Abstract
Infection with HIV can cripple the immune system and lead to AIDS. Hepatitis B virus (HBV) is a hepadnavirus that causes human liver diseases. Both pathogens are major public health problems affecting millions of people worldwide. The polymerases from both viruses are the most common drug target for viral inhibition, sharing common architecture at their active sites. The L-nucleoside drugs emtricitabine and lamivudine are widely used HIV reverse transcriptase (RT) and HBV polymerase (Pol) inhibitors. Nevertheless, structural details of their binding to RT(Pol)/nucleic acid remained unknown until recently. Here, we discuss the implications of these structures, alongside related complexes with L-dNTPs, for the development of novel Lnucleos(t)ide drugs, and prospects for repurposing them.
Keywords: HIV-1 reverse transcriptase, L-nucleoside, hepatitis B virus polymerase, drug resistance, drug development, repurposing
Graphical Abstract
Introduction
HIV/AIDS became a global epidemic during the 1980s, and remains a major threat to global health. The most common drug target for inhibition is HIV-1 RT, which is essential for viral replication. Anti-RT drugs have a central role in highly active antiretroviral therapy (HAART; see Glossary), a treatment regimen that typically includes two nucleos(t)ide RT inhibitors (NRTIs), as well as in pre-exposure prophylaxis (PrEP). With nearly 40 million people with HIV worldwide and about half receiving HAART, this success story was effectively repurposed against HBV, which presents a Pol structurally related to RT, making NRTIs the backbone of HBV treatment. HBV causes inflammation of the liver, triggering several hepatic diseases, and affects ~250 million patients worldwide.1–3
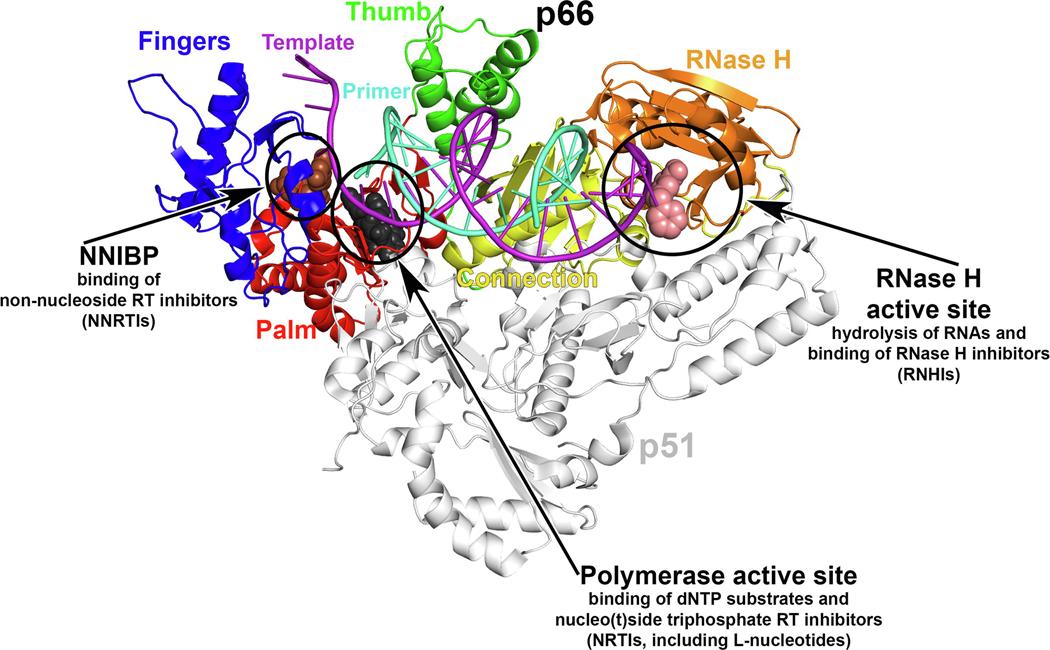
RT functions as an RNA- and DNA-dependent DNA Pol, making a DNA copy of the viral RNA genome by incorporating dNTPs to a growing primer strand. RT is formed by a p66/p51 heterodimer, in which the first monomer bears catalytic activities (Pol and RNase H) and the second provides a stabilizing scaffold (Figure 1a). It can be easily argued that RT is the best understood Pol in terms of structure, mechanisms of catalysis, inhibition, and resistance.2,4–6 By contrast, HBV Pol, which is a multifunctional protein comprising four domains (a terminal protein, spacer, RT, and RNase H), is highly insoluble when expressed as a recombinant protein.7 This has precluded structural studies, and structural homology models based on the moderate similarity of its RT domain to HIV-1 RT (~35% sequence identity) have been the only tool supporting structure-based drug design.8,9
Figure 1.
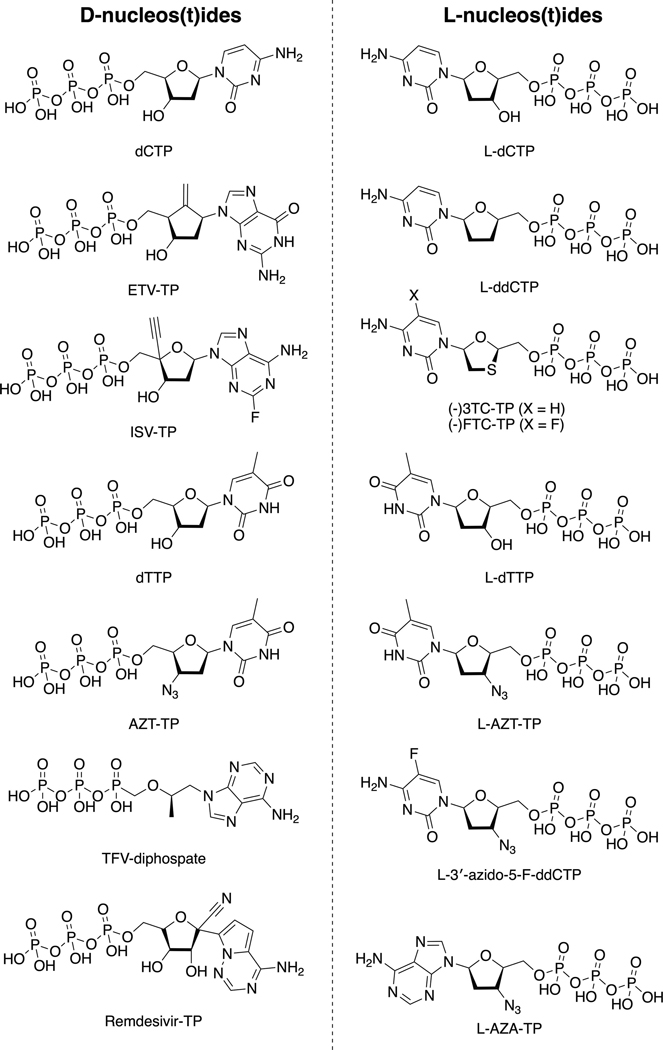
HIV-1 reverse transcriptase (RT) and nucleos(t)ides. (a) HIV-1 RT structure and main binding sites for substrates and inhibitors indicated and described. The structure shown corresponds to RT with bound double-strand (ds)DNA and incoming EFdA-TP [Protein Data Bank (PDB) IDP 5J2M], with template primer, with p66 polymerase fingers, palm, thumb, connection subdomains, and RNase H domain, and with p51 indicated. The NNRTI rilpivirine (brown spheres; PDB ID: 4G1Q) and RNHI 11b (light-pink spheres; PDB ID: 6AOC) are superposed. (b) 2D chemical structures of some of the nucleos(t)ide molecules mentioned in this review, grouped by stereochemistry, as indicated. Abbreviations: (−)-FTC-TP, (−)-2’,3’-dideoxy-5-fluoro-3’thiacytidine-TP; (−)-3TC-TP, (−)-2’,3’-dideoxy-3’-thiacytidine-TP; AZT-TP, 3’-azido-3’-deoxythymidine-TP; dCTP, 2′‐deoxycytidine triphosphate; dTTP, 2’-deoxythymidine TP; ETV-TP, 2-amino-9-[(1S,3R,4S)-4-hydroxy-3-(hydroxymethyl)-2methylidenecyclopentyl]-6,9-dihydro-3H-purin-6-one-TP; ISV-TP, 4’-ethynyl-2-fluoro-2’-deoxyadenosine-TP; L-3ʹ-azido-5-FddCTP, 3’-azido-5-fluoro-2ʹ,3ʹ -dideoxythymidine-TP; L-AZA-TP, L-3’-azido-3’-deoxyadenosine-TP; L-AZT-TP, L-3’-azido-3’deoxythymidine-TP; L-dCTP, L-2′‐deoxycytidine TP; L-ddCTP, L-2′,3′‐dideoxycytidine triphosphate; L-dTTP, L-2′‐deoxythymidine TP; Remdesivir-TP, 1’-cyano-adenosine TP; TFV-diphosphate, [R]-9-[2-(phosphonomethoxy)propyl]adenine diphosphate. Adapted from 2 (a).
Among NRTIs, emtricitabine (FTC) and lamivudine (3TC) are two of the most widely used nucleoside drugs targeting HIV RT and HBV Pol. As with other NRTIs, they are prodrugs administered as nucleosides, but their activity in the body requires conversion to their triphosphate (TP) form. These were discovered during the late 1980s and approved by the US Food and Drug Administration (FDA) between 1997 and 2003. Since then, their biochemical mechanism of action and resistance have been well studied and characterized.10–12 Conversely, structural data of their binding to RT(Pol)/nucleic acid remained elusive until 2019.3,13–15
In this review, we provide an overview of the recent structures of RT/DNA (including wild-type RT, the major drug-resistant mutant M184V, and HBV-associated amino acid substitutions) with the mentioned L-nucleotide drugs, as well as novel structures of RT/DNA with L-dCTP and L-dTTP [the TP form of anti-HBV drug telbivudine (L-dT)16,17] in conjunction with kinetic experiments. These data allow detailed visualization of L-nucleotide binding, incorporation, and resistance. We next discuss the drug development avenues that these structures suggest. Finally, we focus on several recent reports of experiments in cellular and animal models as well as clinical trials indicating that L-nucleosides bear potential for being repurposed for treating cancer, diabetes, and other infectious and autoimmune diseases.
