Abstract
Previous bacterial transport studies have utilized fluorophores which have been shown to adversely affect the physiology of stained cells. This research was undertaken to identify alternative fluorescent stains that do not adversely affect the transport or viability of bacteria. Initial work was performed with a groundwater isolate, Comamonas sp. strain DA001. Potential compounds were first screened to determine staining efficiencies and adverse side effects. 5-(And 6-)-carboxyfluorescein diacetate, succinimidyl ester (CFDA/SE) efficiently stained DA001 without causing undesirable effects on cell adhesion or viability. Members of many other gram-negative and gram-positive bacterial genera were also effectively stained with CFDA/SE. More than 95% of CFDA/SE-stained Comamonas sp. strain DA001 cells incubated in artificial groundwater (under no-growth conditions) remained fluorescent for at least 28 days as determined by epifluorescent microscopy and flow cytometry. No differences in the survival and culturability of CFDA/SE-stained and unstained DA001 cells in groundwater or saturated sediment microcosms were detected. The bright, yellow-green cells were readily distinguished from autofluorescing sediment particles by epifluorescence microscopy. A high throughput method using microplate spectrofluorometry was developed, which had a detection limit of mid-105 CFDA-stained cells/ml; the detection limit for flow cytometry was on the order of 1,000 cells/ml. The results of laboratory-scale bacterial transport experiments performed with intact sediment cores and nondividing DA001 cells revealed good agreement between the aqueous cell concentrations determined by the microplate assay and those determined by other enumeration methods. This research indicates that CFDA/SE is very efficient for labeling cells for bacterial transport experiments and that it may be useful for other microbial ecology research as well.
There is heightened interest in using degradative microbes to bioremediate soil and groundwater contaminated with recalcitrant pollutants, a process known as bioaugmentation. Bioaugmentation has been used successfully to remediate groundwater contaminated with chlorinated solvents (10, 28, 40, 42) and is expected to be useful for other compounds as well. Bioaugmentation requires that effective concentrations of microorganisms be predictably transported to and through contaminated areas of the subsurface. Current tracking technologies are of limited use due to the effects of labeling compounds on cell viability or other properties, high detection limits or interferences from indigenous organisms, or regulatory concerns about the release of genetically modified or antibiotic-resistant microbial strains. Therefore, new methods for tracking viable bacterial cells under both laboratory and field conditions are being developed as part of a major research project examining the physical, chemical, and biological controls on bacterial transport under way under the auspices of the Acceleration Element of the Natural and Accelerated Bioremediation Research Program at the U. S. Department of Energy (DOE) South Oyster site (Oyster, Va.). In order to examine the processes in detail, the movement and distribution of introduced bacteria must be monitored.
Several methods have previously been developed to monitor microbial transport through, and microbial interactions with, porous media during laboratory and field experiments. Selective plate counting has been used with some success to detect injected organisms in downgradient monitoring wells (6, 28, 42), but this method was unsuccessful during a field-scale bacterial transport experiment at the DOE North Oyster, Va., site, where an indigenous strain was injected (7). Plating methods can have detection limits around 100 to 1,000 culturable cells per ml of sample, providing that there is no overgrowth of indigenous bacteria on the plates. Selective plating may allow lower detection limits but requires extensive a priori screening of every potential degradative organism for antibiotic sensitivity and carbon source utilization profile. A major disadvantage of this method is that it does not detect “viable but nonculturable” cells (21, 30, 31, 34, 36, 38, 50), which still may be metabolically active but unable to form colonies on solid media.
Stable isotopes are increasingly being used for field experiments. Bacteria grown on 13C-enriched glucose prior to injection into the surficial, uncontaminated aquifer at the North Oyster, Va., site were detected in downgradient monitoring wells by converting collected particulate organic matter to carbon dioxide and measuring the 13CO2 by Carlo-Erba isotope ratio mass spectrometry (7). However, additional methods were required to unequivocally establish that the 13C represented intact cells of the bacteria injected rather than protozoans or other indigenous microbes which may have incorporated labeled cellular material via predation of live target cells or consumption of dead target cells (W. Holben, University of Montana, personal communication, 1999).
One of the molecular methods available, PCR, has been used to count very low numbers of bacteria in a variety of environments (25, 44, 45, 49, 51, 53). PCR and nucleic acid quantification can usually be performed only in the laboratory, which precludes the use of these methods in the field to obtain near-real-time cell detection and enumeration data. Genetically engineered microorganisms carrying the genes coding for the green fluorescent protein have been developed (11, 12, 29), which has allowed enumeration of these organisms by measuring fluorescence. However, the use of these genetically engineered microorganisms has been restricted to laboratory, greenhouse, and lysimeter experiments because of regulatory concerns.
Labeling cells with fluorescent stains has been employed to examine bacterial attachment to surfaces (13), to count the numbers of total and active cells in a variety of environmental samples (37, 39, 55), and for in situ injection of groundwater bacteria (1, 16–19). One of the stains used, 4′,6-diamino-2-phenylindole (DAPI), specifically binds to nucleic acids (RNA and DNA), which enables it to universally label cells in an organism-independent manner. The binding mechanism adversely affects normal cell function (33), resulting in a loss of viability. The potential effects of this compound on the transport properties of an organism have been shown to be minimal for short-term experiments (24). Another stain, 4-phenyl spiro-furan-2-(3H),1 phthalan-3,3′-dione (Fluram, fluorescamine), forms covalent bonds with free amino groups. Fluram was shown not to effect the adhesion of a Rhodococcus strain to titanium-rich particles (13), but its effects on other species have not been examined. For fluorescent stains to be useful for studying bacterial transport and monitoring bioaugmentation, they must have minimal effects on bacterial adhesion, viability, and metabolic activity, while at the same time they must be retained in the cells for at least several weeks.
New fluorescent compounds which may allow cells to be stained without a loss of activity or viability or changes in adhesive properties have been and continue to be developed. Many of these dyes have been developed specifically for eukaryotic cell staining, but the principles underlying their use makes them applicable to prokaryote staining as well. Some of the newer dyes specifically stain cell membranes, while others cross the membrane and covalently bond to intracellular proteins. In either case, some of these dyes have been shown to be retained in cells for at least 3 to 4 weeks without a loss of cell viability or alterations in cell function or adhesion (20). This work was undertaken to critically evaluate the labeling of bacteria with numerous fluorescent stains and to develop an easy and effective high-throughput detection procedure for quantifying fluorescently labeled cells during in situ bacterial transport experiments.
MATERIALS AND METHODS
Chemicals and media.
The following compounds were purchased from Molecular Probes (Eugene, Oreg.): calcein AM; calcein blue AM; 5-(and 6-)-carboxyfluorescein diacetate, succinimidyl ester (CFDA/SE); 7-amino-4-chloromethylcoumarin (CellTracker Blue CMAC); 5-chloromethylfluorescein diacetate (CellTracker Green) (CMFDA); 4-(4-[dihexadecylamino]styryl)-N-methylpyridinium iodide (DiA); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine-5,5′-disulfonic acid [DiIC18(3)-DS]; 1,6-diphenyl-1,3,5-hexatriene (DPH); Oregon Green 488 (carboxy-DFFDA); 1,1′-dioctadecyl-6,6′-diphenyl-3,3,3′,3′-tetramethylindocarbocyanine chloride (6,6-Ph2-DiIC18); rhodamine B (hexyl ester, chloride); 5-sulfofluorescein diacetate, sodium salt (SFDA); SlowFade Light mounting fluid; 5-(and-6)-chloromethyl SNARF-1 acetate; (1,1′-dioctadecyl - 6,6′ - di(4 - sulfophenyl) - 3,3,3′,3′ - tetramethyl - indocarbocyanine [SP -DiIC18(3)]; 3,3′-dioctadecyl-5,5′-di(4-sulfophenyl)-oxacarbocyanine, sodium salt [SP-DiOC18(3)]; and 5-(and-6)-carboxytetramethylrhodamine, succinimidyl ester (TAMRA/SE). Fluorescamine and 5-cyano-2,3-ditolyl-tetrazolium chloride (CTC) were purchased from Fluka Chemical (Milwaukee, Wis.) and PolySciences (Warrington, Pa.). DAPI and acridine orange (AO) were obtained from Sigma Chemical (Milwaukee, Wis.). R2A agar was purchased from Fisher Scientific (Fair Lawn, N.J.).
