Abstract
The conversion of sulfate to an excess of free sulfide requires stringent reductive conditions. Dissimilatory sulfate reduction is used in nature by sulfate-reducing bacteria for respiration and results in the conversion of sulfate to sulfide. However, this dissimilatory sulfate reduction pathway is inhibited by oxygen and is thus limited to anaerobic environments. As an alternative, we have metabolically engineered a novel aerobic sulfate reduction pathway for the secretion of sulfides. The assimilatory sulfate reduction pathway was redirected to overproduce cysteine, and excess cysteine was converted to sulfide by cysteine desulfhydrase. As a potential application for this pathway, a bacterium was engineered with this pathway and was used to aerobically precipitate cadmium as cadmium sulfide, which was deposited on the cell surface. To maximize sulfide production and cadmium precipitation, the production of cysteine desulfhydrase was modulated to achieve an optimal balance between the production and degradation of cysteine.
Dissimilatory reduction of sulfate to hydrogen sulfide is used by a diverse group of heterotrophic strict anaerobes as a sink for electrons generated during oxidation of a carbon source (2). Industrially, this source of sulfide has been used to precipitate metals in wastewater treatment reactors and has been proposed for stabilization of metals in soils and for formation of metal sulfide “quantum” particles for microelectronics applications (10). For removal of heavy metals from wastewater, addition of hydrogen sulfide (biologically or nonbiologically) can be especially effective because the metal sulfide precipitates are extremely insoluble and stable (14). Biological hydrogen sulfide production could also be used to precipitate and stabilize heavy metals in situ. Previous research on bioprecipitation has predominantly focused on using sulfate-reducing bacteria to produce sulfide and precipitate heavy metals as metal sulfides (7, 15, 16, 18, 19). However, sulfate-reducing bacteria are obligate anaerobes (2) and their application is limited to anaerobic environments.
Sulfide is also produced from sulfate during assimilatory sulfate reduction for the synthesis of cysteine (11) and methionine. Unlike dissimilatory sulfate reduction, assimilatory sulfate reduction is tightly regulated so that little or no excess sulfide is produced and secreted from the cell. Furthermore, assimilatory sulfate reduction operates under many growth conditions, such that the strict anaerobic conditions necessary for dissimilatory sulfate reduction are not required. An aerobic sulfide production pathway could be useful for precipitation and removal or stabilization of heavy metal contaminants, for the formation of metal sulfide quantum particles, or for any other use of sulfide under conditions that are not strictly anaerobic. As a step towards developing such applications, we have redirected the assimilatory sulfate reduction pathway to create an aerobic sulfide production pathway and have shown its use for the bioprecipitation of metals.
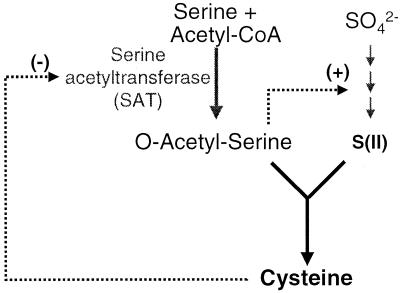
The aerobic sulfide production pathway was engineered by overproducing two unique enzymes in Escherichia coli: a serine acetyltransferase (SAT or CysE) that is insensitive to feedback inhibition by cysteine and a cysteine desulfhydrase from Treponema denticola. In cysteine biosynthesis, SAT catalyzes the acetylation of serine to form O-acetylserine, the final precursor to cysteine (11). In addition, some O-acetylserine is converted to N-acetylserine, which triggers the induction of the sulfate assimilation genes (Fig. 1) (11). Denk and Bock found that a single amino acid change (methionine to isoleucine at position 256) rendered the SAT insensitive to feedback inhibition by cysteine, and production of this mutant enzyme resulted in cysteine overproduction by E. coli (6).
FIG. 1.
Cysteine biosynthesis pathways and regulation of SAT activity.
