymin B. Targeting the spliceosome machinery: A new therapeutic axis in cancer? Biochem Pharmacol. 2021;189:114039. doi: 10.1016/j.bcp.2020.114039. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Luhrmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the survival of motor neuron protein impairs mrna localization and local translation in the growth cone of motor neurons. J Neurosci. 2016;36:3811–3820. doi: 10.1523/JNEUROSCI.2396-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro FP, Alvizi L, Zechi-Ceide RM, Bertola D, Felix TM, de Souza J, Raskin S, Twigg SR, Weiner AM, Armas P, et al. A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet. 2014;94:120–128. doi: 10.1016/j.ajhg.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund C, Kuhne R, Yang H, Park S, Reinherz EL, Wagner G. Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules. EMBO J. 2002;21:5985–5995. doi: 10.1093/emboj/cdf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Matos P, Jordan P. The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA. 2008;14:2538–2549. doi: 10.1261/rna.1253408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignano E, Jachiet V, Fenaux P, Ades L, Fain O, Mekinian A. Autoimmune manifestations associated with myelodysplastic syndromes. Ann Hematol. 2018;97:2015–2023. doi: 10.1007/s00277-018-3472-9. [DOI] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney ML, Golde DW. Myelodysplasia. N Engl J Med. 1999;340:1649–1660. doi: 10.1056/NEJM199905273402107. [DOI] [PubMed] [Google Scholar]
- Heinze M, Kofler M, Freund C. Investigating the functional role of CD2BP2 in T cells. Int Immunol. 2007;19:1313–1318. doi: 10.1093/intimm/dxm100. [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016;30:989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Polaski JT, Taylor J, Castel P, Chen S, Kobayashi S, Hogg SJ, Hayashi Y, Pineda JMB, El Marabti E, et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat Genet. 2021;53(5):707–718. doi: 10.1038/s41588-021-00828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishak CA, Loo Yau H, De Carvalho DD. Spliceosome-targeted therapies induce dsRNA responses. Immunity. 2021;54:11–13. doi: 10.1016/j.immuni.2020.12.012. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Ariizumi Y, Shiga A, Kato T, Tan CF, Sato T, Miki Y, Yokoo M, Fujino T, Koyama A, et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22:4136–4147. doi: 10.1093/hmg/ddt262. [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Teer JK, Cherukuri PF, Hansen NF, Loftus SK, Center NIHIS, Chong K, Mullikin JC, Biesecker LG. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am J Hum Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/S1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gecz J, Sefiani A, Hoffmann K, Moser B, Haas S, et al. Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet. 2003;35:313–315. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- Katsuyama T, Li H, Krishfield SM, Kyttaris VC, Moulton VR. Splicing factor SRSF1 limits IFN-gamma production via RhoH and ameliorates experimental nephritis. Rheumatology (oxford) 2021;60:420–429. doi: 10.1093/rheumatology/keaa300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, Li Y, Chung YR, Micol JB, Murphy ME, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler M, Heuer K, Zech T, Freund C. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J Biol Chem. 2004;279:28292–28297. doi: 10.1074/jbc.M402008200. [DOI] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabo A, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- Komeno Y, Huang YJ, Qiu J, Lin L, Xu Y, Zhou Y, Chen L, Monterroza DD, Li H, DeKelver RC, et al. SRSF2 Is Essential for Hematopoiesis, and Its Myelodysplastic Syndrome-Related Mutations Dysregulate Alternative Pre-mRNA Splicing. Mol Cell Biol. 2015;35:3071–3082. doi: 10.1128/MCB.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, Xu E, Zhang H, Lai M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380:476–484. doi: 10.1016/j.canlet.