Abstract
There is limited knowledge of interspecies interactions in biofilm communities. In this study, Pseudomonas sp. strain GJ1, a 2-chloroethanol (2-CE)-degrading organism, and Pseudomonas putida DMP1, a p-cresol-degrading organism, produced distinct biofilms in response to model mixed waste streams composed of 2-CE and various p-cresol concentrations. The two organisms maintained a commensal relationship, with DMP1 mitigating the inhibitory effects of p-cresol on GJ1. A triple-labeling technique compatible with confocal microscopy was used to investigate the influence of toxicant concentrations on biofilm morphology, species distribution, and exopolysaccharide production. Single-species biofilms of GJ1 shifted from loosely associated cell clusters connected by exopolysaccharide to densely packed structures as the p-cresol concentrations increased, and biofilm formation was severely inhibited at high p-cresol concentrations. In contrast, GJ1 was abundant when associated with DMP1 in a dual-species biofilm at all p-cresol concentrations, although at high p-cresol concentrations it was present only in regions of the biofilm where it was surrounded by DMP1. Evidence in support of a commensal relationship between DMP1 and GJ1 was obtained by comparing GJ1-DMP1 biofilms with dual-species biofilms containing GJ1 and Escherichia coli ATCC 33456, an adhesive strain that does not mineralize p-cresol. Additionally, the data indicated that only tower-like cell structures in the GJ1-DMP1 biofilm produced exopolysaccharide, in contrast to the uniform distribution of EPS in the single-species GJ1 biofilm.
Biofilms of environmental and medical significance frequently consist of diverse populations of microorganisms (4, 12). A range of metabolic interactions have been observed among microorganisms in biofilms, including mutualistic and commensal relationships (16, 26). Moreover, metabolic interactions within biofilms may be facilitated by the spatial arrangement of interacting cells (11, 19, 20, 27). In biofilms that detoxify mixed organic wastes, the metabolic interactions among bacteria could potentially influence biofilm structure and development, since the metabolism of complex organic pollutants often involves multispecies bacterial consortia (10, 23). For example, fluctuating toxicant concentrations could provide selective pressure that alters the species distribution in a biofilm, ultimately influencing biofilm activity.
Another feature of biofilms that may respond to changing toxicant concentrations is the exopolysaccharide (EPS) matrix. EPS is an integral structural and functional component of biofilm systems (6, 24), can account for up to 90% of the organic matter in a biofilm (25), and helps protect organisms in the biofilm community from environmental stresses (1; T. R. Neu, G. Packroff, and J. R. Lawrence, Abstr. 97th Gen. Meet. Am. Soc. Microbiol. 1997, p. 396, 1997). Recent research has characterized the composition and quantity of EPS produced (17, 18), as well as its presence in natural biofilms (8). Few studies have investigated the relationship among EPS, biofilm architecture, and toxicant concentrations.
Recently, scanning confocal laser microscopy has been developed into a powerful tool for elucidating the three-dimensional architecture and species distribution of biofilm systems (5, 22) and may be useful for correlating biofilm structure with interspecies relationships. In this study, a triple-labeling technique that was compatible with confocal microscopy was developed to characterize the response of a dual-species biofilm to two compounds that are commonly found in mixed organic chemical waste. The biofilm consisted of Pseudomonas putida DMP1, a p-cresol-degrading organism, and Pseudomonas sp. strain GJ1, a 2-chloroethanol (2-CE)-degrading organism. The morphology, species distribution, and EPS production of biofilms grown under increasingly inhibitory toxicant concentrations were examined using scanning confocal laser microscopy. The resulting images elucidated the effect of the interspecies metabolic interactions on the organization of the biofilm.
MATERIALS AND METHODS
Strains, plasmids, and media.
Pseudomonas sp. strain GJ1, a 2-CE degrader, and Pseudomonas putida DMP-1, a p-cresol degrader, were described previously (13; Y. T. Wang and M. Qu, Abstr. 65th Annu. Conf. Expos. Water Environ. Fed., p. 63, 1992). Escherichia coli ATCC 33456, a wild-type Escherichia coli strain, was obtained from the American Type Culture Collection (21). Plasmid pSMC21 is pUCP-based cloning vector containing gfpmut2 and the ampicillin, carbenecillin, and kanamycin resistance genes. Plasmid pVLT33 is an RSF-1010-based, broad-host-range plasmid harboring the kanamycin resistance gene. All strains were grown in MMV (13), a minimal medium supplemented with 50 μg of kanamycin ml−1. During all biofilm colonization processes, 0.1% succinate, a substrate that is not inhibitory to either strain, was provided as the sole carbon source for both GJ1 and DMP1. All strains were cultured at ambient temperature (22 ± 1°C).
