Abstract
The ABCD-GENE score was developed to identify patients at risk for diminished antiplatelet effects with clopidogrel after percutaneous coronary intervention (PCI) and utilizes Age, Body mass index, Chronic kidney disease, Diabetes and CYP2C19 GENEtic variants. The objective of this study was to validate the ability of the ABCD-GENE score to predict risk for atherothrombotic events in a diverse, real-world population of clopidogrel-treated PCI patients who received clinical CYP2C19 genotyping to guide antiplatelet therapy. A total of 2341 adult patients who underwent PCI, were genotyped for CYP2C19, and received treatment with clopidogrel across four institutions were included (mean age 64±12 years, 35% female, 20% Black). The primary outcome was major atherothrombotic events, defined as the composite of all-cause death, myocardial infarction, ischemic stroke, stent thrombosis, or revascularization for unstable angina within 12 months following PCI. Major adverse cardiovascular events (MACE), defined as the composite of cardiovascular death, myocardial infarction, ischemic stroke, or stent thrombosis, was assessed as the secondary outcome. Outcomes were compared between patients with an ABCD-GENE score ≥10 versus <10. The risk of major atherothrombotic events was higher in patients with an ABCD-GENE score ≥10 (n=505) versus <10 (n=1836; 24.6 versus 14.7 events per 100 patient-years, adjusted hazard ratio (HR), 1.66; 95% CI, 1.23–2.25; p<0.001). The risk for MACE was also higher among patients with a score ≥10 versus <10 (16.7 versus 10.1 events per 100 patient-years, adjusted HR, 1.59; 95% CI, 1.11–2.30; p=0.013). Our diverse, real-world data demonstrate diminished clopidogrel effectiveness in PCI patients with an ABCD-GENE score ≥10.
Keywords: clopidogrel, CYP2C19, precision medicine, genetic testing, percutaneous coronary intervention
Introduction
Dual antiplatelet therapy with aspirin and a P2Y12 inhibitor is recommended after percutaneous coronary intervention (PCI) to decrease the risk of ischemic events.(1, 2) In clinical trials of patients presenting with an acute coronary syndrome (ACS), in which CYP2C19 genotyping was not performed to guide treatment selection, prasugrel and ticagrelor were superior to clopidogrel in reducing major atherothrombotic events and are thus preferred in this setting.(1–4) However, prasugrel and ticagrelor are associated with increased bleeding risk and higher cost compared with clopidogrel, which in addition to ticagrelor-associated dyspnea, limit their widespread use.(3–6) Thus, clopidogrel remains commonly prescribed in clinical practice, including in patients undergoing PCI following an ACS.(7, 8)
Clopidogrel is a prodrug that relies on the cytochrome P450 2C19 (CYP2C19) enzyme for biotransformation to its pharmacologically active metabolite. The wide variability in clopidogrel-induced antiplatelet effects, which in a considerable number of patients is impaired leading to high on-treatment platelet reactivity (HPR), is due to both genetic determinants(9) and clinical variables.(10–12) Importantly, HPR is a marker of increased risk of thrombotic complications in patients undergoing PCI.(10, 13) Clopidogrel effectiveness is specifically influenced by genetic variability in CYP2C19 that contributes to wide interindividual variability in clopidogrel metabolism and inhibition of platelet aggregation.(14, 15) Approximately 30% of the U.S. population carries at least one CYP2C19 no function allele.(16) Among clopidogrel-treated patients, no function allele carriers have lower concentrations of the active clopidogrel metabolite, higher platelet reactivity and HPR rates, and are at increased risk for cardiovascular events after PCI compared to those without a no function allele.(17, 18) In addition to CYP2C19 genotype, clinical factors, including older age, higher body mass index (BMI), chronic kidney disease (CKD), and diabetes mellitus (DM), contribute to HPR and are associated with reduced clopidogrel effectiveness.(11, 19–23)
The ABCD-GENE (Age, Body mass index, Chronic kidney disease, and Diabetes mellitus, and CYP2C19 GENEtic variants) score includes CYP2C19 genotype and clinical factors independently associated with HPR among clopidogrel-treated patients following PCI.(24) The score was evaluated in two clopidogrel-treated European cohorts, with a score ≥10 predicting both HPR following elective PCI(25) and increased risk for major adverse cardiovascular events (MACE) among patients with myocardial infarction (MI), most of whom underwent PCI.(24, 26) The ABCD-GENE score was subsequently shown to identify patients at increased risk of MACE in post-hoc analyses of predominately European and Asian clinical trial participants receiving clopidogrel after PCI or ischemic stroke.(27, 28) However, validation of the ABCD-GENE score in a more diverse cohort of patients undergoing PCI in real-world clinical practice is necessary to assess the generalizability and potential clinical utility of this risk stratification tool. In addition, only the CYP2C19*2 no function allele was considered in the evaluation of the score. While this is the most common no function allele across populations, there are other less common alleles associated with absence of CYP2C19 function, and the Association for Molecular Pathology recommends that both the CYP2C19*2 and CYP2C19*3 no function alleles should be included in clinical CYP2C19 testing.(29) The objective of this analysis was to validate the association between the ABCD-GENE score and major atherothrombotic events in a diverse real-world population of clopidogrel-treated PCI patients who received clinical CYP2C19 genotyping that included, at a minimum, the *2 and *3 alleles.
Methods
Study Population
The study population included patients who were ≥18 years of age, underwent elective or emergent PCI, received P2Y12 inhibitor therapy, and were genotyped clinically for CYP2C19, as previously described.(30, 31) Four institutions (University of Florida, Gainesville; University of Florida, Jacksonville; University of North Carolina, Chapel Hill; University of Maryland, Baltimore) contributed data for 3,688 patients meeting these criteria, of whom, 2,341 were treated with clopidogrel. Data collection was approved by the Institutional Review Board at each site.
CYP2C19 genotype was determined in a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvement Amendments (CLIA)-licensed laboratory at each institution, and results were reported in the electronic health record, as previously described.(32–34) All sites interrogated the CYP2C19*2, *3, and *17 alleles, with two of four sites testing for additional rare no function alleles (Table S1). Patients carrying a no function allele were assigned the CYP2C19 poor (PM; two no function alleles) or intermediate (IM; one no function allele) metabolizer phenotype, consistent with Clinical Pharmacogenetics Implementation Consortium guidelines.(16) At each institution, alternative antiplatelet therapy with prasugrel or ticagrelor was recommended for CYP2C19 IMs and PMs, in the absence of contraindications, whereas no prescribing recommendations were made for those without a no function allele.(34) The ultimate prescribing decision was left to the clinician’s discretion, and approximately 45% of IMs and PMs were treated with clopidogrel across sites.(30, 31)
Data abstraction
Data were manually abstracted from the electronic health record at each site using a common data collection form, as previously described.(35) Data for eligible patients were abstracted through review of patient encounters or phone interviews, starting with the hospitalization for the index PCI (defined as the PCI performed in association with CYP2C19 genotyping) and including subsequent outpatient encounters and hospitalizations. Patients were followed longitudinally until an outcome of interest occurred, clopidogrel was discontinued, or the prespecified follow-up time of 12 months post-PCI was achieved, whichever occurred first. Patients were included regardless of length of follow-up. Ischemic events were identified from provider-reported diagnoses at each encounter or clinical notes in the event of death. Data collection procedures were approved by the institutional review board at each institution.