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Kurtovic-Kozaric A, Przychodzen B, Singh J, Konarska MM, Clemente MJ, Otrock ZK, Nakashima M, Hsi ED, Yoshida K, Shiraishi Y, et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia. 2015;29:126–136. doi: 10.1038/leu.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Luhrmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5–102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, North K, Kim E, Jang E, Obeng E, Lu SX, Liu B, Inoue D, Yoshimi A, Ki M, et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell. 2018;34:225–241. doi: 10.1016/j.ccell.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol. 2014;32:22–29. doi: 10.1016/j.semcdb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, Pigneux A, Wetzler M, Stuart RK, Erba HP, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM, Gener B, Gillessen-Kaesbach G, et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu T, Jin X, Zhang X, Yuan H, Cheng J, Lee J, Zhang B, Zhang M, Wu J, Wang L, et al. A novel missense SNRNP200 mutation associated with autosomal dominant retinitis pigmentosa in a Chinese family. PLoS ONE. 2012;7:e45464. doi: 10.1371/journal.pone.0045464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KJ, Ansari M, Abou Jamra R, Clarke A, El Chehadeh S, FitzPatrick DR, Greenslade M, Henderson A, Hurst J, Keller K, et al. PUF60 variants cause a syndrome of ID, short stature, microcephaly, coloboma, craniofacial, cardiac, renal and spinal features. Eur J Hum Genet. 2017;25:552–559. doi: 10.1038/ejhg.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z, Wang Z, Luo L, Chen Y, Han G, Wang R, Xiao H, Li X, Hou C, Feng J, et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. 2019;12:1164–1173. doi: 10.1038/s41385-019-0184-y. [DOI] [PubMed] [Google Scholar]
- Lynch DC, Revil T, Schwartzentruber J, Bhoj EJ, Innes AM, Lamont RE, Lemire EG, Chodirker BN, Taylor JP, Zackai EH, et al. Disrupted auto-regulation of the spliceosomal gene SNRPB causes cerebro-costo-mandibular syndrome. Nat Commun. 2014;5:4483. doi: 10.1038/ncomms5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, Sanada M, Grossmann V, Sundaresan J, Shiraishi Y, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SL, Leonidou A, Wai P, Marchio C, Ng CK, Sapino A, Salomon AV, Reis-Filho JS, Weigelt B, Natrajan RC. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235:571–580. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, Elena C, Galli A, Walldin G, Della Porta MG, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, Travaglino E, Groves MJ, Godfrey AL, Ambaglio I, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques F, Tenney J, Duran I, Martin J, Nevarez L, Pogue R, Krakow D, Cohn DH, Li B. Altered mRNA splicing, chondrocyte gene expression and abnormal skeletal development due to SF3B4 mutations in rodriguez acrofacial dysostosis. PLoS Genet. 2016;12:1006307. doi: 10.1371/journal.pgen.1006307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv Exp Med Biol. 2007;623:14–35. doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- Maubaret CG, Vaclavik V, Mukhopadhyay R, Waseem NH, Churchill A, Holder GE, Moore AT, Bhattacharya SS, Webster AR. Autosomal dominant retinitis pigmentosa with intrafamilial variability and incomplete penetrance in two families carrying mutations in PRPF8. Invest Ophthalmol vis Sci. 2011;52:9304–9309. doi: 10.1167/iovs.11-8372. [DOI] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, Kohlmann A, Alpermann T, Yoshida K, Ogawa S, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–3088. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Roifman M, Braunschweig U, Yuen RK, Alexandrova R, Bates A, Reid B, Nalpathamkalam T, Wang Z, Thiruvahindrapuram B, et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman Syndrome by disrupting minor intron splicing. Nat Commun. 2015;6:8718. doi: 10.1038/ncomms9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF) J Biol Chem. 2011;286:4150–4164. doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA (1993) Splicing of precursors to mRNA by the spliceosome. In RNA World, Gesteland RF, Atkins JF, eds. (Cold Spring Harbor, New York, Cold Spring Harbor Labratory Press), pp. 303-357