Growth and toxicity studies.
Pseudomonas sp. strain GJ1 harboring pSMC21 (3) was grown in MMV with the following concentrations of 2-CE (Aldrich, Milwaukee, Wis.): 5, 10, 15, 20, 25, and 30 mM. The specific growth rates of Pseudomonas sp. strain GJ1 harboring pSMC21 in suspended culture were determined. All growth experiments were performed at least twice, and the mean values are reported. P. putida DMP1 harboring pVLT33 (9) was grown in MMV with the following concentrations of p-cresol (Aldrich): 0, 0.2, 0.5, 0.7, 0.9, and 1.9 mM. The suspended culture growth rates of DMP1 were determined.
The effect of p-cresol on the specific growth rate of GJ1 was determined by growing GJ1 in MMV with 20 mM 2-CE and various p-cresol concentrations (0, 0.2, 0.5, 0.7, 0.9, and 1.9 mM). The effect of 2-CE on the specific growth rate of DMP1 was determined by growing DMP1 in MMV with the optimal p-cresol concentration (0.7 mM) and various 2-CE concentrations (0, 5, 10, 15, 20, 25, and 30 mM).
Bench-scale flow cell.
Biofilms were prepared, as described previously, in bench-scale parallel-plate flow cells (reactor volume, of 0.35 ml) (5). A coverglass was glued to a plastic frame using General Electric silicone rubber adhesive sealant RTV 102 (GE Silicones, Waterford, N.Y.). Flow cells were operated in recirculating (start-up) or continuous modes at flow rates of 0.86 or 0.12 ml/min, respectively.
GJ1 and DMP1 dual-species biofilms.
For colonization of the surface, medium containing equal concentrations of exponentially growing GJ1 (harboring pSMC21) and DMP1 (harboring pVLT33) was recirculated through a flow cell for 6 h. Subsequently, sterile medium containing 20 mM 2-CE and either 0.7 or 1.9 mM p-cresol was continuously pumped through the flow cell for 45 h. MMV (15 ml) was pumped through each flow cell to remove cells that were not attached to the coverglass. The biofilms in the flow cells were then stained and imaged as described below.
GJ1 and ATCC 33456 dual-species biofilms.
Exponentially growing E. coli ATCC 33456 harboring pVLT33 was recirculated through a flow cell for 6 h to promote colonization. Exponentially growing Pseudomonas sp. strain GJ1 harboring pSMC21 was then recirculated through the same flow cell for 3 h. After both colonization steps, sterile medium containing 20 mM 2-CE and either 0.7 or 1.9 mM p-cresol was continuously pumped through the flow cell for 45 h. To maintain ATCC 33456, which cannot grow on either 2-CE or p-cresol, 0.002% glucose was added to the medium. The addition of glucose did not influence strain GJ1, which is unable to grow on glucose. The resulting biofilms were rinsed, stained, and imaged.
Single-species biofilms.
Flow cells were colonized as described above with medium containing either GJ1 harboring pSMC21, ATCC 33456 harboring pVLT33, or DMP1 harboring pVLT33. The colonized flow cells were then switched to the continuous-flow conditions described above. Again, ATCC 33456 biofilms were switched to medium containing 0.002% glucose. The GJ1 biofilms were rinsed and stained with calcofluor white, an EPS stain, but not with SYTO 59, prior to imaging. The ATCC 33456 and DMP1 biofilms were rinsed and stained with both SYTO 59 and calcofluor white before imaging.
Staining biofilms with SYTO 59.
After being rinsed, flow cells containing either E. coli ATCC 33456 or P. putida DMP1 were stained with 20 μM SYTO 59 (Molecular Probes, Inc., Eugene, Oreg.), a soluble nucleic acid dye that emits in the red region, for 10 min.
Staining biofilms with calcofluor white.
