SUMMARY
ADP-ribosylation (ADPRylation) is a posttranslational modification of proteins catalyzed by ADP-ribosyl transferase (ART) enzymes, including nuclear PARPs (e.g., PARP1 and PARP2). Historically, studies of ADPRylation and PARPs have focused on DNA damage responses in cancers, but more recent studies have elucidated diverse roles in a broader array of biological processes. Here we summarize the expanding array of molecular mechanisms underlying the biological functions of nuclear PARPs with a focus on PARP1, the founding member of the family. This includes roles in DNA repair, chromatin regulation, gene expression, ribosome biogenesis, and RNA biology. We also present new concepts in PARP1-dependent regulation, including PAR-dependent posttranslational modifications, “ADPR spray,” and PAR-mediated biomolecular condensate formation. Moreover, we review advances in the therapeutic mechanisms of PARP inhibitors (PARPi), as well as progress on the mechanisms of PARPi resistance. Collectively, the recent progress in the field has yielded new insights into the expanding universe of PARP1-mediated molecular and therapeutic mechanisms in a variety of biological processes.
Keywords: ADP-ribosylation, poly (ADP-ribose) polymerase (PARP), PARP inhibitor (PARPi), DNA damage response, DNA replication, chromatin, histone, posttranslational modification (PTM), gene regulation, ribosome biogenesis, RNA biology, biomolecular condensate, therapeutics, therapeutic resistance
eTOC Blurb
Huang and Kraus review the expanding array of molecular mechanisms underlying the biological, pathological, and therapeutic functions of nuclear PARPs and ADP-ribosylation, including roles in DNA repair, chromatin regulation, gene expression, and RNA biology. They also present emerging concepts, including PAR-dependent posttranslational modifications, ADP-ribose spray, and PAR-mediated biomolecular condensate formation.
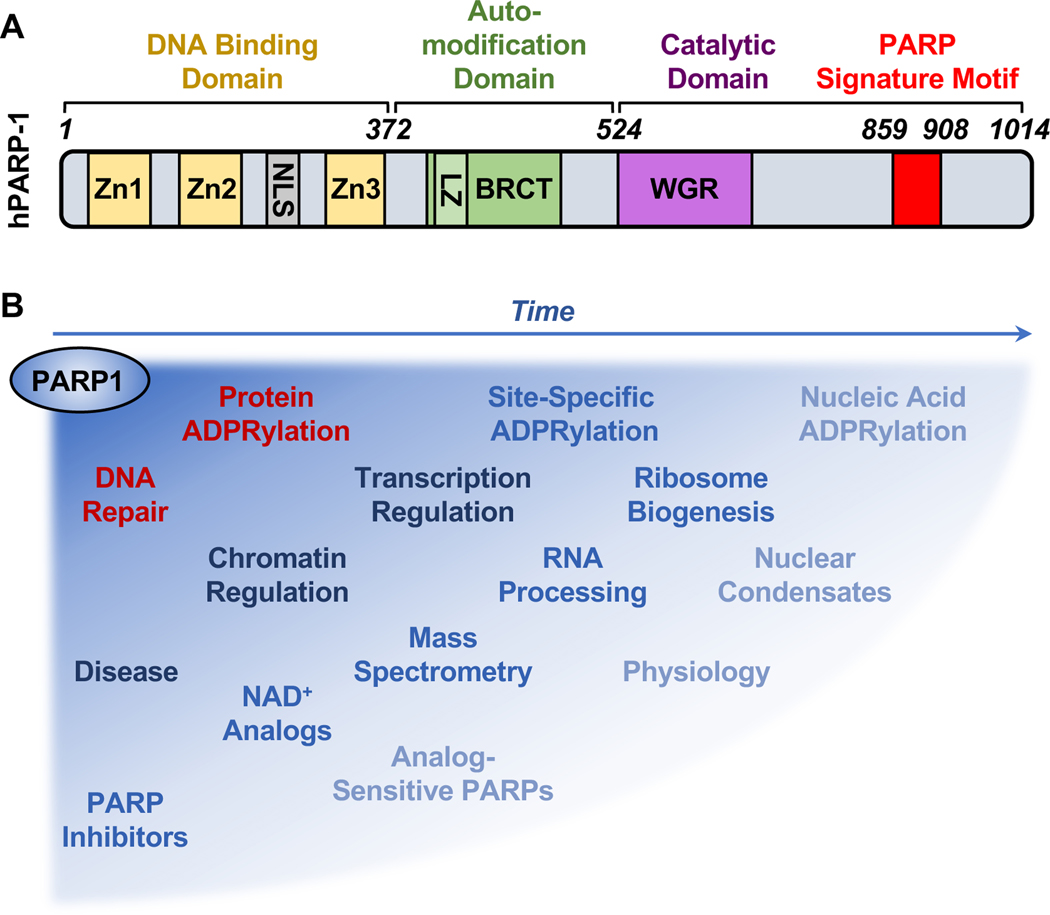
ADP-ribosylation (ADPRylation) is a posttranslational modification of proteins that has received considerable attention recently. This interest has been driven by an increased understanding of the biochemistry, molecular biology, physiology, and pathology of the poly(ADP-ribose) polymerase (PARP) family of enzymes that mediate ADPRylation, as well as the successful therapeutic targeting of PARP1, the founding member of the family (Figure 1A). Although great strides have been made in determining the mechanisms of action of PARP1 in the nucleus, especially with respect to DNA repair, more work is needed to fully understand the broad, multifaceted ways in which PARP1 controls the functions of the genome. In addition, our understanding of the therapeutic mechanisms of PARP1 inhibitors needs some revision as well. This review address both of these topics. The evolution of our understanding of PARP1 and nuclear ADPRylation can be gleaned by comparing the discussion herein with a similarly focused review published by our lab over a decade ago (Krishnakumar and Kraus, 2010a) (Figure 1B).
Figure 1. The expanding universe of PARP1 biology and tools used to investigate its functions.
(A) Structural and functional domains of PARP1, including (1) a DNA binding domain (DBD) containing three zinc finger (Zn) motifs and a nuclear localization signal (NLS), (2) an automodification domain containing a BRCA C-terminus (BRCT) motif, and (3) a catalytic domain containing a tryptophan (W)-glycine (G)-arginine (R) (WGR) motif and the PARP catalytic site signature motif.
(B) Schematic illustrating how the understanding of PARP1 biology, as well as the methods and tools used to study it, has evolved over time from the original focus on protein ADPRylation and DNA repair.
A BRIEF OVERVIEW OF PARPS AND ADPRYLATION
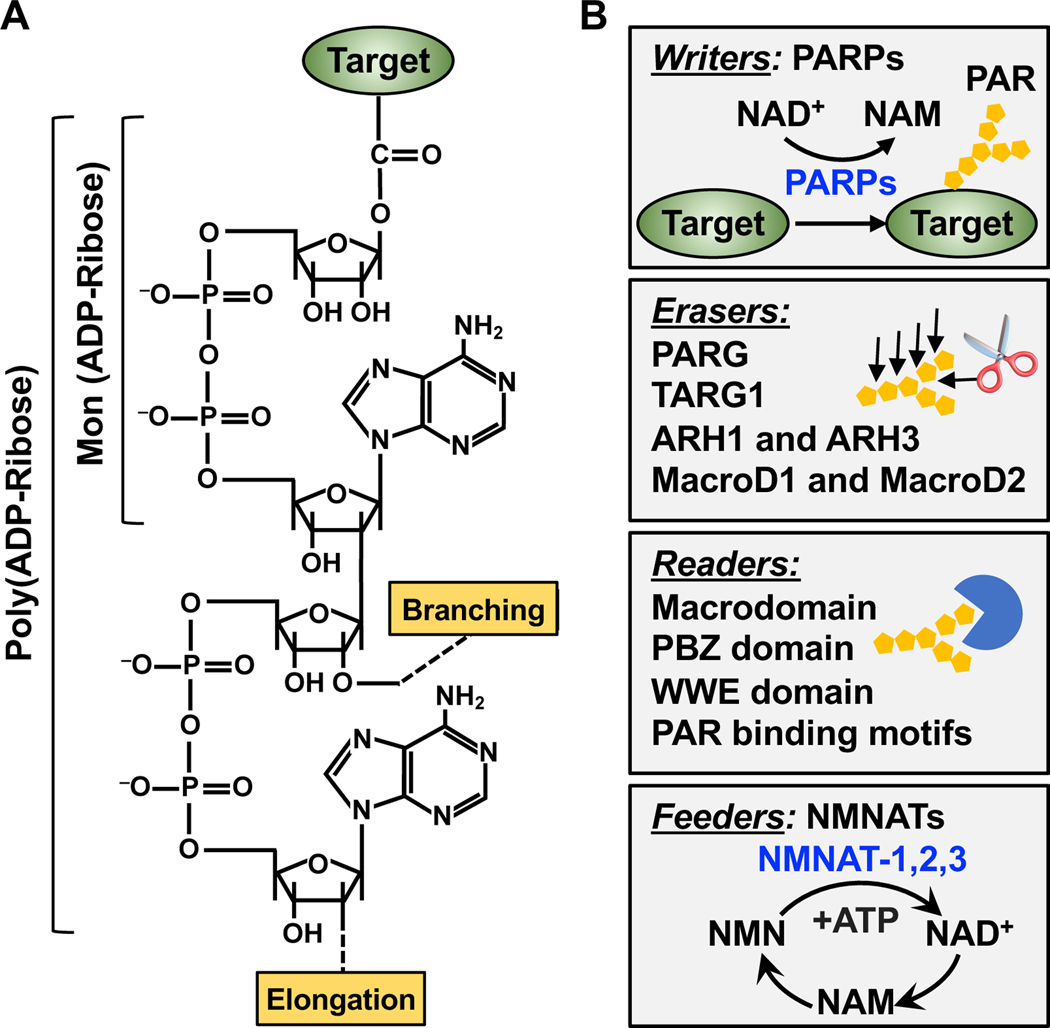
ADPRylation is a post-translational modification (PTM) of proteins first discovered in the 1960s (Chambon et al., 1966; Hasegawa et al., 1967; Reeder et al., 1967). During ADPRylation, ADP-ribose moieties are enzymatically transferred from nicotinamide adenine dinucleotide (NAD+) to a wide variety of substrate proteins (Ryu et al., 2015) and possibly even nucleic acids (Groslambert et al., 2021; Munnur et al., 2019; Weixler et al., 2021). ADPRylation is catalyzed by the PARP family of ADP-ribosyl transferases, leading to the covalent attachment of either a single ADP-ribose unit (monoADPRylation or MARylation) or polymers of ADP-ribose units (polyADPRylation or PARylation) (Ame et al., 2004; Luscher et al., 2021) (Figure 2A). The ‘poly’ modification can be highly heterogeneous, with various numbers of ADP-ribose units, as well as both linear and branched structures (Gupte et al., 2017) (Figure 2A). Interestingly, the entire pathway leading from NAD+ synthesis from nicotinamide mononucleotide (NMN) and ATP, to PARP1 catalytic activity and substrate modification, was observed in the first characterization of ADPRylation using nuclear extracts nearly 60 years ago (Chambon et al., 1963) But, the intimate functional relationship between the nuclear NAD+ synthase nicotinamide mononucleotide adenylyltransferase (NMNAT-1) and PARP1 activity in the nuclear compartment was only recently elucidated (Ryu et al., 2018).
Figure 2. Dynamics of PARP-mediated ADPRylation.
(A) Chemical structures of mono(ADP-ribose) (MAR) and poly(ADP-ribose) (PAR). The positions on the ribose moieties where the PAR polymer is elongated and branched are indicated.
(B) Dynamics of PARP-mediated ADPRylation. PARP family members serve as “writers” of ADPRylation by catalytically transferring ADPR moieties from NAD+ to substrate proteins. The ADPR units can be hydrolyzed and removed by “erasers,” such as PARG, TARG1, ARH1, ARH3, MacroD1, and MacroD2. The ADPR units on target proteins can be recognized by “readers” containing macrodomains, WWE domains, PAR-binding zinc fingers (PBZs), or PAR-binding motifs. The NAD+ required for ADPRylation is supplied through NAD+ synthesis pathways catalyzed by “feeders” - the nicotinamide mononucleotide adenylyl transferases (NMNATs).
ADPRylation is a dynamic and reversible PTM, which is maintained and regulated by a network of “writers” (PARPs), “readers” (ADPR binding domains), and “erasers” (ADP-ribosyl hydrolases) (Gupte et al., 2017) (Figure 2B). The relevance of the readers and erasers to ADPRylation and PARP function are discussed in detail for the examples presented below, as well as elsewhere (Kliza et al., 2021; Palazzo et al., 2017; Pourfarjam et al., 2020; Rack et al., 2020). ADPRylation has been linked to a wide variety of molecular and cellular processes through its ability to alter the properties and functions of substrate proteins, as well as their network of interacting proteins (Gibson and Kraus, 2012). Moreover, some PARPs, such as PARP1, have catalytic-independent functions and can act as structural components in cellular processes (Liu and Kraus, 2017; Luo and Kraus, 2012). Although the historical focus has been on the functions of PARP1 in DNA repair (Figure 3), more recent studies have identified roles for PARP1 in many different cellular processes, from rDNA transcription and ribosome biogenesis, to chromatin modulation, transcription, and RNA processing (Figure 4). All of the processes have the potential to create therapeutic opportunities for PARP inhibitors (PARPi).
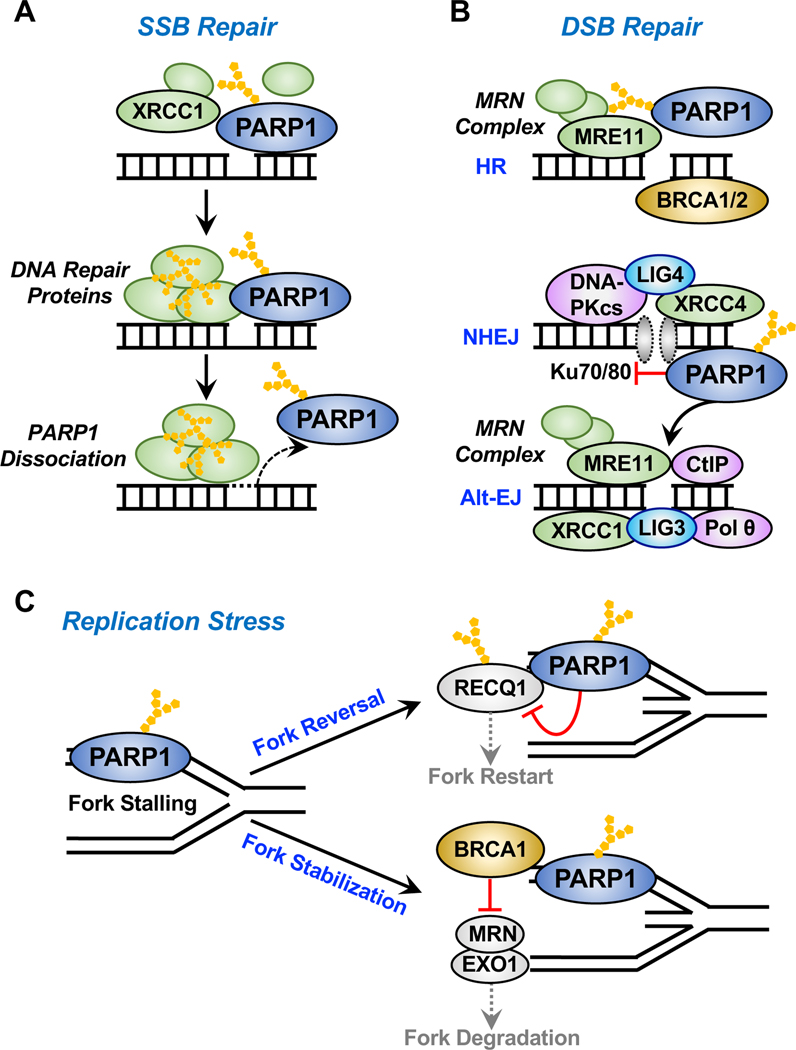
Figure 3. Roles of PARP1 in DNA damage repair.
