Abstract
Eutypa lata is the causal fungal agent of Eutypa dieback, a serious grapevine necrotic disease. The erratic and delayed (1 to 2 months) appearance of characteristic conidia on culture media and the presence of numerous microorganisms in decaying wood make it difficult either to identify or to detect E. lata in grapevine wood samples. We designed six pairs of PCR primers for diagnosis of E. lata. Three primer pairs were derived from ribosomal DNA internal transcribed spacer sequences, and three pairs were derived from randomly amplified polymorphic DNA fragments. The six primer pairs could be used to amplify DNAs extracted from all of the E. lata isolates tested. They did not amplify DNAs from fungi and bacteria representing more than 50 different species of microorganisms associated with grapevine. We developed a simple protocol, leading to a rapid release of DNA, that enabled us to identify E. lata from pure or mixed cultures as well as from grapevine wood samples. Identification of E. lata in wood was achieved within a few hours, instead of the several weeks required for classical cultures on agar medium. We believe that the procedure described here can be adapted to detect other microorganisms involved in woody plant diseases.
Eutypa lata (Pers.: Fr.) Tul et C. Tul. (synonym E. armeniacae Hansf. and M. V. Carter; anamorph Libertella blepharis A. L. Smith) is a worldwide ascomycete pathogen of many perennial plants (3, 7). In grapevine (Vitis vinifera L.), E. lata causes a serious disease named Eutypa dieback that may lead to significant reductions in yield (21). The fungus is also found in vines suffering from the black measles disease, where it acts as a pioneering fungus (16, 20). Eutypa dieback is a chronic disease that develops extremely slowly (4) and has highly variable symptom expression, making diagnosis in vineyards difficult (18). The first noticeable symptoms are stunted spring shoots with small, chlorotic, cupped leaves. These symptoms may be due to phytotoxic compounds produced by the mycelium in the older wood (11, 17). Symptoms appear in a vineyard several years after infection, and their severity is influenced by numerous factors (22). In the wood, infection results in a brown lesion which appears as sectors in cross sections of diseased vines (18). Control is based on the protection of pruning wounds and on the removal of infected parts of the vines (5).
E. lata is usually identified by traditional methods of isolation and culturing (5). E. lata does not produce sexual structures in artificial media, so identification depends primarily on the morphology of the mycelium (5, 6), although serological techniques have also been developed (12, 29). Observation of conidia is often necessary to confirm the identification of E. lata, but their production requires at least 1 to 2 months and they are not produced by all isolates (5). The growth of E. lata on culture media may be slower than that of other saprophytic microorganisms present in grapevine wood. Thus, the fungus may go unobserved on isolation plates, leading to an erroneous diagnosis. In some cases, determining if the fungus is present requires careful subcultures, which is extremely time-consuming and tedious.
PCR with species-specific primers can provide an easy way to detect fungal plant pathogens both in culture and in plant tissues. Assays may be based on amplification of a ribosomal DNA (rDNA) region encompassing internal transcribed spacers (ITS) and 5.8S rDNA (10, 13, 19, 32, 35). The use of PCR primers derived from the sequences of randomly amplified polymorphic DNA (RAPD) fragments was also fruitful (14, 23, 31). Our objective was to develop a reliable assay, based on PCR, for a quick and accurate diagnosis of E. lata. For this purpose, we designed and tested six primer pairs to identify E. lata cultures. Three primer pairs were derived from E. lata rDNA sequences, and three were derived from sequences of RAPD fragments obtained in a previous study (26). Two primer pairs, one per approach, were used to detect the fungus directly in the wood of diseased grapevines.
MATERIALS AND METHODS
Isolates of E. lata.
We used 60 E. lata isolates that originated from different geographic locations and that were obtained from the wood or stromata of grapevines (Table 1). Isolates were stored long-term in distilled water at 4°C, as previously described (25). Some cultures were initiated by transferring mycelial plugs on malt-agar (MA) medium (15 g of Cristomalt [Materne, Fruibourg, France] per liter, 20 g of agar per liter) to petri dishes. Plates were incubated at 22°C for 1 week. About 40 mg of fresh mycelium was harvested per isolate by scraping the surface of the agar with an inoculation loop and transferred to a microcentrifuge tube (2 ml). Cultures were also grown on potato dextrose agar medium (39 g/liter; Difco Laboratories, Detroit, Mich.), from which liquid cultures were initiated to produce mycelia according to the method of Péros et al. (28). Mycelium samples were immediately freeze-dried and stored at 4°C until DNA was extracted.
TABLE 1.
