Abstract
Keloid scarring is a kind of pathological healing manifestation after skin injury and possesses various tumor properties, such as the Warburg effect, epithelial–mesenchymal transition (EMT), expression imbalances of apoptosis-related genes and the presence of stem cells. Abnormal expression of tumor signatures is critical to the initiation and operation of these effects. Although previous experimental studies have recognized the potential value of a single or several tumor biomolecules in keloids, a comprehensive evaluation system for multiple tumor signatures in keloid scarring is still lacking. This paper aims to summarize tumor biomolecules in keloids from the perspectives of liquid biopsy, genetics, proteomics and epigenetics and to investigate their mechanisms of action and feasibility from bench to bedside. Liquid biopsy is suitable for the early screening of people with keloids due to its noninvasive and accurate performance. Epigenetic biomarkers do not require changes in the gene sequence and their reversibility and tissue specificity make them ideal therapeutic targets. Nonetheless, given the ethnic specificity and genetic predisposition of keloids, more large-sample multicenter studies are indispensable for determining the prevalence of these signatures and for establishing diagnostic criteria and therapeutic efficacy estimations based on these molecules.
Keywords: Tumor signatures, Keloid, Proteome, Epigenetics, Exosomes, Tumor biomolecules, Biomarkers
Highlights.
Keloid scarring is a kind of pathological healing manifestation after skin injury and possesses various tumor properties.
Liquid biopsy has the potential to screen for people with keloids due to its non-invasive and accurate properties.
Epigenetic biomarkers do not require changes in the gene sequence, and their reversibility and tissue specificity make them ideal therapeutic targets.
Background
Keloid disorder is a group of ‘tumor-like’ pathological scars that grows invasively beyond the injury boundary, with very few spontaneous degenerative changes [1, 2]. Keloid scarring is an unpreventable consequence of trauma, burns, surgery, or no apparent induction, with fibroblast overgrowth and excessive collagen secretion as the typical pathological features [3]. The pathogenesis of keloids is intricate and involves the comprehensive effects of multiple factors, such as genetic susceptibility, persistent inflammatory response, endocrine factors and race specificity [4]. Keloids generally occur in areas such as the anterior chest and scapular areas, which are intermittently or continuously in high tension in daily activities [5]. The reason may be that mechanical force signals are converted into chemical signals through mechanically sensitive ion channels and G-protein conjugate receptors on the surfaces of cell membranes, affecting downstream pathways such as transforming growth factor beta (TGF-β)/Smad pathways and tumor necrosis factor alpha (TNF-α)/nuclear factor-kappa B (NF-κB) pathways, thereby stimulating fibroblast proliferation and collagen fiber accumulation [6]. There are numerous keloid treatments, including surgery, radiation therapy and drug injections. Nonetheless, due to the lack of clear biomarkers for the early identification and prevention of keloid scarring, the diagnosis and treatment are relatively immature and can merely be implemented after the formation of keloids [4, 7]. Moreover, the undetermined pathogenesis of keloids leads to the failure of etiological treatment, and the recurrence rate of surgical and nonsurgical symptomatic therapy is high [8].
In recent years, with the increasing understanding of keloids, more scholars have detected the tumor-like properties of keloids, such as the Warburg effect, epithelial–mesenchymal transition (EMT), expression imbalances of apoptosis-related genes and the existence of keloid stem cells [9, 10]. Abnormal expression of tumor signatures in keloids is critical to the initiation and operation of these effects. Under the stimulation of trauma, infection or surgery, some tumor-related factors are activated as dominant modulators or signal molecules, inducing a series of reactions, such as the inflammatory response and accumulation of extracellular matrix [11]. Consequently, they have broad prospects in experimental research and clinical application, including early screening, prevention, assessment of aggressiveness and treatment-response prediction in keloids.
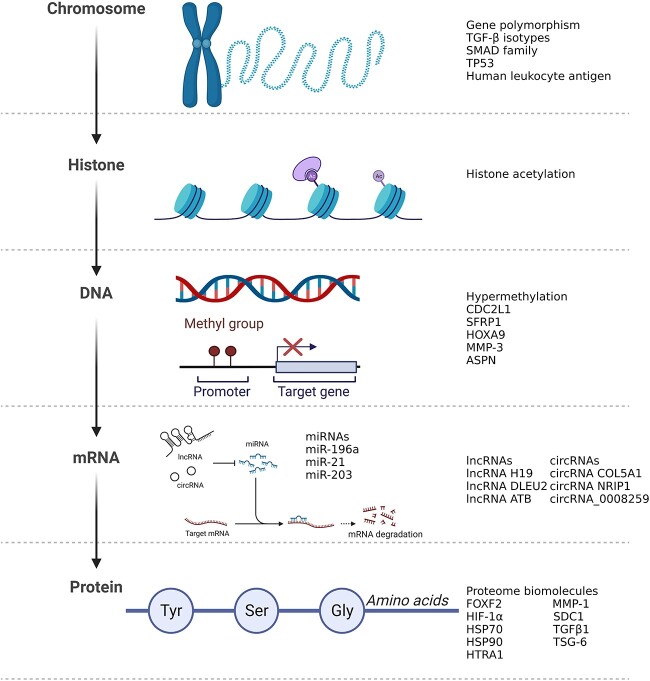
Although the potential value of a single or several tumor signatures in keloids has been acknowledged in many experimental studies [12, 13], it is essential to establish a comprehensive evaluation system that summarizes various tumor signatures in keloids and thoroughly assesses the practicality of these factors in clinical practice. From this perspective, the objective of this article is to summarize the tumor signatures that are abnormally expressed in keloids and to investigate their mechanism of action and their feasibility from bench to bedside (Figure 1).
Figure 1.