L-nucleotides: binding, incorporation, and drug resistance in HIV-1 RT
The first L-nucleoside analogs identified as potential therapeutics were racemic mixtures of the natural D- and the unnatural Lnucleosides, but it was later observed that one of the stereoisomers was usually primarily responsible for the activity. To date, the most relevant L-nucleosides discovered are still 3TC and FTC, which exhibit not only good antiviral properties, but also low toxicity and a good therapeutic index. The latter results from a combination of efficient phosphorylation by host nucleotide kinases, resistance to deamination by nucleoside deaminases, and the low activity of off-target cellular DNA polymerases, especially mitochondrial DNA polymerase Pol γ. The low stereoselectivity of some viral polymerases toward L-nucleosides (such as HIV-1 RT and HBV Pol) and some nucleoside kinases contrasts with the stricter enantioselectivity of nucleoside degradation enzymes. Thus, this suggests that stereoselectivity is a fortuitous event, rather than evolutionarily driven, because none of these enzymes create or alter chiral centers (reviewed in 18–20).
Paradoxically, the first structural studies of binding and incorporation of (−)3TC-TP and (−)FTC-TP (Figure 1b) were done first with bacterial and cellular DNA polymerases.15,20,21 These studies revealed different mechanisms and degree of stereoselectivity by those polymerases, supporting a lack of evolutionary pressure for stereoselectivity. The Arnold laboratory had previously determined crystal structures of the 3TC-resistant mutant HIV-1 RT (M184I) in both the presence and absence of a dsDNA,22 developing a reliable model that explained the mechanism of the main resistance mutation for 3TC and FTC. However, the mechanistic basis of RT stereoselectivity was not completely understood.
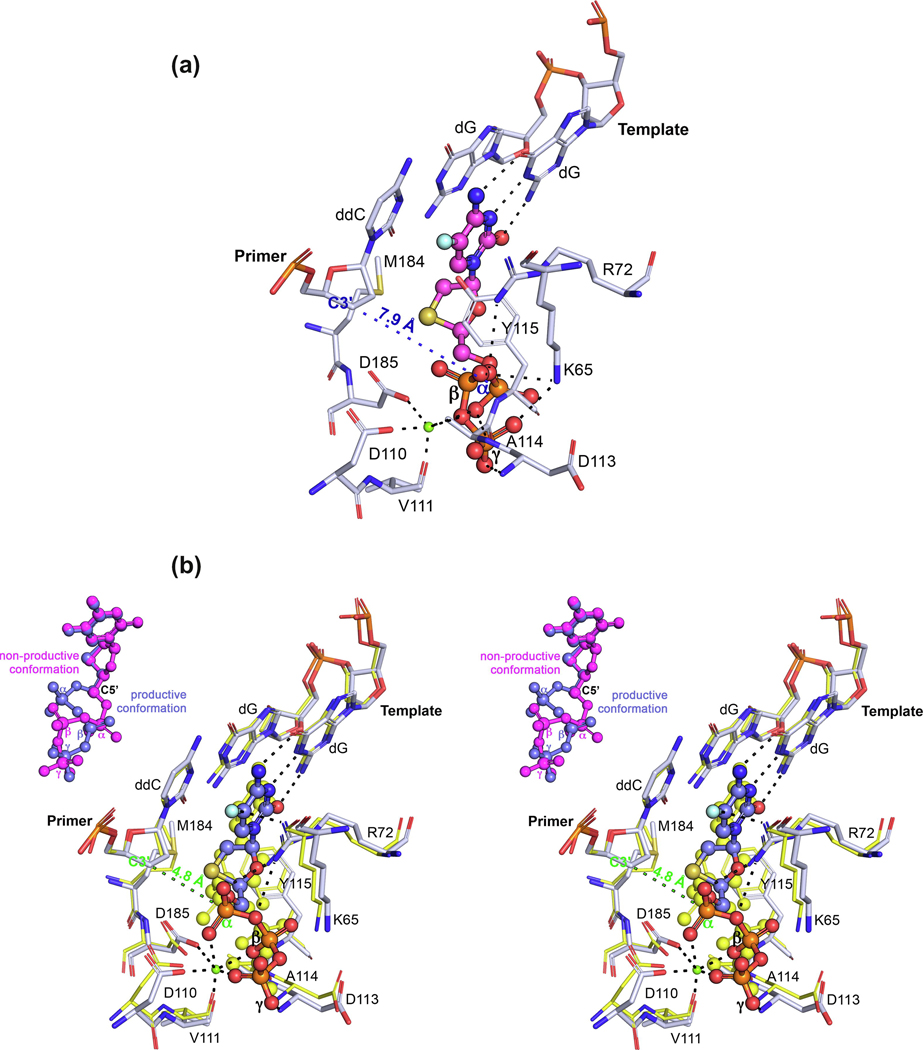
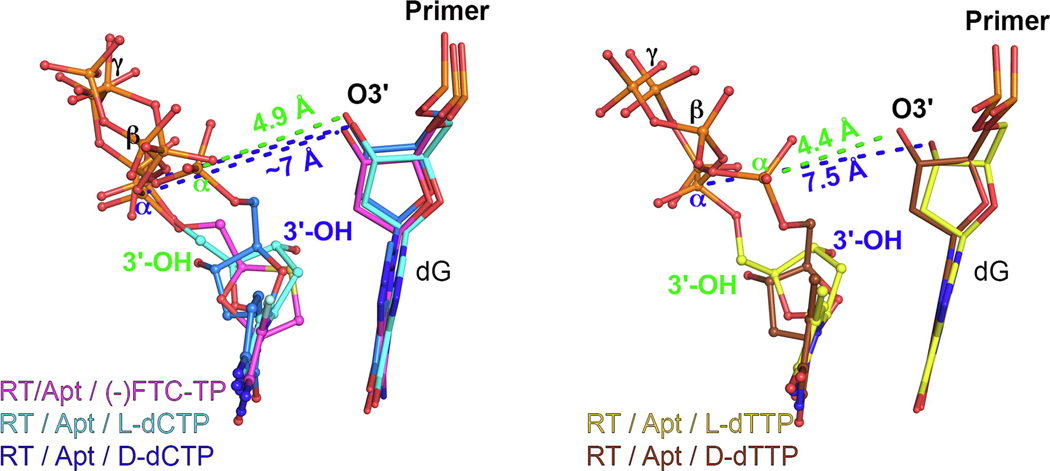
Since 2019, four different articles have been published involving RT/DNA complexes containing either the TP or monophosphate (MP) of 3TC and FTC,13–15 with one of them using a mutant HIV-1 RT as a surrogate for HBV Pol,3 described in the next section. The first set of articles13–15 used a similar approach for the crystallographic studies: an engineered RT protein cross‐linked via a disulfide bridge to the primer strand of a double-strand (ds)DNA, with a dideoxy-terminated primer strand. Both groups reported structures with incoming D-dCTP, (−)FTC‐TP and (+)FTC‐TP (L and D stereoisomers, respectively), and (−)3TC‐TP (L stereoisomer). These crystal structures showed that RT/dsDNA complexes bind L-nucleotides readily, with no steric hindrance that would allow discrimination, in concordance with biochemical and virological data (Figure 2). In most of the structures, the incoming L-nucleotide analogs bind using Watson–Crick base pairing/stacking and a 180° rotation of the sugar ring (compared with D-nucleotides), in a ‘nonproductive’ state, because the distance from the C3ʹ in the deoxyribose ring of the last priming nucleotide [pretranslocation (P)-site] and the α-phosphate (Pα) of the L-nucleotide is ~8 Å. Strikingly, Hung and colleagues,15 in the structures with incoming (−)FTC‐TP and (−)3TC‐TP, observed a second conformation of the analog, a ‘productive’ state, with a C3ʹ−Pα distance of 4.8 Å (Figure 2b), resulting from pivoting of the TP through the C5′ carbon. This means that, if the P-site had a 3ʹ-hydroxy (3ʹ-OH) group, it would be poised for nucleophilic attack to the Pα of the L-nucleotide.
Figure 2.
L-nucleotides: binding, incorporation, and drug resistance in HIV-1 reverse transcriptase (RT). (a) Atomic model of the binding of (−)-FTC-TP (magenta) to HIV-1 RT/DNA (white) in a nonproductive conformation [Protein Data Bank (PDB) IDP 6UJX], with the C3ʹ−Pα distance indicated. (b) Atomic model of the binding of (−)-FTC-TP (violet) to HIV-1 RT/DNA (white) in a productive conformation (PDB ID: 6UJX), with the C3ʹ−Pα distance indicated in green. The previous is superposed with the HIV-1 RT/DNA/dCTP (PDB ID: 6UIT, yellow). An inset is included with the comparison of the two (−)-FTC-TP conformations in PDB ID 6UJX. (c) Superposition of the HIV-1 RT/DNA (gray) with incorporated (−)-FTC-MP (monophosphate, pink, PDB ID: 6WPH) with the HIV-1 RT/DNA bound to the nonproductive (−)-FTC-TP conformer [as in (a)], with C5ʹ atoms indicated for both [C5ʹa and C5ʹb refers to the same atom in the two alternate conformations of (−)-FTC-MP], with an arrow indicating the pivoting movement around this atom. (d) Superposition of the complex depicted in (a) with the complexes indicated (PDB IDs: 6UK0, 6UIR, and 6UIS, respectively, from top to bottom). (e) Superposition of the wild-type and resistance mutant M184V RT/dsDNA/(−)-FTC-TP complexes (as indicated previously), with the steric clash (causing resistance) between the CG1 carbon atom of V184 and the sulfur atom of the oxathiolane ring depicted with spheres (corresponding to their van der Waals volumes). The incoming dNTPs/NRTI-TPs (nucleoside RT inhibitor-TPs) are shown as sphere and stick models [except in (e), for clarity]. The amino acids and nucleic acids, within a distance of 5 Å, are shown as stick models. H‐bond interaction distances within 3.5 Å to the compounds are depicted as black dotted lines. Ca2+ ions are shown as green spheres.
The kinetics experiments reported in the literature14,23 showed that L-nucleotide analogs bind more tightly to RT/nucleic acid compared with their D-nucleotide counterparts and D-dCTP, but that the rate of incorporation is significantly slower. Thus, the structural information allows us to hypothesize two possible scenarios: (i) after long-lived binding of the L-nucleotide in the nonproductive state, chemistry will be preceded by a rearrangement to the productive state; or (ii) the L-nucleotide can bind in either nonproductive or productive states (even more, the nonproductive state could be the majority population), which might decrease the incorporation rate compared with the ‘one-state’ D-nucleotides. Thus, either the L-nucleotide conformational change or binding conformation sampling will be the rate-limiting step, as suggested by pre-steady state kinetic data with lack of a burst.14,23
Although the authors speculate about other possibilities, we hypothesize that the nonproductive state can be stabilized further than the productive state by additional contacts with K65 and the TP moiety. Indeed, Figure 2a highlights two contacts between K65 of RT and the nonproductive state of (−)FTC-TP that are closer than 3.5 Å, whereas, in Figure 2b, such contacts do not exist with the productive state of (−)FTC-TP. Although the K65R mutation is infrequent, it is the other mutation apart from M184V that reduces 3TC or FTC susceptibility (24,25 and the HIV Stanford database; https://hivdb.stanford.edu/dr-summary/resistance-notes/NRTI/). Kinetic experiments show a sharp decrease in the incorporation rate upon K65R mutation, which can be understood by the restricted mobility of the K65R/R72 pair (forming a rigid platform via stacking of their guanidinium groups, as reported previously 24).