An artificial groundwater designated NCAGW and based on the groundwater chemistry of the aerobic Narrow Channel Focus Area (NC) of the South Oyster bacterial transport site was used during this research in place of real groundwater when a sterile, carbon-free medium was needed to eliminate the potential effects of predation, competition, and growth. NCAGW contained (per liter) 70 mg of Ca(NO3)2 · 4H2O, 60 mg of MgSO4 · 7H2O, 60 mg of NaHCO3, 29 mg of CaCl2 · 2H2O, 25 mg of CaSO4 · 2H2O, 10 mg of KNO3, and 0.4 mg of NaH2PO4. During some experiments, a simplified recipe (mNCAGW) was used, which contained (per liter) 80 mg of CaSO4 · 2H2O, 66 mg of NaHCO3, 20 mg of CaCl2 · 2H2O, and 16 mg of KHCO3. The pH of both artificial groundwaters was adjusted to 6.0 with HCl, and sterilization was performed by filtration though 0.2-μm-pore-size cellulose acetate membranes. Phosphate-buffered saline (PBS) (pH 7.4) and phosphate buffer (PB) (pH 7.0) were also sterilized by filtration.
Bacterial strains.
An indigenous aquifer organism, DA001, was isolated from groundwater from the South Oyster Narrow Channel Focus Area. DA001 was identified as a member of a Comamonas species by phylogenetic analysis of its 16S rRNA gene sequence (similarity index [SI] = 0.965). This strain has been used extensively for laboratory experiments performed as part of the DOE bacterial transport research project and will be used for field-scale experiments as well. Therefore, the majority of the fluorescent stain screening was focused on finding compounds which would facilitate tracking this organism. Adhesion-deficient variants of DA001 were selected by using a standardized adhesion assay (9) employing South Oyster site sediment. DA001 was used for screening the fluorescent stains when the adhesion values stabilized around 30 to 40%. Routine growth of the organism was performed by using 0.2% (wt/vol) sodium acetate in a basal salts medium (16) at room temperature.
Additional bacterial strains used during this research included Acinetobacter johnsonii ATCC 17909, Alcaligenes eutrophus ATCC 17909, Arthrobacter globiformis ATCC 8010, Bacillus subtilis ATCC 6051, Cytophaga pectinovora ATCC 19366, Escherichia coli BL, Flavobacterium odoratum ATCC 4651, Klebsiella sp. strain 1PC, Micrococcus luteus ATCC 4698, Pseudomonas cepacia ATCC 25416, Pseudomonas fluorescens ATCC 13525, Pseudomonas putida ATCC 12633, Pseudomonas sp. strains LB300 and To11A, Rahnella aquatilis BFB, Rhodococcus rhodochrous ATCC 13808, Sphingomonas capsulata ATCC 14666, and Streptomyces albus ATCC 3004. Several facultatively iron-reducing bacteria which were isolated from the suboxic South Oyster Focus Area of the South Oyster Bacterial Transport Site were also screened, including strain FER-02, which is most closely related to Paenibacillus polymyxa (SI = 0.905), strain SO-B2, which is most closely related to Enterobacter aerogenes (SI = 0.953), and two unidentified low-G+C-content gram-positive strains, SO-9 and SO-S10. Most of these organisms were routinely grown in R2A broth at room temperature; the only exceptions were M. luteus and F. odoratum, which were grown in tryptic soy broth. Staining was performed as it was with strain DA001; cells were incubated for 48 h at 15°C in NCAGW, washed, and resuspended in NCAGW prior to determination of staining efficiency.
Fluorescent stain screening.
Previously published procedures were followed for staining with SFDA (46), fluorescamine (13), and CTC (37, 39). Initial staining with other compounds was performed by using the manufacturers' recommended protocols for carrier solvents, cell suspension buffers, final compound concentrations, and temperature conditions. The compounds were usually added as concentrated stocks in carrier solvents consisting of dimethyl sulfoxide (DMSO) or dimethyl formamide to achieve final concentrations of 10 to 50 μM, with the carrier solvent present at a concentration of ≤0.2% (vol/vol).
During the first stage of screening, each compound was tested for its ability to efficiently and uniformly stain DA001 cells. Qualitative assessment of staining was performed with a Nikon Labophot microscope equipped with an Hg100W high-intensity light source, an EF-D episcopic-fluorescence attachment, an H-III photomicrographic attachment, and the following illumination blocks: UV-1A (Ex 365/10, Dichroic 400, Em 400), B-2H (Ex 470/20, Dichroic 510, Em 515), and G1A (Ex 510-560 and 546/5, Dichroic 565, Em 570). Staining was assessed quantitatively by epifluorescence microscopy of cells counterstained with either AO (for blue-fluorescing stains) or DAPI (for green- and red-fluorescing stains) after filtration of cells onto black polycarbonate filters (pore size, 0.2 μm; Millipore Corp., Bedford, Mass.). At least 20 fields per polycarbonate filter were examined; the number of AO- or DAPI-positive cells and the number of these cells that were also stained with the test compound were counted by alternating between two epifluorescence filter cubes. The staining efficiency was calculated as follows: % efficiency = (no. of cells stained with test compound)/(no. of cells stained with AO or DAPI) × 100. The efficiency of CFDA/SE staining was validated by flow cytometry (see below).
Stains that exhibited a high staining efficiency (>90%) were used in stage two of the screening process, during which the effects of the stain on the culturability and adhesion of DA001 were evaluated. DA001 was grown and washed as described previously, and the final cell suspension was split into two portions. One portion was amended with the stain in the carrier solvent, while the other received only the carrier solvent. Both portions then underwent the same procedure with respect to incubation temperature, time, etc. After staining was complete, aliquots of both portions were diluted and plated in triplicate onto R2A agar to determine the number of culturable cells. Additional aliquots were tested in triplicate to determine the percentage of cells adhering to Oyster site sediment by using the standardized adhesion assay cited above. Changes in adhesion of ±10% and decreases in culturability of 10 to 20% were deemed acceptable.
The third and final stage of the screening process was to determine the longevity of cell fluorescence and was performed only with those compounds that yielded acceptable results in the previous stages of screening. Cells were stained, washed, and incubated at 15°C in sterile NCAGW with shaking. After 48 to 72 h, the cells were washed twice with NCAGW, resuspended in NCAGW, and incubated again at 15°C with shaking. Over a 3-week period, samples were removed and analyzed for culturable cells by plating onto R2A agar and for staining efficiency as described above. Direct counts of stained cells were determined by epifluorescence microscopy by using established methods (23).
Optimization of CFDA/SE staining protocol and sample handling.
The general staining procedure proved to be adequate for the initial screening. However, there was a desire to minimize the amount of CFDA/SE needed to stain a given number of cells and to optimize the overall staining procedure in order to both reduce the costs associated with labeling cells for field-scale experiments and facilitate scaling-up of the staining procedure. The experiments performed included staining in different buffer solutions (NCAGW, PBS, PB), staining of stationary cultures versus staining of log-phase cultures, staining with different temperature regimens (ambient, 37°C, cycling from 25 to 37°C), and staining with different cell densities (109 and 1010 cells/ml) and with different CFDA/SE concentrations (10, 50, and 100 μM). In general, staining took approximately 3 to 5 h. Specific details are given in the Results and Discussion.
Although samples were kept covered with aluminum foil, exposure to incident fluorescent light from overhead illumination did occur during normal processing and handling. Exposure to incandescent light occurred under some conditions. A series of experiments was conducted to determine the effects of exposure to both types of light, as well as sunlight, on stained cells. A suspension of stained DA001 cells (approximately 5 × 106 cells/ml) in NCAGW was prepared, and 40-ml portions were distributed into four sterile 50-ml polypropylene centrifuge tubes (Corning P/N 25330-50; Corning, Inc., Corning, N.Y.). One tube was wrapped completely with aluminum foil and served as the control. One of the three remaining tubes was allowed to sit uncovered on a laboratory bench exposed to fluorescent light, one was placed 15 cm from a 60-W incandescent light bulb, and one was placed outside in direct sunlight. Aliquots from each tube were taken initially and then periodically for up to 18 h (incandescent treatment only). Total fluorescence and the amount of fluorescence per cell were determined by microplate spectrofluorometry and flow cytometry, respectively.
Microcosm fitness experiments.
