Abstract
DNA nanostructures have proven potential in biomedicine. However, their intracellular interactions—especially cytosolic stability—remain mostly unknown and attempts to discern this are confounded by the complexities of endocytic uptake and entrapment. Here, we bypass the endocytic uptake and evaluate the DNA structural stability directly in live cells. Commonly used DNA structures—crosshairs and a tetrahedron—were labeled with a multistep Förster resonance energy transfer dye cascade and microinjected into the cytosol of transformed and primary cells. Energy transfer loss, as monitored by fluorescence microscopy, reported the structure’s direct time-resolved breakdown in cellula. The results showed rapid degradation of the DNA crosshair within 20 min, while the tetrahedron remained consistently intact for at least 1 h postinjection. Nuclease assays in conjunction with a current understanding of the tetrahedron’s torsional rigidity confirmed its higher stability. Such studies can inform design parameters for future DNA nanostructures where programmable degradation rates may be required.
Keywords: DNA nanotechnology, tetrahedron, cell, cytoplasm, stability, nuclease, FRET, dye, microinjection
Graphical Abstract
The widespread adoption of mRNA-based vaccines against SARS-CoV-2-virus-associated disease confirmed that nucleic acid based therapeutics have arrived.1,2 Extending from the application of these relatively simple oligonucleotides to the complex plethora of structures now afforded by DNA nanotechnology portends future therapeutic roles.3–5 DNA nanostructures can act as designer scaffolds for displaying antigenic proteins where the stoichiometry, spacing, and affinity can be optimized. Indeed, Veneziano showed that B-cell activation could be maximized in mice by as little as five HIV-1 antigens optimally displayed on engineered 40 nm viral-like DNA nanoparticles.6 Such structures can also act as targeted carriers for drug delivery. Here, they are typically functionalized with a targeting moiety such as folic acid or RGD peptidyl repeats for binding to cognate receptors that are upregulated on certain tumorigenic cells. The assemblies are then preferentially taken up into those cells, where they deliver their drug payload.4,5 Functionalized DNA structures can even actively participate in complex theranostic applications, where they functionally mimic a nanoscale robot to sense the presence of targets or receptors on cells via aptamers and other ligands and then reconfigure themselves to release a drug or imaging probe.7,8
For DNA nanotechnology to achieve its full therapeutic potential, many fundamental biological questions must be answered. Since most therapeutics require cellular uptake and metabolism, the uptake and fate of DNA structures needs to be elucidated. DNA nanostructures typically undergo uptake by some form of endocytosis, be it receptor-mediated or a more nonspecific mechanism.3,4 Once inside cells, DNA structures enter the endolysosomal system, where their final trajectory becomes less clear.9,10 While evidence exists for DNA structures undergoing degradation in the harsh lysosomal environment, especially since cells contain endogenous nucleases,11 they are still able to escape from these compartments and reach the cytosol to achieve the desired effect and, if required, furthermore enter the nucleus.3–5,7 Significantly, the stability of designer DNA structures directly within the cellular cytosol is hitherto unknown.9 It is on this last critical question that we focus our attention.
Preliminary information exists on the stability of complex DNA structures in cellular media and lysates.3,12–14 However, these do not reflect a true cytoplasmic environment in terms of protein content, molecular density, and the complex dynamics of an enclosed living biological system that is regulated at the nanoscale. Clearly, the putative pathway that a DNA structure takes on its journey through initial cell membrane interaction, endocytosis, and then escape to the cytosol is complex and will dramatically confound any study of cytoplasmic stability. Here, we describe a methodology that entirely bypasses the initial complexity of the endolysosomal system and allows for direct monitoring of DNA structural stability in the cellular cytosol. Direct cellular microinjection of plasmid DNA monitored with a later fluorescent in situ hybridization readout has provided information on cytosolic integrity,15 but higher-resolution analysis is required to monitor the degradation of more complex DNA architectures. Herein, we embedded a multistep Förster resonance energy transfer (FRET) dye relay into prototypical DNA assemblies. As FRET inherently depends upon the proximity between component donor (D) and acceptor (A) dyes with a one-sixth power dependency on D–A separation,16,17 we follow its presence as a direct reflection of the underlying DNA scaffold’s structural integrity. Following assembly, DNA structures were directly microinjected into the cellular cytosol of different cell types while FRET was monitored quantitatively via confocal fluorescent microscopy in a time-resolved manner in cellula (Figure 1). The results indicate that (i) a DNA tetrahedron design is far more stable in the cytoplasm in comparison to structures with linear double-stranded DNA (dsDNA) “arms”, (ii) the primary cause of cytosolic degradation is endogenous nucleases, and (iii) such studies can inform design parameters for creating DNA therapeutics.
