Abstract
Genome-wide association studies (GWAS) have identified several risk loci for post-traumatic stress disorder (PTSD); however, how they confer PTSD risk remains unclear. We aimed to identify genes that confer PTSD risk through their effects on brain protein abundance to provide new insights into PTSD pathogenesis. To that end, we integrated human brain proteomes with PTSD GWAS results to perform a proteome-wide association study (PWAS) of PTSD, followed by Mendelian randomization, using a discovery and confirmatory study design. Brain proteomes (N=525) were profiled from the dorsolateral prefrontal cortex using mass spectrometry. The Million Veteran Program (MVP) PTSD GWAS (n=186,689) was used for the discovery PWAS, and the Psychiatric Genomics Consortium PTSD GWAS (n=174,659) was used for the confirmatory PWAS. To understand whether genes identified at the protein-level were also evident at the transcript-level, we performed a transcriptome-wide association study (TWAS) using human brain transcriptomes (N=888) and the MVP PTSD GWAS results. We identified 11 genes that contribute to PTSD pathogenesis via their respective cis-regulated brain protein abundance. Seven of 11 genes (64%) replicated in the confirmatory PWAS and 4 of 11 also had their cis-regulated brain mRNA levels associated with PTSD. High confidence level was assigned to 9 of 11 genes after considering evidence from the confirmatory PWAS and TWAS. Most of the identified genes are expressed in other PTSD-relevant brain regions and several are preferentially expressed in excitatory neurons, astrocytes, and oligodendrocyte precursor cells. These genes are novel, promising targets for mechanistic and therapeutic studies to find new treatments for PTSD.
Post-traumatic stress disorder (PTSD) is a mental illness that can manifest after exposure to extremely stressful or life-threatening events. The lifetime prevalence of PTSD is approximately 8% in the general population but can be as high as 31% in veterans or individuals with higher frequency of severe psychological stress or trauma exposure1, 2. PTSD symptoms can be debilitating and last for years3, 4. While current treatments for PTSD have provided clinical improvement for many people, even the most positive assessments of treatment effectiveness would argue that there is a substantial minority who do not improve with treatment5–8. For instance, medications for PTSD tend to have small effect sizes, and only about 20% of the patients achieved loss of PTSD diagnosis after pharmacotherapy6, 7. Likewise, about 60% of the patients continued to be diagnosed with PTSD after undergoing trauma-focused therapies, the first-line therapies for PTSD5, 9. Thus, there is a dire need for effective treatments for PTSD10.
To support the development of new therapeutics, we need a better understanding of the biological mechanisms underlying PTSD. Risk for PTSD is multifactorial, including both genetic and environmental factors (severity of traumatic event, prior stress exposure, lack of social support, coping styles, and others)11. Genetic factors contribute up to 40% of the vulnerability to PTSD12, 13 and is a window to identify novel treatment targets. Furthermore, genetically informed drug targets are more likely to lead to successful FDA approval14. The Million Veteran Program (MVP) and Psychiatric Genomics Consortia (PGC) have made great strides in understanding the genetic architecture of PTSD though genome-wide association studies (GWAS) involving hundreds of thousands of participants of European ancestry15–17. These GWAS have identified several risk loci for PTSD; however, how these loci confer PTSD risk remains unclear. Here, we aimed to identify genetic variants that confer PTSD risk through their effects on brain protein abundance to provide new insights into PTSD pathogenesis.
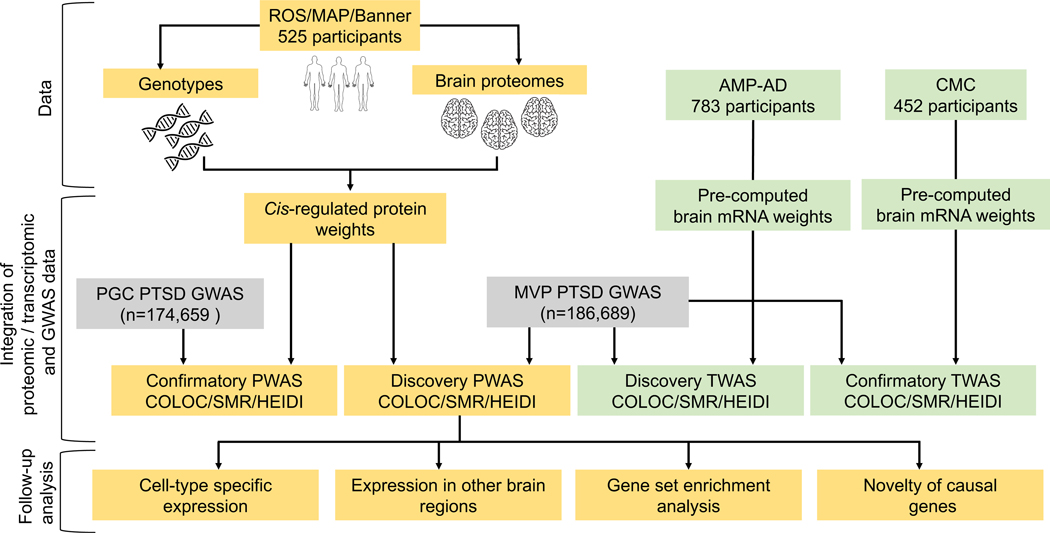
To that end, we employed two independent complementary analytical approaches18, 19 to integrate reference human brain proteomes with PTSD GWAS results using a discovery and confirmatory study design (Figure 1). Specifically, we performed a proteome-wide association study (PWAS) of PTSD followed by Mendelian randomization18, 19 to identify potential causal genes that act via their cis-regulated brain protein abundance to contribute to PTSD pathogenesis. Furthermore, to examine these genes at both the transcript and protein levels, we performed a transcriptome-wide association study (TWAS) of PTSD integrating reference human brain transcriptomes with the PTSD GWAS results. Lastly, we examined the expression of the potential causal genes in other PTSD-relevant brain regions and in specific cell types. We present here the first PWAS of PTSD to identify brain proteins that are promising targets for further mechanistic and therapeutic studies.
Figure 1:
Graphical abstract
RESULTS
Discovery PWAS of PTSD
We performed a discovery PWAS of PTSD by integrating the MVP PTSD GWAS results (n = 186,689 EA participants)17 with reference human brain proteomes (n = 525)20, 21 following the FUSION pipeline19. The integration involves two steps. In the first step, the genetically regulated protein levels, referred to as cis-regulated protein levels, were estimated using the human brain proteomes and corresponding genome-wide genotypes in 525 individuals. Before estimation, the proteomic data underwent quality control and removal of effects of cognitive diagnosis and technical factors. After quality control, the proteomic profiles consisted of 8610 proteins, of which 2179 proteins had significant heritability (i.e., heritability p < 0.01), for which the cis genetic component of protein expression can be estimated. In the second step, the cis-regulated protein levels were integrated with the PTSD GWAS summary association statistics using FUSION19 to perform a PWAS of PTSD.