Illustrations of tumor-associated molecules, giving examples of how these molecules may be involved in gene expression from chromosomes to proteins in keloids. Created with BioRender.com. TGF-β transforming growth factor beta, miRNAs microRNAs, MMP matrix metalloproteinase, ASPN asporin, HSP heat shock protein, HIF-1α hypoxia inducible factor 1 subunit alpha, IL interleukin, VEGF vascular endothelial growth factor
Review
Signatures in blood and body fluids—liquid biopsy
Clinical pathological sampling is ordinarily undertaken by surgical biopsy or needle biopsy, which is an invasive test. ‘Liquid biopsy’, as a novel blood test, is easy to perform, and frequent sampling is allowed. Assessing and analyzing circulating cells, DNA and exosomes released from lesions into the blood assist in diagnosing and clarifying the nature of diseases and locating lesions that may metastasize [14, 15]. Burgeoning liquid biopsy is beneficial for keloid prevention and screening as it avoids the skin injury generated by surgical biopsy, and the diagnostic efficiency is notably enhanced [16].
Interestingly, a recent article based on single-cell sequencing and T-cell subtyping identified serum levels of soluble human leukocyte antigen-E (sHLA-E) as a predictive biomarker and potential therapeutic target for keloids. Its sensitivity and specificity could reach 83.69% and 92.16%, which were validated in 104 patients with keloids, 512 healthy donors and 100 patients with an interfering disease. sHLA-E levels decreased following intralesional therapy of keloids with triamcinolone combined with 5-fluorouracil, and monitoring of sHLA-E levels in keloids could be used to determine the recurrence of keloids and guide clinical treatment [17].
Currently, the progression of liquid biopsy in keloids is principally reflected in the investigation of exosomes. Exosomes are derived from polyvesicles formed by intracellular lysosomal microparticle invagination, which transfer signaling molecules such as proteins, DNA and microRNAs (miRNAs) to mediate cell–cell communication and participate in various biological behaviors, including the immune response, cell migration and differentiation [18, 19]. Exosomes are an indispensable link in the process of fibrosis and may serve as a new research direction in the diagnosis and treatment of keloids [20, 21]. Exosomes derived from human endothelial progenitor cells organize fibroblast angiogenesis and mesenchymal–endothelial transformation by transporting miRNA-133 [22]. Exosomes can stimulate the phenotypic differentiation of normal fibroblasts into myofibroblasts that are conducive to invasion and metastasis by TGF-β/Smad signaling pathways [23, 24]. Mechanical stress is able to influence vascular formation by adjusting the secretion of fibroblast exosomes [25].
It is worth noting that the main significance of exosomes for translational medicine is that they serve as ideal non-invasive diagnostic and prognostic markers for personalized monitoring of disease occurrence and progression and as nucleic acid or drug delivery vectors [26]. Exosomes act as vehicles for cell communication, acquiring their contents from their parental cells, and thus exosomes in body fluids may reflect the characteristics, status and disease evolution of their parental cells. For example, glypican 1 can be used to diagnose pancreatic cancer and determine its stage with 100% sensitivity and specificity [27]. For keloids, exosomes contain signaling molecules that are transported to distant sites to regulate the microenvironment of the fibrosis process. Therefore, monitoring the secretion of specific exosomes is expected to provide early screening for patients with keloids. For example, fibroblasts in keloids secrete significantly more exosomal miRNA-21 than normal skin tissue [28, 29]; thus, the evaluation of exosomes containing specific signaling molecules may aid in differentiating keloids from normal skin tissue.
Due to their excellent biocompatibility, low immunogenicity, first-class membrane permeability and penetration, exosomes have incomparable advantages over artificial drug delivery carriers such as liposomes or lipid-based nanoparticles as nucleic acid or drug delivery carriers [30]. In the treatment of keloids, exosomes secreted by mesenchymal stromal cells are capable of inhibiting inflammation and scar formation in the process of wound healing [31]. Autologous transdermal injection of exosomes derived from fibroblasts regulated the expression of procollagen type I and matrix metalloproteinase 1 (MMP-1) and impacted dermal collagen deposition and degradation [32]. Therefore, inducing specific exosomes or constructing drug vectors based on exosomes to intervene in the process of fibrosis and scar formation may be a new direction for the development of new keloid drugs.
Genetic signatures
The first study concentrating on keloid susceptibility sites came from Marneros et al.’s genome-wide linkage survey of two keloid families (Japanese and African–American) [33]. Genetic predisposition to keloids in the Japanese family was linked to locus D2S1399 on chromosome 2q23, a locus that controls the encoding of TNF-α-induced protein 6 (TNFAIP6). Keloid tendency in the African–American family was found to be linked to locus D7S499 on chromosome 7p11, which encodes epidermal growth factor receptor (EGFR). EFGR is exceptionally widely expressed in ovarian cancer and lung adenocarcinoma, is the focal point of targeted therapy and clinical molecular detection [34] and its expression has been substantiated to be significantly higher in keloids than in normal skin tissue [35].
Regarding gene polymorphisms, previous studies have chiefly investigated TGF-β isotypes (TGF-β1, TGF-β2, TGF-β3), members of the SMAD family (SMAD3, SMAD6, SMAD7), mutations in p53 and human leukocyte antigen (HLA) in keloids [36–38]. In an analysis of p53 gene mutations in keloids, mutations were observed in all keloid samples along with increased cell proliferation and an imbalance in apoptosis that was not observed in other normal skin tissues of the same patients [39]. In another study involving 15 keloid samples, mutations in exons of the p53 gene were discovered in all keloid samples [40]. In addition, the importance of the HLA haplotype in the process of keloid fibrosis has been verified in relevant studies [41]. In studies on the association between the HLA system and keloids, the risk of keloid development in Caucasians was positively correlated with the frequency of the HLA-DRB1*15 phenotype, and the distribution of HLA-DQA1 and DQB1 alleles was enormously disparate between keloid and normal populations in the analysis of Chinese Han keloid patients [42, 43].