Figure 1.
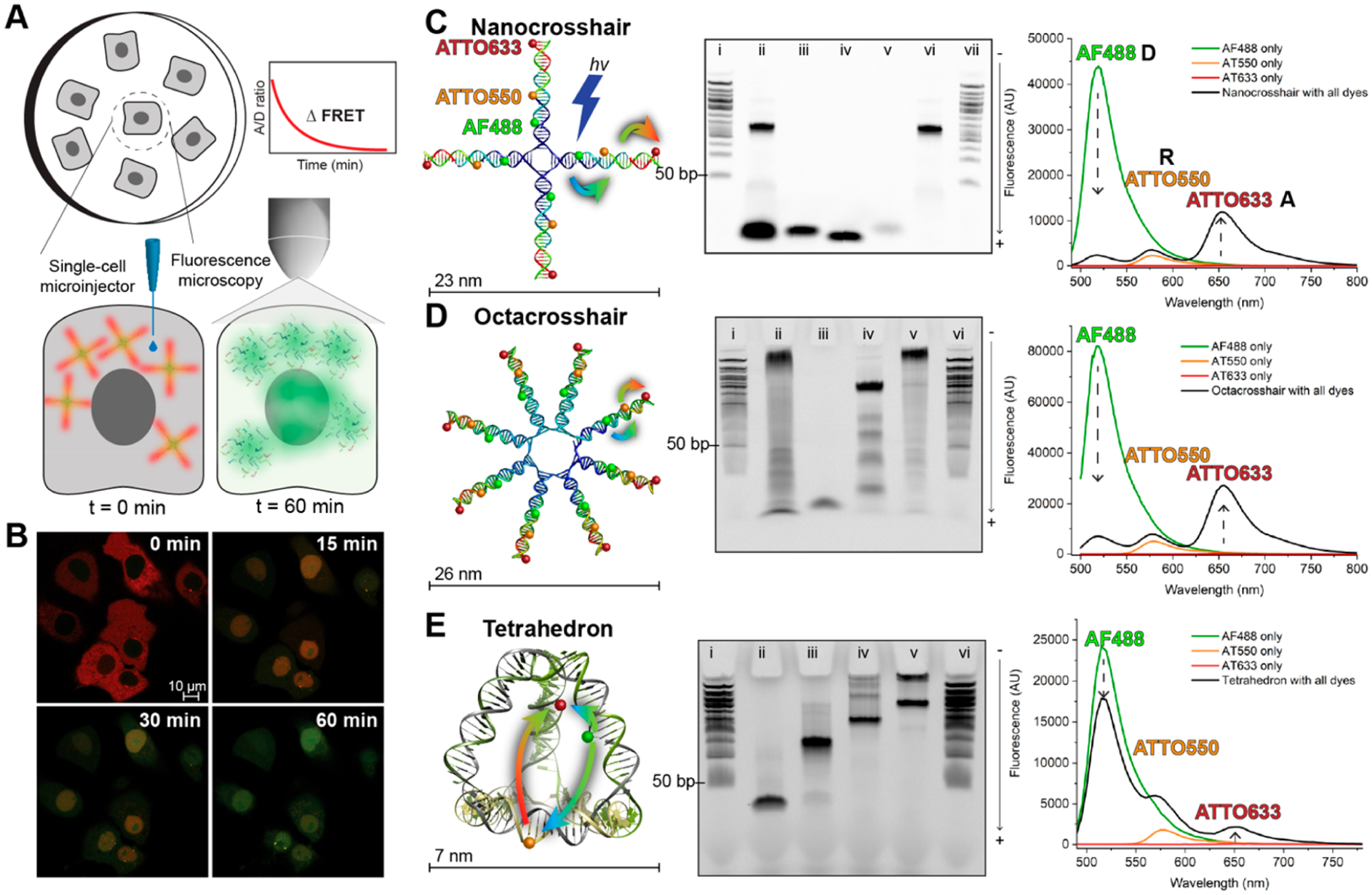
Illustrative summary of the methodology and DNA structures tested for cytosolic stability. (A) Surface-adherent mammalian cells were individually injected with femtoliter volumes of DNA structures in the cytosol. The fluorescence intensity and FRET between the three dyes was measured using confocal microscopy over the course of 60 min. (B) Representative confocal images of COS-1 monkey kidney cells injected with the DNA nanocrosshair at different time points. At t = 0 min, the acceptor fluorescence (red) is high in the cytosolic region of the cells, whereas at t = 60 min the D fluorescence (green) is visible largely in the nuclei. At t = 15 and 30 min, there is intermediate fluorescence representing diminishing FRET. (C–E) Structure of each DNA nanostructure (left), their formation characterization via 10% PAGE (middle), and their fluorescence spectra (right). (C) DNA nanocrosshair with four copies of the three-dye FRET network arranged linearly on each arm. Representative 10% PAGE gel in 1X TBE comprised of the following lanes: (i, vii) 50 bp DNA ladder; (ii) unpurified nanocrosshair; (iii) AF488-labeled strand; (iv) AT550-labeled strand; (v) AT633-labeled strand; (vi) purified nanocrosshair. Steady-state fluorescence spectra of the nanocrosshair with all dyes present versus individual dyes only (excitation wavelength 466 nm). (D) DNA octacrosshair with eight copies of the three-dye FRET network arranged linearly on each arm. Representative 10% PAGE gel in 1X TBE comprised of the following lanes: (i, vi) 50 bp DNA ladder; (ii) unpurified octacrosshair; (iii) AF488 strand; (iv) partially assembled octacrosshair; (v) purified octacrosshair. Steady-state fluorescence spectra of the octacrosshair with all dyes present versus individual dyes only (excitation wavelength 466 nm). (E) DNA tetrahedron with one copy of the three-dye FRET network arranged with one dye per edge. Representative 10% PAGE gel in 1X TBE buffer (0.089 M Tris, 0.089 M boric acid, and 0.002 M EDTA, pH 8.3) comprised of the following lanes: (i, vi) 50 bp DNA ladder; (ii) T1 strand only; (iii) T1 + T2; (iv) T1 + T2 + T3; (v) fully formed tetrahedron. Steady-state fluorescence spectra of the tetrahedron with all dyes present versus individual dyes only (excitation wavelength 466 nm). Colored arrows in the schematics indicate D → R and R → A FRET steps.
Three DNA structures common to biotechnology were employed here.18,19 A nanocrosshair (160 bp) and octacross-hair (320 bp) were designed as four- or eight-arm DNA tiles of ~23 and 26 nm diameter, respectively,20 while the 7 nm tetrahedron (126 bp) originates from the work of Turberfield (Figure 1).21 The plural “crosshairs” designate both the nano- and octacrosshairs. Crosshairs were chosen since they are simple to assemble and primarily consist of linear dsDNA in an open “dendritic” type confirmation. In contrast, the tetrahedron is a far more compact, closed structure and previous reports suggest it to have intrinsic resistance to nuclease activity.3,12 For FRET monitoring, a three-dye FRET cascade consisting of Alexa Fluor 488 (D; AF488), ATTO550 (relay-R; AT550), and ATTO633 (A; AT633) was embedded into the DNA structures. Positioning of the dyes in the crosshairs had the Ds closest to the center, Rs in the middle of each arm, and As at the outer ends. In the tetrahedron, each dye is positioned on one edge. The self-assembly of each structure was confirmed via polyacrylamide gel electrophoresis (PAGE) (Figure 1C–E). For the crosshairs, the formation efficiency was <100%, requiring purification with Amicon centrifugal filtration columns prior to experiments. The tetrahedron showed nearly complete formation efficiency and was utilized without further purification. In solution, the crosshairs manifest an initial AF488 D decrease of ~94% and an estimated anywhere-to-end FRET efficiency (Eae) of 31 ± 4%, while the tetrahedron showed an initial D quenching of 26% and an Eae value of 10 ± 4% (Figures 1C–E). Eae accounts for the direct excitation of both the D and R dyes (see the Supporting Information).22,23 The difference in initial D quench and Eae between structural classes arises since the crosshairs’ dyes can be placed linearly close to each other on the same arm, while the tetrahedron’s architecture dictates that the dyes be placed on different arms to correctly report structural degradation. The FRET efficiency, and therefore structural integrity, is subsequently reported as an A/D ratio. The steady-state A/D ratio for the fully formed structures is 0.1 up to 5 and for ssDNA strands is 0. A list of the DNA sequences and pertinent dye FRET characterization is given in the Supporting Information.