Microcosm experiments were performed to assess the potential effects that CFDA/SE may have on the fitness or survival of the stained organism. A single culture of DA001 was grown and split into two portions. One portion was stained as described above, while the other underwent the same incubation procedure in the absence of CFDA/SE. After the cells were washed and resuspended in mNCAGW, they were incubated at 15°C for 72 h. Both cell solutions were washed and resuspended in fresh mNCAGW prior to addition to aqueous microcosms (50 ml of mNCAGW or Narrow Channel Focus Area [NC] groundwater in 250-ml bottles) and saturated NC sediment microcosms (2 g of wet sediment in 50-ml centrifuge tubes; total liquid volume, 2 ml). Unstained and CFDA/SE stained DA001 cells were added to obtain initial densities of 107 cells per ml or g. The aqueous microcosms were incubated at 15°C with the bottles gently shaken to keep the cells in suspension; the sediment microcosms were incubated statically. Aliquots of the aqueous microcosms were removed over a 3-week period for determination of culturable DA001 concentrations by plating onto R2A agar; CFDA/SE-stained cells were counted by direct microscopic enumeration. Replicates of the sediment microcosms were sacrificed at the same time points and extracted with 18 ml of mNCAGW, and the DA001 concentrations were determined by plating and microscopic examination (CFDA/SE-stained cells only). The indigenous population of DA001 and other microorganisms in the South Oyster sediment is quite small, which results in a low background concentration and allows added DA001 to be enumerated by plating.
Detection methods for CFDA/SE-stained cells.
During this work, alternatives to direct microscopic enumeration of CFDA/SE-stained cells were investigated, particularly alternatives which would facilitate high-throughput, low-cost sample analysis. Discrete sample fluorometry was evaluated by using a model 10-AU-005 field fluorometer rented from Turner Designs (Sunnyvale, Calif.). Ninety-six-well black microplate formats were tested by using a model BF10000 FLUOROCOUNT fluorescence microplate reader (Packard Instrument Company, Meriden, Conn.) and a SPECTRAmax GEMINI dual-scanning microplate spectrofluorometer (Molecular Devices Corporation, Sunnyvale, Calif.). Both of these devices measured the total fluorescence emitted by each well of the microplate and reported the fluorescence intensity in relative fluorescence units. Ninety-six-well black OptiPlate HTRF-96 microtiter plates (P/N 6005407) were purchased from Packard. A flowthrough fluorescence quantification system consisting of a Hitachi A11 high-performance liquid chromatography (HPLC) pump and an AL10 108 position autosampler coupled to an FL-750 HPLC spectrofluorometer (McPherson, Inc., Chelmsford, Mass.) equipped with 6-, 12-, and 24-μl flow cells was also evaluated. NCAGW was the mobile phase, and no column was present between the autosampler and the detector.
Enumeration of CFDA/SE-stained DA001 cells was also done by flow cytometry performed at the Princeton University Flow Cytometry Core Facility. A 1-ml sample was mixed with 10 μl of a solution containing a known concentration of 1.0-μm-diameter carboxylate-modified TransFluorSpheres (Ex 488 nm/Em 645 nm; Molecular Probes, Inc.) prior to being introduced into a FACScan instrument (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). The cell concentration in the sample was determined as described elsewhere (3, 27, 43) by using the TransFluorSpheres to calculate the volume of sample analyzed. The detection limit of the flow cytometer was determined by enumerating CFDA/SE-stained cells that had been serially diluted in NCAGW, NC groundwater, formaldehyde-fixed NC groundwater, and fixed NC groundwater adjusted to pH 8.0. The staining efficiency of CFDA/SE, as determined by epifluorescent microscopy, was verified with a FACS Vantage cell sorter (Becton Dickinson Immunocytometry Systems).
Intact-core bacterial transport experiments.
Intact sediment cores (length, 70 cm; diameter, 7.2 cm) were collected from both the vadose and saturated zones during an excavation at the South Oyster field site in August 1998. The core orientation was parallel to the in situ groundwater flow. Further details regarding the site and core collection can be found elsewhere (7, 14). Intact cores were prepared and operated as described elsewhere (8, 14), with slight modifications to tubing size and configuration as described by DeFlaun et al. (M. DeFlaun, M. Fuller, B. Mailloux, T. Onstott, W. Holben, W. Johnson, P. Zhang, D. Balkwill, and D. White, submitted for publication). The final height of the sediment after core preparation was 50 cm. Initial injections of sodium bromide (50 μg/ml as Br) or [3H]water (approximately 10,000 dpm/ml) were performed as described previously (8, 14). These injections were used to calculate the flow velocity and core porosity, permeability, and dispersivity and served as conservative tracers to which the transport of the bacterial cells was compared.
During experiment 1, 300 ml (equal to one-half of the pore volume [PV] of the sediment) of NC groundwater containing approximately 7 × 107 CFDA/SE-stained DA001 cells was introduced into core NC7-2 (vadose zone core), and this was followed by continuous injection of NC groundwater. The DA001 cell concentrations in the core effluent fractions collected at 20-min intervals were determined by plating, epifluorescent direct counting of CFDA/SE-stained cells, and microplate spectrofluorometry. After 28 PV of groundwater had passed through the sediment, the core was drained, frozen, and split (8). A template consisting of a grid of 5 by 28 squares (each square was 1.6 by 1.6 cm) was used as a guide to subsample the sediment. The sediment under each grid space was transferred to a preweighed plastic centrifuge tube with a metal spatula, the tube was weighed again, and the weight of the tube plus sediment was recorded. The sediment in the tube was thoroughly homogenized with a metal spatula, and the gravimetric water contents of selected samples obtained along the length of the core were determined. All of the samples within the first 15 cm from the influent end of the core and selected samples from the remainder of the core were plated and used for DA001 CFU determination. Briefly, 1.0 g (wet weight) of sediment was placed in 9 ml of PBS, vortexed for 60 s, serially diluted in PBS, and plated onto R2A plates in triplicate. The concentrations of DA001 in selected samples were also determined by epifluorescent direct counting of CFDA/SE-stained cells. The samples used for microscopy were prepared as the samples used for CFU determinations were, except that the sediment suspension was allowed to settle for 300 s before a sample was removed and filtered onto black polycarbonate membranes for direct counting.
Experiment 2 was performed similarly, except that DA001 cells were first labeled with [14C]acetate by the method of McEldowney and Fletcher (26) as described by DeFlaun et al. (8). After 14C labeling, the cells were washed and stained with CFDA/SE. One-half PV containing 9 × 107 cells was introduced into core NC9-2SAT (a saturated zone core); this was followed by continuous injection of NC site groundwater, and effluent fractions were collected at 20-min intervals. DA001 concentrations were determined by plating, direct counting of CFDA/SE-stained cells, and microplate spectrofluorometry, as well as by scintillation counting as described elsewhere (8, 14).
RESULTS AND DISCUSSION
Fluorescent stain screening.
A total of 19 fluorescent stains were evaluated; the results of the screening are presented in Table 1. Only seven (37%) of the stains exhibited a high-efficiency staining of Comamonas sp. strain DA001. This indicates that stains developed primarily for use with mammalian cells do not necessarily work with bacterial cells. However, it was encouraging that only one of the stains, fluorescamine, caused significant reductions in cell culturability and significant increases in cell adhesion to sediment. This effect was most likely due to either the acetone carrier solvent or the fact that the fluorescamine reacted with free amine groups at the cell surface.
TABLE 1.
Screening results for Comamonas sp. strain DA001
| Stain | Color | Efficiency | Undesirable effects on:
|
Well retained | |
|---|---|---|---|---|---|
| Cell culturabilitya | Adhesionb | ||||
| Calcein blue AM | Blue | Low | NDc | ND | ND |
| CMAC | Blue | Moderate | No | No | ND |
| DPH | Blue | Low | ND | ND | ND |
| Fluorescamine | Blue-green | High | Yes | Yes | ND |
| Calcein AM | Green | Low | ND | ND | ND |
| CFDA/SE | Green-yellow | High | No | No | Yes |
| CMFDA | Green | High | No | No | No |
| DiA | Green | High | No | No | No |
| Oregon Green 488 | Green | Low | ND | ND | ND |
| SFDA | Green-yellow | Low | No | ND | ND |
| SP-DiOC18(3) | Green | Low | ND | ND | ND |
| SNARF-1 | Red | Low | ND | ND | ND |
| CTC | Red | Low | ND | ND | ND |
| DiI | Red/orange | High | No | No | ND |
| DiIC18(3)-DS | Red/orange | Moderate | ND | ND | ND |
| 6,6-Ph2-DiIC18(3) | Red/orange | High | No | ND | ND |
| Rhodamine B, hexyl ester | Red/orange | Highd | No | ND | ND |
| SP-DiIC18(3) | Red/orange | Moderated | No | No | ND |
| TAMRA/SE | Red | High | No | No | Yes |
A decrease in culturability compared to the culturability of unstained cells was considered an undesirable effect.