Origins of the 60 isolates of E. lata used in this study
| Isolatea | Geographical origin (country, region) | Yr of isolation | Grape cultivar |
|---|---|---|---|
| BX 1-10b | France, Bordeaux | 1990 | Cabernet Sauvignon |
| 8Db | France | 1991 | Unknown |
| 8Fb | Italy, Verona | 1988 | Unknown |
| Fronsac 88b | France, Fronsac | 1988 | Unknown |
| 91-P-55b | France | 1991 | Unknown |
| 91-P-80b | France | 1991 | Unknown |
| AL2b | France, Traenheim | 1996 | Gewurztraminer |
| SO 14b | France, Saint-Emilion | 1996 | Cabernet Franc |
| SO 22b | France, Montbazillac | 1996 | Sauvignon |
| SO 41b | France, Jurançon | 1996 | Gros Manseng |
| CO 15b | France, St. Laurent de Cognac | 1996 | Ugni Blanc |
| CO 23b | France, Pouillac | 1996 | Ugni Blanc |
| 42-2b | France, Bergerac | 1996 | Unknown |
| PL6b | France, Illats | 1998 | Unknown |
| T-96-1b | Tunisia, Kelibia | 1996 | Muscat d'Italie |
| BE 236 | France, Beauvoisin | 1996 | Carignan |
| AN 238 | France, Antugnac | 1996 | Mauzac |
| LP 388 | France, Les Touches de Périgny | 1996 | Ugni Blanc |
| PE 390 | France, Pérignac | 1996 | Ugni Blanc |
| LC 405 | France, St. Laurent de Cognac | 1996 | Ugni Blanc |
| MT 247 | France, Montreal | 1996 | Cinsaut |
| AS 250 | France, Assignan | 1996 | Grenache |
| LY 252 | France, Leyrac | 1996 | Cinsaut |
| ME 256 | France, Monteils | 1996 | Cinsaut |
| CC 260 | France, Cannes-Clairan | 1996 | Carignan |
| PI 266 | France, Pinet | 1996 | Cinsaut |
| SR 270 | France, St. Christol | 1996 | Grenache |
| BC 279 | France, Beaucaire | 1996 | Cabernet |
| MO 393 | France, Montguyon | 1997 | Ugni Blanc |
| RQ 461 | France, Riquewihr | 1997 | Chasselas |
| GW 463 | France, Gueberschwihr | 1997 | Chasselas |
| OW 465 | France, Obermorschwihr | 1997 | Gewurztraminer |
| OI 467 | France, Ollwiller | 1997 | Pinot |
| BH 470 | France, Bergheim | 1997 | Riesling |
| WZ 471 | France, Wintzenheim | 1997 | Pinot |
| FH 473 | France, Furdenheim | 1997 | Gewurztraminer |
| BV 474 | France, Benardville | 1997 | Auxerrois |
| OB 477 | France, Obernai | 1997 | Riesling |
| WG 479 | France, Wangen | 1997 | Sylvaner |
| GZ 394 | France, Gémozac | 1997 | Ugni Blanc |
| SF 396 | France, St. Ford sur Gironde | 1997 | Ugni Blanc |
| SX 397 | France, Sablonceaux | 1997 | Ugni Blanc |
| CY 398 | France, Le Chay | 1997 | Ugni Blanc |
| SI 400 | France, Saintes | 1997 | Ugni Blanc |
| JO 402 | France, Jonzac | 1997 | Ugni Blanc |
| BI 324 | Italy, Broni | 1995 | Merlot |
| BI 325 | Italy, Broni | 1995 | Trebbiano Toscano |
| PG 343 | Italy, Padenghe | 1995 | Trebbiano di Lugana |
| SB 359 | Italy, S. Benedetto | 1995 | Trebbiano Toscano |
| IM 334 | Italy, Imola | 1996 | Sangiovese |
| LZ 338 | Italy, Lazise | 1996 | Corvina |
| LZ 340 | Italy, Lazise | 1996 | Corvina |
| SG 368 | Italy, S. Giorgio | 1996 | Trebbiano di Lugana |
| SP 372 | Italy, Sommacampagna | 1996 | Trebbiano Toscano |
| TG 374 | Italy, Teglio | 1996 | Chiavennasca |
| PP 496 | Spain, Pacs del Penedes | 1999 | Cabernet Sauvignon |
| PP 497 | Spain, Pacs del Penedes | 1999 | Cabernet Sauvignon |
| PP 498 | Spain, Pacs del Penedes | 1999 | Cabernet Sauvignon |
| IR 512 | Spain, Santa Maria de Mirallés | 1999 | Sauvignon |
| IR 513 | Spain, Santa Maria de Mirallés | 1999 | Sauvignon |
E. lata isolates are available from us upon request.
Isolates used for testing all six PCR primer pairs listed in Table 3.
Microorganisms associated with grapevine.
To verify that PCR primers specifically amplified E. lata DNA, we tested a total of 100 isolates of microorganisms representing more than 50 species of fungi and bacteria (Table 2). All fungal isolates, except isolates of Agaricus bisporus and Armillaria mellea, were collected from grapevines. They either originated from local collections or were isolated in 1998 from grapevine cane surfaces, pruning wounds, pieces of wood, or cuttings. Fungi were grown on MA medium, and bacteria were grown on King's B medium (15). Most bacterial species obtained from the French Collection of Phytopathogenic Bacteria (Institut National de la Recherche Agronomique [INRA], Angers, France) were chosen to be representative of bacterial species that have been previously detected in grapevine (2).
TABLE 2.
