Abstract
Background:
Substance use disorders (SUDs) are associated with disruptions in circadian rhythms. Both human and animal work has shown the integral role for circadian clocks in the modulation of reward behaviors. Interestingly, astrocytes have emerged as key regulators of circadian rhythmicity. However, no studies to date have identified the role of circadian astrocyte function in the nucleus accumbens (NAc), a hub for reward regulation, or determined the importance of these rhythms for reward-related behavior.
Methods:
Using astrocyte-specific RNA-sequencing across time-of-day, we first characterized diurnal variation of the NAc astrocyte transcriptome. We then investigated the functional significance of this circadian regulation through viral-mediated disruption of molecular clock function in NAc astrocytes, followed by assessment of reward-related behaviors, metabolic-related molecular assays, and whole-cell electrophysiology in the NAc.
Results:
Strikingly, ~43% of the astrocyte transcriptome has a diurnal rhythm and key metabolic pathways were enriched among the top rhythmic genes. Moreover, mice with a viral-mediated loss of molecular clock function in NAc astrocytes show a significant increase in locomotor response to novelty, exploratory drive, operant food self-administration and motivation. At the molecular level, these animals also show disrupted metabolic gene expression, along with significant downregulation of both lactate and glutathione levels in the NAc. Importantly, loss of NAc astrocyte clock function also significantly altered glutamatergic signaling onto neighboring medium spiny neurons, alongside upregulated glutamate-related gene expression.
Conclusions:
Taken together, these findings demonstrate a novel role for astrocyte circadian molecular clock function in the regulation of the NAc and reward-related behaviors.
Keywords: Astrocytes, Nucleus Accumbens, Circadian Rhythms, Reward, BMAL1, Metabolic State
Introduction
Substance use disorder (SUD) remains a devastating public health issue in the United States (1, 2) that lacks effective therapeutics (3). A potential treatment target may involve circadian rhythms. While it has long been known that individuals with SUDs have disrupted circadian and sleep/wake rhythms (4, 5), there is a growing body of evidence suggesting a bidirectional relationship between circadian rhythm disruption and substance abuse. Both human and animal studies indicate disruptions to circadian rhythms and sleep/wake may increase SUD vulnerability (6). Specifically, changes at the molecular level of the circadian system may play an essential role in the establishment and/or reinforcement of drug addiction. In turn, exposure to drugs of abuse further disrupts molecular clock function, resulting in a self-perpetuating cycle.
In mammals, circadian rhythms are controlled by a series of transcription-translation feedback loops (TTFLs) that cycle every 24h (7). This ‘molecular clock’ consists of the core circadian transcription factors CLOCK or NPAS2, which form a complex with BMAL1 and drive the expression of the proteins Period (PER) and Cryptochrome (CRY). PER and CRY heterodimerize in the cytosol, translocate into the nucleus, and interact with CLOCK/NPAS2:BMAL1 to inhibit their own transcription. This molecular clock is present in nearly every cell throughout the body and is organized into a synchronous, hierarchical system by a “central clock” in the suprachiasmatic nucleus (SCN) that works to temporally control nearly all physiology and behavior (8, 9). Notably, disrupted rhythmicity through mutations in the circadian molecular clock genes has been associated with altered reward regulation. In humans, variants in BMAL1, PER, and CLOCK are associated with increased drug use and reward dependence (10–17). In mice, mutations in the Per, Clock, or Npas2 genes can significantly alter exploratory drive, cocaine locomotor sensitization, cocaine preference and self-administration/motivation, and alcohol self-administration (18–28). In addition to regulating reward, disruptions to circadian rhythms have also been associated with many other psychiatric disorders often comorbid with substance use (e.g., anxiety, depression, bipolar, etc.) (29–32). While together these studies highlight the role a functional molecular clock plays in regulating reward-related behaviors, most research to date has primarily focused on molecular clock function in neurons specifically, or in whole reward regions, like the nucleus accumbens (NAc) (33, 34). Interestingly, a growing body of work suggests astrocytes play a critical role in both the regulation of circadian rhythms and reward processing (35, 36).
Astrocytes are a highly abundant glial cell type essential for many modulatory and support functions across the central nervous system (37), including regulation of glutamate levels and neurometabolic homeostasis (38–41). Like neurons, astrocytes also contain a circadian molecular clock (42–44), and core astrocyte functions are regulated by clock genes (45–47). Interestingly, several recent studies have demonstrated astrocyte rhythmicity is also important for regulating circadian rhythmicity. SCN astrocytes show anti-phasic rhythmicity relative to SCN neurons, whereby they suppress neuronal activity during the night via regulation of extracellular glutamate (48). This anti-phasic relationship is important regulating SCN circadian timekeeping at both the molecular and behavioral levels (48–50). In addition to regulating circadian rhythms (48), extensive work has shown astrocytic regulation of glutamate in the NAc to be important for reward-regulation and drug addiction vulnerability (51, 35, 52, 53, 34). However, while we know astrocytes regulate circadian rhythms and reward through their roles in the SCN and NAc, respectively, no studies to date have investigated NAc astrocyte rhythmicity in the NAc and/or its potential role in regulating NAc function in the context of reward.
Methods and Materials
See the Supplemental Information for detailed methods and materials.
Animals
Aldh1l1-eGFP/Rpl10a mice were used for characterizing astrocyte rhythmicity (54–56), while Bmal1 “floxed” (BMFL; Bmal1lox) mice were used for loss of rhythmicity experiments (57) (The Jackson Laboratory; Bar Harbor, ME). Both sexes were used throughout. All mice were maintained on a 12:12 light-dark cycle (lights on: 0700 zeitgeber time (ZT) 0; lights off: 1900, ZT12) and provided ad libitum food and water access unless otherwise indicated.
Immunoprecipitation (IP) and RNA extraction from NAc Astrocytes
NAc tissue was collected from Aldh1l1-eGFP/Rpl10a mice across 6 times of day (ZT2,6,10,14,18,&22; 10Male/8Female/ZT). Immunoprecipitation of polyribosomes was processed using a modified protocol described previously (58, 59). Final eluted NAc astrocyte-specific RNA was used for RNA-sequencing.
RNA sequencing and Analyses
Following quality/integrity assessment and library preparation, Total RNA sequencing was performed using the Illumina NextSeq 500 platform (Illumina; San Diego, CA). After pre-processing and filtering of sequencing data, 12,739 genes remained for further analysis. The JTK_CYCLE R package was used to detect circadian rhythmicity of NAc astrocyte-specific genes (60, 61). Ingenuity Pathway Analysis (IPA) (QIAGEN; Hilden, Germany) (62) and Metascape (https://metascape.org/) (63) were used to identify enriched molecular pathways/processes in both the top enriched and top rhythmic gene lists.
RNAscope ® in situ hybridization (ISH)
For qualitative validation of Aldh1l1-eGFP/Rpl10a mice, the RNAscope ISH Fluorescent Multiplex Assay (ACD; Newark, CA) was utilized.
Viral-Placement Surgery and Verification
BMFL mice (8 weeks old) were injected into the NAc with AAV8-GFAP-Cre-GFP or AAV8-GFAP-eGFP viral vectors (UNC Viral Vector Core; Chapel Hill, NC) as previously published (25). Mice were allowed ≥2 weeks to recover before behavioral testing or tissue collection. Viral placement, spread, and specificity were verified using immunofluorescence (IF).
Behavioral Testing
BMFL mice expressing NAc-specific GFAP-Cre or eGFP control were run through a panel of behavioral tests assessing reward-related behaviors at ZT2–6 or ZT14–18.
Gene Expression and Metabolic Assays
In whole NAc tissue from BMFL mice expressing NAc-specific GFAP-Cre or eGFP control, reverse transcriptase quantitative PCR (RT-qPCR) was used to measure astrocyte, metabolic, and glutamate-related gene expression (ZT4–6). Primers provided in Table S1.
Both L(+)-Lactate and glutathione (GSH) levels were also measured utilizing established and validated colorimetric and luminescent assays (64–67), at ZT5 & ZT17.
Electrophysiology Recordings
Whole-cell patch-clamp recordings were conducted at ZT4–6 and ZT16–18 using BMFL NAc-containing sections expressing GFAP-Cre or eGFP control.
Statistical Analyses
GraphPad Prism 9 (GraphPad Software; San Diego, CA) was utilized for all statistical data analyses. All analyses tested for sex as a variable; if a main effect of sex was detected, sexes are displayed separately. Unless otherwise noted, all data are shown as mean ± SEM with α=0.05.
Results
The NAc astrocyte transcriptome is highly rhythmic
Our lab and others have previously demonstrated whole NAc rhythmicity at both the cellular and molecular levels (68, 69); however, no studies to date have investigated NAc astrocyte rhythmicity. To do this, we utilized the Aldh1l1-eGFP/Rpl10a mouse model (70–72), wherein astrocytes exclusively express an eGFP tag on the RpL10a ribosomal subunit that allows for immunoprecipitation and isolation of astrocyte-specific ribo-associated mRNA (Figure 1A). We first isolated astrocyte-specific mRNA from whole NAc tissue harvested across 6 times of day in male and female mice following a previously published protocol (58, 59). Isolated mRNA was then processed for RNA-sequencing analysis, resulting in the detection of 12,739 expressed genes. Notably, cell-type enrichment heatmapping using established cell-type markers (54, 56, 73–75) revealed a high degree of specificity for astrocyte enrichment (Figure 1B). Moreover, the eGFP tag is highly-colocalized with Aldh1l1, a pan-astrocyte marker (54, 55), and not with Vgat, a GABA transporter expressed in NAc MSNs (Figure 1C). Ingenuity Pathway Analysis (62) and Metascape analysis (63) revealed historically astrocyte-associated biological processes and canonical pathways to be enriched among the top 200 expressed genes (Figure 1D), further validating this method and underscoring their potential importance in the NAc. These processes were also highly interconnected, as revealed by an enrichment network analysis (Supplemental Figure S1).
Figure 1. Aldh1l1-eGFP/Rpl10a mice allow for isolation of astrocyte-specific ribo-associated mRNA.
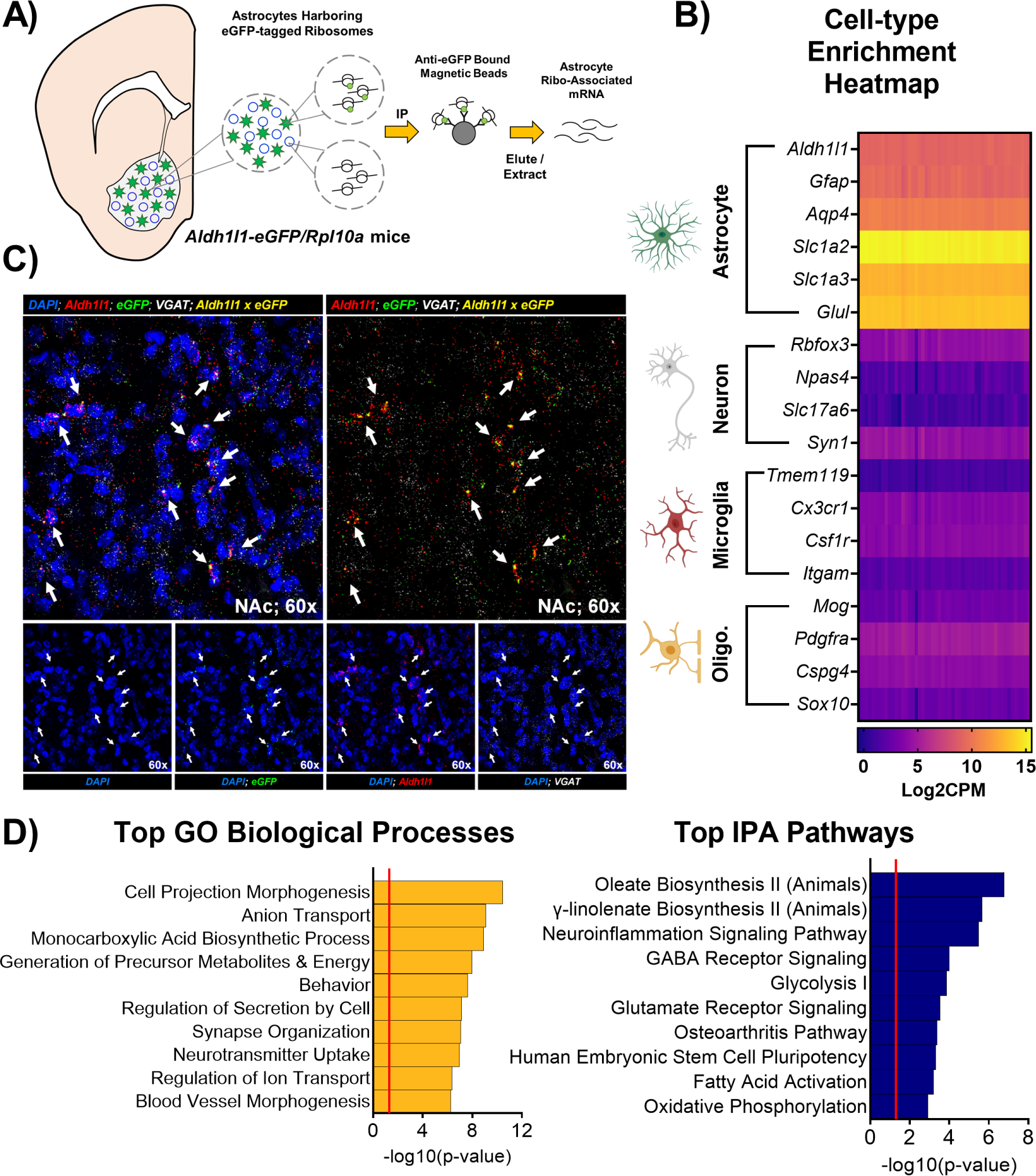
Schematic illustrating the immunoprecipitation (IP) of astrocyte-specific ribo-associated mRNA from whole NAc tissue of Aldh1l1-eGFP/Rpl10a mice that express an eGFP tag on the ribosomal subunit RpL10a. (B) Isolated mRNA is highly specific for astrocytes, as revealed by a cell-type enrichment heatmap generated from plotting Log2CPM values of established cell-type markers from our RNA-sequencing dataset (alternate names: Slc1a2 = EAAT2 or Glt-1; Slc1a3 = EAAT1 or Glast; Glul = GS; Rbfox3 = NeuN; Slc17a6 = Vglut2). Columns are the individual samples, rows are individual genes. Purple indicates low expression, while yelllow indicates peak expression. (C) Moreover, the fluorophore eGFP is expressed specifically in cells expressing the pan-astrocyte marker Aldh1l1, but not in cells expressing the neuronal GABA transporter VGAT, as determined by RNAscope; arrows indicate cells with co-expression of Aldh1l1 and eGFP. (D) Following RNA-seq, Ingenuity Pathway Analysis (62) and Metascape analysis (63) revealed the top canonical pathways and biological processes enriched among the top 200 expressed genes in NAc astrocytes include key metabolic and synaptic neurotransmission functions generally attributed to astrocytes. Notable NAc-astrocyte enriched processes include glycolysis and lactate synthesis, oxidative phosphorylation, glutamate receptor signaling, neurotransmitter uptake (e.g., glutamate), and neuroinflammation signaling. Red line in bar graphs indicates significance threshold of p<0.05 or −log10(p-value) > 1.3.