Study Endpoints
The primary outcome was major atherothrombotic events, defined as the composite of all-cause death, MI, ischemic stroke, stent thrombosis, or revascularization for unstable angina within 12 months following the index PCI. MACE, defined as the composite of cardiovascular death, MI, ischemic stroke, or stent thrombosis, within 12 months post-PCI was assessed as the secondary outcome. All events were independently reviewed and verified by an interventional cardiologist.
Data Analysis
Data were curated and aggregated at the University of Florida, Gainesville. The ABCD-GENE score was calculated from information at the time of index PCI, with four points assigned for age >75 years, four points for body mass index >30 kg/m2, three points for CKD (defined as an estimated glomerular filtration rate of <60 ml/min/1.73m2), three points for DM, six points for IMs, and 24 points for PMs.(24)
Patient characteristics and event rates (per 100 patient-years) for the primary and secondary endpoints were compared between clopidogrel-treated patients with an ABCD-GENE score ≥10 versus <10 using the chi-square test or two-sample t-test as appropriate.(24) Unadjusted hazard ratios (HR) and 95% confidence intervals (CIs) were calculated using Cox proportional hazards regression. Multivariable Cox regression was used to estimate the risk for major atherothrombotic events and MACE, and adjusted HRs and 95% CIs were calculated. The adjusted model included sex, smoking status, index PCI indication (i.e., MI versus unstable angina or elective PCI), PCI strategy (i.e., stent vs percutaneous transluminal coronary angioplasty), and institution.
An exploratory analysis arose based on the observation that 159 patients with an ABCD-GENE score <10 were CYP2C19 IMs. Given known effects of IM status in reducing antiplatelet effects and clinical effectiveness of clopidogrel,(16) the ABCD-GENE score <10 group was stratified into IMs and non-IMs. Median scores were compared between IMs versus non-IMs with a score <10 using the Mann-Whitney U test. The risk for major atherothrombotic events and MACE among CYP2C19 IMs and non-IMs with an ABCD-GENE score <10, compared with the ≥10 group, was evaluated using multivariable Cox regression as described in the primary analysis. Similarly, based on the observation that 216 patients with a score ≥10 did not have a CYP2C19 no function allele, as a second exploratory analysis, we compared risk for major atherothrombotic events and MACE between CYP2C19 IM/PMs versus non-IM/PMs with a score ≥10.
Kaplan-Meier analyses were used to depict the cumulative incidence of major atherothrombotic events and MACE by ABCD-GENE score group during the 12-month follow-up period. P<0.05 was considered statistically significant. All statistical analyses were performed using R statistical software (version 4.1).(36)
Results
Patient Characteristics
Characteristics of the 2,341 patients included in the analysis are shown in Table 1. All were treated with clopidogrel at the time of event or last follow-up. Mean (SD) age was 64±12 years, 817 (35%) were female, 470 (20%) were Black or African American, and 1601 (68%) had an ACS indication for PCI. A total of 448 (19%) patients were no function allele carriers (18% IMs, 1% PMs), and 505 (22%) had an ABCD-GENE score ≥10. Among CYP2C19 IMs, six (1%) had a diplotype that did not include the CYP2C19*2 allele (*1/*3, *1/*4, *1/*8 [n=2], *6/*17, and *8/*17).
Table 1.
Patient Characteristics at the Time of Index Percutaneous Coronary Intervention
| Full Cohort (n=2341) |
ABCD-GENE Score <10 (n=1836) | ABCD-GENE Score ≥10 (n=505) | p value for comparison of ABCD-GENE score <10 vs ≥10 | |
|---|---|---|---|---|
| Age, years | 64.2 ± 12.0 | 63.1 ± 11.7 | 68.2 ± 12.1 | <0.001 |
| Female sex | 817 (34.9) | 609 (33.2) | 208 (41.2) | 0.001 |
| Race | ||||
| White | 1733 (74.0) | 1357 (73.9) | 376 (74.5) | 0.968 |
| Black or African American | 470 (20.1) | 370 (20.2) | 100 (19.8) | |
| Other or Unknown Racec | 138 (5.9) | 109 (5.9) | 29 (5.7) | |
| BMI, kg/m 2 | 29.9 ± 6.6 | 28.9 ± 6.1 | 33.6 ± 7.0 | <0.001 |
| Current smoker | 672 (28.7) | 579 (31.5) | 93 (18.4) | <0.001 |
| PCI Indication a | ||||
| STEMI | 401 (17.1) | 342 (18.6) | 59 (11.7) | 0.17 |
| NSTEMI | 661 (28.2) | 505 (27.5) | 156 (30.9) | |
| UA | 539 (23.0) | 408 (22.2) | 131 (25.9) | |
| Stable CAD/Elective | 740 (31.6) | 581 (31.6) | 159 (31.5) | |
| PCI Strategy b | ||||
| Drug-eluting stent | 2009 (85.8) | 1587 (86.4) | 422 (83.6) | <0.001 |
| Bare metal stent | 261 (11.1) | 210 (11.4) | 51 (10.1) | |
| PTCA | 71 (3.0) | 39 (2.1) | 32 (6.3) | |
| Medical history | ||||
| Hypertension | 1953 (83.4) | 1490 (81.2) | 463 (91.7) | <0.001 |
| Dyslipidemia | 1540 (65.8) | 1167 (63.6) | 373 (73.9) | <0.001 |
| Prior MI | 616 (26.3) | 467 (25.4) | 149 (29.5) | 0.075 |
| Prior Revascularization | 1065 (45.5) | 797 (43.4) | 268 (53.1) | <0.001 |
| Prior Stent | 721 (30.8) | 539 (29.4) | 182 (36.0) | 0.005 |
| Prior CABG | 277 (11.8) | 204 (11.1) | 73 (14.5) | 0.047 |
| Prior PTCA | 67 (2.9) | 54 (2.9) | 13 (2.6) | 0.774 |
| Stroke/TIA | 287 (12.3) | 218 (11.9) | 69 (13.7) | 0.313 |
| PVD | 232 (9.9) | 176 (9.6) | 56 (11.1) | 0.359 |
| Heart failure | 387 (16.5) | 273 (14.9) | 114 (22.6) | <0.001 |
| Atrial fibrillation | 254 (10.9) | 178 (9.7) | 76 (15.0) | 0.001 |
| Gastrointestinal or intracranial hemorrhage | 68 (2.9) | 44 (2.4) | 24 (4.8) | 0.008 |
| Cancer | 120 (5.1) | 99 (5.4) | 21 (4.2) | 0.318 |
| Discharge medication | ||||
| Aspirin | 2288 (97.7) | 1799 (98.0) | 489 (96.8) | 0.17 |
| Statin | 2185 (93.3) | 1717 (93.5) | 468 (92.7) | 0.566 |
| β-blocker | 2007 (85.7) | 1575 (85.8) | 432 (85.5) | 0.948 |
| ACE inhibitor or ARB | 1555 (66.4) | 1213 (66.1) | 342 (67.7) | 0.519 |
| ARA | 99 (4.2) | 77 (4.2) | 22 (4.4) | 0.971 |
| Anticoagulant | 264 (11.3) | 176 (9.6) | 88 (17.4) | <0.001 |
| PPI | 763 (32.6) | 595 (32.4) | 168 (33.3) | 0.755 |
p value for comparison of primary PCI indication categories (i.e., myocardial infarction [STEMI/NSTEMI] vs. unstable angina or Stable CAD/Elective) between groups.
p value for comparison of PCI strategy (i.e., any stent type vs. PTCA) between groups.