Fungal and bacterial species used to verify the specificities of the PCR primers
| Species | No. of isolates tested | Origin(s)a |
|---|---|---|
| Fungi | ||
| Alternaria sp. | 2 | Collection and isolation |
| Agaricus bisporus | 3 | Collection |
| Armillaria mellea | 1 | Collection |
| Aspergillus sp. | 1 | Collection |
| Botrytis cinerea | 10 | Isolation |
| Chaetomium sp. | 3 | Collection and isolation |
| Cladosporium sp. | 1 | Collection |
| Colletotrichum sp. | 1 | Collection |
| Coniothyrium sp. | 2 | Collection and isolation |
| Cytospora sp. | 1 | Collection |
| Epicoccum sp. | 4 | Isolation |
| Fusarium sp. | 4 | Collection and isolation |
| Geotrichum sp. | 1 | Collection |
| Gliocladium sp. | 7 | Collection and isolation |
| Gloesporium ampelophagum | 1 | Collection |
| Guignardia bidwellii | 2 | Collection |
| Nigrospora sp. | 1 | Collection |
| Papularia sp. | 1 | Collection |
| Penicillium sp. | 1 | Collection |
| Pestalotia sp. | 1 | Isolation |
| Phaeoacremonium chlamydosporum | 1 | Collection |
| Phaeoacremonium aleophilum | 1 | Collection |
| Phaeoacremonium sp. | 1 | Collection |
| Phellinus sp. | 3 | Collection |
| Phoma diplodiella | 1 | Collection |
| Phomopsis viticola | 3 | Collection and isolation |
| Plasmopara viticola | 2 | Collection |
| Pullularia pullulans | 2 | Collection |
| Rhodotorula sp. | 1 | Isolation |
| Rhizopus sp. | 1 | Collection |
| Sphaeropsis sp. | 2 | Isolation |
| Stereum hirsutum | 1 | Collection |
| Trichoderma sp. | 1 | Isolation |
| Tritirachium sp. | 1 | Collection |
| Ulocladium atrum | 2 | Collection |
| Uncinula necator | 2 | Collection |
| Bacteria | ||
| Bacillus subtilis | 1 | CFBP 491 |
| Clavibacter michiganensis | 1 | CFBP 2492 |
| Curtobacterium flaccumflasciens pv. flaccumflasciens | 1 | CFBP 3418 |
| Enterobacter agglomerans | 2 | CFBP 1189, CFBP 3845 |
| Enterobacter agglomerans | 2 | Isolation |
| Pseudomonas aeruginosa | 1 | Isolation |
| Pseudomonas cepacia | 2 | Isolation |
| Pseudomonas cichorii | 1 | CFBP 2101 |
| Pseudomonas corrugata | 1 | CFBP 2431 |
| Pseudomonas fluorescens | 3 | Isolation |
| Pseudomonas fluorescens bv. 1 | 1 | CFBP 2123 |
| Pseudomonas putida | 1 | CFBP 2066 |
| Pseudomonas sp. | 4 | Isolation |
| Pseudomonas syringae pv. syringae | 1 | CFBP 3069 |
| Rhodococcus fasciens | 1 | CFBP 2401 |
| Xanthomonas campestris pv. dieffenbachiae | 1 | CFBP 2482 |
| Unidentified | 3 |
Isolates originated either from isolations specially made for this study in 1998 from grapevine material or from different collections, namely, local collections (Bordeaux and Montpellier, France) for fungal isolates (not numbered) and the French Collection of Phytopathogenic Bacteria (INRA, Centre de Recherches d'Angers, Beaucouzé Cédex, France) (numbered bacterial strains). Except for the last-named strains, all isolates are available from us upon request.
DNA extraction.
Fungal DNA was obtained from freeze-dried material according to the method of either Délye et al. (8) or Péros et al. (28). Bacterial DNA was obtained by heating 50-μl aliquots of dense cell suspensions (108 bacteria per ml) at 95°C for 10 min in sterile water. Samples were kept at −20°C until they were used for PCR amplifications.
Primer design.
DNA of E. lata isolate BX 1-10 was amplified by using the universal primers ITS1 and ITS4 (34) at a final concentration of 0.2 μM each. The amplification mix and cycling programs were as previously described (8), except that the annealing temperature was 65°C. The sequence of the PCR product was determined by Eurogentec SA (Seraing, Belgium). Searches for the most closely related sequences were carried out with the EMBL and GenBank databases using the BLAST program (1, 24). We designed three oligonucleotide primer pairs (Table 3) targeting E. lata rDNA and sharing little or no homology with other known rDNA sequences. Primers were synthesized by Eurogentec SA. Three other primer pairs were designed from the sequences of the variable RAPD fragments A10-410, B02-830, and D18-530, obtained in a previous work (26), using the PRIME procedure of the Genetics Computer Group software package (9). These primers were synthesized by Life Technologies (Eragny, France).
TABLE 3.