Cysteine desulfhydrase is an aminotransferase that converts cysteine into pyruvate, ammonia, and hydrogen sulfide. Chu and colleagues discovered an especially active cysteine desulfhydrase from T. denticola (4), a bacterium isolated from a dental patient.
By combining the activities of the mutant SAT and the cysteine desulfhydrase, we created an aerobic sulfide production pathway and demonstrated its use in metal precipitation. Producing the mutant SAT resulted in the overproduction of cysteine, and cysteine desulfhydrase was used to convert excess cysteine to sulfide. The secreted sulfide precipitated cadmium and effectively removed it from solution.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pRock carried the ColE1 origin of replication, the hok/sok stability cassette from the parB locus of plasmid R1, and the lactose-inducible Ptac promoter. The cysteine desulfhydrase gene from T. denticola was inserted into pRock under control of Ptac to form pCysdesulf/LacI2/Rock. Plasmid pBAD33 was constructed by Guzman et al. (9). The mutant SAT gene from pCys2 (6) was amplified and fused with a strong consensus ribosomal binding site and an 8-bp spacer (AGGAGGTTTTTATT) using PCR and cloned into the SacI and HindIII sites of pBAD33 to create pCysE*/AraC. Plasmid pCysE*/AraC was used in this study instead of pCys2 because the p15 origin of replication from pBAD33 allows stable plasmid coexistence with pRock or pCysDesulf/LacI2/Rock. Various combinations of these plasmids were transformed into E. coli DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara,leu)7697 galU galK λ− rpsL nupG] by electroporation.
Cell culture.
E. coli cultures were grown in a defined medium buffered with 3-[N-morpholino]propanesulfonic acid at pH 7.3. The culture medium was prepared as previously described (13) with the following exceptions: 2 mM glycerol phosphate was substituted for inorganic phosphate, and 4 mM K2SO4 was used as the sulfate source. In addition, the medium was supplemented with all amino acids (except for tyrosine, methionine, and cysteine) at concentrations recommended by Wanner (17). When necessary, cysteine was supplemented at a concentration of 500 μM. Between 0 and 100 μM isopropyl β-d-thiogalactopyranoside (IPTG) and 5 mM arabinose were added to induce gene expression. Ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml) were also added to the medium. Cultures used to inoculate metal-containing cultures were grown in medium containing 60 mM glycerol and harvested during exponential growth. The E. coli cultures were then resuspended to an optical density at 600 nm (OD600) of 1.0 in medium containing 30 mM glucose and amended with cadmium chloride as necessary. The mixed culture was obtained by combining 2.5 ml of E. coli transformed with pRock and pCysE*/AraC and 2.5 ml of E. coli transformed with pCysDesulf/LacI2/Rock and pBAD33. Five-milliliter cultures were grown aerobically in culture tubes in an incubated shaker (37°C; 200 rpm). The cultures were analyzed after 12 h of incubation.
Assays.
The acid-labile sulfide content of the cultures was determined by a colorimetric assay reported previously (1). Cysteine was measured by the colorimetric assay using ninhydrin as outlined by Gaitonde (8).
Analysis of cadmium removal.
Cell cultures were centrifuged at 17,000 × g for 2 min. The culture supernatant was filtered with 0.22-μm-pore-size syringe filters, diluted in 10% HNO3, and analyzed for cadmium content on a Perkin-Elmer Optima 3000DV inductively coupled plasma spectrometer.
Preparation of cell lysate.
Late-exponential-phase cells (5 to 10 ml; OD600, >0.6 and <1.2) were centrifuged at 17,000 × g for 10 min and then resuspended in 60 μl of a 10% sucrose–50 mM Tris solution (pH 7.5). To this suspension, 75 μl of lysis buffer (10% sucrose, 300 mM NaCl, 90 mM EDTA, 3-mg/ml lysozyme, 50 mM Tris-HCl [pH 7.5]) was added. Next, the suspension was mixed and incubated on ice for 2 h. The cell suspension was then frozen and thawed five times by cycling between 37°C incubation in a water bath and freezing in liquid nitrogen. Finally, the suspension was sonicated (40% power) for 5 s using a Branson Sonifier.