Initial cellular experiments utilized COS-1 monkey kidney epithelial cells. Their relatively large size and discernible nuclei help prevent off-target injections, while their strong surface adherence prevents cellular displacement and microinjector clogging. Details on the cell culture, microinjection protocol, fluorescence microscopy, and subsequent FRET analysis are given in the Supporting Information. Fluorescence microscopy was realized with multichannel measurements in parallel to record not only the energy transfer between dyes but also their direct excitation (Table S7). Representative confocal images of COS-1 cells microinjected with 1 μM of nanocrosshair or tetrahedron in HEPES with MgCl2 buffer at t = 0 and 60 min are shown in Figure 2A,B, respectively. Different fluorescent channels monitoring AF488 D (green), AT550 R (orange), AT633 A (red), and all merge –initially show even cytosolic dye emission distribution, including especially from the terminal AT633 A, when 488 nm laser light was used to excite the initial D and initiate the FRET cascade. Three other channels measured the direct excitation of AT550 and AT633 at their corresponding excitation wavelengths (Figures S4 and S5). These images also confirmed that the DNA structures are relatively well dispersed throughout the cellular cytosol, unlike 100 nm large plasmid-PEI complexes that have been observed previously to confine into vesicular puncta 24 h after cytosolic injection.24 For the nanocrosshair, after 1 h there was almost no remaining cytosolic fluorescence visible in the red AT663 A channel and there was a severe reduction in the AT550 R channel. From this we conclude that the nanocrosshair underwent significant structural degradation during this time period. The octacrosshair responded to COS-1 cytosolic injections in the same manner when it was injected at a total DNA concentration equal to that of the nanocrosshair, 0.5 μM (Figure S6). For the nanocrosshair, we noted AF448 staining in the nuclei at t = 0 and intensifying nuclear staining of all three dyes at t = 60 min. Dyes being selectively taken up by nuclei along with small dye-labeled DNA fragments is known; these are likely smaller than 5 nm, which concomitantly increases their amenability to cross the nuclear pores.25,26 This suggests that the crosshairs begin degrading immediately upon entering the cytosol. The tetrahedron behaved significantly differently, wherein the nuclei remained mostly clear of fluorescence and no substantial loss in sensitized fluorescence was noted for the R and terminal A channels. For both structures, the colocalization coefficient over time confirmed these observations (Figure S3). We conclude that the tetrahedron is significantly more stable in the COS-1 cytosolic environment over this time period. A small decrease in overall fluorescence intensity across all channels was also noted during the experiments (8–30%; Figure S12), but measurable fluorescence and FRET are observed for up to 5 h (Figure S14). The decreasing fluorescence is hypothesized to be driven by photobleaching and cellular volume changes. Fortunately, ratiometric monitoring of FRET serves to effectively negate this issue.16,17 Control experiments verified our experimental format—these include a ssDNA negative control sample to represent fluorescence from a fully fragmented DNA tetrahedron under the same microinjection conditions (Figures S11–S13 and S15). These controls validated that FRET from the injected tetrahedron was resolvable (even though the FRET is weaker) by our experimental setup and was consistently greater than that of the control ssDNA sample.
Figure 2.
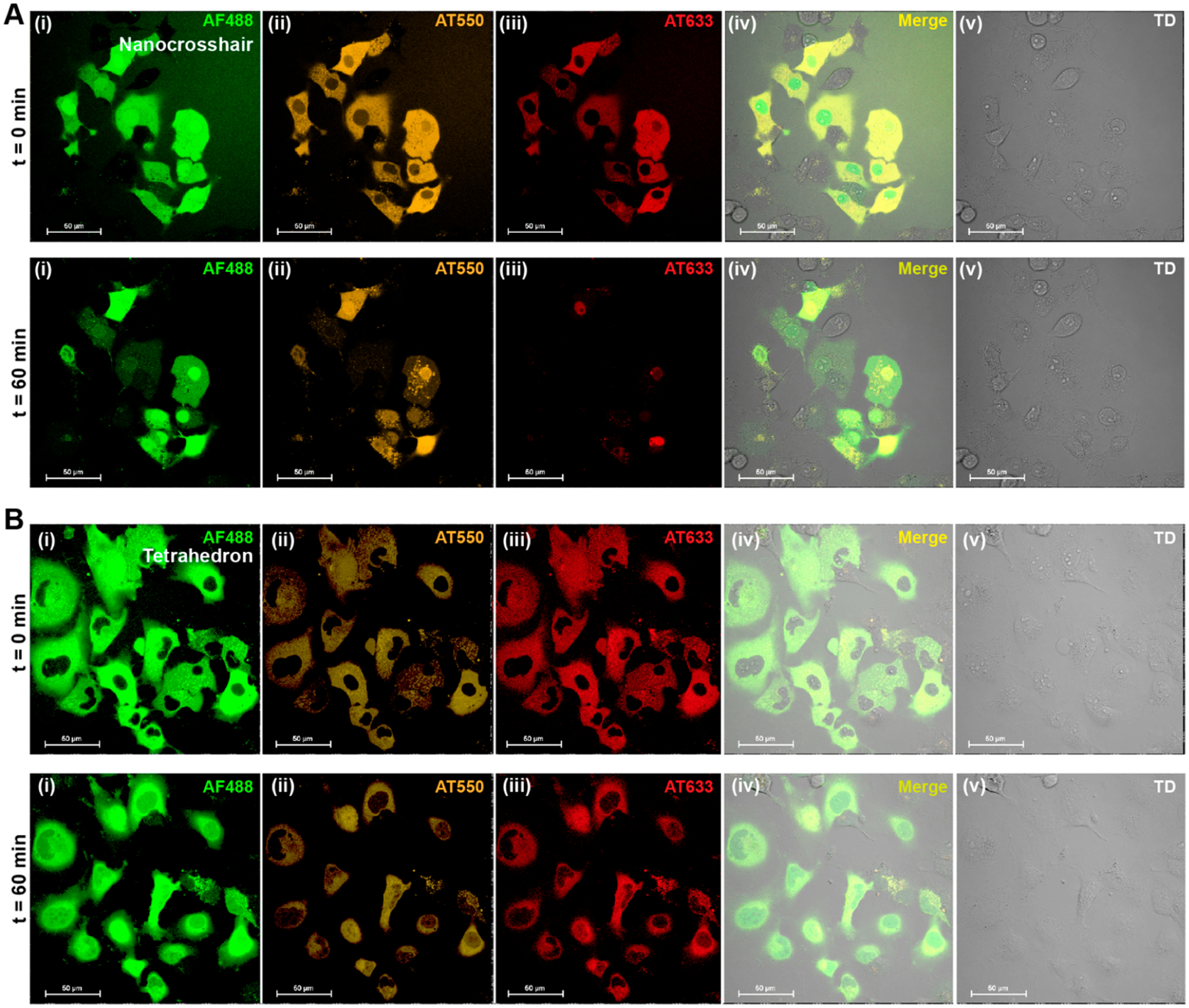
Fluorescence of a cytosol-injected nanocrosshair and tetrahedron. Representative four-channel confocal image as well as merged image of COS-1 cells injected with (A) a nanocrosshair and (B) a tetrahedron. The top panels represent fluorescence scans at t = 0 min, and the bottom scans represent t = 60 min. In each experiment, 1 μM of the DNA structure was injected at roughly 15 fL volume. The injection parameters were 500 hPa injection pressure (Pi), 0.30 s injection time (ti), and 45 hPa compensation pressure (Pc). The confocal imaging channels represented here are (i) AF488 D, (ii) AT550 R, (iii) AT633 A, (iv) merged, and (v) direct transmission (TD). All fluorescence channels were excited at 488 nm. Regions of interest (ROIs) were drawn on individual cells to acquire time-resolved florescence in each channel. Further experimental details can be found in the Supporting Information.
Figure 3 summarizes the degradation rate of the structures in COS-1 cells as a normalized (divided by the value at t = 0) ratio of A/D fluorescence (red/green channel) over time. Non-normalized A/D values for fully formed DNA structures ranged from 0.5 to 8 and for the ssDNA control was at around 0.12 (Figure S13). Changes in normalized A/D from 1 to 0 thus directly correlate with the proportion of degraded structure present. Both crosshairs showed rapid and nearly complete degradation within the first 10 min, suggesting a structural half-life of 3–4 min (Figure 3A). Our initial hypothesis was that the higher DNA density of the octacrosshair (eight arms) would delay its degradation rate. However, the octacrosshair degraded at a nominally higher rate when it was microinjected at total DNA concentration equal to that of the nanocrosshair (Figure 3A,B). One possibility underlying this interesting result may be that of enzymatic “hopping” or “scooting”, a process whereby a nuclease rapidly moves from localized site to site, continuously catalyzing DNA hydrolysis rather than diffusing away to the bulk and then rebinding to this or other substrates between each cleavage step. This modality has been noted for protease activity on nanoparticles displaying multiple peptidyl substrates and is manifested kinetically as higher enzymatic efficiency (i.e., kcat/KM).27,28 The assumption that the primary cause for crosshair degradation in the cytosol is nuclease-driven digestion (vide infra) suggests that the octacrosshair acts as an inherently higher density localized substrate, which functionally enhances the degradation rate versus the nanocrosshair. Reflecting the results of Figure 2B, Figure 3C confirms that the tetrahedron is significantly more robust within the COS-1 cellular cytoplasm with a statistically insignificant loss in average A/D ratio. Since the octacrosshair did not enhance stability, subsequent cellular experiments compared only the nanocrosshair and tetrahedron.