Primers pairs designed for specific amplification of E. lata DNA
| Primer pair | Sequence of primer (5′-3′) | Genomic origin | Expected PCR product size (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| SCA 10A | TAGTGGTGTCAGTGAAAGG | RAPD fragment A10-410a | 350 | 60 |
| SCA 10B | GTGCTAAAGCTTAAAATCCC | RAPD fragment A10-410 | 350 | 60 |
| SCB 02A | AATCGATGTGAGAGATGG | RAPD fragment B02-830a | ||
| SCB 02B | AGGTCAATGATAGCCAAC | RAPD fragment B02-830 | 700 | 60 |
| SCD 18A | GAGTACGTTGGTACAATGG | RAPD fragment D18-530a | ||
| SCD 18B | ACTCTCTCTCGTCTTTTGC | RAPD fragment D18-530 | 450 | 60 |
| Lata 1 | GAGCTACCCTGTAGCCCGCTG | rDNA ITS1 regionb | ||
| Lata 2-1 | CTATCCGGAGATAGGCTCCC | rDNA ITS2 regionb | 302 | 65 |
| Lata 1 | GAGCTACCCTGTAGCCCGCTG | rDNA ITS1 region | ||
| Lata 2-2 | GACGTCAGCCGTGACACACC | rDNA ITS2 region | 385 | 65 |
| Lata 3 | GCCTACCCGCCGGTGGACAC | rDNA ITS1 region | ||
| Lata 2-1 | CTATCCGGAGATAGGCTCCC | rDNA ITS2 region | 281 | 65 |
PCRs.
All PCR amplifications were performed in 25-μl mixes, as previously described (8). Primers were used at a final concentration of 0.2 μM each. The cycling programs consisted of 37 cycles with a 30-s denaturation at 94°C, a 30-s annealing at a specific temperature (Table 3), and a 1-min extension at 72°C. The suitability of each fungal DNA sample for PCR was checked with the universal primers ITS1 and ITS4 (34) at an annealing temperature of 55°C. Aliquots of 5 μl from heated bacterial suspensions were checked using the universal bacterial primers FD1 and RP2 (33) at an annealing temperature of 45°C. Amplified DNA fragments were visualized under UV light after electrophoresis at 100 V on 1% (wt/vol) agarose gels stained with ethidium bromide (0.8 μg/ml) and run in 0.5× Tris-borate-EDTA buffer (Euromédex, Souffelweyersheim, France). Positive and negative amplification controls were included in each experiment.
Rapid recovery of DNA for identification of E. lata.
To identify E. lata from pure or mixed cultures after 1 week of incubation, approximately 0.1 mg (fresh weight) of fungal material was collected by gently scraping the surface of culture with a sterilized loop and placed into 0.5-ml microcentrifuge tubes containing 50 μl of sterile, deionized water. Samples were incubated at 95°C for 15 min and immediately placed on ice. Five-microliter aliquots of the resulting supernatants were used for PCRs. To ensure that E. lata DNA could be readily amplified from cultures where other microorganisms dominated, 45 samples of about 0.1 mg of fresh mycelial material, collected from E. lata cultures highly contaminated by saprophytic microorganisms after routine isolation from grapevine lesions, were analyzed using the primer pairs Lata 1 and Lata 2.2 and SCA 10A and SCA 10B.
Detection of E. lata from grapevine wood.
Eighteen grape plants were collected during the summer of 1999, and cross sections were made in woody parts to look for characteristic brown lesions (5). After surface sterilization by rapid flaming, a piece of necrotic wood, 1 by 2 by 1 cm in size, was taken from close to the margin of the lesion by using pruning shears. Both culturing followed by PCR identification of E. lata and direct detection of E. lata in wood were used to analyze the necrotic wood samples obtained. For culturing of E. lata, 20 small wood chips, 3 by 5 by 5 mm in size, were sliced from a piece of necrotic wood, disinfected in calcium hypochlorite solution (1.8% active chlorine), placed onto MA medium in petri dishes, and incubated for 1 week at 23°C. Plates were assessed visually for the presence of typical mycelia. From each piece of necrotic wood examined, a mixture of mycelia growing on MA medium was analyzed with the PCR primer pair Lata 1 and Lata 2.2.
To detect E. lata directly in the grapevine wood tissues, we cut three thin wood shavings, about 5 by 5 by 0.2 mm in size, from the piece of wood using a sterile scalpel (no. 22; Swann Morton, Sheffield, England) and placed them individually into 1.5-ml microcentrifuge tubes. Samples were incubated at 95°C for 15 min after the addition of 50 μl of sterile, deionized water, and immediately placed on ice. Five microliters of 1:10 and 1:100 dilutions of the resulting supernatant were used for PCR analysis using the Lata 1-Lata 2.2 and SCA 10A-SCA 10B primer pairs.
RESULTS
Primer design for E. lata rDNA regions.
We sequenced a 574-bp-long DNA fragment encompassing E. lata ITS1, ITS2, and the 5.8S rDNA (GenBank accession number AF099911). This sequence did not differ from the sequences of two other strains of this fungus (J. Mugnier, personal communication). The highest similarity (91% identity) was found with the corresponding rDNA regions of the fungus Xylaria cubensis (GenBank accession number AF163032). Primers targeting E. lata ITS regions (Table 3) were designed so that the last four bases at their 3′ termini hybridized with regions specific to the E. lata rDNA sequence.