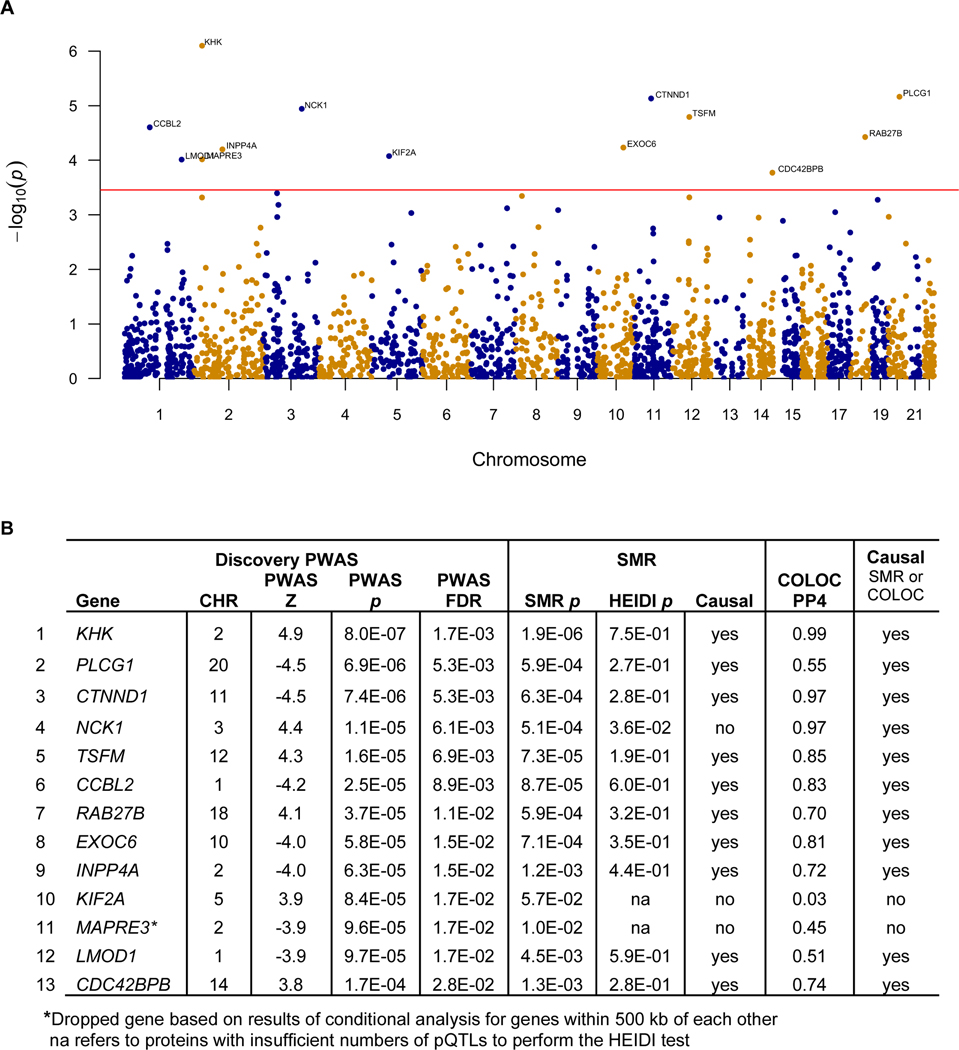
The discovery PWAS of PTSD identified 13 genes whose cis-regulated brain protein levels were associated with PTSD at FDR p <0.05 (Figure 2, Supplementary Table 1). Among these 13 genes, two are located within 500 kb of each other (MAPRE3 and KHK on chromosome 2). Conditional analysis identified KHK in the pair as jointly significant and MAPRE3 was no longer significant and thus was dropped. Therefore, the discovery PWAS identified 12 independent genes whose cis-regulated brain protein levels were associated with PTSD at FDR p <0.05.
Figure 2: Results of the discovery PWAS of PTSD.
2A: Manhattan plot for the discovery PWAS of PTSD, which identified 13 genes whose cis-regulated brain protein abundances were associated with PTSD at FDR p<0.05. 2B: Table for the results of the discovery PWAS of PTSD, followed by Mendelian randomization, HEIDI, and COLOC. Among these 13 genes, 1 was dropped (marked with asterisk) due to not being jointly significant when it was considered with another gene within 500 kb of its location in the conditional analysis. Among the remaining 12 genes, 11 were consistent with being causal based on Mendelian randomization and HEIDI or COLOC.
The association between cis-regulated protein levels and PTSD identified from the PWAS can arise from pleiotropy (SNP affecting both protein abundance and PTSD), causality (SNP affecting protein abundance, which in turn influences PTSD), or linkage disequilibrium (LD, i.e. SNP affecting protein abundance is in LD with SNP affecting PTSD). We tested for the possibilities of pleiotropy/causality versus LD using summary data-level Mendelian randomization (SMR)18 as well as COLOC22. For simplicity, we will refer to pleiotropy or causality as consistent with being causal henceforth. SMR/HEIDI and COLOC results suggest that 11 of the 12 PWAS significant proteins were consistent with being causal (Figure 2, Supplementary Table 2). In sum, we performed a discovery PWAS of PTSD, followed by Mendelian randomization and COLOC and identified 11 genes that modulate their brain protein expression to predispose to PTSD.
Confirmatory PWAS of PTSD
To increase confidence in our PWAS findings, we performed a confirmatory PWAS of PTSD by integrating results from an independent PGC PTSD GWAS (n = 174,659 EA participants)16 with the 525 reference brain proteomes (Supplementary table 3). Focusing on the 11 potential causal genes identified in the discovery PWAS, we performed a meta-analysis of the discovery and confirmatory PWAS for these genes. Replication was defined as having a smaller p-value for association in the meta-analysis than the p-values from both the discovery and confirmation PWAS as well as having consistent directions of association between protein expression and PTSD in the discovery and confirmatory PWAS. We found that 7 of 11 genes replicated (64%; Table 1).
Table 1:
Results of the confirmatory PWAS of PTSD, followed by a meta-analysis of the discovery and confirmatory PWAS for the 11 causal proteins. Seven of the 11 proteins (64%) replicated. Replication was defined has having the meta-analysis p-value smaller than that from both the discovery and replication PWAS and in the same direction of association.
| Discovery PWAS | Confirmatory PWAS | Meta-analysis | Replicated | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Causal | Z | p | FDR | Z | p | p | Direction | ||
|
| ||||||||||
| 1 | KHK | yes | 4.9 | 8.0E-07 | 0.002 | 2.5 | 0.01 | 3.8E-08 | ++ | yes |
| 2 | PLCG1 | yes | -4.5 | 6.9E-06 | 0.005 | -2.6 | 0.01 | 2.0E-07 | -- | yes |
| 3 | CTNND1 | yes | -4.5 | 7.4E-06 | 0.005 | -3.1 | 0.00 | 4.7E-08 | -- | yes |
| 4 | NCK1 | yes | 4.4 | 1.1E-05 | 0.006 | -1.1 | 0.25 | 1.7E-03 | +- | no |
| 5 | TSFM | yes | 4.3 | 1.6E-05 | 0.007 | 0.8 | 0.43 | 4.4E-05 | ++ | no |
| 6 | CCBL2 | yes | -4.2 | 2.5E-05 | 0.009 | -2.8 | 0.01 | 4.7E-07 | -- | yes |
| 7 | RAB27B | yes | 4.1 | 3.8E-05 | 0.012 | -0.7 | 0.47 | 1.7E-03 | +- | no |
| 8 | EXOC6 | yes | -4.0 | 5.9E-05 | 0.015 | -1.3 | 0.18 | 3.7E-05 | -- | yes |
| 9 | INPP4A | yes | -4.0 | 6.3E-05 | 0.015 | -2.2 | 0.03 | 5.3E-06 | -- | yes |
| 10 | LMOD1 | yes | -3.9 | 9.7E-05 | 0.017 | 0.5 | 0.64 | 2.1E-03 | -+ | no |
| 11 | CDC42BPB | yes | 3.8 | 1.7E-04 | 0.028 | 2.1 | 0.04 | 1.7E-05 | ++ | yes |
Examining the potential PTSD-causal genes at the transcript level
Given the central dogma that DNA is transcribed into mRNA, which is translated into protein, we investigated the relationship between brain transcript level and PTSD for these 11 PTSD genes identified herein as potentially causal. To this end, we performed a TWAS of PTSD by integrating the MVP PTSD GWAS results17 with the 888 transcriptomes mainly profiled from the frontal cortex of postmortem brain tissues donated by participants of the Accelerating Medicines Partnership Alzheimer’s Disease (AMP-AD)21, 23. This will be referred to as the TWAS of PTSD. Likewise, we performed a second TWAS using the MVP PTSD GWAS17 and the protein weights estimated from the 452 transcriptomes profiled from the dPFC of the Common Mind Consortium (CMC) participants24 and will refer to this as the CMC-TWAS of PTSD. Among the 11 genes from the discovery PWAS, 2 in the TWAS and 4 in the CMC-TWAS were also associated with PTSD at the transcript level at FDR <0.05 (Table 2). Together, 4 of the 11 transcripts were associated with PTSD in either TWAS at FDR <0.05 (Table 2). In sum, 36% of the PTSD potential causal proteins also have their transcript level associated with PTSD.