After staining of the biofilms with SYTO 59, all flow cells were stained in the dark with 0.025% calcofluor white M2R (Sigma Chemical Co., St. Louis, Mo.) for 1 min. This dye binds to β-linked polysaccharides, such as cellulose and chitin (14). Calcofluor white cannot penetrate intact cell membranes and does not stain viable cells (15). The biofilms were then rinsed with 2 ml of MMV to decrease the background fluorescence. The stained flow cells were imaged using confocal microscopy.
Confocal microscopy.
Confocal microscopy was performed using an MRC-1024 laser-scanning confocal imaging system (Bio-Rad Microsciences, Cambridge, Mass.) equipped with a Diaphot 200 inverted microscope (Nikon, Inc., Tokyo, Japan). All images were obtained with a 20× lens. The 488-nm line from a Kr/Ar laser was used to simultaneously excite both green fluorescent protein (GFP) and SYTO 59. GFP was detected using a standard fluorescein filter set (522/35 band-pass filter), and SYTO 59 emissions were detected using a 605/32 band-pass filter. The 363-nm line from an argon ion UV laser (model ENT 622; Innova Technology/Coherent Enterprises, Santa Clara, Calif.) was used to excite the calcofluor white, which was then detected using a 455/30 band-pass filter. The GFP emissions were directed to the green channel, the SYTO 59 emissions were directed to the red channel, and the calcofluor white emissions were directed to the blue channel. The resulting images were resized and printed using Adobe Photoshop 5.0 (Adobe Systems, Inc., San Jose, Calif.) software.
RESULTS
Growth and toxicity studies.
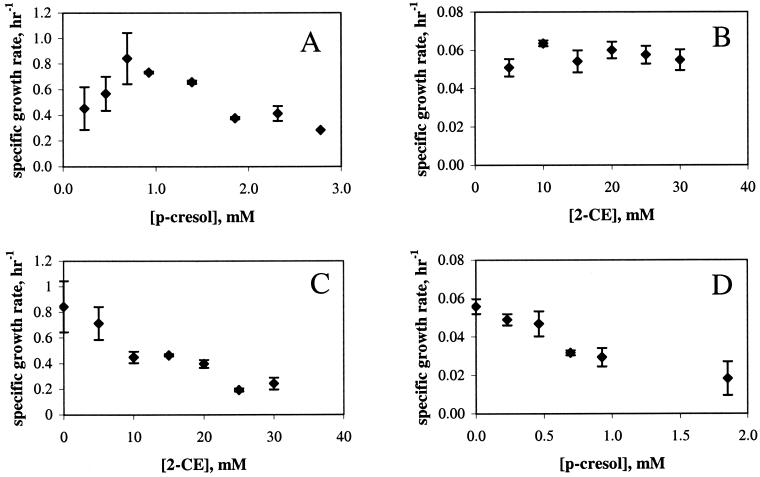
P. putida DMP1 grew optimally at 0.7 mM (75 ppm) p-cresol and experienced substrate inhibition at p-cresol concentrations greater than 0.7 mM (Fig. 1A). In contrast, the growth of Pseudomonas sp. strain GJ1 was unaffected by 2-CE at all concentrations tested and was an order of magnitude slower than that of DMP1 (Fig. 1B). DMP1 was unable to grow on 2-CE (Fig. 1C), and GJ1 was unable to grow on p-cresol (Fig. 1D). Thus, each member of the biofilm had an independent carbon source. For purposes of investigating interactions between GJ1 and DMP1 in a biofilm, 0.7 or 1.9 mM (200 ppm) p-cresol concentrations were selected; 0.7 mM was the optimal concentration for DMP1 growth, while DMP1 substrate inhibition occurred at 1.9 mM p-cresol. A 2-CE concentration of 20 mM (1,600 ppm) was used.
FIG. 1.
Growth and inhibition kinetics. (A and C) P. putida DMP1. (B and D) Pseudomonas sp. strain GJ1. (A) P. putida DMP1 growth on p-cresol. (B) Pseudomonas sp. strain GJ1 growth on 2-CE. (C) Inhibition of P. putida DMP1 growth with p-cresol as the carbon source and 2-CE as the inhibitor. (D) Inhibition of Pseudomonas sp. strain GJ1 growth with: 2-CE as the carbon source and p-cresol as the inhibitor.