Figure 3.
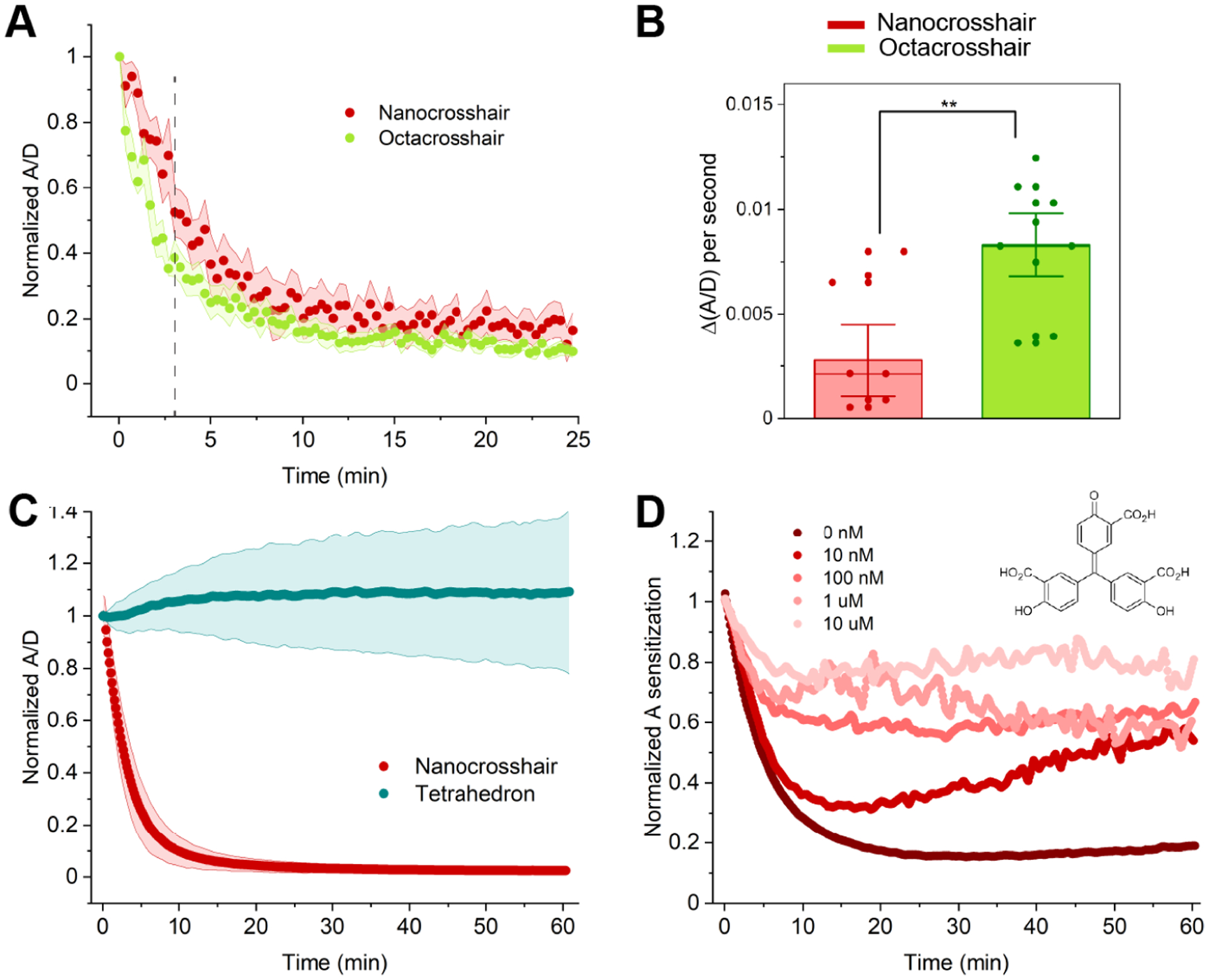
FRET characterization of microinjected DNA nanostructures. (A) Time-resolved change in normalized A/D of nanocrosshair versus octacrosshair. For normalization, all initial A/D fluorescence ratios at t = 0 were set to 1 and subsequent ratios were adjusted to this to put data on the same comparative scale. In each experiment, equimolar amounts of DNA were injected, such that 1 μM of DNA nanocrosshair and 0.5 μM octacrosshair were injected. The injection parameters were 500 hPa injection pressure (Pi), 0.30 s injection time (ti), and 45 hPa compensation pressure (Pc). (B) Rate of ratio change in the initial 200 s from the vertical dashed line in (A). (C) Time-resolved change in normalized A/D in nanocrosshair versus tetrahedron. (nanocrosshair n value, 91 cells; tetrahedron n value, 85 cells). In each experiment, 1 μM of nanocrosshair or tetrahedron was injected. The injection parameters were 500 hPa injection pressure (Pi), 0.30 s injection time (ti), and 45 hPa compensation pressure (Pc). (D) Time-resolved A sensitization change in coinjection of nanocrosshair with varying concentrations of ATA (structure shown in inset) in COS-1 cells (n = 30 cells per ATA concentration). In each experiment, 1 μM of nanocrosshair was prepared with different ATA concentrations and injected. The injection parameters were 500 hPa injection pressure (Pi), 0.30 s injection time (ti), and 45 hPa compensation pressure (Pc). Shaded regions in (A) and (C) represent standard deviations from the mean in each plot.
We next sought confirmatory evidence that cytosolic nuclease activity was responsible for the rapid degradation of the nanocrosshair. Aurintricarboxylic acid (ATA), a known nuclease inhibitor that functions competitively to inhibit protein–DNA interactions, was coinjected with samples to inhibit nuclease activity within the cells.29 A microinjected nanocrosshair supplemented with 10-fold increasing concentrations of ATA (from 10 nM up to 10 μM) revealed a progressively decreasing A sensitization change as a function of increasing ATA concentration, which is exactly the behavior expected for nuclease inhibition (Figure 3D). ATA is known to inhibit hepatitis C viral replicase in vitro and in vivo with IC50 values in the 150–250 nM range, and the current results suggest the nuclease inhibition kinetics here to be on par with that.30 Protein-catalyzed activity in the form of nucleases thus appear to be responsible for the DNA structure degradation seen in the COS-1 cells.
