Abstract
We describe a modified Agrobacterium-mediated method for the efficient transformation of Agaricus bisporus. Salient features of this procedure include cocultivation of Agrobacterium and fruiting body gill tissue and use of a vector with a homologous promoter. This method offers new prospects for the genetic manipulation of this commercially important mushroom species.
We have devised a highly efficient, convenient, and expeditious genetic transformation system for the button mushroom Agaricus bisporus. Our method is based on the Agrobacterium-mediated fungal transformation (agro-transformation) system originally described for the yeast Saccharomyces cerevisiae (1, 2). The unavailability of a practical gene transfer system is the single largest obstacle precluding the use of molecular approaches for the genetic improvement of mushrooms. Despite considerable interest in the development of a transformation scheme (3, 12, 14, 17, 18), no method is in general use today, owing to low efficiency or lack of utility and convenience. Recently, De Groot et al. (7) transformed several fungi, including A. bisporus, using the agro-transformation system. Although this method was more convenient than the existing protoplast-based scheme (17, 18), it suffered from a comparably low efficiency of transformation on A. bisporus. The agro-transformation method described in this paper offers a practical means for exploiting transgenic approaches for the genetic manipulation and improvement of mushrooms.
Fruiting bodies of commercial hybrid strains of A. bisporus (Sylvan 608, Sylvan 130, Amycel U1, Amycel 2500, Lambert 900, and LeLion X22) were grown at The Pennsylvania State University (PSU) mushroom research facilities (13). Vegetative cultures derived from commercial spawn were maintained on potato dextrose yeast agar (5). Genomic DNA was isolated from fruiting bodies (1 to 3 g) and broth cultures (100 mg of mycelium) as described previously (5).
Strains AGL-1 and EHA105 of Agrobacterium tumefaciens were provided by Mark Guiltinan, Department of Horticulture, PSU. Strain GV3850 was from Qiaoling Jin, U.S. Department of Energy Plant Research Laboratory, Michigan State University. Plasmid PCSN44, containing the Escherichia coli hygromycin B phosphotransferase (hph) gene with the Aspergillus nidulans trpC promoter (16), was provided by Seogchan Kang, Department of Plant Pathology, PSU. The binary vector pCAMBIA1300 (CAMBIA, Canberra, Australia) and plasmid PE2113-EGFP with the Aequorea victoria enhanced green fluorescent protein (EGFP) gene and the cauliflower mosaic virus (CaMV) 35S promoter and terminator also were provided by Mark Guiltinan. An ∼280-bp promoter sequence for the glyceraldehyde-3-phosphate dehydrogenase (gpd) gene of A. bisporus (8) was obtained by PCR amplification using either primers gpd-FH (5′-GAAGAAGCTTTAAGAGGTCCGC-3′) and gpd-RK (5′-CAGGTACCGGCGATAAGCTTGTTGTG) or primers gpd-FH and gpd-RC (5′-CAATCGATGGCGATAAGCTTGTTGTG).
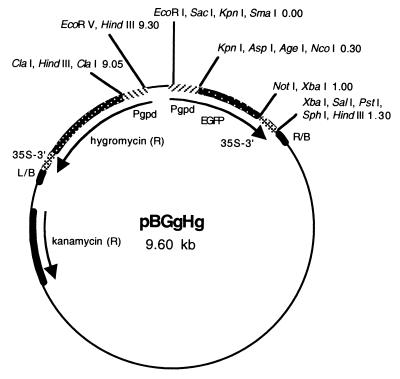
Our binary plasmid vector (9.6 kb), designated pBGgHg, consisted of a pCAMBIA1300 backbone containing the hph and EGFP genes, each of which was joined to the CaMV 35S terminator and controlled by the gpd promoter from A. bisporus (Fig. 1). In order to construct vector pBGgHg, intermediate plasmid pEGFP.g was generated by excising the CaMV 35S promoter from PE2113-EGFP with HindIII and KpnI and inserting the gpd promoter sequence obtained by PCR amplification with primers gpd-FH and gpd-RK containing HindIII and KpnI restriction sites, respectively. Intermediate plasmid pHph.g was designed from PCSN44 by excision of the trpC promoter with HindIII and ClaI and blunt-end ligation to the gpd promoter derived by PCR amplification with primers gpd-FH and gpd-RC. Intermediate plasmid pBHg was made by digesting pCAMBIA1300 with BstXI and XhoI to remove the hph gene and the CaMV 35S promoter and inserting by blunt-end ligation the hph gene and the gpd promoter, which was excised from pHph.g using BamHI. Finally, pBGgHg was constructed by excising the EGFP gene with the gpd promoter from pEGFP.g using EcoRI and HindIII and inserting this fragment by blunt-end ligation at the BamHI site in pBHg.