Proteome signatures
Maintaining proliferation
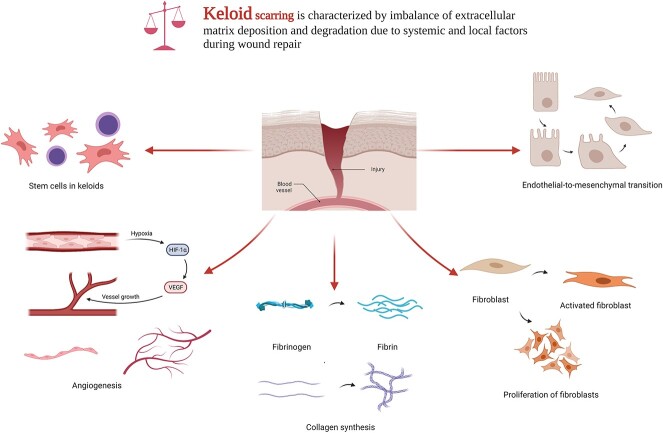
To date, the abnormal expression of one or several tumor biomolecules and their functions in keloids (Table 1) have been reported in several studies, including proliferation, apoptosis, extracellular matrix (ECM) deposition, angiogenesis, inflammatory response and EMT (Figure 2). In signaling fibroblast division and differentiation, keloids are considerably different from normal skin tissues in the silencing or downregulation of apoptotic factors, oncogene activation, and the regulation of classical signaling pathways and stress proteins [44–46].
Table 1.
Tumor-associated protein biomolecules differentially expressed in keloids
| Biomolecules | Abbreviation | Involved physiological function | Expression in keloids | Associated biological processes in keloids | Reference |
|---|---|---|---|---|---|
| Proliferation and apoptosis | |||||
| Catenin (Cadherin-associated protein), beta 1 | β-Catenin | Cell growth Cell adhesion |
Up-regulated | Maintenance of skin fibrosis | [51, 52, 54] |
| B-Cell lymphoma-2 | BCL2 | Programmed cell death | Positive in keratinocytes and fibroblasts | Blocked fibroblast apoptosis | [48] |
| Proto-oncogene C-Fos | C-FOS | Transactivating factor | Positive in dermal fibroblasts | Regulation of fibroblast proliferation | [48] |
| Calcitonin gene-related peptide | CGRP | Vasodilation | Up-regulated | Gene expression of growth factors in fibroblasts | [46] |
| c-Myc transcription factor | cMYC | Cell cycle progression Cellular transformation Apoptosis |
Down-regulated | Apoptosis | [47] |
| Proto-oncogene C-Jun | C-JUN | Transactivating factor | Positive in dermal fibroblasts | Regulation of fibroblast proliferation | [48] |
| Defender against cell death 1 | DAD-1 | Programmed cell death | Down-regulated | Apoptosis | [47] |
| Nineteen Kd interacting protein-3 | NIP3 | Pro-apoptotic factor | Down-regulated | Apoptosis | [47] |
| Tumor protein P53 | TP53 | Cell cycle arrest DNA repair Senescence |
Down-regulated | Decreased fibroblast proliferation | [48, 50] |
| Tumor protein P63 | TP63 | Maintenance of epithelial self-renewal | Up-regulated | Blockade of TP53 activity | [50] |
| Tumor necrosis factor receptor type 1-associated death domain protein | TRADD | Programmed cell death | Down-regulated | Apoptosis | [47] |
| Wingless-type MMTV integration site family, member 5A | Wnt5A | Proliferation Apoptosis Differentiation |
Up-regulated | Proliferation of fibroblasts | [52] |
| Wingless-type MMTV integration site family, member 10A | Wnt10A | Proliferation Apoptosis Differentiation |
Up-regulated | Proliferation of keloid progenitor cells | [53] |
| Collagen deposition and extracellular matrix formation | |||||
| Collagen type I alpha 1 chain | COLIα1 | Type I collagen synthesis | Up-regulated | Collagen synthesis | [71] |
| Heat shock protein 27 | HSP27 | Correct folding of proteins | Up-regulated | Matrix synthesis | [76] |
| Heat shock protein 47 | HSP47 | Collagen biosynthesis | Up-regulated | Synthesis and secretion of collagen | [76] |
| Interleukin 6 | IL-6 | Inflammation Maturation of B cells |
Up-regulated | Increased accumulation of ECM Regulation of fibroblast migration Regulation of matrix metalloproteinase function |
[60, 65] |
| Integrin subunit alpha 2 | ITGα2 | Modulation of collagenase gene expression Organization of newly synthesized ECM |
Up-regulated | Matrix synthesis | [46] |
| Matrix metalloproteinase 2 | MMP-2 | Cleaving components of the ECM Cleaving molecules involved in signal transduction |
Up-regulated | Remodeling of collagen bundle regions Degradation of ECM |
[67, 68] |
| Matrix metalloproteinase 13 | MMP-13 | Breakdown of ECM | Up-regulated | Remodeling of the extracellular matrix Immune response |
[66] |
| Matrix metalloproteinase 19 | MMP-19 | Breakdown of ECM | Up-regulated | Remodeling of the extracellular matrix Chronic inflammation |
[46] |
| Osteopontin | OPN | Cell adhesion Signal transduction |
Up-regulated | Collagen synthesis Fibroproliferation |
[73] |
| Periostin | POSTN | Tissue development Regeneration |
Up-regulated | Collagen synthesis Proliferation of KFB Migration of KFB Invasion of KFB |
[71] |
| S100 Calcium binding protein | S100 | Cell cycle progression Cell differentiation |
Down-regulated | ECM production | [69] |
| Thymic stromal lymphopoietin | TSLP | Immune response Inflammatory response |
Up-regulated | Increased collagen I and collagen III expression | [58] |
| Inflammatory response | |||||
| Chemokine-like factor 1 | CKLF-1 | Immune surveillance Inflammation response |
Up-regulated | Inflammatory response | [60] |
| C-X-C Motif chemokine receptor 4 | CXCR4 | Regulation of cell migration Inflammatory response |
Up-regulated | Inflammatory response | [59, 60] |
| C-X-C Motif chemokine ligand 9 | CXCL9 | Immunoregulatory Inflammation response |
Up-regulated | Inflammatory response | [61] |
| C-X-C Motif chemokine ligand 12 | CXCL12 | Immune surveillance Inflammation response |
Up-regulated | Activation of the inflammatory response | [62] |
| Interleukin 17 | IL-17 | Proinflammatory cytokine | Positive | Inflammation Fibrosis |
[109, 110] |
| Angiogenesis | |||||
| Interleukin 8 | IL-8 | Chemotactic factor | Up-regulated | Increased recruitment of endothelial progenitor cells | [80] |
| Jagged canonical notch ligand 1 | JAG1 | Fibroblast growth factor-induced angiogenesis | Up-regulated | Angiogenesis Enhanced fibroblast activity |
[83] |
| Vascular endothelial growth factor | VEGF | Angiogenesis Endothelial cell growth |
Positive in the basal layer of the epidermis | Angiogenesis | [85, 86] |
| Platelet-derived growth factor | PDGF | Angiogenesis Cell proliferation Differentiation |
Positive | Angiogenesis Fibronectin production |