Cysteine desulfhydrase activity assay.
Cysteine desulfhydrase activity was measured using a colorimetric assay adapted from the method of Chu et al. (5). In a 2-ml microfuge tube, 1 μl of lysate was added to 999 μl of 0.1 mM cysteine in phosphate-buffered saline (8 g of NaCl/liter, 0.2 g of KCl/liter, 1.44 g of Na2HPO4/liter, 0.24 g of KH2PO4/liter [pH 7.4]). The mixture was incubated for 1 h at 37°C. Next, 0.1 ml of 0.02 M N,N-dimethyl-p-phenylenediamine sulfate in 7.2 N HCl and 0.1 ml of 0.3 M FeCl3 in 1.2 N HCl were added, the mixture was vortexed, and color was allowed to develop for 20 min. Samples were then centrifuged for 5 min at 17,000 × g. The supernatant was diluted 1/10 in water, and its absorbance was measured at 670 nm. Calibration standards were prepared by adding sodium sulfide to phosphate-buffered saline at concentrations from 0 to 0.2 mM. The standards also contained 0.1 mM cysteine.
TEM and EDXS.
Cell samples were washed in phosphate buffer solution and fixed overnight in a solution of 2% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.2. Samples were then rinsed with 0.1 M sodium cacodylate (pH 7.2), post-fixed in a solution of 0.5% osmium tetroxide in 0.1 M sodium cacodylate (pH 7.2), and rinsed with deionized water. The samples were then dehydrated in a graded acetone series and embedded in Epon-Araldite resin. Samples 40 nm thick were sectioned using a Reichert Ultracut E microtome and collected on uncoated 300-mesh copper grids. Samples were analyzed by transmission electron microscopy (TEM) and energy-dispersive X-ray spectroscopy (EDXS) using a JEOL 200CX scanning transmission microscope and a Kevex 8000 EDXS system.
RESULTS
Individual components of the pathway.
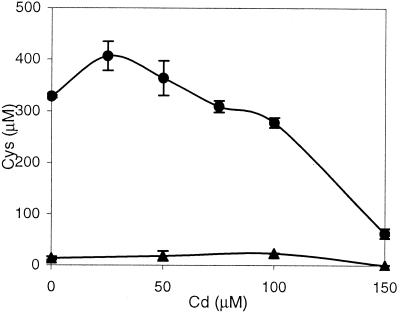
Initial experiments were conducted to confirm the individual activities of the mutant SAT and cysteine desulfhydrase. E. coli producing the mutant SAT secreted cysteine, even in the presence of cadmium (Fig. 2). Using sulfate as the only sulfur source, the engineered E. coli produced 364 μM cysteine in the presence of 50 μM cadmium and 278 μM cysteine in the presence of 100 μM cadmium after 12 h. While optimal cysteine production actually occurred in 25 μM cadmium, production was clearly inhibited at higher cadmium concentrations. The control did not produce any significant amount of cysteine at the cadmium concentrations evaluated.
FIG. 2.
Cysteine production as a function of added cadmium. E. coli DH10B harboring pCysE*/AraC and pRock, which produces the mutant SAT, secreted cysteine into the medium (circles); cysteine production was diminished at higher cadmium concentrations. The control (E. coli harboring pBAD33 and pRock) secreted minimal amounts of cysteine (triangles). Data and error bars represent the means and errors for duplicate experiments.
For evaluation of the cadmium precipitation potential of cysteine desulfhydrase, the enzyme was overproduced in E. coli and the culture medium was supplemented with cysteine. Cultures were inoculated at an OD600 of 1.0, and production of acid-labile sulfide (a measure of sulfide present as a metal sulfide) and removal of aqueous cadmium were measured after 12 h of culturing. Production of free, aqueous sulfide could not be accurately measured because it reacts with cadmium at a near-instantaneous rate and can oxidize under aerobic culture conditions. When the medium was supplemented with 500 μM cysteine and 100 μM Cd, cysteine desulfhydrase-producing E. coli generated 112 μM acid-labile sulfide and removed 99 μM of 100 μM cadmium from solution. The near-unity molar ratio of sulfide to removed cadmium indicated that cadmium was precipitated as cadmium sulfide. In contrast, the control E. coli (no overproduced cysteine desulfhydrase) produced only 60 μM acid-labile sulfide and removed 80 μM cadmium.