Table 2:
Association between PTSD and the cis-regulated mRNA levels of the 11 causal proteins identified in the discovery PWAS. Of note, the discovery PWAS was based on MVP (symptom severity scale), the replication PWAS was based on PGC (case-control study), and the TWASs were based on MVP (symptom severity scale). Novel genes indicate genes not located within 1 Megabase of the MVP PTSD GWAS SNPs with p < 5×10−8.
| Gene | Discovery PWAS | Confirmatory PWAS | TWAS (AMP-AD) | TWAS (CMC) | Novel genes | Confidence level | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1 | CCBL2 | causal | replicated | significant | significant | yes | very high |
| 2 | EXOC6 | causal | replicated | - | significant | yes | very high |
| 3 | INPP4A | causal | replicated | - | - | yes | high |
| 4 | CDC42BPB | causal | replicated | - | - | yes | high |
| 5 | KHK | causal | replicated | - | - | yes | high |
| 6 | PLCG1 | causal | replicated | - | - | yes | high |
| 7 | CTNND1 | causal | replicated | - | - | yes | high |
| 8 | TSFM | causal | no | significant | significant | yes | high |
| 9 | NCK1 | causal | no | - | significant | yes | high |
| 10 | RAB27B | causal | no | - | - | no | moderate |
| 11 | LMOD1 | causal | no | not significant | not significant | yes | low |
“-” indicates genes not present in the TWAS due to not having significant SNP-based heritability. “replicated” indicates that the p-value from the meta-analysis was smaller than those from both the discovery and confirmatory PWAS.
Though not the main focus on this paper, the TWAS of PTSD found 46 brain transcripts associated with PTSD at FDR p<0.05 (Supplementary Table 4). Of those, 26 had evidence consistent with being causal based on SMR and HEIDI (Supplementary Table 4). The CMC-TWAS yielded 40 brain transcripts associated with PTSD at FDR p<0.05 (Supplementary Table 5). Of these, 28 were consistent with being causal based on SMR and HEIDI (Supplementary Table 5). In comparing the TWAS of PTSD (including 6604 genes) with the CMC-TWAS of PTSD (including 5311 genes), we found 3341 genes reported in both TWAS. Among the 46 significant transcripts from the TWAS, 17 were reported in the CMC-TWAS, and 14 / 17 were associated with PTSD at nominal p <0.05 while 8 / 17 associated with PTSD at FDR p<0.05 (adjusted for 5311 transcripts; Supplementary table 6). Among the 40 significant transcripts from the CMC-TWAS, 21 were included in the TWAS, and 17 / 21 were associated with PTSD at p<0.05, while 8 / 21 were associated with PTSD at FDR p<0.05 (Supplementary table 6).
Cell-type specific expression of the potential PTSD causal genes
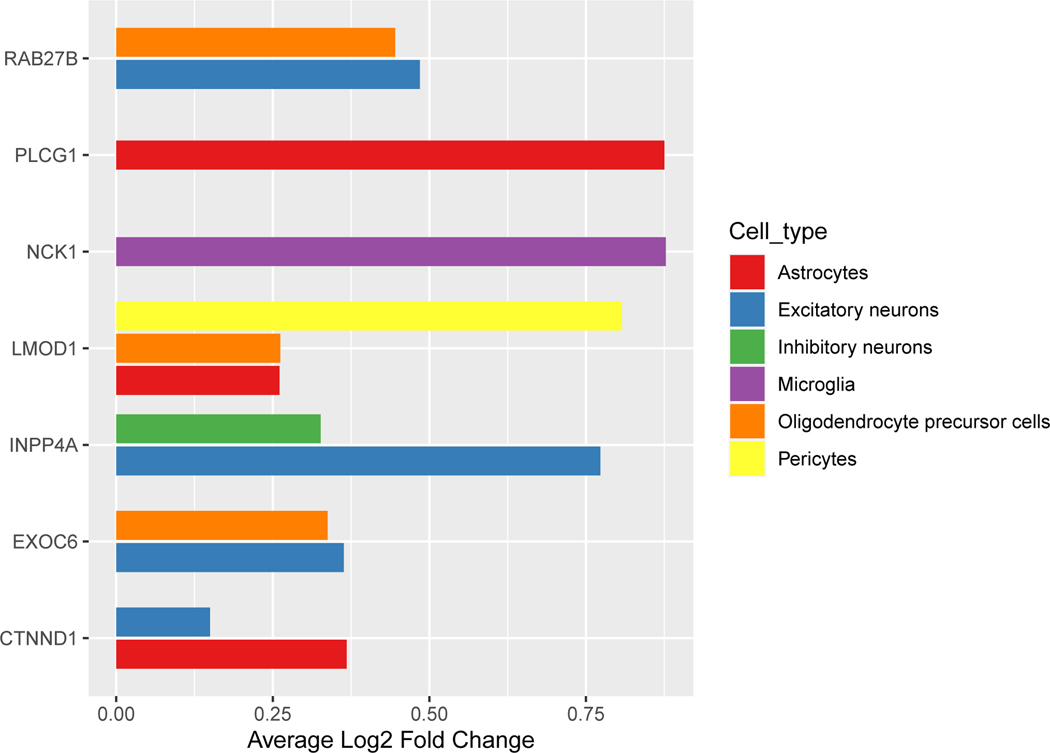
To understand gene activities across cell types, we examined brain cell-type specific expression of these genes using human single cell RNA sequencing data profiled from the dPFC of cognitively normal donors25. Among these 11 genes, 7 were enriched in one or more cell types, including excitatory neurons, inhibitory neurons, astrocytes, microglia, oligodendrocyte precursor cells, and pericytes, while the other 4 did not show evidence of enrichment in a particular cell type (Figure 3, Supplementary table 7). Four genes were highly expressed in excitatory neurons (RAB27B, INPP4A, EXOC6, and CTNND1; Figure 3) and one gene was highly expressed in inhibitory neurons (INPP4A).
Figure 3: Bar graph of single-cell-type enrichment for the 11 potential PTSD causal genes.
Among these genes, seven were enriched in a particular cell type at Bonferroni adjusted p < 0.05 (adjusted for 17,551 genes). Depicted here is the average log fold enrichment for each gene by each brain cell type. Data were from human brain single nuclei RNA-sequencing from the dPFC from Mathys et al, 2019. The x-axis shows the average log fold change for expression of the gene in the particular cell type versus the rest of the other cell types. The y-axis shows the gene name. The full statistics are presented in Supplementary Table 7.