PARP1 binds to sites of DNA damage, is rapidly activated, and catalyzes autoADPRylation, as well as transmodification of local substrate proteins, playing important roles in various DNA repair pathways.
(A) Activated PARP1 at sites of single-strand breaks (SSBs) recruits DNA repair effectors, such as X-ray repair cross-complementing protein 1 (XRCC1), to mediate DNA repair. AutoADPRylation causes the dissociation of PARP1 from the DNA damage site, which is ultimately required for the efficient DNA repair.
(B) In response to double-strand breaks (DSBs), activated PARP1 recruits the MRE11-RAD50NBS1 (MRN) complex and other repair factors to the sites of damage for homologous recombination (HR)-mediated repair. PARP1 also differentially regulates classical and alternative non-homologous end-joining (NHEJ) repair pathways. PARP1 binds to DSBs in direct competition with Ku70/80 proteins, thus inhibiting the classical pathway of NHEJ that utilizes Ku, DNA-PKcs, DNA ligase IV, and XRCC4. Conversely, PARP1 mediates the recruitment of MRN and CtIP to DSBs to promotes alternative NHEJ (Alt-EJ).
(C) PARP1 links DNA damage to DNA replication by regulating DNA replication fork progression during replication stress. PARP1 is activated at the stalled replication forks in response to replication stress. Activated PARP1 may promote replication fork reversal or stabilization by multiple distinct mechanisms, such as (1) inhibiting the ATP-dependent DNA helicase Q1 (RECQ1), which is responsible for restart of the stalled replication fork or (2) inhibiting fork degradation by MRN and EXO1 through the recruitment of BRCA1. Conflicting studies have suggested an active role of PARP1 in replication fork restart through MRE11 based on the prevention or delay of replication restart upon PARP1 depletion, but the evidence directly linking PARP1 and MRE11 during fork restart is limited.
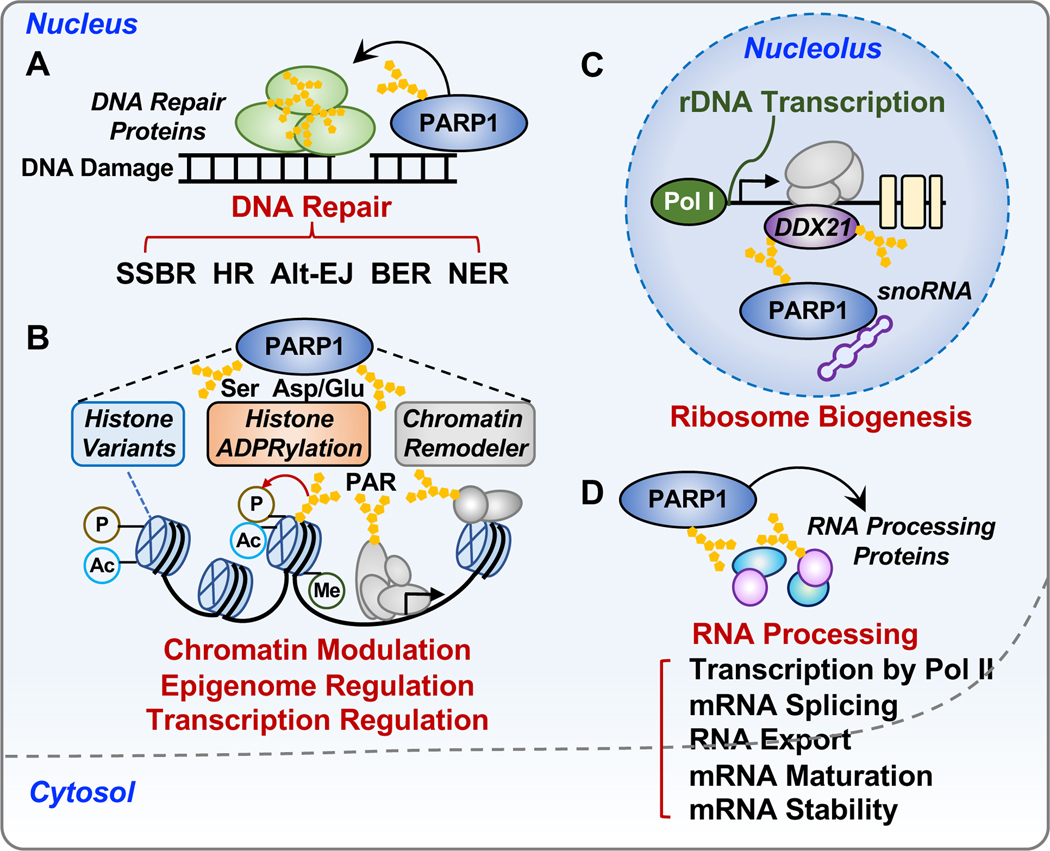
Figure 4. Biological functions of PARP1 in the nucleus.
PARP1 regulates a broad array of processes in the nucleus, such as DNA repair, chromatin regulation, gene expression, ribosome biogenesis, and RNA processing.
(A) Roles of PARP1 in DNA damage repair networks, including single-strand break (SSB) repair, base excision repair (BER), nucleotide excision repair (NER), and DSB repair through both homologous recombination (HR) and alternative end-joining (Alt-EJ) pathways.
(B) Roles of PARP1 in chromatin regulation. PARP1 modulates the structure and function of chromatin through the (1) regulation of chromatin composition, including histone variants and linker histone; (2) regulation of histone ADPRylation and other histone PTMs, including phosphorylation, acetylation and methylation; and (3) regulation of chromatin remodelers.
(C) Roles of PARP1 in ribosome biogenesis. SnoRNA-mediated activation of PARP1 promotes PARylation of DDX21, a DEAD-box RNA helicase in the nucleolus, resulting in the enhanced rDNA transcription and subsequent protein synthesis.
(D) Roles of PARP1 in RNA biology. PARP1 regulates multiple steps of RNA processing by interacting with and modifying RNA binding proteins and RNA processing factors.
The preceding brief overview of PARPs and ADPRylation merely scratches the surface of a vibrant and growing field. In addition, the field is much broader than the specific focus on nuclear PARPs - PARP1 in particular - presented in this review. Several other excellent reviews provide additional details that fill in the gaps not addressed here. In the space below, we focus on a few key aspects of the regulation of nuclear processes by PARP proteins, with a limited set of the many possible examples presented in each case.
NUCLEAR PARPS AND ADPRYLATION IN DNA DAMAGE RESPONSES
Historically, functional studies of PARPs and ADP-ribosylation have focused on DNA damage responses. Among the 17 PARP family members, only PARP1, PARP2, and PARP3, which all localize to the nucleus, are thought to be catalytically activated by damaged DNA and play important roles in cellular responses to DNA damage (Ame et al., 1999; D’Amours et al., 1999; Langelier et al., 2014; Rulten et al., 2011). PARP1, PARP2, and PARP3 shared conserved C-terminal domains: a Trp-Gly-Arg (WGR) domain and a catalytic (Cat) domain; the latter includes two subdomains, a helical domain (HD) and an ADP-ribosyl transferase (ART) domain (Langelier et al., 2014) (Figure 1A). An allosteric change-induced destabilization of the Cat domain is essential for DNA-dependent activation of PARP1, PARP2, and PARP3 (Langelier et al., 2012; Langelier et al., 2014). These allosteric effects on PARP activity allow for regulating responses to DNA damage. Other nucleic acid and protein activators of PARP1 catalytic activity have been identified (Alemasova and Lavrik, 2019; Kim et al., 2019), but their roles in PARP1-mediated DNA and other aspects of PARP1 biology need further characterization.
PARP1 functions in multiple DNA repair pathways.
PARP1, the founding member of the PARP family, is responsible for the vast majority of cellular PARP activity during DNA damage (Dantzer et al., 2006). PARP1 can be activated by various forms of DNA lesions, including single strand breaks (SSBs) (El-Khamisy et al., 2003; Eustermann et al., 2011; Satoh and Lindahl, 1992) (Figure 3A), double strand breaks (DSBs) (Haince et al., 2008; Krishnakumar and Kraus, 2010a; Langelier et al., 2011; 2012) (Figure 3B), and stalled replication forks (Bryant et al., 2009; Min et al., 2013) (Figure 3C). PARP1 can also bind to several non-B DNA structures, which present challenges for the maintenance of genome stability, such as DNA hairpins, cruciforms, and G-quadruplexes (Edwards et al., 2020; Lonskaya et al., 2005; Scheibye-Knudsen et al., 2016). Recent work also suggests that PARP1 is activated by unligated Okazaki fragment intermediates during DNA replication, helping to mediate their repair in S phase cells (Hanzlikova et al., 2018).
PARP1 plays important roles in DNA damage repair networks, including SSB repair, base excision repair (BER), nucleotide excision repair (NER), and DSB repair through both homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways (Audebert et al., 2004; Bryant et al., 2009; Caldecott et al., 1996; Ceccaldi et al., 2016; El-Khamisy et al., 2003; Koczor et al., 2021; Mansour et al., 2010; Masson et al., 1998; Okano et al., 2003; Ray Chaudhuri and Nussenzweig, 2017; Robu et al., 2013; Sallmyr and Tomkinson, 2018; Sugimura et al., 2008; Wang et al., 2006) (Figure 4A). Within these pathways, PARP1 binds to sites of DNA damage, is rapidly activated, and catalyzes autoADPRylation, as well as transmodification of local substrate proteins (Figures 3 and 4A). The latter include DNA repair proteins, histones, and non-histone chromatin-associated proteins. The extent of PARP1 activation functions as a rheostat to facilitate the molecular decision to promote repair of the DNA lesions or drive the cells down one of a number of potential cell death pathways (Luo and Kraus, 2012).
PARP1 links DNA damage to DNA replication.
DSBs are one of the most hazardous DNA lesions for cells. Structural studies have shown that PARP1 binds to DSBs through its zinc fingers and WGR domain, which facilitate its catalytic activation (Langelier et al., 2011; 2012; Langelier et al., 2010; Langelier et al., 2018). Upon binding, PARP1 is robustly activated to catalyze PARylation events at the sites of DSBs. DSBs are repaired by two major pathways, HR or NHEJ, determined by the cell cycle phase and the nature of the DSB ends (Symington and Gautier, 2011). HR is an error-free repair of DSBs using the sister chromatid as a template, and dominates in the S and G2 phases of the cell cycle (Ray Chaudhuri and Nussenzweig, 2017; Symington and Gautier, 2011). Activated PARP1 at the sites of DSBs promotes rapid recruitment of MRE11-RAD50-NBS1 (MRN) complex via a PAR-dependent mechanism for initiation of HR, as well as other critical HR components, including EXO1, BRCA1, and BRCA2 for repair of DSBs through the HR pathway (Haince et al., 2008; Li and Yu, 2013; Slade, 2020; Zhang et al., 2015a; Zhang et al., 2015b) (Figure 3B). PARP1 is also required for late stages of HR to prevent hyper-recombination by controlling the activity of BRCA1 through PARylation (Hu et al., 2014).
Previous studies have suggested that activated PARP1 is associated with DNA replication (Anachkova et al., 1989; Lehmann et al., 1974). Although the specific effects and outcomes remain unresolved, a number of studies have pointed to a role for PARP1 in regulating DNA replication fork progression. PARP1 binds to and is activated by stalled replication forks (Bryant et al., 2009; Min et al., 2013). Evidence from a number of studies indicates that activated PARP1 promotes replication fork reversal and reduces replication fork speed upon replication stress, serving to stabilize the replication fork (Berti et al., 2013; Ray Chaudhuri et al., 2012) (Figure 3C). In this model, PARPi increase replication fork speed by impairing fork reversal, leading to replication stress and DNA damage responses (Genois et al., 2021; Maya-Mendoza et al., 2018; Sugimura et al., 2008) (Figures 3C and 6C). In contrast, several other studies have suggested an active role of PARP1 in replication fork restart through MRE11 based on the prevention or delay of replication restart upon PARP1 depletion (Bryant et al., 2009; Min et al., 2013; Yang et al., 2004), but the evidence directly linking PARP1 and MRE11 in this model is limited. On the other side of the same coin, inhibitors of poly(ADP-ribosyl) glycohydrolase (PARG), which degrades PAR chains, sensitizes cells to radiation-induced DNA damage, and induces persistent fork stalling and replication catastrophe (Houl et al., 2019; Pillay et al., 2019; Ray Chaudhuri et al., 2015). Links among PARP1, PARG, ADPRylation, and DNA replication open exciting new avenues for understanding these processes and the potential to target them therapeutically, as discussed below. Clearly, additional studies are required to resolve the discrepancies.
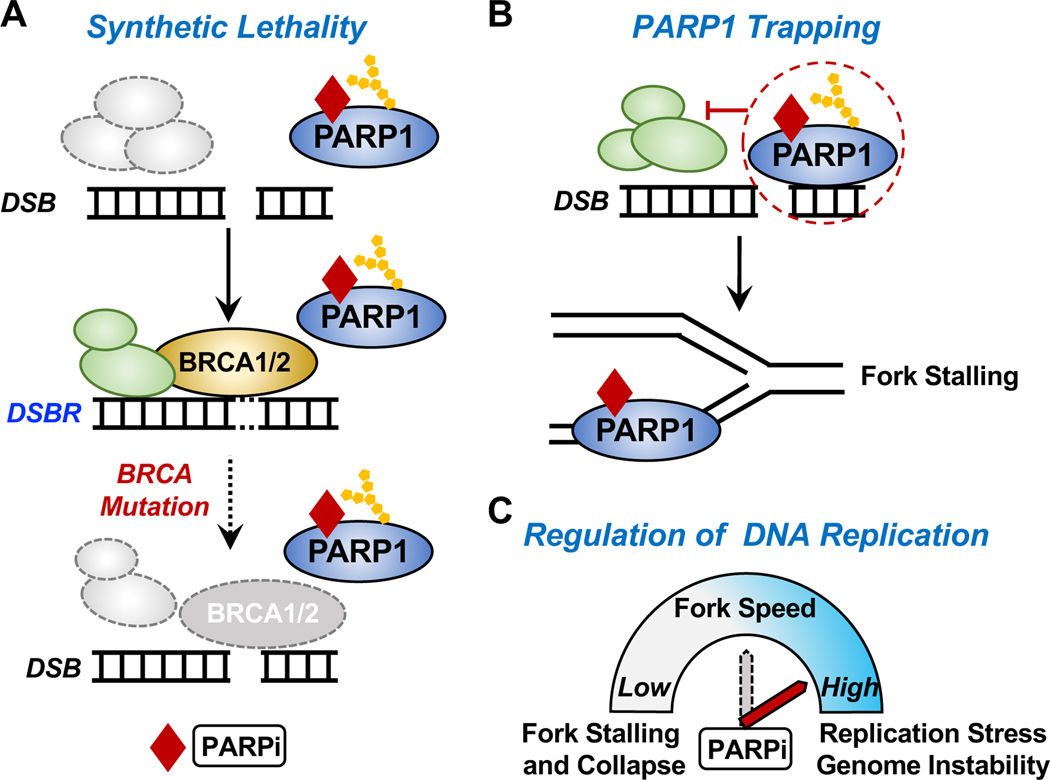
Figure 6. Mechanisms of action of PARP inhibitors.