Assembly of the sulfide production pathway.
We devised two strategies to combine cysteine overproduction with cysteine desulfhydrase activity to create a pathway that converts sulfate to sulfide under aerobic conditions. In the first strategy, both the mutant SAT and cysteine desulfhydrase were produced by a single E. coli strain. The cysteine produced by the E. coli is converted immediately to sulfide by cysteine desulfhydrase. The second strategy utilized a mixed culture containing E. coli overproducing SAT only and E. coli overproducing cysteine desulfhydrase only. The former strain secretes cysteine, and the latter converts the cysteine to sulfide.
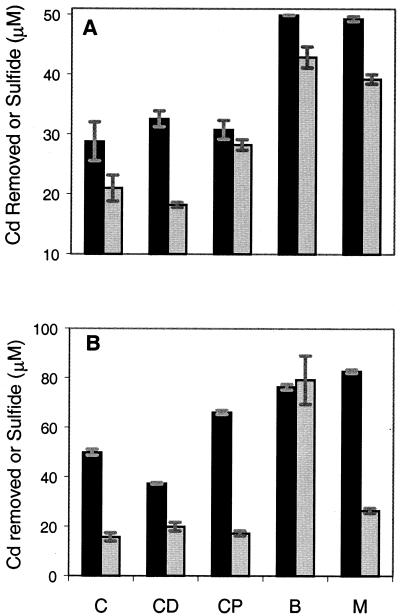
E. coli cultures were initially inoculated at an OD600 of 1.0 and grown aerobically in a defined salts medium with sulfate as the only sulfur source. Cultures were spiked with either 50 or 100 μM cadmium chloride, induced with 100 μM IPTG, and analyzed after 12 h. At a 50 μM concentration of cadmium, E. coli producing both the SAT and the cysteine desulfhydrase produced the greatest amount of acid labile sulfide (42.9 μM) and removed nearly all (99.8%) of the cadmium from solution (Fig. 3A). Although slightly less proficient than the E. coli producing both enzymes, the mixed culture also removed cadmium and produced sulfide. The molar ratios of sulfide to removed cadmium of these cultures were 0.86 and 0.80, respectively, and suggest that a significant amount of cadmium was precipitated as cadmium sulfide. Control cultures producing neither enzyme, producing only cysteine desulfhydrase, and producing only SAT removed significantly less cadmium and yielded less sulfide. Generation of sulfide by the various controls could stem from native cysteine biosynthesis and tryptophan aminotransferase, which has a low level of cysteine desulfhydrase activity (12). Cadmium not precipitated as cadmium sulfide could have been removed through carbonate precipitation, phosphate precipitation, or adsorption to cellular materials. At a 100 μM concentration of cadmium, the E. coli strain producing both enzymes was the only culture to produce a high amount of sulfide, three times as much sulfide as the mixed culture (Fig. 3B). This high level of sulfide production was accompanied by a stoichiometric amount of cadmium removed and strongly suggested that it was being precipitated as cadmium sulfide.
FIG. 3.