Primer design for RAPD fragments.
Sequenced RAPD fragments, A10-410, B02-830, and D18-530, were 410, 830, and 530 bp long, respectively. The PRIME analysis identified only one pair of primers for fragment A10-410. We chose one pair of primers among the 15 and one among the 6 proposed for fragments B02-830 and D18-530, respectively.
PCR amplifications.
Optimal annealing temperatures were 65°C for all primer pairs targeting rDNA and 60°C for all primer pairs derived from RAPD fragments. All primer pairs yielded a single amplicon of the expected size (Fig. 1) when they were used for PCR with DNAs extracted from 15 E. lata isolates (Table 1). The Lata 1-Lata 2.2 and SCA 10A-SCA 10B primer pairs yielded a single amplicon of the predicted size for 45 more E. lata isolates (Table 1).
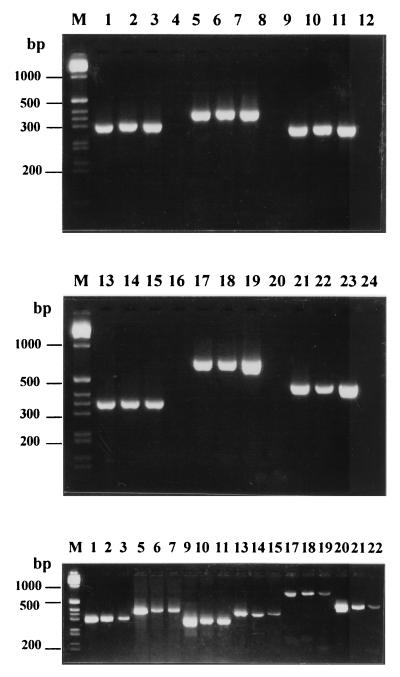
FIG. 1.
PCR products obtained with DNA extracted from E. lata according to a protocol described in reference 8 (top and middle) or by boiling (bottom) were from the following isolates: BX 1-10 (lanes 1, 5, and 9), 8D (lanes 2, 6, and 10), and 8F (lanes 3, 7, and 11). Lanes 4, 8, 12, 16, 20, and 24, H2O negative control (no DNA). Primers used were Lata 1 and Lata 2.1 (lanes 1 to 4), Lata 1 and Lata 2.2 (lanes 5 to 8), Lata 3 and Lata 2.1. (lanes 9 to 12), SCA 10A and SCA 10B (lanes 13 to 16), SCB 02A and SCB 02B (lanes 17 to 20), and SCD 18A and SCD 18B (lanes 21 to 24). Lane M, molecular weight markers (1-kb DNA ladder; Life Technologies).
Specificities of primer pairs.
None of the six primer pairs amplified DNA from any of 100 isolates of microorganisms tested (Table 2). DNA could be amplified from all these samples when we used fungal or bacterial universal primers, with these primers thus serving as positive controls and demonstrating the presence of DNA suitable for PCR (not shown).
Identification of E. lata from crude mycelium extract.
All six primer pairs yielded single PCR products of the expected sizes when E. lata crude DNA was used as the template in the PCR mix (Fig. 1). All 45 mixed mycelial samples, collected from highly contaminated E. lata cultures, also yielded only the expected amplification products.
Double-blind test.
A double-blind test was performed to confirm these results. MA medium cultures were initiated from diseased grapevine wood or from pruning wound material. After 1 week of incubation, 50 mycelial samples were visually assigned to three classes (Table 4): mycelium resembling E. lata (“Ela”), mycelium that might or might not be E. lata (“doubtful”), and mycelium not resembling E. lata (“other”). These samples were subcultured and identified by visual assessment and independently characterized by PCR using the primer pairs Lata 1 and Lata 2.2 and SCA 10A and SCA 10B. Identification using subculturing and PCR were in agreement for all 18 Ela samples and all 17 doubtful samples (Table 4). Both PCR and subculturing identified 2 out of the 18 Ela samples as false positives. None of the 15 samples classified as other was identified as E. lata by subculturing. But E. lata was detected by PCR in two samples classified as other. Subsequent culturing confirmed that these two samples were indeed mixed samples containing minute amounts of E. lata.
TABLE 4.
Double-blind identification of E. lata in 50 mycelium subcultures
| Classa | No. of samples | No. identified as E. lata
|
|||
|---|---|---|---|---|---|
| Visually in subcultures
|
Using PCR
|
||||
| + | − | + | − | ||
| Ela | 18 | 16 | 2 | 16 | 2 |
| Doubtful | 17 | 0 | 17 | 0 | 17 |
| Other | 15 | 0 | 15 | 2 | 13 |
Based on visual assessment in bulk cultures prior to subculturing. Ela, mycelium visually identified as E. lata; doubtful, whitish mycelium resembling E. lata; other, mycelium not resembling E. lata.
Detection of E. lata in wood samples.
