Abstract
Contemporary paediatric clinical trials have improved 5-year event-free survival above 85% and 5-year overall survival above 90% in B-cell acute lymphoblastic leukaemia (ALL) in many study groups, whilst outcomes for T-cell ALL are still lagging behind by 5–10% in most studies. Several factors have contributed to this discrepant outcome. First, patients with T-cell ALL are generally older than those with B-cell ALL and, therefore, have poorer tolerance to chemotherapy, especially dexamethasone and asparaginase, and have increased risk of extramedullary relapse. Second, a higher proportion of patients with B-cell ALL have favourable genetic subtypes (eg, ETV6–RUNX1 and high hyperdiploidy), which confer a superior outcome compared with favourable subtypes of T-cell ALL. Third, T-cell ALL blasts are generally more resistant to conventional chemotherapeutic drugs than are B-cell ALL blasts. Finally, patients with B-cell ALL are more amendable to available targeted therapies, such as Philadelphia chromosome-positive and some Philadelphia chromosome-like ALL cases to ABL-class tyrosine kinase inhibitors, and CD19-positive and CD22-postive B-cell ALL cases to a variety of immunotherapies. Several novel treatments under investigation might narrow the gap in survival between T-cell ALL and B-cell ALL, although novel treatment options for T-cell ALL are limited.
Introduction
Acute lymphoblastic leukaemia (ALL) is the most common paediatric cancer, accounting for nearly a quarter of all cases of paediatric cancer.1 Based on the immunophenotype, ALL cases can be broadly classified as B-cell or T-cell ALL, with T-cell ALL comprising 10–15% of newly diagnosed cases, depending on age range, race, or ethnicity of the population in question. Historically, patients with T-cell ALL have had worse outcomes than patients with B-cell ALL in most clinical trials, with a few exceptions such as the Berlin-Frankfurt-Münster 86 protocol2 and the Dana-Farber Cancer Institute Protocol 85–01.3 These two clinical trials yielded similar results for patients with B-cell ALL and T-cell ALL, a finding attributed to the intensive use of cyclophosphamide, dexamethasone, asparaginase, and anthracycline for both trials. With recent advances in risk-directed treatment, 5-year event-free survival has exceeded 85% and overall survival over 90% for all patients with ALL in many contemporary clinical trials (table 1).4–15 However, the outcome for T-cell ALL is still inferior to that for B-cell ALL in most studies. In this Review, we assess the clinical and biological characteristics, and the treatment components, that potentially accounted for the differences in treatment response and outcome between B-cell ALL and T-cell ALL, before summarising recent developments in biological studies and treatment that could be used to improve outcomes for both B-cell and T-cell ALL.
Table 1:
Treatment results for B-cell ALL and T-cell ALL in major paediatric clinical trials
| Years of study | Patients (n) | Age range, years | Patients with T-cell ALL (%) | Patients who received cranial irradiation (%) | 5-year event-free survival, % (SE or 95% CI) | 5-year overall survival, % (SE or 95% CI) | References | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | B-cell | T-cell | Total | B-cell | T-cell | |||||||
| Associazione Italiana di Ematologia Pediatrica Group-Berlin-Frankfurt-Münster 2000 | 2000–06 | 4741 | 1–18 | 13·2% | 18% | NA | 80·4% (0·9) at 7 years | 75·9% (2·0) at 7 years* | NA | 91·8% (0·5) at 7 years | 80·7% (1·9) at 7 years* | Conter et al (2010)4 and Schrappe et al (2011)5 |
| COG (Children’s Cancer Group, Paediatric Oncology Group, COG) studies† | 2000–05 | 7153 | 0–22 | 7·1% | NA | NA | NA | NA | 90·4% (0·5) | 91·1% (0·5) | 81·6% (2·2) | Hunger et al (2012)6 |
| Dutch COG-10 | 2004–12 | 778 | 1–18 | 14·9% | 0·6% | 87·0% (1·2) | 88·4% (1·3) | 80·0% (3·7) | 91·9% (1·0) | 93·3% (1·0) | 84·4% (3·4) | Pieters et al (2016)7 |
| Dana-Farber Cancer Institute Consortium 05–001 | 2005–10 | 551 | 1–18 | 12·5% | Approximately 23% | 85% (82–88) | 85% (82–88) | 87% (76–93) | 91% (88–93) | 91% (88–93) | 91% (81–96) | Place et al (2015)8 |
| European Organisation for Research and Treatment of Cancer-Children’s Leukemia Group 58 951 | 1998–2008 | 1947 | 0–17 | 15·2% | None | 81·3% (0·9) at 8 years | Dexa: 83·4% (1·4), pred: 82·0% (1·4) at 8 years | Dexa: 71·3% (3·8), pred: 76·7% (3·5) at 8 years | 88·1% (0·8) at 8 years | Dexa: 89·6% (1·2), pred: 90·2% (1·1) at 8 years | Dexa: 74·2% (3·8), pred: 82·1% (3·2) at 8 years | Domenech et al (2014)9 |
| Medical Research Council UK ALL 2003 Trial | 2003–11 | 3126 | 1–24 | 12·4% | 2% | 87·3% (86·1–88·5) | 88·1% (86·6–89·5) | 81·2% (77·3–85·1) | 91·6% (90·6–92·6) | 92·4% (91·4–93·4) | 86·4% (82·9–89·9) | Vora et al (2014)10 and Patrick et al (2014)11 |
| Malaysia-Singapore Trial 2003 | 2002–11 | 556 | 0–18 | 8·8% | Approximately 20% | 80·6% (3·5) at 6 years | 80·7% (3·7) at 6 years | 80·5% (11·6) at 6 years | 88·4% (3·1) at 6 years | NA | NA | Yeoh et al (2012)12 |
| Nordic Society of Paediatric Haematology and Oncology ALL 2008 Trial | 2008–14 | 1509 | 1–17 | 15·3% | None | 85% (1) | 86% (1) | 74% (3) | 91% (1) | NA | NA | Toft et al (2018)13 |
| St Jude Children’s Research Hospital Total XV Trial | 2000–07 | 498 | 1–18 | 15·3% | None | 87·3% (84·4–90·2) | 86·9% (3·1) | 78·4% (7·8) | 93·5% (91·3–95·7) | 94·6% (1·9) | 87·6% (6·3) | Pui et al (2009)14 and Pui etal (2015)15 |
ALL=acute lymphoblastic leukaemia. NA=not available. COG=Children’s Oncology Group. Dexa=dexamethasone. Pred=prednisone.
Excluded 145 patients not evaluable for minimal residual disease study, 18 of whom died or relapsed before day 78 of induction.
Children’s Cancer Group 1961–91; Pediatric Oncology Group 9494/9407/9904/9905/9906; COG AALL00P2/0232.
Clinical differences
Compared with patients with B-cell ALL, those patients with T-cell ALL are generally older; more likely to be male; have higher presenting leucocyte count, haemoglobin concentration, and platelet count; and have a higher frequency of mediastinal mass and blast cells in cerebrospinal fluid due to CNS disease status 2 (<5 leucocytes per μL with blasts), CNS3 (≥5 leucocytes per μL with blasts), or traumatic lumbar puncture with blasts (≥10 erythrocytes per μL with blasts) at diagnosis (table 2). Older age at presentation contributed to the generally poorer outcome of patients with T-cell ALL than that of patients with B-cell ALL. Besides compliance issues, adolescents (10–19 years old) have more treatment-related morbidities and mortality, especially those associated with dexamethasone and asparaginase, such as osteonecrosis, thrombosis, pancreatitis, and liver dysfunction, than do younger children.13 Increased toxicities were attributed to poorer clearance and hence increased exposure to dexamethasone in the older age group, and the potentiated effect of concomitant use of asparaginase on dexamethasone.16 Increased dose intensity of treatment with dexamethasone and asparaginase generally improved the outcome of patients with T-cell ALL.16 However, a randomised study of non-very high-risk patients (ie, did not fit the criteria, defined by the presence of blast count in peripheral blood ≥1 × 109/L at completion of the prephase [day 8], presence of t[9;22], t[4;11], or another MLL rearrangement, near-haploidy [<34 chromosomes], hypodiploidy [35–40 chromosomes], acute undifferentiated leukaemia, minimal residual disease ≥10−2 at completion of induction [day 35], or failure to achieve complete remission or good partial response) found that prolonged treatment with native Escherichia coli asparaginase was unable to improve outcome for patients with T-cell ALL.17
Table 2:
Comparative characteristics of patients with B-cell ALL and T-cell ALL treated in Total 11, 12, 13A, 13B, 14, 15, and 16 Studies at St Jude Children’s Research Hospital, 1984–2017
| Number of patients (%) | p value | ||
|---|---|---|---|
| B-cell ALL (n=1302) | T-cell ALL (n=257) | ||
| Age, years | ·· | ·· | p<0·0001 |
| <1 | 27 (2%) | 3 (1%) | ·· |
| 1–9 | 964 (74%) | 142 (55%) | ·· |
| 10–15 | 235 (18%) | 87 (34%) | ·· |
| 16–18 | 76 (6%) | 25 (10%) | ·· |
| Median | 5·10 | 8·92 | ·· |
| Gender | ·· | ·· | p<0·0001 |
| Male | 703 (54%) | 184 (72%) | ·· |
| Female | 599 (46%) | 73 (28%) | ·· |
| Race | ·· | ·· | p<0·0001 |
| White | 1033 (79%) | 181 (70%) | ·· |
| Black | 182 (14%) | 65 (25%) | ·· |
| Others* | 87 (7%) | 11 (4%) | ·· |
| Leucocyte count, ×109/L | ·· | ·· | p<0·0001 |
| <10 | 641 (49%) | 40 (16%) | ·· |
| 10–24 | 274 (21%) | 38 (15%) | ·· |
| 25–49 | 144 (11%) | 28 (11%) | ·· |
| 50–99 | 133 (10%) | 43 (17%) | ·· |
| ≥100 | 110 (8%) | 108 (42%) | ·· |
| Median | 10·13 | 76·40 | ·· |
| Haemoglobin, g/dL | ·· | ·· | p<0·0001 |
| ≤10 | 1028 (79%) | 119 (46%) | ·· |
| >10 | 274 (21%) | 138 (54%) | ·· |
| Median | 7·7 | 10·5 | ·· |
| Platelet count, ×109/L | p=0·0054 | ||
| ≤100 | 903 (69%) | 155 (60%) | ·· |
| >100 | 399 (31%) | 102 (40%) | ·· |
| Median | 52 | 70 | ·· |
| Mediastinal mass | ·· | ·· | p<0·0001 |
| Absent | 1296 (100%)† | 180 (70%) | ·· |
| Present | 6 (<1%) | 77 (30%) | ·· |
| CNS status‡ | ·· | ·· | p<0·0001 |
| CNS1 | 856 (66%) | 127 (49%) | ·· |
| CNS2 | 342 (26%) | 90 (35%) | ·· |
| CNS3 | 29 (2%) | 17 (7%) | ·· |
| Traumatic lumbar puncture with blasts | 74 (6%) | 23 (9%) | ·· |
ALL=acute lymphoblastic leukaemia.