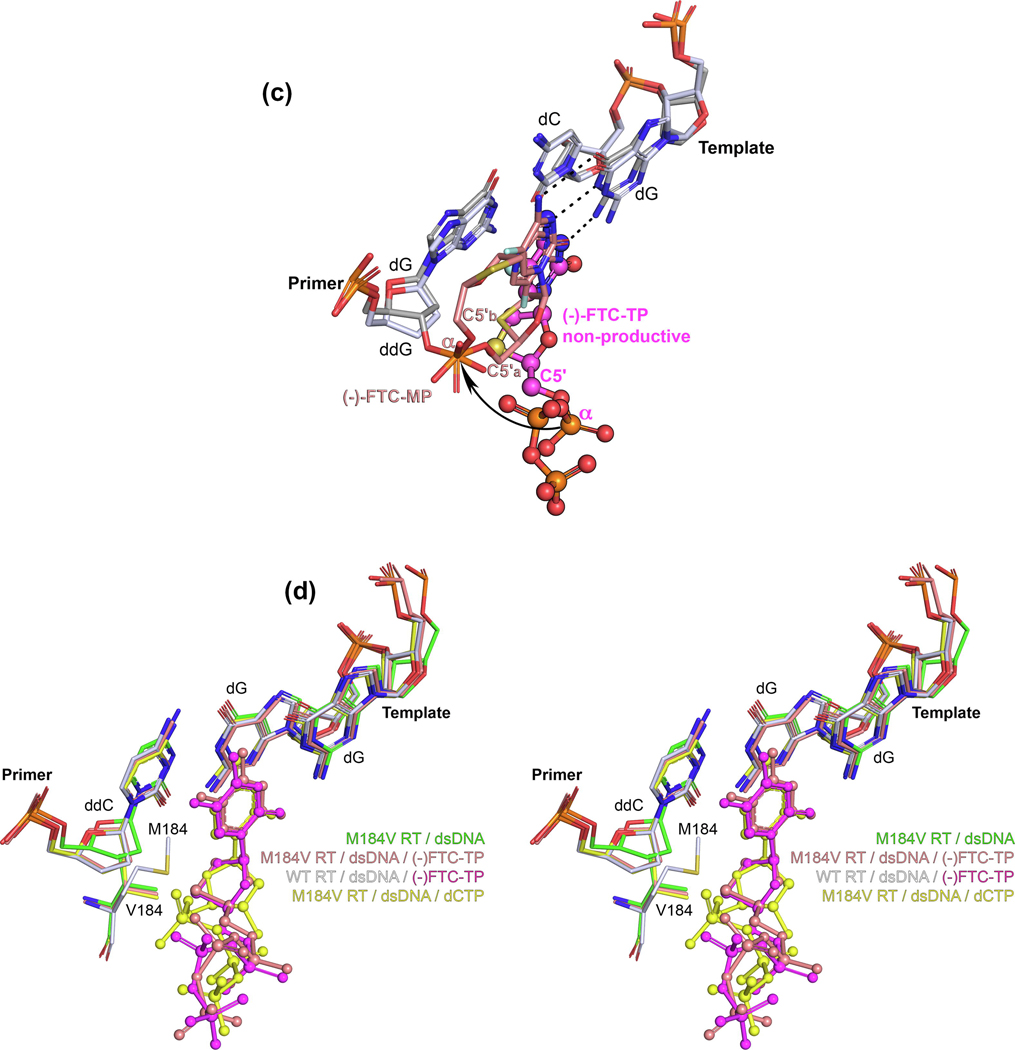
Bertoletti and colleagues recently published a follow-up study13 in which they report the postcatalytic nucleotide (N)-site structure of RT/dsDNA/(−)FTC-MP (FTC incorporated into the primer strand but before translocation to the P-site). They used the same crosslinking strategy as in their previous work, but with a primer strand terminated with dGMP [instead of dideoxy(dd)GMP] and subsequent co-crystallization in the presence of Mg2+ and (−)FTC-TP. The cross-linking between the sixth nucleotide in the primer strand and Q258C blocked translocation and captured the N-site complex. This structure (Figure 2c) permits visualization of the postchemistry step. Even more, a double conformation of the incorporated (−)FTC-MP is observed. Overall, this collection of structures shows the whole sequence of events, from binding to incorporation of (−)FTC-TP (Figures 2a–c). In addition, Bertoletti and colleagues compared the same type of postcatalytic complex but terminated with the D-thymidine analog d4T-MP (Supplementary Data Chart S1 in the supplemental information online), providing a snapshot of the mechanism of how RT develops resistance to d4T via excision (whereas FTC and 3TC are not susceptible to excision), complementing previous biochemical and virological observations.26–28
Returning to the work of Hung and colleagues,15 they also determined structures of M184V RT-DNA, and ternary structures with (−)FTC-TP and dCTP to pinpoint the mechanism by which the M184V mutation discriminates L-nucleotide binding. Interestingly, in the binary M184V RT-DNA complex, they observed a shift of the primer strand as observed previously in a similar M184I RT-DNA complex (Figure 2d).22 However, the ternary complexes with (−)FTC-TP and dCTP have the primer positioned almost identically to the wild-type structures. This reduces the likelihood that primer repositioning is a significant contributing factor to the M184V mutation resistance mechanism. The ternary structures show that the M184V mutation provokes steric hindrance by directly contacting the sulfur atom in the oxathiolane ring of (−)FTC-TP, bound in a nonproductive conformation (Figure 2e).
L-nucleotides: recognition and drug resistance in HBV RT
The intractability of HBV Pol heterologous expression has precluded structural studies, including with the FDA-approved drugs 3TC and FTC. Earlier attempts8 took advantage of the structural homology between the Pol active site of retroviral RTs and Pol. Recently, Yasutake and coworkers translated this strategy from modeling to structure determination. Using HIV-1 RT as a surrogate, they introduce HBV-mimicking mutations to study the detailed interactions between HBV nucleoside inhibitors (NIs) in the active triphosphate form and the HBV-mimicking RT/DNA complexes. First, they used this strategy to understand the structural basis for HBV NI entecavir (Figure 1b).29,30 Recently, they did the same with 3TC,3 and also studied the mechanism of resistance to 3TC of the HBV Pol double-mutant L528M/M552V (note that the authors used the HBV RT domain numbering L180M/M204V, whereas here we use full-length HBV Pol numbering).
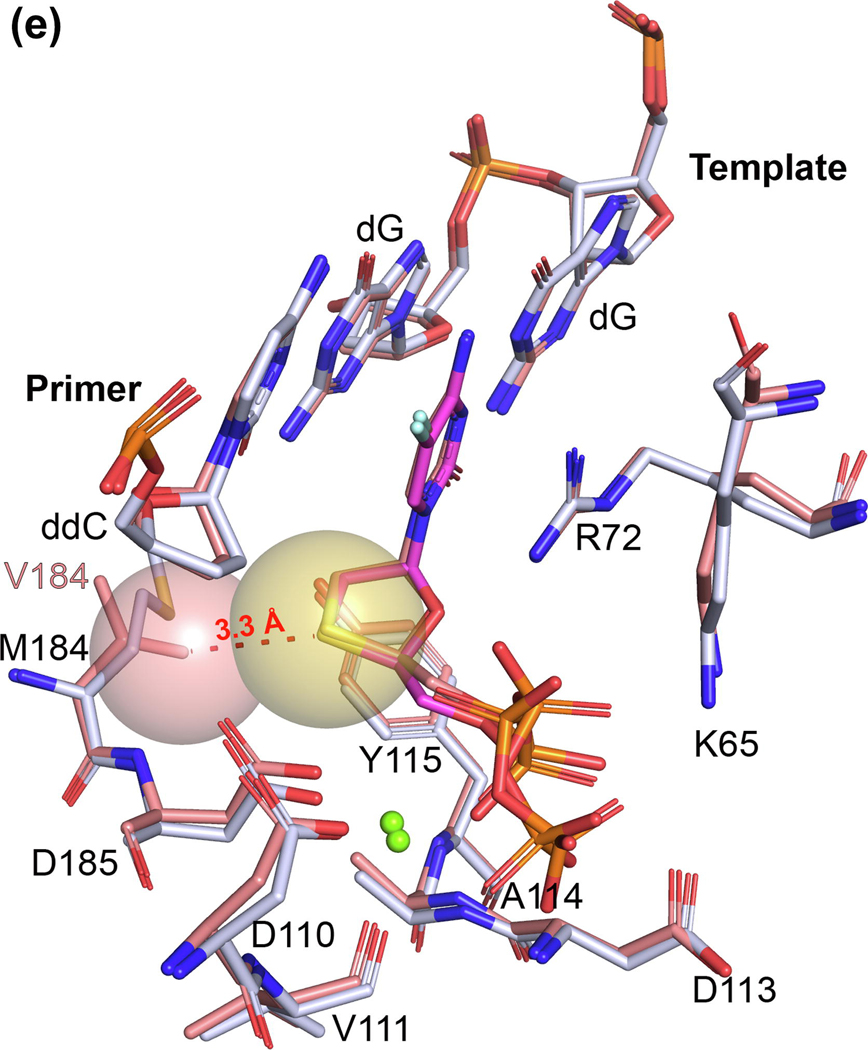
For crystallization of RT/nucleic acid complexes, Yasutake and coworkers used a previously developed 38‐mer hairpin template‐primer DNA aptamer (referred to Apt henceforth; Figure 3a),31,32 which allows crystallization with a 3ʹ-OH terminated primer and subsequent in crystallo soaking with dNTPs and NRTITPs. In this work, they introduced three HBV-mimicking mutations into HIV-1 RT: Y115F/F116Y/Q151M (designated 3M). Despite these mutations, they observed that the incoming (−)3TC‐TP was bound in the nonproductive state, almost identical to the structures determined previously.14,15 In this case, because the primer terminal nucleotide bears the 3ʹ-OH, they found that the 3ʹ-OH primer−L-nucleotide Pα distance was ~7 Å, in concordance with the observed distance of ~8 Å in the structures with dideoxy-terminated primers (Figure 3b).
Figure 3.