We next used JTK_Cycle, a non-parametric statistical algorithm, to identify transcripts with diurnal variation in expression (60). Out of the 12,739 genes we identified, approximately 43.3% display a significant diurnal rhythmicity (Figure 2A). Rank-rank hypergeometric overlap (RRHO) revealed a nearly identical overlap between males and females (Supplemental Figure S2) - thus analyses proceeded with sexes grouped together. Interestingly, through assessing circadian acrophase (time of peak expression), we see the majority of rhythmic genes peak between ZT14-ZT22, coinciding with the mouse’s active phase (Figure 2B). Moreover, pathway enrichment analyses of top rhythmic transcripts revealed circadian pathways/processes among the most enriched (Figure 2C; Supplemental Figure S3). Also, many of the top 20 rhythmic transcripts are known core clock genes and their expression patterns align with established expression relationships (Figure 2D). Interestingly, alongside circadian pathways and processes, important metabolic-relevant functions were also enriched among the top rhythmic genes (Figure 2C). Finally, in an enrichment network analysis of the top rhythmic biological processes, the circadian regulation and glucocorticoid receptor signaling node clusters show a high degree of interconnectivity (Figure 2E; Supplemental Figure S3).
Figure 2. Nucleus accumbens astrocytes are highly rhythmic at the transcriptome level.
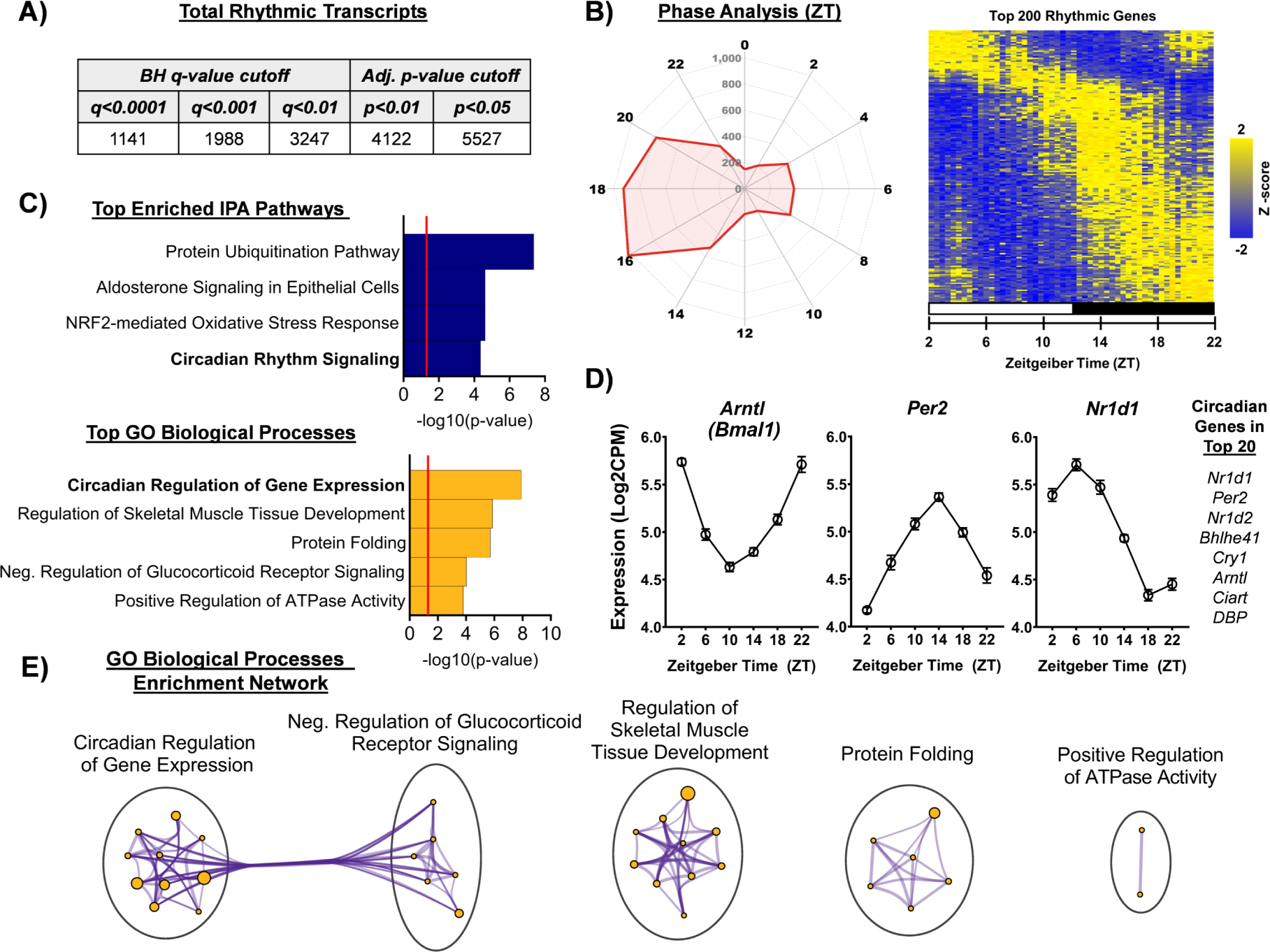
Among the 12,739 genes identified in NAc astrocytes, approximately 43.3% of genes exhibited rhythmic expression across the 6 ZTs sampled (ZT2,6,10,14,18,22; n=4–5 per ZT, JTK_Cycle), or 5,527 genes at the Bonferroni-adjusted p-value cutoff of p<0.05. Even using the most stringent statistical cutoff (Benjamini-Hochberg q-value cutoff of q<0.0001), approximately ~9% of transcripts still display diurnal rhythmicity, or 1,141 genes. (B) Phase analysis through radar plotting of top rhythmic transcripts (p<0.05) and heatmap visualization of the top 200 rhythmic genes reveal a majority of rhythmic transcripts peak between ZT14 and ZT18, coinciding with the dark phase / active phase. The radar plot perimeter axis represents ZT, while the inner axis is the number of genes with peak expression at a given ZT. Heatmap displays Z-transformed expression for each of the top 200 statistically rhythmic genes (as determined by JTK_Cycle), ranked in descending order along the Y-axis (rows) and each subject ordered by ZT along the x-axis (columns). The white bar indicates lights on (ZT2,6,10) and the black bars indicate lights off (ZT14,18,22). (C) IPA pathway analysis and Metascape Gene Ontology (GO) Biological Process analysis revealed both circadian and metabolic-related processes enriched among the top rhythmic genes. Notable pathways/processes include circadian rhythm signaling and circadian regulation of gene expression, as well as NRF2-mediated oxidative stress response, regulation of ATPase activity, and glucocorticoid receptor signaling. Red line in bar graphs indicates significance threshold of p<0.05 or −log10(p-value) > 1.3. (D) Core circadian genes are among the top rhythmic genes and show expression patterns that align with previously established functional relationships; i.e., Arntl (gene that encodes BMAL1) is anti-phasic with Per2 and Nr1d1). (E) Among the top enriched rhythmic GO biological processes, the circadian regulation of gene expression and glucocorticoid receptor signaling node clusters show a high degree of interconnectivity.
Loss of BMAL1 function in NAc astrocytes increases locomotor response to novelty during the day
To investigate the functional importance of NAc astrocyte rhythmicity for reward, Bmal1 floxed mutant mice (Bmal1f/f) were stereotaxically injected into the NAc with an adeno-associated virus (AAV) expressing either Cre recombinase or eGFP control under the Gfap astrocyte-specific promoter (Figure 3A). NAc expression of GFAP-Cre results in loss of astrocyte molecular clock function and rhythmicity due to deletion of BMAL1’s functional domain (57, 49, 76), which IF revealed to be both localized and specific (Figure 3A,B). To assess reward-related behavior, mice were first run through the locomotor response to novelty (LRN) task (ZT4–6 and ZT16–18), a behavior established as a predictor of drug-seeking propensity (77–79). Interestingly, GFAP-Cre mice show a significantly higher LRN during the day, relative to GFAP-eGFP control mice (Figure 3C), measured as total locomotor activity in the first hour of the two-hour task (Figure 3C and inset). Notably, this increased LRN was not due to any differences in habituation or total activity (Figure 3D and inset). Moreover, no significant differences were observed during the night (Figure 3E–G). However, looking across phase, eGFP mice displayed a significant diurnal variation in LRN that is disrupted in GFAP-Cre mice, driven by increased LRN during the day (Supplemental Figure S4).
Figure 3. Loss of BMAL1 function in NAc astrocytes increases locomotor response to novelty in mice during the day.