Asian, Hispanic, and mixed race.
100% due to rounding.
Information not available for one patient with B-cell ALL.
T-cell ALL appears to have a 2–3 times higher incidence in male patients than female patients (table 2), a finding in which the genetic causes are not fully understood. Inactivating somatic mutations and deletions of the X-linked PHF6 tumour suppressor gene have been identified in a cohort of 38% adult and 16% paediatric T-cell ALL samples, and occurred almost exclusively in male patients with T-cell ALL (found in 31·5% of male patients and 2·6% of female patients).18 Because male patients have a single copy of PHF6, loss of function mutation in this gene is much more likely to trigger T-cell ALL leukaemogenesis than in female patients, pointing to PHF6 as a plausibly haplosufficient tumour suppressor gene in this type of leukaemia. Mutational loss of PHF6 has been associated with T-cell ALL development, which is driven by aberrant expression of the homeobox transcription factor oncogenes TLX1 and TLX3.18 PHF6 mutations were not associated with NOTCH1, FBXW7 or PTEN mutations, nor were they associated with overall survival.18
Compared with B-cell ALL, T-cell ALL affects a higher proportion of black patients,6,19 who generally have poorer survival than that of white patients;6 however, with contemporary treatment options, the gap in overall 5-year survival rates has significantly decreased between the two races from 11·0% in 1990–94 to 3·3% in 2000–05, especially among those with T-cell ALL (from 5·0% to 0·02% in one study).6 Through genome-wide association studies (GWAS), 13 ALL risk loci have been identified for B-cell ALL in genes associated with haemapoietic development and tumour suppressor pathways.20 These known risk alleles have different effects on T-cell ALL or B-cell ALL susceptibility. For example, ARID5B risk variant is largely specific to B-cell ALL,21 whereas the CDKN2A/CDKN2B allele is associated with a comparable level of increase in the risk of developing B-cell ALL and T-cell ALL.22 Overall, the genetic basis of T-cell ALL predisposition remains poorly understood.
National Cancer Institute (NCI)/Rome criteria
In a workshop sponsored by the Cancer Therapy Evaluation Program of the US NCI, presenting age and leucocyte count were used to develop a risk stratification schema for B-cell ALL.23 Patients with B-cell ALL were classified into two risk groups: a standard-risk group, comprising two-thirds of patients aged between 1 year and 9 years and presenting a leucocyte count of less than 50 × 109 cells per L; and a high-risk group, comprising the remaining third of patients aged younger than 1 year, aged 10 years and older, or presenting with a leucocyte count ≥50 × 109 cells per L. This NCI criteria is not useful for T-cell ALL. Although high presenting leucocyte count appears to have little effect on event-free survival or overall survival in T-cell ALL, leucocytosis over 1 × 106 cells per μL and the presence of blasts in the cerebrospinal fluid at diagnosis are independent risk factors for CNS relapse in T-cell ALL.14,24
Immunophenotypic differences
Leukaemia develops from transforming events in early haemapoietic progenitors, with subsequent acquisition of multiple alterations in genes involved in survival, differentiation, and proliferation.25 Both normal B and T lymphocytes derive from haemapoietic stem cells in the bone marrow. B lymphocytes mature in the bone marrow through a series of developmental stages identifiable through expression of specific transcription factors and cell surface markers. T-lymphocyte development occurs as early lymphoid progenitors emigrate from the marrow to the thymus. Functional T cells undergo positive selection and autoreactive T cells are elimated (undergo negative selection). Most cases of B-cell ALL originate from clones that are developmentally arrested at the pre–pro-B cell or pro-B cell stages. Except for mature B-cell ALL, developmental stage lacks prognostic or therapeutic relevance.25 T-cell ALL arises from clones at various stages of intrathymic development. The European Group for the Immunological Classification of Leukemia divides T-cell ALL according to developmental stage into pro-T, pre-T or immature, cortical-T, and mature-T (appendix p 1).26 Although useful in some older studies, developmental stage in T-cell ALL also no longer has independent prognostic value with contemporary treatments based on minimum residual disease-based risk stratification.
Early T-cell precursor ALL and mixed-phenotype acute leukaemia
Early T-cell precursor ALL is a subtype of T-cell ALL defined by a specific immunophenotype, with expression of T-lineage and myeloid and early progenitor cell markers (appendix p 1), and represents 10–15% of all T-cell ALL cases.27 Early T-cell precursor ALL arises from a T-cell clone in early lineage development and does not meet the WHO definition of an acute leukaemia of ambiguous lineage.28,29 The genetic alterations found in early T-cell precursor ALL are distinct from those of non-early T-cell precursor ALL and are most similar to those of blasts from T-myeloid mixed-phenotype acute leukaemia.30 Most cases of early T-cell precursor ALL have alterations in transcriptional regulators (>80%), JAK and STAT signalling (>40%), and epigenetic regulation (>80%).30 Notch signalling and cell cycle alterations are less common in early T-cell precursor ALL than in non-early T-cell precursor ALL.
Early studies suggested early T-cell precursor ALL might signify a particularly poor prognosis with high minimum residual disease at the end of remission induction, and was associated with a less than 20% 5-year overall survival.27 Early T-cell precursor ALL tends to be inherently corticosteroid resistant, and thus has a markedly higher induction failure than non-early T-cell precursor ALL. In the Children’s Oncology Group AALL0434 clinical trial, M3 marrow (>25% blast by morphology) was found in 7·8% of patients with early T-cell precursor ALL but in only 1·1% of patients with non-early T-cell precursor ALL at the end of induction.31 Data published in 2016 from multiple groups suggested that early T-cell precursor ALL was associated with event-free survival and overall survival outcomes approaching those of non-early T-cell precursor ALL when the protocol contained more intensive cyclophosphamide therapy.31,32
Mixed-phenotype acute leukaemias, a collection of high-risk subtypes with myeloid and lymphoid features, can be broadly classified into B-myeloid and T-myeloid subtypes.28,29 ZNF384 rearrangement is common in the B-myeloid subtype, and biallelic WT1 alterations are common in the T-myeloid subtype, which shares genomic features with early T-cell precursor ALL, such as RAS and JAK–STAT pathway mutations.28 In a study of mixed-phenotype leukaemia, ALL-type therapy was superior to acute myeloid leukaemia-type therapy, which was preferable only for a minority of patients with CD19-negative leukaemia.29
Genetic differences
Based on leukaemia cell genetic abnormalities, ALL cases can be broadly classified into three categories: low, intermediate, and high risk, although the proportions of patients in each category can differ between T-cell and B-cell ALL (figure, table 3).33,34 Heterogeneity exists in treatment response among patients with the same genetic subtypes, partly due to cooperative mutations and germline genetic variants.20 Nevertheless, the genetic alterations can be combined with clinical variables and response to therapy to risk stratify patients with B-cell ALL into prognostic groups. Although genetic alterations can be used to classify T-cell ALL, their prognostic value is tenuous in the context of minimum residual disease-stratified therapy. Throughout, child or childhood will refer to patients <21 years of age, unless otherwise indicated.
Figure: Estimated frequencies of specific genetic subtypes of childhood ALL.