L-nucleotides: recognition and drug resistance in hepatitis B virus (HBV) reverse transcriptase (RT). (a) 2D diagram of the 38‐mer hairpin template‐primer mimicking DNA aptamer (Apt) co-crystallized with HIV-1 RT used in this and previous studies32 as well as by Yasutake and coworkers.3,29,30 The nucleotide (N)-site and pretranslocation (P)-site are indicated, with the first poised for dNTP/NRTI-TP (plus divalent cation) soaking. (b) Superposition of the HIV-1 RT/DNA/(−)-FTC-TP in a nonproductive conformation [Protein Data Bank (PDB) ID: 6UJX, as indicated in Figure 2 in the main text] with the HBVmimicking HIV-1 RT mutant 3M RT/Apt (in cyan, except the three indicated mutations in violet) in complex with (−)-3TC-TP (in cyan; PDB ID: 6KDJ), with the C3ʹ−Pα and O3ʹ−Pα distances indicated, respectively. (c) Superposition of the 3M RT/Apt(−)3TC-TP complex with the F160/M184 3M cognate complex (in dark violet; PDB ID: 6KDO) representative of the L528M/M552V HBV resistance mutations. Note that the triphosphate moiety of (−)-3TC-TP could not be assigned by the authors and, thus, is not represented. The fingers subdomain electron density is sparse (see main text), most likely moving between open and closed states, as denoted by the arrow and the distances of key residues, K65 and R72, in both instances. (d) Superposition of the 3M RT/Apt(−)-3TC-TP complex with Q151Mc RT/dsDNA complexes (PDB ID: 5TXO and 5TXP), displaying the conserved interaction between F116Y and K73, and illustrating the similarity between the active site of HBV polymerase (Pol) and this HIV-1 RT mutant. Ca2+ ions are shown as green spheres and the water molecule as a red sphere.
Regarding the mechanism of resistance of double-mutant HBV Pol L528M/M552V, the F160M/M184V 3M RT/Apt/(−)3TC‐TP structure showed defined electron density for the cytosine base and oxathiolane moiety but no clear density for the triphosphate and Mg2+. The latter points to a loosely bound (−)3TC‐TP, whereas the oxathiolane deviates from that in the 3M RT/Apt/(−)3TC‐TP complex by ~1 Å. Related to this, Yasutake and coworkers modeled an open conformation of the fingers subdomain. In most structures of RT/nucleic acid with incoming dNTP or NI-TP, the fingers subdomain clamps onto the former via R72 and K65 interactions with the triphosphate moiety.2,6 The electron density for the fingers region is sparse (data not shown), which points to this subdomain opening and closing, further explaining the lack of clear density of the triphosphate moiety and the loose binding of (−)3TC‐TP (Figure 3c). Given that the electron density of V184 is clear (data not shown), it appears that the misaligned oxathiolane avoids the steric clash with the side chain of V184 at the expense of a reduced binding, explaining the resistance to 3TC.
Similarly, the Arnold group recently studied the resistance mutations Q151M and Q151M complex (designated Q151Mc, formed by Q151M and four associated mutations, A62V, V75I, F77L, and F116Y) in HIV-1 RT.33 Q151M might introduce a conformational perturbation at the N-site, and the mutated pocket might exist in multiple conformations. It was found that Q151Mc RT/dsDNA complexed with either dATP (with an additional interaction of the 3ʹ-OH group with the main chain of Y115) or ddATP appeared to compensate for this perturbation by restricting the side chain flexibility of M151 via the F116Y mutation, because Y116 is hydrogenbonded to K73 (Figure 3d).33 In the case of both 3M RT and F160M/M184V 3M RT/Apt complexes, this interaction is conserved via an interfacial water molecule, which is additionally hydrogen-bonded to the main chain of M151 and the side chain of Y146 (Figure 3d).
Therefore, this supports our hypothesis that wild-type HBV Pol has an N-site conformation closely related to that observed in Q151Mc structures, removing conformational heterogeneity and maintaining the polymerization efficiency of both enzymes. The HBV Pol L528M/M552V mutant (equivalent to the surrogate F160M/M184V 3M RT) maintains the overall conformation seen by the wildtype HBV Pol surrogate, but both mutations impair efficient binding of (−)3TC‐TP, also lacking the 3ʹ-OH interaction present in the wild-type and Q151M mutants in HIV-1 RT/dsDNA/dATP complexes.
A word of caution is necessary, given the distant homology outside the Pol active site between HBV Pol and HIV RT, as well as the differences in the Pol active sites of both. Thus, other scenarios that cannot be captured by the surrogate structural model are possible. Indeed, Yasutake et al.3 complemented this structural work with antiviral activity measurements of 3TC and other NRTIs against different HBV-mimicking RT mutants, including that used for crystallization. The surrogate model appears to appropriately explain the inhibition and resistance profile for 3TC and ETV (good HBV NIs), but does not work as well for tenofovir (TFV, Figure 1b) or islatravir (ISV, a 4ʹ-ethynyl NRTI in Phase III clinical trials; Figure 1b). TFV is not affected by M184V in HIV-1 or M552V in HBV, but the 3M RT mutant antiviral activity is significantly affected by introducing the M184V mutation. In addition, ISV still holds remarkable antiviral activity against the 3M RT, albeit in reality is a moderate HBV NI.3 Although imperfect, in the absence of HBV Pol structures, this approach can be useful (in combination with antiviral activities, as done by Yasutake and coworkers) for HBV structure-based drug design.
L-dNTPs: binding to HIV-1 RT and implications for the development of novel L-nucleotide analogs
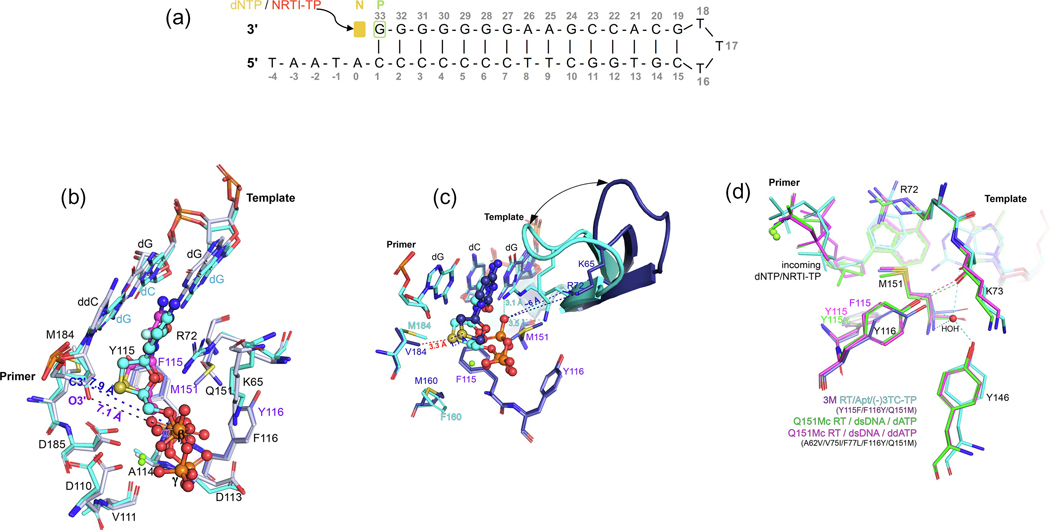
Concurrently, we determined additional structures related to the structures reported above: (i) structures of HIV-1 RT/Apt in complex with incoming (−)FTC‐TP and D and L stereoisomer pairs of dCTP and dTTP; and (ii) the corresponding kinetic data. These complement the previous studies and provide avenues for the design of novel L-nucleoside analogs, discussed herein. Similar to that of Yasutake and colleagues, our crystallographic system differs in that the protein is wild-type HIV-1 RT; it has been crystallized in a different space group and our complexes include the catalytically inactive ion Ca2+ (see methods in the supplemental information online). The incoming (−)FTC‐TP, L-dCTP and L-dTTP are all bound to RT/Apt in the nonproductive state described previously: with the L-sugar engaged in Watson–Crick base pairing/stacking, and with a ~7 Å 3ʹ-OH primer−L-nucleotide Pα distance3 (Figures 4a–c). Notably, all of the structures discussed here, including the postincorporation structure by Bertoletti and colleagues, present a closed conformation of the fingers subdomain, which latches onto the triphosphate moiety of the incoming nucleotide (Figure 3c). In all of these ternary complexes, metal-chelating and triphosphate interactions are very similar (Figures 2–4).
Figure 4.
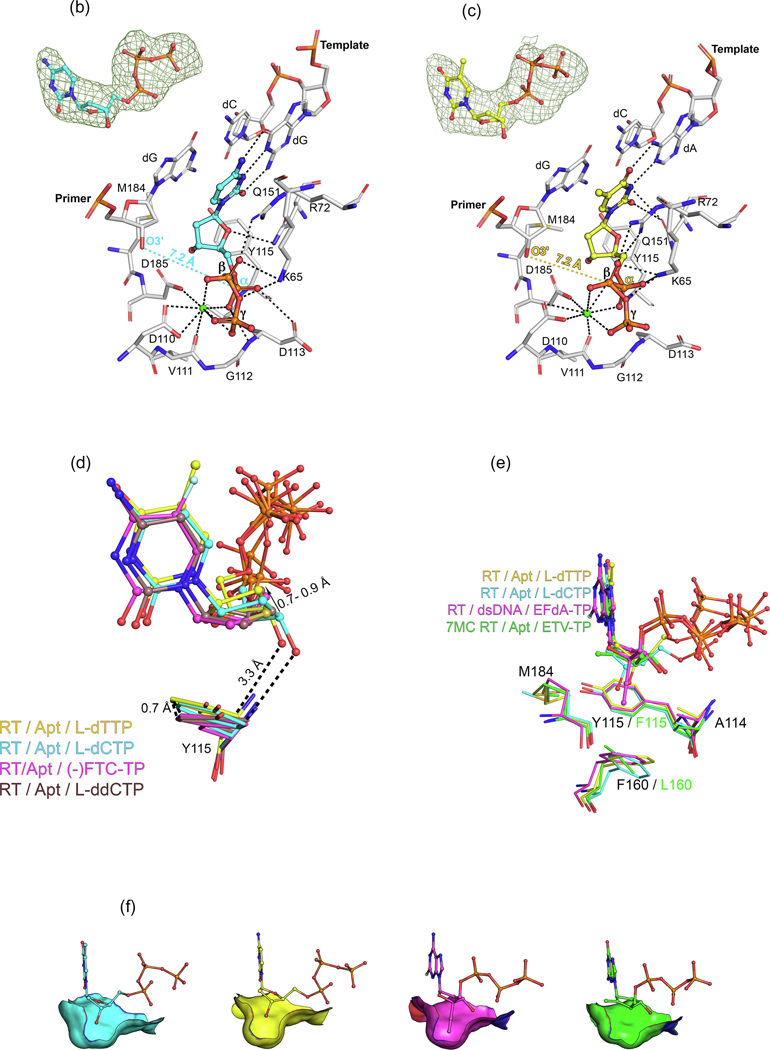
L-dNTPs: binding to HIV-1 reverse transcriptase (RT) and implications for development of novel L-nucleotide analogs. (a) Comparison between the positioning of bound D- and L-dNTPs [and (−)-FTC-TP] to HIV-1 RT/Apt, with respect to the pretranslocation (P)-site nucleotide, with O3ʹ−Pα distances indicated in green for D-dNTPs and in blue for L-dNTPs [and ()-FTC-TP]. The Protein Data Bank (PDB) IDs of these new structures can be found in the ‘Accession Numbers’ section in the main text and in Table S3 in the supplemental information online. (b) Atomic model of the binding of L-dCTP (cyan) to HIV-1 RT/Apt (white), with the O3ʹ−Pα distance indicated in cyan, and inset with the difference polder Fo – Fc map (green mesh, 5σ). (c) Atomic model of the binding of L-dTTP (yellow) to HIV-1 RT/Apt (white), with the O3ʹ−Pα distance indicated in yellow, and inset with the difference polder Fo – Fc map (green mesh, 4.5σ). (d) Detail of the correlated positioning of L-dNTPs and LddNTPs and Y115, an active site residue known as the ‘steric gate’ excluding rNTP binding. The presence of the L-3ʹ-OH group triggers a slight repositioning of L-dNTPs compared with L-ddNTPs (PDB ID: 6P1X for the complex with L-ddCTP, the rest are included as aforementioned). (e) Side view of the preformed pocket in which the 4ʹ-ethynyl substituent of ISV-TP binds (PDB ID: 5J2M), superposed onto both RT/Apt/L-dNTP complexes and the 7MC RT/Apt/ETV-TP complex (PDB ID: 6IKA). Colors as indicated in the legend, with common residues indicated in black. (f) The same subpocket described in (e) is shown with the residues represented as accessible surface area (ASA, https://en.wikipedia.org/wiki/Accessible_surface_area) and with all the complexes compared side-to-side. It illustrates the optimal fit of the 4ʹ-ethynyl substituent, the suboptimal fit of the L-3ʹ-OH substituent, and the lack of binding in the case of ETV-TP. The incoming L-dNTPs are shown as sphere-and-stick models. The amino acids and nucleic acids, within a distance of 5 Å, are shown as stick models. H‐bond interaction distances within 3.5 Å to the compounds are depicted as black-dotted lines. Ca2+ ions are shown as green spheres.