FIG. 1.
Organization of binary vector pBGgHg. pBGgHg is 9.6 kb in size and consists of a pCAMBIA1300 backbone containing the kanamycin resistance (R) gene and the right border (R/B) and left border (L/B) sequences of Agrobacterium T-DNA. The hygromycin resistance and EGFP genes are located between the border sequences, and each is joined to the A. bisporus glyceraldehyde-3-phosphate dehydrogenase promoter (Pgpd) and the cauliflower mosaic virus terminator (35S-3′). Shown are restriction enzyme sites with map distances in kilobases.
Southern blot analysis was carried out with a 32P-labeled ∼1-kb fragment of the hph gene as a probe and SacI-digested genomic DNA. SacI does not cut within the hph gene. PCR analysis was done (4) using primers gpd-FH and hph-R (5′-GGCGACCTCGTATTGGGAATC-3′), which defined an ∼970-bp sequence spanning the gpd promoter and the hph gene.
Aside from the aforementioned modifications with regard to the plasmid vector, we used the agro-transformation procedure of Bundock et al. (1) as extended by De Groot et al. (7), except that fruiting body tissue instead of basidiospores was cocultivated with A. tumefaciens. This appears to have a major impact on transformation efficiency. We selected fruiting bodies that were near maturity but without exposed gills. Using a scalpel, the veil was cut from the fruiting body and the exposed gill tissue was aseptically excised and sectioned into 2 to 5-mm square pieces.
For transformation experiments, Agrobacterium was grown in 5 ml of minimal medium containing kanamycin at 50 μg/ml for 2 days at 28°C. One milliliter of the fresh culture was transferred to 100 ml of minimal medium with kanamycin and grown overnight at 28°C to an optical density at 600 nm of 0.5 to 0.8. Bacteria were collected by centrifugation and resuspended in induction medium containing 200 μM acetosyringone to an optical density at 600 nm of 0.5. In order to preinduce the virulence of A. tumefaciens, the bacterial suspension was incubated for 3 to 6 h at room temperature with gyratory shaking at 100 rpm. Fruiting body gill tissue pieces were vacuum infiltrated with the suspension of induced bacteria until the air had been completely purged. The evacuated tissue was transferred to a piece of sterile 3MM Whatman filter paper overlaid on cocultivation medium and incubated for 3 days at room temperature. Tissue pieces were transferred to selection medium (SM) containing hygromycin at 30 μg/ml and maintained at room temperature. For final selection, colonies growing from the tissue pieces were transferred to SM containing hygromycin at 50 μg/ml. Each experiment included a nontransformed control consisting of either tissue pieces that were vacuum infiltrated with induction medium alone or infiltrated with noninduced bacteria.
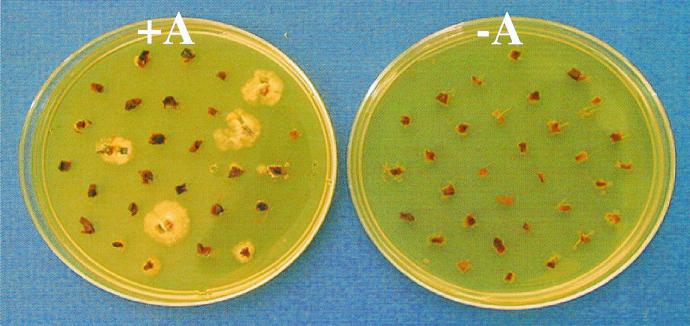
Hygromycin-resistant colonies appeared at the margins of the tissue pieces after 9 to 14 days on SM with hygromycin at 30 μg/ml (Fig. 2). Our optimal protocol typically provided 30 to 40% efficiency of transformation (percentage of tissue pieces regenerating colonies on hygromycin medium). This is an order of magnitude higher than the floral dip agro-transformation procedure for Arabidopsis thaliana (6) and 7 orders of magnitude higher (i.e., ∼0.00003%) than the reported agro-transformation method for A. bisporus using basidiospores (7). The lower efficiency of the previous A. bisporus protocol was probably due to the inherently low germination rate of the spores and the use of a heterologous promoter. Our procedure offers higher effective efficiency and greater convenience than the original method and is more expeditious considering the time expended in spore printing and the several weeks required for spore germination and selection.
FIG. 2.
Selection of putative hygromycin-resistant transformants of A. bisporus. Pieces of fruiting body gill tissue were cocultivated with (+A) and without (−A) A. tumefaciens strain AGL-1 carrying the vector pBGgHg containing the A. bisporus gpd promoter and hph gene construct. Shown is the appearance of the cultures after 2 weeks on SM with hygromycin at 30 μg/ml.