An increase in adhesion of stained cells compared to adhesion of unstained cells was considered undesirable.
ND, not determined.
Fluorescence faded quickly.
None of the blue stains examined were found to be acceptable (Table 1). Some of the red stains yielded promising results; the most notable were DiI, SP-DiIC18(3), and TAMRA/SE, which are currently undergoing further testing (Table 1). The ability to reduce CTC, which results in intense red intracellular crystals, has been proposed as a widely applicable means to assess the percentage of bacterial cells in a given sample which are metabolically active (2, 37, 39, 52, 54). It has been reported that staining conditions can significantly affect the reduction of CTC to CT-formazan by bacteria (35, 41, 48). However, even after several modifications to the CTC staining protocol were evaluated (pH 6 versus pH 8; 5 mM PB versus 50 mM PB; presence and absence of 10% R2A broth), no uptake and intracellular deposition of CT-formazan crystals by DA001 were observed. DA001 is a member of the genus Comamonas, whose members are likely to constitute a significant proportion of many terrestrial and aquatic environmental microbial communities. The inability of CTC to stain DA001 cells was rather surprising but confirms the findings of others (48) that CTC staining should not be regarded as a general and universal measure of metabolic activity.
Of the seven green stains that were evaluated, CFDA/SE, CMFDA, and DiA met all the requirements of the first and second stages of the screening analysis (Table 1). However, during the third stage of testing, both CMFDA and DiA were lost by cells after only 2 to 3 days. CFDA/SE was able to label cells without compromising cell viability or altering cell adhesion characteristics. A high percentage of cells also remained fluorescent for at least 3 weeks during incubation in mNCAGW at 15°C after staining with 10, 50, and 100 μM CFDA/SE (data not shown). Additional work using flow cytometry and epifluorescence microscopy has shown that CFDA/SE-stained cells remain fluorescent in formaldehyde-fixed groundwater samples for up to 3 months (data not shown).
CFDA/SE has been used extensively in mammalian cell studies to track nondividing cells for up to 6 months and growing cells through at least eight divisions (32). However, this stain has been used only on a limited basis for labeling prokaryotes, mostly in the area of food microbiology. Breeuwer et al. (4) used CFDA/SE to allow for continuous measurement of intracellular pH in Lactococcus lactis, while Bunthof et al. (5) used it to assess the viability of the same organism incubated under a variety of different stress conditions. Uerkert et al. (47) combined the use of CFDA/SE with flow cytometry to monitor cell division and cell injury of Lactobacillus plantarum.
All the strains screened were able to cleave the two acetates of CFDA/SE, as evident from the fluorescence of the supernatant during the staining procedure. However, the resulting fluorophore either was unable to react with or was not retained by the cells of some species (Table 2). Both Breeuwer et al. (4) and Uerkert et al. (47) reported difficulty staining E. coli and other gram-negative bacteria with CFDA/SE. They attributed this to the inability of the compound to pass through the outer membrane of the gram-negative cell wall and stated that EDTA improved uptake. The present research indicated that CFDA/SE was able to efficiently stain E. coli BL cells without requiring any pretreatment, although the fluorescence was very faint. Addition of EDTA (5 mM) during staining of DA001 actually resulted in lower fluorescence per cell compared to cells not exposed to EDTA (data not shown). CFDA/SE stained members of most gram-negative genera quite well; the notable exceptions were the five pseudomonads. Most of the gram-positive strains were stained very efficiently and brightly; the only exception was S. albus cells. This further indicates that the inability of CFDA/SE to stain members of a given genus is not simply a function of cell wall organization. One intriguing aspect of this screening was the frequent observation that a cell showing bright green CFDA/SE fluorescence emitted no blue fluorescence when it was counterstained with DAPI. Whether this was due to poor uptake or rapid loss of DAPI from the cells is not known, but it may have some implications for previous bacterial transport studies which utilized DAPI as the cell-labeling compound (18, 24). Although the longevity of CFDA/SE staining has not been determined for these other bacterial strains, the strains that were stained remained stained for at least 72 h in NCAGW at 15°C. The fact that a broad range of organisms were efficiently stained indicates that the CFDA/SE labeling method may be widely applicable for other microbial ecology research. However, due to the variability encountered during this research with respect to the staining procedure and the organism being stained, optimization of CFDA/SE staining should be performed on a case-by-case basis.
TABLE 2.
CFDA/SE staining efficiency for different bacteria
| Organism | Staining efficiency (%) | Comments |
|---|---|---|
| Gram-negative organisms | ||
| Acinetobacter johnsonii | 98 ± 4 | Moderately bright staining but high background fluorescence |
| Alcaligenes eutrophus | 61 ± 27 | Faint to moderately bright staining |
| Comamonas sp. strain DA001 | 99 ± 5 | |
| Cytophaga pectinovora | 100 ± 0 | Some CFDA/SE-positive cells are not DAPI positive |
| Enterobacter aerogenes | 98 ± 5 | |
| Erwinia herbicola | 98 ± 5 | Some very faint staining |
| Escherichia coli BL | 100 ± 0 | Some very faint staining; some CFDA/SE-positive cells are not DAPI positive |
| Flavobacterium odoratum | 100 ± 0 | Very good staining, some variability |
| Klebsiella sp. strain 1PC | 100 ± 0 | Bright staining |
| Pseudomonas cepacia | 100 ± 0 | Faint staining but still visible |
| Pseudomonas fluorescens | <10 | Very faint staining |
| Pseudomonas putida | <10 | |
| Pseudomonas sp. strain LB300 | <10 | Very faint staining |
| Pseudomonas sp. strain To11A | 100 ± 0 | Faint staining but still visible |
| Rahnella aquatilis BFB | 100 ± 0 | Moderately bright staining; some CFDA/SE-positive cells are not DAPI positive |
| Sphingomonas capsulata | 100 ± 0 | Even staining; some CFDA/SE-positive cells are not DAPI positive |
| Gram-positive organisms | ||
| Arthrobacter globiformis | 100 ± 0 | Very good staining; cells are more visible with CFDA/SE than with DAPI |
| Bacillus subtilis | 100 ± 0 | Some variable staining and high background fluorescence |
| Micrococcus luteus | 100 ± 0 | Very bright staining; some CFDA/SE-stained cells are not stained with DAPI |
| Paenibacillus polymyxa FER-02 | 100 ± 0 | Uniform; some CFDA/SE-stained cells are not stained with DAPI |
| Rhodococcus rhodochrous | 99 ± 5 | Generally uniform staining |
| Streptomyces albus | <10 | Very few cells are stained |
| Unidentified low-G+C-content strain SO-B2 | 100 ± 0 | Very bright staining |
| Unidentified low-G+C-content strain SO-9 | 100 ± 0 | |
| SO-S10 | 100 ± 0 |
Microcosm fitness experiments.