The most remarkable feature of the RT/Apt/L-dCTP and L-dTTP structures compared with the D stereoisomer pair and the cognate (−)FTC-TP structure is the positioning of the sugar moiety. The presence of the 3ʹ-OH group pushes the L-deoxyribose ring (in both L-dCTP and L-dTTP) upward compared with (−)FTC-TP and L-ddCTP (the latter reported by Bertoletti and colleagues14), with a concurrent analogous shift of Y115, a residue that is known to act as a ‘gatekeeper’ or ‘steric gate’ that impedes NTPs from being properly bound and incorporated.34,35 This is likely due to the weak polar contact between the 3ʹ-OH group and the meta-carbon (Cε2 atom) in Y115 (Figure 4d).
We have also determined pre-steady state kinetics of the same complexes (Table S1 in the supplemental information online). Although literature data pertaining to the natural dNTPs and (−)FTC-TP are clear, there are contradictory data on L-dNTPs, which rely only on steady-state kinetics.36–39 Using template-primer substrates, we observed a slow RT-catalyzed incorporation of both L-dNTPs (data not shown), hindering the use of a pre-steady state approach because of nucleic acid fall-off from RT. To circumvent this, we used the tightly-binding Apt, with which RT falls off much less than does a dsDNA or RNA-DNA template-primer (for detailed methods, see the supplemental information online).32,40 The slow dissociation rate of RT on the Apt allowed the use of extended time points that are required to observe L-dNTP addition under pre-steady state conditions (Figures S1 and S2 in the supplemental information online). It was recently shown that Apt, containing two 2′-O-methyl modified nucleotides in the template region near the Pol active site, behaves more similarly to an RNA/DNA template-primer than to a dsDNA.40 This helps to rationalize the finding that these kinetic data are more similar to those reported using RNA/DNA template-primer than with dsDNA23 (Table S1 in the supplemental information online).
As previously observed for (−)FTC-TP and (−)3TC-TP 23, the binding of L-dCTP to RT is on the same order of magnitude to D-dCTP (1.5-fold worse KD), whereas L-dTTP presents a 9.5-fold worse KD. In addition, in a similar fashion, the incorporation rate (kpol) is affected by the L stereochemistry in L-dNTPs, and is an order of magnitude higher than that for (−)FTC-TP, resulting in a drop of catalytic efficiency of over 104-fold. Therefore, our kinetics data show that, even though RT has a relaxed stereoselectivity for L-nucleotide drugs, its D-stereoselectivity for L-dNTPs is similar to all Pol reported (Table S2 in the supplemental information online). Nevertheless, as discussed by Vyas and colleagues20 and herein, D-stereoselectivity is case dependent. The five different DNA Pols analyzed by combined structural and kinetics-based approaches show unique mechanisms, reinforcing that this is due to randomness rather than to an evolutionary pressure. Indeed, in a world essentially comprising only D-nucleosid(t)es, a nonstereoselective enzyme might not have a reduced fitness because no selective pressure exists.
Thus, in general, L-dNTPs and L-nucleotide drugs are able to bind readily to Pols, via chelation, base pairing, stacking, and electrostatic contacts with the triphosphate moiety, but the required conformational change or sampling needed for incorporation severely reduces the rate and, hence, the efficiency (Figures 2b,c, and 4a). Our results and the recent structural work with HIV-1 RT are in good agreement with the kinetic model proposed by Kellinger and Johnson.41 By fluorescently labeling RT in the fingers subdomain, they take into account RT dynamics and kinetics simultaneously. In essence, they observe for D-nucleotides and that the fingers closure provides tight binding and optimal alignment for catalysis. Meanwhile, for L-nucleotides, there remains tight binding, but the geometry is not optimal for catalysis, requiring a rearrangement and/or sampling and, consequently, leading to a slower incorporation rate. This results in a longer half-life of the ternary complex for L-nucleotides.
The previous model coincides with our observations in crystallo with our RT/Apt crystal system (data not shown), when we soaked with: (i) D-dCTP (and Mg2+), full incorporation occurred in ~30 min; (ii) (−)FTC-TP (and Mn2+), full incorporation occurred in ~2 days; and (iii) L-dCTP (and Mn2+), partial incorporation appeared to have occurred in ~2 days. Thus, Kellinger and Johnson speculated whether in vivo L-NRTI-TPs could act as competitive inhibitors, in addition to chain terminators.41
Based on all of these findings, we hypothesize that L-nucleotides with the appropriate 3ʹ-substituent could be strong RT competitive inhibitors, similar to nucleotide-competing reverse transcriptase inhibitors (NcRTIs), a different class of chemotype that inhibit RT by competing with dNTP incorporation.2 As shown by Sohl and colleagues,21 human mitochondrial DNA Pol γ discriminates against the 5-fluorine (F) of (−)FTC-TP (but not RT), explaining the lower toxicity of FTC over 3TC. Thus, any new derivative should bear the F substituent. Interestingly, a superposition of the RT/Apt/L-dCTP complex with RT/dsDNA/islatravir-TP (ISV-TP)42 (Figures 4e,f) showed good overlap between the 3ʹ-OH in the L-nucleotide and the 4’-ethynyl group of ISV-TP. Recent work42,43 reported that the ethynyl group binds optimally to the preformed subpocket delimitated by A114, Y115, F160, M184, and catalytic D185 in RT. Other substituents (ethyl, methyl, cyano, or azido) can provide good binding and inhibition in the case of D-nucleotides.9,43–45
It is then conceivable that one of the previous substituents placed at the 3′ position of an L-nucleotide could provide tight binding and competitive inhibition toward RT. Hence, the competitive inhibition mechanism of such compounds would be of a translocation-defective RT inhibitor, similarly to ISV-TP. In fact, L-AZT-TP, the L-stereoisomer of 3′-azido-3′-deoxythymidine (active form of the drug AZT), was shown to be moderately active against RT, with a Ki of 2 μM (~250-fold less inhibitory than D-AZT-TP).46 Although L-AZT did not show HIV antiviral activity, L-3′-azido-5-FddC showed strong antiviral activity against both HIV and HBV.47 It is reasonable that the triphosphate form of L-3′-azido-5-FddC, bearing both the L-3ʹ substituent and the 5-F as in (−)-FTC-TP, might inhibit (at least in HIV) viral replication through RT competitive inhibition.
Previous work reported that L-nucleosides bearing only a 3′-OH group specifically showed antiviral activity against HBV but not against HIV.47 This indicates that L-dC and L-dT might not be efficiently phosphorylated in lymphocytes as in hepatocytes and/or that their triphosphate metabolites are poor inhibitors (the second is supported by our data). Overall, L-nucleosides as L-dT might work in HBV because of a combination of: (i) better phosphorylation in hepatic cells; also supported by the fact that an L-AZA prodrug showed strong HIV and HBV antiviral activity (whereas L-AZA was devoid of activity)48; and (ii) better inhibition of Pol activity, given that HBV Pol presents a Phe in the position of Y115 and a Leu in place of F160, with this pocket likely being more shallow,30 potentially better accommodating the 3′-OH group (Figure 4f). The different overall architecture of HBV Pol in relation to RT might also induce a slightly different arrangement of the active site, facilitating the 3′-OH group interaction.
In summary, these novel structures along with kinetics data complement previous findings and provide interesting insights for the development of novel L-nucleos(t)ide analogs, targeting both HIV-1 and HBV.