[85] |
| Endothelial–mesenchymal transition | |||||
| Interleukin 18 | IL-18 | Proinflammatory cytokine | Up-regulated | Epithelial–mesenchymal interaction Increased collagen expression Increased secretion of profibrotic cytokines |
[98] |
| Phosphatase and tensin homolog deleted on chromosome ten | PTEN | Tumor suppressor | Positive in keloid keratinocytes | EMT transition | [99, 100] |
| Signal transducer and activator of transcription 3 | STAT3 | Cell growth Apoptosis |
Up-regulated | Epithelial–mesenchymal interaction Cell proliferation Collagen production |
[101] |
| Wingless-type MMTV integration site family, member 3A | Wnt3A | Regulation of cell fate Patterning during embryogenesis |
Up-regulated | EMT transition | [102] |
| Stem cells in keloids | |||||
| Kruppel-like factor 4 | KLF4 | Differentiation of epithelial cells | Positive in keloid-derived precursor cells | Development of individualized induced pluripotent stem cells from fibroblasts | [107] |
| Lin-28 homolog | LIN28 | Developmental timing | Positive in keloid-derived precursor cells | Development of individualized induced pluripotent stem cells from fibroblasts | [107] |
| Octamer-binding protein 4 | 4-Oct | Embryonic development Stem cell pluripotency |
Positive in keloid-derived precursor cells | Development of individualized induced pluripotent stem cells from fibroblasts | [107] |
| Sry-box transcription factor 2 | SOX2 | Embryonic development Determination of cell fate |
Positive in keloid-derived precursor cells | Development of individualized induced pluripotent stem cells from fibroblasts | [107] |
KFB keloid fibroblast cells, EMT endothelial–mesenchymal transition, ECM extracellular matrix
Figure 2.
Key pathological processes involved in the development of keloids by proteomic molecules. Created with BioRender.com. HIF-1α hypoxia inducible factor 1 subunit alpha, VEGF vascular endothelial growth factor
In studies of multiple apoptosis-related factors in keloids, the expression levels of TRADD, TSG-6 and NIP3, which are participants of programmed cell death and apoptosis, were downregulated [47]. Immunohistochemical staining of keloids revealed strong staining of the oncogenes cMYC and Bcl-2 (negative regulators of programmed cell death), negative staining of TP53 (a negative regulator of cell proliferation), and positive staining for the transcription factors c-Jun and c-Fos, which activate protooncogenes and facilitate sustained fibroblast proliferation signals [48–50]. β-Catenin, Wnt5α and Wnt10α are activated, and their expression levels are positively connected with fibroblast proliferation through the classical Wnt/β-catenin signaling pathway [51–54]. HTRA1, a member of the serine protease family induced by heat shock, has the ability to recognize misfolded proteins and degrade a variety of substrates, including ECM [55]. Due to the protease activity of HTRA1, it has been confirmed to be involved in autophagy, apoptosis and the metastasis of malignant tumors. Active keloid lesions express more HTRA1 than normal skin and boost keloid formation during wound healing by involving the cell proliferation process and specific remodeling of the extracellular matrix. Knockdown of HTRA1 in keloid inhibits keloid cell proliferation and exogenous supplementation of HTRA1 in culture medium counteracts this process. [56].
Deposition of ECM
Histologically, keloids microscopically present as spiral collagenous fibers, dense collagenous nodules accumulated by fibroblasts and collagenous fibers, diffuse microvascular injury and degeneration or necrosis of fibroblasts [57]. Excessive collagen deposition, or fibrosis, is another focus of research into the pathological mechanisms of keloids [58]. Proteins that regulate the process of fibrosis are primarily concentrated in three groups, namely, cytokines involved in the inflammatory response, components that constitute the extracellular matrix and collagen-processing proteins [59–62].
The cytokine interleukin 6 (IL-6) has been reported to be conducive to the occurrence of a variety of diseases with fibrotic processes, such as scleroderma, cardiac hyperplasia and pulmonary fibrosis [63]. The expression of IL-6 stimulates the synthesis of type I collagen by inducing inflammatory responses and mediating the TGF-β/Smad pathway [64]. Defects in IL-6 delay the re-epithelialization of wound healing and inhibit MMP expression [60, 65]. Activation of MMPs is associated with the invasion ability of various malignant tumors [66]. The roles of MMP-1 and MMP-2 involve accelerating the lysis of ECM and the remodeling of collagen bundles, intensifying the migration ability of fibroblasts and creating conditions for the migration of fibroblasts to the surrounding normal skin [67, 68]. S100, a protein secreted by keratinocytes in inflammatory diseases such as bronchitis and rheumatoid arthritis, has been verified to be down-regulated in keloids. In S100A7- and S100A15-treated fibroblasts, the expression levels of TGF-β subtype molecules, type I collagen, type III collagen and fibronectin-1 decreased [69].
Periostin is expressed in the dermis and basement membrane during embryonic development and is gradually degraded after birth [70]. Nevertheless, in keloids, periostin is upregulated and regulates the fibrosis process and keratinocyte proliferation and is involved in maintaining the mechanical properties of keloids [71]. Osteopontin (OPN), a noncollagenous bone matrix protein, is predominantly involved in the proliferation of fibroblasts and the occurrence and metastasis of epithelial-derived tumors [72]. The expression of OPN in various cells is low under physiological conditions, whereas OPN positivity in the epidermis and fibroblasts was noted by immunohistochemical staining of keloid [73].