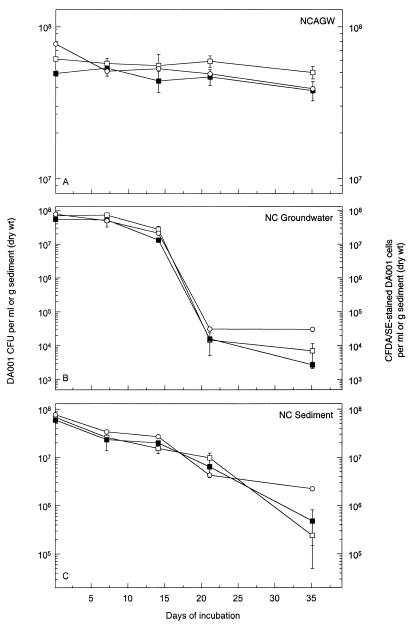
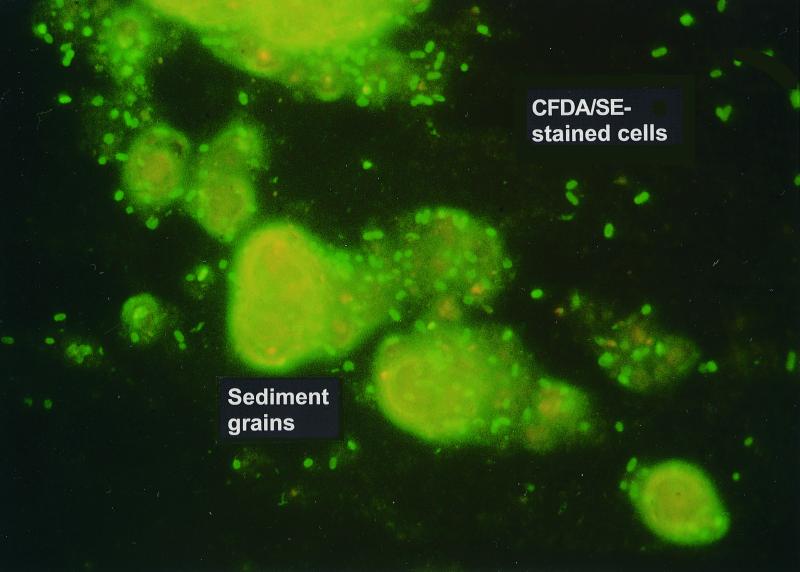
The survival and culturability of unstained and CFDA/SE-stained DA001 cells were virtually identical in mNCAGW, NC groundwater, and NC sediment microcosms (Fig. 1). Survival was substantial in the NC sediment, possibly due to the protection from predation that the sediment afforded compared to NC groundwater. Cells remained fluorescent in all matrices for up to 35 days. The culturable cell densities in the different microcosms were mirrored by the concentrations of CFDA/SE-stained cells over time (Fig. 1), indicating that fluorescence is likely lost shortly after cells become nonviable. Given this, and the fact that neither the NC groundwater microcosms nor the NC sediment microcosms exhibited a detectable increase in culturable cells, it is reasonable to assume that the introduced DA001 cells remained in a nongrowth state throughout the incubation and did not utilize any labile carbon present. If growth did occur, the number of culturable cells would be expected to increase, while the number of detectable CFDA-stained cells would be expected to decrease due to dilution of the intracellular concentration of the fluor with each cell division. An exciting result was the ability to enumerate CFDA/SE-stained cells in the sediment microcosms by epifluorescence microscopy, even in the presence of brightly autofluorescing mineral grains (Fig. 2).
FIG. 1.
Comparison of survival of unstained and CFDA/SE-stained DA001 cells as determined by plate counting of unstained (□) and CFDA/SE-stained (■) cells and by epifluorescent direct counting (○) in NCAGW microcosms (A), NC groundwater microcosms (B), and NC sediment microcosms (C). The error bars represent 1 standard deviation of the mean.
FIG. 2.
Micrograph of CFDA/SE-stained DA001 cells in the presence of autofluorescing NC sediment grains.
Optimization of CFDA/SE staining and sample processing.
Initial optimization was performed by using only epifluorescence microscopy to qualitatively assess the effects of different protocols on the efficiency and brightness of CFDA/SE staining. Staining was performed by using 1.0-ml (total volume) samples containing 1 × 109 DA001 cells/ml in 2.0-ml amber microcentrifuge tubes shaken vigorously (300 to 600 rpm), with a final CFDA/SE concentration of 100 μM (delivered in 2 μl of DMSO). Staining was most effective when the cells were suspended in PBS instead of 5 or 50 mM PB or mNCAGW. Staining in basal salts medium during growth on acetate (same cell and CFDA concentrations) yielded uniformly faintly fluorescent cells. Incubation at 35 to 41°C yielded better staining than incubation at room temperature. Using a CFDA/SE solution which had been repeatedly frozen and thawed instead of a freshly prepared solution did not affect the quality of staining in any medium. During microscopic examination, cells stained in PBS appeared to contain very bright yellow-green intracellular inclusions; the fluorescence usually dispersed throughout the entire cell after incubation in NCAGW at 15°C for a few days. The use of SlowFade Light mounting fluid proved to be very effective for preventing fading of cell fluorescence during observation and counting.
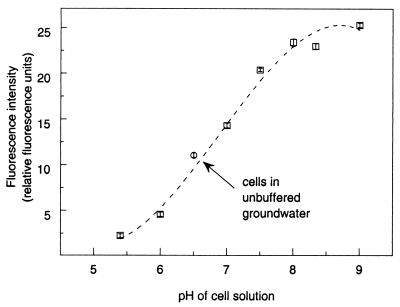
Microplate spectrofluorometry allowed further refinement of the staining and sample analysis protocols by allowing the total fluorescence of cells stained by different procedures to be quantified. The best staining of DA001 cells occurred when a suspension containing 1 × 1010 cells/ml was incubated with stirring at 37 to 42°C in the presence of 100 μM CFDA/SE (Table 3). Higher or lower concentrations of cells or CFDA/SE resulted in less efficient staining; shaking and incubation at room temperature also produced less-fluorescent cells. The fluorescence intensity of the cells was directly proportional to the pH of the solution in which the cells were suspended, with a shift from pH 6 to pH 8 resulting in fivefold-higher cell fluorescence (Fig. 3).
TABLE 3.
Effects of cell density, CFDA/SE concentration, and incubation procedure on fluorescence intensity of stained Comamonas sp. strain DA001 cellsa
| Expt | Mixing conditions | Cell density (cells/ml) | CFDA/SE final concn (μM) | Fluorescence intensity (relative fluorescence units)b |
|---|---|---|---|---|
| A | Stirred, 42°C | 1.0 × 109 | 100 | 30.2 ± 0.3 |
| 1.0 × 1010 | 100 | 72.7 ± 1.6 | ||
| A | Shaken, 35–42°C | 1.0 × 1010 | 50 | 25.6 ± 0.5 |
| 100 | 38.5 ± 0.4 | |||
| 2.5 × 1010 | 50 | 15.9 ± 0.5 | ||
| 100 | 25.2 ± 0.5 | |||
| B | Stirred, room temp | 1.0 × 1010 | 100 | 47.4 ± 0.9 |
| Stirred, 40°C | 1.0 × 1010 | 100 | 82.7 ± 1.9 |
All cells were stained in PBS, washed, incubated at 15°C for 48 h, washed, and resuspended in NCAGW to a final optical density at 550 nm of 0.003 for experiment A and in Oyster groundwater to a final optical density at 550 nm of 0.005 for experiment B prior to microplate spectrofluorimetry. Experiment B samples were also adjusted to pH 8.0 (0.01 mM phosphates).
Measured with the SPECTRAmax GEMINI scanning microplate spectrofluorometer.
FIG. 3.
Fluorescence intensities of suspensions of CFDA/SE-stained DA001 cells (approximately 5 × 106 cells/ml) buffered to different pH values, as measured with the SPECTRAmax GEMINI scanning microplate spectrofluorometer. The error bars represent 1 standard deviation of the mean based on eight analysis replicates.
Based on these experiments, a standard staining protocol was established. Cells were grown in appropriate media to the early stationary phase, harvested by centrifugation, washed twice in PBS, and resuspended in PBS to a concentration of 1 × 1010 cells/ml in a 50-ml centrifuge tube. CFDA/SE (50 mM in DMSO) was added to a final concentration of 100 μM. A small stir bar was added, and the tube was placed in a water bath at 25°C with a stir plate positioned under the bath to allow vigorous mixing of the solution during staining. The temperature of the water bath was cycled between 25 and 37°C five times, and the total staining time was approximately 2.5 h. The stained cells were then harvested by centrifugation, washed twice in NCAGW, and resuspended in a final volume of NCAGW equal to the original culture volume. Cells were incubated at 15°C for 48 to 72 h with shaking, harvested by centrifugation, washed twice in NCAGW, and resuspended in fresh NCAGW before use in any experiments.
Product literature available from Molecular Probes regarding CFDA/SE indicated that all solutions containing the compound needed to be protected from light to avoid fluorescence fading (20). Cells stained with CFDA/SE proved to be very photostable during laboratory exposure to incandescent and fluorescent light; there was no detectable decrease in the total fluorescence of the cell solution as determined by microplate spectrofluorometry or in the fluorescence per cell as determined by flow cytometry (data not shown). However, exposure to direct sunlight led to significant losses (>20%) of both total and per-cell fluorescence within 10 min (data not shown). Care should therefore be taken to avoid exposure of samples containing CFDA/SE-stained cells to sunlight.