Sulfide production (light bars) and cadmium removal (dark bars) by the following cultures: C, E. coli producing neither cysteine desulfhydrase nor the mutant SAT (pBAD33 and pRock); CD, E. coli producing only cysteine desulfhydrase (pBAD33 and pCysDesulf/LacI2/Rock); CP, E. coli producing only the mutant SAT (pCysE*/AraC and pRock) for cysteine production; B, E. coli producing both the mutant SAT and the cysteine desulfhydrase; and M, a mixed culture containing E. coli producing only cysteine desulfhydrase (pBAD33 and pCysDesulf/LacI2/Rock) and E. coli producing only the mutant SAT (pCysE*/AraC and pRock). (A) Acid-labile sulfide production and cadmium removal by cultures initially containing 50 μM cadmium. (B) Acid-labile sulfide production and cadmium removal by cultures initially containing 100 μM cadmium. In all cultures, gene expression was fully induced, and cadmium removal and sulfide production were measured 12 h after inoculation of cultures. Data and error bars represent the means and errors for duplicate experiments.
Toxicity of cysteine desulfhydrase.
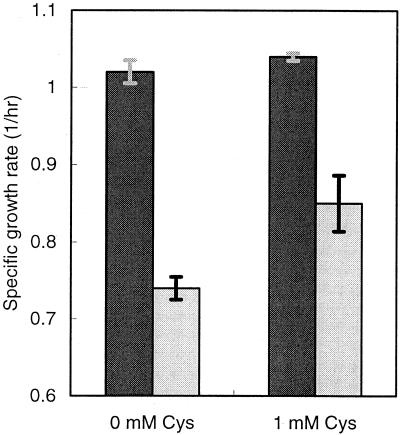
The growth of cells expressing cysteine desulfhydrase was impaired in the absence of exogenous cysteine (Fig. 4). When induced by 100 μM IPTG, E. coli expressing the cysteine desulfhydrase gene grew at a considerably slower rate (0.74 h−1) than the control producing no cysteine desulfhydrase (1.02 h−1). Because minimal amounts of sulfide would be produced in the absence of an abundant source of cysteine, the inhibition of growth can be attributed to the toxicity of cysteine desulfhydrase, not the generation of sulfide. Supplementing cultures with cysteine (1 mM) partially alleviated the toxicity of cysteine desulfhydrase and resulted in an increase in the growth rate of the E. coli producing cysteine desulfhydrase.
FIG. 4.
Effect of cysteine desulfhydrase on the growth rate with and without additional cysteine. Growth rates of E. coli producing cysteine desulfhydrase (light bars) and of the control, not producing cysteine desulfhydrase (dark bars), are shown.
Optimization of the sulfide production pathway.
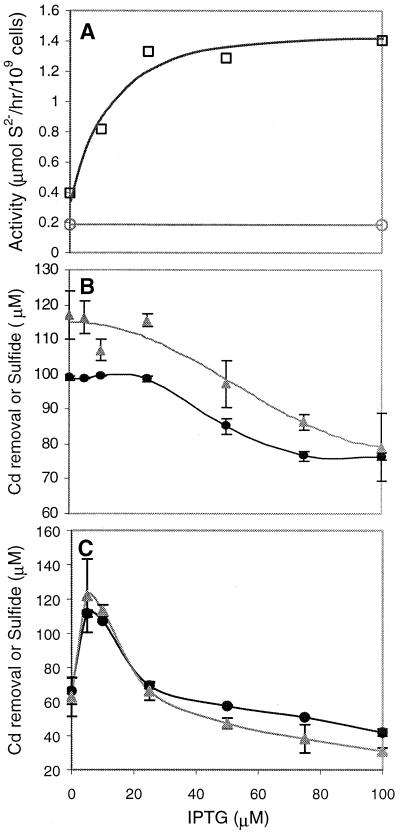
Varying the concentration of the inducer (IPTG) caused the expression of the cysteine desulfhydrase gene to be optimized, so that the toxicity of cysteine desulfhydrase was minimized and sulfide production and cadmium removal were maximized. Cysteine desulfhydrase activity was effectively modulated at inducer concentrations of between 0 and 100 μM IPTG (Fig. 5A). At a 100 μM concentration of cadmium and low induction levels (≤25 μM IPTG), the E. coli producing both enzymes produced the highest amounts of sulfide and removed cadmium almost completely (Fig. 5B), demonstrating that even the lowest achievable expression of cysteine desulfhydrase was sufficient for removal of 100 μM cadmium.