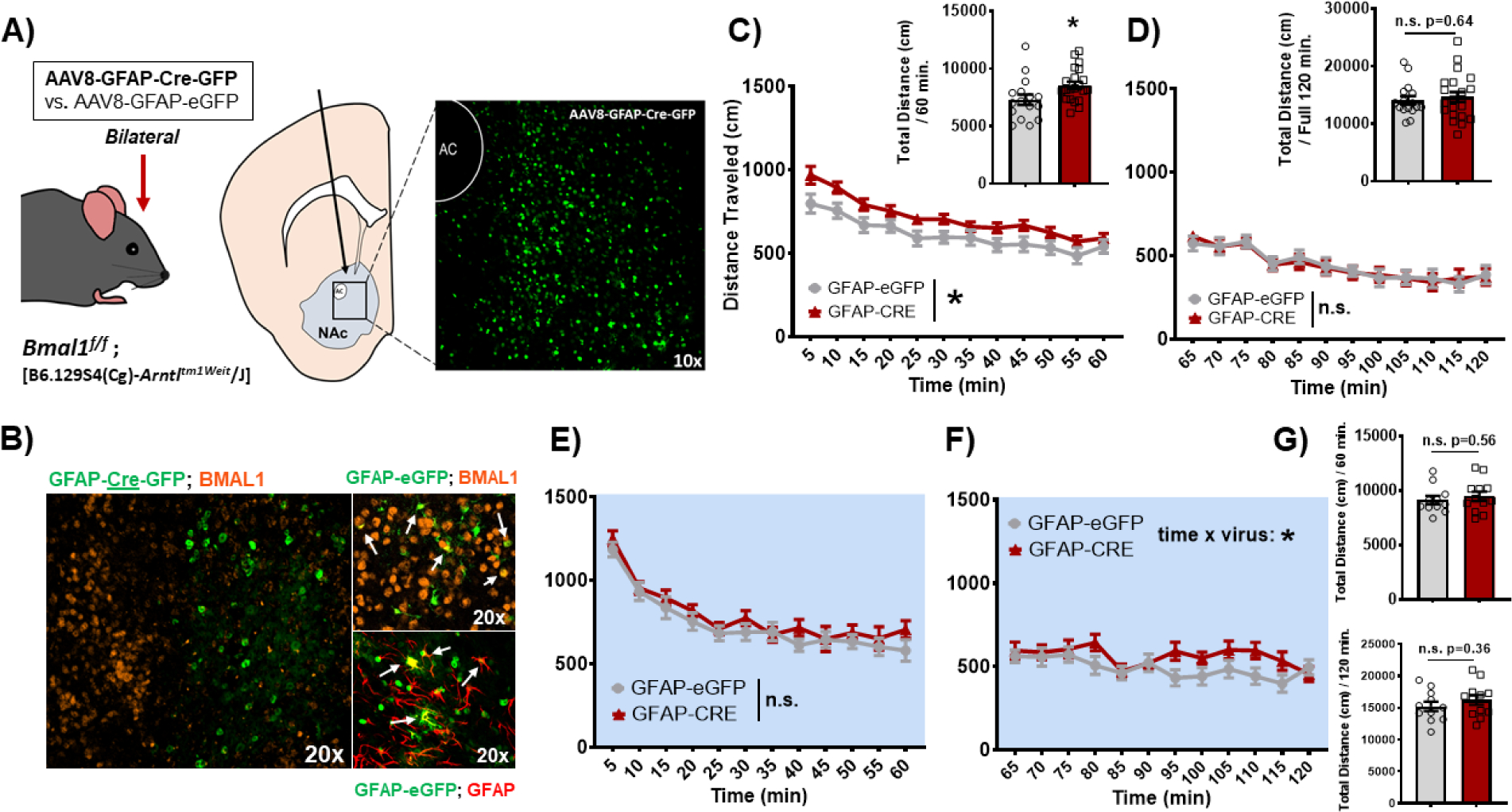
(A) Schematic illustrating the bilateral stereotaxic injection of either AAV8-GFAP-Cre-GFP or GFAP-eGFP virus into the NAc of Bmal1 floxed mutant mice; injection at AP: +1.5, ML: ± 1.5, and DV: −4.4 microns resulted in a region-specific expression of the virus in the NAc. (B) GFAP-Cre-GFP virus resulted in a NAc-specific loss of BMAL1 detection by immunofluorescence (IF), specifically in GFAP+ cells (i.e., astrocytes). Arrows indicate cells with co-expression. (C) In the locomotor motor response to novelty (LRN) task, mice with a loss of BMAL1 function in NAc astrocytes (GFAP-Cre) show a significantly greater LRN relative to control mice during the day (Virus: F(1, 36) = 5.68, * p=0.02; Time: F(11, 396) = 40.41, p<0.0001; Time × Virus: F(11, 396) = 0.92, p=0.51; inset, t(35)=2.32, * p=0.02). (D) This increase in LRN is not driven by differences in habituation or overall locomotor activity across the full task (inset) Virus: F(1, 36) = 0.0003, n.s. p=0.98; Time: F(11, 396) = 16.66, p<0.0001; Time × Virus: F(11, 396) = 0.26, p=0.99; inset, t(35) = 0.46, p=0.64. (E) However, during the night, mice show no significant differences in locomotor response to novelty (Virus: F(1, 22) = 1.017, p=0.32; Time: F(11, 242) = 44.68, p<0.0001; Time × Virus: F(11, 242) = 0.62, p=0.8), (F) habituation (Virus: F(1, 22) = 2.19, p=0.15; Time: F(11, 242) = 3.28, p=0.0003; Time × Virus: F(11, 242) = 1.91, p=0.03), or (G) overall locomotor differences (top: t(21)=0.58, p=0.56; bottom: t(21)=0.91, p=0.36). White background indicates behavior run during the day (ZT 2–6). Blue background indicates behavior run during the night (ZT 14–18). Mean ± SEM; n=12–22; * p<0.05, n.s. = not significant.
Loss of BMAL1 function in NAc astrocytes increases exploratory drive behavior during the day
Alongside LRN, increased exploratory drive and novelty-seeking behavior are also established predictors of vulnerability for drug-seeking and addiction-like behaviors in rodent models (77, 80, 78, 81–83). Given this association, mice were then run through a battery of exploratory drive / anxiety-related behavioral tasks, including open field (OF), light-dark box test (L/D), elevated plus maze (EPM), and novelty suppressed feeding (NSF), either during the day or at night. Strikingly, across all 4 tasks, mice with disrupted NAc astrocyte molecular clock function exhibit significantly elevated exploratory drive during the day, relative to controls (Figure 4A–D). Following z-normalization of the 4 behaviors, a method previously shown to increase sensitivity, reliability, and robustness of behavioral phenotyping (84), exploratory drive was found to be significantly greater in GFAP-Cre mice during the day (Figure 4E; top) but not at night (bottom) – further underscoring the phenotype. Interestingly, looking across phase, eGFP mice display a significant diurnal variation in exploratory drive that is disrupted or attenuated in GFAP-Cre mice, driven by effects during the day (Supplemental Figure S5). Finally, this phenotype is not attributed to locomotor differences across tasks or motivation to eat in NSF (Supplemental Figure S6).
Figure 4. Loss of BMAL1 function in NAc astrocytes increases exploratory drive in mice during the day.
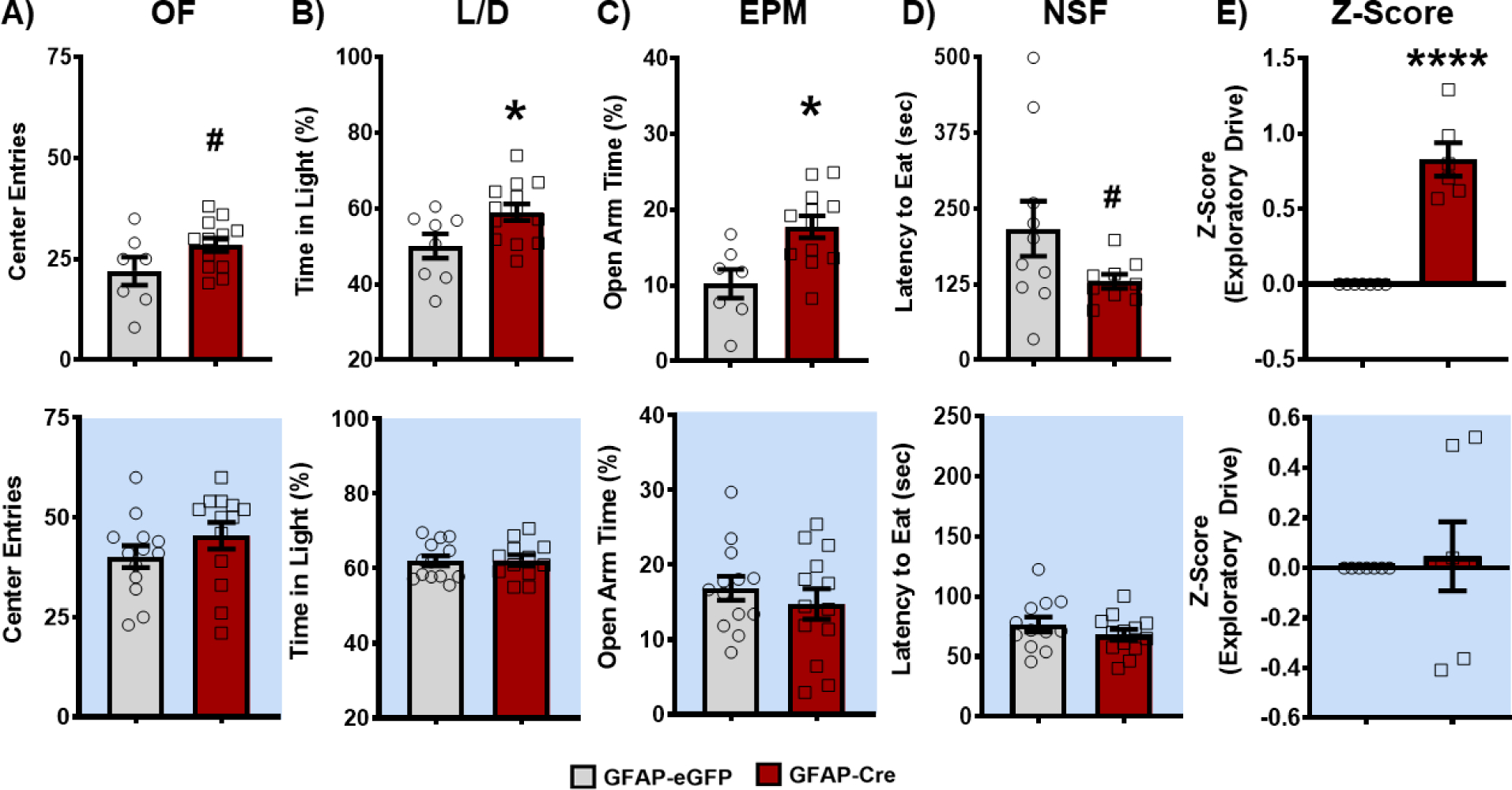
Both BMFL mice expressing NAc-specific GFAP-Cre or eGFP control were run through a panel of behavioral assays testing exploratory drive. (A) In the open field (OF) task, GFAP-Cre mice show an increased number of center entries relative to eGFP controls during the day (top: t(18) = 1.91; # p=0.07), but not during the night (bottom: t(24) = 1.21; p=0.24). (B) In the light-dark box test (L/D), GFAP-Cre mice spend a significantly greater percentage of time in the brightly lit chamber of the arena during the day (top: t(19) = 2.37; * p=0.02), but not during the night (bottom: t(23) = 0.09; p=0.92). (C) GFAP-Cre mice also spent significantly more time in the open arms of the elevated plus maze (EPM) during the day (top: t(18) = 2.35; * p=0.03), but not during the night (bottom: t(24) = 0.81; p=0.43). (D) Following an overnight food restriction, GFAP-Cre mice had a shorter latency to eat food in the novelty suppressed feeding (NSF) task during the day (top: t(22) = 1.9; # p=0.06), but not during the night (bottom: t(23) = 1.15; p=0.26). (E) Across the behavioral panel, Z-normalization of the 4 behavioral tasks revealed a loss of BMAL1 function in NAc astrocytes significantly increases exploratory drive behavior relative to control mice during the day (top: t(11) = 8.14, **** p<0.0001), but not during the night (bottom: t(12) = 0.34; p=0.74). White background indicates behavior run during the day (ZT 2–6). Blue background indicates behavior run during the night (ZT 14–18). Mean ± SEM; n=7–13; # p≤0.07, * p<0.05, **** p<0.0001.
Loss of BMAL1 function in NAc astrocytes increases food self-administration and motivation during the day
While LRN and exploratory drive have been established as reward-relevant behaviors, we next investigated reward specifically. Mice were trained either during the day or at night in an operant food self-administration task in which mice discriminate between two levers (active versus inactive) to receive a food pellet. Mice were first trained on a fixed ratio 1 (FR1) schedule whereby 1 active lever press is rewarded with 1 food pellet, and inactive lever presses result in no pellets (Figure 5A). On the FR1 schedule, all mice successfully acquired the task (criteria: ≥ 25 pellets for 3 sessions) both during the day (Figure 5B) and at night (Figure 5D). While no significant differences were detected between virus groups across phase (Figure 5B,D), a main effect of sex was detected in that male mice exhibit a higher response rate than females, both during the day (Figure 5B) and at night (Figure 5D); this finding is consistent with previous literature showing sex differences in operant appetitive learning (85–87). Moreover, looking across phase, while male mice did not show diurnal variation in responding, female eGFP and GFAP-Cre mice both show a significant diurnal variation in food self-administration at FR1 that was enhanced in the GFAP-Cre group (Supplemental Figure S7). Following FR1 training, food self-administration motivation was tested through the escalation of progressively more demanding FR schedules. Strikingly, across progressively harder schedules (FR3, FR5, and FR10), male GFAP-Cre mice (but not females) demonstrate a robust increase in daytime food self-administration despite the increasingly more demanding schedules (Figure 5C; Supplemental Figure S8A), but not at the night (Figure 5E; Supplemental Figure S8B). Finally, much like at FR1, female eGFP and GFAP-Cre mice both show a highly significant diurnal variation in food self-administration motivation (Supplemental Figure S7), but not males.
Figure 5. Loss of BMAL1 function in NAc astrocytes increases operant food self-administration and motivation during the day.