Expression of potential PTSD causal genes in other brain regions
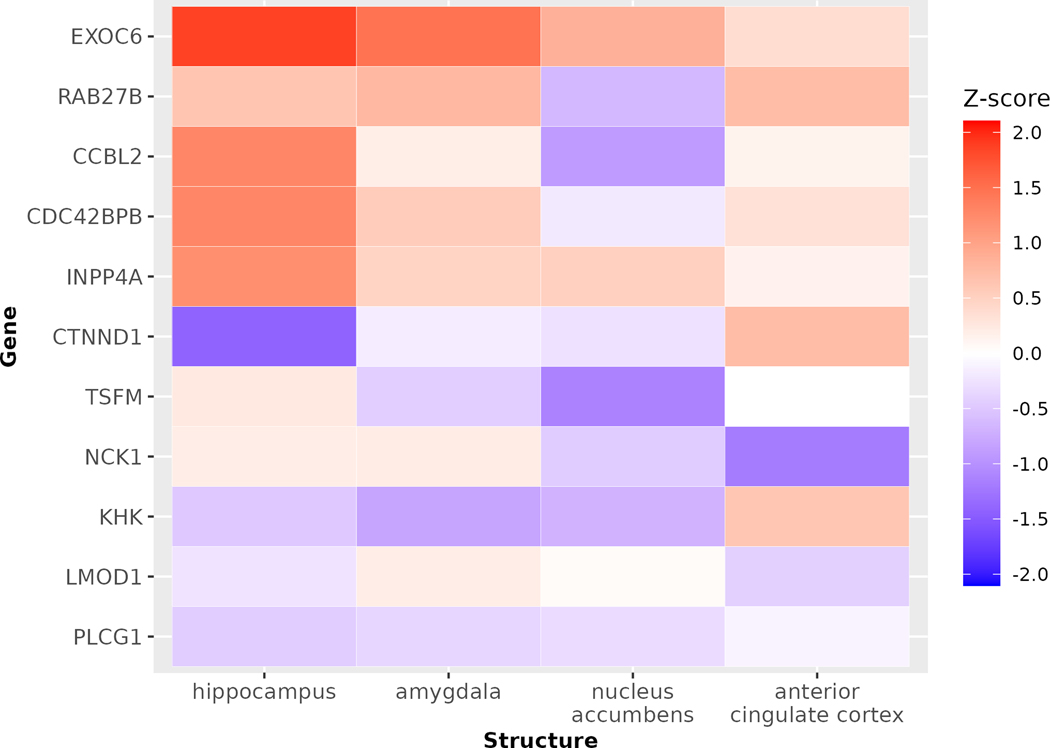
Next, we asked whether these 11 genes are expressed in other PTSD-relevant brain regions such as amygdala, hippocampus, anterior cingulate cortex, medial prefrontal cortex, and nucleus accumbens26. To this end, we used RNA microarray data from the Allen Brain Atlas27, which have been normalized to allow for comparison across genes. The microarray data were not available for medial prefrontal cortex. Among the 11 potential causal genes, 7 (64%) are highly expressed or expressed in the hippocampus, 8 (73%) are highly expressed or expressed in the amygdala, 7 (64%) are highly or lowly expressed in the anterior cingulate cortex, and 2 (14%) are highly expressed in the nucleus accumbens (Figure 4). These observations suggest that the majority of the 11 potential PTSD causal genes are expressed in PTSD-relevant brain regions.
Figure 4: Expression of the 11 potential PTSD causal genes in different PTSD-relevant brain regions.
We accessed microarray expression data from 6 neurotypical adults from the Allen Brain Atlas data portal. The data have been normalized to allow comparison across genes. We focused on expression for the 11 genes of interest in four PTSD-relevant regions (amygdala, anterior cingulate cortex, hippocampus, and nucleus accumbens). The medial prefrontal cortex is also considered PTSD-relevant, but microarray data were unavailable for this structure. Each gene was represented on the microarray by at least two probes. For this heatmap, we selected the single most abundant probe per gene and averaged the expression Z-scores across samples for each gene and brain structure. The higher Z score indicates higher expression in that brain region.
Protein-protein interaction network analysis
In examining the connectivity and biological pathways for the 11 potential PTSD causal proteins, we performed network-based genomic analysis using GeNets28. Based on protein-protein interaction, we found one protein community including NCK1 and PLCG1 (Supplementary Figure 1). A community is defined as a set of proteins that are more connected to each other than they are to other proteins28. GeNets also enables gene set enrichment analysis using the canonical pathways from gene sets in the MSigDB29. We found that these 11 genes were enriched in angiogenesis and vasculogenesis (VEGFR1 pathway), B-cell receptor activation, axon guidance (Netrin pathway), vesicular trafficking (ERRB1 receptor proximal pathway and ARF6 trafficking pathway), cell-cell adhesion (E-Cadherin stabilization pathway), insulin pathway, and metabolism of inositol phosphate (Supplementary Figure 1). Interestingly, 4 proteins were enriched in several pathways - CTNND1, EXOC6, NCK1, and PLCG1.
Novelty of these PTSD causal genes
To determine the novelty of these 11 potentially causal PTSD genes, we determined the lowest p-values for the single nucleotide polymorphisms (SNPs) within 1Mb window of these genes using the summary statistics from the MVP PTSD GWAS17. We found that one gene was located within 1 Mb of a genome-wide significant GWAS signal (RAB27B), while the other 10 genes were not, suggesting that these 10 are novel genes not implicated in PTSD by the latest GWAS (Table 2, Supplementary table 8). The p values for these 11 genes range from 3.7×10−5 to 1.6×10−7. These findings are consistent with those from prior TWAS studies that found risk genes in regions without genome-wide significant p values24, 30, 31. Furthermore, these 11 genes point to specific brain proteins that they likely act through to predispose to PTSD.
PTSD and MDD have shared potential causal genes
PTSD and MDD commonly co-occur, with a comorbidity rate of approximately 50%32. Interestingly, we observed a moderate genetic correlation between PTSD and MDD (correlation of 0.6, p = 2.6 × 10−146) using the MVP PTSD GWAS17 and the latest depression GWAS33, suggesting a substantial shared genetic basis between PTSD and MDD. A natural question that arises is whether there are shared causal genes between PTSD and MDD. In a prior study, we identified 25 potential depression causal genes acting via their cis-regulated brain protein levels20. Here, we found that PTSD and MDD have 2 shared potential causal genes - RAB27B (Z = 4.1, FDR p=0.012 for PTSD; Z = 5.7, FDR p=7.1×10−6 for MDD) and CTNND1 (Z = −4.5, FDR p=0.005 for PTSD; Z = −4.8, FDR p=5.3×10−4 for MDD). These genes predispose to PTSD and MDD via their cis-regulated brain protein levels in consistent directions of association; higher cis-regulated brain protein level of RAB27B and lower cis-regulated brain protein level of CTNND1 were associated with higher risk for PTSD and MDD.
Level of evidence for the potential PTSD causal genes
We assigned a level of confidence for the 11 potential causal genes identified in the discovery PWAS based on evidence for replication in the confirmatory PWAS and results from the two TWAS. The transcriptomes used in the TWAS were profiled with a different technology from mass spectrometry and from brain tissues of donors mostly independent from donors of brains for the proteomes, hence, the TWAS provided an independent layer of validation for the PWAS findings. Taking into consideration findings from the confirmatory PWAS and both TWAS, we had low confidence in LMOD1 as it did not replicate in the confirmatory PWAS or in either TWAS (Table 2) and moderate confidence in RAB27B because it did not replicate in the confirmatory PWAS and was not reported in the TWAS due to lack of SNP-based heritability (Table 2). We had high confidence in 7 genes (INPP4A, CDC42BPB, KHK, PLCG1, and CTNND1; Table 2) because they replicated in the confirmatory PWAS and in TSFM and NCK1 because they were also associated with PTSD at the transcript level in both TWAS (Table 2). Finally, we had very high confidence in CCBL2 and EXOC6 because they replicated in the confirmatory PWAS and were also associated with PTSD at the transcript level in both TWAS (Table 2). In sum, among the 11 potentially causal genes, we have high level of confidence in 9 of them.