Both substrates inhibited the growth of the organism not able to utilize it as a carbon source. DMP1 growth was inhibited by the presence of 2-CE (Fig. 1C). At 20 mM 2-CE, the 2-CE concentration used for all biofilm studies, the planktonic specific growth rate of DMP1 was diminished to one-fourth of that with no 2-CE. GJ1 growth was also inhibited by the presence of p-cresol in the medium (Fig. 1D). At 0.7 or 1.9 mM p-cresol, the conditions investigated in this study, the specific growth rate decreased to two-thirds and one-third of that with no p-cresol, respectively. Since DMP1 grew substantially faster than GJ1 under all planktonic conditions, the inhibition of DMP1 by 2-CE was not an important variable and was not explored in this study.
Biofilm morphology, species distribution, and EPS production.
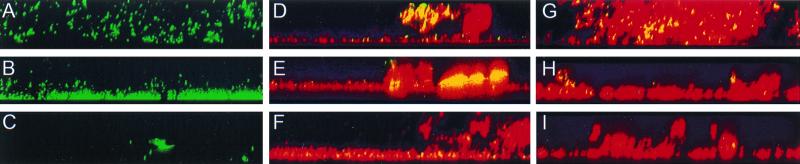
Vertical sections of GJ1 single-species biofilms (Fig. 2A to C), GJ1-DMP1 dual-species biofilms (Fig. 2D to F), and GJ1-ATCC 33456 dual-species biofilms (Fig. 2G to I) were obtained using confocal microscopy. These biofilms were cultured in medium containing succinate (Fig. 2A, D, and G), 20 mM 2-CE and 0.7 mM p-cresol (Fig. 2B, E, and H), or 20 mM 2-CE and 1.9 mM p-cresol (Fig. 2C, F, and I). In all images, green or yellow cells correspond to GJ1 and red cells correspond to either DMP1 (Fig. 2D to F) or ATCC 33456 (Fig. 2G to I). Multiple images were collected for each set of experimental conditions; a representative image is presented in all cases.
FIG. 2.
Vertical sections of biofilms. (A to C) Pseudomonas sp. strain GJ1 (green) monoculture biofilms. (D to F) Pseudomonas sp. strain GJ1 (yellow) and P. putida DMP1 (red) coculture biofilms. (G to I) Pseudomonas sp. strain GJ1 (yellow) and E. coli ATCC 33456 (red) coculture biofilms. (A, D, and G) Cells were grown on succinate. (B, E, and H) Cells were grown on 20 mM 2-CE and 0.7 mM p-cresol. (C, F, and I) Cells were grown on 20 mM 2-CE and 1.9 mM p-cresol. Magnification, ×200.
A distinct change was observed in the GJ1 single-species biofilm morphology as the p-cresol concentration increased (Fig. 2, top to bottom). When GJ1 biofilms were cultured in medium with succinate (in the absence of p-cresol), a biofilm consisting of loosely associated cell clusters formed (Fig. 2A). These cell clusters were connected by EPS (data not shown). When the GJ1 biofilm was developed in medium containing 20 mM 2-CE and 0.7 mM p-cresol, a morphological change resulted in a biofilm that was dense and attached to the substratum (Fig. 2B). A more dramatic change in morphology resulted when the GJ1 biofilm was developed in medium containing 20 mM 2-CE and 1.9 mM p-cresol (Fig. 2C). Under these conditions, only a few clusters of GJ1 were able to survive. In contrast, DMP1 single-species biofilms were dense and closely attached to the substratum under all p-cresol concentrations (data not shown).
Qualitatively, both the morphology and species distribution of the dual-species GJ1-DMP1 biofilms were influenced by the p-cresol concentration. When the GJ1-DMP1 biofilms were cultivated on succinate, the two species were intermingled in the biofilm (Fig. 2D). Under these conditions, yellow GJ1 cells were found near the biofilm/bulk-fluid interface. In contrast to the case where a GJ1 biofilm was developed on succinate alone, the biofilm containing both GJ1 and DMP1 formed a tightly packed biofilm, resembling that of a DMP1 single-species biofilm (data not shown). When the biofilm was developed in medium containing 20 mM 2-CE and 0.7 mM p-cresol, the yellow GJ1 cells were present only between layers of the red DMP1 cells (Fig. 2E). Similarly, GJ1 was found only deep within the biofilm when the biofilm was developed in 20 mM 2-CE and 1.9 mM p-cresol (Fig. 2F). There was also a decline in the GJ1 population when the p-cresol concentration was increased to 1.9 mM. When the medium contained either 0.7 or 1.9 mM p-cresol, GJ1 was no longer evident near the biofilm/bulk-fluid interface.