The choice of promoter, strain of A. tumefaciens, and type of fruiting body tissue were critical for optimal transformation efficiency. In two experiments comparing various promoter constructs with the hph gene, the A. bisporus gpd, A. nidulans trpC, and CaMV 35S promoters provided average transformation efficiencies of 15, 1, and 0%, respectively. Of the three bacterial strains examined, AGL-1 and EHA105, but not GV3850, transformed A. bisporus, each averaging ∼23% efficiency in two experiments. Also, in a replicated experiment, higher efficiencies were obtained with gill tissue (64%) than with the fleshy tissue derived from the fruiting body cap and stem (9%). Our standard protocol entailed the use of plasmid vector pBGgHg, bacterial strain AGL-1, and gill tissue.
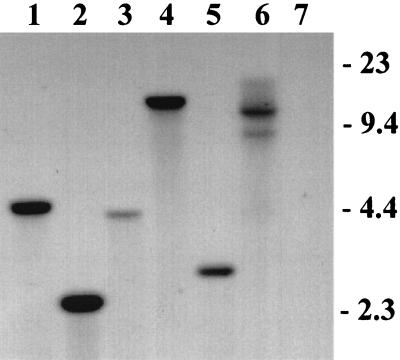
Southern blot analyses confirmed that the hph gene was integrated into the genome of A. bisporus (Fig. 3). We detected no false positives by Southern blot analysis or PCR amplification (Fig. 4) among 37 antibiotic-resistant cultures. Many of the transformed cultures appeared to have a single integrated copy of the gene at random sites, although some showed evidence of up to four integration events. However, a precise determination of copy number would require an analysis of homokaryotic lines derived by single-spore culture.
FIG. 3.
Southern blot analysis of DNA isolated from putative hygromycin-resistant transformants of A. bisporus. Genomic DNA (5 to 10 μg) was isolated from broth cultures, digested with SacI, and probed with an ∼1-kb 32P-labeled hph gene sequence. Lanes: 1 to 6, DNA isolated from putative transformants AT1 to AT6, respectively; 7, DNA isolated from nontransformed A. bisporus. The positions of molecular DNA size markers (kilobases) are shown on the right.
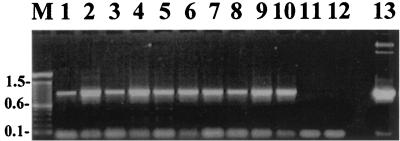
FIG. 4.
PCR analysis of DNA isolated from putative hygromycin-resistant transformants of A. bisporus. PCR amplification was carried out on genomic DNA using primers (gpd-FH and hph-R) defining an ∼970-bp sequence spanning the A. bisporus gpd promoter and the hph gene. Lanes: 1 to 10, DNA isolated from putative transformants AT3, AT4, AT9, AT10, AT11, AT12, AT16, AT19, AT24, and AT31, respectively; 11, negative control with water; 12, DNA isolated from nontransformed A. bisporus; 13, positive control with vector pBGgHg; M, DNA molecular size markers (kilobases).
Southern blot analysis established that the EGFP gene also was incorporated into the genome, but we were not able to confirm GFP fluorescence in an examination of three transformants. With the available data, we cannot draw conclusions about the expression of this transgene in A. bisporus but note that it has been problematic in other organisms owing to aberrant mRNA processing (9) and codon preference (15, 19).
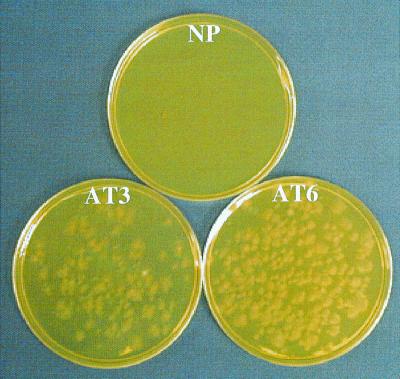
We can regenerate transgenic vegetative cultures in less than 2 weeks and produce mature fruiting bodies 8 weeks later under controlled environmental conditions. We have cropped 30 hygromycin-resistant transgenic mushroom lines, and all have borne fruiting bodies. After final selection for several weeks with hygromycin at 50 μg/ml, we typically maintained hygromycin-resistant cultures for weeks to months on a medium without antibiotic selection. For cropping trials, these cultures were grown without selection for an additional 8 weeks on rye grain, compost, and peat. The antibiotic resistance trait was stably maintained to the extent that it was expressed by the developing fruiting bodies and basidiospores (Fig. 5). In one study, all of the tissue pieces sampled from 120 randomly selected fruiting bodies representing six transgenic lines were found to inherit the trait as determined by antibiotic selection.