Wood chips, placed on MA medium, developed a whitish mycelium identified as E. lata by visual assessment of 17 of 18 grapevine lesions investigated (Table 5). However, mycelial growth of E. lata was not observed from more than half of the wood chips examined from five grapevines (grapevines 13 to 17). No E. lata mycelial growth was clearly observed from wood chips from grapevine 18. PCR analysis of mycelial mixtures collected from one wood chip per lesion, however, showed that E. lata was present in all 18 grapevines investigated. Direct PCR analysis of thin wood shavings, using primer pairs Lata 1 and Lata 2.2 and SCA 10A and SCA 10B, indicated the presence of E. lata in all 18 grapevine lesions examined (Fig. 2). The rate of successful detection of E. lata depended upon the primer pair and the dilution used. For grapevines from which E. lata grew from less than half of the wood chips (grapevines 13 to 18) (Table 5), the frequency of successful amplifications of E. lata from wood shavings was higher when we used the rDNA-targeting primers Lata 1 and Lata 2.2. With both primer pairs, more positive results were obtained with 1:10-diluted templates than with 1:100-diluted templates.
TABLE 5.
E. lata diagnosis of 18 diseased grapevines: comparison of results of isolation, PCR identification from crude mycelium after isolation, and direct PCR detection from wood material
| Grapevine | Visual assessment of E lata myceliuma | PCR identification from culturesb | Direct PCR detection from wood shavings with the primer pairc:
|
|||
|---|---|---|---|---|---|---|
| Lata 1- Lata 2.2
|
SCA 10A- SCA 10B
|
|||||
| 1/10 dilution | 1/100 dilution | 1/10 dilution | 1/100 dilution | |||
| 1 | 14 | 1 | 3 | 3 | 3 | 3 |
| 2 | 12 | 1 | 3 | 3 | 3 | 3 |
| 3 | 16 | 1 | 3 | 3 | 3 | 3 |
| 4 | 15 | 1 | 3 | 3 | 3 | 3 |
| 5 | 15 | 1 | 3 | 3 | 3 | 3 |
| 6 | 18 | 1 | 3 | 3 | 3 | 3 |
| 7 | 15 | 1 | 3 | 3 | 3 | 3 |
| 8 | 15 | 1 | 3 | 3 | 3 | 2 |
| 9 | 16 | 1 | 3 | 3 | 3 | 2 |
| 10 | 19 | 1 | 3 | 3 | 3 | 2 |
| 11 | 18 | 1 | 3 | 3 | 3 | 2 |
| 12 | 14 | 1 | 3 | 3 | 2 | 2 |
| 13 | 9 | 1 | 3 | 3 | 2 | 1 |
| 14 | 7 | 1 | 3 | 3 | 2 | 1 |
| 15 | 6 | 1 | 3 | 1 | 1 | 1 |
| 16 | 5 | 1 | 3 | 1 | 1 | 1 |
| 17 | 3 | 1 | 3 | 1 | 1 | 0 |
| 18 | 0 | 1 | 2 | 0 | 1 | 0 |
Number of wood chips from which the development of E. lata was observed per 20 wood chips deposited onto culture medium.
Number of mycelial samples where E. lata was identified per one sample collected from the culture medium per lesion.
Number of wood shavings where E. lata was detected per three wood shavings analyzed.
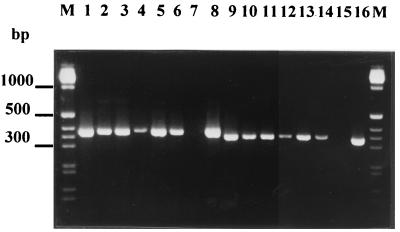
FIG. 2.
PCR detection of E. lata in wood samples from necrotic lesions. Amplifications were performed from 5-μl aliquots of 10-fold and 100-fold dilutions of supernatants obtained after boiling three wood shavings separately in sterile water (lanes 1 to 2 and 9 to 10, lanes 3 to 4 and 11 to 12, and lanes 5 to 6 and 13 to 14; odd-numbered lanes, 10-fold dilutions; even-numbered lanes, 100-fold dilutions). Primers used were Lata 1 and Lata 2.2 (lanes 1 to 8) and SCA 10A and SCA 10B (lanes 9 to 16). Lanes 7 and 15, H2O negative control (no DNA); lanes 8 and 16, E. lata positive control (DNA extracted from isolate BX 1-10); lane M, molecular weight markers (1-kb DNA ladder; Life Technologies).
DISCUSSION
Our objective was to develop PCR assays for rapid identification and detection of E. lata from grapevine wood. We used two primer design approaches and identified a set of three primer pairs based on E. lata rDNA sequences and another set of three primer pairs from RAPD fragment sequences. The Lata 1-Lata 2.2 and SCA 10A-SCA 10B primer pairs could amplify DNA from all 60 E. lata isolates tested, and none of the six primer pairs amplified DNA from any of the 96 isolates of microorganisms associated with grapevine. Consequently all six PCR primer pairs are potentially useful for specific identification of E. lata.
From mycelium cultures, our PCR assay enables, in less than 5 h, unambiguous identification of E. lata among the whitish mycelia obtained from wood chips deposited, thus avoiding cumbersome subcultures of the fungus and the need for visual comparison with reference cultures. PCR-assisted identification of E. lata cultures may be performed as soon as there is sufficient fungal material on MA medium (3 to 7 days) without the need to wait for the uncertain formation of characteristic conidia (1 to 2 months). No specialized mycological training is required, and the reagents and equipment needed are common in most molecular biology laboratories.