Detailed breakdown includes patients of the following ancestries: American Indian or Alaska Native (n=29), Asian (n=14), mixed ancestry (n=2), Native Hawaiian or Other Pacific Islander (n=2), or unknown (n=91).
Data are presented as number (%) or mean ± SD.
ACE, angiotensin converting enzyme; ARA, aldosterone receptor antagonist; ARB, angiotensin receptor blocker; BMI, body mass index; CABG, coronary artery bypass grafting; IM, CYP2C19 intermediate metabolizer; MI, myocardial infarction; PCI, percutaneous coronary intervention; PPI, proton pump inhibitor; PTCA, percutaneous transluminal coronary angioplasty; PVD, peripheral vascular disease; STEMI, ST-segment elevation myocardial infarction; TIA, transient ischemic attack
Apart from the expected differences in ABCD-GENE score components, there also were significant differences in sex, smoking status, PCI strategy (i.e., any stent type versus percutaneous transluminal coronary angioplasty), medical history, and anticoagulant use between clopidogrel-treated patients with an ABCD-GENE score ≥10 versus <10 (Table 1). All clinical components of the ABCD-GENE score were more prevalent in the ≥10 versus the <10 group (Table 2). Among those with an ABCD-GENE score ≥10, 269 (53%) and 20 (4%) patients were CYP2C19 IMs and PMs compared with 159 (9%) and 0 patients with a score <10, respectively (Table 2).
Table 2.
Prevalence of the clinical and genetic components of the ABCD-GENE score for clopidogrel-treated patients
| Full cohort (n=2341) |
ABCD-GENE <10 (n=1836) |
ABCD-GENE ≥10 (n=505) |
p value for comparison of ABCD-GENE score <10 vs ≥10 | |
|---|---|---|---|---|
| ABCD-GENE Score | 6 [3 – 8] | 4 [3 – 7] | 11 [10 – 13] | <0.001 |
| Age >75 years | 412 (18%) | 239 (13%) | 173 (34%) | <0.001 |
| BMI >30 kg/m2 | 1051 (45%) | 658 (36%) | 393 (78%) | <0.001 |
| CKD | 670 (29%) | 353 (19%) | 317 (63%) | <0.001 |
| Diabetes | 987 (42%) | 641 (35%) | 346 (69%) | <0.001 |
| CYP2C19 Genetic Test | ||||
| 2 LOF alleles (PM) | 20 (1%) | 0 | 20 (4%) | <0.001a |
| 1 LOF allele (IM) | 428 (18%) | 159 (9%) | 269 (53%) | |
| 0 LOF alleles (NM, RM, UM) | 1893 (81%) | 1677 (91%) | 216 (43%) |
p value for comparison of ≥1 LOF alleles vs 0 LOF alleles between groups.
Data are presented as median [IQR] or number (%).
BMI, body mass index; CKD, chronic kidney disease defined as an estimated glomerular filtration rate of < 60 ml/min/1.73m2; IM, CYP2C19 intermediate metabolizer; NM, CYP2C19 normal metabolizer; PM, CYP2C19 poor metabolizer; RM, CYP2C19 rapid metabolizer; UM, CYP2C19 ultra-rapid metabolizer; LOF, loss-of-function.
Clinical outcomes
Median [IQR] follow-up time after PCI was 229 [37–346] days for patients with an ABCD-GENE score <10 and 182 [29–330] days in those with a score ≥10. The rate of major atherothrombotic events was higher in patients with an ABCD-GENE score ≥10 versus <10 (24.6 versus 14.7 per 100 patient-years; adjusted hazard ratio [HR], 1.66; 95% CI, 1.23 – 2.25; p<0.001) (Table 3, Figure 1A). Patients with an ABCD-GENE score ≥10 also had a higher rate of MACE compared to patients with a score <10 (event rate 16.7 versus 10.1 per 100 patient-years; adjusted HR, 1.59; 95% CI, 1.11 – 2.30; p=0.013) (Table 3, Figure 1B).
Table 3.
Association of ABCD-GENE Score with Ischemic Outcomes within 1-Year After Percutaneous Coronary Intervention
| No. of events | Event rate per 100 person-years | Unadjusted HR | Adjusted HRa | |
|---|---|---|---|---|
| Major atherothrombotic events | ||||
| ABCD-GENE score <10 (n=1836) |
150 | 14.7 (95% CI, 12.3 – 17.0) |
Reference | Reference |
| ABCD-GENE score ≥10 (n=505) |
62 | 24.6 (95% CI, 18.5 – 30.8) |
1.63 (95% CI, 1.21 – 2.19) p=0.001 |
1.66 (95% CI, 1.23 – 2.25) p<0.001 |
| MACE | ||||
| ABCD-GENE score <10 (n=1836) |
103 | 10.1 (95% CI, 8.1 – 12.0) |
Reference | Reference |
| ABCD-GENE score ≥10 (n=505) |
42 | 16.7 (95% CI, 11.6 – 21.7) |
1.60 (95% CI, 1.12 – 2.29) p=0.011 |
1.59 (95% CI, 1.11 – 2.30) p=0.013 |
Model adjusted for: sex, smoking status, percutaneous coronary intervention (PCI) indication (i.e., MI vs. UA or elective PCI), PCI strategy (i.e., stent versus percutaneous transluminal coronary angioplasty), and institution
ABCD-GENE (Age, oBesity, Chronic kidney disease, Diabetes, CYP2C19 GENEtic variants) score; MACE, major adverse cardiovascular events. Major atherothrombotic events defined as the composite of death, myocardial infarction (MI), ischemic stroke, stent thrombosis (ST), or unstable (UA) requiring revascularization. MACE defined as the composite of cardiovascular death, MI, ST, or ischemic stroke.
Figure 1. Time-to-Event Curves for Atherothrombotic Events.