Repurposing L-nucleoside drugs in cancer and diabetes
Identification of novel drugs is a costly and resource intensive endeavor, which makes drug (as well as know-how and technology) repurposing a clearly advantageous ‘shortcut’. A recent and prominent example of this process is the identification of remdesivir, a nucleoside prodrug the active form of which is a nucleoside-TP (Figure 1b) that inhibits the viral RNA-dependent RNA polymerase (RdRp) from Ebola virus (EBOV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).49,50 Initially, remdesivir was developed by Gilead to target HCV and respiratory syncytial virus (RSV), but was later repurposed because it did not work against these viruses.49 Another recent and impressive example has been the ‘reuse’ of a quantitative mass spectrometry-based proteomic approach developed initially for mapping virus–host interactions in HIV for SARS-CoV-2 and coronaviruses in general. This has yielded several potential avenues for drug repurposing,51 including plitidepsin, an inhibitor of the host protein eEF1A developed initially to treat multiple myeloma, which has successfully undergone a Phase I/II clinical study for the treatment of Coronavirus 2019 (COVID-19; see Supplementary Data Chart S1 in the supplemental information online).52
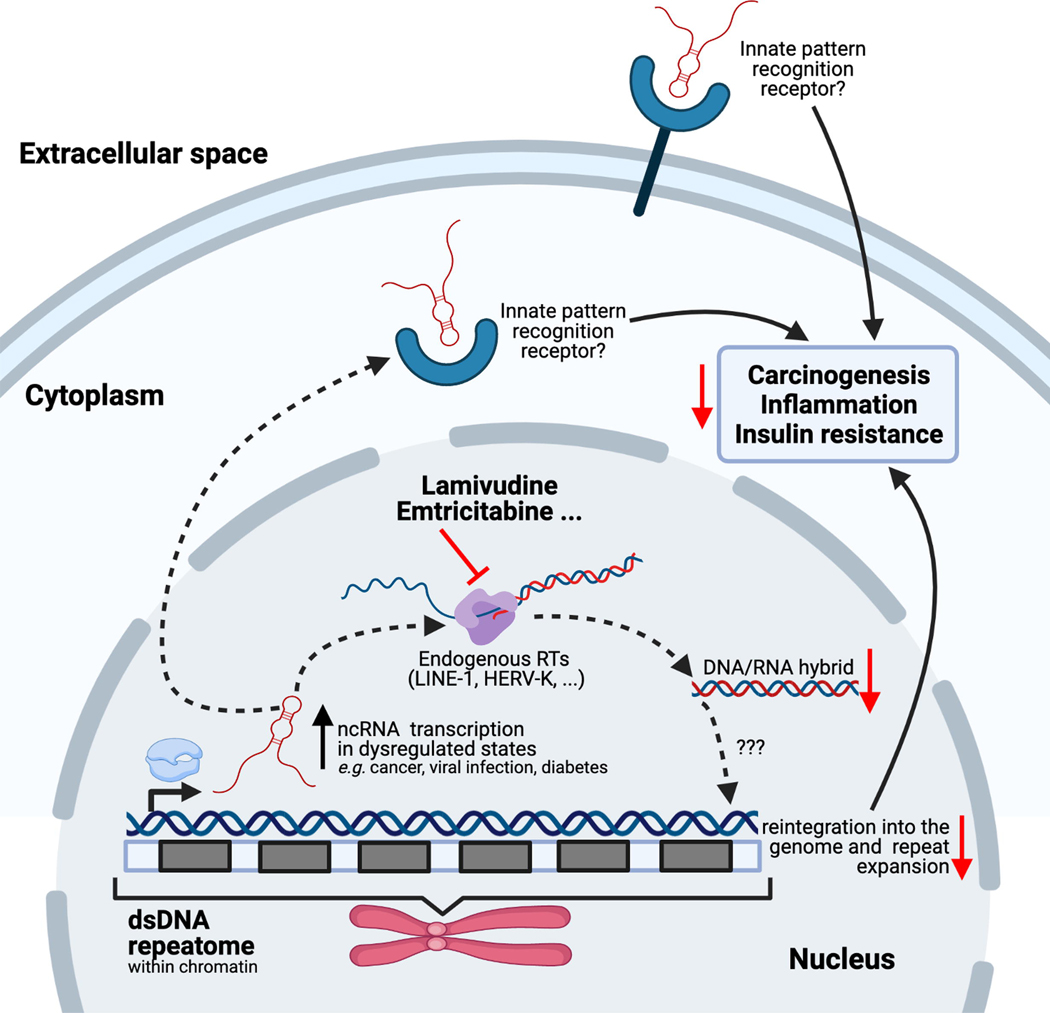
Regarding L-nucleoside drugs (successfully repurposed for HBV treatment as detailed above), recent findings strongly support potential repurposing for treating certain cancers related to their anti-RT activity. Repurposing NRTIs and nucleotide analogs to treat cancer by inhibition of human DNA Pols (especially those involved in DNA repair) is an intriguing avenue. Given that it is out the scope of this review, we refer the reader to several studies and reviews for more detail.53–56 It has been recognized that (at least some) tumors mimic viruses.57 The dysregulated genetic and epigenetic landscape of tumors (e.g., p53 mutation or DNA methylation) promotes transcription of RNA silenced in normal cells, such as the RNA repetitive elements dubbed the ‘repeatome’, which constitutes about one-third of the human genome.58,59 Some of these repeatome RNAs, including pericentromeric satellite repeats HSATII, come from ancient retroviruses integrated into the human genome (most of them lacking functionality). In some cancers, these repeats are reverse transcribed and reintegrated into the genome, resulting in their expansion. Given that these RNA repeats resemble viral sequences, it is possible that this is a mechanism tumor cells use to escape innate immunity57,60 (Figure 5). Blocking this endogenous RT activity with NRTIs that normally target HIV stopped cancer progression in cellular and xenograft cancer models.60,61 A recent Phase II trial of lamivudine (3TC) in p53 mutant metastatic colorectal cancer showed an association of clinical benefit in patients treated with 3TC that correlated with lower HSATII repeat RNA levels.62 However, the identity of this endogenous RT activity in cancer cells and whether it comes from multiple sources are unknown.
Figure 5.
Repurposing L-nucleoside drugs in cancer and diabetes. In cells in dysregulated states (as in cancer, viral infection, or diabetes), the normally silent ‘repeatome’ expands via a multistep process that involves upregulated transcription of noncoding RNAs (ncRNAs), reverse transcription by endogenous reverse transcriptases [RTs, such as long interspersed nuclear elements (LINE-1) or human endogenous retrovirus K (HERV-K), encoded in retrotransposons that form part of the same repeatome] and reintegration of the expanded repeats through an unknown mechanism. The current understanding is that nucleos(t)ide RT inhibitors (NRTIs), such as the L-nucleotides lamivudine or emtricitabine, can inhibit the repeat expansion, which correlates with carcinogenesis, inflammation, or insulin resistance, and could provide a better prognosis for these pathologies. Some of the ncRNA transcripts might also be detected by innate pattern recognition receptors, because of their similarity to viral sequences, inducing a proinflammatory immune response. Although the overall process is full of outstanding questions, it is clear that NRTIs can block the pernicious effects triggered by these repeat expansions through inhibition of reverse transcription, but there is evidence showing they can work through additional RT-independent inhibition mechanisms. Created with BioRender (BioRender.com). Adapted from 60. See 57,60,61,65,67,68 for detailed information.
RT activity in mammalian cells is primarily due to retrotransposons, mainly long interspersed nuclear elements (LINEs) and human endogenous retrovirus (HERV) family members, as well as human telomerase reverse transcriptase (hTERT).63,64 Given that recombinant expression has proven challenging, specific probes (e.g., NRTIs) are necessary to identify the sources of mammalian RT activity and better characterize their action in pathologies.65 Banuelos-Sanchez and colleagues recently developed an assay in mammalian cells to screen for specific inhibitors of LINE-1 retrotransposons, which represent ~17% of the human genome.65 Among the 30 nucleotide inhibitors and several NNRTIs screened, the newly developed D-nucleoside inhibitor GBS-149 (Supplementary Data Chart S1 in the supplemental information online), (−)FTC and (−)3TC were found to be nontoxic, mammalian-specific LINE-1 inhibitors. Banuelos-Sanchez and colleagues also discussed in detail other studies in which NRTIs have been tested against retrotransposons, and we refer the reader to it for a more complete analysis of the state of the art for this field.65 Additionally, (−)FTC and (−)3TC have been shown to inhibit HERV-K, which might be the only active HERV member in humans (although, in general, HERVs are inactive). Despite acting on both LINE-1 and HERV-K, the NRTI inhibition profiles appear to differ.65 Regarding hTERT, the NRTIs AZT and TFV block hTERT activity, but (−)FTC and (−)3TC show no inhibition.66
In the case of type 2 diabetes mellitus (T2DM), a recent study revealed that NRTIs could be repurposed to reduce ‘inflammasome’ activation (Figure 5).67 A thorough analysis of several health insurance databases in the USA showed that patients with AIDS or HBV treated with NRTIs present a 33% lower risk of T2DM incidence. Moreover, lamivudine has been shown to improve insulin sensitivity and reduce inflammasome activation in diabetic human cells and mice fed a high-fat diet. The underlying mechanism is again linked to repeatome RNA transcripts. In this case, it might be due to elevated short interspersed nuclear element (SINE) RNA transcripts, which are known activators of the inflammasome. SINEs are able to hijack the endogenous RT activity of LINE-1 and blocking this activity with repurposed NRTIs holds promise for reducing incidence of T2DM. However, NNRTIs and other non-RT HIV inhibitors are not associated with a reduced frequency of T2DM. It is also not clear whether the NRTI effect is due to direct inhibition of endogenous RT activity or if it is independent.68 In fact, mounting evidence shows that the NRTI effect on inflammasome inhibition in age-related macular disease69–71 results from direct inhibition of endogenous LINE-1 RT activity69 and from independent mechanisms, as shown by use of alkylated NRTIs (not inhibiting RT activity).70 Hence, both mechanisms might contribute to NRTImediated diabetes prevention.
Concluding remarks
L-nucleosides have revolutionized AIDS and HBV treatment through exploitation of the relaxed stereoselectivity of viral RTs compared with cellular DNA Pols, also making them less toxic. Their discovery was possible through medicinal chemistry, and virological and biochemical approaches, but the detailed molecular mechanisms remained elusive until 2019. A combined structural and kinetics approach with several DNA Pols, and now with HIV-1 RT and a surrogate HBV Pol model, allows us to further rationalize their mechanism of action and to devise novel L-nucleoside drugs. Additionally, their favorable bioavailability and biosafety profile makes them candidates for repurposing them in cancer and T2DM, largely driven by the mimicry between tumor and diabetic cells and viruses. In addition, other pathologies linked to RNA repeats, particularly neurological disorders, could be next in line for this approach.65 Lastly, although (−)3TC and (−)FTC are not effective against viral RdRps,72 bacterial DNA-dependent DNA and RNA Pols could be inhibited by L-nucleosides, as studies with T7 RNA polymerase73 and Dpo456 (Table S2 in the supplemental information online) suggest.
Supplementary Material
Acknowledgments
The authors thank members of the Arnold group for helpful discussions. We also thank the many investigators in the fields of nucleoside inhibitors, HIV, and HBV for their continuing contributions toward understanding the magnificent RT enzyme, as it pertains to structure, function, inhibition, and resistance, making it one of the most important drug targets in medicine. We also want to apologize to those investigators whose papers have not been cited due to space limitations. This work was supported by NIH R01 AI027690 and the HIVE Center Grant U54 AI150472 (both to E.A.), and R01AI150480 to J.J.D.