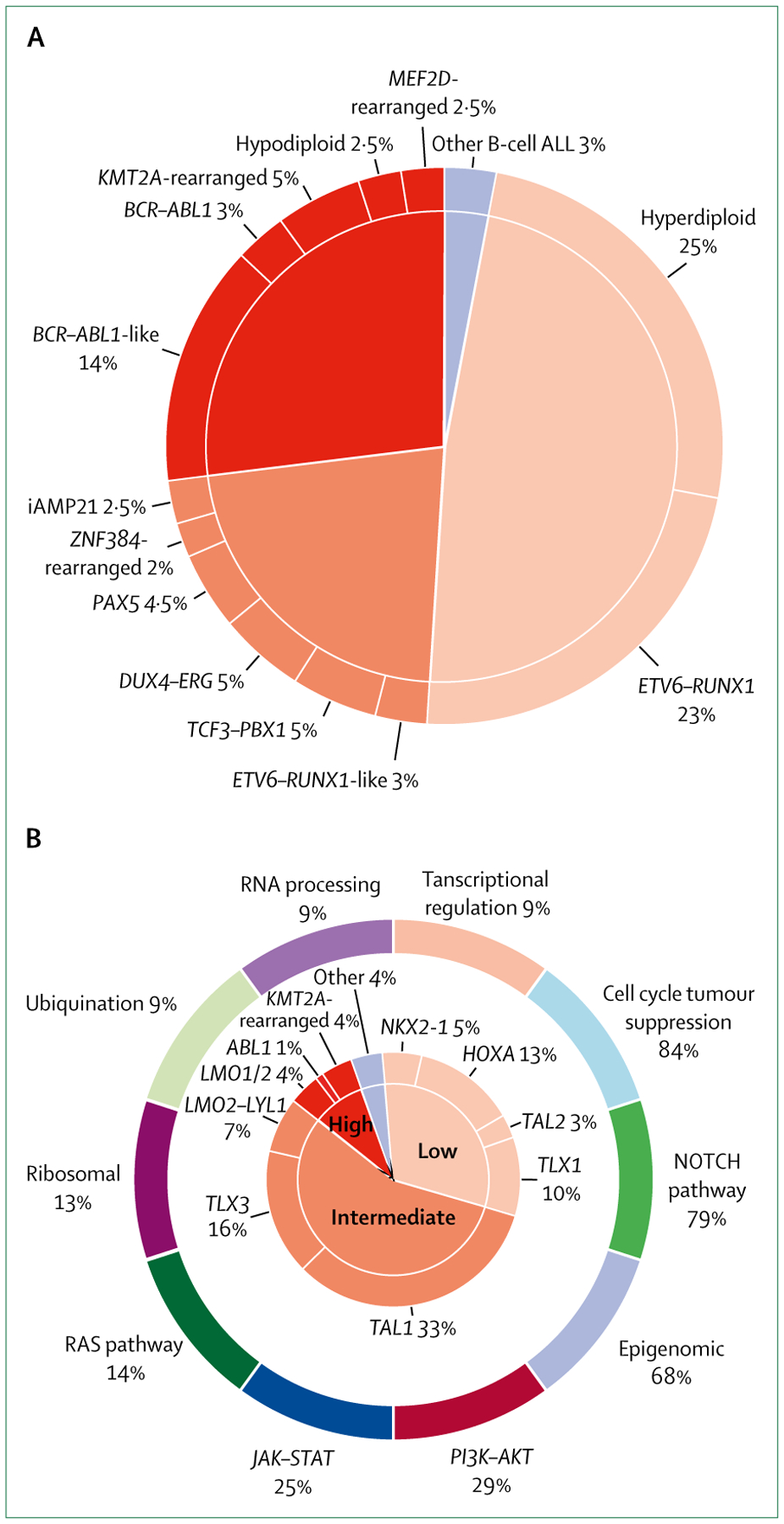
Pie charts represent patients with B-cell ALL treated in the St Jude Total Therapy study XV33 with modifications to account for discoveries of novel genetic abnormalities (A), and patients with T-cell ALL who were studied as part of the Therapeutically Applicable Research to Generate Effective Treatments initiative and treated in Children’s Oncology Group studies (B).34 T-cell ALL cases were divided according to dysregulation of targetable functional pathways (outer ring). Subtypes are grouped into low-risk, intermediate-risk, and high-risk categories on the basis of 5-year survival rates: over 90%, 70–90%, and less than 70% in B-cell ALL (overall survival) and T-cell ALL (event-free survival). The outcome (event-free survival) for patients treated in the AALL0434 trial,35 used for the T-cell ALL risk grouping in this figure, is superior to that in most other published studies. ALL=acute lymphoblastic leukaemia.
Table 3:
Prognostic indicators for patients with B-cell ALL and T-cell ALL
| Patients with B-cell ALL | Patients with T-cell ALL | |
|---|---|---|
| Clinical | ||
| Favourable | Age 1–10 years and leucocyte count <50 × 109/L, Caucasian or Asian, CNS1 disease status | CNS1 disease status |
| Intermediate | Age ≥10 years, leucocyte count 50–100 × 109/L, black race, CNS2 disease status, testicular leukaemia | CNS2 or CNS3 disease status, testicular leukaemia |
| Unfavourable | Age <1 year, leucocyte count >100 × 109/L, native American or Hispanic race, CNS3 disease status | Leucocyte count >200 × 109/L* |
| Biological | ||
| Favourable | ETV6-RUNX1-type, hyperdiploidy (51–67 chromosomes; DNA index ≥1·16) | NOTCH1 mutation, FBXW7 mutation |
| Intermediate | ETV6-RUNX1-like, TCF3-PBX1, DUX4-ERG, PAX5, ZNF384-rearranged, iAMP21 | Early T-cell precursor ALL |
| Unfavourable | BCR-ABL1-type, BCR-ABL1-like, KMT2A-rearranged, MEF2D-rearranged, hypodiploid (<44 chromosomes) | RAS mutation, PTEN mutation, lack of biallelic TRG rearrangements |
| Response to chemotherapy (minimal residual disease) | ||
| Favourable | Day 8 blood minimal residual disease <10−4, day 15 or day 19 bone marrow minimal residual disease <10−2, end of induction bone marrow minimal residual disease <10−4 | End of induction bone marrow minimal residual disease <10−4 |
| Intermediate | End of induction bone marrow minimal residual disease from 10−4 to 10−2 | End of induction bone marrow minimal residual disease ≥10−4 and end of consolidation minimal residual disease ≥10−4 and <10−3 |
| Unfavourable | End of induction bone marrow >1% blasts | End of consolidation bone marrow minimal residual disease ≥10−3 |
ALL=acute lymphoblastic leukaemia.
Not consistent.
B-cell ALL genetic alterations
ETV6–RUNX fusion (also known as TEL-AML1)
ETV6–RUNX1 is the most common genetic fusion in childhood B-cell ALL, occurring in 20–25% of patients.36 The translocation, t(12;21)(p13;q22), is cryptic cyto-genetically, but is identifiable by reverse transcription-PCR or florescence in-situ hybridisation (FISH). Studies evaluating cord blood, monozygotic twins, and neonatal blood spots of children with ALL show that ETV6–RUNX1 fusion is an initiating event that usually occurs in utero, leading to a preleukaemic state with a postnatal latency period of up to 14 years.37 ETV6–RUNX1 is not sufficient for leukaemogenesis and additional alterations of genes involved in the cell cycle and B-cell lineage differentiation, such as CDKN2A or PAX5, or chromosomal gains or losses, are needed for leukaemic transformation.37 Most studies show that ETV6–RUNX1 is an independent predictor of favourable outcome, which has led many cooperative groups to reduce treatment intensity in this subgroup of patients,38 with the exception of those with persistent minimum residual disease who have worse outcomes.39 By use of RNA sequencing, a new subtype of ETV6–RUNX1-like ALL was identified with the same gene-expression profile as that of ALL with ETV6–RUNX1 and coexisting ETV6 and IKZF1 alteration, but without the fusion transcript.40 These cases appeared to have a low-to-intermediate risk prognosis, with two cases of relapse among the ten cases studied.