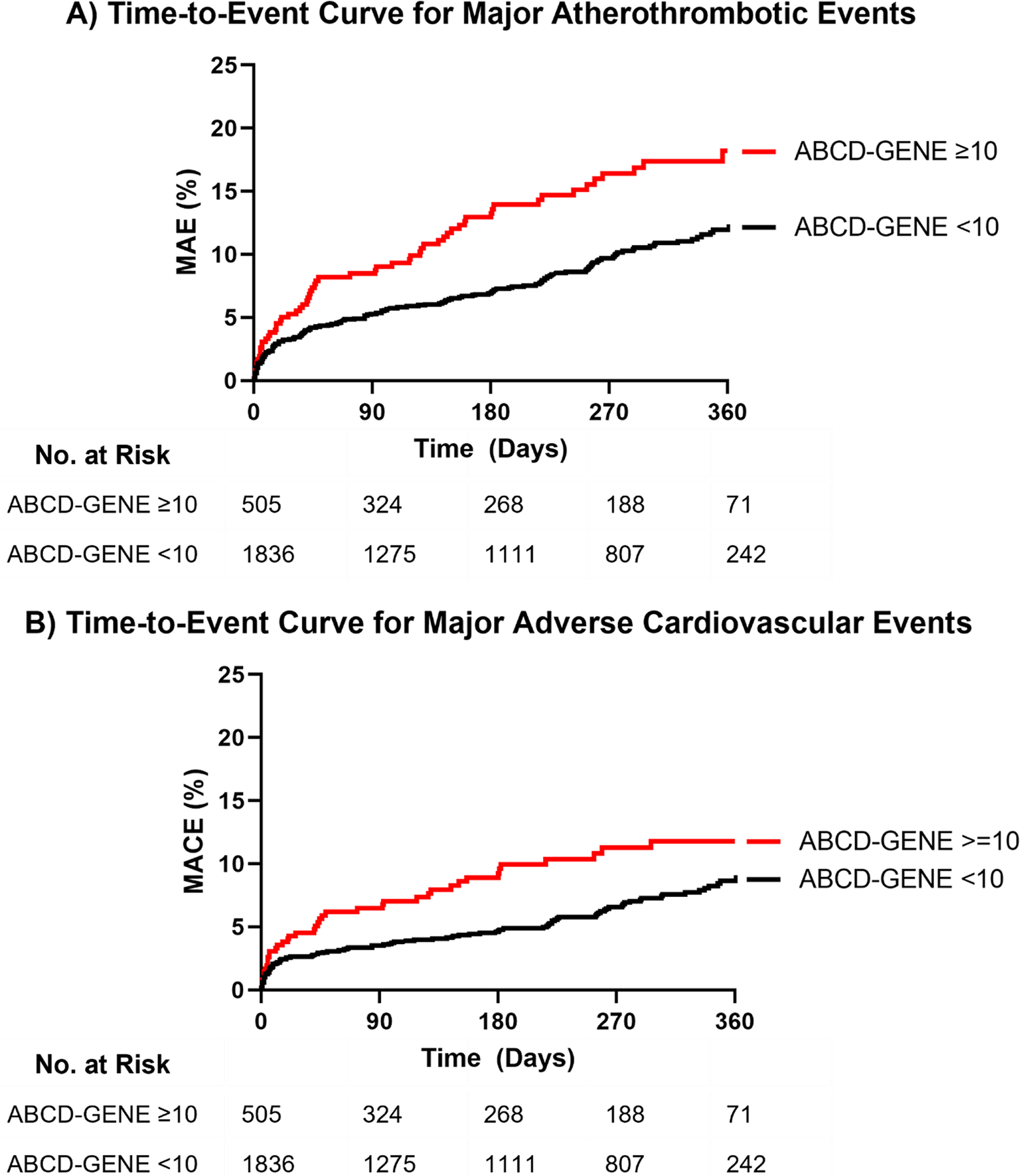
A) Major atherothrombotic events (MAE), defined as the first occurrence of death, myocardial infarction (MI), ischemic stroke, stent thrombosis (ST), or unstable angina requiring revascularization at 1-year post-PCI. Adjusted hazard ratio (HR) 1.66 (95% CI, 1.23 – 2.25), p<0.001. B) Major adverse cardiovascular events (MACE), defined as the first occurrence of cardiovascular death, MI, ST, or ischemic stroke at 1-year post-PCI. Adjusted HR 1.59 (95% CI, 1.11 – 2.30), p=0.013. The tails of the Kaplan-Meier curves in Panels A and B were truncated at 360 days post-PCI, after which time less than 10% of each stratum were available for follow-up. In Panel A, n=4 events were observed in the ABCD-GENE score <10 group after day 360, when only 71 (4% of patients with a score <10) patients were still in follow-up. In Panel B, n=1 event occurred after day 360 in the ABCD-GENE score <10 group. No cardiovascular events occurred after day 360 for the ABCD-GENE score ≥10 group when 16 (3%) patients were available for follow-up.
Characteristics of patients with an ABCD-GENE score <10 stratified by IM status are shown in Table S2. Median [IQR] ABCD-GENE score was greater among CYP2C19 IMs versus non-IMs with a score <10 (6 [IQR, 6–9] versus 4 [IQR, 0–7]; p<0.001). Kaplan-Meier curves for major atherothrombotic events are depicted for patients with an ABCD-GENE score ≥10 and <10, stratified by CYP2C19 IM status, in Figure 2. The major atherothrombotic event rate was lower among non-IMs with an ABCD-GENE score <10 versus patients with a score ≥10 (adjusted HR, 0.58; 95% CI, 0.43–0.79; p<0.001). In contrast, there was no significant difference in the major atherothrombotic event rate between IMs with a score <10 and patients with a score ≥10 (adjusted HR, 1.03; 95% CI, 0.59–1.81; p=0.917) (Table 4). Similar results were observed for MACE among non-IMs with a score <10 (adjusted HR, 0.61; 95% CI, 0.42–0.88; p=0.008) and IMs with a score <10 (adjusted HR, 0.97; 95% CI, 0.48–1.95; p=0.925) compared to those with a score ≥10 (Table 4; Figure S1). In the subset of patients with an ABCD-GENE score ≥10 (n=505), major atherothrombotic event and MACE rates were not significantly different between IM/PMs versus non-IM/PMs (Table S3).
Figure 2. Time-to-Event Curve for Major Atherothrombotic Events Stratified by ABCD-GENE Score and CYP2C19 IM Status.