(A) Synthetic lethality. The simultaneous inhibition of PARP1 activity in DNA repair by PARP inhibitors (PARPi) and loss of BRCA1/2 functionality (e.g., through genetic lesions) leads to the failure of DNA damage repair and subsequent cell death.
(B) PARPi-induced PARP1 “trapping.” Some PARPi “trap” PARP1 on DNA, thus preventing autoPARylation of PARP1 and release from the site of damaged DNA. This interferes with the catalytic cycle of PARP1 and efficient DNA damage repair. PARP1 trapping acts as a replication barrier, which impairs the progression of DNA replication forks and leads to fork stalling.
(C) Regulation of DNA replication progression by PARPi. Activated PARP1 promotes replication fork reversal and reduces replication fork speed upon replication stress (see Figure 3C). In this regard, PARPi increases replication fork speed, leading to replication stress and genome instability.
Roles for PARP2 and PARP3 in DNA repair pathways.
PARP2, another nuclear ‘polyenzyme’, and PARP3, a nuclear mono(ADP-ribosyl) transferase (MART) are also activated by DNA breaks (Ame et al., 1999; Langelier et al., 2014; Rulten et al., 2011). PARP2 and PARP3 are preferentially activated by DNA with 5′ phosphorylated ends (Langelier et al., 2014). The functions of PARP2 in DNA damage repair partially overlap with PARP1. PARP2 contributes to BER by promoting recruitment of XRCC1 and polynucleotide kinase-phosphatase (PNKP) to sites of SSBs (Hanzlikova et al., 2017; Sukhanova et al., 2019) and promotes HR in response to DSBs (Fouquin et al., 2017). PARP2 is also likely to have unique function in DNA damage repair. PARP2 is recruited to sites of DNA damage, is activated by PARP1 in a PARylation-dependent manner, and catalyzes branched PARylation at the damage site (Chen et al., 2018). PARP3 has been shown to contribute preferentially to DSB repair (Beck et al., 2014; Rulten et al., 2011). PARP3 is stimulated by DSBs and functions together with the PAR-binding protein aprataxin- and PNK-like factor (APLF) to accelerate NHEJ in response to DSBs (Boehler et al., 2011; Rulten et al., 2011). In addition, based on biochemical and molecular assays, certain types of complex DNA breaks can be ADPRylated by both PARP2 and PARP3 (Belousova et al., 2018; Munnur and Ahel, 2017; Zarkovic et al., 2018). The potential role of DNA ADPRylation in DNA damage response in cells is an exciting possible mechanism that requires additional investigation.
NUCLEAR PARPS AND ADPRYLATION IN THE REGULATION OF CHROMATIN AND GENE EXPRESSION
Chromatin, which is comprised of repeating arrays of nucleosomes (i.e., histones plus genomic DNA), is modulated in response to intrinsic and extrinsic signals by changes in its composition and posttranslational modifications (PTMs), as well as the actions of ATP-dependent chromatin remodeling complexes. An effect on chromatin structure was one of the first cellular functions defined for PARP1, driven in part by the ADPRylation of histone proteins (Kraus and Lis, 2003; Krishnakumar and Kraus, 2010a). This was demonstrated clearly and dramatically by studies in Drosophila with polytene chromosomes, which decondense in a PARP1- and PAR-dependent manner upon heat shock or ecdysone exposure (Tulin and Spradling, 2003). Nuclear PARPs have been implicated in the structure, compaction, and function of chromatin, which are essential for transcriptional regulation and DNA damage repair (Krishnakumar and Kraus, 2010a). As described in detail below, PARP1 may act locally at regulatory elements. In addition, PARP1 and PARylation contribute to the regulation of higher order three-dimensional chromatin structures through the chromatin architecture protein CCCTC-binding factor (CTCF) (Zlatanova and Caiafa, 2009). In this regard, PARP1 and PARylation regulate CTCF-dependent chromatin insulation and the stabilization of CTCF binding and chromatin structure (Lupey-Green et al., 2018; Yu et al., 2004). The interaction between PARP1 and CTCF is reciprocal since CTCF can also induce PARP1 activation (Guastafierro et al., 2008; Zhao et al., 2015). Oscillations in PARP1-CTCF interactions were shown to regulate transcription of circadian genes by modulating their recruitment to nuclear periphery through changes in chromosome topology (Zhao et al., 2015). In the text below, we review the role of PARP1 in the regulation of chromatin.
Nuclear PARPs and histone ADPRylation.
Nuclear PARPs modulate the structure and function of chromatin through histone ADPRylation (Figure 4B). All core histone proteins H2A, H2B, H3, and H4, as well as linker histone H1, were identified decades ago as substrates for ADPRylation (Boulikas, 1988; Huletsky et al., 1985). Early studies suggested that PARP-mediated histone ADPRylation promotes chromatin decompaction (de Murcia et al., 1986; Poirier et al., 1982; Tulin and Spradling, 2003). Recent advances in chemical biology and mass spectrometry have led to the identification of specific sites of ADPRylation on histones (Gibson et al., 2016; Hendriks et al., 2019; Hottiger, 2011; Huang et al., 2020a; Karch et al., 2017; Leidecker et al., 2016). This has led to a greater understanding of the molecular mechanisms and functional roles of site-specific histone ADPRylation.
During DNA damage responses, histone ADPRylation occurs predominantly on serine residues, which requires the accessory factor histone PARylation factor 1 (HPF1) for efficient transmodification of histones (Bartlett et al., 2018; Bonfiglio et al., 2017; Gibbs-Seymour et al., 2016; Hendriks et al., 2021; Langelier et al., 2021; Palazzo et al., 2018). Structural studies have shown that HPF1 completes the active sites of PARP1 and PARP2 to promote histone ADPRylation (Suskiewicz et al., 2020). Moreover, the PARP2-HPF1 complex bridges two nucleosomes and aligns the broken DNA in a position suitable for ligation (Bilokapic et al., 2020). This induces structural alterations in PARP2 that signal DNA break recognition to the catalytic domain, leading to HPF1 binding and PARP2 activation.
ADPRylation of histones at serines, as well as acidic residues, plays a key role in various aspects of DNA repair. For example, in the soil-dwelling amoebae Dictyostelium, ADPRylation of the histone variant H3b at Ser10 and Ser28 maintains genome stability by preventing premature mitotic entry with unresolved DNA damage, thus maintaining genome stability (Brustel et al., 2022). ADPRylation of the histone variant H2AX at Glu141, which occurs in response to oxidative DNA damage, facilitates BER repair, but also suppresses the H2AXγ-mediated DSB response (Chen et al., 2021). PARP3 is also activated by nucleosomes containing nicked DNA, directing MARylation on Glu2 of histone H2B (Grundy et al., 2016). These are undoubtedly just a few of the many examples linking site-specific histone ADPRylation to DNA repair.
A recent study from our lab highlighted the physiological significance of histone H2B ADPRylation in adipogenesis and obesity, as well as the functional consequences of crosstalk between histone ADPRylation and other histone PTMs (Huang et al., 2020a) (Figure 4B). Mechanistically, this study identified a subset of snoRNAs as physiological activators of PARP1 and characterized NMNAT1 as a factor promoting PARP1 catalytic activity toward specific Glu and Asp residues on histone proteins (Huang et al., 2020a). Both in vitro and in vivo assays demonstrated that ADPRylation of H2B Glu35 inhibits phosphorylation of H2B Ser36, thereby attenuating adipogenic gene expression and increased fat mass in cultured adipocytes and mice (Huang et al., 2020a). Together, recent findings on HPF1 and NMNAT1 suggest distinct mechanisms for “specificity factors” that can direct PARP1 activity to different substrate residues in different biological and cellular contexts. This concept might also be expanded generally to other PARP family members, such as PARP7, which has dual specificity for Cys and Asp/Glu (Palavalli Parsons et al., 2021; Rodriguez et al., 2021).
Additional examples of crosstalk between ADPRylation and other histone modification occur during gene regulation and DNA repair. For example, H3 Ser10 ADPRylation and H3 Lys9 acetylation are mutually exclusive in response to DNA damage (Bartlett et al., 2018; Liszczak et al., 2018). H3 Ser10 ADPRylation was also recently shown to block H3 Lys9 methylation study (Hananya et al., 2021). In addition, ADPRylation of H2AX at Glu141 inhibits phosphorylation at Ser139 during DNA repair (Chen et al., 2021). Moreover, PARP1 prevents the demethylation of H3K4me3 by blocking the histone demethylase KDM5B through PARylation, leading to the opening of promoter chromatin (Krishnakumar and Kraus, 2010b) (Figure 4B).
Regulation of chromatin structure and composition by PARP1.
Studies in Drosophila have shown that PARP1 activation can induce large-scale chromatin alterations in response to heat shock or ecdysone treatment (Petesch and Lis, 2008; 2012; Tulin and Spradling, 2003). For some genes (e.g., Hsp70), this involves a PARP1- and transcription-dependent loss of nucleosomes over the body of the gene (Petesch and Lis, 2008). PARP1 is associated with the 5’-end of the Hsp70 gene, and its enzymatic activity is rapidly induced by heat shock (Petesch and Lis, 2012). This activation promotes PARP redistribution and PAR accumulation throughout the gene in a manner dependent on the transcription factor HSF and the acetylation of H2A Lys5 at the 5’-end of the gene by Tip60 (Petesch and Lis, 2012). The activation and spread of PARP1 is required for the rapid, signal-induced nucleosome loss over the Hsp70 gene (Petesch and Lis, 2012).
Although the linker histone H1 was one of the first chromatin substrates identified for PARP1, the functional interplay between PARP1 and H1 may be more dependent on structural, rather than catalytic, aspects of PARP1 function. Like H1, PARP1 binds at the dyad axis of the nucleosome where the linker DNA exits the nucleosome (Kim et al., 2004). Given the overlapping binding sites, PARP1 can effectively compete with H1 for binding and exclude H1 from nucleosomes. In cells, PARP1 binding to chromatin and catalytic activity is enriched at promoters (Gibson et al., 2016; Krishnakumar et al., 2008). PARP1 localization to promoters allows PARP1 to clear H1, promote an open chromatin architecture, and allow transcription to proceed (Krishnakumar et al., 2008; Krishnakumar and Kraus, 2010b). These effects occur independently of PARP1 catalytic activity. Together, these results highlight the dual roles (i.e., catalytic-dependent and -independent) roles of PARP1 in the regulation of chromatin structure and composition.
Functional interplay between PARP1 and histone variants.
In addition to the canonical core histones, a broader set of histone variants can alter the composition and function of chromatin (Martire and Banaszynski, 2020), including the functions of PARP1 within chromatin. The H2A variant H2Av, the Drosophila homolog of the mammalian histone H2A variants H2Az and H2Ax, facilities PARP1 binding to active chromatin. PARP1 binds to H2Av-containing nucleosomes via a hydrophobic patch formed by histone H4. Histone H4 can trigger activation of PARP1 catalytic activity through the C-terminal Cat domain of PARP1 (Pinnola et al., 2007; Thomas et al., 2019). Phosphorylation of H2Av activates PARP1 by increasing the accessibility of H4 (Thomas et al., 2014), resulting in chromatin decondensation and transcriptional activation of heat shock-induced genes (Kotova et al., 2011). Another H2A variant, macroH2A1.1, cooperates with the activated PARP1 through its PAR-binding macrodomain, resulting in chromatin rearrangement and gene regulation through histone H2B acetylation (Chen et al., 2014; Timinszky et al., 2009). In addition, the H2A variant macroH2A1.2 and PAR binding by a non-canonical PAR-binding region unique to H3K4 histone demethylase KDM5A allow recruitment of KDM5A to sites of DNA damage to facilitate homology-directed repair and repression of transcription at DNA breaks (Kumbhar et al., 2021). Notably, while recruitment of KDM5A is promoted by PAR binding (Kumbhar et al., 2021), recruitment of the KDM5B demethylase is blocked by PARylation (Krishnakumar and Kraus, 2010b). The deposition of the H3 variant H3.3 in chromatin at sites of DNA damage is promoted by activated PARP1 through the chromatin remodeler CHD2 to facilitate NHEJ (Luijsterburg et al., 2016). These are undoubtedly just a few of the examples of how the incorporation of histone variants impacts chromatin-dependent regulation by PARP1 (Figure 4B).
Regulation of ATP-dependent chromatin remodeling complexes by PARP1.
Nuclear PARPs regulate ATP-dependent chromatin remodelers in a PAR-dependent manner, leading to changes of chromatin compaction. ALC1, a chromatin remodeling enzyme, is rapidly recruited and allosterically activated at sites of DNA damage by activated PARP1 via the PAR-binding macrodomain of ALC1. This drives chromatin relaxation for DNA damage repair (Ahel et al., 2009; Gottschalk et al., 2009; Ooi et al., 2021; Singh et al., 2017; Wang et al., 2021a). Chromatin relaxation induced by ALC1 upon PAR binding promotes recruitment of other chromatin remodelers, such as CHD3 and CHD4 (Smith et al., 2018). PARP1 plays multiple roles in ALC1dependent nucleosome remodeling (Bacic et al., 2021; Ooi et al., 2021). Nucleosome remodeling by ALC1 is required for the release of DNA-damage-recruited PARP2 from chromatin (Blessing et al., 2020) and is also involved in the timely removal of PARP1 from DNA lesions (Juhasz et al., 2020). Site-specific ADPRylation of histones (e.g., at H2B Ser6 or H3 Ser10) converts nucleosomes into robust substrates for remodeling by ALC1 (Mohapatra et al., 2021).
APLF, a histone chaperone and DNA repair factor, is recruited to the sites of DNA damage by activated PARP1 via its PAR-binding zinc-finger (PBZ) domain (Mehrotra et al., 2011). Activated PARP1 also promotes the recruitment of SMARCA5/SNF2H, the catalytic subunit of ISWI chromatin remodeling complexes, to the sites of DNA damage (Smeenk et al., 2013). In contrast, PARylation of bromodomain-containing protein 7 (BRD7), a component of SWI/SNF chromatin remodeling complex, promotes its degradation through the PAR binding E3 ubiquitin ligase RNF146 (Hu et al., 2019) (Figure 4B).