Aneuploidy
Entire chromosome losses or gains (aneuploidy) are frequently identified in ALL blasts. High hyperdiploidy (51–67 chromosomes; DNA index ≥1·16) is very common in B-cell ALL, being identified in 20–30% of cases.33,36 Non-random gains in chromosomes 4, 10, 14, 17, and 21 are most common, and high hyperdiploidy is associated with a favourable prognosis. Similar to ETV6–RUNX1, initiating events often occur prenatally.41 The cause of high hyperdiploidy is poorly understood; however, GWAS have shown an association between 10q21.2 mutations and high hyperdiploidy.21 The 10q21 risk locus for high hyperdiploid ALL is mediated by the single nucleotide polymorphism rs7090445, which interacts with the ARID5B transcription start site and affects RUNX3 binding, leading to abnormal lymphocyte development and clonal expansion.42
Hypodiploid ALL (<44 chromosomes) is uncommon (2–3% of cases) and associated with inferior outcome compared with hyperdiploid ALL. Hypodiploid ALL is heterogeneous, comprising several subgroups of differing biology and prognosis.43 Near-haploid ALL (25–29 chromosomes) is characterised by genetic alterations affecting RAS and receptor tyrosine kinase signalling, and a high frequency of IKZF3 alterations.43 Low-hypodiploid cases (32–39 chromosomes) frequently have alterations in TP53, IKZF2, and RB1. TP53 alterations have been identified in 90% of patients with low hypodiploid ALL, with approximately 50% of these cases having germline TP53 alterations.43 Patients with germline TP53 alterations are at increased risk of relapse and secondary cancer development.44 Thus, all patients with low hypodiploid ALL should be tested for Li-Fraumeni Syndrome, a hereditary cancer predisposition syndrome. Patients with high-hypodiploid ALL with 44 chromosomes have been found to have better survival outcomes than those with 40–43 chromosomes, who in turn had better outcomes than those patients who had near haploidy or low haploidy.45 Data have shown that hypodiploid patients with negative minimum residual disease at the end of remission induction therapy can be treated with chemotherapy alone.45
BCR–ABL1 fusion gene (also known as the Philadelphia chromosome translocation) and Philadelphia chromosome-like ALL
Rearrangement of t(9;22)(q34;q11.2) resulting in the Philadelphia chromosome and BCR–ABL1 oncogene fusion occurs in approximately 3% of childhood ALL and approximately 25% of adult cases of ALL. Before the development of tyrosine kinase inhibitors (TKIs), Philadelphia chromosome-positive B-cell ALL was a marker of poor survival, associated with an overall survival of less than 50%.46 More recent trials combining the TKIs imatinib or dasatinib with multiagent chemotherapy have shown markedly improved outcomes, with 5-year overall survival approaching 75%.46 Alterations in IKZF1 are present in approximately 80% of Philadelphia chromosome-positive B-cell ALL cases and might be associated with chemotherapy resistance and inferior outcomes compared with Philadelphia chromosome-positive ALL without IKZF1 mutations, although more research is needed to confirm this association.47
Philadelphia chromosome-like ALL is characterised by an activated kinase gene expression profile resembling that of Philadelphia chromosome-positive ALL with a high frequency of IKZF1 alterations, but is BCR–ABL1-negative. Patients with Philadelphia chromosome-like ALL represent approximately 10% of those with childhood NCI standard risk, about 15% of childhood NCI high risk, about 20% of adolescent cases of ALL, and approximately 27% of young adult cases of ALL.48,49 Philadelphia chromosome-like ALL is heterogeneous and associated with a wide range of genetic alterations that are responsive to different tyrosine kinase or signal pathway inhibitors (appendix p 2). CRLF2 rearrangements leading to activation of PI3K/AKT/mTOR and JAK–STAT signalling are common in Philadelphia chromosome-like ALL, especially in older patients and in Native American and Hispanic or Latino populations.47 Approximately 50% of patients with CRLF2-rearranged Philadelphia chromosome-like ALL harbour JAK2 or JAK1 mutations.48,49 Among the patients without CRLF2 rearrangements, 15–20% have rearrangements in ABL1, ABL2, CSF1R, platelet-derived growth factor receptors (PDGFR) alpha and beta, and might respond to ABL-class TKIs. 10–15% of patients without CRLF2 rearrangements have lesions that activate JAK–STAT signalling, including JAK2 fusions or truncating rearrangements in erythropoietin receptor.48,49 Although most Philadelphia chromosome-like ALL cases require intensive treatment, those with negative minimum residual disease at the end of induction therapy have excellent outcomes (as defined in each citation), even when treated with low-intensity chemotherapy.48,49
TCF3–PBX1 and HLF fusion genes
TCF3 (formerly E2A) encodes an E protein that is a basic helix-loop-helix transcription factor involved in B-lymphocyte and T-lymphocyte development.36,50 For B-cell ALL, t(1;19)(q23;p13.3) with TCF3–PBX1 fusion occurs in 4% of children and t(17;19)(q22;p13.3) with TCF3–HLF fusion occurs in less than 1% of children.47 Once associated with poor prognosis, ALL with TCF3–PBX1 is now considered a low-to-intermediate risk genotype with contemporary minimum residual disease-stratified treatment. TCF3–HLF, by contrast, is associated with very poor survival outcomes.47
Other B-cell ALL genetic alterations
iAMP21 was first described in 2003, after detection of multiple copies of RUNX1 during testing by FISH for ETV6–RUNX1, and was associated with poor outcomes.51 iAMP21 occurs in approximately 2% of childhood B-cell ALL and arises from complex structural rearrangements of chromosome 21, including breakage–fusion–bridge cycles followed by chromothripsis. Contemporary intensified therapy has significantly improved its prognosis.52
DUX4 overexpression as a consequence of IGH–DUX4 fusions occurs in approximately 5% of patients with B-ALL.53 The rearrangements lead to loss of function of ERG, which can also be seen with ERG–DUX4 fusions. Despite a high frequency of IKZF1 deletions, IGH–DUX4 has favourable prognosis.40 MEF2D and ZNF384 rearrangements have been identified from unique gene expression signatures.54 Each appear to be represented in approximately 3% of childhood cases of B-cell ALL on the basis of initial reports, but more data are needed to establish actual prevalence. MEF2D rearrangements might result in inferior outcomes and ZNF384 rearrangements in intermediate outcomes.20 More recently, RNA sequencing has identified new subtypes, driven by the lymphoid transcriptional factor gene PAX5, which affected 4–5% of patients and appeared to relate to an intermediate prognosis.55
T-cell ALL genetic alterations
T-cell ALL is more genetically diverse than B-cell ALL, and no genetic alterations have been identified that reproducibly and independently predict outcome. In a comprehensive genomic analysis as part of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative, 106 putative driver genes were identified among 264 children and young adults with T-cell ALL.34
Grouping by transcription factor oncogenes
T-cell ALL can be divided into biological subgroups either by gene expression profiling or by mutated functional pathways (figure).34 T-cell ALL can be segregated by increased expression of various transcription factors, including TAL1, TLX1 (also known as HOX11), TLX3, LMO1, LMO2, MEF2C, and HOXA.56 The increased expression is often the result of a chromosomal rearrangement that juxtaposes the T-cell receptor and the proto-oncogenes that encode the transcription factors. Alternative mechanisms include translocations that rearrange the proto-oncogenes with other partners; mutations or insertions in non-coding regions that can activate the proto-oncogenes; or alterations such as duplications that lead to amplification of the transcription factors.57 TAL1 can be activated through multiple mechanisms, such as by mutations in the TAL1 structural loop that create superenhancers. Other transcription factor genes, including MYB, MED12, and MYCN, can be altered by rearrangements, mutations, or amplifications.
Grouping by dysregulation of targetable functional pathways
T-cell ALL can also be divided into subgroups on the basis of genomic alterations that lead to dysregulation of potentially targetable functional pathways (figure).34 Importantly, alterations in these pathways are not mutually exclusive, and T-cell ALL blasts commonly have genomic alterations leading to dysregulation of multiple pathways. A strong correlation has been identified between the type and frequency of genetic alterations, the developmental stage of T-cell ALL blasts, and different T-cell ALL subgroups. For example, NRAS and FLT3 mutations are associated with immature T-cell ALL, JAK3 and STAT5B mutations with HOXA1-deregulated ALL, PTPN2 mutations with TLX1-deregulated ALL, and PIK3R1 and PTEN mutations with TAL1-deregulated ALL.34
The most common signalling pathway abnormally activated in T-cell ALL is that mediated by the Notch1 pathway (70–80% of patients).34 Notch1 signalling is crucial in T-cell lineage commitment, activation, and proliferation, and can be dysregulated through multiple mechanisms. Activating mutations in NOTCH1 or loss of function of the negative regulator FBXW7 leading to aberrant activation of the NOTCH pathway occur in 65–70% of T-cell ALL cases, often co-occurring with loss of the CDKN2A locus, which encodes tumour suppressors p16INK4A and p14ARF.56 Notch1 can also be activated as a result of dysregulation of other pathways, including the c-myc and PI3K/AKT/mTOR pathways.
PI3K/AKT/mTOR signalling, which has important roles in cell death, metabolism, and proliferation, is abnormally activated in T-cell ALL in approximately 30% of patients, most commonly from loss of PTEN, resulting in AKT activation.31 PTEN loss can occur because of PTEN deletions or mutations. Additionally, PI3K/AKT/mTOR can be activated by mutations in PI3KCA, PIK3R1, IL7R, or AKT, or secondarily from activation of Notch, Ras, or JAK–STAT.31 Abnormal activation of JAK–STAT (approximately 25% of patients) and RAS (approximately 15% of patients) is also relatively frequent in T-cell ALL, most commonly in early T-cell precursor ALL.34 JAK–STAT can be activated through JAK pathway gene fusions such as TEL–JAK2, through mutations in IL7R, JAK1, JAK3, STAT5, or through deletions in PTPN2.31 RAS can be activated through mutations in NRAS, FLT3, B-Raf proto-oncogene, serine/threonine kinase (BRAF), and KRAS.31 Both JAK–STAT and RAS can also be indirectly activated through dysregulation of other signalling pathways including PI3K/AKT/mTOR.
In addition to signalling pathway dysregulation, recurrent alterations in genes regulating cell cycle progression or tumour suppression, including in CDKN2A, CDKN2B, cyclin D3 (CCND3), and RB1, are common (>80% of patients).34 Epigenetic regulators, including CREB binding protein (CREBBP) and PHF6, are commonly mutated in T-cell ALL (>60% of patients), especially in early T-cell precursor ALL and the LMO2/LYL1 and TLX3 subgroups.34
Other genetic abnormalities in T-cell ALL
A number of rare fusions including NUP214-ABL1, SET nuclear proto-oncogene-NUP214, STMN1-SP1, and EML-ABL1 can be seen in T-cell ALL.34 Absence of ABD is prevalent in patients with T-cell ALL who do not respond to induction therapy.56 As ABD is characteristic of early thymocyte precursors in normal lymphocyte development, there might be overlap between ABD and early T-cell precursor ALL.