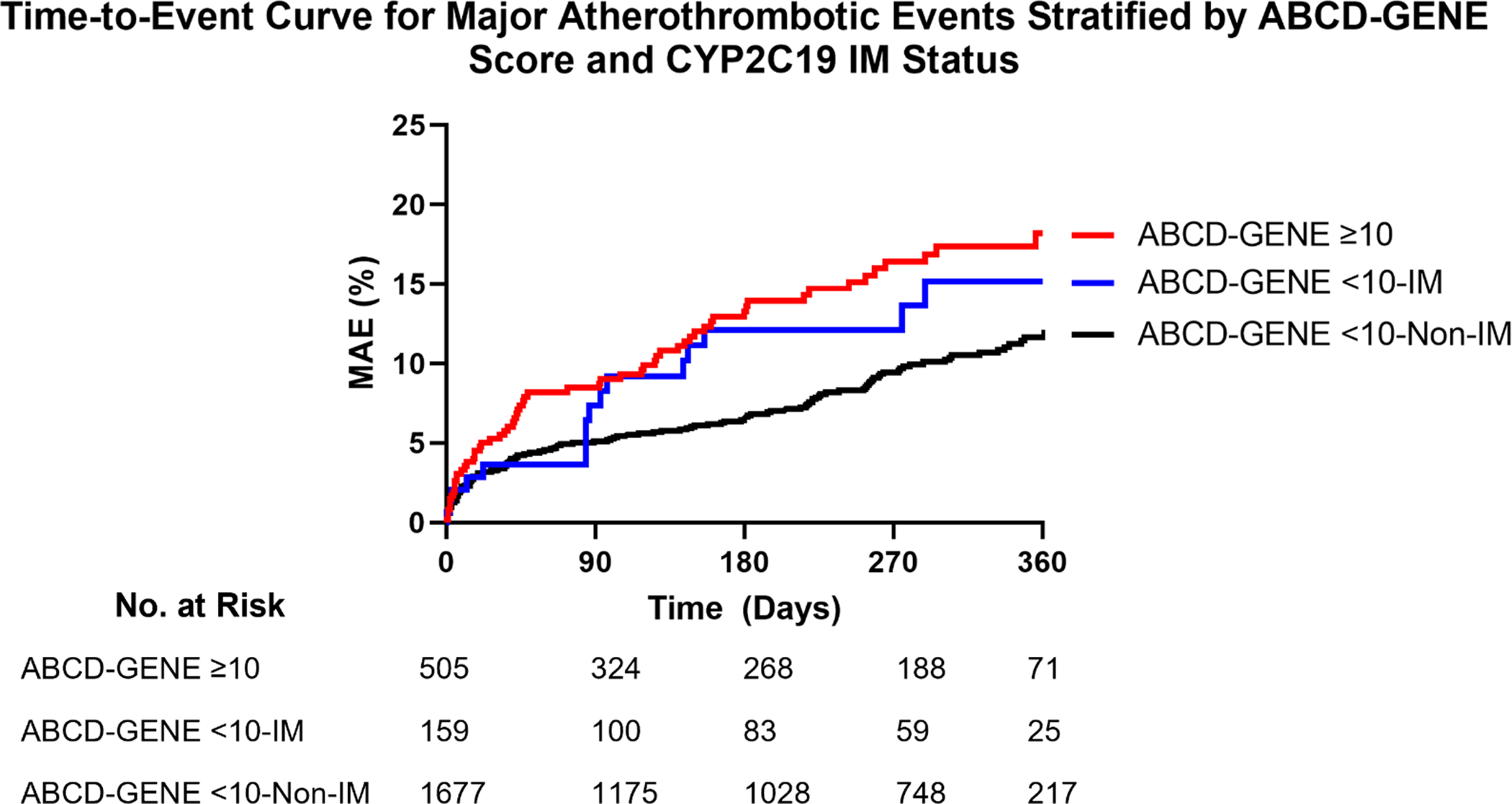
Major atherothrombotic events (MAE), defined as the first occurrence of death, myocardial infarction, ischemic stroke, stent thrombosis, or unstable angina requiring revascularization at 1-year post-PCI. Adjusted hazard ratio (HR) for ABCD-GENE score <10-Non-IM versus ≥10 = 0.58 (95% CI, 0.43–0.79), p<0.001. Adjusted HR for ABCD-GENE score <10-IM versus ≥10 = 1.03 (95% CI, 0.59–1.81), p=0.917. IM: CYP2C19 intermediate metabolizer. The tails of the Kaplan-Meier curve were truncated at 360 days post-PCI, after which time less than 10% of each stratum were available for follow-up. In the ABCD-GENE <10-Non-IM group, n=4 events were observed after day 360, when only 64 patients (4%) were still in follow-up. No MAE occurred after day 360 for the ABCD-GENE score ≥10 or score <10-IM groups when 16 (3%) and 7 (4%) patients were available for follow-up, respectively.
Table 4.
Association Between ABCD-GENE Score and Major Atherothrombotic Events with CYP2C19 Intermediate Metabolizers (IMs) and non-IMs within 1-Year After Percutaneous Coronary Intervention
| No. of events | Event rate per 100 person-years | Unadjusted HR | Adjusted HRa | |
|---|---|---|---|---|
| Major atherothrombotic events | ||||
| ABCD-GENE score ≥10 (n=505) |
62 | 24.6 (95% CI, 18.5 – 30.8) |
Reference | Reference |
| ABCD-GENE score <10: IMs (n=159) |
16 | 20.1 (95% CI, 10.3 – 30.0) |
0.82 (95% CI, 0.47–1.42) P=0.482 |
1.03 (95% CI, 0.59–1.81) P=0.917 |
| ABCD-GENE score <10: Non-IMs (n=1677) |
134 | 14.2 (95% CI, 11.8 – 16.6) |
0.60 (95% CI, 0.44–0.81) P<0.001 |
0.58 (95% CI, 0.43–0.79) P<0.001 |
| MACE | ||||
| ABCD-GENE score ≥10 (n=505) |
42 | 16.7 (95% CI, 11.6 – 21.7) |
Reference | Reference |
| ABCD-GENE score <10: IMs (n=159) |
10 | 12.6 (95% CI, 4.8 – 20.4) |
0.76 (95% CI, 0.38–1.51) P=0.431 |
0.97 (95% CI, 0.48–1.95) P=0.925 |
| ABCD-GENE score <10: Non-IMs (n=1677) |
93 | 9.9 (95% CI, 7.9 – 11.9) |
0.61 (95% CI, 0.43–0.88) P=0.009 |
0.61 (95% CI, 0.42–0.88) P=0.008 |
Model adjusted for: sex, smoking status, percutaneous coronary intervention (PCI) indication (i.e., MI vs. UA or elective PCI), PCI strategy (i.e., stent versus percutaneous transluminal coronary angioplasty), and institution
ABCD-GENE (Age, oBesity, Chronic kidney disease, Diabetes, CYP2C19 GENEtic variants); IM: CYP2C19 intermediate metabolizer; MACE, major adverse cardiovascular events. Non-CYP2C19 IMs consists of CYP2C19: normal, rapid, and ultra-rapid metabolizers. Major atherothrombotic events defined as the composite of death, myocardial infarction (MI), ischemic stroke, stent thrombosis (ST), or unstable angina (UA) requiring revascularization. MACE defined as the composite of cardiovascular death, MI, ST, or ischemic stroke.