PARP1 AND RIBOSOME BIOGENESIS
A substantial amount of PARP1 and PARP2 is localized in the absence of genotoxic stress to the nucleolus, which is the site of ribosome biogenesis (Desnoyers et al., 1996; Guetg et al., 2012; Kim et al., 2019; Meder et al., 2005; Rancourt and Satoh, 2009). Emerging evidence has indicated that PARP1 plays an important role in ribosome biogenesis, including ribosomal RNA (rRNA) transcription, pre-rRNA processing, and pre-ribosome assembly in the nucleolus (Kim et al., 2020). Two types of nucleolar RNAs, pRNA (promoter associated RNA) and snoRNA (small nucleolus RNA), were shown to interact with PARP1 and mediate the effects of PARP1 and PARylation on rRNA transcription (Guetg et al., 2012; Kim et al., 2019). PARP1 interacts with TIP5, a component of the chromatin remodeling complex NoRC, through pRNA to establish transcriptionally inactive ribosomal DNA (rDNA) chromatin, thus silencing rRNA transcription (Guetg et al., 2012). In contrast, snoRNA-mediated activation of PARP1 promotes PARylation of DDX21, a DEAD-box RNA helicase in the nucleolus, resulting in enhanced rRNA transcription, increased ribosome assembly, and subsequent protein synthesis (Kim et al., 2019) (Figure 4C). The opposing effects of PARP1 on rRNA transcription shown by these two studies suggest that the range of RNA types bound by PARP1 and the functional consequences of PARP1-RNA interactions may vary in different biological contexts.
PARP1 enzymatic activity is also required for the proper localization of nucleolar proteins to the proximity of precursor rRNA, thus controlling rRNA processing and subsequent ribosome assembly (Boamah et al., 2012). PARP1 and PARylation also regulate the nucleolar-nucleoplasmic shuttling of several DNA damage repair factors in response to genotoxic stress (Veith et al., 2019). In addition, an ADP-ribose hydrolase, TARG1, controls rRNA transcription when localized to transcriptionally active nucleoli. Activated PARP1 and PARP2 induce the relocalization of TARG1 to the nucleoplasm through PAR binding in response to DNA damage, thus inhibiting the nucleolar function of TARG1 in ribosome biogenesis (Butepage et al., 2018). Interestingly, homozygous mutations leading to defects in TARG1 cause severe neurodegeneration in humans (Sharifi et al., 2013).
Given the important roles of PARP1 in ribosome biogenesis, the localization of PARP1 and associated nucleolar proteins may represent key biomarkers for related diseases. In breast cancer cells, increased nucleolar localization of DDX21 by activated PARP1 drives ribosome biogenesis and cancer cell proliferation (Kim et al., 2019). In neurons, PARP1-mediated PARylation of DNA methyltransferase 1 (DNMT1) inhibits DNMT1’s activity for the methylation of rDNA. In Alzheimer’s disease, mislocalization of PARP1 from the nucleolus decreases PARylation of DNMT1, thus leading to the hypermethylation of rDNA and impaired rRNA transcription (Zeng et al., 2016). Together, these results identify a broad array of functions for PARP1 in the nucleolus, including rRNA transcription, pre-rRNA processing, and ribosome assembly, some of which may be useful targets for therapeutic interventions.
NUCLEAR PARPS AND RNA BIOLOGY
A growing body of work has described important roles for PARPs and ADPRylation in RNA biology (Eleazer and Fondufe-Mittendorf, 2020; Kim et al., 2020). In fact, proteomic studies have consistently identified RNA processing and splicing factors as the most highly enriched class of nuclear PARP substrates (Figure 4D), even more than DNA repair, chromatin, and transcription proteins (Buch-Larsen et al., 2020; Gibson et al., 2016; Jungmichel et al., 2013; Larsen et al., 2018; Zhang et al., 2013; Zhen and Yu, 2018).
Binding of PARP1 and PARP2 to RNAs and activation of their catalytic activities.
Recent studies have identified various RNA species, including mRNAs and non-coding RNA, that bind to nuclear PARPs (Guetg et al., 2012; Kim et al., 2019; Melikishvili et al., 2017). The interactions with PARP1 are mediated through the amino-terminal nucleic acid binding domain. Zinc fingers 1 and 2 (Zn1 and Zn2) are required for PARP1 binding to DNA, while Zn3 is important for PARP1 binding to RNA, as deletion of Zn1 and Zn2 enhances PARP1 binding to RNA (Guetg et al., 2012; Huambachano et al., 2011; Melikishvili et al., 2017). Importantly, PARP1 has been shown to be specifically activated by RNA in vitro, as well as in various cellular and biological contexts, independent of DNA damage (Huang et al., 2020a; Huang et al., 2020b; Kim et al., 2019; Melikishvili et al., 2017).
RNA-dependent activation of PARP1 may require the WGR domain (Huambachano et al., 2011), but the details of the required structural and functional determinants in PARP1 and the interacting RNAs have not been well defined. Deletion of individual domain of PARP1 seems to decrease RNA binding to a similar extent, but none of those mutants completely abrogate its RNA binding ability in vitro (Melikishvili et al., 2017). CRISPR-Cas9 mediated deletion of selected snoRNAs decreases cellular levels of PARylation in 3T3-L1 preadipocytes and promotes their differentiation into mature adipocytes, implicating the interaction between PARP1 and RNA in physiological processes (Huang et al., 2020a). RNA, including rRNA, has also been reported as a potent activator of PARP2 in vitro and in vivo (Chen et al., 2018; Leger et al., 2014). The SAP (SAF-A/B, Acinus, and PIAS) and WGR domains of PARP2 are required for RNA binding and catalytic activation (Leger et al., 2014).
Functional outcomes of PARP1 in RNA biology.
PARP1 and PARylation play key roles in many steps in gene regulation, including ribosome biogenesis, transcription by RNA polymerase II (Pol II), and RNA splicing (Figure 4D). Transcription elongation by Pol II is a highly regulated process (Jonkers and Lis, 2015). In addition to the chromatin-dependent mechanisms of transcriptional regulation noted above, PARP1 also regulates transcription elongation through the negative elongation factor NELF, which acts to restrict transcriptional elongation and stimulate promoter-proximal pausing by Pol II (Gibson et al., 2016). PARP1 PARylates and inhibits the NELF-E subunit of the NELF complex, which promotes the release of Pol II into productive elongation (Gibson et al., 2016). This may have important consequences for signal-regulated gene expression in cancers (Gadad et al., 2021).
A growing body of work has identified PARP1 as an important regulator of mRNA splicing. PARP1 binding to nucleosomes can promote changes in the chromatin state that regulate Pol II kinetics, thus affecting co-transcriptional alternative splicing (Eleazer and Fondufe-Mittendorf, 2020; Kim et al., 2020; Matveeva et al., 2019a). Moreover, PARP1 can regulate RNA binding proteins (RBPs) and RNA processing factors in alternative splicing through covalent PARylation events, non-covalent PAR binding, and protein-protein interaction. Proteomic approaches have identified various RBPs and splicing factors among PARP1 substrates, the PARP1 interactome, and the PAR-binding proteome (Isabelle et al., 2010; Jungmichel et al., 2013; Teloni and Altmeyer, 2016; Zhen and Yu, 2018). PARP1-mediated PARylation is important for regulating the activity of splicing factors, but scaffolds formed through the interaction between PARP1 and chromatin or regulatory factors are also critical for splicing regulation (Eleazer and Fondufe-Mittendorf, 2020). PARP1 and PARylation are also involved in the regulation of mRNA stabilization and degradation through an array of mechanisms, which are also related to their roles in splicing regulation (Ke et al., 2017; Matveeva et al., 2019b). PARP1 inhibits polyadenylation of mRNA through PARylation of poly(A) polymerases (PAP), impacting the stability and maturation of mRNA (Di Giammartino et al., 2013). Additionally, PARP1 can regulate mRNA translation by PARylating Myef2, which binds to the mRNAs of protein-coding genes in oligodendroglial cells through their 3’ untranslated regions (UTRs) (Wang et al., 2021b). As the subset of results presented here illustrate, PARP1 can impact the production, stability, and processing of various types of RNAs (Figure 4D).
ADDITIONAL CONCEPTS AND MECHANISMS OF PARP1-DEPENDENT REGULATION
In the sections above, we summarized the role of nuclear PARPs in several key nuclear processes, focusing on the actions of PARP1. In this section, we summarize several concepts that underlie the mechanisms of PARP1-dependent regulation.
PAR-dependent post-translational modifications, including PAR-dependent PARylation.
The discovery of ADPR binding domains (ARBDs), including macrodomains and WWE domains, have pointed to new possibilities for PAR-dependent interactions, including those that drive protein substrates to their cognate enzymes (Kumbhar et al., 2021; Palazzo et al., 2017; Rack et al., 2020; Teloni and Altmeyer, 2016). Examples include (1) “PAR-dependent PARylation,” where a substrate protein interacts with automodified PARP1 through PAR, leading to subsequent PARylation of the substrate protein by PARP1 and (2) other “PAR-dependent post-translational modifications,” where a PARP PARylates a substrate protein, which promotes PAR-dependent interactions with a modifying enzyme, leading to post-translational modification of the PARP substrate protein by the modifying enzyme (Figure 5A). The latter was first demonstrated for PAR-dependent ubiquitylation, where the initial PARylation of a substrate protein (i.e., 3BP2) by a PARP (i.e., Tankyrase) was required for subsequent ubiquitylation by a PAR-binding E3 ubiquitin ligase (i.e., RNF146) (Gibson and Kraus, 2012; Guettler et al., 2011; Levaot et al., 2011). More recently, PARP1-mediated PARylation of Stat1α at Asp721 in the transactivation domain was shown to promote subsequent interferon gamma-mediated transactivation through phosphorylation at Ser727 in macrophages, although the molecular details of the phosphorylation event were not elucidated (Gupte et al., 2021).
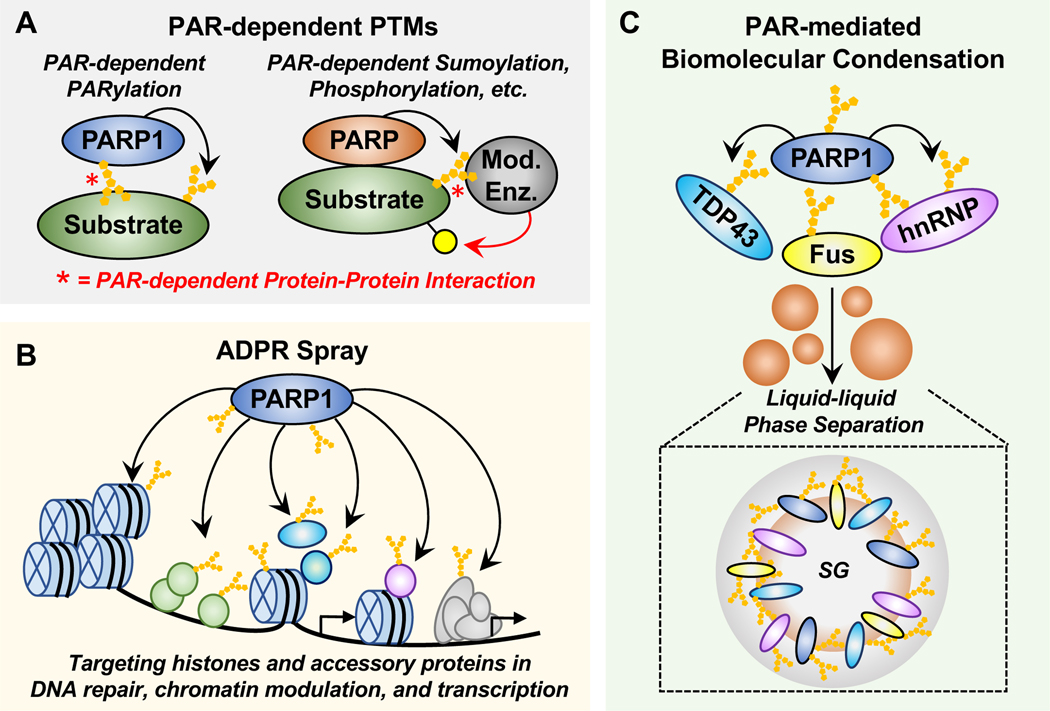
Figure 5. Emerging concepts and mechanisms of PARP1-dependent regulation.
(A) PAR-dependent PTMs. PAR-dependent protein-protein interactions can drive substrates to their cognate modifying enzymes. Examples include (1) “PAR-dependent PARylation,” where a substrate protein interacts with automodified PARP1 through PAR, leading to subsequent PARylation of the substrate protein by PARP1 and (2) other “PAR-dependent post-translational modifications,” where a PARP (e.g., PARP1, PARP5) interacts with and PARylates a substrate protein, which promotes PAR-dependent interactions with a modifying enzyme, leading to posttranslational modification (e.g., ubiquitylation, phosphorylation) of the PARP substrate protein by that modifying enzyme.
(B) ADPR spray. ADPRylation events mediated by nuclear PARP1 may drive a high density “ADPR spray” across key histone and accessory proteins in processes including DNA repair, chromatin modulation, and transcription.
(C) PAR-mediated biomolecular condensate formation and function. Free or protein-linked PAR may regulate the formation or dissociation of biomolecular condensates. In this example, PAR facilitates the phase separation of FUS, TDP43, and hnRNPA1 to form stress granules (SG) in the pathogenesis of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
More recently, a number of examples of “PAR-dependent PARylation,” in which PAR-dependent interactions drive substrates to automodified PARP1 for subsequent ADPRylation, have been characterized. For example, p53 binds to automodified PARP1 through specific non-covalent interactions between PAR and the C-terminal domain of p53, leading to its covalent ADPRylation by PARP1 (Fischbach et al., 2018) (Figure 5A). Likewise, the RNA-helicase DDX21 binds to automodified PARP1 through specific non-covalent interactions between PAR and the C-terminal RNA recognition motif of DDX21, leading to its covalent ADPRylation by PARP1 (Kim et al., 2019). Interestingly, as exemplified by DDX21, a number of RNA binding domains also bind PAR (Teloni and Altmeyer, 2016), suggesting a broader array of protein modules available to drive PAR-dependent interactions. The similarities among the chemical compositions and structures of PAR, RNA, and DNA suggest that promiscuity in binding these macromolecules by various proteins is probably common.
“ADPR spray.”
Nuclear enzymes that mediate PTMs, including those deposited on histones and chromatin-associated proteins, engage a variety of targets whose simultaneous modification may serve common functions in a particular process or pathway. In 2012, Psakhye and Jentsch proposed a model for multisite sumoylation of several colocalized proteins in the same pathway by SUMO ligase, which they called “SUMO spray” (Psakhye and Jentsch, 2012). In 2018, Weinert et al. proposed an “acetyl spray” mechanism for the acetyltransferases p300 and CBP, which drives high density acetylation across multiple targets in chromatin that creates a dispersed interface for interactions or functions driven by p300/CBP-mediated acetylation (Weinert et al., 2018). We suggest that this is also a useful paradigm for thinking about ADPRylation events mediated by nuclear PARPs, which may drive a high density “ADPR spray” across key histone and accessory proteins in processes, including DNA repair, chromatin modulation, and transcription. Further work is needed to substantiate this idea, but it could help to explain the diversity of PARP substrates in each process (Figure 5B).
PAR-mediated biomolecular condensate formation and function.
Biomolecular condensates are a class organelles and organelle subdomains that lack a bounding membrane (Sabari, 2020). They can form in cells through a range of processes, including the phase separation of proteins, RNA, and other biopolymers into colloidal emulsions, liquid crystals, solid crystals, or aggregates. Recent studies have shown that PAR mediates liquid-liquid phase separation to form biomolecular condensates in a variety of biological processes and pathological diseases (Leung, 2020). The structure and properties of PAR – a nucleic acid-like negatively charged polymer – is well suited for this purpose. PAR-mediated condensates have been linked to cancer, viral infection, and neurodegeneration (Leung, 2020). Moreover, PAR-mediated condensates may be useful targets of PARPi, which can modulate their formation and dynamics, as well as the trafficking of their components.