To understand why the crosshairs degrade intracellularly while the tetrahedron remains more stable, the structures were assayed against three distinct nucleases in a fluorescence plate assay. Figure 4A shows the tetrahedron and crosshairs on the same size scale as crystallographically derived models of DNase I (substrate, nonspecific dsDNA; source, human), Exonuclease I (ExoI, substrate 3′-end only of ssDNA, E. coli), and Exonuclease III (ExoIII, substrate 3′-ends only of dsDNA with overhangs, E. coli). A control-labeled single-crosshair arm (single arm) was included to test the advantage (or lack thereof) potentially provided by the crosshairs’ full shape and structure in the digestion assays. The tetrahedron appears resistant to structural degradation in the presence of DNase I and ExoI, while the crosshairs and the single arm show a rapid loss of structural integrity (Figure 4B). Only ExoIII seemed to have an effect on tetrahedron integrity.31 This pattern is highlighted in Figure 4C, which focuses on the rate of A/D ratio change over the first 5 min of the assay (the initial slope region in the curves). The high rate of tetrahedron digestion by ExoIII may again reflect the accelerated-continuous digestion modality on the highly localized substrate described above. Figure 4D compares initial nuclease digestion rates for the nanocrosshair and single arm in the presence/absence of ATA. Here, significant inhibition of degradation rates by ATA serves to confirm the results of Figure 3D, where this inhibitor was used intracellularly. Even though ExoI and ExoIII may not be present in COS-1 cells, examining their DNA binding sites in comparison to the DNA structures (Figure 4A) provides insight into any potential steric advantage that the tetrahedron possesses. The ExoI DNA binding site is the most sterically complex, where the enzyme would need to wrap around substrate DNA and potentially distort its linearity. Although the tetrahedron vertices contain one ss A nucleotide that could act as a substrate for ExoI, the tetrahedron is known to be resistant to physical distortions due to its short edge length, vertices, and torsional rigidity.3,12–14 In contrast, DNase I and ExoIII binding sites are less complex and could potentially act on all the structures. The crosshairs present multiple freely accessible ds termini that probably breathe considerably and become susceptible to nuclease binding and subsequent digestion.32 Results from assaying the single arm in Figure 4C serve to support this latter notion. From this we conclude that the tetrahedron architecture provides some intrinsic protection against nucleases exhibiting DNase I and ExoI type activity. In contrast, ExoIII, which targets ds ends of linear DNA, is active on the tetrahedron, suggesting that some such sites are available within this structure. Tetrahedron resistance to degradation has been observed in other physiologically relevant environments, including cell lysates.3,12,33 It should be noted that the number of strand termini in the crosshairs (1 of 3′- or 5′-terminus on average per 5 bp) is different from that in the tetrahedron (1 of 3′- or 5′-terminus on average per 15 bp), yet the hydrolysis of the tetrahedron seems to be enzyme-specific, suggesting that the shape (and not number of strand termini) could influence the susceptibility to digestion.
Figure 4.
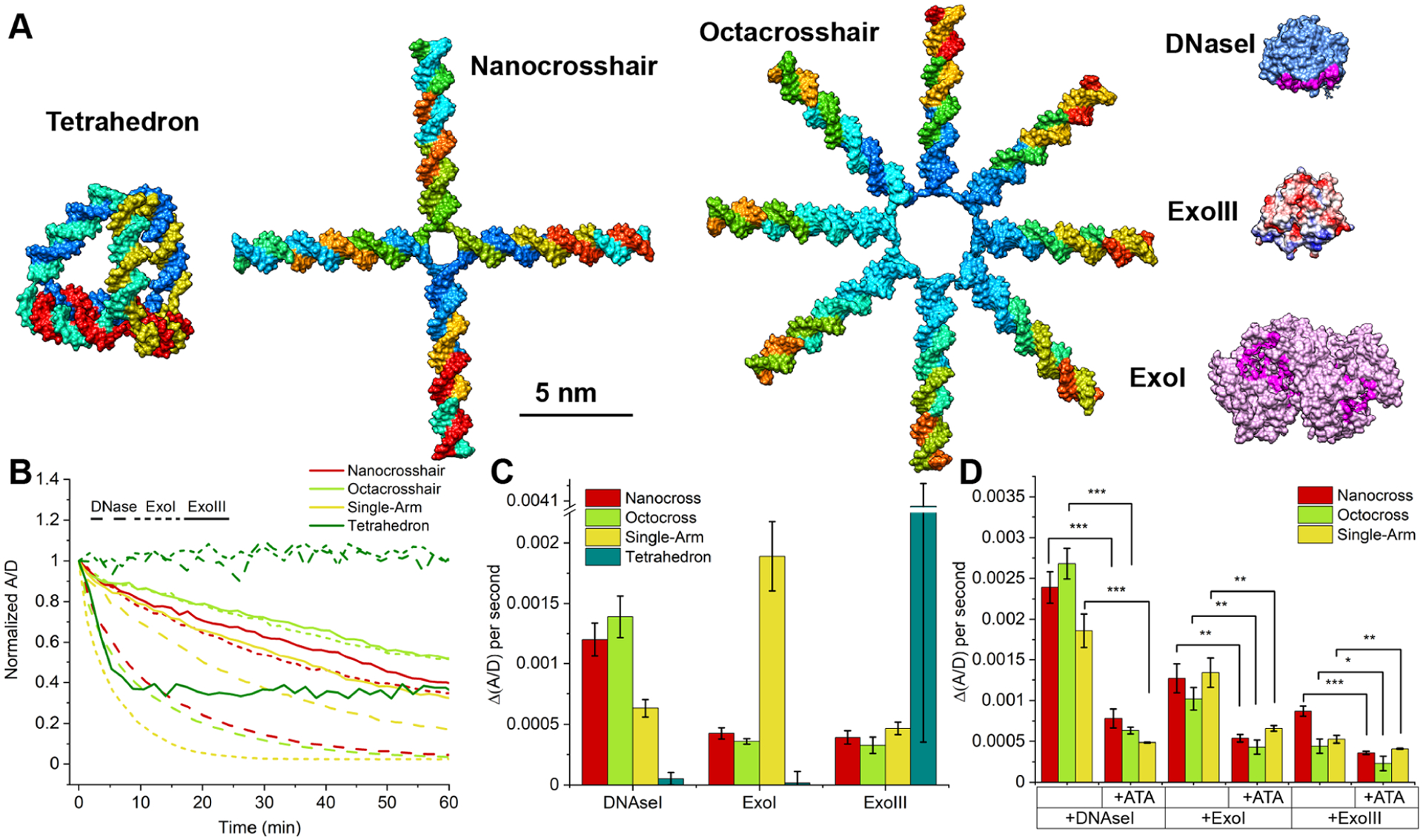
DNA nanostructure stability evaluation using nucleases. (A) Structural comparison of the three DNA nanostructures with common nucleases (DNase I, Exonuclease I, and Exonuclease III) all rendered on the same size scale. The DNA binding site of each nuclease is shown; DNase I (PDB access number 1dnk.pdb) and ExoI (4js5.pdb) binding sites are highlighted in magenta, while ExoIII (1ako.pdb) is highlighted in blue (highlighting its polar region). (B) Time-resolved changes in normalized A/D when the DNA structures were subjected to nuclease digestion in solution: DNase I at 2 units/mL; Exonuclease III at 500 units/mL; Exonuclease I at 50 units/mL. (C) Rate of initial fluorescence change in the presence of different nucleases at t = 5 min. (D) Rate of initial fluorescence change with and without ATA present (1 mg/mL), in the same manner as for (C) at t = 5 min. ***p < 0.0005; **p < 0.005; *p < 0.05 (n = 3).