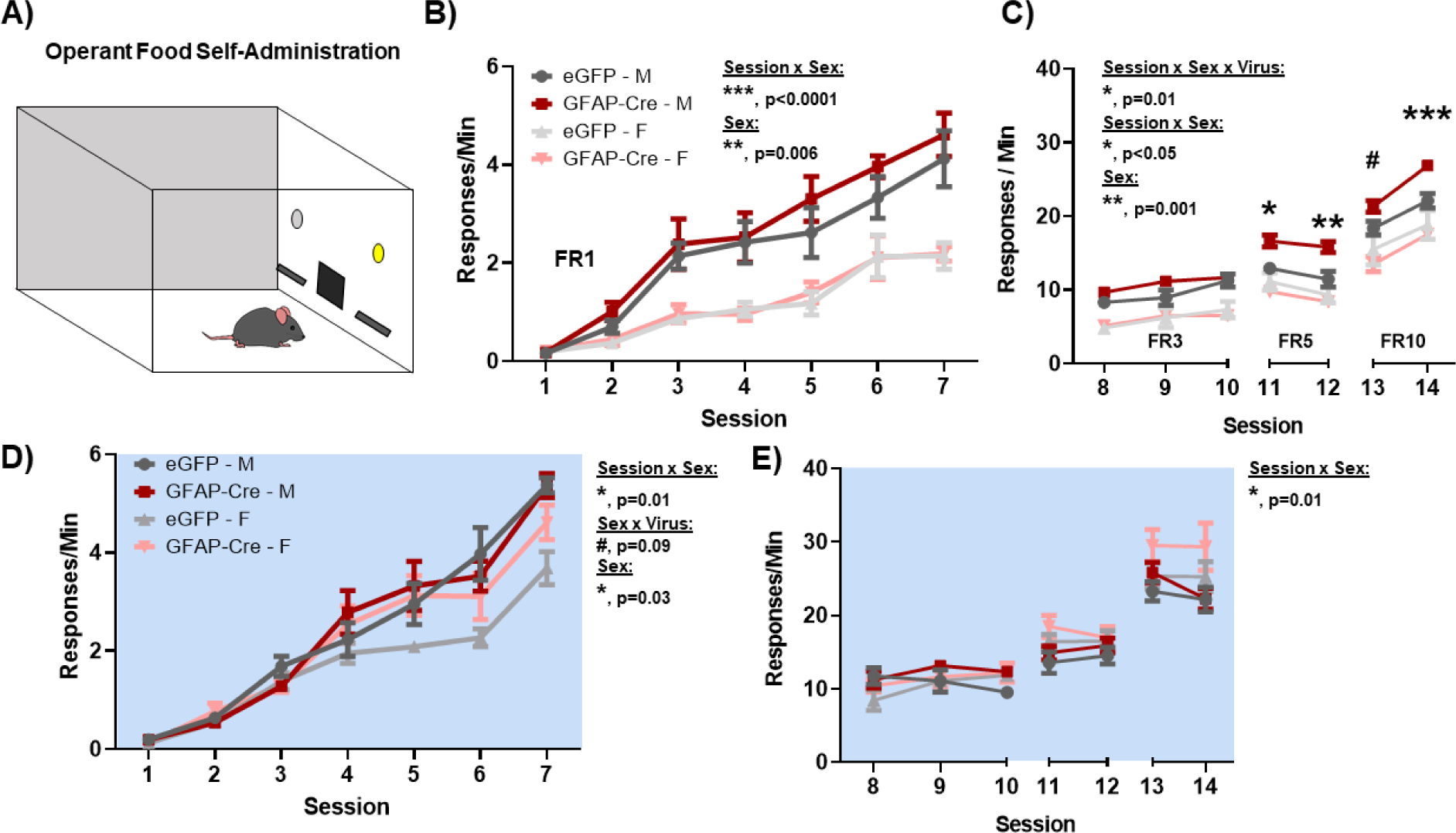
To assess natural reward and reward-motivation, both BMFL mice expressing NAc-specific GFAP-Cre or eGFP control were trained to self-administer food pellets in an operant food self-administration task both during the day and at night. (A) Mice were trained to discriminate between an active and an inactive lever, whereby active lever pressing resulted in a fixed ratio (FR) of food pellets. (B) On an FR1 schedule (i.e., 1 lever press : 1 pellet), GFAP-Cre mice successfully learn to self-administer food (Main effect of Session: F(6, 60) = 59.63, p<0.0001), but show no significant differences relative to controls (Virus: F(1, 10) = 1.68, p=0.22). In both virus groups, males show a significantly greater response rate relative to females (Sex: F(0.4, 4) = 37.11, ** p=0.006; Session × Sex: F(3.5, 35.2) = 8.52, *** p<0.0001). (C) To test reward motivation, mice were tested across increasingly difficult FR schedules (i.e., FR3, 3 presses : 1 pellet; FR5, 5 presses : 1 pellet; FR10, 10 presses : 1 pellet). GFAP-Cre male mice not only show a robust maintenance, but also a significant increase in food self-administration rate across FR schedules during the day, relative to control counterparts (Session × Sex × Virus: F(6, 59) = 2.96, * p=0.01; Sex × Virus: F(1, 10) = 5.09, * p<0.05; Sex: (F(0.6, 6) = 34.92, ** p=0.001). No effect of the virus was seen in female mice. (D) During the night, GFAP-Cre mice also successfully learn to self-administer food (Main effect of Session: F(7, 70) = 164.4, p<0.0001), with a main effect of sex (Sex: F(0.37, 3.7) = 15.01, * p=0.03; Session × Sex: F(2.8, 25.96) = 3.55, * p=0.03; Sex × Virus: F(1, 10) = 5.05, * p=0.04), but no significant effects of virus (Virus: F(1, 10) = 2.29, p=0.16). (E) During the night, no significant differences are seen in motivation as mice are tested across increasing FR schedules (Session × Sex × Virus: F(6, 58) = 1.1, p=0.37; Sex × Virus: F(1, 10) = 0.09, p=0.76 ; Sex: F(0.37, 3.67) = 2.17, p=0.17; Session × Sex: F(2.5, 24.32) = 4.75, * p=0.01). White background indicates behavior run during the day (ZT 2–6). Blue background indicates behavior run during the night (ZT 14–18). Mean ± SEM; n = 6; # p=0.07, * p<0.05, ** p<0.01, *** p=0.0001.
Loss of BMAL1 function in NAc astrocytes disrupts metabolic homeostasis
Given that astrocytes play a crucial role in regulating cellular metabolic state, we next wanted to investigate how disrupting NAc astrocyte molecular clock function affects this key function. Utilizing RT-qPCR in GFAP-Cre or eGFP whole NAc tissue, we first assessed expression of key astrocyte and metabolic-state relevant genes during the day (ZT4–6; i.e., time of behavioral phenotype) (Figure 6A). No significant difference was seen in Aldh1l1 expression. However, GFAP-Cre mice exhibit a significant upregulation of Gfap and Nrf2, established markers of astrocyte reactivity and oxidative stress induced anti-oxidant response, respectively (88, 89). Moreover, GFAP-Cre mice exhibited a significant downregulation of Gclc, Pgc1α, Ldha, Mct1, and Mct2, all genes relevant for glutathione (GSH) production, mitochondrial function, and lactate synthesis/shuttling, respectively.
Figure 6. Loss of BMAL1 function in NAc astrocytes disrupts NAc metabolic homeostasis.
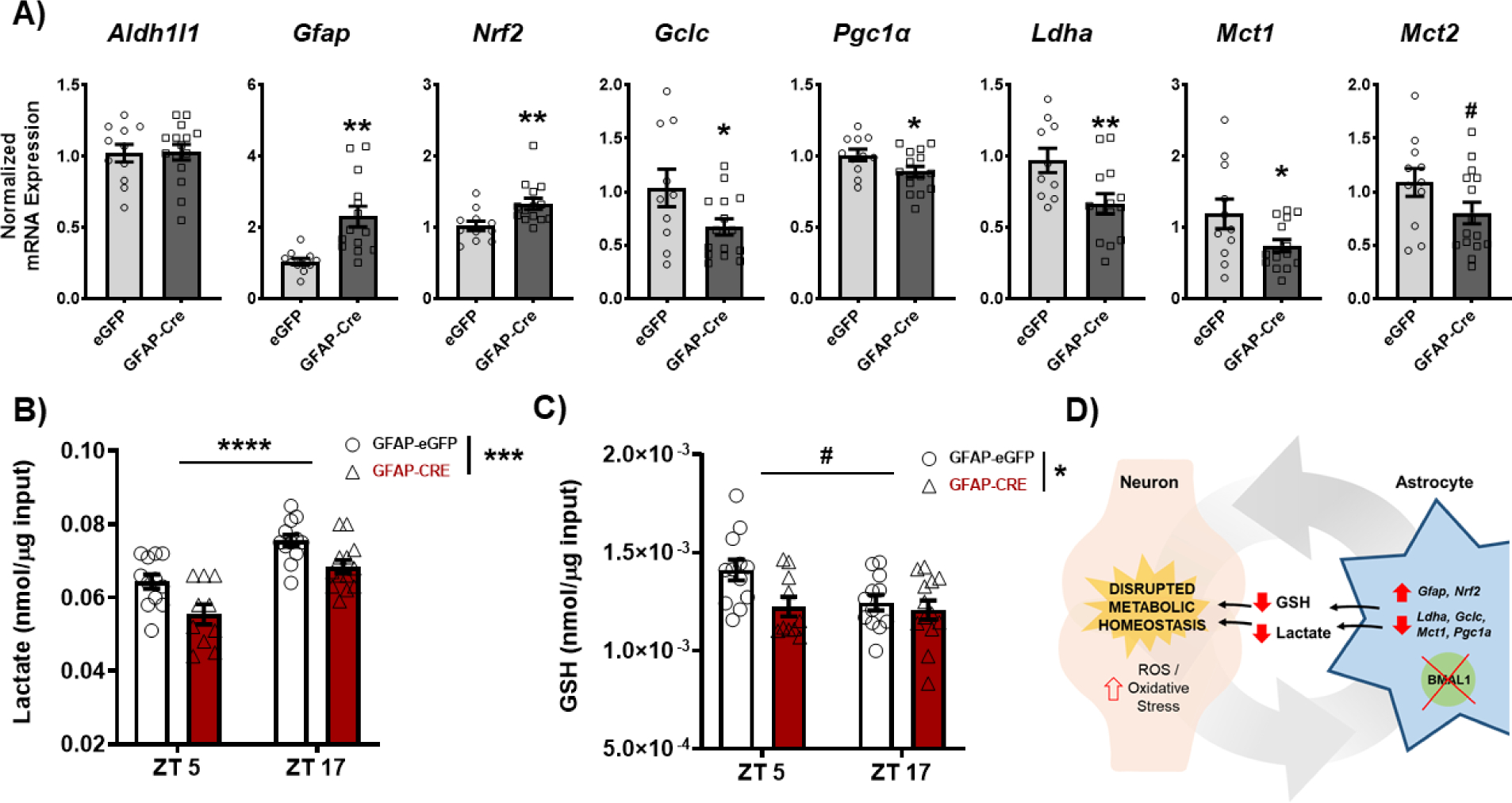
NAc tissue from both BMFL mice expressing NAc-specific GFAP-Cre or eGFP control was processed through a range of molecular assays assessing factors relevant for regulation of cellular metabolic state. (A) Through reverse transcriptase quantitative PCR (RT-qPCR), a panel of metabolic genes was assessed in the whole NAc of GFAP-Cre and eGFP control mice (ZT4–6). While there were no differences in the astrocyte marker Aldh1l1 (t(24) = 0.09, p=0.92), GFAP-Cre mice showed significantly elevated expression of Gfap (t(23) = 3.74, ** p=0.001) and Nrf2 (t(23) = 2.87, ** p=0.008), markers of astrocyte activation and oxidative stress antioxidant response. GFAP-Cre mice also showed significant reductions in genes relevant for glutathione (GSH) production (Gclc; t(23) = 2.15, * p<0.05), mitochondrial biogenesis (Pgc1a; t(24) = 2.13, * p<0.05), and lactate synthesis and shuttling (Ldha, t(22) = 2.75, * p=0.01; Mct1, t(24) = 2.18, * p<0.05; & Mct2, t(24) = 1.77, # p=0.08). (B) Moreover, loss of BMAL1 function in NAc astrocytes significantly reduces the concentration of lactate (Time: F(1, 43) = 35.76, ****p<0.0001; Virus: F(1, 43) = 15.79, *** p=0.0003; Interaction: F (1, 43) = 0.20, p=0.65) and (C) GSH (Time: F(1, 43) = 3.56, # p=0.06; Virus: F1, 43) = 5.31, * p=0.02; Interaction: F(1, 43) = 2.36, p=0.13) in the whole NAc across time of day. (D) The effects on key metabolic genes, lactate, and GSH are hypothesized to disrupt neurometabolic homeostasis, potentially leading to the downstream effects on NAc-regulated behavior. Gene expression normalized to 18s. Mean± SEM. N=10–15. # p=0.06; * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
We next wanted to determine the concentration of lactate and GSH in the NAc following disruption of NAc astrocyte molecular clock function. Astrocytes synthesize and shuttle both lactate and GSH to neurons to help support their metabolic demands and prevent oxidative stress, respectively (90, 40, 41). To measure lactate, NAc tissue from GFAP-Cre mice was run through an L(+)-Lactate colorimetric enzyme-linked immunosorbent assay (ELISA). Alongside the reductions in lactate-related gene expression (Figure 6A), GFAP-Cre mice exhibited a significant reduction in lactate concentration across time of day, relative to controls (Figure 6B). Interestingly, NAc lactate concentration exhibits diurnal variation in eGFP controls which was disrupted in GFAP-Cre mice. Alongside lactate measures, NAc tissue from GFAP-Cre mice was also run through a GSH luminescent ELISA. Much like lactate levels, GFAP-Cre mice exhibit significantly reduced GSH concentration across time of day, relative to controls (Figure 6C). Notably, control mice also exhibit diurnal variation in NAc GSH concentration that is significantly reduced in GFAP-Cre mice.