From diseased lesions, conventional culture-based diagnosis generally requires the excision of 10 to 20 small wood chips and 1 to 2 months of subculture. Using our PCR-based protocol, only 1 day and at most three wood samples are required for E. lata diagnosis. We detected E. lata in wood samples by using 1:10 and 1:100 dilutions of supernatants obtained by boiling thin wood shavings. This success may be due to the relative lack of PCR inhibitors released from necrotic wood tissues and/or to the dilution of extraction material (36). Primer pair Lata 1 and Lata 2.2 was more efficient than primer pair SCA 10A and SCA 10B (Table 5) in detecting E. lata in wood samples that were probably less heavily colonized. This difference may be due to the high copy number of rDNA targeted by primers Lata 1 and Lata 2.2 (34).
Most PCR procedures used for diagnostic purposes are based upon lengthy extraction protocols that include steps with phenol or other specific buffers (see, e.g., references 10, 14, 19, and 33). Successful amplication of DNA recovered from boiled mycelium has already been reported by others (13, 30). However, no PCR assay enabling the detection of microorganisms directly in woody plant material has yet been reported. Quick diagnosis of E. lata in grapevine necrotic tissues will be particularly useful in identifying the origin of dieback of vines that do not show characteristic symptoms of Eutypa dieback during the growing season. Rapid methods to detect E. lata from grapevine wood material also will be of great use in the study of the epidemiology of E. lata disease (e.g., wound receptivity). Further work is necessary to verify that our PCR detection assay can reliably detect the fungus in nonnecrotic material. If so, then PCR detection of E. lata could be used to monitor the development and to trace the movement of the fungus in grapevine tissues, to assess infections in situ after a short incubation period, or to compare either the levels of pathogenicity of isolates or the levels of resistance of cultivars. Our PCR assay may also be adapted to study the nature and the complexity of the fungal community present in the decaying wood of grapevine (27). Similar procedures may also be used to detect and/or study microorganisms associated with wood cankers or diseases in other perennials.
In conclusion, the assays we developed are specific and do not require complicated preparation of samples for either diagnostic cultures or DNA extraction and purification. Thus, our assays are significantly faster than other methods used to date and can be combined with conventional isolation procedures to contribute to a robust diagnosis of E. lata. We are confident that PCR-based detection procedures derived from the one described here will be helpful to study microflora present within woody plant parts.
ACKNOWLEDGMENTS
This work was supported in part by grants from the European Union (FAIR contract no. 8001 CT95-0654) and from the Conseil Interprofessionnel du Vin de Bordeaux (CIVB).
We thank F. Jailloux, P. Larignon, B. Lung, and J. Roudet (INRA, URSV, Villenave d'Ornon, France) for providing isolates of fungi from their collections; L. Gardan and S. Belouin (INRA, Pathologie, Angers, France) for donating bacterial strains; J. Mugnier for supplying partial E. lata rDNA sequences; S. Giry Laterrière for participating in the blind test; and F. Lahogue, D. Roche, E. Laveau, C. Leppert, and J. M. Limiñana for technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bell C R, Dickie G A, Harvey W L G, Chan J W Y F. Endophytic bacteria in grapevine. Can J Microbiol. 1995;41:46–53. [Google Scholar]
- 3.Bolay A, Carter M V. Newly recorded hosts of Eutypa lata (=E. armeniacae) in Australia. Plant Prot Q. 1985;1:10–12. [Google Scholar]
- 4.Carter M V. Eutypa dieback (“Dying arm”) disease of vines—progress towards control. Aust Grapegrower Winemaker. 1978;172:27–28. [Google Scholar]
- 5.Carter M V. The status of Eutypa lata as a pathogen. International Mycological Institute, Phytopathological Paper 32. Wallingford, England: CAB International; 1991. [Google Scholar]
- 6.Carter M V. Wood and root diseases caused by fungi. Eutypa dieback. In: Pearson R C, Goheen A C, editors. Compendium of grape diseases. 3rd ed. St. Paul, Minn: APS Press; 1994. pp. 32–34. [Google Scholar]
- 7.Carter M V, Bolay A, Rappaz F. An annotated host list and bibliography of Eutypa armeniacae. Rev Plant Pathol. 1983;62:251–258. [Google Scholar]
- 8.Délye C, Corio-Costet M F, Laigret F. A RAPD assay for strain typing of the biotrophic grape powdery mildew fungus Uncinula necator using DNA extracted from the mycelium. Exp Mycol. 1995;19:234–237. [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faggian R, Bulman S R, Lawrie A C, Porter I J. Specific polymerase chain reaction primers for the detection of Plasmodiophora brassicae in soil and water. Phytopathology. 1999;89:392–397. doi: 10.1094/PHYTO.1999.89.5.392. [DOI] [PubMed] [Google Scholar]