Discussion
The ABCD-GENE score is a recently developed tool that integrates clinical factors and CYP2C19 genotype to identify PCI patients at risk for diminished clopidogrel antiplatelet effects and clinical effectiveness.(24) The score was originally derived using platelet reactivity data from a randomized, controlled trial(37) and associated with clinical outcomes in two European registries, POPULAR (Do Platelet Function Assays Predict Clinical Outcomes in Clopidogrel-Pretreated Patients Undergoing Elective PCI)(25) and FAST-MI (French Registry of Acute ST-Elevation and Non–ST-Elevation Myocardial Infarction),(26) which were conducted in the Netherlands and France, respectively. An ABCD-GENE score ≥10 was associated with diminished clopidogrel response and increased risk of MACE versus a score <10.(24) The score was further evaluated in Asian patients for its association with HPR with clopidogrel after PCI and its association with recurrent stroke with clopidogrel plus aspirin versus aspirin alone following minor stroke or transient ischemic attack.(28, 38) Most recently, the predictive value of the ABCD-GENE score in identifying patients at increased risk of ischemic events was shown in a post-hoc analysis of data from predominately White or Asian participants in a clinical trial of genotype-guided therapy after PCI.(27) Similar to findings from European registries and post-hoc analysis of clinical trial data, we found that the risk of major atherothrombotic events and MACE was approximately 1.6-fold higher in clopidogrel-treated PCI patients with an ABCD-GENE score ≥10 compared to those with a score <10 in a diverse, real-world clinical setting. Moreover, the increased risk for MACE observed herein among those with an ABCD-GENE score ≥10 versus <10 (adjusted HR, 1.59; 95% CI, 1.11 – 2.30) was comparable to the risk for MACE reported in the FAST-MI registry (adjusted HR, 1.48; 95% CI, 1.16 – 1.90).(24)
Our findings help increase the generalizability of the ABCD-GENE score given the real-world study population who underwent emergent or elective PCI; had a high comorbidity burden, especially in regard to prevalence of obesity, CKD, DM, and history of MI,(26) and were diverse in ancestry. The population reported herein was from the United States, and 20% were Black or African American. Further, the patients included in our analysis received CYP2C19 testing as part of their clinical care to help guide post-PCI antiplatelet therapy and thus represent patients in whom the score would be applied to in practice. All patients in the current analysis were genotyped for both the CYP2C19*2 and *3 no function alleles at a minimum, as recommended per current guidelines; although detection of the *3 and other no function alleles only resulted in 1% of additional patients classified as IM or PM. These data add to the evidence of ABCD-GENE score associations with outcomes during clopidogrel treatment after PCI and demonstrate diminished clopidogrel effectiveness in a real-world population of PCI patients with a score ≥10.(24, 28)
Given evidence that CYP2C19 genotype influences the effectiveness of clopidogrel, but not prasugrel or ticagrelor, pharmacogenetic guidelines recommend prasugrel or ticagrelor for CYP2C19 IMs and PMs in the absence of contraindications.(16) Similarly, the Food and Drug Administration-approved clopidogrel labeling recommends alternative therapy in PMs, with recommendations for alternative therapy expanded to IMs in their Table of Pharmacogenetic Associations.(39, 40) Clinical trials and observational studies have examined outcomes with guided approaches to the selection of P2Y12 inhibitor therapy, using either genetic or platelet function testing.(35, 41–43) Recent large-scale meta-analyses of these data have shown improved outcomes, including better safety and efficacy profiles, with a guided approach to P2Y12 inhibitor therapy selection compared with a non-guided approach.(44, 45) Importantly, outcomes varied depending on whether P2Y12 inhibitor therapy was escalated from clopidogrel to alternative therapy with prasugrel or ticagrelor in no function allele carriers or de-escalated from prasugrel or ticagrelor to clopidogrel in patients without a no function allele. Compared to a non-guided treatment approach, an escalation approach was associated with a reduction in ischemic events without any increase in bleeding, while a de-escalation approach was associated with a reduction in bleeding, without any increase in ischemic events.(45) These findings suggest a guided approach can be used to better balance the risks and benefits of P2Y12 inhibitors and facilitate the selection of antiplatelet therapy post-PCI.
Importantly, CYP2C19 genotype is only one factor explaining the inter-patient variability in clopidogrel response.(9) Advanced age, obesity, CKD, and DM have also been associated with diminished antiplatelet effects of clopidogrel. Specifically, the SENIOR-PLATELET study showed that clopidogrel-treated patients age 75 years or older were more likely to have high residual platelet reactivity compared with patients younger than 75 years.(23) Similarly, elevated BMI has been associated with an attenuated clopidogrel response.(22, 46, 47) A meta-analysis of clopidogrel-treated patients determined that patients with CKD were more likely to develop HPR and were at increased risk for cardiovascular events compared to patients without CKD.(19) Diabetes is also associated with significantly reduced clopidogrel active metabolite formation and increased platelet reactivity.(20, 21) Although the mechanism underlying these effects remains unclear, suppression of hepatic CYP2C expression and activity in the setting of chronic inflammatory conditions (such as advanced age, obesity, CKD, and diabetes) could contribute to reduced clopidogrel active metabolite formation.(48–50) The ABCD-GENE score combines these independent risk factors for clopidogrel non-response. With the increasing uptake of CYP2C19 testing in clinical practice,(34) the ABCD-GENE score represents a feasible means of more precisely predicting clopidogrel response following PCI.
While all patients in our cohort were genotyped clinically, and prasugrel or ticagrelor was recommended in patients with a no function allele, 19% of no function allele carriers (IM, 18%; PM, 1%) remained on clopidogrel. Most of these patients were IMs, suggesting that physicians may be less likely to heed recommendations to avoid clopidogrel in IMs compared to PMs, perhaps because the clopidogrel labeling only addresses PMs. In an exploratory analysis from this project that focused on the 159 CYP2C19 IMs with an ABCD-GENE score <10, the risk for major atherothrombotic events and MACE was not different between IMs with an ABCD-GENE score <10 and anyone with a score ≥10. These data suggest that patients with a no function allele are at increased risk for ischemic events with clopidogrel even if their ABCD-GENE score is <10. A previous analysis showed that when the ABCD-GENE score is considered as a continuous variable, each one-point increase in the score increases the relative risk of MACE by approximately 4%.(24) The actual score was higher in the IM group, and we cannot rule out that this contributed to our findings. In addition, an exploratory analysis within the 505 patients with a score ≥10 showed that major atherothrombotic and MACE event rates were not significantly different between CYP2C19 IM/PMs versus non-IM/PMs, suggesting that patients without a no function allele are at increased risk for ischemic events with clopidogrel if their ABCD-GENE score is ≥10. Taken together, data from the current analysis add to the ABCD-GENE score literature and suggest that patients with at least one no function allele or an ABCD-GENE score ≥10 are at elevated risk of diminished clopidogrel clinical effectiveness. Given this and the small sample size, these data should be interpreted with caution, and further study of clopidogrel response among IMs with a score <10 and among non-IM/PMs with a score ≥10 is warranted.
This study is not without limitations. Notably, the pharmacodynamic outcome, HPR, was not assessed in our study, and thus we could not examine the ability of the ABCD-GENE score to predict HPR with clopidogrel in our population or determine whether HPR mediated the observed increased risk for ischemic events in patients with a score ≥10. Both the primary outcome and the more restrictive MACE definition were consistent with the results published in the original ABCD-GENE score manuscript,(24) which increased confidence in the validity of the current results using real-world data. In addition, data on the severity and duration of various comorbid conditions, such as diabetes and hyperlipidemia, and laboratory parameters, such as glucose, cholesterol, C-reactive protein, and liver enzymes were not collected, and the potential impact on ischemic outcomes was not evaluated. Finally, events were identified based on electronic health record review, and events that occurred in other health systems may have been missed.
In summary, our real-world outcomes data extend the association between the ABCD-GENE score and cardiovascular events with clopidogrel treatment to a diverse, real-world U.S. population who underwent PCI and received clinical CYP2C19 genotyping. Within this population, an ABCD-GENE score ≥10 was associated with an increased risk for atherothrombotic events compared with a score <10. Our data suggest the ABCD-GENE score may be used in patients undergoing PCI to identify poor responders to clopidogrel. Whether treatment with an alternative P2Y12 inhibitor would improve outcomes in patients with a high score remains to be determined. Given the significant body of evidence on factors contributing to poor clopidogrel response informing recent advances in guided selection of DAPT for patients undergoing PCI, the ABCD-GENE score represents a practical approach to integrate both clinical and genetic predictors of clopidogrel non-response to identify those patients at risk for diminished clopidogrel clinical effectiveness.