- Nguyen HD, Leong WY, Li W, Reddy PNG, Sullivan JD, Walter MJ, Zou L, Graubert TA. Spliceosome mutations induce R Loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 2018;78:5363–5374. doi: 10.1158/0008-5472.CAN-17-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishanian TG, Waldman T. Interaction of the BMPR-IA tumor suppressor with a developmentally relevant splicing factor. Biochem Biophys Res Commun. 2004;323:91–97. doi: 10.1016/j.bbrc.2004.08.060. [DOI] [PubMed] [Google Scholar]
- Nishizawa K, Freund C, Li J, Wagner G, Reinherz EL. Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc Natl Acad Sci U S A. 1998;95:14897–14902. doi: 10.1073/pnas.95.25.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoyatleva T, Tang Y, Rafalska I, Stamm S. Pre-mRNA missplicing as a cause of human disease. Prog Mol Subcell Biol. 2006;44:27–46. doi: 10.1007/978-3-540-34449-0_2. [DOI] [PubMed] [Google Scholar]
- O'Connor BP, Danhorn T, De Arras L, Flatley BR, Marcus RA, Farias-Hesson E, Leach SM, Alper S. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015;11:1004932. doi: 10.1371/journal.pgen.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29:909–917. doi: 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano L, Caira M. Risks for infection in patients with myelodysplasia and acute leukemia. Curr Opin Infect Dis. 2012;25:612–618. doi: 10.1097/QCO.0b013e328358b000. [DOI] [PubMed] [Google Scholar]
- Paine I, Posey JE, Grochowski CM, Jhangiani SN, Rosenheck S, Kleyner R, Marmorale T, Yoon M, Wang K, Robison R, et al. Paralog Studies Augment Gene Discovery: DDX and DHX Genes. Am J Hum Genet. 2019;105:302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternack SM, Refke M, Paknia E, Hennies HC, Franz T, Schafer N, Fryer A, van Steensel M, Sweeney E, Just M, et al. Mutations in SNRPE, which encodes a core protein of the spliceosome, cause autosomal-dominant hypotrichosis simplex. Am J Hum Genet. 2013;92:81–87. doi: 10.1016/j.ajhg.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Paz S, Ritchie A, Mauer C, Caputi M. The RNA binding protein SRSF1 is a master switch of gene expression and regulation in the immune system. Cytokine Growth Factor Rev. 2021;57:19–26. doi: 10.1016/j.cytogfr.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellagatti A, Boultwood J. The molecular pathogenesis of the myelodysplastic syndromes. Eur J Haematol. 2015;95:3–15. doi: 10.1111/ejh.12515. [DOI] [PubMed] [Google Scholar]
- Petit F, Escande F, Jourdain AS, Porchet N, Amiel J, Doray B, Delrue MA, Flori E, Kim CA, Marlin S, et al. Nager syndrome: confirmation of SF3B4 haploinsufficiency as the major cause. Clin Genet. 2014;86:246–251. doi: 10.1111/cge.12259. [DOI] [PubMed] [Google Scholar]
- Pollyea DA, Harris C, Rabe JL, Hedin BR, De Arras L, Katz S, Wheeler E, Bejar R, Walter MJ, Jordan CT, et al. Myelodysplastic syndrome-associated spliceosome gene mutations enhance innate immune signaling. Haematologica. 2019;104:e388–e392. doi: 10.3324/haematol.2018.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, Singh J, Padgett RA, Gu X, Phillips JG, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Wang F, Yu G, Wang D, Yao Y, You M, Liu J, Liu J, Sun Z, Ji C et al (2021) SRSF1 serves as a critical posttranscriptional regulator at the late stage of thymocyte development. Sci Adv 7:eabf0753 [DOI] [PMC free article] [PubMed]