Genetic abnormalities seen in B-cell ALL and T-cell ALL
Although the majority of genetic lesions in B-cell ALL and T-cell ALL are mutually exclusive, some lesions can be found in both, including KMT2A rearrangement and BCR–ABL1. Somatic translocations involving KMT2A (formerly MLL) on 11q23 occur in 2–5% of childhood cases of ALL and approximately 75% of infant patients with ALL.47,58 KMT2A encodes a methyltransferase that is important in haemapoietic stem cell development. KMT2A rearrangements are strong drivers of leukaemia development, and require very few cooperating genetic alterations to induce leukaemia formation.58 Infants with KMT2A rearrangement have a particularly poor prognosis, especially those younger than 90 days old at diagnosis.58 The prognostic significance of KMT2A rearrangement in children aged 1 year or older with B-cell ALL varies according to the fusion partner, and more than 100 different partners have been reported.47,58 Older children (≥1 year) with KMT2A rearrangement have more somatic mutations (mean of 6·5 mutations per patient vs 1·3 mutations per patient in infants), have frequent mutations in epigenetic regulators, and have an intermediate prognosis.47,58 KMT2A rearrangement also occurs in 5–10% of T-cell ALL cases. Similar to B-cell ALL, prognosis varies based on fusion partners.36 As described earlier, BCR–ABL1 fusions are common in B-cell ALL, and BCR–ABL1 is considerably more common than other ABL1 fusions. In T-cell ALL, ABL1 can form fusions with BCR, NUP214, and EML1;34 however, these fusions are not common.
Differences in biology and prognosis
A number of biological factors account for the slightly inferior outcome of T-cell ALL than that of B-cell ALL. First, the proportion of favourable (low-risk) genetic subtypes is higher in B-cell ALL than in T-cell ALL (figure). Second, many of the specific genetic subtypes of B-cell ALL have clear prognostic and therapeutic implications and benefit from the availability of risk-stratified therapy and targeted therapeutics. With contemporary risk-stratified treatment, approximately 50% of patients with B-cell ALL have low-risk genetic subtypes, ETV6–RUNX1 and high hyperdiploidy, with 5-year overall survival of more than 90%;7,14 approximately 30% of patients have high-risk ALL, such as Philadelphia chromosome-positive, Philadelphia chromosome-like, hypodiploid, KMT2A-rearranged ALL, and MEF2D-rearranged ALL, with 5-year overall survival of less than 70%;20,45,49 and the remaining patients with intermediate-risk ALL have 5-year overall survival between 70% and 90%.20,55,59 Risk-adapted treatment allows intensification of therapy for unfavourable biological lesions and reduction in therapy intensity for favourable biological lesions. In this regard, intensification of therapy in patients with iAMP21,52 and the addition of ABL-class TKIs for all Philadelphia chromosome-positive B-ALL and approximately 10–15% of patients with Philadelphia chromosome-like ALL with ABL-class fusion transcripts, have significantly improved outcome.46,47,49
In contrast to B-cell ALL, no consensus genetic classification with prognostic or therapeutic implications has been reached for T-cell ALL, as most genetic alterations in T-cell ALL are not absolutely predictive of treatment outcome.36 The majority of relapses among patients with T-cell ALL actually occur in those classified to have favourable (low-risk) leukaemia based on minimum residual disease, and the prognosis for relapsed T-cell ALL is poor.36 Moreover, the interpretation of genetic testing is not simple, and the apparent prognostic implications of many alterations vary between studies. For example, activation of the Notch1 pathway by NOTCH1 or FBXW7 mutations has been associated with a favourable prognosis in some studies, whereas alterations in PTEN, partly through its modulatory effect of NOTCH1, have been associated with an unfavourable prognosis.60–63 In the UKALL 2003 trial,62 for example, neither PTEN nor K-RAS or N-RAS genotype significantly influenced the clinical outcome, whereas in the French Acute Lymphoblastic Leukemia (FRALLE) 2000 T trial,64 both PTEN and K-RAS or N-RAS mutations were associated with poor outcome. Additional studies are required to clarify these discordant results.
The TARGET study34 identified several putative prognostic genomic alterations that required validation. In a univariate analysis, a number of lesions were associated with increased risk of relapse, including alterations in AKT1, MLLT10, PTEN, and CNOT3. Nevertheless, because of the small number of patients who relapsed in this study (n=20), the analysis was not powered to determine if any alterations could predict event-free survival or overall survival.
Unlike B-cell ALL, precision medicine approaches for T-cell ALL are lagging behind. As the biology of T-cell ALL is now better understood, future trials in T-cell ALL should be able to rationally and selectively incorporate new drugs into their treatment design. Notably, both JAK and ABL1 kinases are often activated in T-cell ALL, and might be amendable through treatment with JAK and ABL1 inhibitors.56
Differences in treatment responses
Minimum residual disease measurement accounts for leukaemic cell genetics and microenvironment, host pharmacogenetics, and treatment efficacy. Minimal residual disease can be measured by different techniques that can quantify leukaemic blasts, including flow cytometry, PCR, and next-generation sequencing. It is usually calculated as the number of leukaemic blasts relative to the number of nucleated cells.36 As such, minimum residual disease is considered the most important independent prognostic indicator for ALL,15 and the effect of its assessment has been consistent across treatment regimens, methods and timings of minimum residual disease assessment, various cutoff values, and immunophenotypic or genotypic subtypes of leukaemia.65 For example, although IKZF1 deletion has been associated with poor outcomes in many studies of B-cell ALL, two studies have shown that patients with IKZF1 deletion and negativity for minimum residual disease (≤10−4) at the end of induction therapy actually have highly favourable outcomes, even when treated with low-intensity treatment for standard-risk ALL.66–68
In general, minimum residual disease measurement at two timepoints should be used in concert with information on genetic abnormalities to improve its predictive accuracy and value.59 Early measurement (table 3) can identify patients with highly curable and drug-sensitive leukaemia, who exhibit negativity for minimum residual disease after treatment with only three or four chemotherapeutic drugs.59 Patients with favourable genetic subtypes of B-cell ALL (eg, ETV6–RUNX1-type, high hyperdiploidy) who exhibit negativity for minimum residual disease at a very early timepoint (eg, day 8 or 15 of remission induction therapy), especially when confirmed by a highly sensitive minimum residual disease assay, such as next-generation sequencing, might benefit from reduction of treatment intensity.68 By contrast, high-level minimum residual disease (eg, 10−2) at the end of induction therapy or persistent leukaemia after consolidation therapy (after exposure to seven or more drugs) generally predicts a poor prognosis.68 In this regard, minimum residual disease is particularly useful for identifying patients with T-cell ALL with persistent minimum residual disease after remission induction and early intensification therapies who might benefit from more intensive chemotherapy or allogeneic transplantation.59,68
With recent improvements in treatment and risk stratification, subgroups of patients with T-cell ALL might now be predicted to have better survival outcomes.64,69 For example, 30% of patients with T-cell ALL with NOTCH1 or FBXW7 mutations and germline RAS or PTEN mutations, a leucocyte count less than 200 × 109/L at diagnosis, and minimum residual disease less than 10−4 at the end of induction who were treated on the FRALLE 2000 T protocol had a remarkable 5-year disease-free survival of 98·3%.64 In the Associazione Italiana di Ematologia Pediatrica Group (AIEOP)–Berlin-Frankfurt-Munster (BFM) ALL 2000 study,5 patients with T-cell ALL who were negative for minimum residual disease (≤10−4) at day 78, irrespective of minimum residual disease status at day 33, had similar cumulative risk of relapse as those who achieved negativity for minimum residual disease at day 33 (8·5% vs 7·5% relapse), whereas those positive for minimum residual disease at day 78 had a high cumulative risk of relapse that increased with higher minimum residual disease values: 26·3% for less than 10−3, 33·0% for 10−3, and 44·7% for more than 10−3. One explanation is that the minimum residual disease status at day 78 reflects the response to induction consolidation protocol 1B, comprising two courses of cyclophosphamide, cytarabine, and mercaptopurine, a combination that is particularly important for T-cell ALL. Despite high minimum residual disease values at the early timepoint, patients with early T-cell precursor ALL had intermediate-to-favourable outcomes with treatment regimens containing protocol 1B.5,31,32 Similarly, patients with high hyperdiploid B-cell ALL who had poor response to remission induction might be cured with chemotherapy featuring consolidation treatment with high-dose methotrexate and mercaptopurine, a combination particularly effective for this genotype.70 To this end, one study found that patients who had decreasing minimum residual disease between the endpoint of induction therapy and week 17 of continuation treatment (25 weeks after the completion of remission induction) could be treated with chemotherapy alone.15
Differential sensitivity to cytotoxic chemotherapy
Previously, most cooperative groups treated de novo B-cell ALL and T-cell ALL for 2–3 years with the same chemotherapy backbone. With time it became apparent that B-cell ALL and T-cell ALL blasts had different sensitivities to many conventional cytotoxic chemotherapeutics, including hydrocortisone, daunorubicin, asparaginase, cytarabine, and methotexate.71 Although some study groups use B-cell ALL-focused and T-cell ALL-focused treatment approaches, the same 12–15 cytotoxic drugs are typically used for both, and the primary differences are in the dosing and scheduling of the drugs. All of the differences in therapies for B-cell ALL and T-cell ALL are beyond the scope of this Review; however, we will highlight a few examples.