The heat shock protein (HSP) family acts as a molecular chaperone to prevent stress-induced protein aggregation, and multiple HSP molecules have specific abnormal expression profiles in a variety of tumors [74]. For example, HSP27 can be a predictor of the progression of endometrial carcinomas [75]. As the HSP family is involved in the folding and synthesis of collagen in the endoplasmic reticulum, members of the HSP family are markedly increased in keloids and maintain the correct folding of collagen [76]. HSP47 directly or indirectly engages in collagen synthesis and secretion through the TGF-β/Smad pathway or as a molecular chaperone of collagen [77].
Angiogenesis
The continuous supply of nutrients is indispensable for tumor proliferation and metastasis. Accordingly, the expression of multiple angiogenesis regulators is associated with the degree of malignancy and the aggressiveness and prognosis of a variety of tumors and this phenomenon is similar in keloids [78, 79]. The cytokine IL-8 has been ascertained to amplify endothelial progenitor cell recruitment in keloids [80], and JAG1 may boost angiogenesis induced by fibroblast growth factor (FGF) through the Notch signaling pathway [81–83]. As the main angiogenic mediators, platelet-derive growth factor (PDGF) and vascular endothelial growth factor (VEGF) are expressed in myofibroblasts, vascular endothelial cells and vascular pericytes in keloids, and maintain the vascular density required for nutrient transport and metabolism by stimulating the proliferation of vascular endothelial cells [84–86]. PDGF synthesized and secreted by fibroblasts may encourage chemotaxis and the division of microvascular smooth muscle cells through paracrine action, which may be the molecular pathological basis of microvascular distortion and hyperplastic occlusion [87, 88]. Disruption of nutrient supply follows hyperplastic occlusion of blood vessels, hinders the regeneration of transitional structures between the dermis and the epidermis, and is ultimately manifested histologically as the loss of dermal papilla, basement membrane and skin appendages [89].
Energy metabolism
3D images of keloids reveal the extensive distribution of occluded microvessels, possibly due to excessive proliferation of endothelial cells protruding into the lumen and compression of surrounding fibroblasts and collagen. The vascular distribution in the center is sparser and the lumen is more flattened, suggesting a more hypoxic environment in the center of the keloid [87, 90]. Moreover, keloid energy metabolism is similar to that of tumors, with much higher lactic acid levels than those of normal skin, sustaining continuous anaerobic glycolysis [91, 92]. Hypoxia inducible factor 1 (HIF-1) induced under hypoxia promotes the expression of the inflammatory factor IL-6 by activating the TLR4/MYD88/NF-κB pathway [93]. Likewise, it elevates the secretion of connective tissue growth factor and supports the transformation of dermal fibroblasts into myofibroblasts by activating the TGF-β/Smad signaling pathway [94, 95].
EMT transition
EMT is a biological process in which epithelial cells undergo biochemical adjustments to acquire a mesenchymal phenotype, forfeit polarity and acquire invasiveness. EMT is ubiquitous and enables cells to achieve higher antiapoptotic competence [96–98]. In keloids, the tumor suppressor phosphatase and tensin homolog (PTEN) and Akt signaling pathways have been reported to participate in keratinocyte and fibroblast phenotypic modifications and the acquisition of stemness [99–101]. Treating human dermis microvascular endothelial cells with Wnt-3α increases the expression of vimentin, a characteristic protein of mesenchymal cells [102]. Activated HIF-1α in hypoxic environments is instrumental in the EMT process and migration capacity of keratinocytes, which can be reversed by hyperbaric oxygen therapy [103, 104].
Stem cells in keloids
Fibroblasts from normal skin do not have cloning capability. However, a cloning culture of keloid fibroblasts demonstrated that the self-renewal ability of keloid fibroblasts was much higher than that of normal fibroblasts, and they were able to continuously expand outwards, which might be related to keloid stem cells [105, 106]. Keloid stem cells mostly have two phenotypes: mesenchymal stem cells and hematopoietic stem cells [105, 106].
Immunohistochemical staining of the embryonic stem cell markers OCT4 and SOX2 is positive in keloid-associated lymphoid tissue [107]. OCT4 is a crucial transcription factor in embryonic stem cells that maintains the self-renewal and undifferentiated state of embryonic stem cells by activating pluripotency-related genes and inhibiting the expression of differentiation-related genes [108]. In addition, the increased expression of the IL-17/IL-6 axis in the inflammatory response is partly responsible for the formation of self-renewing and multicompetent derived precursors in keloids [109, 110].
Clinical translation
Advances in transcriptomics, proteomics and experimental techniques have facilitated better identification of molecular signatures of keloids and understanding of keloid pathogenesis, and certain biomolecules that have been identified exhibit some potential for clinical application in the scientific translation of proteomics. Bagabir et al. revealed strong immunoreactivity of syndecan-1 in the reticular dermis of keloids by immunohistochemical staining, whereas staining was completely absent or slight in hyperplastic scars, normal scars, healthy skin and dermatofibrosarcoma protuberans, thus recommending syndecan-1 as a diagnostic marker to differentiate keloids and avoid misdiagnosis of dermatofibrosarcoma protuberans [111]. Syndecan-1, a heparan sulfate proteoglycan, is a principal component of the ECM and mediates cell adhesion and migration. Syndecan-1 on the cell surface restricts cell migration and enhances the binding force between cells and the ECM. The loss of expression of syndecan-1 is a marker of the transformation of epithelial cells into mesenchymal cells [112]. Knockdown of Syndecan-1 resulted in a markedly diminished proliferation of keloid fibroblasts and reduced expression of components of the extracellular matrix [113].