Supplementary Material
Table S1. CYP2C19 Genotyping Platform and Alleles Detected at Each Institution
Table S2. Comparison of Patient Characteristics at the Time of Index Percutaneous Coronary Intervention Between Patients with an ABCD-GENE score <10, stratified by CYP2C19 intermediate metabolizer status, and those with a score ≥10
Table S3. Association of ABCD-GENE Score with Ischemic Outcomes within 1-Year after Percutaneous Coronary Intervention Among Patients with an ABCD-GENE Score ≥10 (n=505) Stratified by CYP2C19 IM/PM Status
Figure S1. Time-to-Event Curve for Major Adverse Cardiovascular Events Stratified by ABCD-GENE Score and CYP2C19 IM Status. Major adverse cardiovascular events (MACE), defined as the first occurrence of cardiovascular death, myocardial infarction, ischemic stroke, or stent thrombosis at 1-year post-PCI. Adjusted hazard ratio (HR) for ABCD-GENE score <10: non-IM versus ≥10 = 0.61 (95% CI, 0.42–0.88), p=0.008. Adjusted HR for ABCD-GENE score <10: IM versus ≥10 = 0.97 (95% CI, 0.48–1.95), p=0.925. IM: CYP2C19 intermediate metabolizer. The tails of the Kaplan-Meier curve were truncated at 360 days post-PCI, after which time less than 10% of each stratum were available for follow-up. In the ABCD-GENE <10-Non-IM group, n=1 event was observed after day 360, when only 65 patients (4%) were still in follow-up. No MACE occurred after day 360 for the ABCD-GENE score ≥10 or score <10-IM groups when 17 (3%) and 7 (4%) patients were available for follow-up, respectively.
Study Highlights.
What is the current knowledge on the topic?
The ABCD-GENE score was developed using platelet function data and includes five independent predictors of high on-treatment platelet reactivity to predict clopidogrel non-response after percutaneous coronary intervention (PCI). While the score is predictive of clopidogrel non-response in predominately European and Asian patients and clinical trial participants, its ability to predict outcomes in a diverse, high-risk, real-world population is unknown.
What question did this study address?
The objective of this study was to validate the association between the ABCD-GENE score and risk for major atherothrombotic events in a diverse real-world population of clopidogrel-treated PCI patients.
What does this study add to our knowledge?
These data extend the association between an ABCD-GENE score ≥10 and increased risk for adverse atherothrombotic events among patients receiving clopidogrel after PCI to a diverse real-world population.
How might this change clinical pharmacology or translational science?
These data support use of the ABCD-GENE score for patients undergoing PCI to more precisely predict risk for poor response to clopidogrel.
Funding:
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute (1R01HL149752), National Human Genome Research Institute (U01HG007269), and National Center for Advancing Translational Sciences (UL1TR001427). Spartan Bioscience Inc. (Ottawa, ON) provided the genotyping platforms and kits for initial testing at the UF Health Jacksonville site. C.D.T. was supported in part by the National Human Genome Research Institute (T32 HG008958). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of Interest:
C.D.T. - None; F.F. has received consulting fees or honoraria from AstraZeneca, Bayer, and Sanofi; his institution has received research grants from PLx Pharma, and the Scott R. MacKenzie Foundation.; E.C.K. - None; J.S.R. - None; M.W. - None; R.D.A. - None; A.L.D. - None; Y.G. - None; M.N.G. - None; R.A.K. - None; N.K. - None; J.G.M. - None; C.W.M. - None; I.R.M. - None; P.S. - None; A.L.B. - None; J.A.J. is on the United Health Group Scientific Advisory Board.; G.A.S. - None; A.G.W has received consulting fees from Arbor Pharmaceuticals and Genentech Inc. Her institution has received funding for research for which she served as principal investigator from Merck, Sharpe and Dohme.; D.J.A. has received consulting fees or honoraria from Abbott, Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Boehringer Ingelheim, Bristol Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Haemonetics, Janssen, Merck, PhaseBio, PLx Pharma, Pfizer, and Sanofi; his institution has received research grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli Lilly, Gilead, Idorsia, Janssen, Matsutani Chemical Industry Co, Merck, Novartis, Osprey Medical, Renal Guard Solutions, and the Scott R. MacKenzie Foundation.; C.R.L. - None; L.H.C. - None
References:
- (1).Levine GN et al. 2016 ACC/AHA guideline focused update on duration of dual antiplatelet therapy in patients with coronary artery disease: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Thorac Cardiovasc Surg. 152, 1243–75 (2016). [DOI] [PubMed] [Google Scholar]
- (2).Collet JP et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur Heart J. 42, 1289–367 (2021). [DOI] [PubMed] [Google Scholar]
- (3).Wiviott SD et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 357, 2001–15 (2007). [DOI] [PubMed] [Google Scholar]
- (4).Wallentin L et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 361, 1045–57 (2009). [DOI] [PubMed] [Google Scholar]
- (5).Khalid U et al. Prescription Patterns of Clopidogrel, Prasugrel, and Ticagrelor After Percutaneous Coronary Intervention With Stent Implantation (from the NCDR PINNACLE Registry). Am J Cardiol. 124, 1807–12 (2019). [DOI] [PubMed] [Google Scholar]
- (6).Zanchin T et al. Frequency, Reasons, and Impact of Premature Ticagrelor Discontinuation in Patients Undergoing Coronary Revascularization in Routine Clinical Practice: Results From the Bern Percutaneous Coronary Intervention Registry. Circ Cardiovasc Interv. 11, e006132 (2018). [DOI] [PubMed] [Google Scholar]
- (7).Basra SS et al. Ticagrelor Use in Acute Myocardial Infarction: Insights From the National Cardiovascular Data Registry. J Am Heart Assoc. 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dayoub EJ et al. Trends in Platelet Adenosine Diphosphate P2Y12 Receptor Inhibitor Use and Adherence Among Antiplatelet-Naive Patients After Percutaneous Coronary Intervention, 2008–2016. JAMA Intern Med. 178, 943–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shuldiner AR et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. Jama. 302, 849–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Aradi D et al. Bleeding and stent thrombosis on P2Y12-inhibitors: collaborative analysis on the role of platelet reactivity for risk stratification after percutaneous coronary intervention. Eur Heart J. 36, 1762–71 (2015). [DOI] [PubMed] [Google Scholar]
- (11).Angiolillo DJ et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 49, 1505–16 (2007). [DOI] [PubMed] [Google Scholar]
- (12).Sibbing D et al. Updated Expert Consensus Statement on Platelet Function and Genetic Testing for Guiding P2Y12 Receptor Inhibitor Treatment in Percutaneous Coronary Intervention. JACC Cardiovasc Interv. 12, 1521–37 (2019). [DOI] [PubMed] [Google Scholar]
- (13).Stone GW et al. Platelet reactivity and clinical outcomes after coronary artery implantation of drug-eluting stents (ADAPT-DES): a prospective multicentre registry study. Lancet. 382, 614–23 (2013). [DOI] [PubMed] [Google Scholar]
- (14).Mega JL et al. Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. Jama. 306, 2221–8 (2011). [DOI] [PubMed] [Google Scholar]
- (15).Varenhorst C et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J. 30, 1744–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee CR et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin Pharmacol Ther. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mega JL et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. Jama. 304, 1821–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mega JL et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 360, 354–62 (2009). [DOI] [PubMed] [Google Scholar]