Stained-cell detection methods.
The Turner Designs fluorometer was able to detect CFDA/SE-stained cells, and the lower detection limit was approximately 1 × 104 cells/ml when the instrument's 25-ml discrete sample chamber was used. However, the instrument was somewhat awkward to set up and operate, and the data capture capabilities were somewhat limited. The use of an HPLC fluorescence detector coupled with small-volume flow cells did not yield consistent results, and several instrument problems precluded further development.
Both the Packard FLUOROCOUNT and Molecular Devices SPECTRAmax GEMINI fluorescence microplate readers were similar with respect to the lower detection limit (approximately 105 cells/ml) and sample analysis time (<1 min per 96-well plate). The major advantage of the GEMINI instrument was the fact that it was a scanning spectrofluorometer that employed dual monochrometers rather than filters. This allowed the excitation and emission wavelengths to be manipulated to optimize the sensitivity for any fluorescent compound under any conditions and also eliminated the need to buy a new filter set for each new fluorescent compound being screened. The optimized spectrofluorometer settings for CFDA/SE-stained cells were as follows: Ex 495 nm, Em 538 nm, cutoff 530 nm. The detection limit for the SPECTRAmax GEMINI instrument was lowered by 0.33 to 0.5 order of magnitude by adjusting the pH of the samples to 8.0 with potassium phosphate buffer (pH 8.0) (final concentration, 0.01 mM) prior to analysis.
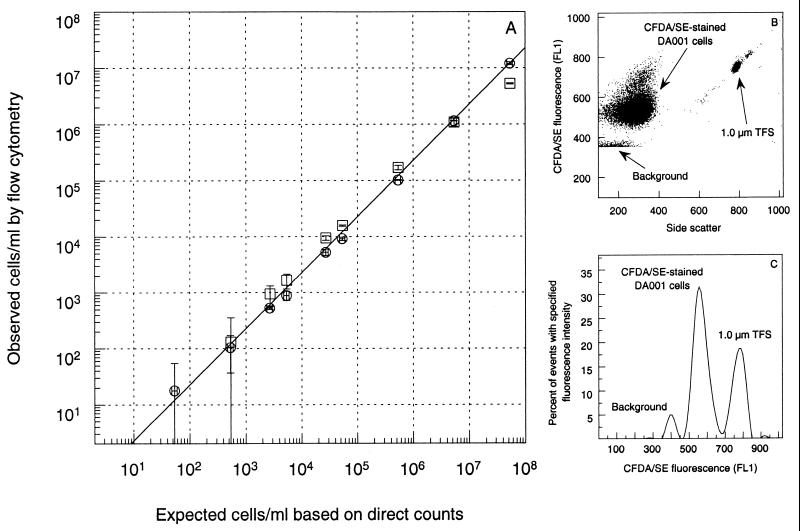
Flow cytometry also proved to be effective for analyzing and enumerating CFDA/SE-stained DA001 cells. When the FACS Vantage instrument was used to count cells stained with both DAPI and CFDA/SE (data not shown), the staining efficiency was determined to be >95%. A dilution series of CFDA/SE-stained cells in NC groundwater and fixed, pH-adjusted NC groundwater indicated that the practical lower detection limit was just slightly greater than 1,000 cells/ml (Fig. 4A); dilutions in NCAGW and fixed NC groundwater gave similar results (data not shown). The absolute lower detection limit is determined by the amount of sample that is analyzed, as well as any background fluorescence, but theoretically it is 1 cell/ml. Figures 4B and C present flow cytometry results for CFDA/SE-stained cells in NC groundwater, which clearly illustrate that the stained cells can be distinguished from indigenous bacterial cells of similar density (i.e., equivalent side scatter values), as well as from the TransFluorSpheres added to the sample for enumeration purposes. This second point is important, because although the strongest fluorescence emission from the TransFluorSpheres is in the red range (645 nm), a significant amount of green fluorescence is emitted during resonance energy transfer among the three dyes in the microspheres.
FIG. 4.
(A) Expected and observed CFDA/SE-stained DA001 cell concentrations in NC groundwater (pH 6.0) (□) and NC groundwater preserved with 1% (wt/vol) formaldehyde and buffered to pH 8 (○), as determined by epifluorescence microscopy and flow cytometry. The x-axis error bars represent the calculated error based on three replicate direct microscopic counts of the initial cell solution used to prepare each dilution series. The y-axis error bars represent the error associated with the mean from three replicate flow cytometry analyses after the mean from three replicate flow cytometry analyses of the blank had been subtracted to account for background fluorescence. (B and C) Flow cytometric dot plot (B) and histogram (C) of CFDA/SE-stained DA001 cells in formaldehyde-fixed NC groundwater. FL1 is the optical filter in the flow cytometer which allows the sample fluorescence emitted at 530 ± 30 nm to be quantified. TFS, TransFluorSpheres.
Intact-core experiments.
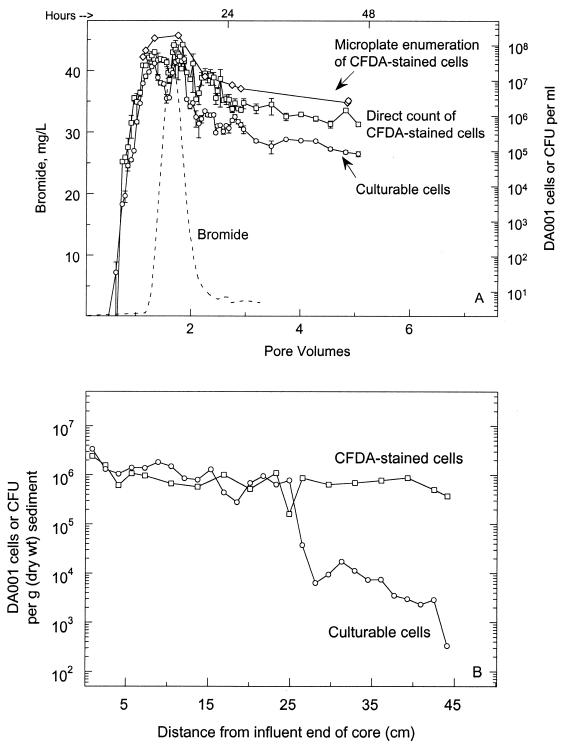
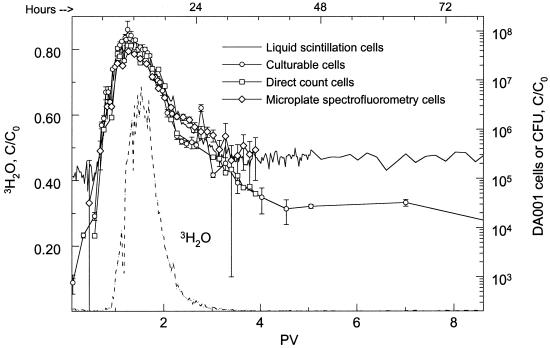
Transport of DA001 cells through intact sediment cores was similar regardless of the tracking method used (Fig. 5A and 6). Plate counting of DA001 CFU and liquid scintillation analysis of 14C-labeled DA001 cells yielded the same results as direct microscopic counting and microplate spectrofluorometry of CFDA/SE-stained cells. During the later time points of experiment 2, the CFDA/SE direct counts matched the CFU very closely, while cell concentrations based on microplate spectrofluorometry and liquid scintillation paralleled each other. In both experiments, cells passed through the cores significantly faster than the conservative tracers passed through, and the peak concentration was observed to elute from the cores sooner for the cells than for the tracers. This is seen more clearly in Fig. 6 than in Fig. 5A because of the multiple peaks in the core NC7-2 cell breakthrough curve.
FIG. 5.
(A) Concentrations of CFDA/SE-stained DA001 cells in the effluent of intact sediment core NC7-2, as determined by plate counting (○), epifluorescent direct counting (□), and microplate spectrofluorometry (◊). Breakthrough of the conservative bromide tracer is also shown (–––). The initial concentrations of DA001 and Br injected into the core were 7 × 107 cells/ml and 50 μg/ml, respectively. The error bars represent 1 standard deviation of the mean. (B) Sediment concentration profile for CFDA/SE-stained DA001 cells along the length of intact core NC7-2, as determined by plate counting (○) and epifluorescent direct counting (□).
FIG. 6.