DISCUSSION
In this study we sought to identify brain proteins that predispose to PTSD to find new treatment targets. To this end, we integrated the latest PTSD GWAS results with reference human brain proteomes and transcriptomes, followed by Mendelian randomization, using a discovery and confirmatory study design. In light of the range of severity of PTSD symptoms, we focused on PTSD symptom severity as the outcome in the discovery dataset and validated the identified proteins in the replication dataset with the case/control design as the outcome. We identified 11 potential causal PTSD genes that act via their cis-regulated brain proteins to contribute to PTSD pathogenesis. Notably, 7 / 11 genes (64%) replicated in a confirmatory PWAS, and 4 / 11 genes also had their cis-regulated brain mRNA levels associated with PTSD, providing an additional layer of confirmation.
Only 36% of the potential PTSD causal proteins also have their transcript levels associated with PTSD, suggesting that brain transcripts may not be a good proxy for brain proteins in many genes, and vice versa, consistent with observations from prior studies34, 35. This is likely due to the layers of regulation after transcription, including microRNA regulation, translational regulation, and post-translational regulation. Interestingly, when we compared our TWAS and PWAS findings with the differentially expressed mRNAs in dPFC in PTSD from a published transcriptome-wide differential expression analysis36, 1 / 11 (9%) of our PWAS-significant genes (LMOD1) and 2 / 42 (5%) of our TWAS-significant genes (WNT3 and DPYSL5) were differentially expressed in the dPFC in PTSD at FDR p<0.05. Since proteins are the main functional components of cells and biological processes, these findings highlight that studying measured brain proteomes directly offer additional important insights.
Notably, we found 2 potential causal genes that are shared between PTSD and MDD, RAB27B and CTNND1. They are associated with both PTSD and MDD, which are comorbid in 50% of the PTSD patients32. Treatments that target these shared genes may benefit patients with comorbid PTSD and depression and should be prioritized for further mechanistic and therapeutic studies.
Three previous studies included a TWAS of PTSD17, 36, 37 and our TWAS findings are very consistent with these published results. For instance, Huckins and colleagues performed a TWAS of PTSD integrating the GTEx transcriptomes and PGC PTSD GWAS results37. Focusing on their results in the dPFC, the brain region we studied here, we found that 6 of our 46 TWAS-significant genes were associated with PTSD in Huckins’ TWAS at nominal p<0.05 and in consistent direction of association (SLC35F6, DPYSL5, PET112, B3GNT1, CCBL2, LRRC37A2, Supplementary table 9). Likewise, Girgenti and colleagues performed a TWAS of PTSD using the MVP PTSD GWAS and GTEx transcriptomes from brain (cortical and CNS tissue) and other non-CNS tissues and published their significant findings36, which we used to compared with our TWAS results. Five of their 17 cortical tissue-TWAS-significant genes36 were present in our TWAS, and all 5 were significant in our TWAS at FDR p<0.05 (Supplementary table 10). We could not find information on direction of association in Girgenti et al’s publication to compare with ours. Similarly, Stein and colleagues performed a TWAS of PTSD integrating the MVP PTSD GWAS with GTEx transcriptomes and found 10 significant genes in their TWAS in the dPFC17. Five of these 10 genes were reported in our TWAS and all 5 were significant in our TWAS at FDR p<0.05 and with consistent direction of association (LRRC37A4P, ARL17A, RBM6, LRRC37A2, RNF123, Supplementary table 11). Taken together, our TWAS findings were highly consistent with those of the published TWAS for the commonly reported genes - 100% consistent with Girgenti et al cortical tissue-TWAS36 and 100% consistent with Stein et al dPFC-TWAS17.
We also compared our PWAS findings with results from the published TWAS. Among our 11 causal proteins from the PWAS, one was associated with PTSD in the Huckins’ TWAS37 at nominal p<0.05 and in consistent direction (CCBL2; Supplementary table 9). One of the 10 TWAS-significant genes in the dPFC in Stein et al study17 was present in our PWAS, but its protein level was not associated with PTSD in our PWAS (RNF123). Among Girgenti et al’s TWAS-significant genes36, 4 genes were present in our PWAS, and 2 / 4 were associated with PTSD in our PWAS at p <0.05 (MON1A and PLCD3) but none was significant at FDR p <0.05 (Supplementary table 12). Again, we were unable to find the direction of association in Girganti et al published study to compare with ours. As expected, findings from our TWAS and the published TWAS were more consistent with each other than between our PWAS and the published TWAS, as the TWAS and PWAS can provide independent information.
Based on published literature, these 11 genes are involved in several biological processes, including vesicular trafficking (EXOC638, RAB27B39), signal transduction (PLCG1, CTNND1, NCK138), neurotransmission, maintenance of excitatory neurons, and immune / inflammation (CCBL240, 41). Specifically, CCBL2 is a transcriptional co-activator and plays an important and dose-dependent role in neurodevelopment and maintenance of excitatory forebrain neurons42. Furthermore, CCBL2 is also known as kynurenine aminotransferase 3 (KAT3), which transaminates kynurenine to form kynurenic acid. Thus, CCBL2 is an important factor in the kynurenine pathway, which interfaces between the immune / inflammatory response and serotoninergic and glutamatergic neurotransmission40, 41. Other processes these genes are involved in include sugar metabolism (KHK38), cell-cell adhesion (CTNND138), cell migration (LMOD1, CDC42BPB38), cell survival (INPP4A43), and mitochondrial function (TSFM38). Among these genes, CTNND1 was associated with five psychiatric disorders (ADHD, autism spectrum disorder, major depressive disorder, obsessive compulsive disorder, and schizophrenia) in GWAS44 and is differentially expressed in the brain in PTSD social-stress mouse model45. Drug compounds targeting these 14 genes have not been studied in clinical trials when we checked the Open Targets database46.
Our findings should be interpreted in light of the study’s limitations. First, we could examine only 2179 brain proteins in the PWAS. This is because SNP-based heritability estimates for proteins depend on the power for the study, which is dependent on the number of reference brain proteomes. Currently, we had 525 human brain proteomes and future studies with a larger number of human brain proteomes can have more power to identify heritable proteins and thus more proteins will be examined in the PWAS. Second, we do not have measured brain proteins in post-mortem brains of subjects with PTSD to further validate our findings and this should be the focus of future studies. Nevertheless, we have a confirmatory PWAS and two TWAS to provide multiple layers of validation for our findings. Third, we were limited to studies of European-ancestry subjects due to the power of the available GWAS. Fourth, the proportion of participants with PTSD also having MDD in the two published PTSD GWAS that we used the summary statistics of is unknown. Hence, it is possible that the identified genes for PTSD may contribute to MDD as well. However, this appears to be limited as we only found 2 common causal genes between PTSD and MDD. Fifth, PTSD is a heterogeneous syndrome that has overlapping symptoms with MDD or anxiety disorders; hence interpretation of the findings should take this into consideration. Sixth, the PTSD GWAS we used in the discovery PWAS focused on the continuous PTSD phenotype (i.e., PTSD symptom severity) while the PTSD GWAS used in the confirmatory PWAS focused on the binary PTSD phenotype (i.e., diagnosis of PTSD) as these were the largest available GWAS. However, this is mitigated by the high phenotypic correlation (r = 0.86) and very high genetic correlation (rg = 0.97) between the binary and continuous phenotype of PTSD17, suggesting that they are likely influenced by the same genetic basis.
This study has several strengths. First, this is the first PWAS of PTSD to the best of our knowledge. We note that among the 7 potentially causal genes identified in the discovery PWAS and replicated in the confirmatory PWAS, only 2 (29%) showed association at the transcript level in the TWAS. This highlights the important insights afforded by studying measured brain proteins. Second, we used a discovery and confirmatory study design to enhance the level of confidence in our findings. Third, we performed two TWAS of PTSD using 888 transcriptomes from the AMP-AD in the first TWAS and 452 brain transcriptomes from the CommonMind Consortium in the second TWAS. These TWAS illuminate the association at the transcript level and provide an additional layer of validation for the PWAS findings. Fourth, this is the first study that performed both a PWAS and TWAS to gain more insights in the action of genes at both the transcript and protein level.