- (19).Wu Y, Song Y, Pan Y, Gong Y & Zhou Y High on-clopidogrel platelet reactivity and chronic kidney disease: a meta-analysis of literature studies. Scand Cardiovasc J. 53, 55–61 (2019). [DOI] [PubMed] [Google Scholar]
- (20).Angiolillo DJ et al. Platelet function profiles in patients with type 2 diabetes and coronary artery disease on combined aspirin and clopidogrel treatment. Diabetes. 54, 2430–5 (2005). [DOI] [PubMed] [Google Scholar]
- (21).Angiolillo DJ et al. Impaired responsiveness to the platelet P2Y12 receptor antagonist clopidogrel in patients with type 2 diabetes and coronary artery disease. J Am Coll Cardiol. 64, 1005–14 (2014). [DOI] [PubMed] [Google Scholar]
- (22).Angiolillo DJ et al. Platelet aggregation according to body mass index in patients undergoing coronary stenting: should clopidogrel loading-dose be weight adjusted? J Invasive Cardiol. 16, 169–74 (2004). [PubMed] [Google Scholar]
- (23).Silvain J et al. High on-thienopyridine platelet reactivity in elderly coronary patients: the SENIOR-PLATELET study. Eur Heart J. 33, 1241–9 (2012). [DOI] [PubMed] [Google Scholar]
- (24).Angiolillo DJ et al. Derivation, Validation, and Prognostic Utility of a Prediction Rule for Nonresponse to Clopidogrel: The ABCD-GENE Score. JACC Cardiovasc Interv. 13, 606–17 (2020). [DOI] [PubMed] [Google Scholar]
- (25).Breet NJ et al. Comparison of Platelet Function Tests in Predicting Clinical Outcome in Patients Undergoing Coronary Stent Implantation. JAMA. 303, 754–62 (2010). [DOI] [PubMed] [Google Scholar]
- (26).Simon T et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 360, 363–75 (2009). [DOI] [PubMed] [Google Scholar]
- (27).Capodanno D et al. ABCD-GENE Score and Clinical Outcomes Following Percutaneous Coronary Intervention: Insights from the TAILOR-PCI Trial. J Am Heart Assoc. 11, e024156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dai L et al. Application of Age, Body Mass Index, Chronic Kidney Disease, Diabetes, and Genotyping Score for Efficacy of Clopidogrel: Secondary Analysis of the CHANCE Trial. Stroke. Strokeaha120033049 (2021). [DOI] [PubMed] [Google Scholar]
- (29).Pratt VM et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn. 20, 269–76 (2018). [DOI] [PubMed] [Google Scholar]
- (30).Lee CR et al. Impact of the CYP2C19*17 Allele on Outcomes in Patients Receiving Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. Clin Pharmacol Ther. 109, 705–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Beitelshees AL et al. CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention in Diverse Clinical Settings. J Am Heart Assoc. e024159 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cavallari LH et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med. 16, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lee CR et al. Clinical Outcomes and Sustainability of Using CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. Circ Genom Precis Med. 11, e002069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Empey PE et al. Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther. 104, 664–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cavallari LH et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv. 11, 181–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).R Core Team. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2021). [Google Scholar]
- (37).Price MJ et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA. 305, 1097–105 (2011). [DOI] [PubMed] [Google Scholar]
- (38).Saito Y et al. Validation of the ABCD-GENE score to identify high platelet reactivity in east Asian patients undergoing percutaneous coronary intervention. Int J Cardiol. 327, 15–8 (2021). [DOI] [PubMed] [Google Scholar]
- (39).U.S. Food & Drug Administration. Table of Pharmacogenetic Associations. <https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations> (2021). Accessed February 3, 2022.
- (40).Plavix (clopidogrel) [package insert]. Bridgewater, NJ: Sanofi-aventis US, LLC; 2021. [Google Scholar]
- (41).Sibbing D et al. Guided de-escalation of antiplatelet treatment in patients with acute coronary syndrome undergoing percutaneous coronary intervention (TROPICAL-ACS): a randomised, open-label, multicentre trial. Lancet. 390, 1747–57 (2017). [DOI] [PubMed] [Google Scholar]
- (42).Claassens DMF et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N Engl J Med. 381, 1621–31 (2019). [DOI] [PubMed] [Google Scholar]
- (43).Pereira NL et al. Effect of Genotype-Guided Oral P2Y12 Inhibitor Selection vs Conventional Clopidogrel Therapy on Ischemic Outcomes After Percutaneous Coronary Intervention: The TAILOR-PCI Randomized Clinical Trial. Jama. 324, 761–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Galli M et al. Comparative effects of guided vs. potent P2Y12 inhibitor therapy in acute coronary syndrome: a network meta-analysis of 61 898 patients from 15 randomized trials. Eur Heart J. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Galli M et al. Guided versus standard antiplatelet therapy in patients undergoing percutaneous coronary intervention: a systematic review and meta-analysis. Lancet. 397, 1470–83 (2021). [DOI] [PubMed] [Google Scholar]
- (46).Cuisset T et al. Relationship between aspirin and clopidogrel responses in acute coronary syndrome and clinical predictors of non response. Thromb Res. 123, 597–603 (2009). [DOI] [PubMed] [Google Scholar]
- (47).Nardin M et al. Body Mass Index and Platelet Reactivity During Dual Antiplatelet Therapy With Clopidogrel or Ticagrelor. J Cardiovasc Pharmacol. 66, 364–70 (2015). [DOI] [PubMed] [Google Scholar]
- (48).Morgan ET Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther. 85, 434–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sun Y et al. Clopidogrel Resistance in a Murine Model of Diet-Induced Obesity Is Mediated by the Interleukin-1 Receptor and Overcome With DT-678. Arterioscler Thromb Vasc Biol. 40, 1533–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jiang LP et al. Is platelet responsiveness to clopidogrel attenuated in overweight or obese patients and why? A reverse translational study in mice. Br J Pharmacol. 179, 46–64 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. CYP2C19 Genotyping Platform and Alleles Detected at Each Institution
Table S2. Comparison of Patient Characteristics at the Time of Index Percutaneous Coronary Intervention Between Patients with an ABCD-GENE score <10, stratified by CYP2C19 intermediate metabolizer status, and those with a score ≥10
Table S3. Association of ABCD-GENE Score with Ischemic Outcomes within 1-Year after Percutaneous Coronary Intervention Among Patients with an ABCD-GENE Score ≥10 (n=505) Stratified by CYP2C19 IM/PM Status
Figure S1. Time-to-Event Curve for Major Adverse Cardiovascular Events Stratified by ABCD-GENE Score and CYP2C19 IM Status. Major adverse cardiovascular events (MACE), defined as the first occurrence of cardiovascular death, myocardial infarction, ischemic stroke, or stent thrombosis at 1-year post-PCI. Adjusted hazard ratio (HR) for ABCD-GENE score <10: non-IM versus ≥10 = 0.61 (95% CI, 0.42–0.88), p=0.008. Adjusted HR for ABCD-GENE score <10: IM versus ≥10 = 0.97 (95% CI, 0.48–1.95), p=0.925. IM: CYP2C19 intermediate metabolizer. The tails of the Kaplan-Meier curve were truncated at 360 days post-PCI, after which time less than 10% of each stratum were available for follow-up. In the ABCD-GENE <10-Non-IM group, n=1 event was observed after day 360, when only 65 patients (4%) were still in follow-up. No MACE occurred after day 360 for the ABCD-GENE score ≥10 or score <10-IM groups when 17 (3%) and 7 (4%) patients were available for follow-up, respectively.