Glossary
- Highly active antiretroviral therapy (HAART)
also called combination antiretroviral therapy (cART, or simply ART); uses ‘cocktails’ of antiretroviral drugs, targeting the viral proteins RT, protease (RT), and integrase (IN), as well as entry and fusion inhibitors. Patients with HIV taking the HAART regimen as prescribed can maintain an undetectable viral load and have very low risk of transmitting HIV. HAART usually includes two NRTIs and often an NNRTI. Among the ~30 FDA-approved HIV drugs and combinations, about half contain RT inhibitors, with five approved NRTIs (including L-nucleosides lamivudine and emtricitabine, see Figure 1a in the main text) and five approved NNRTIs.
- Inflammasome
innate immune response related to several diseases, such as ageing, diabetes, cancer, or viral infections. Inflammasomes are part of a superfamily of pattern recognition receptors (PRRs) and act as cytoplasmatic sensors triggering inflammation. They are multiprotein complexes that enhance inflammatory responses by promoting the production and secretion of key cytokines.
- Nucleotide (N)-site and pretranslocation (P)-site
initial step in nucleotide incorporation is the binding of the incoming dNTP at the N-site. In processive DNA synthesis, the nucleic acid substrate must translocate to the P-site relative to RT to make the N-site available to bind the next incoming dNTP.
- Nucleoside
organic molecule formed by a nucleobase linked to a sugar (usually ribose or 2’-deoxyribose). Besides the natural nucleosides, with D (right-handed) stereochemistry, several nucleoside analogs are used as antiviral or anticancer agents. They present different variations and substitutions in either the nucleobase and/or the sugar ring. These compounds are activated in the cells by being phosphorylated into (deoxy)nucleoside triphosphates [(d)NTPs]. They are administered as nucleosides because charged nucleotides cannot easily cross cell membranes. Among them, L-nucleoside analogs comply with all these requirements and additionally present an inverse L (left-handed) stereochemistry.
- Nucleoside reverse transcriptase inhibitor (NRTI)
drug class that blocks the Pol activity of the HIV-1 RT and HBV Pol RT domains. They are analogs of the naturally occurring nucleosides lacking (or with a modified) 3’-hydroxyl (OH) group, indispensable for the next incoming dNTP incorporation (through nucleophilic attack of a 3ʹ-OH to the Pα of the dNTP). Thus, they act as chain terminators. Some NRTIs, such as ETV or ISV (Figure 1a in the main text), bear a 3ʹ-OH group, allowing further incorporation of dNTPs, acting via delayed chain termination. In opposition to NRTIs, which are orthosteric inhibitors binding at the Pol active site, non-NRTIs (NNRTIs) are allosteric, binding ~10 Å away from the active site (Figure 1b in the main text). NNRTIs provoke thumb subdomain hyperextension, and displacement of the 3ʹ-OH group of the primer terminus from a catalytically poised to an inactive configuration.
- Nucleotide
building block of DNA and RNA; an organic molecule compriisng a nucleoside and a phosphate.
- Pre-exposure prophylaxis (PrEP)
treatment for patients who are at very high risk of getting HIV; a pill is taken daily to prevent infection. Standard therapy comprises a daily dose that always include an L-nucleoside drug and tenofovir. Studies show that daily use of PrEP reduces the risk of getting HIV from sex by ~99%, and by 74% among injecting drug users.
- Retrotransposons
genetic elements that copy and paste themselves into different genomic locations by converting RNA back into DNA via reverse transcription using an RNA transposition intermediate. Different types of retrotransposon carry out this process by distinct mechanisms. Long terminal repeat (LTR) retrotransposons and non-LTR retrotransposons use element-encoded enzymes to mediate their mobility. In addition, the endonuclease and reverse transcriptase activities of non-LTR retrotransposons also have a central role in mobilizing non-autonomous short interspersed elements (SINEs), certain classes of non-coding RNA and mRNA encoding pseudogenes (defective genes).
- Therapeutic index
quantitative measurement of the relative safety of a drug. It is the ratio of the dose of a therapeutic agent that causes the therapeutic effect to the dose that causes toxicity.
Biographies
Author biographies

Francesc Xavier Ruiz
Francesc Xavier Ruiz is an assistant research professor at the Center for Advanced Biotechnology and Medicine (CABM) at Rutgers University (USA). He pursued undergraduate and graduate studies in biochemistry at Universitat Autònoma de Barcelona in Spain, followed by a postdoctoral stay at Institut de Génétique et de Biologie Moléculaire et Cellulaire (France) in the lab of Alberto Podjarny. During this period, he studied the structure, function, and inhibition of human aldo-keto reductases involved in the first step of retinoic acid biosynthesis. In 2015, Dr Ruiz joined the lab of Eddy Arnold. His current research focuses on the structure and function of human and viral proteins, especially HIV-1 reverse transcriptase (RT), with an emphasis on catalysis and inhibition for drug discovery.

Anthony Hoang
Anthony Hoang is currently a PhD candidate in chemical, physical, and structural biology at Baylor College of Medicine (USA). He received his BA in molecular biology and biochemistry at Rutgers University under the guidance of Eddy Arnold, conducting research on HIV-1 RT structure determination of nucleoside inhibitor complexes. His current research focuses on characterizing the structure, function, and inhibition of anion exchange transporters.

Eddy Arnold
Eddy Arnold has been a CABM Faculty Member since 1987. He is on the Rutgers University Board of Governors and is a distinguished professor of chemistry and chemical biology. He pursued undergraduate and graduate studies in organic chemistry and crystallography at Cornell University with Jon Clardy, and performed postdoctoral research at Purdue University, with the late Michael G. Rossmann, obtaining the first structure of an animal virus (human common cold virus). He has been a central contributor to the study of HIV RT structure and function, solving multiple HIV-1 RT structures in complex with a variety of antiviral drugs and model segments of the HIV genome. In collaboration with the late Paul Janssen, his laboratory’s structure-based drug design effort was instrumental in the discovery and development of two US Food and Drug Administration (FDA)-approved anti-HIV drugs.
Footnotes
Declaration of interests
The authors declare no conflicts of interest.
Teaser: Despite the longtime use of L-nucleoside drugs in AIDS and HBV treatment, structural details of recognition by HIV-1 reverse transcriptase were only recently unveiled. These structures provide a guide for drug development and repurposing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Menendez-Arias L, Sebastian-Martin A, Alvarez M. Viral reverse transcriptases. Virus Res. 2017; 234: 153–176. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz FX, Arnold E. Evolving understanding of HIV-1 reverse transcriptase structure, function, inhibition, and resistance. Curr Opin Struct Biol. 2020; 61: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasutake Y, Hattori SI, Tamura N, Matsuda K, Kohgo S, Maeda K, et al. Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine. Sci Rep. 2020; 10(1): 3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das K, Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 1. Curr Opin Virol. 2013; 3(2): 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das K, Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 2. Curr Opin Virol. 2013; 3(2): 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009; 385(3): 693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vörös J, Urbanek A, Rautureau GJ, O’Connor M, Fisher HC, Ashcroft AE, et al. Large-scale production and structural and biophysical characterizations of the human hepatitis B virus polymerase. J Virol. 2014; 88(5): 2584–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, et al. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J Virol. 2001; 75(10): 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higashi-Kuwata N, Hayashi S, Das D, Kohgo S, Murakami S, Hattori SI, et al. CMCdG, a novel nucleoside analog with favorable safety features, exerts potent activity against wild-type and entecavir-resistant hepatitis B virus. Antimicrob Agents Chemother. 2019; 63(4): e02143–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liotta DC, Painter GR. Discovery and development of the anti-human immunodeficiency virus drug, emtricitabine (Emtriva, FTC). Acc Chem Res. 2016; 49(10): 2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinazi RF, Chu CK, Peck A, McMillan A, Mathis R, Cannon D, et al. Activities of the four optical isomers of 2’,3’-dideoxy-3’-thiacytidine (BCH-189) against human immunodeficiency virus type 1 in human lymphocytes. Antimicrob Agents Chemother. 1992; 36(3): 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schinazi RF, McMillan A, Cannon D, Mathis R, Lloyd RM, Peck A, et al. Selective inhibition of human immunodeficiency viruses by racemates and enantiomers of cis-5-fluoro-1-[2-(hydroxymethyl)-1,3-oxathiolan-5-yl]cytosine. Antimicrob Agents Chemother. 1992; 36(11): 2423–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertoletti N, Chan AH, Schinazi RF, Anderson KS. Post-catalytic complexes with emtricitabine or stavudine and HIV-1 reverse transcriptase reveal new mechanistic insights for nucleotide incorporation and drug resistance. Molecules. 2020; 25(20): 4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoletti N, Chan AH, Schinazi RF, Yin YW, Anderson KS. Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: first crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine. Protein Sci. 2019; 28(9): 1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung M, Tokarsky EJ, Lagpacan L, Zhang L, Suo Z, Lansdon EB. Elucidating molecular interactions of L-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance. Commun Biol. 2019; 2: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen J.King of the pills. Science. 2015; 348(6235): 622–625. [DOI] [PubMed] [Google Scholar]
- 17.Menendez-Arias L, Alvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr Opin Virol. 2014; 8: 1–9. [DOI] [PubMed] [Google Scholar]
- 18.Maury G.The enantioselectivity of enzymes involved in current antiviral therapy using nucleoside analogues: a new strategy? Antivir Chem Chemother. 2000; 11(3): 165–189. [DOI] [PubMed] [Google Scholar]
- 19.Seley-Radtke KL, Yates MK. The evolution of nucleoside analogue antivirals: a review for chemists and non-chemists. Part 1: early structural modifications to the nucleoside scaffold. Antiviral Res. 2018; 154: 66–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vyas R, Reed AJ, Raper AT, Zahurancik WJ, Wallenmeyer PC, Suo Z. Structural basis for the D-stereoselectivity of human DNA polymerase beta. Nucleic Acids Res. 2017; 45(10): 6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohl CD, Szymanski MR, Mislak AC, Shumate CK, Amiralaei S, Schinazi RF, et al. Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma. Proc Natl Acad Sci U S A. 2015; 112(28): 8596–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarafianos SG, Das K, Clark AD Jr, Ding J, Boyer PL, Hughes SH, et al. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with b-branched amino acids. Proc Natl Acad Sci U S A. 1999; 96(18): 10027–10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray AS, Murakami E, Peterson CN, Shi J, Schinazi RF, Anderson KS. Interactions of enantiomers of 2’,3’-didehydro-2’,3’-dideoxy-fluorocytidine with wild type and M184V mutant HIV-1 reverse transcriptase. Antiviral Res. 2002; 56(3): 189–205. [DOI] [PubMed] [Google Scholar]