FIG. 5.
Optimization of cysteine desulfhydrase activity for cadmium removal. (A) Cysteine desulfhydrase activity as a function of the inducer concentration in E. coli producing only cysteine desulfhydrase (squares) and the control (circles). (B and C) Optimization of sulfide production (triangles) and cadmium removal (circles) by varying cysteine desulfhydrase activity in E. coli producing both the mutant serine acetyl transferase and cysteine desulfhydrase at 100 μM cadmium (B) and 125 μM cadmium (C). Cadmium removal and sulfide production were measured 12 h after inoculation of cultures. Data and error bars represent the means and errors for duplicate experiments.
At a 125 μM concentration of cadmium, cysteine desulfhydrase was optimally induced at a 5 μM concentration of IPTG, and the engineered E. coli produced the greatest amount of sulfide and removed the most cadmium (98% of the initial amount) from solution (Fig. 5C). In this optimal case, there was a high enough level of cysteine desulfhydrase activity to produce a sufficient quantity of sulfide but a low enough level of activity to minimize the burden of overproducing cysteine desulfhydrase.
Visualization and analysis of precipitation.
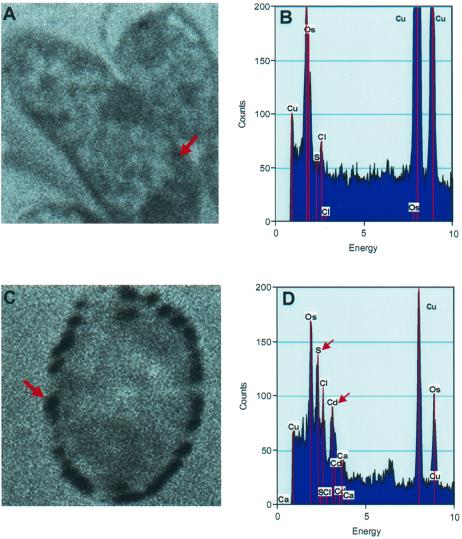
The E. coli producing both enzymes in the cytoplasm most effectively precipitated cadmium (50 μM) from solution and was analyzed by TEM and EDXS, an analytical method for determining elemental composition. Electron microscopy revealed dense granules on the cell wall (Fig. 6C), and EDXS revealed that these granules were an accumulation of both cadmium and sulfur (Fig. 6D), indicating that the cadmium precipitated on the cell wall as cadmium sulfide. No granules or accumulation of cadmium and sulfur were detected in the control producing neither enzyme (Fig. 6A and B).
FIG. 6.
TEM and EDXS analysis of E. coli grown in 50 μM cadmium. (A) TEM analysis of the control E. coli producing neither cysteine desulfhydrase nor the mutant SAT did not show precipitation of cadmium on the cell wall. (B) An EDXS spectrum of the control showed no accumulation of cadmium on the cell wall. (C) TEM analysis of the E. coli producing both cysteine desulfhydrase and the mutant SAT in the cytoplasm revealed dense granules on the cell wall. The arrow points to the spot at which the EDXS spectrum was taken. (D) An EDXS spectrum of the E. coli producing both enzymes showed that the dense granules were an accumulation of cadmium and sulfur, indicating that cadmium precipitated as cadmium sulfide on the cell wall. Copper and osmium peaks were due to the copper grid and the osmium tetraoxide fixative.
DISCUSSION
In this work, E. coli was metabolically engineered to convert sulfate via cysteine to sulfide under aerobic conditions. Unlike the sulfide generation pathway of sulfate-reducing bacteria, this novel pathway is not inhibited by oxygen and is not limited to anaerobic applications. The creation of this metabolic pathway diverges from the commonly accepted notion that biological sulfide generation must occur under anaerobic conditions and suggests a potential, although unprecedented, mechanism for production of aerobic sulfide from sulfate by naturally occurring organisms. Moreover, since the pathway is not coupled to anaerobic respiration, this sulfate reduction pathway could be expressed in a variety of organisms. As such, this engineered sulfide generation pathway is a possible alternative to anaerobic sulfate reduction. While sulfate-reducing bacteria have robust characteristics that for certain applications may be superior to the system presented here, there may be certain aerobic applications for which sulfate-reducing organisms would not survive and aerobic organisms expressing such a pathway would be desired.