- 11.Fallot J, Deswarte C, Dalmayrac S, Colrat S, Roustan J P. L'Eutypiose de la vigne: isolement d'une molécule synthétisée par Eutypa lata et toxique pour la vigne. C R Acad Sci Ser III. 1997;320:149–158. [Google Scholar]
- 12.Francki R I B, Carter M V. The serological properties of Eutypa armeniacae mycelium and ascospores. Aust J Biol Sci. 1970;23:713–716. [Google Scholar]
- 13.Hamelin R C, Bourassa M, Rail J, Dusabenyagasani M, Jacobi V, Laflamme G. PCR detection of Gremmeniella abietina, the causal agent of Scleroderris canker of pine. Mycol Res. 2000;104:527–532. [Google Scholar]
- 14.Kelly A G, Bainbridge B W, Heale J B, Peréz-Artes E, Jiménez-Diaz R M. In planta-polymerase-chain-reaction detection of the wilt-inducing pathotype of Fusarium oxysporum f. sp. ciceris in chickpea (Cicer arietinum L.) Physiol Mol Plant Pathol. 1998;52:397–409. [Google Scholar]
- 15.King E O, Ward M K, Rainey D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 16.Larignon P, Dubos B. Fungi associated with Esca disease. Eur J Plant Pathol. 1997;103:147–157. [Google Scholar]
- 17.Mauro M C, Vaillant V, Tey-Ruhl P, Mathieu Y, Fallot J. In vitro study of the relationship between Vitis vinifera and Eutypa lata (Pers.: Fr.) Tul. I. Demonstration of toxic compounds secreted by the fungus. Am J Enol Viticult. 1988;39:200–204. [Google Scholar]
- 18.Moller W J, Kasimatis A N. Dieback of grapevines caused by Eutypa armeniacae. Plant Dis Rep. 1978;62:254–258. [Google Scholar]
- 19.Moricca S, Ragazzi A, Kasuga T, Mitchelson K R. Detection of Fusarium oxysporum f. sp. vasinfectum in cotton tissue by polymerase chain reaction. Plant Pathol. 1998;47:486–494. [Google Scholar]
- 20.Mugnai L, Graniti A, Surico G. Esca (black measles) and brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis. 1999;83:404–418. doi: 10.1094/PDIS.1999.83.5.404. [DOI] [PubMed] [Google Scholar]
- 21.Munkvold G P, Duthie J A, Marois J J. Reductions in yield and vegetative growth of grapevines due to Eutypa dieback. Phytopathology. 1994;84:186–192. [Google Scholar]
- 22.Munkvold G P, Marois J J. Factors associated with variation in susceptibility of grapevine pruning wounds to infection by Eutypa lata. Phytopathology. 1995;85:249–256. [Google Scholar]
- 23.Paran I, Michelmore R W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet. 1993;85:985–993. doi: 10.1007/BF00215038. [DOI] [PubMed] [Google Scholar]
- 24.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Péros J-P, Berger G. A rapid method to assess the aggressiveness of E. lata isolates and the susceptibility of grapevine cultivars to Euypa dieback. Agronomie. 1994;14:515–523. [Google Scholar]
- 26.Péros J-P, Berger G, Lahogue F. Variation in pathogenicity and genetic structure in the Eutypa lata population of a single vineyard. Phytopathology. 1997;87:799–806. doi: 10.1094/PHYTO.1997.87.8.799. [DOI] [PubMed] [Google Scholar]
- 27.Péros J-P, Jamaux-Despréaux I, Berger G, Gerba D. The potential importance of diversity in Eutypa lata and co-colonising fungi in explaining variation in development of grapevine dieback. Mycol Res. 1999;103:1385–1390. [Google Scholar]
- 28.Péros J-P, This P, Confuron Y, Chacon H. Comparison by isoenzyme and RAPD analysis of some isolates of the grapevine dieback fungus, Eutypa lata. Am J Enol Viticult. 1996;47:49–56. [Google Scholar]
- 29.Price T V. Serological identification of Eutypa armeniacae. Aust J Biol Sci. 1973;26:389–394. [Google Scholar]
- 30.Rollo F, Salvi R, Torchia P. Highly sensitive and fast detection of Phoma tracheiphila by polymerase chain reaction. Appl Microbiol Biotechnol. 1990;32:572–576. doi: 10.1007/BF00173730. [DOI] [PubMed] [Google Scholar]
- 31.Tilsala-Timisjärvi A, Alatossava T. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl Environ Microbiol. 1998;64:4816–4819. doi: 10.1128/aem.64.12.4816-4819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward E, Adams M J. Analysis of ribosomal DNA sequences of Polymyxa species and related fungi and the development of genus- and species-specific PCR primers. Mycol Res. 1998;102:965–974. [Google Scholar]
- 33.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Innis M A, Gelfrand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 35.Willits D A, Sherwood J E. Polymerase chain reaction detection of Ustilago hordei in leaves of susceptible and resistant barley varieties. Phytopathology. 1998;89:212–217. doi: 10.1094/PHYTO.1999.89.3.212. [DOI] [PubMed] [Google Scholar]
- 36.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]