Concentrations of 14C-labeled DA001 cells stained with CFDA/SE in the effluent of intact sediment core NC9-2SAT, as determined by liquid scintillation counting (——), plate counting (○), epifluorescent direct counting (□), and microplate spectrofluorometry (◊). Breakthrough of the conservative 3H2O tracer is also shown (–––). The initial concentration of DA001 injected into the core was 9 × 107 cells/ml. The error bars represent 1 standard deviation of the mean.
The distribution of DA001 cells remaining in the sediment of core NC7-2 is shown in Fig. 5B. There is agreement between the plate count and direct microscopic count data for approximately one-half the length of the core, and the profile is roughly flat over this interval. At 25 cm from the influent end of the core, the number of CFU rapidly decreases, whereas the direct counts of CFDA/SE-stained cells remain relatively unchanged. The reason for this is not yet known. The distribution of DA001 in the sediment based on the direct count data, however, is more representative than the distribution based on the CFU data when the data are compared to the sediment distribution of DA001 in other intact cores (H. Dong, T. Onstott, M. DeFlaun, M. Fuller, S. Streger, R. Rothmel, and B. Mailloux, submitted for publication). Therefore, it is assumed that in this case the direct count data represent the actual sediment distribution of DA001 more realistically than the CFU data do.
Conclusions.
This research resulted in the development of a long-term fluorescent staining technique for nondividing cells which has no apparent effects on cell viability, adhesion, and transport characteristics. The stained cells were quantifiable by epifluorescence microscopy, microplate spectrofluorometry, and flow cytometry. Cells stained with CFDA/SE could be distinguished from autofluorescing sediment particles by microscopy, allowing cell densities in sediment samples to be determined. Transport of DA001 cells determined by enumeration of CFDA/SE-stained cells was identical to DA001 transport determined by more established methods. It is believed that the utility of CFDA/SE for tracking microorganisms will become limited if the stained cells begin to grow, since this will result in approximately twofold dilution of the fluorescent moiety with each successive cell division. Additional work with this technique that will be reported elsewhere includes field-scale bacterial transport experiments using CFDA/SE-stained DA001 cells with near-real-time microplate spectrofluorometry enumeration (M. Fuller, B. Mailloux, P. Zhang, S. Vainberg, W. Johnson, T. Onstott, and M. DeFlaun, submitted for publication); experiments examining protozoan predation dynamics using CFDA/SE-stained cells under both laboratory and field conditions; experiments coupling the staining procedure with an immunomagnetic separation-concentration (ferrographic) method to reduce the lower detection limit to 5 to 10 cells/ml (22); and several more intact-core bacterial transport experiments.
This bacterial tracking technique has applications outside the field of bacterial transport. We believe that it may prove to be useful in public health microbiology, allowing the survival and movement of pathogen and pathogen surrogates to be monitored in terrestrial, aquatic, and even food-processing environments. The technique may also be useful for studying infection and colonization by pathogens in vivo using animal models.
ACKNOWLEDGMENTS
We acknowledge the support of the DOE Office of Energy Research Natural and Accelerated Bioremediation Research Program Assessment Element (grant DE-FG02-98ER62712).
We also acknowledge Frank Wobber, the program manager for the grant, for his leadership and Tim Griffin of Golder Associates for his excellent management of the field site operations. Access to the field site was granted by The Nature Conservancy. We also thank Kate Gillespie, a research associate who assisted with the experiments, and Andrew Beavis, Princeton University Flow Cytometry Core Facility Manager, for his assistance with flow cytometric analyses.
REFERENCES
- 1.Bales R, Li S, Maguire K, Yahya M, Gerba C, Harvey R. Virus and bacteria transport in a sandy aquifer, Cape Cod, MA. Ground Water. 1995;33:653–661. [Google Scholar]
- 2.Bhupathiraju V, Hernandez M, Landfear D, Alvarez-Cohen L. Application of tetrazolium dye as an indicator of viability in anaerobic bacteria. J Microbiol Methods. 1999;37:231–243. doi: 10.1016/s0167-7012(99)00069-x. [DOI] [PubMed] [Google Scholar]
- 3.Blaß S, Lennartz K. A simple method for counting small numbers of cells by flow cytometry. Z Naturforsh Teil C Biochem Biophys Biol Virol. 1991;46:1130–1133. [PubMed] [Google Scholar]
- 4.Breeuwer P, Drocourt J-L, Rombouts F, Abee T. A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl Environ Microbiol. 1996;62:178–183. doi: 10.1128/aem.62.1.178-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunthof C J, van den Braak S, Breeuwer P, Rombouts F M, Abee T. Rapid fluorescence assessment of the viability of stressed Lactococcus lactis. Appl Environ Microbiol. 1999;65:3681–3689. doi: 10.1128/aem.65.8.3681-3689.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champ D, Schroeter J. Bacterial transport in fractured rock—a field-scale tracer test at the Chalk River Nuclear Laboratories. Water Sci Technol. 1988;20:81–87. [Google Scholar]
- 7.DeFlaun M, Murray C, Holben W, Scheibe T, Mills A, Ginn T, Griffin T, Majer E, Wilson J. Preliminary observations on bacterial transport in a coastal plain aquifer. FEMS Microbiol Rev. 1997;20:473–487. [Google Scholar]
- 8.DeFlaun M, Oppenheimer S, Streger S, Condee C, Fletcher M. Alterations in adhesion, transport, and membrane characteristics in an adhesion-deficient pseudomonad. Appl Environ Microbiol. 1999;65:759–765. doi: 10.1128/aem.65.2.759-765.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFlaun M, Tanzer A, McAteer A, Marshall B, Levy S. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl Environ Microbiol. 1990;56:112–119. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dybas M, Barcelona M, Bezborodnikov S, Davies S, Forney L, Heuer H, Kawka O, Mayotte T, Sepulveda-Torres L, Smalla K, Sneathen M, Tiedje J, Voice T, Wiggert D, Witt M, Criddle C. Pilot-scale evaluation of bioaugmentation for in situ remediation of a carbon tetrachloride-contaminated aquifer. Environ Sci Technol. 1998;32:3598–3611. [Google Scholar]
- 11.Errampalli D, Leung K, Cassidy M, Kostrzynska M, Blears M, Lee H, Trevors J. Applications of the green fluorescent protein as a molecular marker in environmental microorganisms. J Microbiol Methods. 1999;35:187–199. doi: 10.1016/s0167-7012(99)00024-x. [DOI] [PubMed] [Google Scholar]
- 12.Errampalli D, Okamura H, Lee H, Trevors J, van Elsas J. Green fluorescent protein as a marker to monitor survival of phenanthrene-mineralizing Pseudomonas sp. UG14Gr in creosote-containing soil. FEMS Microbiol Ecol. 1998;26:181–191. [Google Scholar]
- 13.Fleminger G, Shabtai Y. Direct and rapid analysis of the adhesion of bacteria to solid surfaces: interaction of fluorescently labeled Rhodococcus strain GIN-1 (NCIMB 40340) cells with titanium-rich particles. Appl Environ Microbiol. 1995;61:4357–4361. doi: 10.1128/aem.61.12.4357-4361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller, M., H. Dong, B. Mailloux, T. Onstott, and M. DeFlaun. Examining bacterial transport in intact cores from Oyster, Virginia: effect of sedimentary facies type on bacterial breakthrough and retention. Water Resour. Res., in press.