We next examined the tetrahedron performance in several other cell lines to see if its cytosolic stability would hold true beyond COS-1 cells. The same type of microinjection experiments were performed in primary dermal fibroblast cells and human astrocyte cells to compare against other transformed cell lines, including human A549 adenocarcinoma alveolar basal epithelial cells and HeLa adenocarcinoma cervical epithelial cells (Figures S7–S10). As seen in Figure 5A–E, the tetrahedron consistently demonstrates similar stability profiles across all cell lines, while nanocrosshair instability is again confirmed by microinjection into the HeLa cells (Figure 5F). Along with A/D ratio, the change in A sensitization (when exciting the D) divided by A fluorescence upon direct excitation is also shown. The latter provides additional confidence in the results, since the tetrahedron does display a lower FRET Eae value in comparison to the crosshairs and, as noted above, the samples also undergo some loss of overall fluorescence intensity over time in cells during the experiments (Figure S12). Although the two data plots do not directly superimpose over each other, the net results and conclusions remain the same.
Figure 5.
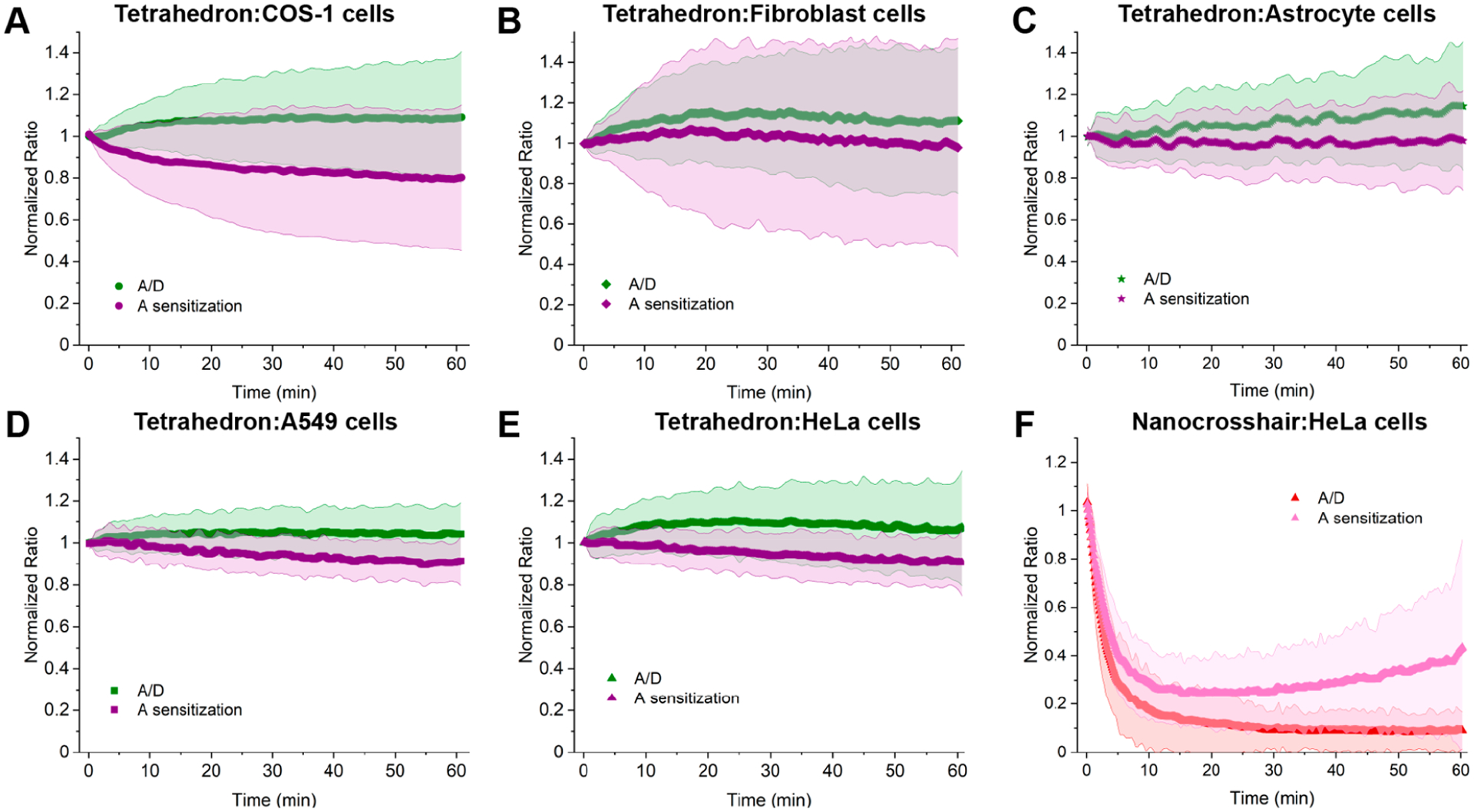
Cytosolic behavior of the DNA tetrahedron and nanocrosshair in different cell types. Change in FRET for cytosolically injected DNA tetrahedron in (A) monkey kidney COS-1 cells, (B) human fibroblast cells, (C) human astrocyte cells, (D) human lung A549 cells, and (E) human cervical HeLa cells. Human fibroblast and astrocyte cells are primary cell types, while the others are all transformed cell lines. For the tetrahedron, no statistical difference in response is seen between primary and transformed cells. (F) Nanocrosshair injected in human cervical HeLa cells. The shaded regions represent the standard deviation from the average (the number of cells in each graph is given in Table S6).
Several hundred genes encoding putative nucleases are present in the human genome, including DNase I. Beyond their role in cellular processes, it is believed that some function secondarily as part of the innate immune system and help destroy pathogenic nucleic acids during infection.34 This is perhaps why exogenous linear DNAs/RNAs are typically not stable or well tolerated within human cells. For DNA nanostructures to achieve viable roles as therapeutics, in situ diagnostics, and even complex theranostic devices, a full understanding of their pathway into a cell and then to their ultimate site of action is needed.9 The complexities of endosomal uptake into cells continue to hamper attempts toward this understanding. Beyond harboring its own nucleases, the endolysosomal system manifests further complicating factors such as pH changes and recycling activity.35,36 The methodology demonstrated here bypasses this issue and reports directly on the DNA nanostructure stability within the cellular cytosol; this may be relevant to chemo- and electrotransfection studies as well.37 This is not to say that understanding DNA stability following endocytic uptake and later endosomal escape is not just as critically important, rather, that the endogenous nuclease activity and complex lifecycle of these compartments make the subsequent contribution of cytosolic degradation hard to separately parse in this joint context. Hopefully, it is only a matter of time before DNA nanostructures are paired with agents to promote endosomal release, e.g. drugs or peptides, which may help negate this issue somewhat.38–40 In terms of applying this method during initial endocytic uptake, receptor concentration into endosomes and the nanoscale confines of such compartments suggest that this may be quite challenging due to highly localized assembly concentrations which can induce intra-assembly solution-phase FRET.16,41
As an initial investigation, this study already informs the value of design rules needed for creating DNA-based therapeutics. A preliminary evaluation suggests that data can be collected for at least 5 h (Figure S14), which is promising. If a DNA nanostructure is intended to deliver a drug or mRNA to the cytosol, then lower stability is desirable and simpler tile-based structures may suffice. Such a carrier could have good clearance from the cell via exocytosis after delivering the payload.42 One interesting corollary of this may be that denser-linear structures may degrade even more quickly (octa- vs nanocrosshair data; Figures 3A,B). In contrast, if the structure’s purpose requires longer stability, slower drug release kinetics, or shuttling through the cytosol to the nucleus, then more stable assemblies such as the tetrahedron may be warranted. Combining this approach with in-solution nuclease assays also provides useful insight toward what type of nuclease susceptibility different DNA structures do have. The ability of DNA to shuttle from the cytosol into the nucleus can also be evaluated through this technology.43 Determining the cytosolic stability of other types of DNA nanostructures across a broad range of sizes and complexities only requires judicious placement of dyes into their structure for assaying with this generalized approach. This methodology may further be amenable to provide data on related processes in the cellular cytosol: for example, whether proteins or other molecules displayed on the DNA structure itself interact with or are substrates for other cytosolic processes. Further utility includes confirming the stability of structures incorporating non-canonical nuclease-resistant nucleic acids such as peptide nucleic acids (PNAs) or those with phosphorothioate internucleotide linkages.3,5,12,14,44 Although this methodology only provides data on one aspect of the DNA nanostructure lifecycle within cells, its application can nevertheless help contribute to improving the design for in vivo biomedical applications. Given its non-natural structure, tetrahedron-based designs may represent an excellent place to start.12,19,45,46
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the U.S. Naval Research Laboratory (NRL), the Office of Naval Research, and the NRL’s Nanoscience Institute for programmatic funding. D.M. was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number K99EB030013. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.2c00917.