Optimization of the engineered pathway by varying gene expression was essential for maximal production of sulfide and, in the cadmium precipitation example, maximal removal of cadmium from solution. Cysteine production requires a significant amount of energy, in the form of ATP, and reducing equivalents, in the form of NADPH. Hence, production of cysteine at levels higher than are necessary for protein synthesis robs the cell of precursors that would allow faster growth. However, it was not necessary to vary the expression of the mutant SAT gene, since preliminary growth studies indicated that overproduction of the mutant SAT (at least at the levels attainable with this plasmid-promoter combination) did not have a significant effect on the growth of the E. coli (data not shown).
In contrast, cysteine desulfhydrase was found to be toxic to cell growth. Supplementation of the medium with cysteine partially relieved the metabolic burden of the overproduced cysteine desulfhydrase. This indicated that cysteine desulfhydrase converts cysteine that could be used for protein synthesis to sulfide, thereby reducing the growth rate of a cell producing that protein. This burden is significantly greater than what one observes in producing a nonmetabolic enzyme at similar levels (3) and made cysteine desulfhydrase production a key parameter in optimizing the pathway.
In addition, we demonstrated that the sulfide secreted by the cell could be used to precipitate cadmium in a complex of cadmium and sulfur, most likely cadmium sulfide, on the cell wall. While the culture producing both enzymes removed 99.8% of cadmium with an initial 50 μM concentration in solution, at a 100 μM initial concentration of cadmium, a significant amount of cadmium (23.6% of the initial concentration) remained in solution. Because E. coli producing only cysteine desulfhydrase exhibited enough enzymatic activity to produce more than 100 μM sulfide and because E. coli producing only the mutant SAT secreted more than enough cysteine to produce 100 μM sulfide, it appears that the metabolic burdens of producing both enzymes acted together to impair sulfide production. Although this combined burden was not large enough to preclude the nearly complete removal of 50 μM cadmium from solution, it is possible that the increased toxicity of 100 μM cadmium inhibited the cells to such an extent that the metabolic burden prevented complete removal.
It is interesting to note the sensitivity of the engineered E. coli to the concentration of cadmium and production of cysteine desulfhydrase. At low, tolerable concentrations of cadmium (50 μM), the metabolic burden of producing cysteine desulfhydrase at the maximal induction level does not prevent the cells from efficiently removing cadmium and producing sulfide (Fig. 3A, column B). However, at more toxic concentrations of cadmium (100 and 125 μM), the toxic effects of cysteine desulfhydrase became evident and high induction levels adversely affected sulfide production and cadmium removal. This result emphasizes further the importance of optimizing production of SAT and cysteine desulfhydrase.
This study represents an important step towards the development of an aerobic sulfate reduction pathway for the precipitation of heavy metals and is the first report of an organism genetically engineered to precipitate a heavy metal as a metal sulfide. However, in its current stage of development, this system is not an immediately practical means of remediating metal-polluted environments. Clearly, if the engineered pathway were to be employed for heavy metal removal from, or stabilization in, a contaminated environment, a more environmentally relevant organism (such as a pseudomonad or a gram-positive microorganism) would be utilized. Furthermore, additional steps would be needed to make such a genetically engineered organism competitive in a heterogeneous microbial environment. For example, the organism might be engineered with mechanisms enabling it to tolerate toxic concentrations of heavy metals, thus giving it a competitive advantage over indigenous organisms.
ACKNOWLEDGMENTS
We thank August Bock (Lehrstul fur Microbiologie der Universitat Munchen, Munich, Germany) for plasmid pCys2, Jon Beckwith (Harvard Medical School, Boston, Mass.) for plasmid pBAD33, and Chuck Echer (National Center for Electron Microscopy, Lawrence Berkeley Laboratory, Berkeley, Calif.) for assistance with electron microscopy and EDXS.