FIG. 5.
Expression of the hygromycin resistance trait in first-generation basidiospores produced from transgenic cultures of A. bisporus. Basidiospores from two transgenic lines (AT3 and AT6) and the nontransformed parental strain (NP) were plated on SM with hygromycin at 50 μg/ml. The viability of the basidiospores of the parental strain was established on SM without antibiotic (data not shown).
Classical breeding of A. bisporus has been difficult, relating primarily to the predominantly secondarily homothallic life cycle of this fungus (11). Therefore, it is not surprising that the cultivated strains display extremely limited genetic variation (10). The transformation technique described herein provides a practical method for using transgenic technology in the genetic improvement of this commercially important mushroom and represents an important tool for the molecular genetic analysis of biological processes in this species.
Acknowledgments
We are grateful to Cheng Lu and Qing Shen for technical assistance.
This research was supported in part by a grant from Sylvan Inc.
REFERENCES
- 1.Bundock P, Den Dulk-Ras A, Beijersbergen A, Hooykaas P J J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bundock P, Hooykaas P J J. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc Natl Acad Sci USA. 1996;93:15272–15275. doi: 10.1073/pnas.93.26.15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Challen M P, Elliott T J. Evaluation of the 5-fluoroindole resistance marker for mushroom transformation. Cult Mush Newsl. 1994;2:13–20. [Google Scholar]
- 4.Chen X, Romaine C P, Ospina-Giraldo M D, Royse D J. A PCR-based test for the identification of Trichoderma harzianum biotypes 2 and 4 inciting the worldwide green mold epidemic in cultivated Agaricus bisporus. Appl Microbiol Biotechnol. 1999;52:246–250. [Google Scholar]
- 5.Chen X, Romaine C P, Tan Q, Schlagnhaufer B, Ospina-Giraldo M D, Royse D J, Huff D R. PCR-based genotyping of epidemic and preepidemic Trichoderma isolates associated with green mold of Agaricus bisporus. Appl Environ Microbiol. 1999;65:2674–2678. doi: 10.1128/aem.65.6.2674-2678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clough S J, Bent A F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 7.De Groot M J A, Bundock P, Hooykaas P J J, Beijersbergen A G M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 8.Harmsen M C, Schuren F H J, Moukha S M, Van Zuilen C M, Punt P J, Wessels J G H. Sequence analysis of the glyceraldehyde 3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr Genet. 1992;22:447–454. doi: 10.1007/BF00326409. [DOI] [PubMed] [Google Scholar]
- 9.Haseloff J, Siemering K R, Prasher D C, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerrigan R W, Horgen P A, Anderson J B. The California population of Agaricus bisporus comprises at least two ancestral elements. Syst Bot. 1993;18:123–136. [Google Scholar]
- 11.Kerrigan R W, Royer J C, Baller L M, Kohli Q, Horgen P A, Anderson J B. Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics. 1992;133:225–236. doi: 10.1093/genetics/133.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li A, Horgen P A. Attempts to develop a transformation system in Agaricus bisporus, using particle bombardment and several other novel approaches. Cult Mush Newsl. 1993;1:11–16. [Google Scholar]
- 13.Romaine C P, Schlagnhaufer B. Characteristics of a hydrated alginate-based delivery system for cultivation of the button mushroom. Appl Environ Microbiol. 1992;58:3060–3066. doi: 10.1128/aem.58.9.3060-3066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Royer J C, Horgen P A. Towards a transformation system for Agaricus bisporus. In: Van Griensven L J L D, editor. Genetics and breeding of Agaricus. Wageningen, The Netherlands: Pudoc; 1991. pp. 135–139. [Google Scholar]
- 15.Spellig T, Bottin A, Kahmann R. Green fluorescent protein (gfp) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- 16.Staben C, Jensen B, Singer M, Pollock J, Schechtman M, Kinsey J, Selker E. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet Newsl. 1989;36:79–81. [Google Scholar]
- 17.van der Rhee M D, Graca P M A, Huizing H J, Mooibroek H. Transformation of the cultivated mushroom, Agaricus bisporus, to hygromycin B resistance. Mol Gen Genet. 1996;250:252–258. doi: 10.1007/BF02174382. [DOI] [PubMed] [Google Scholar]
- 18.van der Rhee M D, Werten M O, Huizing H J, Mooibroek H. Highly efficient homologous integration via tandem exo-beta-1,3-glucanase genes in the common mushroom, Agaricus bisporus. Curr Genet. 1996;30:166–173. doi: 10.1007/s002940050116. [DOI] [PubMed] [Google Scholar]
- 19.Zolotukin S, Potter M, Hauswirth W W, Guy J, Muzycka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]