Several biomolecules such as forkhead box F2 (FOXF2), HIF-1α and TSG-6 are expected to be potential targets for keloid treatment (Table 2). Stevenson et al. performed transcriptome and metabolome sequencing analysis of keloid promoters and observed significant differences in both methylation and expression of FOXF2 in keloids, and knockdown of the transcriptional regulator FOXF2 significantly suppressed the expression of extracellular matrix-related genes and collagen I production [114]. Keloid fibroblasts treated with inhibitors targeting HSP90 exhibited a dose-dependent reduction in proliferative capacity and cell migration, and a similar phenomenon was observed with interference with HSP70 expression [76, 115, 116]. Tan et al. found that the expression level of TSG-6 within keloid lesions was significantly reduced compared to the dermis of normal skin [117], and Li et al. induced growth inhibition and G2/M phase block of keloid fibroblasts and activation of the mitochondrial apoptotic pathway by lentiviral transfection of TSG-6 in keloids [118, 119]. Hypoxia triggers HIF-1α protein accumulation and EMT processes in keloids [104, 120]. Hyperbaric oxygen therapy, which increases tissue oxygenation, alleviates the inflammatory process, regulates HIF-1α expression and reduces the recurrence rate of keloids after surgical excision and radiotherapy [121]. 2-Methoxyestradiol, which aims at HIF-1α, assists keloid fibroblasts to increase their sensitivity to radiotherapy [122]. Resveratrol, by targeting HIF-1α, reverses the effects of hypoxia on keloid and induces the onset of apoptosis [123].
Table 2.
Potential proteomic biomarkers in keloids
| Biomarker | Abbreviation | Involved physiological function | Expression in keloids | Associated biological processes in keloids | Role in keloids | Reference |
|---|---|---|---|---|---|---|
| Forkhead Box F2 | FOXF2 | Cell proliferation Cell invasion |
Up-regulated | Maintenance of extracellular matrix-related gene expression | Involvement in the pathogenesis of keloids Potential therapeutic target |
[114] |
| Hypoxia inducible factor 1 subunit alpha | HIF-1α | Energy metabolism Angiogenesis |
Up-regulated | Metabolic adaptation to hypoxia | Involvement in the pathogenesis of keloids | [93, 120] |
| Heat shock protein 70 | HSP70 | Correct folding of proteins | Up-regulated | HSP70 knockdown decreases collagen production in KFB | Potential therapeutic target | [76, 115] |
| Heat shock protein 90 | HSP90 | Cell cycle control Signal transduction |
Up-regulated | Regulation of apoptosis, proliferation and migration of fibroblasts Heat shock protein 90 inhibitor induces apoptosis and reduces cell migration in keloid fibroblasts |
Potential therapeutic target | [116] |
| High-temperature requirement A serine peptidase 1 | HTRA1 | Cell growth | Up-regulated | Acceleration of cell proliferation Remodeling of keloid-specific ECM |
Involvement in the pathogenesis of keloids | [56] |
| Matrix metalloproteinase 1 | MMP-1 | Breakdown of ECM | Up-regulated | Regulation of fibroblast migration Regulation of matrix metalloproteinase function |
Involvement in the pathogenesis of keloids | [68] |
| Syndecan-1 | SDC1 | Cell binding Cell signaling Cytoskeletal organization |
Up-regulated | Matrix synthesis | Potential molecular diagnostic biomarker | [111, 113] |
| Transforming growth factor β-1 | TGF-β1 | Cell growth Cell differentiation |
Up-regulated | Collagen synthesis Fibroproliferation |
Involvement in the pathogenesis of keloids | [38, 71] |
| TNF-α-stimulated Gene-6 | TSG-6 | ECM stability Cell migration |
Down-regulated | Induction of apoptosis in KFB Remodeling of the extracellular matrix Inflammation |
Potential therapeutic target | [117–119] |
KFB keloid fibroblast cells, EMT endothelial–mesenchymal transition, ECM extracellular matrix
Epigenetic signatures
miRNAs
Ubiquitously expressed in eukaryotes, miRNAs are a class of endogenous single-stranded RNAs with a length of ~21–23 nucleotides, characterized by fine regulation of oncogenes and their specific expression in tissue [124, 125]. miRNAs (Table 3) have been proven to foster or curb keloid progression by influencing cell proliferation and invasion, EMT processes, ECM deposition and classical signaling pathway transduction [126–138].
Table 3.
Abnormal expression and biological processes of tumor-associated microRNAs in keloids
| miRNA | miRNA expression | Potential value | Target genes | Main associated tumors | References |
|---|---|---|---|---|---|
| miR-1-3p | Down-regulated | Induced cell apoptosis Suppression of proliferation and migration |
TM4SF1 | Oral squamous cell carcinoma Prostate cancer |
[126] |
| miR-1224-5p | Down-regulated | Suppressed cell proliferation, migration and invasion | SMAD3 | Rectal cancer Lung cancer |
[150] |
| miR-124-3p | Down-regulated | Promoted cell apoptosis Inhibited fibroblast-induced angiogenesis Inhibited proliferation and migration |
TGF-βR1 SMAD5 |
Prostate cancer Gastric cancer Colorectal cancer |
[165, 166] |
| miR-133a-3p | Down-regulated | Inhibited fibrosis and proliferation | IRF5 | Colorectal cancer Prostate cancer |
[127] |
| miR-138-5p | Down-regulated | Induced cell apoptosis | CDK6 | Colorectal cancer Breast cancer |
[143] |
| miR-141-3p | Down-regulated | Induced cell apoptosis Suppression of proliferation and migration |
GAB1 | Rectal cancer Renal cell carcinoma |
[128] |
| miR-152-3p | Up-regulated | Increased cell proliferation and invasion Increased type I collagen, type III collagen and fibronectin production |
FOXF1 | Colorectal cancer Prostate cancer |
[129] |
| miR-152-5p | Down-regulated | Inhibited proliferation Reduced migration Promoted apoptosis |
SMAD3 | Liver cancer Gastric cancer |
[151] |
| miR-181a | Up-regulated | Inhibited apoptosis Enhanced keloid fibroblast DNA synthesis and proliferation |
PHLPP2 | Lung cancer Ovarian cancer |
[142] |
| miR-188-5p | Down-regulated | Inhibited cell proliferation Suppressed DNA synthesis Suppression of migration and invasion |
MMP-2 MMP-9 |
Breast cancer Gastric cancer |
[130] |