In summary, we identified 11 genes for which we provided varying levels of support that they contribute to PTSD pathogenesis via their cis-regulated brain protein abundance. These are promising targets for further mechanistic and therapeutic studies to find new effective treatment for PTSD.
METHODS
Human brain proteomic and genetic data
The reference human brain proteomes were profiled from the dorsolateral prefrontal cortex (dPFC) of post-mortem brain samples donated by participants of European descent of the Religious Orders Study and Rush Memory and Aging Project (ROS/MAP)47 and Banner Sun Health Research Institute (Banner) and have been described in detail previously20, 21, 48, 49. ROS/MAP participants provided informed consent, signed an Anatomic Gift Act, and a repository consent to allow their data and biospecimens to be repurposed. The studies were approved by an Institutional Review Board of Rush University Medical Center. All Banner participants or their legal representatives signed an informed consent and the study was approved by the Banner Sun Health Research Institute Institutional Review Board.
Proteomic profiling was performed using isobaric tandem mass tag (TMT) peptide labeling and analyzed by liquid chromatography coupled to mass spectrometry as described in detail previously20, 21, 48, 49. Prior to TMT labeling, samples were randomized by age, sex, post-mortem interval, cognitive diagnosis, and pathologies to prevent batch effect. Peptides from each individual sample and the global internal standard were labeled using the TMT 10-plex kit (ThermoFisher). High pH fractionation was performed as previously described with slight modifications50. Database searches and protein quantification have been described in detail here21, 48. Briefly, all raw files were analyzed using the Proteome Discoverer suite (version 2.3 ThermoFisher Scientific) and MS2 spectra were searched against the canonical UniProtKB Human proteome database downloaded in April 2015 for Banner proteomes and in February 2019 for ROS/MAP proteomes. Percolator was used to filter peptide spectral matches (PSM) and peptides to a false discovery rate (FDR) of less than 1%. Following spectral assignment, peptides were assembled into proteins, which were further filtered based on the combined probabilities of their constituent peptides to a final FDR of 1%. In cases of redundancy, shared peptides were assigned to the protein sequence based on parsimony. Reporter ions were quantified from MS2 or MS3 scans using an integration tolerance of 20 ppm with the most confident centroid setting.
The quality control of the proteomes has been described in detail previously20, 21, 48. Briefly, for each batch, the GIS were used to check for proteins outside of the 95% confidence interval and set to missing. Proteomic analysis in ROS/MAP proteomes identified 12,691 proteins and in Banner proteomes identified 11,518 proteins. Proteins with missing values in more than 50% of the subjects were excluded. Each protein abundance was then scaled by a sample-specific total protein abundance to remove effects of protein loading differences, and then log2 transformed. Outlier samples were identified and removed through iterative principal component analysis (PCA). In each iteration, samples more than four standard deviations from the mean of either the first or second principal component were removed, and principal components were recalculated for the next iteration. We used regression to remove effects of protein batch, MS2 versus MS3 reporter quantitation mode, sex, age at death, postmortem interval, study (ROS vs. MAP), and the final clinical diagnosis of cognitive status from the proteomic profile before estimating the protein weights. After this step, we generated a combined set of ROS/MAP and Banner proteomic profiles by scaling to the mean and one unit of standard deviation.
Genotyping for subjects with proteomic data (and of European ancestry) was generated by either whole genome sequencing (WGS) and/or genome-wide genotyping by either Illumina OmniQuad Express or Affymetrix GeneChip 6.0 platforms for ROS/MAP subjects51, 52. We prioritized using WGS over genotyping where available and performed quality control of WGS and genotyping data separately using Plink53. Individuals from Banner were genotyped using Affymetrix Precision Medicine Array using DNA extracted from brain using Qiagen GenePure kit. We excluded individuals with genotyping missing rate >5%, variants with Hardy Weinberg equilibrium p-value < 1E-08, variants with missing genotype rate >5%, variants with minor allele frequency <1%, and variants that are not single nucleotide polymorphisms (SNPs). Genotyping for each individual was imputed to the 1000 Genome Project Phase 354 using the Michigan Imputation Server55 and SNPs with imputation > 0.3 were retained. Genotyping was filtered to include 1,190,321 HapMap SNPs from the 489 individuals of European descent from the 1000 Genomes Project19, which is provided by FUSION19 and often referred to as the linkage disequilibrium (LD) reference panel. We used KING to remove individuals who were estimated to be closer than second degree kinship56. The sample outliers in population structures were identified and removed by EIGENSTRAT. After quality control, 525 subjects with both proteomic and genetic data were included in the analyses.
Human brain transcriptomic and genetic data
The 888 reference brain transcriptomes were profiled from post-mortem brain samples of 783 individuals of European descent recruited by the ROS/MAP, Mayo, and Mount Sinai Brain Bank studies47, 57, 58. The transcriptomes were mainly profiled from the dPFC and also from frontal cortex, temporal cortex, inferior frontal gyrus, superior temporal gyrus, and perirhinal gyrus. Alignment, quality control, and normalization of RNA-sequencing data have been described in detail before20, 21, 59. Briefly, BAM files were converted to FASTQ format using Picard v.2.2.4, followed by alignment of reads to GRCh38 reference genome using STAR v.2.460. Gene-level counts were computed using STAR. Genes with < 1 count per million in at least 50% of the samples and with missing gene length and percent GC content were removed. Outlier samples were removed. Effects of batch, sex, post-mortem interval, age at death, final diagnosis of cognitive status, and brain region were regressed from these transcriptomes. After quality control, 13,650 mRNAs remained and were considered for the TWAS. Genome-wide genotypes were generated as previously described47, 57, 58, and quality control of the genotypes was the same as was described above.
Additionally, we used the mRNA weights calculated from the 452 transcriptomes profiled from the dPFC from the CommonMind Consortium participants provided here (gusevlab.org/projects/fusion24) to perform the second TWAS of PTSD.
PTSD GWAS summary statistics
For the discovery PWAS, we used the summary statistics from the MVP PTSD GWAS17, which included 186,689 participants of European descent. We used the summary statistics of the GWAS of PTSD using the PCL total score17. For the confirmatory PWAS, we use the summary statistics from the PGC-PTSD GWAS, which used the PTSD case control phenotype and included 174,659 participants of European ancestry16.
Statistical approach
For the PWAS, we used FUSION (downloaded from http://gusevlab.org/projects/fusion on May 2, 2019)19 to estimate protein weights. Briefly, we estimated SNP-based heritability for each gene using protein data. For proteins with significant SNP-based heritability (heritability p-value <0.01), we used FUSION to compute the effect of SNPs on protein abundance using multiple predictive models (top1, blup, lasso, enet, bslmm)19, and the most predictive model was selected. Likewise, for mRNA with significant SNP-based heritability (heritability p-value <0.01), we used FUSION with modifications to estimate mRNA weights. The intention of our modifications of FUSION was to generate the most predictive models of cortical brain transcript expression that were not unduly influenced by either a single individual or a particular brain region. This approach was described and evaluated in detail by Gockley et al23 and is summarized here. First, we used the flag -scale 1 to handle pre-scaled expression values, as expression was scaled across individual brain regions before filtering for matched genotype and combining across brain regions. Second, we ensured that brain expression data from the same individual were included in the same cross validation set and that no fold differed in size by more than 5% from any other fold, and all individuals were only included once in testing the transcript expression models. This cross-fold validation schema prevents information leakage between training and validation folds during model assessment. For both PWAS and TWAS, FUSION was used to combine the genetic effect of PTSD (i.e., PTSD GWAS Z-score) with the mRNA or protein expression weights by calculating the linear sum of for the independent SNPs at the locus to perform a PWAS or TWAS19. The HLA region was excluded due to its complex LD structure.