The U.S. Department of Energy NABIR program supported this research.
REFERENCES
- 1.Aiking H, Kok K, van Heerikhuizen H, van't Riet J. Adaptation to cadmium by Klebsiella aerogenes growing in continuous culture proceeds mainly via formation of cadmium sulfide. Appl Environ Microbiol. 1982;44:938–944. doi: 10.1128/aem.44.4.938-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton L L, Tomei F A. Characteristics and activities of sulfate-reducing bacteria. In: Barton L L, editor. Sulfate-reducing bacteria. New York, N.Y: Plenum Press; 1995. pp. 1–32. [Google Scholar]
- 3.Carrier T, Jones K L, Keasling J D. mRNA stability and plasmid copy number effects on gene expression from an inducible promoter system. Biotechnol Bioeng. 1998;59:666–672. doi: 10.1002/(sici)1097-0290(19980920)59:6<666::aid-bit2>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 4.Chu L, Burgum A, Kolodrubetz D, Holt S C. The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases. Infect Immun. 1995;63:4448–4455. doi: 10.1128/iai.63.11.4448-4455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu L, Ebersole J L, Kurzban G P, Holt S C. Cystalysin, a 46-kilodalton cysteine desulfhydrase from Treponema denticola, with hemolytic and hemoxidative activities. Infect Immun. 1997;65:3231–3238. doi: 10.1128/iai.65.8.3231-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denk D, Bock A. L-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J Gen Microbiol. 1987;133:515–525. doi: 10.1099/00221287-133-3-515. [DOI] [PubMed] [Google Scholar]
- 7.Fortin D, Southam G, Beveridge T J. Nickel sulfide, Iron-nickel sulfide and iron sulfide precipitation by a newly isolated Desulfotomaculum species and its relation to nickel resistance. FEMS Microbiol Ecol. 1994;14:121–132. [Google Scholar]
- 8.Gaitonde M K. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes J D, Richardson D J, Saed S, Evans-Gowing R, Russell D A, Sodeau J R. Cadmium-specific formation of metal sulfide ‘Q-particles’ by Klebsiella pneumoniae. Microbiology. 1997;143:2521–2530. doi: 10.1099/00221287-143-8-2521. [DOI] [PubMed] [Google Scholar]
- 11.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, editor. Escherichia coli and Salmonella cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 514–527. [Google Scholar]
- 12.McFall E, Newman E B. Amino acids as carbon sources. In: Neidhardt F C, editor. Escherichia coli and Salmonella cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 358–379. [Google Scholar]
- 13.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters R W, Ku Y, Bhattacharyya D. Evaluation of recent treatment techniques for removal of heavy metals from industrial wastewaters. Am Inst Chem Eng Symp Ser. 1985;91:165–203. [Google Scholar]
- 15.Peyton B M, Truex M J, Toth J J. Biogenic sulfide precipitation of heavy metals to recycle electroplating and electronics manufacturing process streams PNWD-2315. Batelle; 1995. [Google Scholar]
- 16.Smith A. Harnessing nature to clean up Doñana—scientists recommend a biological solution. Strasbourg, France: European Science Foundation; 1999. [Google Scholar]
- 17.Wanner B. Physiological regulation of a decontrolled lac operon. J Bacteriol. 1977;130:212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White C, Gadd G M. Accumulation and effects of cadmium on sulphate-reducing bacterial biofilms. Microbiology. 1998;144:1407–1415. doi: 10.1099/00221287-144-5-1407. [DOI] [PubMed] [Google Scholar]
- 19.White C, Gadd G M. Mixed sulphate-reducing bacterial cultures for bioprecipitation of toxic metals: factorial and response-surface analysis of the effects of dilution rate, sulphate and subrate concentration. Microbiology. 1996;142:2197–2205. [Google Scholar]