Loss of BMAL1 function in NAc astrocytes alters excitatory synaptic transmission onto neighboring MSNs
To investigate the impact that astrocyte molecular clock disruption may have on NAc synaptic transmission, we performed whole-cell recordings from the principal medium spiny neurons (MSNs) in the NAc from both GFAP-Cre and eGFP control mice during the day and night (Figure 7A). Following the disruption of NAc astrocyte molecular clock function, the evoked AMPA/NMDA EPSC ratio was significantly decreased in GFAP-Cre mice during the day (Figure 7B), which was accompanied by a daytime increase in AMPA (but not NMDA) receptor EPSC decay kinetics (Figure 7C,D); i.e., suggesting postsynaptic glutamate receptor changes, particularly that of AMPA receptors. Furthermore, the evoked EPSCs exhibited similar paired-pulse ratios across phases (Figure 7E), suggesting the presynaptic release probabilities of mixed glutamatergic inputs were grossly unaltered under basal transmission. Following a train of stimuli, the slow EPSC tail also showed similar decay kinetics across phases (Figure 7F), suggesting comparable glutamate clearance between groups. Importantly, no significant differences in any of these parameters were detected at night (Figure 7G,H,I,K; Supplemental Figure S9), though a trending decrease in paired-pulse ratio was observed (Figure 7J). Interestingly, alongside these electrophysiological changes, GFAP-Cre mice also show a significant upregulation of glutamate-related gene expression in the whole NAc during the day – with upregulation of nearly all AMPA receptor subunits (Figure 8).
Figure 7. Loss of BMAL1 function in the NAc alters excitatory synaptic transmission onto neighboring MSNs during the day.
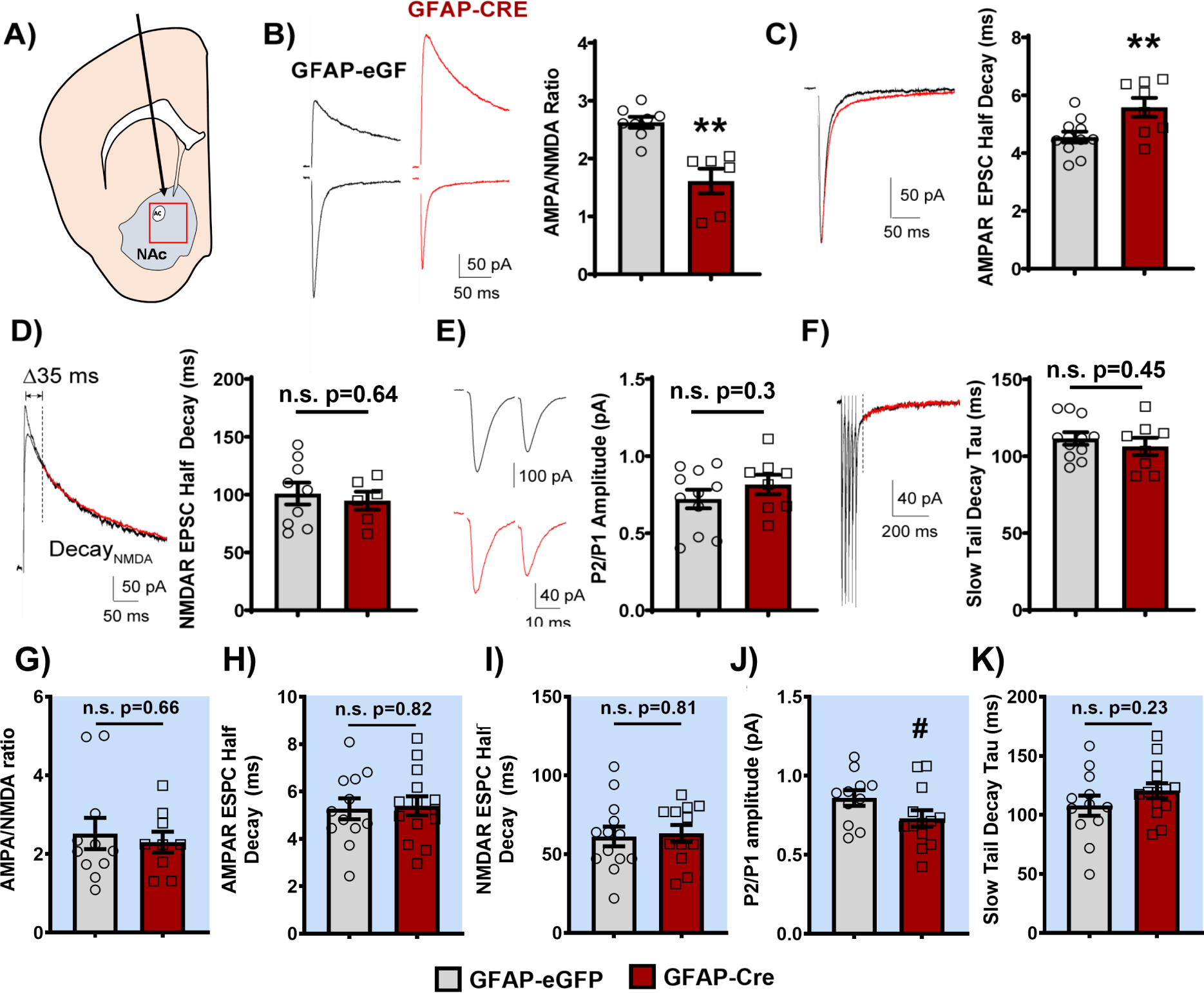
Slice electrophysiology was used to assess the effects of loss of BMAL1 function in NAc astrocytes on glutamatergic signaling in neighboring MSNs during the light and dark phases. (A) Location of stereotaxic injection of AAV8-GFAP-eGFP or AAV8-GFAP-Cre-GFP viral vectors and region targeted for slice electrophysiology experiments. (B) Representative traces (left) and quantification (right) of the AMPA/NMDA ratio. GFAP-Cre mice had a significantly lower AMPA/NMDA ratio compared to GFAP-eGFP controls (t(12) = 4.753, ** p<0.01, unpaired t-test). (C) Representative traces (left) and quantification showing that GFAP-Cre mice have slower AMPA decay kinetics, as measured by AMPA receptor half decay, compared to GFAP-eGFP controls (t(17) = 2.903, ** p<0.01, unpaired t-test). (D) However, there was no difference in NMDA receptor kinetics between GFAP-Cre mice and controls (representative traces, left; quantification, right; t(13) = 0.469, p=0.647, unpaired t-test). (E) Both GFAP-Cre and eGFP mice displayed a similar paired pulse ratio (P2/P1; i.e., EPSC2/EPSC1, measured at 20 ms ISI; t(17) = 1.063, p=0.303, unpaired t-test; representative traces, left; quantification, right). (F) Moreover, there was no difference in the tau of the slow tail decay after a train of high frequency extracellular stimulation (5 × 0.5 ms 50μA pulses at 50Hz) between GFAP-Cre and eGFP mice (measured from 35 ms following the peak of the last AMPA receptor EPSC; t(17) = 0.769, p=0.453, unpaired t-test; representative traces, left; quantification, right). Notably, when recording at night, no significant differences were seen in (G) AMPA/NMDA ratio (t(18) = 0.4396, p=0.665, unpaired t-test), (H) AMPA decay kinetics (t(24) = 0.2177, p=0.829, unpaired t-test), (I) NMDA decay kinetics (t(23) = 0.2336, p=0.817, unpaired t-test), (J) paired pulse ratio (t(22) = 1.781, # p=0.088, unpaired t-test), though trending, nor (K) tau of the slow tail decay (t(24) = 1.225, p=0.2323, unpaired t-test) between GFAP-Cre and eGFP controls. Red traces indicate GFAP-Cre group. Recordings performed ZT4–6 and ZT16–18. Blue background indicates behavior run during the night (ZT 14–18). Mean± SEM. n=6–14. ** p<0.01. n.s. = not significant.
Figure 8. Loss of BMAL1 function in NAc astrocytes upregulates NAc glutamate-related gene expression during the day.
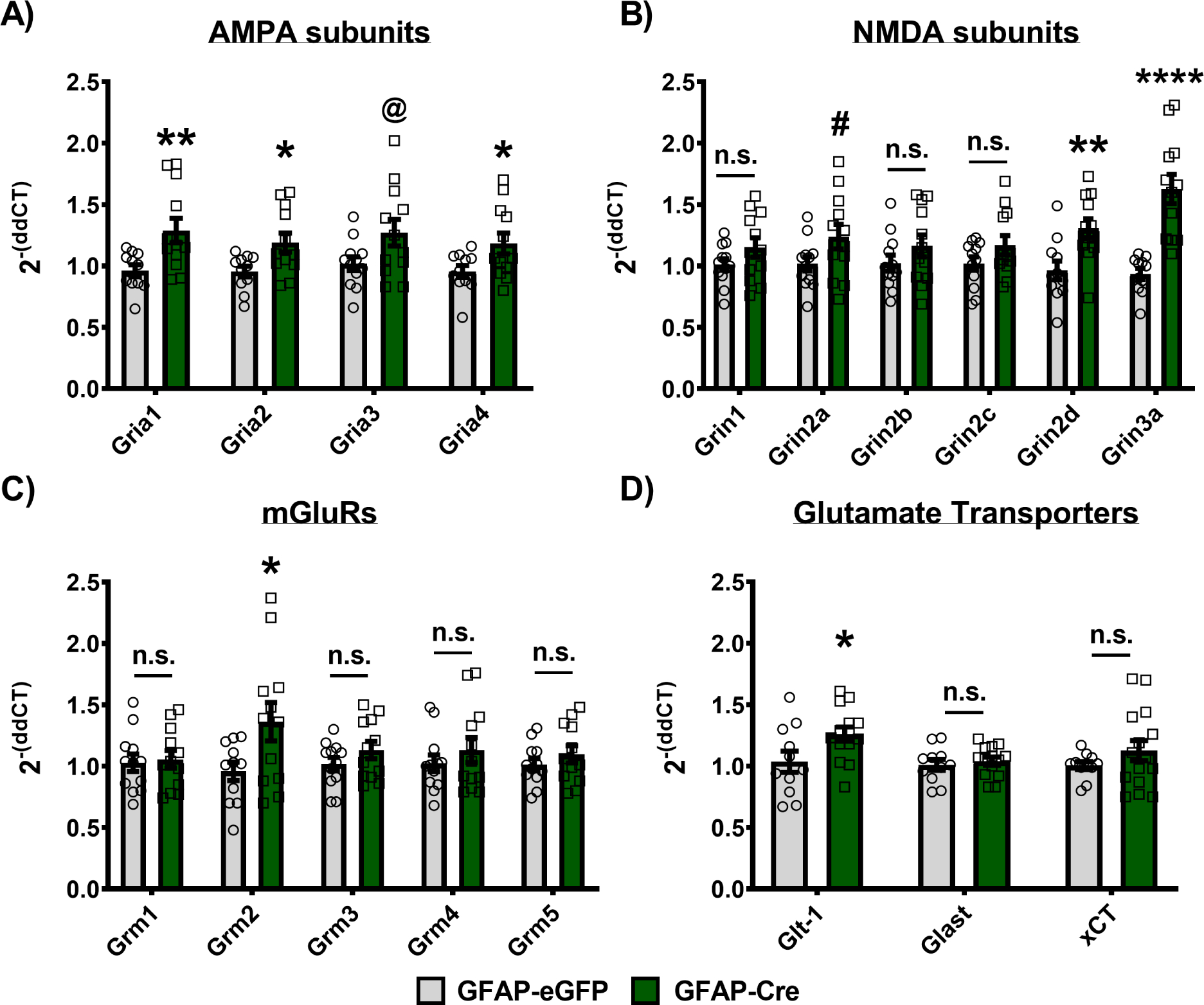
NAc tissue from BMFL mice expressing NAc-specific GFAP-Cre or eGFP control was processed through RT-qPCR assessing daytime whole NAc gene expression of AMPA receptor subunits, NMDA receptor subunits, mGluR receptors, and astrocyte-specific glutamate transporters (ZT4–6). (A) GFAP-Cre mice show significantly upregulated expression of nearly all AMPA receptor subunits, including Gria1 (t(21) = 2.874, ** p<0.01, unpaired t-test), Gria2 (t(21) = 2.583, * p=0.017), Gria4 (t(21) = 2.267, * p<0.05), and a trending increase in Gria3 (t(22) = 2.072, @ p=0.05), all relative to eGFP controls. (B) Looking at NMDA receptor subunit gene expression, only Grin2d (t(21) = 3.156, ** p=0.0048) and Grin3a (t(21) = 5.246, **** p<0.0001) show significant upregulation, with Grin2a (t(22) = 1.92, # p=0.06) showing a trending increase, relative to controls. No significant differences between GFAP-Cre and eGFP control were detected for Grin1 (t(22) = 1.491, p=0.15), Grin2b (t(22) = 1.276, p=0.21), and Grin2c (t(22) = 1.555, p=0.13), while Grin3b was not detectable at all in our whole NAc tissue samples (data not shown). (C) In addition to AMPA and NMDA receptor subunits, we also investigated the expression of the G-protein-coupled mGluR glutamate receptors. GFAP-Cre significantly increased Grm2 relative to eGFP controls (t(21) = 2.257, * p<0.05), but no significant differences were detected in Grm1 (t(22) = 0.2711, p=0.78), Grm3 (t(22) = 1.295, p=0.2), Grm4 (t(22) = 0.8764, p=0.39), or Grm5 (t(22) = 0.9804, p=0.33) expression in the NAc. (D) Finally, expression of astrocyte-specific glutamate transporters was also assessed in the whole NAc of GFAP-Cre and eGFP control mice. Glt-1 (i.e., SLC1A2 or EAAT2) was significantly upregulated in GFAP-Cre mice relative to controls (t(23) = 2.225, * p<0.05), while no significant differences were seen with Glast (i.e., SLC1A3 or EAAT1; t(23) = 0.5574, p=0.58) or the xCT cystine-glutamate antiporter (i.e., SLC7A11; t(24) = 0.2397, p=0.23) expression. Gene expression normalized to 18s. Mean± SEM. n=11–15. # p=0.06; @ p=0.05, * p<0.05, ** p<0.01, **** p<0.0001. n.s. = not significant.