For example, PAR facilitates phase separation at DNA damage repair foci in cells and in vitro. FUS, a nuclear RNA-binding protein, is recruited to the DNA damage sites by binding to PARP1-generated PAR to form damaged DNA-rich compartments for DNA repair (Singatulina et al., 2019). PAR-dependent recruitment of FUS and other FET family members (e.g., EWS and TAF15) to sites of DNA damage sites leads to condensate formation (Altmeyer et al., 2015). These condensates selectively compartmentalize γH2AX and MDC1, but exclude 53BP1 as a downstream effector in DNA damage signaling (Altmeyer et al., 2015), which forms separate condensates (Kilic et al., 2019). Mutations in the FUS nuclear localization sequence induce FUS cytoplasmic mislocalization, which in turn impairs DNA damage signaling, leading to neurodegeneration and FUS aggregate formation (Naumann et al., 2018). PARylation promotes FUS recruitment to DNA damage sites and counteracts FUS cytoplasmic mislocalization. In addition, PAR, several PARPs, and PARG are found in cytoplasmic stress granules and regulate dynamics of stress granules assembly (Catara et al., 2017; Isabelle et al., 2012; Leung et al., 2011). PAR-binding via the PAR-binding motifs of the RNA-binding proteins TDP-43 and hnRNPA1 mediates phase separation, as well as the association of these proteins with stress granules in the pathogenesis of ALS and FTD by promoting aberrant protein aggregation (Duan et al., 2019; McGurk et al., 2018a; McGurk et al., 2018b) (Figure 5C). In vitro analyses have demonstrated liquid-liquid phase separation of FUS, TDP43, and hnRNPA1 through direct interactions with PAR (Altmeyer et al., 2015; Duan et al., 2019; McGurk et al., 2018b; Patel et al., 2015; Singatulina et al., 2019) (Figure 5C). PARP5a and 5b inhibitors have been shown to mitigate TDP-43-associated neurodegeneration in vivo (McGurk et al., 2018a), providing potential therapeutic strategies for treatment of these diseases. Moreover, PAR-dependent condensate formation may be tied to NAD+ biosynthesis in cell (Watanabe et al., 2021). Further exploration in this exciting area will undoubtedly uncover additional aspects of PAR-mediated condensate formation, its regulation, and its function.
MOLECULAR AND THERAPEUTIC MECHANISMS OF PARP INHIBITORS
PARPi are a class of anti-cancer agents, which compete with NAD+ for the catalytic domain of PARP proteins (Steffen et al., 2013; Wahlberg et al., 2012). FDA-approved PARPi are an effective treatment against ovarian, breast, pancreatic and prostate cancers, and may have clinical utility for other cancer types as well (Slade, 2020). These include Olaparib (2014; Lynparza, AstraZeneca), Rucaparib (2016; Rubraca, Clovis Oncology), Niraparib (2017; Zejula, Tesaro), and Talazoparib (2018; Talzenna, Pfizer) (Rose et al., 2020). PARP1 is considered the major, but not exclusive, target for the FDA approved PARPi. Given the conserved structures of the NAD+ binding pockets among the PARP family members, these PARPi can also inhibit the activity of other PARPs, such as PARP2, PARP3, PARP5a, and PARP5b (Steffen et al., 2013; Wahlberg et al., 2012). The past 15 years has witnessed the initial determination of the underlying mechanisms driving the therapeutic effects of PARPi related to the functions of PARP1 in DNA repair, as well as the exploration of a range of other mechanisms that may underlie the therapeutic potential of PARPi (Kim et al., 2021).
Synthetic lethality between PARPi and defects in BRCA1 and BRCA2.
A major mechanism through which PARPi are thought to exert their therapeutic actions is synthetic lethality. Synthetic lethality occurs when a combination of deficiencies (e.g., inactivating mutations in a gene, inhibition of enzyme activity by a drug) leads to cell death, whereas each deficiency alone does not. Synthetic lethality between PARPi and inactivating mutations or deletions of the BRCA1 or BRCA2 genes was first reported in 2005 (Bryant et al., 2005; Farmer et al., 2005). The BRCA1 and BRCA2 proteins encoded by these genes are required for efficient repair of double strand DNA breaks through a HR-dependent pathway. Persistent SSBs caused by PARP1 inhibition promote the collapse of replication forks and trigger HR for repair (Bryant et al., 2005; Farmer et al., 2005). In cancers with BRCA1/2 defects, treatment with PARP inhibition drives synthetic lethality with the genetic lesions, leading to cell death (Lord and Ashworth, 2017) (Figure 6A).
Beyond specific defects in BRCA1/2, other deficiencies in HR (HRD) can lead to “BRCAness” (i.e., HR repair defects sharing similar molecular features with BRCA1/2 mutations) can also lead to synthetic lethality with PARPi (Lord and Ashworth, 2016). Recent studies, however, have reported that PARPi may have significant clinical benefits in patients in the absence of BRCA1/2 defects or HRD, suggesting therapeutic potential beyond DNA repair mechanisms (Frizzell and Kraus, 2009; Ledermann et al., 2014; Lheureux et al., 2017; Scott et al., 2015). In addition, synthetic lethality was also observed between PARPi and deficiencies in chromatin remodelers, such as ALC1 and PBRM1(Chabanon et al., 2021; Hewitt et al., 2021; Juhasz et al., 2020; Verma et al., 2021). In this regard, Olaparib, Rucaparib and Niraparib have been approved for the treatment of cancers in some settings independent of BRCA status (Rose et al., 2020).
PARPi-induced PARP1 “trapping.”
PARP1 is recruited to DNA breaks to initiate the repair process. PARP1 automodification, which plays an important role in promoting the release of PARP1 from DNA repair sites and preventing the retention of chromatin-associated PARP1, is required for the resolution of DNA damage and restoration of cellular function (Prokhorova et al., 2021a). PARPi inhibit PARP1 automodification and block the dissociation of PARP1 from damaged DNA, thereby establishing a stable complex of PARP1 with DNA in a process called PARP1 “trapping” (Lord and Ashworth, 2017; Murai et al., 2012; Zandarashvili et al., 2020). Trapped PARP1 is highly toxic to the cell. The PARPi–PARP1–DNA complexes that form at lesions inhibit the catalytic cycle of PARP1 in DNA damage signaling by preventing the release of PARP1. PARP1 trapping leads to the accumulation of unrepaired SSBs, which impair progression of DNA replication forks and ultimately cause the formation of DNA DSBs (Murai et al., 2012; Murai and Pommier, 2019) (Figure 6B). The methods to detect PARP1 trapping, however, are imprecise (e.g., centrifugation-based chromatin retention assays) and the molecular mechanisms of PARP1 trapping are not clear.
A recent study has illustrated how the relationships between PARP1 trapping and the efficacy of PARPi are not always straightforward. Zandarashvili et al. have explored the mechanisms of PARP1 retention at DNA breaks by different PARP inhibitors using hydrogen/deuterium exchange mass spectrometry (HXMS) combined with X-ray structures (Zandarashvili et al., 2020). Their work showed that the allosteric regulatory helical domain (HD) in PARP1 is subject to different degrees of stabilization/destabilization by different PARPi bound in the NAD+-binding site, resulting in different affinities of PARP1 for DNA breaks (Zandarashvili et al., 2020). Thus, different PARPi can drive distinct allosteric changes in PARP1 that modulate PARP1 trapping, ultimately determining PARPi potency. To drive this point home, they showed that structural modifications of Veliparib, which favors PARP1 release by stabilizing the HD, convert it to an allosteric pro-retention compound that promotes increased trapping and cytotoxicity (Zandarashvili et al., 2020). Further studies are needed to fully understand the molecular mechanisms and clinical significance of PARPi-mediated PARP1 trapping.
Recent studies suggest that inhibition of PARP trapping activity can confer resistance to PARPi (Gogola et al., 2019; Pettitt et al., 2018). In this regard, a PARP1 mutation (R591C) commonly found in samples from PARPi-resistant tumors, is associated with diminished trapping of PARP1 on DNA and PARPi resistance. This finding suggests that PARP1 mutations can decrease DNA trapping and induce PARPi resistance. Moreover, loss of PARG in PARPi-treated cells may promote an accumulation of PAR, thereby reducing PARP1 trapping on DNA, rescuing PARP1-dependent DNA damage signaling, and leading to PARPi resistance (Gogola et al., 2019).
PARPi-induced dysregulation of replication forks.
Early studies provided initial links between PARP1 and DNA replication. For example, PARP activity in newly replicated chromatin was observed to be two to three times greater than in unreplicated chromatin (Anachkova et al., 1989), a result that was recently confirmed (Hanzlikova et al., 2018). Inhibition of PARP1 is toxic in BRCA1- and BRCA2-tumors, which may be due to impaired replication fork protection (Schlacher et al., 2012) and the persistence of unrepaired collapsed forks (Bryant et al., 2005; Farmer et al., 2005). But, conclusions about the specific role and effects of PARP1 in DNA replication have been mixed, with some reports suggesting little to no effect (Anachkova et al., 1989). Recent studies have pointed an important role for PARP1 in promoting replication fork reversal and reducing replication fork speed upon replication stress (Berti et al., 2013; Ray Chaudhuri et al., 2012) (Figure 3C). In this regard, PARPi have been shown to increase replication fork speed, leading to replication stress and genome instability (Genois et al., 2021; Maya-Mendoza et al., 2018; Sugimura et al., 2008) (Figure 6C). However, conflicting results (Bryant et al., 2009; Min et al., 2013; Yang et al., 2004) indicate that further investigation will be required to clearly delineate the therapeutic mechanisms of PARPi with respect to DNA replication.
Emerging evidence suggests that restoration of HR competency may contribute to PARPi resistance. Combinations of PARPi and inhibitors of cell cycle checkpoint proteins (e.g., ATR, CHK1, and WEE1) may provide a therapeutic opportunity to enhance the efficacy of PARPi and circumvent resistance (Haynes et al., 2018; Min et al., 2013). A proposed model is that inhibition of ATR, CHK1 and WEE1 in PARPi-treated cells will reverse G2 arrest and subsequent DNA repair (Haynes et al., 2018). Moreover, since sensitivity to PARGi and PARPi are mutually exclusive in some cell contexts, they may complement each other therapeutically (Pillay et al., 2019). In this regard, PARGi have been explored as an alternate therapeutic option to PARPi and they may be useful in the context of PARPi resistance. PARGi have demonstrated efficacy in preclinical models by sensitizing cells to radiation-induced DNA damage, suppressing replication fork progression, promoting replication catastrophe, and inhibiting cancer cell survival (Houl et al., 2019; Pillay et al., 2019). Interestingly, a recent study has shown that unrestrained PARylation, resulting from depletion of both PARG and ARH3, is highly toxic to cells by disrupting the chromatin modifications (e.g., H3K9ac and H3K27ac) and transcription. In this regard, ARH3 deficiency sensitizes cancer cells to PARGi and renders cancer cells resistant to PARPi (Prokhorova et al., 2021a).
PARPi sensitivity and chromatin regulation.
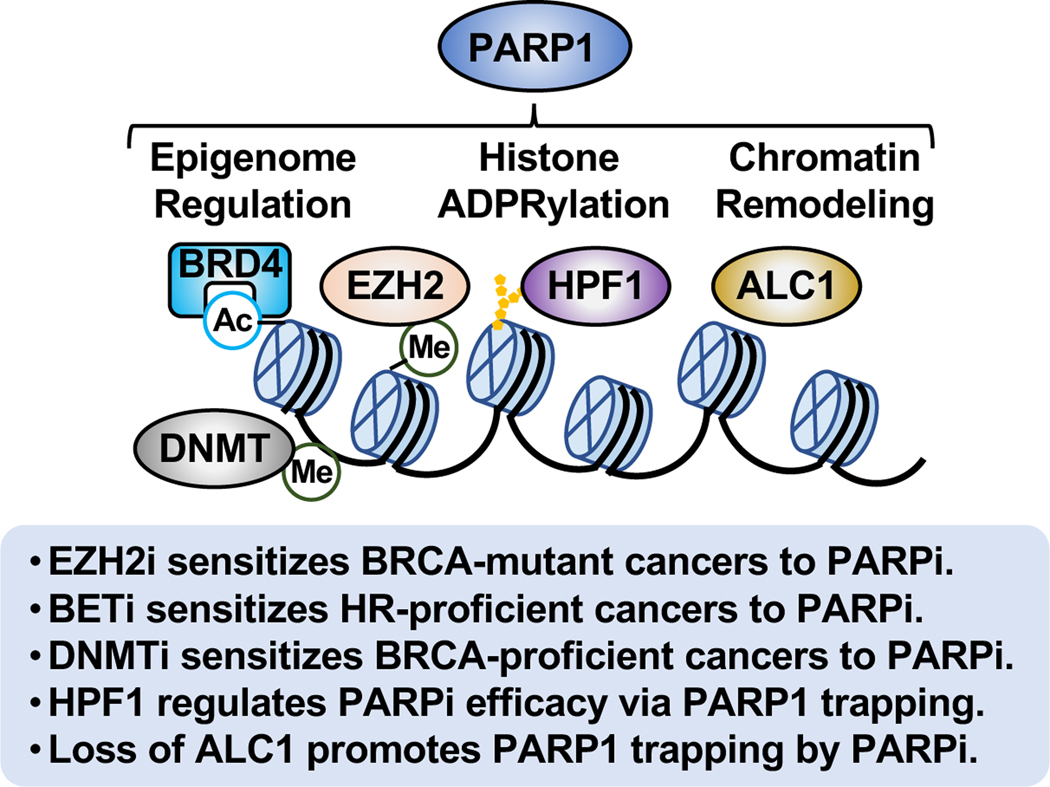
PARP1 plays a number of distinct roles in chromatin regulation, which may impact the sensitivity and efficacy of PARPi. This may involve effects on epigenomic modifications, including histone ADPRylation, as well as chromatin remodeling (Figure 7).
Figure 7. PARPi sensitivity and chromatin regulation.
The multifaceted roles of PARP1 in chromatin regulation impact the sensitivity and efficacy of PARPi. This may involve effects of PARP1 on epigenome regulation, histone ADPRylation, and chromatin remodeling.
PARP1-mediated ADPRylation is functionally linked to other epigenomic modifications, such as methylation, acetylation, and phosphorylation. The histone methyltransferase EZH2 mediates the responses of breast and ovarian cancer cells to PARPi by regulating histone methylation, although the underlying mechanisms have not been clearly resolved. Upon DNA damage, PARP1 PARylates EZH2, then induces dissociation of the PRC2 complex and subsequent EZH2 degradation, thus impairing EZH2-mediated histone H3 trimethylation and gene silencing (Yamaguchi et al., 2018). EZH2 inhibitor sensitizes BRCA-mutant breast cancer cells to PARPi (Yamaguchi et al., 2018). It also sensitizes HR-proficient ovarian cancer cells with high levels of CARM1 to PARPi by increasing NHEJ activity and chromosomal instabilities (Karakashev et al., 2020). Consistent with these findings, inhibition of MUC-1, which is responsible for the nuclear localization of EZH2 and BMI1 to form complexes with PARP1, downregulates EZH2-mediated H3K27 trimethylation and BMI1-mediated H2A ubiquitylation, thus synergistically sensitizing both BRCA-mutant and -proficient triple negative breast cancer cells to PARPi (Yamamoto et al., 2019). Conversely, EZH2 has been reported to localize at stalled replication forks and promote H3K27 trimethylation, mediating replication fork degradation by recruiting the nuclease MUS81. Inhibition of EZH2 has been shown to promote cancer cells to acquire PARPi resistance by promoting replication fork stabilization in BRCA-mutant cancer cells (Rondinelli et al., 2017).