| miR-194-3p | Down-regulated | Inhibited proliferation and migration | RUNX2 | Breast cancer | [131] |
| miR-194-5p | Down-regulated | Inhibited the aggressive phenotypes of keloid fibroblasts | NR2F2 | Pancreatic cancer Colorectal cancer |
[132] |
| miR-196a | Down-regulated | Inhibited expression of type I and III collagens | COLIα1 COLIIIα3 |
Pancreatic cancer Breast cancer |
[133] |
| miR-196b-5p | Down-regulated | Suppressed cell viability, migration and extracellular matrix production | FGF2 | Non-small cell lung cancer Breast cancer |
[134] |
| miR-199a-5p | Down-regulated | Regulation of cell cycle Restrained proliferation |
N/A | Thyroid cancer Lung cancer |
[139] |
| miR-200c | Down-regulated | Suppressed autocrine secretion of TGF-β2 | ZNF217 | Ovarian cancer Breast cancer |
[153] |
| miR-203 | Down-regulated | Induced apoptosis Suppressed proliferation, migration, invasion and ECM production |
SMAD5 EGR1 FGF2 |
Prostate cancer Ovarian cancer |
[135, 136] |
| miR-204 | Down-regulated | N/A | ITGβ5 | Gastric cancer Hepatocellular cancer |
[167] |
| miR-205 | Down-regulated | Induced cell apoptosis Suppression of proliferation and invasion |
N/A | Thyroid cancer Cervical cancer |
[168] |
| miR-205-5p | Down-regulated | Inhibited glycolysis Accelerated apoptosis Inhibited proliferation, migration, invasion and ECM accumulation |
FOXM1 VEGF |
Endometrial cancer Breast cancer |
[149] |
| miR-21 | Up-regulated | Inhibited activation of the caspase-8 egulation of mitochondria-mediated apoptotic signaling pathway Promoted cell proliferation Promoted fibrosis |
FasL SMAD7 PTEN |
Colon cancer Lung cancer |
[28, 29, 154] |
| miR-214-5p | Down-regulated | Induced cell apoptosis Suppression of proliferation and migration |
TM4SF1 | Esophageal cancer Prostate cancer |
[126] |
| miR-21-5p | Up-regulated | Stemness Epithelial–mesenchymal transition |
PTEN | Gastric cancer Lung cancer |
[99] |
| miR-217 | Down-regulated | Inhibited cell proliferation Induced apoptosis |
FN | Prostate cancer Colon cancer |
[144] |
| miR-2392 | Down-regulated | Regulation of epithelial–mesenchymal transition and autophagy | ZEB2 | Gastric cancer | [155] |
| miR-28-5p | Up-regulated | Promoted ECM deposition and cell proliferation | MC1R | Prostate cancer Colorectal cancer |
[164] |
| miR-29a | Down-regulated | Inhibited viability, proliferation, migration and invasion | COLIα1 | Cervical cancer Breast cancer |
[145] |
| miR-30a-5p | Down-regulated | Induced cell apoptosis | BCL2 | Colon cancer Ewing tumor |
[12] |
| miR-31 | Up-regulated | Regulation of cell cycle Restrained apoptosis |
HIF-1α | Lung cancer Breast cancer |
[141] |
| miR-3141 | Down-regulated | Suppressed keloid fibroblast proliferation and migration Promoted cell apoptosis |
SAMD3 | Osteosarcoma | [146] |
| miR-370-3p | Up-regulated | N/A | EGFR | Melanoma Breast cancer |
[167] |
| miR-424-3p | Up-regulated | Enhanced the ability of cell proliferation, migration and collagen secretion Reduced apoptosis |
SMAD7 | Ovarian cancer | [147] |
| miR-4328 | Down-regulated | Induced cell apoptosis Suppressed proliferation and metastasis |
BCL2 | Lung cancer | [137] |
| miR-4417 | Down-regulated | Induced cell apoptosis Suppression of migration and invasion |
CCND1 | Prostate cancer Triple-negative breast cancer |
[138] |
| miR-637 | Down-regulated | Suppressed proliferation and metastasis | SMAD3 | Breast cancer Prostate cancer |
[152] |
| miR-7-5p | Down-regulated | Repressed proliferation, migration and extracellular matrix deposition Promoted cell apoptosis |
EPAC1 | Thyroid cancer Glioma |
[171] |
| miR-769-5p | Down-regulated | Inhibited proliferation, migration and invasion Suppressed extracellular matrix deposition |
EIF3A | Pancreatic cancer Prostate cancer |
[163] |
| miR-96 | Up-regulated | Increased type I and III collagen production | SMAD7 | Breast cancer Urothelial carcinoma |
[148] |
N/A not available, miR microRNA,TM4SF1 transmembrane 4 L six family member 1, SMAD3 SMAD family member 3, TGF-βR1 transforming growth factor beta receptor 1, IRF5 interferon regulatory factor 5, CDK6 cyclin-dependent kinase 6, GAB1 growth factor receptor-bound protein 2-associated binding protein 1, FOXF1 forkhead box F1, PHLPP2 PH domain and leucine-rich repeat protein phosphatase 2, MMP-2 matrix metallopeptidase 2, RUNX2 RUNX family transcription factor 2, NR2F2 nuclear receptor subfamily 2 group F member 2, COLIα1 collagen type I alpha 1 chain, FGF2 fibroblast growth factor 2, ZNF217 zinc finger protein 217, EGR1 early growth response 1, ITGβ5 integrin subunit beta 5, VEGF vascular endothelial growth factor, FasL Fas ligand, PTEN phosphatase and tensin homolog, FN fibronectin, ZEB2 zinc finger E-box binding homeobox 2, MC1R melanocortin 1 receptor, BCL2 B-cell lymphoma-2, HIF-1α hypoxia inducible factor 1 subunit alpha inhibitor, EGFR epidermal growth factor receptor, CCND1 cyclinD1, EPAC1 exchange protein directly activated by CAMP 1, EIF3A eukaryotic translation Initiation factor 3 subunit A
Downregulation of miR-199a-5p, miR-203, miR-217, miR-29a, miR-3141 and miR-138-5p and upregulation of miR-181a, miR-31, miR-96 and miR-424-3p are instrumental in the regulation of specific target genes involved in cell cycle regulation and strengthen the viability, proliferation, invasiveness and resistance to apoptosis of fibroblasts or keratinocytes [136, 139–148]. miR-205-5p regulates FOXM1, a transcription activator of cell proliferation, to influence the proliferation, glucose consumption, lactic acid production and other glycolysis processes of fibroblasts and participates in the PI3K/Akt pathway to impede cell migration [149]. Multiple miRNAs, including miR-1224-5p, miR-152-5p and miR-637, have been determined to act on members of the SMAD family and the TGFβ-Smad pathway, thereby manipulating the production of type I collagen, type III collagen and fibronectin, as well as fibroblast-induced vascular production [150–152]. In addition, miR-200c adjusts the expression of the protein-coding gene ZNF217 and controls the autocrine activity of TGF-β2 [153].