Discussion
In the present study, we demonstrate for the first time that astrocytes in the NAc have strong molecular rhythms, and this circadian function is important for overall NAc function. Using RNA-sequencing across 6 times of day, we reveal approximately 43.3% of detected genes in the NAc astrocyte transcriptome show significant diurnal variation, with circadian and metabolic processes as the top enriched pathways/processes among these rhythmic genes. Furthermore, we demonstrate mice with genetic disruption of NAc astrocyte molecular clock function have significantly altered reward-related behavior, including phase-dependent increases in exploratory drive, as well as increases in food self-administration and motivation. Expanding on the behavioral phenotype, these mice also have disrupted NAc metabolic homeostasis, as revealed by downregulation of lactate and GSH, as well as altered metabolic-related gene expression. Finally, through electrophysiological whole-cell recordings, we provide evidence that disrupting NAc astrocyte molecular clock function also alters excitatory synaptic transmission onto neighboring MSNs via decreased AMPA receptor signaling, alongside upregulated glutamate-related gene expression. Taken together, these novel findings underscore the functional significance of NAc astrocytes and the degree to which their circadian regulation is essential for NAc function and NAc regulated behaviors.
Of particular interest, a potential mechanism underling our GFAP-Cre phenotype may be the uncoupling of circadian regulation of neurons from astrocytes. Astrocytes in the SCN play an integral role in regulating neuronal rhythmicity through circadian controlled uptake/release of glutamate and modulation of GABAergic tone and receptor signaling (91, 48). Moreover, disrupting circadian astrocyte function in the SCN causes a lengthening of the circadian period (i.e., an inability to keep rhythms consolidated to ~24 hours) in both SCN molecular rhythms and overall behavior rhythms (49). Notably, much like the SCN, the NAc consists primarily of GABAergic neurons, and NAc astrocytes play a crucial role maintain glutamate homeostasis (92, 93, 35). Similar to how SCN astrocytic regulation of extracellular glutamate in a diurnal manner is important for regulating circadian rhythms (48), astrocytic regulation of glutamate in the NAc is thought to contribute to reward-regulation and drug addiction vulnerability (51, 35, 52, 53, 34). This is further supported here through pathway analyses revealing glutamate and GABA receptor signaling, and neurotransmitter uptake (glutamate) pathways/processes enriched among NAc astrocytes’ top 200 expressed genes. Likewise, we find disrupting NAc astrocyte circadian function alters AMPA receptor signaling on neighboring MSNs by slowing the decay kinetics and reducing their response relative to that of NMDA receptors, alongside increasing AMPA subunit and specific NMDA, mGluR, and glutamate transporter mRNA levels (perhaps compensatory). Interestingly, previous work in our lab has shown mice with a global functional mutation in CLOCK also have a decreased AMPA/NMDA ratio in NAc MSNs and exhibit some of the same behaviors described here (21, 94). While not investigated, it is possible these previously reported Clock phenotypes are being driven by a loss of CLOCK function in astrocytes. This can be determined in future studies. Taken together, further investigation is needed into how disrupting NAc astrocyte molecular clock function affects glutamate signaling and/or its effects on neuronal rhythmicity, in the context of reward.
In addition to regulating glutamate levels, astrocytes play a central role in the maintenance of neuronal metabolic homeostasis, including synthesis and shuttling of both lactate and GSH (38–41). The astrocyte-to-neuron lactate shuttle (ANLS) hypothesis proposes that increased neuronal activity and subsequent glutamate uptake by astrocytes triggers astrocytic intake of glucose from blood capillaries, increasing glycolysis-mediated production of lactate (a primary CNS energy substrate), and increasing shuttling of lactate to neurons, via monocarboxylate transporters (MCTs), to sustain their energetic needs (95–98). Lactate synthesis and shuttling are known to be regulated by the molecular clock and show diurnal variation (99–104). Interestingly, here we demonstrate disrupting NAc astrocyte clock function leads to a significant downregulation of NAc lactate levels across time of day, as well as genes important for lactate synthesis (Ldha) and shuttling (Mct1 and Mct2). This is of particular interest for reward regulation given the interconnectivity of circadian rhythms, neuronal metabolism, and drug reward (105). While studies in the amygdala find that blocking lactate shuttling attenuates cocaine reward (106–108), in the NAc, the opposite appears to be true. In a recent study, blocking glucocorticoid-mediated lactate release (i.e., decreased lactate) significantly increased morphine preference that could be normalized by restoring lactate (109). This model aligns with our findings. Interestingly, not only is there extensive literature suggesting a dynamic interaction between the circadian system and glucocorticoids in the context of reward (110, 111), but also here we show in NAc astrocytes glucocorticoid receptor signaling is an enriched rhythmic pathway with high interconnectivity with the circadian regulation pathway. This may partly explain our molecular findings, in that glucocorticoid receptor signaling regulates both lactate production and NRF2 pathway activation (112, 113, 109). However, future investigation is needed into this relationship and how it may underly our model.
Moreover, much like the ANLS hypothesis, activation of the NRF2 antioxidant signaling pathway in astrocytes is also coupled with neuronal glutamatergic neurotransmission – whereby glutamate triggers its activation (114, 115, 40, 116). NRF2 is also activated directly by the presence of reactive oxygen species (ROS), inducing expression of GSH enzymes and many other antioxidants (117–119) to remove ROS that would otherwise damage the cell, also known as oxidative stress (120). Importantly, here we show monocarboxylic acid biosynthesis (i.e., lactate), glycolysis, oxidative phosphorylation, and generation of precursor metabolites/energy to be enriched among NAc astrocytes top expressed genes – all processes known to generate ROS and potential oxidative stress. Moreover, circadian analysis revealed NRF2-mediated oxidative stress response and ATPase Activity to be enriched among the top rhythmic transcripts. Further supporting these enriched processes, disrupting NAc astrocyte molecular clock function significantly downregulates NAc GSH levels, as well as related genes, and both Gfap and Nrf2 expression are significantly upregulated, known markers of astrocyte-reactivity and oxidative stress-induced antioxidant response. Given the growing association between oxidative stress and SUDs and the therapeutic potential of antioxidants that act on astrocytes (e.g., n-acetylcysteine, a GSH precursor) (121, 122), future studies will investigate this relationship as it relates circadian reward function in the NAc.
Finally, throughout our studies, males and females were grouped together when no sex differences were statistically detected. Interestingly, only one sex difference was observed, wherein disrupting NAc astrocyte molecular clock function produced a significant increase in light-phase food self-administration and motivation in male mice. This is particularly intriguing given our recent findings that Npas2 mutant mice show a phase- and sex-specific increase in cocaine self-administration, where Npas2 mutation increased dark-phase self-administration behaviors in female mice (28). Notably, ovariectomy normalizes the phenotype, suggesting circulating hormones to be underlying this sex-specific effect (28). While not investigated here, it may be interesting to explore the role of circulating hormones in the sex-effects seen in the current study. Additionally, these nearly opposing phenotypes may be driven by differential circadian effects of neurons versus astrocytes in the NAc – given that NPAS2 is primarily enriched in D1-MSNs (123, 25). Though our RRHO analysis revealed a nearly identical overlap in NAc astrocyte rhythmic transcripts between sexes, this sequencing analysis was limited to just astrocytes and in non-experimentally manipulated mice. Future studies will investigate sex-specific transcriptional changes in both astrocytes and neurons that may arise following disruption of NAc astrocyte molecular clock function.
Collectively, the results of this study demonstrate for the first time that NAc astrocytes exhibit a high degree of diurnal rhythmicity at the transcriptome level, with metabolic relevant pathways/processes enriched among the top rhythmic genes. Moreover, our data reveal that this circadian astrocyte function is not only important for the maintenance of metabolic homeostasis, but also for glutamatergic signaling to MSNs, and downstream regulation of NAc-regulated behaviors. While this circadian NAc astrocyte function most obviously has implications in the context of reward-regulation, as shown here, but also has implications across other psychiatric disorders known to involve the NAc. Ultimately, these data further our understanding of the relationship between circadian rhythms, metabolic state, and reward circuity, and reveal a fundamental role of astrocyte rhythmicity in regulating NAc function.
Supplementary Material
KEY RESOURCES TABLE.
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | Rabbit anti-GFP (IP) | Abcam | ab290; RRID:AB_303395 | |
| Antibody | Chicken anti-GFP (IF) | Abcam | ab13970; RRID:AB_300798 | |
| Antibody | Rabbit anti-BMAL1 (IF) | Novus Biologicals | NB100–2288; RRID:AB_10000794 | |
| Antibody | Mouse anti-GFAP (IF) | Novus Biologicals | NBP1–05197; RRID:AB_1555288 | |
| Bacterial or Viral Strain | AAV8-GFAP-eGFP | University of North Carolina Viral Vector Core | RRID:SCR_002448 | |
| Bacterial or Viral Strain | AAV8-GFAP-Cre-GFP | University of North Carolina Viral Vector Core | RRID:SCR_002448 | |
| Chemical Compound or Drug | RNasin® Plus ribonuclease inhibitor | Promega | cat# N2615 | |
| Chemical Compound or Drug | EDTA-free protein inhibitor cocktail | Roche through Sigma-Aldrich | cat# 11873580001 | |
| Commercial Assay Or Kit | RNAscope in situ hybridization Fluorescent Multiplex Assay | Advanced Cell Diagnostics | cat# 320850 | |
| Commercial Assay Or Kit | RNeasy Plus Mini Kit | QIAGEN | cat# 74134 | |
| Commercial Assay Or Kit | iScript cDNA synthesis kit | Bio-Rad | cat# 1708891 | |
| Commercial Assay Or Kit | SsoAdvanced Universal SYBR Green Supermix kit | Bio-Rad | cat# 1725272 | |
| Commercial Assay Or Kit | EZNA Micro Elute Total RNA kit | Omega Bio-Tek | cat# R6831–02 | |
| Commercial Assay Or Kit | Qubit RNA High Sensitivity assay | Invitrogen | cat# Q32852; RRID:SCR_018095 | |
| Commercial Assay Or Kit | Agilent RNA 6000 Pico Kit | Agilent | cat# 5067–1513; RRID:SCR_018043 | |
| Commercial Assay Or Kit | TruSeq Stranded Total RNA Kit | Illumina | cat# 20020599 | |
| Commercial Assay Or Kit | BioVision Lactate Colorimetric/Fluorometric Assay | BioVision Inc. | cat# K607 | |
| Commercial Assay Or Kit | GSH-Glo™ Glutathione Luminescence Assay | Promega | cat# V6912 | |
| Organism/Strain | Mouse: Bmal1lox (BMFL); male & female | The Jackson Laboratory | JAX Stock No: 007668 | B6.129S4-Arntltm1Weit/J |
| Organism/Strain | Mouse: Aldh1l1-eGFP/Rpl10a; male & female | The Jackson Laboratory | JAX Stock No: 007668; RRID:IMSR_JAX:030247 | B6;FVB-Tg(Aldh1l1-EGFP/Rpl10a)JD133Htz/J |
| Sequence-Based Reagent | Primers for RT-qPCR, see Table S1 | This paper | N/A | |
| Sequence-Based Reagent | Aldh1l1 RNAscope Catalog Probe | Advanced Cell Diagnostics | cat# 405891 | |
| Sequence-Based Reagent | eGFP RNAscope Catalog Probe | Advanced Cell Diagnostics | cat# 400281-c3 | |
| Sequence-Based Reagent | Slc32a1 RNAscope Catalog Probe | Advanced Cell Diagnostics | cat# 319191-c2 | Vgat |
| Software; Algorithm | Graphpad Prism 9 Statistical Software | Graphpad Software | RRID:SCR_002798 | |
| Software; Algorithm | Ethovision XT 13 Behavioral Software | Noldus Information Tech Inc | RRID:SCR_000441 | |
| Software; Algorithm | Med-PC software suite (operant behavior) | Med-Associates Inc | RRID:SCR_012156 | |
| Software; Algorithm | CFX Maestro Software (Real Time qPCR) | Bio-Rad | N/A | |
| Software; Algorithm | Rstudio | https://www.rstudio.com/ | RRID:SCR_000432 | |
| Software; Algorithm | FastQC v0.11.7 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 | |
| Software; Algorithm | HISAT2 v2.1 | http://daehwankimlab.github.io/hisat2/ | RRID:SCR_015530 | |
| Software; Algorithm | Bioconductor edgeR package in R | https://bioconductor.org/packages/release/bioc/html/edgeR.html | RRID:SCR_012802 | |
| Software; Algorithm | JTK_CYCLE package in R (rhythmicity detection) | DOI: 10.1177/0748730410379711 | RRID:SCR_017962 | |
| Software; Algorithm | DisplayR online data anaylsis and visualization software | https://www.displayr.com | N/A | |
| Software; Algorithm | Cytoscape v3.1.2 interaction network bioinformatics software | https://cytoscape.org/ | RRID:SCR_003032 | |
| Software; Algorithm | Ingenuity Pathway Analysis (IPA) | QIAGEN | RRID:SCR_008653 | |
| Software; Algorithm | Metascape (Bioinformatics Database) | https://metascape.org | RRID:SCR_016620 | |
| Software; Algorithm | Olympus Fluoview FV10-ASW 4.2 viewer software | Olympus | RRID:SCR_014215 | |
| Software; Algorithm | pCLAMP 11 electrophysiology software suite | Molecular Devices | RRID:SCR_011323 | |
| Other | QIAshredder homogenization columns | QIAGEN | cat# 79656 | |
| Other | VECTASHIELD mountig medium plus DAPI | Vector Laboratories | RRID:AB_2336788 | |
| Other | Dynabeads Protein G for Immunoprecipitation | Invitrogen | cat# 10009D | |
| Other | ImmEdge® hydrophobic barrier | Vector Laboratories | RRID:AB_2336517 | |
| Other | ProLong™ Gold Antifade Mountant | Invitrogen | cat# P36930 |
Acknowledgments
We would like to thank Ioannis H Migias and Allison Cerwensky for their role in animal care and genotyping. Thank you to Dr. William MacDonald and the University of Pittsburgh Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh for their expertise in Illumina TruSeq Total RNA library preparation and next-generation RNA sequencing. We thank Dr. Mary Kay Lobo for the RiboTag mice along with very helpful experimental advice. Data analysis was performed using Ingenuity Pathway Analysis software licensed through the Molecular Biology Information Service of the Health Sciences Library System at the University of Pittsburgh. This work was funded and supported by the National Institutes of Health (NIH): R01DA039865 (PI: McClung CA), K02DA042886 (PI: McClung CA), P50DA039841 (PI: McClung CA), P50 DA046346 (PI: McClung), R01 MH106460 (PI: McClung), R21DA037636 (PI: McClung), R33DA041872 (Pis Logan RW and McClung CA), K01MH128763 (PI: Ketchesin KD) & T32 NS007433–18 (PI: Sved AF).