- 15.Hareland W A, Crawford R L, Chapman P J, Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975;121:272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey R. In situ and laboratory methods to study subsurface microbial transport. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 586–599. [Google Scholar]
- 17.Harvey R, Garabedian S. Use of colloid filtration theory in modeling movement of bacteria through a contaminated sandy aquifer. Environ Sci Technol. 1991;25:178–185. [Google Scholar]
- 18.Harvey R, George L, Smith R, LeBlanc D. Transport of microspheres and indigenous bacteria through a sandy aquifer: results of natural- and forced-gradient tracer experiments. Environ Sci Technol. 1989;23:51–56. [Google Scholar]
- 19.Harvey R, Kinner N, MacDonald D, Metge D, Bunn A. Role of physical heterogeneity in the interpretation of small-scale laboratory and field observations of bacteria, microbial-sized microspheres, and bromide transport through aquifer sediments. Water Resour Res. 1993;29:2713–2721. [Google Scholar]
- 20.Haugland R. Handbook of fluorescent probes and research chemicals. 6th ed. Eugene, Oreg: Molecular Probes, Inc.; 1996. [Google Scholar]
- 21.Heidelberg J F, Shahamat M, Levin M, Rahman I, Stelma G, Grim C, Colwell R R. Effect of aerosolization on culturability and viability of gram-negative bacteria. Appl Environ Microbiol. 1997;63:3585–3588. doi: 10.1128/aem.63.9.3585-3588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, W., P. Zhang, M. Fuller, B. Mailloux, T. Griffin, T. Onstott, and M. DeFlaun. Ferrographic tracking of bacterial transport in the field at Oyster, VA. Environ. Sci. Technol. in press. [DOI] [PubMed]
- 23.Kepner R L, Jr, Pratt J R. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol Rev. 1994;58:603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Küçükçolak E, Koopman B, Bitton G, Farrah S. Validity of fluorochrome-stained bacteria as tracers of short-term microbial transport through porous media. J Contam Hydrol. 1998;31:349–357. [Google Scholar]
- 25.Leser T D, Boye M, Hendriksen N B. Survival and activity of Pseudomonas sp. strain B13(FR1) in a marine microcosm determined by quantitative PCR and an rRNA-targeting probe and its effect on the indigenous bacterioplankton. Appl Environ Microbiol. 1995;61:1201–1207. doi: 10.1128/aem.61.4.1201-1207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McEldowney S, Fletcher M. Effect of growth conditions and surface characteristics of aquatic bacteria on their attachment to solid surfaces. J Gen Microbiol. 1986;132:513–523. [Google Scholar]
- 27.Molema G, Mesander G, Kroesen B, Helfrich W, Meijer D, de Leij L. Analysis of in vitro lymphocyte adhesion and transendothelial migration by fluorescent-beads-based flow cytometric cell counting. Cytometry. 1998;32:37–43. doi: 10.1002/(sici)1097-0320(19980501)32:1<37::aid-cyto5>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 28.Nelson M J, Kinsella J, Montoya T. In situ biodegradation of TCE contaminated groundwater. Environ Prog. 1990;9:190–196. [Google Scholar]
- 29.Normander B, Hendriksen N B, Nybroe O. Green fluorescent protein-marked Pseudomonas fluorescens: localization, viability, and activity in the natural barley rhizosphere. Appl Environ Microbiol. 1999;65:4646–4651. doi: 10.1128/aem.65.10.4646-4651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver J. Formation of viable but nonculturable cells. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 239–272. [Google Scholar]
- 31.Oliver J D, Hite F, McDougald D, Andon N L, Simpson L M. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl Environ Microbiol. 1995;61:2624–2630. doi: 10.1128/aem.61.7.2624-2630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parish C. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol Cell Biol. 1999;77:499–508. doi: 10.1046/j.1440-1711.1999.00877.x. [DOI] [PubMed] [Google Scholar]
- 33.Parolin C, Montecucco A, Ciarrocchi G, Pedrali-Noy G, Valisena S, Palumbo M, Palu G. The effect of the minor groove binding agent DAPI (2-aminodiphenyl-indole) on DNA-directed enzymes: an attempt to explain inhibition of plasmid expression in Escherichia coli. FEMS Microbiol Lett. 1990;68:341–346. doi: 10.1111/j.1574-6968.1990.tb13962.x. [DOI] [PubMed] [Google Scholar]
- 34.Pommepuy M, Butin M, Derrien A, Gourmelon M, Colwell R R, Cormier M. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl Environ Microbiol. 1996;62:4621–4626. doi: 10.1128/aem.62.12.4621-4626.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyle B, Broadway S, McFetters G. Factors affecting the determination of respiratory activity on the basis of cyanoditolyl tetrazolium chloride reduction with membrane filtration. Appl Environ Microbiol. 1995;61:4304–4309. doi: 10.1128/aem.61.12.4304-4309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman I, Shahamat M, Kirchman P A, Russek-Cohen E, Colwell R R. Methionine uptake and cytopathogenicity of viable but nonculturable Shigella dysenteriae type 1. Appl Environ Microbiol. 1994;60:3573–3578. doi: 10.1128/aem.60.10.3573-3578.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez G G, Phipps D, Ishiguro K, Ridgway H F. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roszak D, Colwell R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;41:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaule G, Flemming H-C, Ridgway H F. Use of 5-cyano-2,3-ditolyl tetrazolium chloride for quantifying planktonic and sessile respiring bacteria in drinking water. Appl Environ Microbiol. 1993;59:3850–3857. doi: 10.1128/aem.59.11.3850-3857.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semprini L, McCarty P. Comparison between model simulations and field results for in-situ biorestoration of chlorinated aliphatics: part 2. Cometabolic transformations. Ground Water. 1992;30:37–44. [Google Scholar]
- 41.Smith J, McFeters G. Effects of substrates and phosphate on INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride) and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli. J Appl Bacteriol. 1996;80:209–215. doi: 10.1111/j.1365-2672.1996.tb03212.x. [DOI] [PubMed] [Google Scholar]
- 42.Steffan R, Sperry K, Walsh M, Vainberg S, Condee C. Field-scale evaluation of in situ bioaugmentation for remediation of chlorinated solvents in groundwater. Environ Sci Technol. 1999;33:2771–2781. [Google Scholar]
- 43.Stewart C, Steinkamp J. Quantitation of cell concentration using the flow cytometer. Cytometry. 1982;2:238–243. doi: 10.1002/cyto.990020407. [DOI] [PubMed] [Google Scholar]
- 44.Thiem S M, Krumme M L, Smith R L, Tiedje J M. Use of molecular techniques to evaluate the survival of a microorganism injected into an aquifer. Appl Environ Microbiol. 1994;60:1059–1067. doi: 10.1128/aem.60.4.1059-1067.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai Y L, Olson B H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuji T, Kawasaki Y, Takeshima S, Sekiya T, Tanaka S. A new fluorescent staining assay for visualizing living microorganisms in soil. Appl Environ Microbiol. 1995;61:3415–3421. doi: 10.1128/aem.61.9.3415-3421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueckert J, von-Caron N, Bos A, ter Steeg P. Flow cytometric analysis of Lactobacillus plantarum to monitor lag times, cell division and injury. Lett Appl Microbiol. 1997;25:295–299. doi: 10.1046/j.1472-765x.1997.00225.x. [DOI] [PubMed] [Google Scholar]
- 48.Ullrich S, Karrasch B, Hoppe H-G, Jeskulke K, Mehrens M. Toxic effects on bacterial metabolism of the redox dye 5-cyano-2,3-ditolyl tetrazolium chloride. Appl Environ Microbiol. 1996;62:4587–4593. doi: 10.1128/aem.62.12.4587-4593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Elsas J, Rosada A, Wolters A, Moore E, Karlson U. Quantitative detection of Sphingomonas chlorophenolica in soil via competitive polymerase chain reaction. J Appl Microbiol. 1998;85:463–471. doi: 10.1046/j.1365-2672.1998.853509.x. [DOI] [PubMed] [Google Scholar]
- 50.van Overbeek L S, Eberl L, Givskov M, Molin S, van Elsas J D. Survival of, and induced stress resistance in, carbon-starved Pseudomonas fluorescens cells residing in soil. Appl Environ Microbiol. 1995;61:4202–4208. doi: 10.1128/aem.61.12.4202-4208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe K, Yamamoto S, Hino S, Harayama S. Population dynamics of phenol-degrading bacteria in activated sludge determined by gyrB-targeted quantitative PCR. Appl Environ Microbiol. 1998;64:1203–1209. doi: 10.1128/aem.64.4.1203-1209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winding A, Binnerup S, Sørensen J. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl Environ Microbiol. 1994;60:2869–2875. doi: 10.1128/aem.60.8.2869-2875.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winkler J, Timmis K, Snyder R. Tracking the response of Burkholderia cepacia G4 5223-PR1 in aquifer microcosms. Appl Environ Microbiol. 1995;61:448–455. doi: 10.1128/aem.61.2.448-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu F, McFeters G. Rapid in situ assessment of physiological activities in bacterial biofilms using fluorescent probes. J Microbiol Methods. 1994;20:1–10. doi: 10.1016/0167-7012(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 55.Yu W, Dodds W, Banks M, Skalsky J, Strauss E. Optimal staining and sample storage time for direct microscopic enumeration of total and active bacteria in soil with two fluorescent dyes. Appl Environ Microbiol. 1995;61:3367–3372. doi: 10.1128/aem.61.9.3367-3372.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]