- Quemener AM, Bachelot L, Forestier A, Donnou-Fournet E, Gilot D, Galibert MD. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip Rev RNA. 2020;11:1594. doi: 10.1002/wrna.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, Ramsay AJ, Bea S, Pinyol M, Martinez-Trillos A, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Krainer AR, Abdel-Wahab O. SnapShot: Splicing Alterations in Cancer. Cell. 2020;180:208–208. doi: 10.1016/j.cell.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber S, Stettler J, Filosa G, Colombo M, Jutzi D, Lenzken SC, Schweingruber C, Bruggmann R, Bachi A, Barabino SM, et al. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 2016;35:1504–1521. doi: 10.15252/embj.201593791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 2006;27:644–653. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzickova S, Stanek D. Mutations in spliceosomal proteins and retina degeneration. RNA Biol. 2017;14:544–552. doi: 10.1080/15476286.2016.1191735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Rebel VI. Acquired mutations that affect pre-mRNA splicing in hematologic malignancies and solid tumors. J Natl Cancer Inst. 2013;105:1540–1549. doi: 10.1093/jnci/djt257. [DOI] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyen E, Singh B, Minana B, Pages A, Mateo F, Pujana MA, Valcarcel J, Eyras E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016;26:732–744. doi: 10.1101/gr.199935.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JL, Murphy MS, Kantheti U, Burnett JM, Hahn MG, Dorrity TJ, Bacas CJ, Mattice EB, Corpuz KD, Barker BR. Polyglutamine binding protein 1 (PQBP1) inhibits innate immune responses to cytosolic DNA. Mol Immunol. 2018;99:182–190. doi: 10.1016/j.molimm.2018.05.014. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol. 2017;18:655–670. doi: 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- Shi Y. The spliceosome: a protein-directed metalloribozyme. J Mol Biol. 2017;429:2640–2653. doi: 10.1016/j.jmb.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, Ndonwi M, Wadugu B, Duncavage EJ, Okeyo-Owuor T, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak K, Jiang Z, Charloteaux B, Mayer A, Habraken Y, Tharun L, Klein S, Xu X, Duong HQ, Vislovukh A, et al. The X-linked trichothiodystrophy-causing gene RNF113A links the spliceosome to cell survival upon DNA damage. Nat Commun. 2020;11:1270. doi: 10.1038/s41467-020-15003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhar SK, Clark J, Gill S, Hamoudi R, Crew AJ, Gwilliam R, Ross M, Linehan WM, Birdsall S, Shipley J, et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–1338. doi: 10.1093/hmg/5.9.1333. [DOI] [PubMed] [Google Scholar]
- Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, Watabe K, Lu Z, Mo YY. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016;7:e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Choudhary GS, Pellagatti A, Choi K, Bolanos LC, Bhagat TD, Gordon-Mitchell S, Von Ahrens D, Pradhan K, Steeples V, et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat Cell Biol. 2019;21:640–650. doi: 10.1038/s41556-019-0314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitali P, Aartsma-Rus A. Splice modulating therapies for human disease. Cell. 2012;148:1085–1088. doi: 10.1016/j.cell.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starczynowski DT, Karsan A. Deregulation of innate immune signaling in myelodysplastic syndromes is associated with deletion of chromosome arm 5q. Cell Cycle. 2010;9:855–856. doi: 10.4161/cc.9.5.11156. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Karsan A. Innate immune signaling in the myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24:343–359. doi: 10.1016/j.hoc.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kumar SA, Shuai S, Diaz-Navarro A, Gutierrez-Fernandez A, De Antonellis P, Cavalli FMG, Juraschka K, Farooq H, Shibahara I, et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature. 2019;574:707–711. doi: 10.1038/s41586-019-1650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2011;88:643–649. doi: 10.1016/j.ajhg.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Thibault P, Abou Elela S, Klinck R, Berson EL, Chabot B, Rivolta C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Lee SC. Mutations in spliceosome genes and therapeutic opportunities in myeloid malignancies. Genes Chromosomes Cancer. 