Other inhibitors of chromatin and genomic modulators also impact PARPi sensitivity. For example, BET inhibitors, which block interactions between BET proteins (e.g., BRD4) and acetylated histones and transcription factors, sensitizes HR-proficient cancers to PARPi by impairing transcription of the BRCA1 and RAD51 genes, which encode proteins essential for HR (Yang et al., 2017). Furthermore, dysregulation of DNA methyltransferases (DNMTs), which causes gene expression changes associated with DNA methylation abnormalities, has been linked to cancers. Combination of DNMT inhibitor and PARPi enhances PARPi trapping, thus sensitizing breast and ovarian cancer cells to PARPi independent of BRCA mutations (Muvarak et al., 2016; Pulliam et al., 2018). Undoubtedly, additional links between the enzymes that control epigenomic modifications and the sensitivity of PARPi are waiting to be discovered.
Emerging evidence indicates that PARPi sensitivity can be mediated through or modulated by histone ADPRylation. Histone PARylation factor HPF1 is essential for PARP1- and PARP2-mediated histone PARylation in response to DNA damage (Bartlett et al., 2018; Bonfiglio et al., 2017; Gibbs-Seymour et al., 2016; Hendriks et al., 2021; Langelier et al., 2021; Palazzo et al., 2018). Moreover, HPF1-dependent PARP1 automodification counters PARP1 trapping and contributes to PARPi tolerance (Prokhorova et al., 2021b). As such, loss of HPF1 sensitizes cancer cells to DNA damaging agents and PARPi (Gibbs-Seymour et al., 2016; Prokhorova et al., 2021b). HPF1 mediates increased affinity of some PARPi (such as Olaparib) for PARP1 and the PARP1/nucleosome complex, suggesting a role for HPF1 in regulating the efficacy of PARP inhibitors through PARP1 trapping (Rudolph et al., 2021). However, HPF1-mediated affinity of different PARPi for PARP1 does not directly reflect the mechanisms of PARP1 trapping and cytotoxicity induced by each PARPi in vivo (Rudolph et al., 2021). Moreover, PARP1 allostery may play a critical role in PARPi-induced PARP1 trapping and may also be involved in HPF1mediated affinity of PARP1 for different PARPi (Zandarashvili et al., 2020). Further investigation of HPF1 as a predictive biomarker for response to PARPi are needed. Interestingly, many oncohistone mutations in a broad array of cancers occur at Asp, Glu, and Ser residues (Nacev et al., 2019), which may be sites of PARP1-mediated ADPRylation. Mutation of these sites in cancers suggests an attempt by cancer cells to circumvent regulatory mechanisms (e.g., ADPRylation) operating through those sites. This possibility requires further exploration.
PARPi sensitivity may also be affected by the action of ATP-dependent chromatin remodeling enzyme, such as ALC1. The chromatin remodeler ALC1 acts as an oncogene, which correlates with poor prognosis for multiple cancer types (Cheng et al., 2013). ALC1 modulates PARPi sensitivity in both HR-deficient and proficient cells (Hewitt et al., 2021; Juhasz et al., 2020; Verma et al., 2021). In response to DNA damage, ALC1 is recruited to DNA lesions by activated PARP1 for chromatin relaxation (Ahel et al., 2009; Gottschalk et al., 2009; Singh et al., 2017). It is also responsible for the timely removal of inactive PARP1 from DNA lesions (Juhasz et al., 2020). Loss of ALC1 reduces chromatin accessibility, increases replication-associated DNA damage, promotes PARPi-induced PARP1 trapping, and interferes with the binding of both HR and NHEJ repair factors to the sites of DNA damage, leading to cell death (Hewitt et al., 2021; Juhasz et al., 2020; Verma et al., 2021). Overexpression of ALC1, observed in many cancer types, may reduce PARPi sensitivity in cancer cells (Juhasz et al., 2020). Development of new therapeutic agents targeting ALC1 that can synergize with the cytotoxic potential of PARPi may be useful in these cancers.
CONCLUSIONS AND PERSPECTIVES
Our understanding of the diverse roles of PARP1 (and PARP2) in a variety of biological processes has been greatly expanded by recent studies that have ventured beyond the traditional roles of PARP1 in DNA damage responses. These studies have begun to reveal PARP1-mediated molecular mechanisms in chromatin regulation, gene expression, ribosome biogenesis, and RNA biology, opening a broader array of mechanisms that may mediated therapeutic outcomes (Figure 4). New concepts underlying PARP1-mediated molecular functions, such as PAR-dependent posttranslational modifications, PAR-mediated condensate formation, and “ADPR spray,” which help to explain the multifaceted roles of PARP1 in physiology and disease (Figure 5).
Knowledge of the biological roles of PARP1 in DNA repair pathways led to the development of PARPi for the treatment of various cancers, especially breast and ovarian cancers with BRCA1/2 mutations or HR-mediated DNA repair deficiencies. But, recent studies suggest potential clinical benefits of PARPi in patients through mechanisms beyond DNA repair. However, these mechanisms remain elusive, limiting our understanding of potential targets for PARPi, predictive biomarkers for PARPi efficacy, and molecular mechanisms underlying PARPi resistance. Recent advances have shown that PARPi-induced dysregulation of DNA replication forks contributes to the therapeutic actions of PARPi (Figure 6). Additional studies exploring the diverse functions of PARP1 in distinct biological processes beyond DNA repair processes will be important to strengthen the rationale for extending the use of PARPi in a broader array of diseases (Figure 4).
In addition to understanding the therapeutic mechanisms of PARPi, elucidating the mechanisms that underlie resistance to PARPi will be important to ensure the therapeutic potential of PARPi. Activity-altering mutations in PARP1, as well as mutation of specific sites of ADPRylation site on PARP1 substrates, might have potential as predictive biomarkers of tumor responses to PARPi in different cancer patients. The diverse roles of PARP1 in chromatin regulation have suggested new strategies for using PARPi in combination with other epigenome-regulating drugs to induce significant tumor regression (Figure 7). Further studies of the molecular mechanisms of action of PARPi are needed, along with validation of additional biomarkers for the prediction of PARPi efficacy.
Given the remaining expansive unexplored knowledge space for the nuclear PARPs, we expect a similarly focused review a decade from now to include a plethora of new and unexpected concepts and results.
ACKNOWLEDGEMENTS
The authors would like to thank members of the Kraus lab for critical comments and review of this manuscript.
FUNDING
The PARP- and ADPRylation-related research in the Kraus lab is supported by grants from the NIH/NIDDK (R01 DK069710), NIH/NCI (R01 CA229487, R01 CA251943), U.S. DOD Ovarian Cancer Research Program (OCRP) (W81XWH-20-OCRP-IIRA), and the Cancer Prevention and Research Institute of Texas (CPRIT) (RP190236), as well funds from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment, to W.L.K.
DECLARATION OF INTERESTS
W.L.K. is a founder of Ribon Therapeutics, Inc. and a founder, consultant, and SAB member of ARase Therapeutics, Inc. He is also a co-holder of U.S. Patent 9,599,606 covering a set of ADP-ribose detection reagents, which have been licensed to and is sold by EMD Millipore. D.H. has nothing to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. (2009). Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243. 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemasova EE, and Lavrik OI (2019). Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res 47, 3811–3827. 10.1093/nar/gkz120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M., Neelsen KJ., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask MD., Streicher W., Jungmichel S., Nielsen ML., and Luka J. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 6, 8088. 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, and de Murcia G. (1999). PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274, 17860–17868. 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, and de Murcia G. (2004). The PARP superfamily. Bioessays 26, 882893. 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Anachkova B, Russev G, and Poirier GG (1989). DNA replication and poly(ADP-ribosyl)ation of chromatin. Cytobios 58, 19–28. [PubMed] [Google Scholar]
- Audebert M, Salles B, and Calsou P. (2004). Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem 279, 55117–55126. 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- Bacic L, Gaullier G, Sabantsev A, Lehmann LC, Brackmann K, Dimakou D, Halic M, Hewitt G, Boulton SJ, and Deindl S. (2021). Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome. Elife 10. 10.7554/eLife.71420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett E, Bonfiglio JJ, Prokhorova E, Colby T, Zobel F, Ahel I, and Matic I. (2018). Interplay of histone marks with serine ADP-ribosylation. Cell Rep 24, 3488–3502 e3485. 10.1016/j.celrep.2018.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Boehler C, Guirouilh Barbat J, Bonnet ME, Illuzzi G, Ronde P, Gauthier LR, Magroun N, Rajendran A, Lopez BS, et al. (2014). PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res 42, 5616–5632. 10.1093/nar/gku174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousova EA, Ishchenko capital A C, and Lavrik OI (2018). DNA is a new target of PARP3. Sci Rep 8, 4176. 10.1038/s41598-018-22673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, et al. (2013). Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20, 347–354. 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Suskiewicz MJ, Ahel I, and Halic M. (2020). Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin. Nature 585, 609–613. 10.1038/s41586-020-2725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing C., Mandemaker IK., Gonzalez-Leal C., Preisser J., Schomburg A., and Ladurner AG. (2020). The oncogenic helicase ALC1 regulates PARP inhibitor potency by trapping PARP2 at DNA breaks. Mol Cell 80, 862–875 e866. 10.1016/j.molcel.2020.10.009. [DOI] [PubMed] [Google Scholar]
- Boamah EK, Kotova E, Garabedian M, Jarnik M, and Tulin AV (2012). Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet 8, e1002442. 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, SanglierCianferani S, Smith S, Schreiber V, Boussin F, and Dantzer F. (2011). Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A 108, 2783–2788. 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, and Matic I. (2017). Serine ADP-ribosylation depends on HPF1. Mol Cell 65, 932–940 e936. 10.1016/j.molcel.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1988). At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J 7, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustel J, Muramoto T, Fumimoto K, Ellins J, Pears CJ, and Lakin ND (2022). Linking DNA repair and cell cycle progression through serine ADP-ribosylation of histones. Nat Commun 13, 185. 10.1038/s41467-021-27867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, and Helleday T. (2009). PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28, 2601–2615. 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, and Helleday T. (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917. 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Buch-Larsen SC, Hendriks IA, Lodge JM, Rykaer M, Furtwangler B, Shishkova E, Westphall MS, Coon JJ, and Nielsen ML (2020). Mapping physiological ADP-ribosylation using activated ion electron transfer dissociation. Cell Rep 32, 108176. 10.1016/j.celrep.2020.108176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butepage M, Preisinger C, von Kriegsheim A, Scheufen A, Lausberg E, Li J, Kappes F, Feederle R, Ernst S, Eckei L, et al. (2018). Nucleolar-nucleoplasmic shuttling of TARG1 and its control by DNA damage-induced poly-ADP-ribosylation and by nucleolar transcription. Sci Rep 8, 6748. 10.1038/s41598-018-25137-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, and Shall S. (1996). XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res 24, 4387–4394. 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catara G, Grimaldi G, Schembri L, Spano D, Turacchio G, Lo Monte M, Beccari AR, Valente C, and Corda D. (2017). PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci Rep 7, 14035. 10.1038/s41598-017-14156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, and D’Andrea AD (2016). Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26, 52–64. 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon RM, Morel D, Eychenne T, Colmet-Daage L, Bajrami I, Dorvault N, Garrido M, Meisenberg C, Lamb A, Ngo C, et al. (2021). PBRM1 deficiency confers synthetic lethality to DNA repair inhibitors in cancer. Cancer Res 81, 2888–2902. 10.1158/00085472.CAN-21-0628. [DOI] [PubMed] [Google Scholar]
- Chambon P, Weill JD, Doly J, Strosser MT, and Mandel P. (1966). On the formation of a novel adenylic compound by enzymatic extracts of liver nuclei. Biochem Biophys Res Commun 26, 638–643. [Google Scholar]
- Chambon P, Weill JD, and Mandel P. (1963). Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 11, 39–43. 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, and Gamble MJ (2014). MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat Struct Mol Biol 21, 981–989. 10.1038/nsmb.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Bian C, Wang X, Liu X, Ahmad Kassab M, Yu Y, and Yu X. (2021). ADPribosylation of histone variant H2AX promotes base excision repair. EMBO J 40, e104542. 10.15252/embj.2020104542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Kassab MA, Dantzer F, and Yu X. (2018). PARP2 mediates branched poly ADPribosylation in response to DNA damage. Nat Commun 9, 3233. 10.1038/s41467-018-05588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Su Y, and Xu F. (2013). CHD1L: a novel oncogene. Mol Cancer 12, 170. 10.1186/1476-4598-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, and Poirier GG (1999). Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342 ( Pt 2), 249–268. [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, and de Murcia G. (2006). Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol 409, 493–510. 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, and Poirier GG (1986). Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem 261, 7011–7017. [PubMed] [Google Scholar]
- Desnoyers S, Kaufmann SH, and Poirier GG (1996). Alteration of the nucleolar localization of poly(ADP-ribose) polymerase upon treatment with transcription inhibitors. Exp Cell Res 227, 146–153. 10.1006/excr.1996.0259. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, and Manley JL (2013). PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell 49, 7–17. 10.1016/j.molcel.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, et al. (2019). PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 29, 233–247. 10.1038/s41422-019-0141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AD, Marecki JC, Byrd AK, Gao J, and Raney KD (2020). G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 10.1093/nar/gkaa1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy SF, Masutani M, Suzuki H, and Caldecott KW (2003). A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res 31, 5526–5533. 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleazer R, and Fondufe-Mittendorf YN (2020). The multifaceted role of PARP1 in RNA biogenesis. Wiley Interdiscip Rev RNA, e12607. 10.1002/wrna.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Videler H, Yang JC, Cole PT, Gruszka D, Veprintsev D, and Neuhaus D. (2011). The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J Mol Biol 407, 149–170. 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. (2005). Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921. 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fischbach A, Kruger A, Hampp S, Assmann G, Rank L, Hufnagel M, Stockl MT, Fischer JMF, Veith S, Rossatti P, et al. (2018). The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res 46, 804–822. 10.1093/nar/gkx1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquin A, Guirouilh-Barbat J, Lopez B, Hall J, Amor-Gueret M, and Pennaneach V. (2017). PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucleic Acids Res 45, 1232512339. 10.1093/nar/gkx881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell KM, and Kraus WL (2009). PARP inhibitors and the treatment of breast cancer: beyond BRCA1/2? Breast Cancer Res 11, 111. 10.1186/bcr2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadad SS, Camacho CV, Malladi V, Hutti CR, Nagari A, and Kraus WL (2021). PARP1 regulates estrogen-dependent gene expression in estrogen receptor alpha-positive breast cancer cells. Mol Cancer Res. 10.1158/1541-7786.MCR-21-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genois MM, Gagne JP, Yasuhara T, Jackson J, Saxena S, Langelier MF, Ahel I, Bedford MT, Pascal JM, Vindigni A, et al. (2021). CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol Cell 81, 784–800 e788. 10.1016/j.molcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs-Seymour I, Fontana P, Rack JGM, and Ahel I. (2016). HPF1/C4orf27 is a PARP-1interacting protein that regulates PARP-1 ADP-ribosylation activity. Mol Cell 62, 432–442. 10.1016/j.molcel.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, and Kraus WL (2012). New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13, 411–424. 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, and Kraus WL (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353, 45–50. 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogola E, Duarte AA, de Ruiter JR, Wiegant WW, Schmid JA, de Bruijn R, James DI, Llobet SG, Vis DJ, Annunziato S, et al. (2019). Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 35, 950–952. 10.1016/j.ccell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, and Conaway RC (2009). Poly(ADPribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A 106, 13770–13774. 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groslambert J, Prokhorova E, and Ahel I. (2021). ADP-ribosylation of DNA and RNA. DNA Repair (Amst) 105, 103144. 10.1016/j.dnarep.2021.103144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy GJ, Polo LM, Zeng Z, Rulten SL, Hoch NC, Paomephan P, Xu Y, Sweet SM, Thorne AW, Oliver AW, et al. (2016). PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B(Glu2). Nat Commun 7, 12404. 10.1038/ncomms12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, and Caiafa P. (2008). CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem 283, 21873–21880. 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, and Santoro R. (2012). Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell 45, 790–800. 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, and Sicheri F. (2011). Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell 147, 1340–1354. 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Gupte R, Liu Z, and Kraus WL (2017). PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 31, 101–126. 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R, Nandu T, and Kraus WL (2021). Nuclear ADP-ribosylation drives IFNgamma-dependent STAT1alpha enhancer formation in macrophages. Nat Commun 12, 3931. 10.1038/s41467-021-24225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, and Poirier GG (2008). PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283, 1197–1208. 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hananya N, Daley SK, Bagert JD, and Muir TW (2021). Synthesis of ADP-ribosylated histones reveals site-specific impacts on chromatin structure and function. J Am Chem Soc 143, 10847–10852. 10.1021/jacs.1c05429. [DOI] [PubMed] [Google Scholar]