- 24.Das K, Bandwar RP, White KL, Feng JY, Sarafianos SG, Tuske S, et al. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J Biol Chem. 2009; 284(50): 35092–35100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, et al. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem. 2004; 279(1): 509–516. [DOI] [PubMed] [Google Scholar]
- 26.Acosta-Hoyos AJ, Scott WA. The Role of nucleotide excision by reverse transcriptase in HIV drug resistance. Viruses. 2010; 2(2): 372–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J Virol. 2002; 76(18): 9143–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matamoros T, Nevot M, Martinez MA, Menendez-Arias L. Thymidine analogue resistance suppression by V75I of HIV-1 reverse transcriptase: effects of substituting valine 75 on stavudine excision and discrimination. J Biol Chem. 2009; 284(47): 32792–32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasutake Y, Hattori SI, Hayashi H, Matsuda K, Tamura N, Kohgo S, et al. HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir. Sci Rep. 2018; 8(1): 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasutake Y, Hattori SI, Tamura N, Matsuda K, Kohgo S, Maeda K, et al. Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4’-modified nucleoside RT inhibitors. Biochem Biophys Res Commun. 2019; 509(4): 943–948. [DOI] [PubMed] [Google Scholar]
- 31.Das K, Balzarini J, Miller MT, Maguire AR, DeStefano JJ, Arnold E. Conformational states of HIV-1 reverse transcriptase for nucleotide incorporation vs pyrophosphorolysis-binding of foscarnet. ACS Chem Biol. 2016; 11(8): 2158–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MT, Tuske S, Das K, DeStefano JJ, Arnold E. Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer. Protein Sci. 2016; 25(1): 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das K, Martinez SE, Arnold E. Structural insights into HIV reverse transcriptase mutations Q151M and Q151M complex that confer multinucleoside drug resistance. Antimicrob Agents Chemother. 2017; 61(6): e00224–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000; 275(26): 19759–19767. [DOI] [PubMed] [Google Scholar]
- 35.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci U S A. 1997; 94(2): 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Focher F, Maga G, Bendiscioli A, Capobianco M, Colonna F, Garbesi A, et al. Stereospecificity of human DNA polymerases alpha, beta, gamma, delta and epsilon, HIV-reverse transcriptase, HSV-1 DNA polymerase, calf thymus terminal transferase and Escherichia coli DNA polymerase I in recognizing D- and L-thymidine 5’-triphosphate as substrate. Nucleic Acids Res. 1995; 23(15): 2840–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maga G, Amacker M, Hübscher U, Gosselin G, Imbach JL, Mathé C,et al. Molecular basis for the enantioselectivity of HIV-1 reverse transcriptase: role of the 3’hydroxyl group of the L-(beta)-ribose in chiral discrimination between D- and L-enantiomers of deoxy- and dideoxy-nucleoside triphosphate analogs. Nucleic Acids Res. 1999; 27(4): 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semizarov DG, Arzumanov AA, Dyatkina NB, Meyer A, Vichier-Guerre S, Gosselin G, et al. Stereoisomers of deoxynucleoside 5’-triphosphates as substrates for template-dependent and -independent DNA polymerases. J Biol Chem. 1997; 272(14): 9556–9560. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi T, Iwanami N, Shudo K, Saneyoshi M. Chiral discrimination of enantiomeric 2’-deoxythymidine 5’-triphosphate by HIV-1 reverse transcriptase and eukaryotic DNA polymerases. Biochem Biophys Res Commun. 1994; 200(2): 1023–1027. [DOI] [PubMed] [Google Scholar]
- 40.Tuske S, Zheng J, Olson ED, Ruiz FX, Pascal BD, Hoang A, et al. Integrative structural biology studies of HIV-1 reverse transcriptase binding to a high-affinity DNA aptamer. Current Research in Structural Biology. 2020; 2: 116–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kellinger MW, Johnson KA. Nucleotide-dependent conformational change governs specificity and analog discrimination by HIV reverse transcriptase. Proc Natl Acad Sci U S A. 2010; 107(17): 7734–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, et al. Structural basis of HIV inhibition by translocation-defective RT inhibitor 4’-ethynyl2-fluoro-2’-deoxyadenosine (EFdA). Proc Natl Acad Sci U S A. 2016; 113(33): 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takamatsu Y, Das D, Kohgo S, Hayashi H, Delino NS, Sarafianos SG, et al. The high genetic barrier of EFdA/MK-8591 stems from strong interactions with the active site of drug-resistant HIV-1 reverse transcriptase. Cell Chem Biol. 2018; 25(10): 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer PL, Julias JG, Ambrose Z, Siddiqui MA, Marquez VE, Hughes SH. The nucleoside analogs 4’C-methyl thymidine and 4’C-ethyl thymidine block DNA synthesis by wild-type HIV-1 RT and excision proficient NRTI resistant RT variants. J Mol Biol. 2007; 371(4): 873–882. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, Hu W, Wang S, Pan Z, Tao L, Guo X, et al. Synthesis of new 2’-deoxy-2’-fluoro-4’-azido nucleoside analogues as potent anti-HIV agents. Eur J Med Chem. 2011; 46(9): 4178–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faraj A, El Alaoui AM, Gosselin G, Imbach JL, Morrow C, Sommadossi JP. Effects of beta-L-3’-azido-3’-deoxythymidine 5’-triphosphate on host and viral DNA polymerases. Antiviral Res. 2000; 47(2): 97–102. [DOI] [PubMed] [Google Scholar]
- 47.Bryant ML, Bridges EG, Placidi L, Faraj A, Loi AG, Pierra C, et al. Antiviral L-nucleosides specific for hepatitis B virus infection. Antimicrob Agents Chemother. 2001; 45(1): 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang HW, Detorio M, Herman BD, Solomon S, Bassit L, Nettles JH, et al. Synthesis, antiviral activity, cytotoxicity and cellular pharmacology of l-3’-azido-2’,3’dideoxypurine nucleosides. Eur J Med Chem. 2011; 46(9): 3832–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilead Sciences. Development of remdesivir: fact sheet. www.gilead.com/-/media/gilead-corporate/files/pdfs/covid-19/gilead_rdv-development-fact-sheet2020.pdf. [Accessed February 17, 2022].
- 50.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020; 295(20): 6785–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020; 370(6521): eabe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White KM, Rosales R, Yildiz S, Kehrer T, Miorin L, Moreno E, et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021; 371(6532): 926–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berdis AJ. Inhibiting DNA polymerases as a therapeutic intervention against cancer. Front Mol Biosci. 2017; 4: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown JA, Pack LR, Fowler JD, Suo Z. Pre-steady-state kinetic analysis of the incorporation of anti-HIV nucleotide analogs catalyzed by human X- and Y-family DNA polymerases. Antimicrob Agents Chemother. 2011; 55(1): 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crespan E, Garbelli A, Amoroso A, Maga G. Exploiting the nucleotide substrate specificity of repair DNA polymerases to develop novel anticancer agents. Molecules. 2011; 16(9): 7994–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaur V, Vyas R, Fowler JD, Efthimiopoulos G, Feng JY, Suo Z. Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Yfamily DNA polymerase. Nucleic Acids Res. 2014; 42(15): 9984–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenbaum BD. Innate immune driven evolution via immunostimulatory RNA: Viruses that mimic hosts, tumors that mimic viruses. Current Opinion in Systems Biology. 2017; 1: 137–142. [Google Scholar]
- 58.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001; 409(6822): 860–921. [DOI] [PubMed] [Google Scholar]
- 59.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol Spectr. 2015; 3(2): MDNA3–0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Younger ST, Rinn JL. Silent pericentromeric repeats speak out. Proc Natl Acad Sci U S A. 2015; 112(49): 15008–15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bersani F, Lee E, Kharchenko PV, Xu AW, Liu M, Xega K, et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc Natl Acad Sci U S A. 2015; 112(49): 15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parikh AR, Rajurkar M, Van Seventer EE, Gemma AJ, Allen JN, Blaszkowsky SJ, et al. Phase II study of lamivudine in p53 mutant metastatic colorectal cancer (mCRC). Journal of Clinical Oncology. 2020; 38(4_suppl): 149. [Google Scholar]
- 63.Belfort M, Curcio MJ, Lue NF. Telomerase and retrotransposons: reverse transcriptases that shaped genomes. Proc Natl Acad Sci U S A. 2011; 108(51): 20304–20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanciano S, Cristofari G. Measuring and interpreting transposable element expression. Nat Rev Genet. 2020; 21(12): 721–736. [DOI] [PubMed] [Google Scholar]
- 65.Banuelos-Sanchez G, Sanchez L, Benitez-Guijarro M, Sanchez-Carnerero V, Salvador-Palomeque C, Tristan-Ramos P, et al. Synthesis and characterization of specific reverse transcriptase inhibitors for mammalian LINE-1 retrotransposons. Cell Chem Biol. 2019; 26(8): 1095–1109. [DOI] [PubMed] [Google Scholar]
- 66.Leeansyah E, Cameron PU, Solomon A, Tennakoon S, Velayudham P, Gouillou M, et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: a potential factor contributing to HIV-associated accelerated aging. J Infect Dis. 2013; 207(7): 1157–1165. [DOI] [PubMed] [Google Scholar]
- 67.Ambati J, Magagnoli J, Leung H, Wang SB, Andrews CA, Fu D, et al. Repurposing anti-inflammasome NRTIs for improving insulin sensitivity and reducing type 2 diabetes development. Nat Commun. 2020; 11(1): 4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fowler BJ, Gelfand BD, Kim Y, Kerur N, Tarallo V, Hirano Y, et al. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science. 2014; 346(6212): 1000–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukuda S, Varshney A, Fowler BJ, Wang SB, Narendran S, Ambati K, et al. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc Natl Acad Sci U S A. 2021; 118(6): e2022751118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendran S, Pereira F, Yerramothu P, Apicella I, Wang SB, Ambati K, et al. Nucleoside reverse transcriptase inhibitors and Kamuvudines inhibit amyloid-beta induced retinal pigmented epithelium degeneration. Signal Transduct Target Ther. 2021; 6(1): 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamada K, Kaneko H, Shimizu H, Suzumura A, Namba R, Takayama K, et al. Lamivudine inhibits Alu RNA-induced retinal pigment epithelium degeneration via anti-inflammatory and anti-senescence activities. Transl Vis Sci Technol. 2020; 9(8): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chien M, Anderson TK, Jockusch S, Tao C, Li X, Kumar S, et al. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J Proteome Res. 2020; 19(11): 4690–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Q, Ke Y, Kan Y, Tang X, Li X, He Y, et al. Compatibility and fidelity of mirror-image thymidine in transcription events by T7 RNA polymerase. Mol Ther Nucleic Acids. 2020; 21: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.