To determine if the apparent protection of GJ1 from p-cresol by DMP1 was due to the specific degradative ability of DMP1, GJ1 was grown in dual-species biofilms with E. coli ATCC 33456, which can form biofilms but is unable to degrade p-cresol. When cultured on succinate, the GJ1-ATCC 33456 biofilm had both species intermingled (Fig. 2G), which was similar to the GJ1-DMP1 biofilm grown under the same conditions (Fig. 2D). When the GJ1-ATCC 33456 biofilm was cultured in 20 mM 2-CE and either 0.7 or 1.9 mM p-cresol, only a few yellow clusters of GJ1 cells were present (Fig. 2H and 2I), significantly fewer than in the GJ1-DMP1 biofilm.
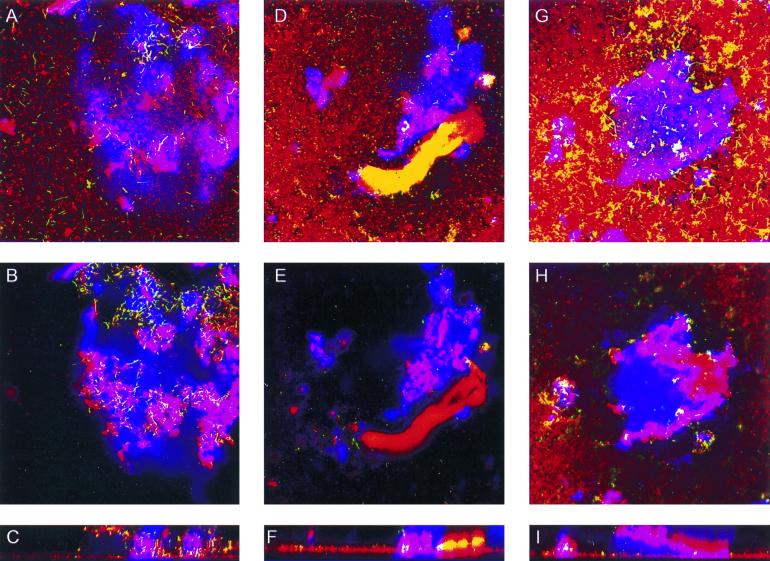
When cultured on succinate, GJ1 and DMP1 were homogeneously distributed in horizontal sections of the biofilm at both 15 and 30 μm above the substratum (Fig. 3A to C). When exposed to 0.7 mM p-cresol, large clusters of GJ1 cells were concentrated beneath DMP1 in the cell towers and in the lower layers of the biofilm (Fig. 3D to F). After exposure to 1.9 mM p-cresol, GJ1 was present only in small clusters in the lower layers of the biofilm (Fig. 3G to I). At 15 μm above the substratum, there were distinct clusters of yellow GJ1 cells homogeneously interspersed with clusters of red DMP1 cells (Fig. 3G).
FIG. 3.
Horizontal and vertical sections of Pseudomonas sp. strain GJ1 (yellow) and P. putida DMP1 (red) coculture biofilms with exopolysaccharides (blue). (A to C) Cells grown in succinate. (D to F) Cells grown on 20 mM 2-CE and 0.7 mM p-cresol. (G to I) Cells grown on 20 mM 2-CE and 1.9 mM p-cresol. Magnification, ×200.
To examine the effect of the organic compounds on EPS formation, GJ1-DMP1 dual-species biofilms were stained with calcofluor white (Fig. 3). In these images, red corresponds to DMP1 cells, yellow corresponds to GJ1 cells, and blue corresponds to EPS. Magenta regions, resulting from the overlap of red and blue pixels, represent DMP1 cells embedded in EPS. Similarly, white regions, resulting from the overlap of yellow and blue pixels, represent GJ1 cells covered in EPS.