- Hanzlikova H, Gittens W, Krejcikova K, Zeng Z, and Caldecott KW (2017). Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res 45, 2546–2557. 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzlikova H, Kalasova I, Demin AA, Pennicott LE, Cihlarova Z, and Caldecott KW (2018). The importance of poly(ADP-Ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol Cell 71, 319–331 e313. 10.1016/j.molcel.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa S, Fujimura S, Shimizu Y, and Sugimura R. (1967). The polymerization of adenosine 5’-diphosphate-ribose moiety of NAD by nuclear enzyme. II. Properties of the reaction product. Biochim Biophys Acta 149, 369–376. [PubMed] [Google Scholar]
- Haynes B, Murai J, and Lee JM (2018). Restored replication fork stabilization, a mechanism of PARP inhibitor resistance, can be overcome by cell cycle checkpoint inhibition. Cancer Treat Rev 71, 1–7. 10.1016/j.ctrv.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Buch-Larsen SC, Prokhorova E, Elsborg JD, Rebak A, Zhu K, Ahel D, Lukas C, Ahel I, and Nielsen ML (2021). The regulatory landscape of the human HPF1- and ARH3-dependent ADP-ribosylome. Nat Commun 12, 5893. 10.1038/s41467-021-26172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Larsen SC, and Nielsen ML (2019). An advanced strategy for comprehensive profiling of ADP-ribosylation sites using mass spectrometry-based proteomics. Mol Cell Proteomics 18, 1010–1026. 10.1074/mcp.TIR119.001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G., Borel V., Segura-Bayona S., Takaki T., Ruis P., Bellelli R., Lehmann LC., Sommerova L., Vancevska A., Tomas-Loba A., et al. (2021). Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol Cell 81, 767–783 e711. 10.1016/j.molcel.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO (2011). ADP-ribosylation of histones by ARTD1: an additional module of the histone code? FEBS Lett 585, 1595–1599. 10.1016/j.febslet.2011.03.031. [DOI] [PubMed] [Google Scholar]
- Houl JH, Ye Z, Brosey CA, Balapiti-Modarage LPF, Namjoshi S, Bacolla A, Laverty D, Walker BL, Pourfarjam Y, Warden LS, et al. (2019). Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death. Nat Commun 10, 5654. 10.1038/s41467-019-13508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Wu W, Li Y, Lin L, Chen D, Yan H, Xiao X, Chen H, Chen Z, Zhang Y, et al. (2019). Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep 20. 10.15252/embr.201846166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Petit SA, Ficarro SB, Toomire KJ, Xie A, Lim E, Cao SA, Park E, Eck MJ, Scully R, et al. (2014). PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov 4, 1430–1447. 10.1158/21598290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huambachano O, Herrera F, Rancourt A, and Satoh MS (2011). Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J Biol Chem 286, 7149–7160. 10.1074/jbc.M110.175190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Camacho CV, Setlem R, Ryu KW, Parameswaran B, Gupta RK, and Kraus WL (2020a). Functional interplay between histone H2B ADP-ribosylation and phosphorylation controls adipogenesis. Mol Cell 79, 934–949 e914. 10.1016/j.molcel.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Kim DS, and Kraus WL (2020b). Specific binding of snoRNAs to PARP-1 promotes NAD(+)-dependent catalytic activation. Biochemistry 59, 1559–1564. 10.1021/acs.biochem.0c00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huletsky A., Niedergang C., Frechette A., Aubin R., Gaudreau A., and Poirier GG. (1985). Sequential ADP-ribosylation pattern of nucleosomal histones. ADP-ribosylation of nucleosomal histones. Eur J Biochem 146, 277–285. 10.1111/j.1432-1033.1985.tb08650.x. [DOI] [PubMed] [Google Scholar]
- Isabelle M, Gagne JP, Gallouzi IE, and Poirier GG (2012). Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci 125, 4555–4566. 10.1242/jcs.106963. [DOI] [PubMed] [Google Scholar]
- Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, Hendzel MJ, and Poirier GG (2010). Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci 8, 22. 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, and Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16, 167–177. 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz S, Smith R, Schauer T, Spekhardt D, Mamar H, Zentout S, Chapuis C, Huet S, and Timinszky G. (2020). The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci Adv 6. 10.1126/sciadv.abb8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, and Nielsen ML (2013). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell 52, 272–285. 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- Karakashev S, Fukumoto T, Zhao B, Lin J, Wu S, Fatkhutdinov N, Park PH, Semenova G, Jean S, Cadungog MG, et al. (2020). EZH2 inhibition sensitizes CARM1-high, homologous recombination proficient ovarian cancers to PARP inhibition. Cancer Cell 37, 157167 e156. 10.1016/j.ccell.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch KR, Langelier MF, Pascal JM, and Garcia BA (2017). The nucleosomal surface is the main target of histone ADP-ribosylation in response to DNA damage. Mol Biosyst 13, 26602671. 10.1039/c7mb00498b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian X, Ba X, Boldogh I, and Zeng X. (2017). PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nat Commun 8, 14632. 10.1038/ncomms14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, and Altmeyer M. (2019). Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J 38, e101379. 10.15252/embj.2018101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Camacho CV, and Kraus WL (2021). Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp Mol Med 53, 42–51. 10.1038/s12276021-00557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS., Camacho CV., Nagari A., Malladi VS., Challa S., and Kraus WL. (2019). Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol Cell 75, 1270–1285 e1214. 10.1016/j.molcel.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Challa S, Jones A, and Kraus WL (2020). PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev 34, 302–320. 10.1101/gad.334433.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, and Kraus WL (2004). NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814. 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kliza KW, Liu Q, Roosenboom LWM, Jansen P, Filippov DV, and Vermeulen M. (2021). Reading ADP-ribosylation signaling using chemical biology and interaction proteomics. Mol Cell 81, 4552–4567 e4558. 10.1016/j.molcel.2021.08.037. [DOI] [PubMed] [Google Scholar]
- Koczor CA, Saville KM, Andrews JF, Clark J, Fang Q, Li J, Al-Rahahleh RQ, Ibrahim M, McClellan S, Makarov MV, et al. (2021). Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD(+)/SIRT6 axis. Cell Rep 37, 109917. 10.1016/j.celrep.2021.109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, and Tulin AV (2011). Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc Natl Acad Sci U S A 108, 6205–6210. 10.1073/pnas.1019644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, and Lis JT (2003). PARP goes transcription. Cell 113, 677–683. 10.1016/s00928674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, and Kraus WL (2008). Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 319, 819–821. 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, and Kraus WL (2010a). The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39, 8–24. 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, and Kraus WL (2010b). PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39, 736–749. 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhar R, Sanchez A, Perren J, Gong F, Corujo D, Medina F, Devanathan SK, Xhemalce B, Matouschek A, Buschbeck M, et al. (2021). Poly(ADP-ribose) binding and macroH2A mediate recruitment and functions of KDM5A at DNA lesions. J Cell Biol 220. 10.1083/jcb.202006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Billur R, Sverzhinsky A, Black BE, and Pascal JM (2021). HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat Commun 12, 6675. 10.1038/s41467-021-27043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, and Pascal JM (2011). Crystal structures of poly(ADPribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem 286, 10690–10701. 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, and Pascal JM (2012). Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732. 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Riccio AA, and Pascal JM (2014). PARP-2 and PARP-3 are selectively activated by 5’ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res 42, 7762–7775. 10.1093/nar/gku474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Ruhl DD, Planck JL, Kraus WL, and Pascal JM (2010). The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem 285, 18877–18887. 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Zandarashvili L, Aguiar PM, Black BE, and Pascal JM (2018). NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun 9, 844. 10.1038/s41467-018-03234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SC, Hendriks IA, Lyon D, Jensen LJ, and Nielsen ML (2018). Systems-wide analysis of serine ADP-ribosylation reveals widespread occurrence and site-specific overlap with phosphorylation. Cell Rep 24, 2493–2505 e2494. 10.1016/j.celrep.2018.07.083. [DOI] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott CL, Meier W, Shapira-Frommer R, Safra T, et al. (2014). Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 15, 852–861. 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- Leger K, Bar D, Savic N, Santoro R, and Hottiger MO (2014). ARTD2 activity is stimulated by RNA. Nucleic Acids Res 42, 5072–5082. 10.1093/nar/gku131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Shall S, and Whish WJ (1974). The relationship between cell growth, macromolecular synthesis and poly ADP-ribose polymerase in lymphoid cells. Exp Cell Res 83, 63–72. 10.1016/0014-4827(74)90688-0. [DOI] [PubMed] [Google Scholar]
- Leidecker O, Bonfiglio JJ, Colby T, Zhang Q, Atanassov I, Zaja R, Palazzo L, Stockum A, Ahel I, and Matic I. (2016). Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat Chem Biol 12, 998–1000. 10.1038/nchembio.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, and Chang P. (2011). Poly(ADPribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42, 489499. 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL (2020). Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol 30, 370–383. 10.1016/j.tcb.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, et al. (2011). Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell 147, 1324–1339. 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lheureux S, Lai Z, Dougherty BA, Runswick S, Hodgson DR, Timms KM, Lanchbury JS, Kaye S, Gourley C, Bowtell D, et al. (2017). Long-term responders on Olaparib maintenance in high-grade serous ovarian cancer: Clinical and molecular characterization. Clin Cancer Res 23, 4086–4094. 10.1158/1078-0432.CCR-16-2615. [DOI] [PubMed] [Google Scholar]
- Li M, and Yu X. (2013). Function of BRCA1 in the DNA damage response is mediated by ADPribosylation. Cancer Cell 23, 693–704. 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszczak G, Diehl KL, Dann GP, and Muir TW (2018). Acetylation blocks DNA damageinduced chromatin ADP-ribosylation. Nat Chem Biol 14, 837–840. 10.1038/s41589-018-0097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Kraus WL (2017). Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol Cell 65, 589–603 e589. 10.1016/j.molcel.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, and Soldatenkov VA (2005). Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem 280, 17076–17083. 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- Lord CJ, and Ashworth A. (2016). BRCAness revisited. Nat Rev Cancer 16, 110–120. 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- Lord CJ, and Ashworth A. (2017). PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158. 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, and van Attikum H. (2016). PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Nonhomologous End-Joining. Mol Cell 61, 547–562. 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, and Kraus WL (2012). On PAR with PARP: cellular stress signaling through poly(ADPribose) and PARP-1. Genes Dev 26, 417–432. 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupey-Green LN, Caruso LB, Madzo J, Martin KA, Tan Y, Hulse M, and Tempera I. (2018). PARP1 Stabilizes CTCF Binding and Chromatin Structure To Maintain Epstein-Barr Virus Latency Type. J Virol 92. 10.1128/JVI.00755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Ahel I, Altmeyer M, Ashworth A, Bai P, Chang P, Cohen M, Corda D, Dantzer F, Daugherty MD, et al. (2021). ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 10.1111/febs.16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour WY, Rhein T, and Dahm-Daphi J. (2010). The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res 38, 6065–6077. 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martire S, and Banaszynski LA (2020). The roles of histone variants in fine-tuning chromatin organization and function. Nat Rev Mol Cell Biol 21, 522–541. 10.1038/s41580-020-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, and de Murcia G. (1998). XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol 18, 3563–3571. 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Al-Tinawi QMH, Rouchka EC, and Fondufe-Mittendorf YN (2019a). Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and co-transcriptional splicing. Epigenetics Chromatin 12, 15. 10.1186/s13072-0190261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Mathbout LF, and Fondufe-Mittendorf YN (2019b). PARP1 is a versatile factor in the regulation of mRNA stability and decay. Sci Rep 9, 3722. 10.1038/s41598-01939969-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, and Bartek J. (2018). High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284. 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, and Bonini NM (2018a). Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol Cell 71, 703–717 e709. 10.1016/j.molcel.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L, Gomes E, Guo L, Shorter J, and Bonini NM (2018b). Poly(ADP-ribose) engages the TDP-43 nuclear-localization sequence to regulate granulo-filamentous aggregation. Biochemistry 57, 6923–6926. 10.1021/acs.biochem.8b00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, and Schreiber V. (2005). PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci 118, 211222. 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, and Ahel I. (2011). DNA repair factor APLF is a histone chaperone. Mol Cell 41, 46–55. 10.1016/j.molcel.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikishvili M., Chariker JH., Rouchka EC., and Fondufe-Mittendorf YN. (2017). Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov 3, 17043. 10.1038/celldisc.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W, Bruhn C, Grigaravicius P, Zhou ZW, Li F, Kruger A, Siddeek B, Greulich KO, Popp O, Meisezahl C, et al. (2013). Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat Commun 4, 2993. 10.1038/ncomms3993. [DOI] [PubMed] [Google Scholar]