Footnotes
Conflict of Interest
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hedegaard H, Miniño AM, Warner M (2020): Drug overdose deaths in the united states, 1999–2018. NCHS Data Brief [PubMed] [Google Scholar]
- 2.National Institute on Drug Abuse (2020): Costs of Substance Abuse | National Institute on Drug Abuse (NIDA) https://www.drugabuse.gov/drug-topics/trends-statistics/costs-substance-abuse
- 3.National Institute on Drug Abuse (2020): Treatment and Recovery Drugs, Brains, and Behavior: The Science of Addiction. https://www.drugabuse.gov/publications/drugs-brains-behavior-science-addiction/treatment-recovery
- 4.Hasler BP, Smith LJ, Cousins JC, Bootzin RR (2012): Circadian rhythms, sleep, and substance abuse. Sleep Med. Rev. 16(1): 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angarita GA, Emadi N, Hodges S, Morgan PT (2016): Sleep abnormalities associated with alcohol, cannabis, cocaine, and opiate use: a comprehensive review. Addict Sci Clin Pract. 11(1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logan RW, Williams WP, McClung CA (2014): Circadian rhythms and addiction: mechanistic insights and future directions. Behav. Neurosci. 128(3): 387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR (2002): Coordination of circadian timing in mammals. Nature. 418(6901): 935–941. [DOI] [PubMed] [Google Scholar]
- 8.Partch CL, Green CB, Takahashi JS (2014): Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 24(2): 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi JS (2017): Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18(3): 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, et al. (2005): The clock gene per2 influences the glutamatergic system and modulates alcohol consumption. Nat. Med. 11(1): 35–42. [DOI] [PubMed] [Google Scholar]
- 11.Comasco E, Nordquist N, Göktürk C, Aslund C, Hallman J, Oreland L, et al. (2010): The clock gene per2 and sleep problems: association with alcohol consumption among swedish adolescents. Ups. J. Med. Sci. 115(1): 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovanen L, Saarikoski ST, Haukka J, Pirkola S, Aromaa A, Lönnqvist J, et al. (2010): Circadian clock gene polymorphisms in alcohol use disorders and alcohol consumption. Alcohol Alcohol. 45(4): 303–311. [DOI] [PubMed] [Google Scholar]
- 13.Sjöholm LK, Kovanen L, Saarikoski ST, Schalling M, Lavebratt C, Partonen T (2010): Clock is suggested to associate with comorbid alcohol use and depressive disorders. J Circadian Rhythms. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong L, Bilbao A, Laucht M, Henriksson R, Yakovleva T, Ridinger M, et al. (2011): Effects of the circadian rhythm gene period 1 (per1) on psychosocial stress-induced alcohol drinking. Am. J. Psychiatry. 168(10): 1090–1098. [DOI] [PubMed] [Google Scholar]
- 15.Shumay E, Fowler JS, Wang GJ, Logan J, Alia-Klein N, Goldstein RZ, et al. (2012): Repeat variation in the human per2 gene as a new genetic marker associated with cocaine addiction and brain dopamine d2 receptor availability. Transl. Psychiatry. 2: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuchimine S, Yasui-Furukori N, Kaneda A, Kaneko S (2013): The clock c3111t polymorphism is associated with reward dependence in healthy japanese subjects. Neuropsychobiology. 67(1): 1–5. [DOI] [PubMed] [Google Scholar]
- 17.Bi J, Gelernter J, Sun J, Kranzler HR (2014): Comparing the utility of homogeneous subtypes of cocaine use and related behaviors with dsm-iv cocaine dependence as traits for genetic association analysis. Am. J. Med. Genet. B, Neuropsychiatr. Genet. 165B(2): 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abarca C, Albrecht U, Spanagel R (2002): Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc. Natl. Acad. Sci. USA. 99(13): 9026–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, et al. (2005): Regulation of dopaminergic transmission and cocaine reward by the clock gene. Proc. Natl. Acad. Sci. USA. 102(26): 9377–9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker-Krail DD, Parekh PK, Ketchesin KD, Yamaguchi S, Yoshino J, Hildebrand MA, et al. (2022): Circadian transcription factor npas2 and the nad+-dependent deacetylase sirt1 interact in the mouse nucleus accumbens and regulate reward. Eur. J. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, et al. (2007): Mania-like behavior induced by disruption of clock. Proc. Natl. Acad. Sci. USA. 104(15): 6406–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozburn AR, Larson EB, Self DW, McClung CA (2012): Cocaine self-administration behaviors in clockδ19 mice. Psychopharmacology. 223(2): 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozburn AR, Falcon E, Mukherjee S, Gillman A, Arey R, Spencer S, et al. (2013): The role of clock in ethanol-related behaviors. Neuropsychopharmacology. 38(12): 2393–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner JR, Ecke LE, Briand LA, Haydon PG, Blendy JA (2013): Cocaine-related behaviors in mice with deficient gliotransmission. Psychopharmacology. 226(1): 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozburn AR, Falcon E, Twaddle A, Nugent AL, Gillman AG, Spencer SM, et al. (2015): Direct regulation of diurnal drd3 expression and cocaine reward by npas2. Biol. Psychiatry. 77(5): 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozburn AR, Kern J, Parekh PK, Logan RW, Liu Z, Falcon E, et al. (2017): Npas2 regulation of anxiety-like behavior and gabaa receptors. Front. Mol. Neurosci. 10: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parekh PK, Logan RW, Ketchesin KD, Becker-Krail D, Shelton MA, Hildebrand MA, et al. (2019): Cell-type-specific regulation of nucleus accumbens synaptic plasticity and cocaine reward sensitivity by the circadian protein, npas2. J. Neurosci. 39(24): 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePoy LM, Becker-Krail DD, Zong W, Petersen K, Shah NM, Brandon JH, et al. (2020): Circadian- and sex-dependent increases in intravenous cocaine self-administration in npas2 mutant mice. J. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClung CA (2013): How might circadian rhythms control mood? let me count the ways... Biol. Psychiatry. 74(4): 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logan RW, McClung CA (2019): Rhythms of life: circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 20(1): 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketchesin KD, Becker-Krail D, McClung CA (2020): Mood-related central and peripheral clocks. Eur. J. Neurosci. 51(1): 326–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker WH, Walton JC, DeVries AC, Nelson RJ (2020): Circadian rhythm disruption and mental health. Transl. Psychiatry. 10(1): 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mannella F, Gurney K, Baldassarre G (2013): The nucleus accumbens as a nexus between values and goals in goal-directed behavior: a review and a new hypothesis. Front. Behav. Neurosci. 7: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scofield MD, Heinsbroek JA, Gipson CD, Kupchik YM, Spencer S, Smith ACW, et al. (2016): The nucleus accumbens: mechanisms of addiction across drug classes reflect the importance of glutamate homeostasis. Pharmacol. Rev. 68(3): 816–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scofield MD, Kalivas PW (2014): Astrocytic dysfunction and addiction: consequences of impaired glutamate homeostasis. Neuroscientist. 20(6): 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim R, Healey KL, Sepulveda-Orengo MT, Reissner KJ (2018): Astroglial correlates of neuropsychiatric disease: from astrocytopathy to astrogliosis. Prog. Neuropsychopharmacol. Biol. Psychiatry. 87(Pt A): 126–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sofroniew MV, Vinters HV (2010): Astrocytes: biology and pathology. Acta Neuropathol. 119(1): 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danbolt NC (2001): Glutamate uptake. Prog. Neurobiol. 65(1): 1–105. [DOI] [PubMed] [Google Scholar]
- 39.Malarkey EB, Parpura V (2008): Mechanisms of glutamate release from astrocytes. Neurochem. Int. 52(1–2): 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolaños JP (2016): Bioenergetics and redox adaptations of astrocytes to neuronal activity. J. Neurochem. 139 Suppl 2: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonçalves C-A, Rodrigues L, Bobermin LD, Zanotto C, Vizuete A, Quincozes-Santos A, et al. (2018): Glycolysis-derived compounds from astrocytes that modulate synaptic communication. Front. Neurosci. 12: 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prolo LM, Takahashi JS, Herzog ED (2005): Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25(2): 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yagita K, Yamanaka I, Emoto N, Kawakami K, Shimada S (2010): Real-time monitoring of circadian clock oscillations in primary cultures of mammalian cells using tol2 transposon-mediated gene transfer strategy. BMC Biotechnol. 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi-Castañeda D, Ortega A (2016): Clock genes in glia cells: a rhythmic history. ASN Neuro. 8(5): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gwak YS, Kang J, Leem JW, Hulsebosch CE (2007): Spinal ampa receptor inhibition attenuates mechanical allodynia and neuronal hyperexcitability following spinal cord injury in rats. J. Neurosci. Res. 85(11): 2352–2359. [DOI] [PubMed] [Google Scholar]
- 46.Beaulé C, Swanstrom A, Leone MJ, Herzog ED (2009): Circadian modulation of gene expression, but not glutamate uptake, in mouse and rat cortical astrocytes. PLoS One. 4(10): e7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marpegan L, Swanstrom AE, Chung K, Simon T, Haydon PG, Khan SK, et al. (2011): Circadian regulation of atp release in astrocytes. J. Neurosci. 31(23): 8342–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH (2017): Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron. 93(6): 1420–1435.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tso CF, Simon T, Greenlaw AC, Puri T, Mieda M, Herzog ED (2017): Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 27(7): 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brancaccio M, Edwards MD, Patton AP, Smyllie NJ, Chesham JE, Maywood ES, et al. (2019): Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science. 363(6423): 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalivas PW (2009): The glutamate homeostasis hypothesis of addiction. Nat. Rev. Neurosci. 10(8): 561–572. [DOI] [PubMed] [Google Scholar]
- 52.Reissner KJ, Gipson CD, Tran PK, Knackstedt LA, Scofield MD, Kalivas PW (2015): Glutamate transporter glt-1 mediates n-acetylcysteine inhibition of cocaine reinstatement. Addict Biol. 20(2): 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scofield MD, Boger HA, Smith RJ, Li H, Haydon PG, Kalivas PW (2015): Gq-dreadd selectively initiates glial glutamate release and inhibits cue-induced cocaine seeking. Biol. Psychiatry. 78(7): 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. (2008): A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28(1): 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, et al. (2011): Molecular comparison of glt1+ and aldh1l1+ astrocytes in vivo in astroglial reporter mice. Glia. 59(2): 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. (2014): An rna-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34(36): 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storch K-F, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, et al. (2007): Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 130(4): 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS (2009): Cell-type-specific isolation of ribosome-associated mrna from complex tissues. Proc. Natl. Acad. Sci. USA. 106(33): 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, et al. (2015): Opposing role for egr3 in nucleus accumbens cell subtypes in cocaine action. J. Neurosci. 35(20): 7927–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes ME, Hogenesch JB, Kornacker K (2010): Jtk_cycle: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 25(5): 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu G, Zhu J, Yu J, Zhou L, Huang JZ, Zhang Z (2014): Evaluation of five methods for genome-wide circadian gene identification. J. Biol. Rhythms. 29(4): 231–242. [DOI] [PubMed] [Google Scholar]