A number of cooperative group trials have compared dexamethasone with prednisone in B-cell ALL and T-cell ALL. Patients in the AIEOP-BFM ALL 2000 trial were randomly assigned to receive dexamethasone (10 mg/m2 per day) or prednisone (60 mg/m2 per day) for 3 weeks (plus tapering of doses over 9 days) during induction therapy after completing a 7-day prednisone prophase.72 Dexamethasone treatment resulted in a lower overall 5-year cumulative risk of relapse (10·8% vs 15·6% for prednisone), with the largest effect observed in reducing extramedullary relapse, but also resulted in higher treatment-related mortality (2·5%) than did prednisone treatment (0·9%). Patients with T-cell ALL with a good response to prednisone (defined as <1·0 × 109/L blasts in blood samples after a 7-day prednisone prophase and one intrathecal methotrexate) had increased event-free survival and overall survival in the dexamethasone treatment group. By contrast, patients with B-cell ALL with a good response to prednisone had superior event-free survival in the dexamethasone treatment group but similar overall survival in both groups, as the ability to salvage patients after relapse was lower in the dexamethasone treatment group. Overall and event-free survival for patients with T-cell ALL or B-cell ALL who had poor response to prednisone prophase was the same in both groups. Investigators concluded that dexamethasone treatment benefited patients with T-cell ALL who had a good response to prednisone. Other cooperative group trials, such as Medical Research Council-ALL97,73 Japanese L95–14,74 Dana-Farber Cancer Institute 91–01P,75 and the Children’s Cancer Group-192276 trials also found higher (albeit non-significant in some of the trials) treatment-related mortality with dexamethasone than with prednisone. Some patients with B-cell ALL, however, can benefit from dexamethasone treatment despite the associated toxicity. In the COG AALL0232 trial,77 comparing dexamethasone treatment for 14 days against prednisone treatment for 28 days during induction therapy for high-risk B-cell ALL, dexamethasone treatment improved outcome in children younger than 10 years of age but was associated with higher risk of osteonecrosis and no survival benefit in older patients. Because of the differential effects of the two drugs in terms of efficacy and toxicity for patients with B-cell ALL or T-cell ALL in different age groups, no consensus is available on how to use these two types of steroid treatment.
A number of cooperative groups have previously treated and continue to treat patients with lower risk ALL with less intensive therapy. The Children’s Oncology Group and Medical Research Council often treat patients with lower risk ALL with a three-drug induction therapy regimen followed by a 4-week oral mercaptopurine-based consolidation, and patients at higher risk with a four-drug induction and more intensive BFM-style consolidation.11 Although this approach has been successful for B-cell ALL, patients with lower risk T-cell ALL do not do as well with the less intensive approach. In the UKALL 2003 trial,11 rapid responder patients with T-cell ALL with NCI high-risk features treated with a four-drug induction and BFM-style consolidation had superior outcomes than those with NCI standard-risk features who were treated with a three-drug induction and oral mercaptopurine-based consolidation.
Methotrexate and asparaginase are both effective drugs in ALL and are used in different schedules and intensities on different backbones. The Dana Faber Cancer Institute ALL Consortium has treated patients with asparaginase-intensive regimens for more than 20 years, showing outcomes in T-cell ALL that are similar to, if not better than, B-cell ALL.8 Additional studies from the Medical Research Council, Pediatric Oncology Group, and St Jude Children’s Research have also shown that additional asparaginase might improve outcomes in T-cell ALL.78 The Children’s Oncology Group has completed two phase 3 clinical trials (AALL0232 and AALL0434), which randomly assigned patients to receive high-dose methotrexate or Capizzi-style escalating methotrexate with pegaspargase.69,77 In AALL0232, high-dose methotrexate was found to have superior efficacy in patients with high-risk B-cell ALL.77 By contrast, in AALL0434, Capizzi-style treatment was found to have superior efficacy in patients with T-cell ALL.69 Reductions in CNS disease and haematological relapses were seen in both trials in the superior group. Arguably, the increased sensitivity to pegaspargase in T-cell ALL compared with B-cell ALL might explain the different results. Of note, in AALL0434, more than 90% of patients with T-cell ALL received prophylactic cranial irradiation. The Capizzi-style group received prophylactic cranial irradiation approximately 5 months earlier in the treatment regimen than did the high-dose methotrexate group, which could have contributed to the improved event-free survival in this group. Regardless, multiple studies, including a comprehensive meta-analysis of 16 623 patients from ten international cooperative groups, have suggested that prophylactic cranial irradiation does not improve survival in patients with T-cell ALL receiving intensified CNS-directed and systemic chemotherapy.79
Availability of novel drugs for relapsed and high-risk disease
Over the past 5 years, a shift has occurred in the treatment of patients with relapsed and refractory B-cell ALL, and a number of targeted therapies and immunotherapies have shown remarkable efficacy, and improved overall survival for this disease. Unfortunately, neither targeted therapies nor immunotherapies have been successful in the treatment of T-cell ALL.
The use of TKIs, including imatinib and dasatinib, has markedly improved survival for patients with Philadelphia chromosome-positive B-cell ALL.46,47 Children with Philadelphia chromosome-like B-cell ALL might also benefit from targeted therapy. Preclinical studies have shown promising results in treating Philadelphia chromosome-like ALL with targeted inhibitors, including TKIs and JAK–STAT inhibitors, leading a number of early-phase clinical trials to integrate targeted drugs into chemotherapy backbones.47,48 Anecdotal case reports have shown impressive activity of targeted therapy in refractory patients, including in the use of imatinib or dasatinib for PDGFRB-rearranged Philadelphia chromosome-like ALL.47,48 For both Philadelphia chromosome-positive and Philadelphia chromosome-like ALL, clinical grade testing in the USA has made it easier to identify these patients and allocate them to precision medicine approaches. In the Children’s Oncology Group and St Jude Children’s Research Hospital trials, patients with B-cell ALL with Philadelphia chromosome-like ALL and lesions that are targetable (appendix p 2) are eligible to participate in trials integrating dasatinib for patients with ABL-class fusion (NCT02883049), or integrating ruxolitinib for those with JAK–STAT involvement (NCT02723994), or both (NCT03117751).
Defining subgroups of T-cell ALL that might benefit from a precision medicine approach has been difficult because it is more biologically heterogeneous than B-cell ALL. One subset of T-cell ALL that might benefit from targeted therapy is early T-cell precursor ALL, which frequently has dysregulated JAK–STAT signalling, and has responded well to JAK–STAT inhibitors in preclinical studies.31 Based on these promising results, St Jude Children’s Research Hospital is integrating ruxolitinib into front-line therapy for patients with early T-cell precursor ALL (NCT03117751). Additionally, anecdotal reports have suggested efficacy of TKIs in T-cell ALL with ABL-class fusion in preclinical and clinical studies.80 Nelarabine is an antimetabolite that has shown particular activity in T-cell ALL in preclinical studies. The Children’s Oncology Group trial AALL0434 randomly assigned patients with intermediate and high-risk T-cell ALL to be treated with or without six 5-day courses of nelarabine. The 4-year disease-free survival was significantly improved for those randomly assigned to receive nelarabine.35 Follow-up of this study is needed to establish if nelarabine improves CNS disease control in T-cell ALL.
Certain types of immunotherapy or targeted therapy have shown efficacy in patients with B-cell ALL: inotuzumab ozogamicin, an anti-CD22 antibody conjugated to a calicheamicin-class cytotoxic drug; blinatumomab, a bispecific T-cell engager that links CD3-positive T cells with CD19-positive B-lineage blast cells; and a number of treatments based around CD19-targeted chimeric antigen receptor (CAR)-modified T cells, including tisagenlecleucel and axicabtagene ciloleucel.81 These drugs have been quickly incorporated into the front-line trials of B-cell ALL. Unfortunately, translating immunotherapies into a T-cell ALL setting has considerable problems, including the theoretical risk of so-called fratricide by T-cell-targeted clones and the risk of severe immunodeficiency from elimination of normal T lymphocytes. Despite these issues, preclinical studies of T-cell ALL have indicated efficacy with the anti-CD38 monoclonal antibody daratumumab and with CAR-T cells targeting a number of antigens, including CD2, CD5, and CD7.82,83 Early-phase trials are currently ongoing (NCT03081910 and NCT03384654). Table 4 summarises novel drugs and targets of interest for B-cell ALL and T-cell ALL.