2019;58:889–902. doi: 10.1002/gcc.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale S, Lotti F, Saieva L, Van Meerbeke JP, Crawford TO, Sumner CJ, Mentis GZ, Pellizzoni L. SMN is essential for the biogenesis of U7 small nuclear ribonucleoprotein and 3'-end formation of histone mRNAs. Cell Rep. 2013;5:1187–1195. doi: 10.1016/j.celrep.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley M, Lynch D, Bernier F, Parboosingh J, Bhoj E, Zackai E, Calder A, Itasaki N, Wakeling E, Scott R, et al. Cerebro-costo-mandibular syndrome: Clinical, radiological, and genetic findings. Am J Med Genet A. 2016;170A:1115–1126. doi: 10.1002/ajmg.a.37587. [DOI] [PubMed] [Google Scholar]
- Towns KV, Kipioti A, Long V, McKibbin M, Maubaret C, Vaclavik V, Ehsani P, Springell K, Kamal M, Ramesar RS, et al. Prognosis for splicing factor PRPF8 retinitis pigmentosa, novel mutations and correlation between human and yeast phenotypes. Hum Mutat. 2010;31:E1361–1376. doi: 10.1002/humu.21236. [DOI] [PubMed] [Google Scholar]
- Tremblay N, Baril M, Chatel-Chaix L, Es-Saad S, Park AY, Koenekoop RK, Lamarre D. Spliceosome SNRNP200 promotes viral RNA sensing and IRF3 activation of antiviral response. PLoS Pathog. 2016;12:e1005772. doi: 10.1371/journal.ppat.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Niemela EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA. 2013;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Will CL, Grote M, Luhrmann R, Frilander MJ. The U11–48K protein contacts the 5' splice site of U12-type introns and the U11–59K protein. Mol Cell Biol. 2008;28:3548–3560. doi: 10.1128/MCB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Taga Y, Yoshimoto R, Mayeda A, Hattori S, Ogawa-Goto K. Component of splicing factor SF3b plays a key role in translational control of polyribosomes on the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2019;116:9340–9349. doi: 10.1073/pnas.1901742116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski LM, Leclair N, Anczukow O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA. 2018;9:1476. doi: 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, et al. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol. 2013;33:396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduci L, Simili M, Rizzo M, Mercatanti A, Evangelista M, Mariani L, Rainaldi G, Pitto L. MicroRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis. J Biol Chem. 2010;285:39551–39563. doi: 10.1074/jbc.M110.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A, Zeidler S, Breedveld G, Overbeek L, Huigh D, Koster L, van der Linde H, de Esch C, Severijnen LA, de Vries BBA, et al. CXorf56, a dendritic neuronal protein, identified as a new candidate gene for X-linked intellectual disability. Eur J Hum Genet. 2018;26:552–560. doi: 10.1038/s41431-017-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma B, Akinyi MV, Norppa AJ, Frilander MJ. Minor spliceosome and disease. Semin Cell Dev Biol. 2018;79:103–112. doi: 10.1016/j.semcdb.2017.09.036. [DOI] [PubMed] [Google Scholar]
- Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, O'Keefe C, Rogers HJ, Sekeres MA, Maciejewski JP, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- Visconte V, Rogers HJ, Singh J, Barnard J, Bupathi M, Traina F, McMahon J, Makishima H, Szpurka H, Jankowska A, et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 2012;120:3173–3186. doi: 10.1182/blood-2012-05-430876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8:375–381. doi: 10.1016/S1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Shionyu M, Kimura T, Kimata K, Watanabe H. Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. J Biol Chem. 2007;282:20728–20738. doi: 10.1074/jbc.M703292200. [DOI] [PubMed] [Google Scholar]
- Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, Abaan OD, Davis SR, Kreitman RJ, Meltzer PS. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D, Gener B, Gonzalez MJ, Seland S, Fischer S, Hehr U, Kuechler A, Hoefsloot LH, de Leeuw N, Gillessen-Kaesbach G, et al. Microcephaly, microtia, preauricular tags, choanal atresia and developmental delay in three unrelated patients: a mandibulofacial dysostosis distinct from Treacher Collins syndrome. Am J Med Genet A. 2009;149A:837–843. doi: 10.1002/ajmg.a.32747. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Newman WG, Wieland T, Berulava T, Kaffe M, Falkenstein D, Beetz C, Graf E, Schwarzmayr T, Douzgou S, et al. Compound heterozygosity of low-frequency promoter deletions and rare loss-of-function mutations in TXNL4A causes Burn-McKeown syndrome. Am J Hum Genet. 2014;95:698–707. doi: 10.1016/j.ajhg.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7):a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R (2006) Spliceosome structure and function. In The RNA World, Gesteland RF et al, eds. (Cold Spring Harbor, New York, Cold Spring Harbor Laboratory Press), pp. 369–400
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Xiong F, Liu HH, Duan CY, Zhang BK, Wei G, Zhang Y, Li S. Arabidopsis JANUS Regulates Embryonic Pattern Formation through Pol II-Mediated Transcription of WOX2 and PIN7. iScience. 2019;19:1179–1188. doi: 10.1016/j.isci.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Xie YA, Abouzeid H, Gordon CT, Fiorentino A, Sun Z, Lehman A, Osman IS, Dharmat R, Riveiro-Alvarez R, et al. Mutations in the spliceosome component CWC27 cause retinal degeneration with or without additional developmental anomalies. Am J Hum Genet. 2017;100:592–604. doi: 10.1016/j.ajhg.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip BH, Dolatshad H, Roy S, Pellagatti A, Boultwood J. Impact of splicing factor mutations on pre-mrna splicing in the myelodysplastic syndromes. Curr Pharm Des. 2016;22:2333–2344. doi: 10.2174/1381612822666160226132112. [DOI] [PubMed] [Google Scholar]
- Yoh SM, Schneider M, Seifried J, Soonthornvacharin S, Akleh RE, Olivieri KC, De Jesus PD, Ruan C, de Castro E, Ruiz PA, et al. PQBP1 Is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell. 2015;161:1293–1305. doi: 10.1016/j.cell.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ogawa S. Splicing factor mutations and cancer. Wiley Interdiscip Rev RNA. 2014;5:445–459. doi: 10.1002/wrna.1222. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kim JH, Carver K, Su Y, Weremowicz S, Mulvey L, Yamamoto S, Brennan C, Mei S, Long H, et al. CLK2 Is an Oncogenic Kinase and Splicing Regulator in Breast Cancer. Cancer Res. 2015;75:1516–1526. doi: 10.1158/0008-5472.CAN-14-2443. [DOI] [PubMed] [Google Scholar]
- Zhang D, Hu Q, Liu X, Ji Y, Chao HP, Liu Y, Tracz A, Kirk J, Buonamici S, Zhu P, et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat Commun. 2020;11:2089. doi: 10.1038/s41467-020-15815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yue T, Choi JH, Nair-Gill E, Zhong X, Wang KW, Zhan X, Li X, Choi M, Tang M, et al. Syndromic immune disorder caused by a viable hypomorphic allele of spliceosome component Snrnp40. Nat Immunol. 2019;20:1322–1334. doi: 10.1038/s41590-019-0464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, Rabadan R, Raza A, Mukherjee S, Manley JL. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112:E4726–4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wei JS, Li SQ, Badgett TC, Song YK, Agarwal S, Coarfa C, Tolman C, Hurd L, Liao H, et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett. 2016;371:214–224. doi: 10.1016/j.canlet.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, Hricik T, Heguy A, Hedvat C, Gonen M, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou N, Huang J, Ho TT, Zhu Z, Qiu Z, Zhou X, Bai C, Wu F, Xu M, et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget. 2016;7:15481–15491. doi: 10.18632/oncotarget.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Lu S, Zhou X, Zhang X, Zhao K, Larsson C. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet. 2006;119:617–623. doi: 10.1007/s00439-006-0168-3. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]