Summary data-based Mendelian Randomization18 (SMR, downloaded from https://cnsgenomics.com/software/smr on May 22, 2019) was used to test whether the PWAS-significant proteins from the FUSION approach mediate the association between genetic variants and PTSD. We used Plink53 to estimate protein quantitative trait loci (pQTL) in the proteomic dataset by linear regression. Next, we used the pQTL results and the MVP PTSD GWAS summary statistics to perform SMR18. We used the conservative unadjusted p-value ≤ 0.05 from Heterogeneity in Dependent Instrument (HEIDI) to suggest that presence of linkage likely influences the main SMR findings. For the colocalization test, we used the COLOC software22 to estimate the posterior probability of the protein and PTSD sharing a causal variant (PP4), and the posterior probability of the protein and PTSD not sharing a causal variant using the marginal association statistics.
A hypothesis-driven meta-analysis of the discovery and confirmatory PWAS was performed using METAL61. We declared as replicated for genes with meta-analysis p-values lower than the p-values of the discovery dataset and had the same direction of association in both datasets.
Using human brain single-cell RNA-sequencing data from 24 cognitively normal donors profiled from the dPFC from Mathys et al25, we examined the cell-type specific expression of the 11 potential PTSD causal genes. First, we performed data preprocessing and transformation using the Seurat package version 3.1.262. Genes were removed if they had fewer than 3 counts in a cell, and cells were removed if they had unique feature counts over 2,500 or less than 200. The RNA counts were normalized and scaled using the NormalizeData and ScaleData functions. We then focused on the 6 main cell types provided by Mathys et al using FindMarkers function: excitatory neuron, inhibitory neuron, astrocyte, microglia, oligodendrocyte, and pericyte. For the 11 PTSD causal genes, we performed differential expression analysis to compare their expression levels in one cell type versus the rest of the other cell types using Wilcoxon Rank Sum test to determine if they are highly expressed in a particular cell type. For multiple testing correction, we used Bonferroni adjustment for all 17,551 genes.
We accessed RNA microarray data from 6 neurotypical adults from the Allen Brain Atlas data portal27. The data have been normalized to allow comparison across genes. We focused on expression for the 11 genes of interest in four PTSD-relevant regions - amygdala, anterior cingulate cortex, hippocampus, and nucleus accumbens. The medial prefrontal cortex is also considered PTSD-relevant, but microarray data were unavailable for this structure. Each gene was represented on the microarray by at least two probes. For this heatmap, we selected the single most abundant probe per gene and averaged the expression Z-scores across samples for each gene and brain structure.
We used GeNets, a web platform for network-based genomic analyses, to investigate networks based on protein-protein interaction (PPI) among the 11 causal proteins28. GeNets used the PPI information from the InWeb3 database, a curated and computationally derived PPI network of 420,000 PPIs of high and lower probability interactions63. GeNets implements an algorithm originally presented in Clauset et al64 that identifies so-called communities in a set of genes. A community is a set of genes that are more connected to one another than they are to other groups of genes. Additionally, GeNets enables gene set enrichment analysis on genes within a PPI network using the canonical pathways from 2199 gene sets in the MSigDB29. GeNets applied a hypergeometric test to obtain p-value for the gene set enrichment and used Bonferroni correction to adjust for multiple hypothesis testing.
We used linkage disequilibrium score regression (LDSC)65 to estimate genetic correlation between major depressive disorder and PTSD using the summary statistics from the latest depression GWAS33 and the MVP PTSD GWAS17.
Supplementary Material
Acknowledgements
We are grateful to the participants of the ROS, MAP, Mayo, Mount Sinai Brain Bank, and Banner Sun Health Research Institute Brain and Body Donation Program for their time and participation. We thank the Psychiatric Genomics Consortium PTSD working group for making the summary statistics from the published PTSD GWAS in Nievergelt et al 2019 available. We thank Jiaqi Liu for her assistance with creating the Figure 3.
The authors thank Million Veteran Program (MVP) staff, researchers, and volunteers, who have contributed to MVP, and especially participants who previously served their country in the military and now generously agreed to enroll in the study. (See https://www.research.va.gov/mvp/ for more details)
The following NIH grants supported this work: P30 AG066511 (A.I.L.), P50 AG025688 (A.I.L.), R01 AG015819 (D.A.B.), R01 AG017917 (D.A.B.), R01 AG056533 (T.S.W., A.P.W.), VA 1IK4 BX005219 (A.P.W), I01 BX005686 (A.P.W). T.S.W is also supported by R56 AG060757, R56 AG062256, RF1 AG057470. N.T.S is also supported by R01 AG053960, R01 AG057911, R01 AG061800. D.A.B is also supported by RC2 AG036547, U01 AG046152, U01 AG061356. A.I.L is also supported by U01 AG046161, U01 AG061357. A.P.W is also supported by U01 MH115484, VA I01 BX003853. CMN, MBS, KCK, KJR were supported by R01MH106595. The Brain and Body Donation Program has been supported by NIH, the Arizona Department of Health Services, the Arizona Biomedical Research Commission and the Michael J. Fox Foundation for Parkinson’s Research.
The views expressed in this work do not necessarily represent the views of the Veterans Administration or the United States Government.
Footnotes
Data Availability
-MVP PTSD GWAS summary statistics:
dbGAP (phs001672.v6.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v6.p1)
-PGC PTSD GWAS summary statistics: publicly available on the PGC website (https://www.med.unc.edu/pgc/download-results/) under for the benefit of the wider biomedical community.
-Proteomic datasets: genotypes, protein expression levels, expression covariates:
https://www.synapse.org/#!Synapse:syn24872746
-AMP-AD datasets: genotypes, transcript expression levels, expression covariates
https://adknowledgeportal.synapse.org/Explore/Studies/DetailsPage?Study=syn22313785
-CMC datasets: We downloaded the transcript weights from http://gusevlab.org/projects/fusion/
https://data.broadinstitute.org/alkesgroup/FUSION/WGT/CMC.BRAIN.RNASEQ.tar.bz2
-Sc-RNAseq datasets: https://www.synapse.org/#!Synapse:syn21589957
-Allen brain atlas https://human.brain-map.org/microarray/search
Competing interests
The authors declare no competing interest.