Table 4:
Novel therapeutic approaches for patients with B-cell ALL and T-cell ALL
| Examples of available drugs | Potential use | |
|---|---|---|
| IL7-JAK-STAT-cytokine receptor-like factor 2 | ||
| JAK inhibitors | Ruxolitinib, tofacitinib, and peficitinib | Early T-cell precursor and Ph-like |
| STAT inhibitors | Pimozide | Early T-cell precursor and Ph-like |
| PI3K/AKT/mTOR | ||
| PI3K inhibitors | Idelalisib | B-cell ALL and T-cell ALL |
| mTOR inhibitors | Sirolimus, everolimus, and temsirolimus | B-cell ALL and T-cell ALL |
| AKT inhibitors | MK-2206, ipatasertib, and afuresertib | B-cell ALL and T-cell ALL |
| mTOR complex 1 or 2 inhibitors | Sapanisertib and vistusertib | B-cell ALL and T-cell ALL |
| PI3K or mTOR dual inhibitors | Dactolisib and gedatolisib | B-cell ALL and T-cell ALL |
| Cell cycle regulation | ||
| CDK4 or CDK6 inhibitors | Ribociclib and palbociclib | B-cell ALL and T-cell ALL |
| Pan-CDK inhibitors | Roniciclib | B-cell ALL and T-cell ALL |
| Proteasome | ||
| Proteasome inhibitors | Bortezomib, ixazomib, and carfilzomib | B-cell ALL and T-cell ALL |
| Neddylation inhibitors | Pevonedistat | B-cell ALL and T-cell ALL |
| Deubiquitinating enzyme inhibitors | In development, preclinical only | B-cell ALL and T-cell ALL |
| E3 ubiquitin ligase inhibitors | In development, preclinical only | B-cell ALL and T-cell ALL |
| MAPK-RAS | ||
| MAPK kinase inhibitors | Trametinib, selumetinib, and cobimetinib | B-cell ALL and T-cell ALL |
| Farynesyltranferase inhibitors | Tipifarnib | B-cell ALL and T-cell ALL |
| Notch receptors | ||
| γ-secretase inhibitors | Crenigacestat and BMS906024 | T-cell ALL |
| Soluble notch proteins | In development, preclinical only | T-cell ALL |
| Mastermind inhibiting peptides | In development, preclinical only | T-cell ALL |
| Apoptotic machinery | ||
| BCL2 inhibitors | Venetoclax | B-cell ALL and T-cell ALL |
| BCL2-like 1 and BCL2 inhibitors | Navitoclax | B-cell ALL and T-cell ALL |
| Epigenetic | ||
| Demethylating agents | Decitabine and 5-azacitidine | B-cell ALL and T-cell ALL |
| Histone deacetylase inhibitors | Romidepsin and vorinostat | B-cell ALL and T-cell ALL |
| Bromodomain containing 4 inhibitors | JQ1, birabresib, and CPI203 | B-cell ALL and T-cell ALL |
| DOT1-like histone lysine methyltransferase inhibitors | Pinometostat | B-cell ALL and T-cell ALL |
| Isocitrate dehydrogenase 1 and 2 inhibitors | AG-120 | B-cell ALL and T-cell ALL |
| Tyrosine kinase inhibitors | ||
| ABL-class inhibitors | Dasatinib, imatinib, and nilotinib | ABL1-translocated B-cell ALL |
| Bruton tyrosine kinase inhibitors | Ibrutinib | ABL1-translocated B-cell ALL |
| Newer cytotoxics | ||
| Antimetabolites | Nelarabine | T-cell ALL |
| Alkylators | Bendamustine | B-cell ALL and T-cell ALL |
| Folate analogue | Pralatrexate | B-cell ALL and T-cell ALL |
| Vincas | Liposomal vincristine | B-cell ALL and T-cell ALL |
| Monoclonal antibodies | ||
| CD20 | Rituximab | B-cell ALL |
| CD22 | Inotuzumab and epratuzumab | B-cell ALL |
| CD25 | Basiliximab | T-cell ALL |
| CD38 | Daratumumab and isatuximab | B-cell ALL and T-cell ALL |
| CD52 | Alemtuzumab | B-cell ALL and T-cell ALL |
| Bispecific T-cell engagers | ||
| CD19 | Blinatumomab | B-cell ALL |
| Chimeric antigen receptors | ||
| CD2, CD5, and CD7 | Preclinical and early-phase trials | T-cell ALL |
| CD19 | Tisagenlecleucel and axicabtagene | B-cell ALL |
| CD22 | Early-phase trials | B-cell ALL |
| CD38 | Early-phase trials | T-cell ALL |
| Dual targeting or multiple antigens | Early-phase trials | B-cell ALL |
ALL=acute lymphoblastic leukaemia. Ph=Philadelphia chromosome positive.
Conclusions
Although patients with T-cell ALL have historically fared worse than those with B-cell ALL, the difference in outcome has narrowed with modern chemotherapy approaches. Improved understanding of ALL biology and the integration of novel therapies are continuing to influence the field and will hopefully continue to improve survival outcomes for both T-cell and B-cell ALL. For T-cell ALL in particular, better methods are needed to identify patients at diagnosis who are likely to relapse, and those who might benefit from individualised therapy.
Supplementary Material
Search strategy and selection criteria
We searched MEDLINE and PubMed for articles published in English from Jan 1, 2008, to Dec 1, 2018, using the search terms “pediatric T-cell acute lymphoblastic leukemia” and “pediatric B-cell acute lymphoblastic leukemia”. Because of the limitation of available references, we preferentially cite review articles.
Acknowledgments
This work was supported by US Department of Health & Human Services, NCI American Lebanese Syrian Associated Charities, and the Leukemia and Lymphoma Society.
Declaration of interests
C-HP reports grants from the National Cancer Institute (NCI; CA021765, P50GM115279, and R01CA036401), and support for patient care and research at his institution from the American Lebanese Syrian Associated Charities, during the writing of this Review. C-HP is on the advisory board of Adaptive Biotechnologies and has received honoraria from Amgen Biotechnology company for giving lectures. DTT reports a grant from the NCI (R01193776), a grant from The Leukemia and Lymphoma Society (MCG 6561-18), and a grant from The American Cancer Society (RSG-14-022-01-CDD), during the conduct of the study. DTT has served on advisory boards for Amgen Biotechnology and La Roche, outside of the submitted work. C-HP is a professor and DTT is a research scholar of the American Cancer Society.
Contributor Information
David T Teachey, Hematology and Oncology, Children’s Hospital of Philadelphia, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Ching-Hon Pui, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin 2010; 64: 83–103. [DOI] [PubMed] [Google Scholar]
- 2.Reiter A, Schrappe M, Ludwig WD, et al. Chemotherapy in 998 unselected childhood acute lymphoblastic leukemia patients. Results and conclusions of the multicenter trial ALL-BFM 86. Blood 1994; 84: 3122–33. [PubMed] [Google Scholar]
- 3.Schorin MA, Blattner S, Gelber RD, et al. Treatment of childhood acute lymphoblastic leukemia: results of Dana-Farber Cancer Institute/Children’s Hospital Acute Lymphoblastic Leukemia Consortium Protocol 85–01. J Clin Oncol 1994; 12: 740–47. [DOI] [PubMed] [Google Scholar]
- 4.Conter V, Bartram CR, Valsecchi MG, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 2010; 115: 3206–14. [DOI] [PubMed] [Google Scholar]
- 5.Schrappe M, Valsecchi MG, Bartram CR, et al. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood 2011; 118: 2077–84. [DOI] [PubMed] [Google Scholar]
- 6.Hunger SP, Lu X, Devidas M, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the Children’s Oncology Group. J Clin Oncol 2012; 30: 1663–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pieters R, de Groot-Kruseman H, Van der Velden V, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol 2016; 34: 2591–601. [DOI] [PubMed] [Google Scholar]
- 8.Place AE, Stevenson KE, Vrooman LM, et al. Intravenous pegylated asparaginase versus intramuscular native Escherichia coli l-asparaginase in newly diagnosed childhood acute lymphoblastic leukaemia (DFCI 05–001): a randomised, open-label phase 3 trial. Lancet Oncol 2015; 16: 1677–90. [DOI] [PubMed] [Google Scholar]
- 9.Domenech C, Suciu S, De Moerloose B, et al. Dexamethasone (6 mg/m2/day) and prednisolone (60 mg/m2/day) were equally effective as induction therapy for childhood acute lymphoblastic leukemia in the EORTC CLG 58951 randomized trial. Haematologica 2014; 99: 1220–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vora A, Goulden N, Mitchell C, et al. Augmented post-remission therapy for a minimal residual disease-defined high-risk subgroup of children and young people with clinical standard-risk and intermediate-risk acute lymphoblastic leukaemia (UKALL 2003): a randomised controlled trial. Lancet Oncol 2014; 15: 809–18. [DOI] [PubMed] [Google Scholar]
- 11.Patrick K, Wade R, Goulden N, et al. Improved outcome for children and young people with T-acute lymphoblastic leukaemia: results of the UKALL 2003 trial. Blood 2014; 124: 3702. [Google Scholar]
- 12.Yeoh AE, Ariffin H, Chai EL, et al. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: results from the Malaysia-Singapore acute lymphoblastic leukemia 2003 study. J Clin Oncol 2012; 30: 2384–92. [DOI] [PubMed] [Google Scholar]
- 13.Toft N, Birgens H, Abrahamsson J, et al. Results of NOPHO ALL2008 treatment for patients aged 1–45 years with acute lymphoblastic leukemia. Leukemia 2018; 32: 606–15. [DOI] [PubMed] [Google Scholar]
- 14.Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009; 360: 2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pui C-H, Pei D, Coustan-Smith E, et al. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol 2015; 16: 465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawedia JD, Liu C, Pei D, et al. Dexamethasone exposure and asparaginase antibodies affect relapse risk in acute lymphoblastic leukemia. Blood 2012; 119: 1658–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondelaers V, Suciu S, De Moerloose B, et al. Prolonged versus standard native E. coli asparaginase therapy in childhood acute lymphoblastic leukemia and non-Hodgkin lymphoma: final results of the EORTC-CLG randomized phase III trial 58951. Haematologica 2017; 102: 1727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Vlierberghe P, Palomero T, Khiabanian H, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 2010; 42: 338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA 2003; 290: 2001–07. [DOI] [PubMed] [Google Scholar]
- 20.Pui CH, Nichols KE, Yang JJ. Somatic and germline genomics in pediatric acute lymphoblastic leukemia. Nat Rev Clin Oncol 2018; published online Dec 13. DOI: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 21.Trevino LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 2009; 41: 1001–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherborne AL, Hosking FJ, Prasad RB, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet 2010; 42: 492–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith M, Arthur D, Camitta B, et al. Uniform approach to risk classification and treatment assignment for children with acute lymphoblastic leukemia. J Clin Oncol 1996; 14: 18–24. [DOI] [PubMed] [Google Scholar]
- 24.Pui C-H, Howard SC. Current management and challenges of malignant disease in the CNS in paediatric leukaemia. Lancet Oncol 2008; 9: 257–68. [DOI] [PubMed] [Google Scholar]
- 25.Campos-Sanchez E, Toboso-Navasa A, Romero-Camarero I, Barajas-Diego M, Sanchez-Garcia I, Cobaleda C. Acute lymphoblastic leukemia and developmental biology: a crucial interrelationship. Cell Cycle 2011; 10: 3473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia 1995; 9: 1783–86. [PubMed] [Google Scholar]