Full description of the materials and methods, experimental details, FRET analysis, and representative confocal data from control experiments (PDF )
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.2c00917
The authors declare no competing financial interest.
Contributor Information
Divita Mathur, College of Science, George Mason University, Fairfax, Virginia 22030, United States; Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States;.
Katherine E. Rogers, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States; Fischell Department of Bioengineering, University of Maryland, College Park, Maryland 20742, United States
Sebastián A. Díaz, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States;.
Megan E. Muroski, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States; American Society of Engineering Education, Washington, DC 20036, United States
William P. Klein, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States; National Research Council, Washington, DC 20001, United States
Okhil K. Nag, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States;.
Kwahun Lee, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States; American Society of Engineering Education, Washington, DC 20036, United States;.
Lauren D. Field, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States; National Research Council, Washington, DC 20001, United States
James B. Delehanty, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States;.
Igor L. Medintz, Center for Bio/Molecular Science and Engineering Code 6900, US Naval Research Laboratory, Washington, DC 20375, United States;.
REFERENCES
- (1).Chauhan G; Madou MJ; Kalra S; Chopra V; Ghosh D; Martinez-Chapa SO Nanotechnology for Covid-19: Therapeutics and Vaccine Research. ACS Nano 2020, 14 (7), 7760–7782. [DOI] [PubMed] [Google Scholar]
- (2).Le TK; Paris C; Khan KS; Robson F; Ng WL; Rocchi P Nucleic Acid-Based Technologies Targeting Coronaviruses. Trends Biochem. Sci 2021, 46 (5), 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mathur D; Medintz IL The Growing Development of DNA Nanostructures for Potential Healthcare-Related Applications. Adv. Healthc. Mater 2019, 8 (9), No. 1801546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hu Q; Li H; Wang L; Gu H; Fan C DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev 2019, 119 (10), 6459–6506. [DOI] [PubMed] [Google Scholar]
- (5).Keller A; Linko V Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem., Int. Ed 2020, 59 (37), 15818–15833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Veneziano R; Moyer TJ; Stone MB; Wamhoff EC; Read BJ; Mukherjee S; Shepherd TR; Das J; Schief WR; Irvine DJ; Bathe M Role of Nanoscale Antigen Organization on B-Cell Activation Probed Using DNA Origami. Nat. Nanotechnol 2020, 15 (8), 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bi S; Dong Y; Jia X; Chen M; Zhong H; Ji B Self-Assembled Multifunctional DNA Nanospheres for Biosensing and Drug Delivery into Specific Target Cells. Nanoscale 2015, 7 (16), 7361–7. [DOI] [PubMed] [Google Scholar]
- (8).Douglas SM; Bachelet I; Church GM A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335 (6070), 831–4. [DOI] [PubMed] [Google Scholar]
- (9).Green CM; Mathur D; Medintz IL Understanding the Fate of DNA Nanostructures inside the Cell. J. Mater. Chem. B 2020, 8 (29), 6170–6178. [DOI] [PubMed] [Google Scholar]
- (10).Okholm AH; Kjems J DNA Nanovehicles and the Biological Barriers. Adv. Drug Delivery Rev 2016, 106, 183–191. [DOI] [PubMed] [Google Scholar]
- (11).Evans CJ; Aguilera RJ DNase II: Genes, Enzymes and Function. Gene 2003, 322, 1–15. [DOI] [PubMed] [Google Scholar]
- (12).Chandrasekaran AR Nuclease Resistance of DNA Nanostructures. Nat. Rev. Chem 2021, 5 (4), 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chandrasekaran AR; Vilcapoma J; Dey P; Wong-Deyrup SW; Dey BK; Halvorsen K Exceptional Nuclease Resistance of Paranemic Crossover (PX) DNA and Crossover-Dependent Bio-stability of DNA Motifs. J. Am. Chem. Soc 2020, 142 (14), 6814–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Keum JW; Bermudez H Enhanced Resistance of DNA Nanostructures to Enzymatic Digestion. Chem. Commun 2009, 45, 7036–7038. [DOI] [PubMed] [Google Scholar]
- (15).Lechardeur D; Sohn KJ; Haardt M; Joshi PB; Monck M; Graham RW; Beatty B; Squire J; O’Brodovich H; Lukacs GL Metabolic Instability of Plasmid DNA in the Cytosol: A Potential Barrier to Gene Transfer. Gene Ther. 1999, 6 (4), 482–97. [DOI] [PubMed] [Google Scholar]
- (16).Algar WR; Hildebrandt N; Vogel SS; Medintz IL FRET as a Biomolecular Research Tool - Understanding Its Potential While Avoiding Pitfalls. Nat. Methods 2019, 16 (9), 815–829. [DOI] [PubMed] [Google Scholar]
- (17).Medintz IL; Hildebrandt N FRET-Förster Resonance Energy Transfer: From Theory to Applications; Wiley: 2013. [Google Scholar]
- (18).Dong Y; Yao C; Zhu Y; Yang L; Luo D; Yang D DNA Functional Materials Assembled from Branched DNA: Design, Synthesis, and Applications. Chem. Rev 2020, 120 (17), 9420–9481. [DOI] [PubMed] [Google Scholar]
- (19).Xie N; Liu S; Yang X; He X; Huang J; Wang K DNA Tetrahedron Nanostructures for Biological Applications: Biosensors and Drug Delivery. Analyst 2017, 142 (18), 3322–3332. [DOI] [PubMed] [Google Scholar]
- (20).Klein WP; Díaz SA; Buckhout-White S; Melinger JS; Cunningham PD; Goldman ER; Ancona MG; Kuang W; Medintz IL Utilizing HomoFRET to Extend DNA-Scaffolded Photonic Networks and Increase Light-Harvesting Capability. Adv. Opt. Mater 2018, 6 (1), 1700679–91. [Google Scholar]
- (21).Erben CM; Goodman RP; Turberfield AJ Single-Molecule Protein Encapsulation in a Rigid DNA Cage. Angew. Chem., Int. Ed 2006, 45 (44), 7414–7. [DOI] [PubMed] [Google Scholar]