- 62.Krämer A, Green J, Pollard J, Tugendreich S (2014): Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 30(4): 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. (2019): Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10(1): 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haskew-Layton RE, Payappilly JB, Smirnova NA, Ma TC, Chan KK, Murphy TH, et al. (2010): Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an nrf2-independent pathway. Proc. Natl. Acad. Sci. USA. 107(40): 17385–17390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baxter PS, Bell KFS, Hasel P, Kaindl AM, Fricker M, Thomson D, et al. (2015): Synaptic nmda receptor activity is coupled to the transcriptional control of the glutathione system. Nat. Commun. 6: 6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lerchundi R, Fernández-Moncada I, Contreras-Baeza Y, Sotelo-Hitschfeld T, Mächler P, Wyss MT, et al. (2015): Nh4(+) triggers the release of astrocytic lactate via mitochondrial pyruvate shunting. Proc. Natl. Acad. Sci. USA. 112(35): 11090–11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muraleedharan R, Gawali MV, Tiwari D, Sukumaran A, Oatman N, Anderson J, et al. (2020): Ampk-regulated astrocytic lactate shuttle plays a non-cell-autonomous role in neuronal survival. Cell Rep. 32(9): 108092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Logan RW, Edgar N, Gillman AG, Hoffman D, Zhu X, McClung CA (2015): Chronic stress induces brain region-specific alterations of molecular rhythms that correlate with depression-like behavior in mice. Biol. Psychiatry. 78(4): 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brami-Cherrier K, Lewis RG, Cervantes M, Liu Y, Tognini P, Baldi P, et al. (2020): Cocaine-mediated circadian reprogramming in the striatum through dopamine d2r and pparγ activation. Nat. Commun. 11(1): 4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, et al. (2008): Application of a translational profiling approach for the comparative analysis of cns cell types. Cell. 135(4): 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakers K, Lake AM, Khazanchi R, Ouwenga R, Vasek MJ, Dani A, et al. (2017): Astrocytes locally translate transcripts in their peripheral processes. Proc. Natl. Acad. Sci. USA. 114(19): E3830–E3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sapkota D, Lake AM, Yang W, Yang C, Wesseling H, Guise A, et al. (2019): Cell-type-specific profiling of alternative translation identifies regulated protein isoform variation in the mouse brain. Cell Rep. 26(3): 594–607.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clarke LE, Liddelow SA, Chakraborty C, Münch AE, Heiman M, Barres BA (2018): Normal aging induces a1-like astrocyte reactivity. Proc. Natl. Acad. Sci. USA. 115(8): E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, et al. (2018): Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 8(1): 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan J, Ma N, Yu B, Zhang W, Wan J (2020): Transcriptomic profiling of microglia and astrocytes throughout aging. J. Neuroinflammation. 17(1): 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lananna BV, Nadarajah CJ, Izumo M, Cedeño MR, Xiong DD, Dimitry J, et al. (2018): Cell-autonomous regulation of astrocyte activation by the circadian clock protein bmal1. Cell Rep. 25(1): 1–9.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stead JDH, Clinton S, Neal C, Schneider J, Jama A, Miller S, et al. (2006): Selective breeding for divergence in novelty-seeking traits: heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet. 36(5): 697–712. [DOI] [PubMed] [Google Scholar]
- 78.Flagel SB, Robinson TE, Clark JJ, Clinton SM, Watson SJ, Seeman P, et al. (2010): An animal model of genetic vulnerability to behavioral disinhibition and responsiveness to reward-related cues: implications for addiction. Neuropsychopharmacology. 35(2): 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Z, Blandino P, Yuan Q, Shen P-H, Hodgkinson CA, Virkkunen M, et al. (2019): Exploratory locomotion, a predictor of addiction vulnerability, is oligogenic in rats selected for this phenotype. Proc. Natl. Acad. Sci. USA. 116(26): 13107–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bush DEA, Vaccarino FJ (2007): Individual differences in elevated plus-maze exploration predicted progressive-ratio cocaine self-administration break points in wistar rats. Psychopharmacology. 194(2): 211–219. [DOI] [PubMed] [Google Scholar]
- 81.Flagel SB, Waselus M, Clinton SM, Watson SJ, Akil H (2014): Antecedents and consequences of drug abuse in rats selectively bred for high and low response to novelty. Neuropharmacology. 76 Pt B: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dickson PE, Ndukum J, Wilcox T, Clark J, Roy B, Zhang L, et al. (2015): Association of novelty-related behaviors and intravenous cocaine self-administration in diversity outbred mice. Psychopharmacology. 232(6): 1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wingo T, Nesil T, Choi J-S, Li MD (2016): Novelty seeking and drug addiction in humans and animals: from behavior to molecules. J Neuroimmune Pharmacol. 11(3): 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guilloux J-P, Seney M, Edgar N, Sibille E (2011): Integrated behavioral z-scoring increases the sensitivity and reliability of behavioral phenotyping in mice: relevance to emotionality and sex. J. Neurosci. Methods. 197(1): 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mishima N, Higashitani F, Teraoka K, Yoshioka R (1986): Sex differences in appetitive learning of mice. Physiol. Behav. 37(2): 263–268. [DOI] [PubMed] [Google Scholar]
- 86.van Haaren F, van Hest A, Heinsbroek RP (1990): Behavioral differences between male and female rats: effects of gonadal hormones on learning and memory. Neurosci. Biobehav. Rev. 14(1): 23–33. [DOI] [PubMed] [Google Scholar]
- 87.McDowell AL, Heath KM, Garraghty PE (2013): The effects of sex and chronic restraint on instrumental learning in rats. Neurosci. J. 2013: 893126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liddelow SA, Barres BA (2017): Reactive astrocytes: production, function, and therapeutic potential. Immunity. 46(6): 957–967. [DOI] [PubMed] [Google Scholar]
- 89.Vomund S, Schäfer A, Parnham MJ, Brüne B, von Knethen A (2017): Nrf2, the master regulator of anti-oxidative responses. Int. J. Mol. Sci. 18(12): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bélanger M, Allaman I, Magistretti PJ (2011): Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 14(6): 724–738. [DOI] [PubMed] [Google Scholar]
- 91.Barca-Mayo O, Pons-Espinal M, Follert P, Armirotti A, Berdondini L, De Pietri Tonelli D (2017): Astrocyte deletion of bmal1 alters daily locomotor activity and cognitive functions via gaba signalling. Nat. Commun. 8: 14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fellin T, D’Ascenzo M, Haydon PG (2007): Astrocytes control neuronal excitability in the nucleus accumbens. ScientificWorldJournal. 7: 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parpura V, Verkhratsky A (2012): Astrocytes revisited: concise historic outlook on glutamate homeostasis and signaling. Croat Med J. 53(6): 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parekh PK, Becker-Krail D, Sundaravelu P, Ishigaki S, Okado H, Sobue G, et al. (2018): Altered glua1 (gria1) function and accumbal synaptic plasticity in the clockδ19 model of bipolar mania. Biol. Psychiatry. 84(11): 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pellerin L, Magistretti PJ (1994): Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. USA. 91(22): 10625–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Magistretti PJ, Pellerin L (1996): Cellular bases of brain energy metabolism and their relevance to functional brain imaging: evidence for a prominent role of astrocytes. Cereb. Cortex. 6(1): 50–61. [DOI] [PubMed] [Google Scholar]
- 97.Pellerin L, Magistretti PJ (2012): Sweet sixteen for anls. J. Cereb. Blood Flow Metab. 32(7): 1152–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Magistretti PJ, Allaman I (2018): Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 19(4): 235–249. [DOI] [PubMed] [Google Scholar]
- 99.Ahlersová E, Ahlers I, Toropila M, Smajda B, Datelinka I (1981): Circadian rhythm of the lactate and pyruvate concentration in rat liver and blood. Physiol Bohemoslov. 30(3): 213–220. [PubMed] [Google Scholar]
- 100.Rutter J, Reick M, Wu LC, McKnight SL (2001): Regulation of clock and npas2 dna binding by the redox state of nad cofactors. Science. 293(5529): 510–514. [DOI] [PubMed] [Google Scholar]
- 101.Naylor E, Aillon DV, Barrett BS, Wilson GS, Johnson DA, Johnson DA, et al. (2012): Lactate as a biomarker for sleep. Sleep. 35(9): 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henriksson E, Huber A-L, Soto EK, Kriebs A, Vaughan ME, Duglan D, et al. (2017): The liver circadian clock modulates biochemical and physiological responses to metformin. J. Biol. Rhythms. 32(4): 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Milićević N, Ten Brink JB, Ten Asbroek ALMA, Bergen AA, Felder-Schmittbuhl M-P (2020): The circadian clock regulates rpe-mediated lactate transport via slc16a1 (mct1). Exp. Eye Res. 190: 107861. [DOI] [PubMed] [Google Scholar]
- 104.Wallace NK, Pollard F, Savenkova M, Karatsoreos IN (2020): Effect of aging on daily rhythms of lactate metabolism in the medial prefrontal cortex of male mice. Neuroscience [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Freyberg Z, Logan RW (2018): The intertwined roles of circadian rhythms and neuronal metabolism fueling drug reward and addiction. Curr. Opin. Physiol. 5: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boury-Jamot B, Carrard A, Martin JL, Halfon O, Magistretti PJ, Boutrel B (2016): Disrupting astrocyte-neuron lactate transfer persistently reduces conditioned responses to cocaine. Mol. Psychiatry. 21(8): 1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boury-Jamot B, Halfon O, Magistretti PJ, Boutrel B (2016): Lactate release from astrocytes to neurons contributes to cocaine memory formation. Bioessays. 38(12): 1266–1273. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Y, Xue Y, Meng S, Luo Y, Liang J, Li J, et al. (2016): Inhibition of lactate transport erases drug memory and prevents drug relapse. Biol. Psychiatry. 79(11): 928–939. [DOI] [PubMed] [Google Scholar]
- 109.Skupio U, Tertil M, Bilecki W, Barut J, Korostynski M, Golda S, et al. (2020): Astrocytes determine conditioned response to morphine via glucocorticoid receptor-dependent regulation of lactate release. Neuropsychopharmacology. 45(2): 404–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perreau-Lenz S, Spanagel R (2015): Clock genes × stress × reward interactions in alcohol and substance use disorders. Alcohol. 49(4): 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Becker-Krail D, McClung C (2016): Implications of circadian rhythm and stress in addiction vulnerability. [version 1; peer review: 2 approved]. F1000Res. 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Allaman I, Pellerin L, Magistretti PJ (2004): Glucocorticoids modulate neurotransmitter-induced glycogen metabolism in cultured cortical astrocytes. J. Neurochem. 88(4): 900–908. [DOI] [PubMed] [Google Scholar]
- 113.Alam MM, Okazaki K, Nguyen LTT, Ota N, Kitamura H, Murakami S, et al. (2017): Glucocorticoid receptor signaling represses the antioxidant response by inhibiting histone acetylation mediated by the transcriptional activator nrf2. J. Biol. Chem. 292(18): 7519–7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Habas A, Hahn J, Wang X, Margeta M (2013): Neuronal activity regulates astrocytic nrf2 signaling. Proc. Natl. Acad. Sci. USA. 110(45): 18291–18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jimenez-Blasco D, Santofimia-Castaño P, Gonzalez A, Almeida A, Bolaños JP (2015): Astrocyte nmda receptors’ activity sustains neuronal survival through a cdk5-nrf2 pathway. Cell Death Differ. 22(11): 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McGann JC, Mandel G (2018): Neuronal activity induces glutathione metabolism gene expression in astrocytes. Glia. 66(9): 2024–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma Q (2013): Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 53: 401–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baxter PS, Hardingham GE (2016): Adaptive regulation of the brain’s antioxidant defences by neurons and astrocytes. Free Radic. Biol. Med. 100: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tu W, Wang H, Li S, Liu Q, Sha H (2019): The anti-inflammatory and anti-oxidant mechanisms of the keap1/nrf2/are signaling pathway in chronic diseases. Aging Dis. 10(3): 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schieber M, Chandel NS (2014): Ros function in redox signaling and oxidative stress. Curr. Biol. 24(10): R453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berríos-Cárcamo P, Quezada M, Quintanilla ME, Morales P, Ezquer M, Herrera-Marschitz M, et al. (2020): Oxidative stress and neuroinflammation as a pivot in drug abuse. a focus on the therapeutic potential of antioxidant and anti-inflammatory agents and biomolecules. Antioxidants (Basel). 9(9): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pavlek LR, Dillard J, Rogers LK (2020): The role of oxidative stress in toxicities due to drugs of abuse. Curr. Opin. Toxicol 20–21: 29–35. [Google Scholar]
- 123.Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, et al. (2000): Impaired cued [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