- 27.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009; 10: 147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander TB, Gu Z, Iacobucci I, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018; 562: 373–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hrusak O, de Haas V, Stancikova J, et al. International cooperative study identifies treatment strategy in childhood ambiguous lineage leukemia. Blood 2018; 132: 264–76. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012; 481: 157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raetz E, Teachey D. T-cell acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program 2016; 1: 580–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conter V, Valsecchi MG, Buldini B, et al. Early T-cell precursor acute lymphoblastic leukaemia in children treated in AIEOP centres with AIEOP-BFM protocols: a retrospective analysis. Lancet Haematol 2016; 3: e80–86. [DOI] [PubMed] [Google Scholar]
- 33.Pui CH, Yang JJ, Hunger SP, et al. Childhood acute lymphoblastic leukemia: progress through collaboration. J Clin Oncol 2015; 33: 2938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017; 49: 1211–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunsmore KP, Winter S, Devidas M, et al. COG AALL0434: a randomized trial testing nelarabine in newly diagnosed t-cell malignancy. J Clin Oncol 2018; 36 (suppl 15): 10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teachey DT, Hunger SP. Predicting relapse risk in childhood acute lymphoblastic leukaemia. Br J Haematol 2013; 162: 606–20. [DOI] [PubMed] [Google Scholar]
- 37.Greaves M A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer 2018; 18: 471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison CJ, Haas O, Harbott J, et al. Detection of prognostically relevant genetic abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: recommendations from the Biology and Diagnosis Committee of the International Berlin-Frankfurt-Munster study group. Br J Haematol 2010; 151: 132–42. [DOI] [PubMed] [Google Scholar]
- 39.Borowitz MJ, Devidas M, Hunger SP, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood 2008; 111: 5477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun 2016; 7: 11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panzer-Grumayer ER, Fasching K, Panzer S, et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood 2002; 100: 347–49. [DOI] [PubMed] [Google Scholar]
- 42.Studd JB, Vijayakrishnan J, Yang M, Migliorini G, Paulsson K, Houlston RS. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat Commun 2017; 8: 14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013; 45: 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian M, Cao X, Devidas M, et al. TP53 germline variations influence the predisposition and prognosis of B-cell acute lymphoblastic leukemia in children. J Clin Oncol 2018; 36: 591–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pui CH, Rebora P, Schrappe M, et al. Outcome of children with hypodiploid acute lymphoblastic leukemia: a retrospective multi-national study. J Clin Oncol 2019; published online Jan 18. DOI: 10.1200/JCO.18.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultz KR, Carroll A, Heerema NA, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia 2014; 28: 1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tasian SK, Hunger SP. Genomic characterization of paediatric acute lymphoblastic leukaemia: an opportunity for precision medicine therapeutics. Br J Haematol 2017; 176: 867–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood 2017; 130: 2064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pui CH, Roberts KG, Yang JJ, Mullighan CG. Philadelphia chromosome-like acute lymphoblastic leukemia. Clin Lymphoma Myeloma Leuk 2017; 17: 464–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki N, Miyazaki K, Nagamachi A, et al. Identification of Zfp521/ZNF521 as a cooperative gene for E2A-HLF to develop acute B-lineage leukemia. Oncogene 2010; 29: 1963–75. [DOI] [PubMed] [Google Scholar]
- 51.Harrison CJ. Blood spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood 2015; 125: 1383–86. [DOI] [PubMed] [Google Scholar]
- 52.Moorman AV, Robinson H, Schwab C, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol 2013; 31: 3389–96. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, McCastlain K, Yoshihara H, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 2016; 48: 1481–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu YF, Wang BY, Zhang WN, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 2016; 8: 173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu Z, Churchman M, Roberts K, et al. PAX5-driven subtypes of B-cell acute lymphoblastic leukemia. Nat Genet 2019; published online Jan 14. DOI: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood 2017; 129: 1113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivera-Reyes A, Hayer KE, Bassing CH. Genomic alterations of non-coding regions underlie human cancer: lessons from T-ALL. Trends Mol Med 2016; 22: 1035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 2015; 47: 330–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pui CH, Pei D, Raimondi SC, et al. Clinical impact of minimal residual disease in children with different subtypes of acute lymphoblastic leukemia treated with response-adapted therapy. Leukemia 2017; 31: 333–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuurbier L, Petricoin EF 3rd, Vuerhard MJ, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica 2012; 97: 1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandapalli OR, Zimmermann M, Kox C, et al. NOTCH1 activation clinically antagonizes the unfavorable effect of PTEN inactivation in BFM-treated children with precursor T-cell acute lymphoblastic leukemia. Haematologica 2013; 98: 928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jenkinson S, Kirkwood AA, Goulden N, Vora A, Linch DC, Gale RE. Impact of PTEN abnormalities on outcome in pediatric patients with T-cell acute lymphoblastic leukemia treated on the MRC UKALL2003 trial. Leukemia 2016; 30: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paganin M, Grillo MF, Silvestri D, et al. The presence of mutated and deleted PTEN is associated with an increased risk of relapse in childhood T cell acute lymphoblastic leukaemia treated with AIEOP-BFM ALL protocols. Br J Haematol 2018; 182: 705–11. [DOI] [PubMed] [Google Scholar]
- 64.Petit A, Trinquand A, Chevret S, et al. Oncogenetic mutations combined with MRD improve outcome prediction in pediatric T-cell acute lymphoblastic leukemia. Blood 2018; 131: 289–300. [DOI] [PubMed] [Google Scholar]
- 65.Berry DA, Zhou S, Higley H, et al. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis. JAMA Oncol 2017; 3: e170580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stanulla M, Dagdan E, Zaliova M, et al. IKZF1plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol 2018; 36: 1240–49. [DOI] [PubMed] [Google Scholar]
- 67.Yeoh AEJ, Lu Y, Chin WHN, et al. Intensifying treatment of childhood B-lymphoblastic leukemia with IKZF1 deletion reduces relapse and improves overall survival: results of Malaysia-Singapore ALL 2010 study. J Clin Oncol 2018; 36: 2726–35. [DOI] [PubMed] [Google Scholar]
- 68.Pui CH, Campana D. Minimal residual disease in pediatric ALL. Oncotarget 2017; 8: 78251–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter SS, Dunsmore KP, Devidas M, et al. Improved survival for children and young adults with T-lineage acute lymphoblastic leukemia: results from the Children’s Oncology Group AALL0434 methotrexate randomization. J Clin Oncol 2018; 36: 2926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schrappe M, Hunger SP, Pui CH, et al. Outcomes after induction failure in childhood acute lymphoblastic leukemia. N Engl J Med 2012; 366: 1371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pieters R, Kaspers GJ, van Wering ER, et al. Cellular drug resistance profiles that might explain the prognostic value of immunophenotype and age in childhood acute lymphoblastic leukemia. Leukemia 1993; 7: 392–97. [PubMed] [Google Scholar]
- 72.Moricke A, Zimmermann M, Valsecchi MG, et al. Dexamethasone vs prednisone in induction treatment of pediatric ALL: results of the randomized trial AIEOP-BFM ALL 2000. Blood 2016; 127: 2101–12. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell CD, Richards SM, Kinsey SE, et al. Benefit of dexamethasone compared with prednisolone for childhood acute lymphoblastic leukaemia: results of the UK Medical Research Council ALL97 randomized trial. Br J Haematol 2005; 129: 734–45. [DOI] [PubMed] [Google Scholar]
- 74.Igarashi S, Manabe A, Ohara A, et al. No advantage of dexamethasone over prednisolone for the outcome of standard- and intermediate-risk childhood acute lymphoblastic leukemia in the Tokyo Children’s Cancer Study Group L95–14 protocol. J Clin Oncol 2005; 23: 6489–98. [DOI] [PubMed] [Google Scholar]
- 75.Hurwitz CA, Silverman LB, Schorin MA, et al. Substituting dexamethasone for prednisone complicates remission induction in children with acute lymphoblastic leukemia. Cancer 2000; 88: 1964–69. [PubMed] [Google Scholar]
- 76.Bostrom BC, Sensel MR, Sather HN, et al. Dexamethasone versus prednisone and daily oral versus weekly intravenous mercaptopurine for patients with standard-risk acute lymphoblastic leukemia: a report from the Children’s Cancer Group. Blood 2003; 101: 3809–17. [DOI] [PubMed] [Google Scholar]
- 77.Larsen EC, Devidas M, Chen S, et al. Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: a report from Children’s Oncology Group Study AALL0232. J Clin Oncol 2016; 34: 2380–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egler RA, Ahuja SP, Matloub Y. L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. J Pharmacol Pharmacother 2016; 7: 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vora A, Andreano A, Pui CH, et al. Influence of cranial radiotherapy on outcome in children with acute lymphoblastic leukemia treated with contemporary therapy. J Clin Oncol 2016; 34: 919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roti G, Stegmaier K. New approaches to target T-ALL. Front Oncol 2014; 4: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teachey DT, Hunger SP. Acute lymphoblastic leukaemia in 2017: immunotherapy for ALL takes the world by storm. Nat Rev Clin Oncol 2018; 15: 69–70. [DOI] [PubMed] [Google Scholar]
- 82.Gomes-Silva D, Srinivasan M, Sharma S, et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood 2017; 130: 285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bride KL, Vincent TL, Im SY, et al. Preclinical efficacy of daratumumab in T-cell acute lymphoblastic leukemia. Blood 2018; 131: 995–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