- (22).Mathur D; Samanta A; Ancona MG; Diaz SA; Kim Y; Melinger JS; Goldman ER; Sadowski JP; Ong LL; Yin P; Medintz IL Understanding Forster Resonance Energy Transfer in the Sheet Regime with DNA Brick-Based Dye Networks. ACS Nano 2021, 15 (10), 16452–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Klein WP; Rolczynski BS; Oliver SM; Zadegan R; Buckhout-White S; Ancona MG; Cunningham PD; Melinger JS; Vora PM; Kuang W; Medintz IL; Díaz SA DNA Origami Chromophore Scaffold Exploiting HomoFRET Energy Transport to Create Molecular Photonic Wires. ACS Appl. Nano Mater 2020, 3 (4), 3323–3336. [Google Scholar]
- (24).Remaut K; Oorschot V; Braeckmans K; Klumperman J; De Smedt SC Lysosomal Capturing of Cytoplasmic Injected Nano-particles by Autophagy: An Additional Barrier to Non Viral Gene Delivery. J. Controlled Release 2014, 195, 29–36. [DOI] [PubMed] [Google Scholar]
- (25).Salman H; Zbaida D; Rabin Y; Chatenay D; Elbaum M Kinetics and Mechanism of DNA Uptake into the Cell Nucleus. Proc. Natl. Acad. Sci. U.S.A 2001, 98 (13), 7247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lacroix A; Vengut-Climent E; de Rochambeau D; Sleiman HF Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale. ACS Cent. Sci 2019, 5 (5), 882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Algar WR; Malonoski A; Deschamps JR; Blanco-Canosa JB; Susumu K; Stewart MH; Johnson BJ; Dawson PE; Medintz IL Proteolytic Activity at Quantum Dot-Conjugates: Kinetic Analysis Reveals Enhanced Enzyme Activity and Localized Interfacial ″Hopping″. Nano Lett. 2012, 12 (7), 3793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dennis AM; Delehanty JB; Medintz IL Emerging Physicochemical Phenomena Along with New Opportunities at the Biomolecular-Nanoparticle Interface. J. Phys. Chem. Lett 2016, 7 (11), 2139–50. [DOI] [PubMed] [Google Scholar]
- (29).Walther W; Stein U; Siegel R; Fichtner I; Schlag PM Use of the Nuclease Inhibitor Aurintricarboxylic Acid (ATA) for Improved Non-Viral Intratumoral in Vivo Gene Transfer by Jet-Injection. J. Gene Med 2005, 7 (4), 477–85. [DOI] [PubMed] [Google Scholar]
- (30).Chen Y; Bopda-Waffo A; Basu A; Krishnan R; Silberstein E; Taylor DR; Talele TT; Arora P; Kaushik-Basu N Characterization of Aurintricarboxylic Acid as a Potent Hepatitis C Virus Replicase Inhibitor. Antivir. Chem. Chemother 2009, 20, 19–36. [DOI] [PubMed] [Google Scholar]
- (31).Ozhalici-Unal H; Armitage BA Fluorescent DNA Nanotags Based on a Self-Assembled DNA Tetrahedron. ACS Nano 2009, 3 (2), 425–33. [DOI] [PubMed] [Google Scholar]
- (32).von Hippel PH; Johnson NP; Marcus AH Fifty Years of DNA ″Breathing″: Reflections on Old and New Approaches. Biopolymers 2013, 99 (12), 923–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mei Q; Wei X; Su F; Liu Y; Youngbull C; Johnson R; Lindsay S; Yan H; Meldrum D Stability of DNA Origami Nanoarrays in Cell Lysate. Nano Lett. 2011, 11 (4), 1477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hartmann G Nucleic Acid Immunity. In Advances in Immunology; Academic Press: 2017; pp 121–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Duncan R; Richardson SC Endocytosis and Intracellular Trafficking as Gateways for Nanomedicine Delivery: Opportunities and Challenges. Mol. Pharmaceutics 2012, 9 (9), 2380–402. [DOI] [PubMed] [Google Scholar]
- (36).Hu YB; Dammer EB; Ren RJ; Wang G The Endosomal-Lysosomal System: From Acidification and Cargo Sorting to Neurodegeneration. Transl. Neurodegener 2015, 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chopra A; Krishnan S; Simmel FC Electrotransfection of Polyamine Folded DNA Origami Structures. Nano Lett. 2016, 16 (10), 6683–6690. [DOI] [PubMed] [Google Scholar]
- (38).Boeneman K; Delehanty JB; Blanco-Canosa JB; Susumu K; Stewart MH; Oh E; Huston AL; Dawson G; Ingale S; Walters R; Domowicz M; Deschamps JR; Algar WR; Dimaggio S; Manono J; Spillmann CM; Thompson D; Jennings TL; Dawson PE; Medintz IL Selecting Improved Peptidyl Motifs for Cytosolic Delivery of Disparate Protein and Nanoparticle Materials. ACS Nano 2013, 7 (5), 3778–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Smith SA; Selby LI; Johnston APR; Such GK The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjugate Chem. 2019, 30 (2), 263–272. [DOI] [PubMed] [Google Scholar]
- (40).Wan Y; Moyle PM; Toth I Endosome Escape Strategies for Improving the Efficacy of Oligonucleotide Delivery Systems. Curr. Med. Chem 2015, 22 (29), 3326–46. [DOI] [PubMed] [Google Scholar]
- (41).Fu M; Dai L; Jiang Q; Tang Y; Zhang X; Ding B; Li J Observation of Intracellular Interactions between DNA Origami and Lysosomes by the Fluorescence Localization Method. Chem. Commun 2016, 52 (59), 9240–2. [DOI] [PubMed] [Google Scholar]
- (42).Wu XA; Choi CH; Zhang C; Hao L; Mirkin CA Intracellular Fate of Spherical Nucleic Acid Nanoparticle Conjugates. J. Am. Chem. Soc 2014, 136 (21), 7726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Luther DC; Jeon T; Goswami R; Nagaraj H; Kim D; Lee YW; Rotello VM Protein Delivery: If Your GFP (or Other Small Protein) Is in the Cytosol, It Will Also Be in the Nucleus. Bioconjugate Chem. 2021, 32 (5), 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wilson C; Keefe AD Building Oligonucleotide Therapeutics Using Non-Natural Chemistries. Curr. Opin. Chem. Biol 2006, 10 (6), 607–14. [DOI] [PubMed] [Google Scholar]
- (45).Copp W; Pontarelli A; Wilds CJ Recent Advances of DNA Tetrahedra for Therapeutic Delivery and Biosensing. Chembiochem 2021, 22 (13), 2237–2246. [DOI] [PubMed] [Google Scholar]
- (46).Shi S; Li Y; Zhang T; Xiao D; Tian T; Chen T; Zhang Y; Li X; Lin Y Biological Effect of Differently Sized Tetrahedral Framework Nucleic Acids: Endocytosis, Proliferation, Migration, and Biodistribution. ACS Appl. Mater. Interfaces 2021, 13 (48), 57067–57074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