- Mohapatra J, Tashiro K, Beckner RL, Sierra J, Kilgore JA, Williams NS, and Liszczak G. (2021). Serine ADP-ribosylation marks nucleosomes for ALC1-dependent chromatin remodeling. Elife 10. 10.7554/eLife.71502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, and Ahel I. (2017). Reversible mono-ADP-ribosylation of DNA breaks. FEBS J 284, 4002–4016. 10.1111/febs.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, Bartlett E, Mikolcevic P, Kirby IT, Rack JGM, Mikoc A, Cohen MS, and Ahel I. (2019). Reversible ADP-ribosylation of RNA. Nucleic Acids Res 47, 5658–5669. 10.1093/nar/gkz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, and Pommier Y. (2012). Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 72, 5588–5599. 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, and Pommier Y. (2019). PARP trapping beyond homologous recombination and platinum sensitivity in cancers. Annual Review of Cancer Biology 3, 131–150. 10.1146/annurevcancerbio-030518-055914. [DOI] [Google Scholar]
- Muvarak NE, Chowdhury K, Xia L, Robert C, Choi EY, Cai Y, Bellani M, Zou Y, Singh ZN, Duong VH, et al. (2016). Enhancing the Cytotoxic Effects of PARP Inhibitors with DNA Demethylating Agents - A Potential Therapy for Cancer. Cancer Cell 30, 637–650. 10.1016/j.ccell.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, Kundra R, Schultz N, Muir TW, and Allis CD (2019). The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567, 473–478. 10.1038/s41586-019-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M., Pal A., Goswami A., Lojewski X., Japtok J., Vehlow A., Naujock M., Gunther R., Jin M., Stanslowsky N., et al. (2018). Impaired DNA damage response signaling by FUSNLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun 9, 335. 10.1038/s41467-017-02299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, and Yasui A. (2003). Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23, 3974–3981. 10.1128/mcb.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Sato S, Tomomori-Sato C, Zhang Y, Wen Z, Banks CAS, Washburn MP, Unruh JR, Florens L, Conaway RC, and Conaway JW (2021). Multiple roles for PARP1 in ALC1-dependent nucleosome remodeling. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2107277118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palavalli Parsons LH, Challa S, Gibson BA, Nandu T, Stokes MS, Huang D, Lea JS, and Kraus WL (2021). Identification of PARP-7 substrates reveals a role for MARylation in microtubule control in ovarian cancer cells. Elife 10. 10.7554/eLife.60481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, and Ahel I. (2018). Serine is the major residue for ADP-ribosylation upon DNA damage. Elife 7. 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Mikoc A, and Ahel I. (2017). ADP-ribosylation: new facets of an ancient modification. FEBS J 284, 2932–2946. 10.1111/febs.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, and Lis JT (2008). Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134, 74–84. 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, and Lis JT (2012). Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell 45, 64–74. 10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt SJ, Krastev DB, Brandsma I, Drean A, Song F, Aleksandrov R, Harrell MI, Menon M, Brough R, Campbell J, et al. (2018). Genome-wide and high-density CRISPRCas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 9, 1849. 10.1038/s41467-018-03917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay N., Tighe A., Nelson L., Littler S., Coulson-Gilmer C., Bah N., Golder A., Bakker B., Spierings DCJ., James DI., et al. (2019). DNA replication vulnerabilities render ovarian cancer cells sensitive to poly(ADP-Ribose) glycohydrolase inhibitors. Cancer Cell 35, 519–533 e518. 10.1016/j.ccell.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Naumova N, Shah M, and Tulin AV (2007). Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem 282, 32511–32519. 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, and Mandel P. (1982). Poly(ADPribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A 79, 3423–3427. 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourfarjam Y, Kasson S, Tran L, Ho C, Lim S, and Kim IK (2020). PARG has a robust endo-glycohydrolase activity that releases protein-free poly(ADP-ribose) chains. Biochem Biophys Res Commun 527, 818–823. 10.1016/j.bbrc.2020.04.120. [DOI] [PubMed] [Google Scholar]
- Prokhorova E, Agnew T, Wondisford AR, Tellier M, Kaminski N, Beijer D, Holder J, Groslambert J, Suskiewicz MJ, Zhu K, et al. (2021a). Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol Cell 81, 2640–2655 e2648. 10.1016/j.molcel.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova E, Zobel F, Smith R, Zentout S, Gibbs-Seymour I, Schutzenhofer K, Peters A, Groslambert J, Zorzini V, Agnew T, et al. (2021b). Serine-linked PARP1 automodification controls PARP inhibitor response. Nat Commun 12, 4055. 10.1038/s41467-02124361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, and Jentsch S. (2012). Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Pulliam N., Fang F., Ozes AR., Tang J., Adewuyi A., Kee H., Lyons J., Baylin SB., Matei D., Nakshatri H., et al. (2018). An effective epigenetic-PARP inhibitor combination therapy for breast and ovarian cancers independent of BRCA mutations. Clin Cancer Res 24, 3163–3175. 10.1158/1078-0432.CCR-18-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack JGM, Palazzo L, and Ahel I. (2020). (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev 34, 263–284. 10.1101/gad.334631.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancourt A, and Satoh MS (2009). Delocalization of nucleolar poly(ADP-ribose) polymerase1 to the nucleoplasm and its novel link to cellular sensitivity to DNA damage. DNA Repair (Amst) 8, 286–297. 10.1016/j.dnarep.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Ahuja AK, Herrador R, and Lopes M. (2015). Poly(ADP-ribosyl) glycohydrolase prevents the accumulation of unusual replication structures during unperturbed S phase. Mol Cell Biol 35, 856–865. 10.1128/MCB.01077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, and Lopes M. (2012). Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19, 417–423. 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A, and Nussenzweig A. (2017). The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 18, 610–621. 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH, Ueda K, Honjo T, Nishizuka Y, and Hayaishi O. (1967). Studies on the polymer of adenosine diphosphate ribose. II. Characterization of the polymer. J Biol Chem 242, 31723179. [PubMed] [Google Scholar]
- Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, and Shah GM (2013). Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci U S A 110, 1658–1663. 10.1073/pnas.1209507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KM, Buch-Larsen SC, Kirby IT, Siordia IR, Hutin D, Rasmussen M, Grant DM, David LL, Matthews J, Nielsen ML, and Cohen MS (2021). Chemical genetics and proteome-wide site mapping reveal cysteine MARylation by PARP-7 on immune-relevant protein targets. Elife 10. 10.7554/eLife.60480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B., Gogola E., Yucel H., Duarte AA., van de Ven M., van der Sluij R., Konstantinopoulos PA., Jonkers J., Ceccaldi R., Rottenberg S., and D’Andrea AD. (2017). EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol 19, 1371–1378. 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- Rose M, Burgess JT, O’Byrne K, Richard DJ, and Bolderson E. (2020). PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front Cell Dev Biol 8, 564601. 10.3389/fcell.2020.564601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Roberts G, and Luger K. (2021). Histone Parylation factor 1 contributes to the inhibition of PARP1 by cancer drugs. Nat Commun 12, 736. 10.1038/s41467-021-20998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-SanMartin B, and Caldecott KW (2011). PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell 41, 33–45. 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Ryu KW, Kim DS, and Kraus WL (2015). New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem Rev 115, 2453–2481. 10.1021/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, and Kraus WL (2018). Metabolic regulation of transcription through compartmentalized NAD(+) biosynthesis. Science 360. 10.1126/science.aan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR (2020). Biomolecular condensates and gene activation in development and disease. Dev Cell 55, 84–96. 10.1016/j.devcel.2020.09.005. [DOI] [PubMed] [Google Scholar]
- Sallmyr A, and Tomkinson AE (2018). Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J Biol Chem 293, 10536–10546. 10.1074/jbc.TM117.000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh MS, and Lindahl T. (1992). Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358. 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Tseng A, Borch Jensen M, Scheibye-Alsing K, Fang EF, Iyama T, Bharti SK, Marosi K, Froetscher L, Kassahun H, et al. (2016). Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA. Proc Natl Acad Sci U S A 113, 12502–12507. 10.1073/pnas.1610198113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M. (2012). A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116. 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Swisher EM, and Kaufmann SH (2015). Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J Clin Oncol 33, 1397–1406. 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, et al. (2013). Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J 32, 1225–1237. 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, Bouhss A, Lavrik OI, and Pastre D. (2019). PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep 27, 1809–1821 e1805. 10.1016/j.celrep.2019.04.031. [DOI] [PubMed] [Google Scholar]
- Singh HR, Nardozza AP, Moller IR, Knobloch G, Kistemaker HAV, Hassler M, Harrer N, Blessing C, Eustermann S, Kotthoff C, et al. (2017). A poly-ADP-ribose trigger releases the auto-inhibition of a chromatin remodeling oncogene. Mol Cell 68, 860–871 e867. 10.1016/j.molcel.2017.11.019. [DOI] [PubMed] [Google Scholar]
- Slade D. (2020). PARP and PARG inhibitors in cancer treatment. Genes Dev 34, 360–394. 10.1101/gad.334516.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, Romeijn RJ, Pastink A, Mailand N, Vermeulen W, and van Attikum H. (2013). Poly(ADPribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J Cell Sci 126, 889–903. 10.1242/jcs.109413. [DOI] [PubMed] [Google Scholar]
- Smith R, Sellou H, Chapuis C, Huet S, and Timinszky G. (2018). CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADPribose)-dependent chromatin relaxation. Nucleic Acids Res 46, 6087–6098. 10.1093/nar/gky334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen JD, Brody JR, Armen RS, and Pascal JM (2013). Structural Implications for Selective Targeting of PARPs. Front Oncol 3, 301. 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K., Takebayashi S., Taguchi H., Takeda S., and Okumura K. (2008). PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol 183, 1203–1212. 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova MV, Hamon L, Kutuzov MM, Joshi V, Abrakhi S, Dobra I, Curmi PA, Pastre D, and Lavrik OI (2019). A Single-Molecule Atomic Force Microscopy Study of PARP1 and PARP2 Recognition of Base Excision Repair DNA Intermediates. J Mol Biol 431, 2655–2673. 10.1016/j.jmb.2019.05.028. [DOI] [PubMed] [Google Scholar]
- Suskiewicz MJ, Zobel F, Ogden TEH, Fontana P, Ariza A, Yang JC, Zhu K, Bracken L, Hawthorne WJ, Ahel D, et al. (2020). HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602. 10.1038/s41586-020-2013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, and Gautier J. (2011). Double-strand break end resection and repair pathway choice. Annu Rev Genet 45, 247–271. 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Teloni F, and Altmeyer M. (2016). Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res 44, 993–1006. 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ji Y, Wu C, Datz H, Boyle C, MacLeod B, Patel S, Ampofo M, Currie M, Harbin J, et al. (2019). Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc Natl Acad Sci U S A 116, 9941–9946. 10.1073/pnas.1901183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CJ, Kotova E, Andrake M, Adolf-Bryfogle J, Glaser R, Regnard C, and Tulin AV (2014). Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly(ADP-ribosyl)ation. Mol Cell 53, 831–842. 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. (2009). A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16, 923–929. 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tulin A, and Spradling A. (2003). Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299, 560–562. 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Veith S., Schink A., Engbrecht M., Mack M., Rank L., Rossatti P., Hakobyan M., Goly D., Hefele T., Frensch M., et al. (2019). PARP1 regulates DNA damage-induced nucleolar-nucleoplasmic shuttling of WRN and XRCC1 in a toxicant and protein-specific manner. Sci Rep 9, 10075. 10.1038/s41598-019-46358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Zhou Y, Cao Z, Deraska PV, Deb M, Arai E, Li W, Shao Y, Puentes L, Li Y, et al. (2021). ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat Cell Biol 23, 160–171. 10.1038/s41556-020-00624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, et al. (2012). Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol 30, 283–288. 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen K, and Chen Z. (2021a). Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome. Nat Commun 12, 4057. 10.1038/s41467-02124320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, and Iliakis G. (2006). PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34, 6170–6182. 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Zhang S, Kim B, Hull VL, Xu J, Prabhu P, Gregory M, MartinezCerdeno V, Zhan X, et al. (2021b). PARP1-mediated PARylation activity is essential for oligodendroglial differentiation and CNS myelination. Cell Rep 37, 109695. 10.1016/j.celrep.2021.109695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Morishita K, Zhou X, Shiizaki S, Uchiyama Y, Koike M, Naguro I, and Ichijo H. (2021). Cells recognize osmotic stress through liquid-liquid phase separation lubricated with poly(ADP-ribose). Nat Commun 12, 1353. 10.1038/s41467-021-21614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Narita T, Satpathy S, Srinivasan B, Hansen BK, Scholz C, Hamilton WB, Zucconi BE, Wang WW, Liu WR, et al. (2018). Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell 174, 231–244 e212. 10.1016/j.cell.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixler L, Scharinger K, Momoh J, Luscher B, Feijs KLH, and Zaja R. (2021). ADPribosylation of RNA and DNA: from in vitro characterization to in vivo function. Nucleic Acids Res 49, 3634–3650. 10.1093/nar/gkab136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Du Y, Nakai K, Ding M, Chang SS, Hsu JL, Yao J, Wei Y, Nie L, Jiao S, et al. (2018). EZH2 contributes to the response to PARP inhibitors through its PARPmediated poly-ADP ribosylation in breast cancer. Oncogene 37, 208–217. 10.1038/onc.2017.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Jin C., Hata T., Yasumizu Y., Zhang Y., Hong D., Maeda T., Miyo M., Hiraki M., Suzuki Y., et al. (2019). MUC1-C Integrates Chromatin Remodeling and PARP1 Activity in the DNA Damage Response of Triple-Negative Breast Cancer Cells. Cancer Res 79, 20312041. 10.1158/0008-5472.CAN-18-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang Y, Shan W, Hu Z, Yuan J, Pi J, Wang Y, Fan L, Tang Z, Li C, et al. (2017). Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci Transl Med 9. 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Cortes U, Patnaik S, Jasin M, and Wang ZQ (2004). Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23, 3872–3882. 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. (2004). Poly(ADP-ribosyl)ation regulates CTCFdependent chromatin insulation. Nat Genet 36, 1105–1110. 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Zandarashvili L, Langelier MF, Velagapudi UK, Hancock MA, Steffen JD, Billur R, Hannan ZM, Wicks AJ, Krastev DB, Pettitt SJ, et al. (2020). Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368. 10.1126/science.aax6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, Biard D, Gasparutto D, Lavrik OI, and Ishchenko AA (2018). Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADPribosylation. Nucleic Acids Res 46, 2417–2431. 10.1093/nar/gkx1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Libien J, Shaik F, Wolk J, and Hernandez AI (2016). Nucleolar PARP-1 expression is decreased in Alzheimer’s disease: Consequences for epigenetic regulation of rDNA and cognition. Neural Plast 2016, 8987928. 10.1155/2016/8987928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Shi J, Bian C, and Yu X. (2015a). Poly(ADP-Ribose) mediates the BRCA2dependent early DNA damage response. Cell Rep 13, 678–689. 10.1016/j.celrep.2015.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Shi J., Chen SH., Bian C., and Yu X. (2015b). The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res 43, 10782–10794. 10.1093/nar/gkv939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, and Yu Y. (2013). Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods 10, 981–984. 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millan-Arino L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS, et al. (2015). PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell 59, 984–997. 10.1016/j.molcel.2015.07.019. [DOI] [PubMed] [Google Scholar]
- Zhen Y, and Yu Y. (2018). Proteomic analysis of the downstream signaling network of PARP1. Biochemistry 57, 429–440. 10.1021/acs.biochem.7b01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, and Caiafa P. (2009). CTCF and its protein partners: divide and rule? J Cell Sci 122, 1275–1284. 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]