REFERENCES
- 1.Bromet E, Sonnega A, Kessler RC. Risk factors for DSM-III-R posttraumatic stress disorder: findings from the National Comorbidity Survey. American journal of epidemiology 1998; 147(4): 353–361. [DOI] [PubMed] [Google Scholar]
- 2.Richardson LK, Frueh BC, Acierno R. Prevalence estimates of combat-related post-traumatic stress disorder: critical review. The Australian and New Zealand journal of psychiatry 2010; 44(1): 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boe HJ, Holgersen KH, Holen A. Mental health outcomes and predictors of chronic disorders after the North Sea oil rig disaster: 27-year longitudinal follow-up study. J Nerv Ment Dis 2011; 199(1): 49–54. [DOI] [PubMed] [Google Scholar]
- 4.Hull AM, Alexander DA, Klein S. Survivors of the Piper Alpha oil platform disaster: long-term follow-up study. The British Journal of Psychiatry 2002; 181(5): 433–438. [DOI] [PubMed] [Google Scholar]
- 5.Steenkamp MM, Litz BT, Hoge CW, Marmar CR. Psychotherapy for Military-Related PTSD: A Review of Randomized Clinical Trials. Jama 2015; 314(5): 489–500. [DOI] [PubMed] [Google Scholar]
- 6.Hoskins M, Pearce J, Bethell A, Dankova L, Barbui C, Tol WA et al. Pharmacotherapy for post-traumatic stress disorder: systematic review and meta-analysis. The British journal of psychiatry : the journal of mental science 2015; 206(2): 93–100. [DOI] [PubMed] [Google Scholar]
- 7.Shiner B, Westgate CL, Gui J, Maguen S, Young-Xu Y, Schnurr PP et al. A Retrospective Comparative Effectiveness Study of Medications for Posttraumatic Stress Disorder in Routine Practice. The Journal of clinical psychiatry 2018; 79(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis C, Roberts NP, Andrew M, Starling E, Bisson JI. Psychological therapies for post-traumatic stress disorder in adults: systematic review and meta-analysis. European journal of psychotraumatology 2020; 11(1): 1729633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steenkamp MM, Litz BT, Marmar CR. First-line Psychotherapies for Military-Related PTSD. Jama 2020; 323(7): 656–657. [DOI] [PubMed] [Google Scholar]
- 10.Krystal JH, Davis LL, Neylan TC, M AR, Schnurr PP, Stein MB et al. It Is Time to Address the Crisis in the Pharmacotherapy of Posttraumatic Stress Disorder: A Consensus Statement of the PTSD Psychopharmacology Working Group. Biological psychiatry 2017; 82(7): e51–e59. [DOI] [PubMed] [Google Scholar]
- 11.Jovanovic T, Ressler KJ. How the neurocircuitry and genetics of fear inhibition may inform our understanding of PTSD. The American journal of psychiatry 2010; 167(6): 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. The American journal of psychiatry 2002; 159(10): 1675–1681. [DOI] [PubMed] [Google Scholar]
- 13.Sartor CE, Grant JD, Lynskey MT, McCutcheon VV, Waldron M, Statham DJ et al. Common heritable contributions to low-risk trauma, high-risk trauma, posttraumatic stress disorder, and major depression. Archives of general psychiatry 2012; 69(3): 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS genetics 2019; 15(12): e1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelernter J, Sun N, Polimanti R, Pietrzak R, Levey DF, Bryois J et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nature neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen CY, Choi KW et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun 2019; 10(1): 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein MB, Levey DF, Cheng Z, Wendt FR, Harrington K, Pathak GA et al. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nature genetics 2021; 53(2): 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature genetics 2016; 48(5): 481–487. [DOI] [PubMed] [Google Scholar]
- 19.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW et al. Integrative approaches for large-scale transcriptome-wide association studies. Nature genetics 2016; 48(3): 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wingo TS, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM et al. Brain proteome-wide association study implicates novel proteins in depression pathogenesis. Nature neuroscience 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wingo AP, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nature genetics 2021; 53(2): 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS genetics 2014; 10(5): e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gockley J, Montgomery KS, Poehlman WL, Wiley JC, Liu Y, Gerasimov E et al. Multi-tissue neocortical transcriptome-wide association study implicates 8 genes across 6 genomic loci in Alzheimer’s disease. Genome medicine 2021; 13(1): 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nature genetics 2018; 50(4): 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019; 570(7761): 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenster RJ, Lebois LAM, Ressler KJ, Suh J. Brain circuit dysfunction in post-traumatic stress disorder: from mouse to man. Nature reviews Neuroscience 2018; 19(9): 535–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489(7416): 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Kim A, Rosenbluh J, Horn H, Greenfeld L, An D et al. GeNets: a unified web platform for network-based genomic analyses. Nature methods 2018; 15(7): 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardinas AF et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nature genetics 2019; 51(4): 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj T, Li YI, Wong G, Humphrey J, Wang M, Ramdhani S et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nature genetics 2018; 50(11): 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rytwinski NK, Scur MD, Feeny NC, Youngstrom EA. The co-occurrence of major depressive disorder among individuals with posttraumatic stress disorder: a meta-analysis. Journal of traumatic stress 2013; 26(3): 299–309. [DOI] [PubMed] [Google Scholar]
- 33.Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature neuroscience 2019; 22(3): 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N et al. Cell type- and brain region-resolved mouse brain proteome. Nature neuroscience 2015; 18(12): 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature reviews Genetics 2012; 13(4): 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girgenti MJ, Wang J, Ji D, Cruz DA, Stein MB, Gelernter J et al. Transcriptomic organization of the human brain in post-traumatic stress disorder. Nature neuroscience 2021; 24(1): 24–33. [DOI] [PubMed] [Google Scholar]
- 37.Huckins LM, Chatzinakos C, Breen MS, Hartmann J, Klengel T, da Silva Almeida AC et al. Analysis of Genetically Regulated Gene Expression Identifies a Prefrontal PTSD Gene, SNRNP35, Specific to Military Cohorts. Cell reports 2020; 31(9): 107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Current protocols in bioinformatics 2016; 54: 1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- 39.Chen D, Guo J, Miki T, Tachibana M, Gahl WA. Molecular cloning and characterization of rab27a and rab27b, novel human rab proteins shared by melanocytes and platelets. Biochemical and molecular medicine 1997; 60(1): 27–37. [DOI] [PubMed] [Google Scholar]
- 40.Kim YK, Jeon SW. Neuroinflammation and the Immune-Kynurenine Pathway in Anxiety Disorders. Current neuropharmacology 2018; 16(5): 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadriu B, Farmer CA, Yuan P, Park LT, Deng ZD, Moaddel R et al. The kynurenine pathway and bipolar disorder: intersection of the monoaminergic and glutamatergic systems and immune response. Molecular psychiatry 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipinski M, Muñoz-Viana R, Del Blanco B, Marquez-Galera A, Medrano-Relinque J, Caramés JM et al. KAT3-dependent acetylation of cell type-specific genes maintains neuronal identity in the adult mouse brain. Nat Commun 2020; 11(1): 2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhuri R, Khanna K, Koundinya D, Pattnaik B, Vatsa D, Agrawal A et al. Novel nuclear translocation of inositol polyphosphate 4-phosphatase is associated with cell cycle, proliferation and survival. Biochimica et biophysica acta Molecular cell research 2018. [DOI] [PubMed] [Google Scholar]
- 44.Relationships Genomic, Loci Novel, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019; 179(7): 1469–1482.e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muhie S, Gautam A, Chakraborty N, Hoke A, Meyerhoff J, Hammamieh R et al. Molecular indicators of stress-induced neuroinflammation in a mouse model simulating features of post-traumatic stress disorder. Translational psychiatry 2017; 7(5): e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N et al. Open Targets Platform: new developments and updates two years on. Nucleic acids research 2019; 47(D1): D1056–d1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer’s disease : JAD 2018; 64(s1): S161–s189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wingo AP, Fan W, Duong DM, Gerasimov ES, Dammer EB, Liu Y et al. Shared proteomic effects of cerebral atherosclerosis and Alzheimer’s disease on the human brain. Nature neuroscience 2020; 23(6): 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nature medicine 2020; 26(5): 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nature protocols 2018; 13(7): 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiology of aging 2012; 33(5): 1017.e1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Scientific data 2018; 5: 180142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American journal of human genetics 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491(7422): 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A et al. Next-generation genotype imputation service and methods. Nature genetics 2016; 48(10): 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics (Oxford, England) 2010; 26(22): 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Scientific data 2016; 3: 160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Scientific data 2018; 5: 180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wan YW, Al-Ouran R, Mangleburg CG, Perumal TM, Lee TV, Allison K et al. Meta-Analysis of the Alzheimer’s Disease Human Brain Transcriptome and Functional Dissection in Mouse Models. Cell reports 2020; 32(2): 107908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics (Oxford, England) 2010; 26(17): 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 2018; 36(5): 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nature biotechnology 2007; 25(3): 309–316. [DOI] [PubMed] [Google Scholar]
- 64.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Physical review E, Statistical, nonlinear, and soft matter physics 2004; 70(6 Pt 2): 066111. [DOI] [PubMed] [Google Scholar]
- 65.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics 2015; 47(3): 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.