The EPS, which was produced by both GJ1 and DMP1 in single-species biofilms (data not shown), was formed in similar quantities in the dual-species biofilms under all p-cresol concentrations tested. The EPS was formed only by cells located in tower- or mushroom-shaped clusters, where there was a high cell density (Fig. 3C, F, and I).
DISCUSSION
The complementary metabolic activities of Pseudomonas sp. strain GJ1 and P. putida DMP1 were combined in a biofilm to produce a single functional system in which both strains were maintained. The planktonic growth and toxicity data (Fig. 1) obtained for the two pseudomonads suggests that GJ1 had a limited chance of survival in the presence of p-cresol. It was also evident that a single strain could not effectively remediate a mixed waste stream containing 2-CE and p-cresol.
Figures 2 and 3 demonstrate that GJ1 benefited from the close interactions with DMP1 in a dual-species biofilm. In the presence of p-cresol, GJ1 was present in the greatest proportion when it was paired with DMP1 in a biofilm. The fact that GJ1 was almost eliminated from single-species (Fig. 2C) or GJ1-ATCC 33456 dual-species (Fig. 2I) biofilms at a p-cresol concentration of 1.9 mM implies that it benefited from being coupled with DMP1, in addition to the nonspecific resistance to toxicants that is associated with biofilms (1). GJ1 depended on DMP1 to mineralize p-cresol and thus detoxify the medium in its vicinity.
The morphological changes in the GJ1-DMP1 biofilms which resulted from exposure to p-cresol caused GJ1 and DMP1 to become increasingly intermingled. A p-cresol concentration of 0.7 mM resulted in GJ1 being surrounded by DMP1 in the z direction (Fig. 2E and 3D to F). At 1.9 mM p-cresol, organizational changes were seen in the x-, y-, and z-directions (Fig. 2F and 3G to I). The contact between strains GJ1 and DMP1 was increased, with GJ1 becoming dispersed throughout the biofilm in the xy plane (Fig. 3G). This arrangement appears to have enhanced the survival of GJ1, possibly because the local concentration of p-cresol in its vicinity was reduced. Similarly, Nielsen et al. recently found that a biofilm of Burkholderia sp. strain LB400 and Pseudomonas sp. strain B13 (FR1) capable of mineralizing 3-chlorobiphenyl altered its morphology in response to changing growth substrates (19).
EPS production by the GJ1-DMP1 coculture biofilms was unaffected by the toxicant concentration. Rather, it appeared to be influenced by biofilm morphology. EPS was found only in tall vertical structures in the biofilm, such as tower- or mushroom-shaped clusters (Fig. 3C, F, and I), suggesting that there were different physiological interactions among the bacteria in the structures, in contrast to other regions of the biofilm. Since EPS was found only in the densely populated vertical clusters, it is possible that its production was controlled by quorum-sensing signals. Davies et al. have shown that quorum sensing plays an important role in P. aeruginosa biofilm structure and development (7); additionally, interspecies cell signaling has been demonstrated for a variety of microorganisms (2).
In summary, the organization and morphology of a dual-species biofilm changed in response to the selective pressure of increasing toxicant concentration. The arrangement of cells in the GJ1-DMP1 biofilm facilitated the survival of strain GJ1, which otherwise would have been eliminated from the biofilm. The commensal relationship between the two strains illustrates how metabolic cooperativity may be essential for maintaining multispecies microbial consortia for biological treatment of mixed organic compounds. Furthermore, an understanding of the mutual or commensal relationships in mixed-culture biofilms could aid in the design of multispecies biofilms for the biosynthesis of specialty chemicals or the biodegradation of xenobiotics that cannot be metabolized by a single organism.
ACKNOWLEDGMENTS
We thank Carolyn Larabell at Lawrence Berkeley National Laboratory for use of the confocal microscope, George O'Toole for his contribution of pSMC21, and Thomas Neu for his advice on EPS staining. We also thank Frank Liao, UC Berkeley, for his assistance with experiments and Eric Granlund, UC Berkeley College of Chemistry machine shop, for his help in designing and constructing the flow cells.
This work was supported by the National Science Foundation (BES-9814088) and the Office of Naval Research (N00014-99-1-0182).
REFERENCES
- 1.Allison D G, Gilbert P. Modification by surface association of antimicrobial susceptibility by bacterial populations. J Ind Microbiol. 1995;15:311–317. doi: 10.1007/BF01569985. [DOI] [PubMed] [Google Scholar]
- 2.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Cowan S E, Gilbert E, Khlebnikov A, Keasling J D. Dual labeling with green fluorescent protein for confocal microscopy. Appl Environ Microbiol. 2000;66:413–418. doi: 10.1128/aem.66.1.413-418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danese P N, Pratt L A, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decho A W, Kawaguchi T. Confocal imaging of in situ natural microbial communities and their extracellular polymeric secretions using nanoplast(R) resin. BioTechniques. 1999;27:1246–1252. [PubMed] [Google Scholar]
- 9.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 10.Field J A, Stams A J M, Kato M, Schraa G. Enhanced biodegradation of aromatic pollutants in cocultures of anaerobic and aerobic bacterial consortia. Antonie Leeuwenhoek. 1995;67:47–77. doi: 10.1007/BF00872195. [DOI] [PubMed] [Google Scholar]
- 11.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M, De Vos W M. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol. 1996;62:1656–1663. doi: 10.1128/aem.62.5.1656-1663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James G A, Beaudette L, Costerton J W. Interspecies bacterial interactions in biofilms. J Ind Microbiol. 1995;15:257–262. [Google Scholar]
- 13.Janssen D B, Scheper A, Witholt B. Biodegradation of 2-chloroethanol and 1,2-dichloroethane by pure bacterial cultures. Prog Ind Microbiol. 1984;20:169–178. [Google Scholar]
- 14.Maeda H, Ishida N. Specificity of binding of hexopyranosyl polysaccharides with fluorescent brightener. J Biochem. 1967;62:276–278. doi: 10.1093/oxfordjournals.jbchem.a128660. [DOI] [PubMed] [Google Scholar]
- 15.Mason D J, Lopez-Amoros R, Allman R, Stark J M, Lloyd D. The ability of membrane potential dyes and calcofluor white to distinguish between viable and non-viable bacteria. J Appl Bacteriol. 1995;78:309–315. doi: 10.1111/j.1365-2672.1995.tb05031.x. [DOI] [PubMed] [Google Scholar]
- 16.Moller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neu T R, Lawrence J R. Lectin-binding analysis in biofilm systems. Methods Enzymol. 1999;310:145–152. doi: 10.1016/s0076-6879(99)10012-0. [DOI] [PubMed] [Google Scholar]
- 18.Neu T R, Marshall K C. Bacterial polymers: physicochemical aspects of their interactions with surfaces. J Biomater Appl. 1990;5:107–133. doi: 10.1177/088532829000500203. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen A T, Toker-Nielsen T, Barken K B, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- 20.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amamm R, Schleifer K-H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen H, Wang Y-T. Simultaneous chromium reduction and phenol degradation in a coculture of Escherichia coli ATCC 33456 and Pseudomonas putida DMP-1. Appl Environ Microbiol. 1995;61:2754–2758. doi: 10.1128/aem.61.7.2754-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart P S, Murga R, Srinivasan R, de Beer D. Biofilm structural heterogeneity visualized by three microscopic methods. Water Res. 1995;29:2006–2009. [Google Scholar]
- 23.Vanginkel C G. Complete degradation of xenobiotic surfactants by consortia of aerobic microorganisms. Biodegradation. 1996;7:151–164. doi: 10.1007/BF00114627. [DOI] [PubMed] [Google Scholar]
- 24.Watnick P I, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–595. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wingender J, Flemming H C. Autoaggregation of microorganisms: flocs and biofilms. In: Winter J, editor. Biotechnology. 2nd ed. IIa. Weinheim, Germany: Wiley-VCH Verlag GmbH; 1999. pp. 65–83. [Google Scholar]
- 26.Wolfaardt G, Lawrence J, Robarts R, Caldwell S, Caldwell D. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–466. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfaardt G M, Lawrence J R, Robarts R D, Caldwell D E. The role of interactions, sessile growth, and nutrient amendments on the degradative efficiency of a microbial consortium. Can J Microbiol. 1994;40:331–340. doi: 10.1139/m94-055. [DOI] [PubMed] [Google Scholar]