Fas ligand (FasL) is expressed on the surface and in the cytoplasm of a variety of tumor cells, and miR-21 has been shown to block mitochondrial-mediated apoptosis by inhibiting FasL activation [154]. miR-21-5p modulates the expression of vimentin in keratinocytes, induces the EMT phenotype and reinforces cell cloning ability and stemness [99]. miR-2392 is vastly downregulated in keloids and has an interaction site with zinc finger E-box binding homeobox 2 (ZEB2); silencing ZEB2 can reverse the effects of the miR-2392 inhibitor on apoptosis, invasion and EMT processes [155].
Long noncoding RNAs (IncRNAs)
As a class of RNA molecules longer than 200 nucleotides, lncRNAs are regulatory molecules in biological processes, including gene expression, gene imprinting, cell division, cell differentiation and plasmatic nuclear transfer [156, 157]. lncRNAs can be employed as cis- or trans-regulators to regulate histone modification, chromatin remodeling and RNA metabolism at the epigenetic level [158]. High-throughput sequencing of tumor tissues and normal tissues shows that abnormal expression of various lncRNAs is closely linked to the occurrence and development of tumors [159, 160]. lncRNAs may be involved in almost all human cancers, and their expression is correlated with the prognosis and metastasis of tumors. Additionally, the secondary structure established by the combination of lncRNAs and specific proteins is anticipated to be an important means to intervene in tumor occurrence and growth [161].
lncRNA DLEU2, lncRNA H19 and lncRNA LINC01116 have been described to act as specific miRNA sponges (Table 4), promoting the proliferation of fibroblasts and enhancing cell migration [136, 162, 163]. lncRNA LINC00937 hinders cell proliferation by the miR-28-5p/MC1R axis in keloids, and its expression is inhibited in fibroblasts [164]. lncRNA HOXA11-AS is upregulated in keloids, and miR-124-3p, as its downstream effector molecule, engages in TGFβR1-mediated angiogenesis and SMAD5-mediated collagen synthesis [165, 166]. lncRNA HOXA11-AS has been demonstrated to support the proliferation of fibroblasts and glycolysis through the miR-205-5p/FOXM1 axis [149]. The expression of lncRNA ATB is positively associated with the self-secretion level of TGF-β2, which is accomplished by downregulating tumor-suppressive miR-200c and subsequently acting on zinc finger protein 217 (ZNF217), an agitator associated with TGF-β [153]. In keloid RNA sequencing and miRNA sequencing, a total of 319 lncRNAs were identified, and two pairs of competing endogenous RNA networks regulating the actin cytoskeleton were constructed: lnc-GLB1L-1/miR-370-3p/EGFR and lnc-CASP9–3/miR-204/ITGB5 [167].
Table 4.
Overview of tumor-associated long non-coding RNAs in keloid scars
| lncRNA | lncRNA expression | Potential value | Target miRNA | Main associated tumors | References |
|---|---|---|---|---|---|
| lncRNA ATB | Up-regulated | Increased autocrine secretion of TGF-β2 | miR-200c | Gastric cancer Colorectal cancer |
[153] |
| lncRNA CACNA1G-AS1 | Up-regulated | Promoted proliferation and invasion Suppressed apoptosis |
miR-205 | Ovarian cancer Colorectal cancer |
[168] |
| lncRNA DLEU2 | Up-regulated | Promoted proliferation and differentiation Suppressed apoptosis |
miR-30b-5p miR-30a-5p |
Pancreatic cancer Lipoma |
[162] |
| lncRNA H19 | Up-regulated | Intensified migration and invasion Increased extracellular matrix deposition |
miR-769-5p miR-29a |
Breast cancer Colorectal cancer |
[145, 163] |
| lncRNA HOXA11-AS | Up-regulated | Promoted fibroblast-induced angiogenesis Promoted glycolysis Inhibited apoptosis Intensified migration and invasion Promoted proliferation and ECM accumulation |
miR-205-5p miR-124-3p |
Glioma Gastric cancer |
[149, 165] |
| lncRNA LINC00937 | Down-regulated | Repressed extracellular matrix deposition Suppressed cell proliferation |
miR-28-5p | Cutaneous melanoma | [164] |
| lncRNA LINC01116 | Up-regulated | Intensified migration and invasion Promoted proliferation and ECM accumulation Suppressed apoptosis |
miR-203 miR-3141 |
Prostate cancer | [136, 146] |
miR microRNA, lncRNA long non-coding RNA, TGF-β2 transforming growth factor beta 2, ECM extracellular matrix
Almost all of these lncRNAs with abnormal expression in keloids have been extensively scrutinized in tumors, and their mechanisms have been confirmed in multiple cancers [160, 161, 168]. It should be acknowledged that there is still a large number of lncRNAs identified in tumors or fibrotic diseases that have not been evaluated in keloids. Furthermore, some lncRNAs have prognostic and diagnostic value in tumors [160, 161], although there is still a lack of relevant studies on whether lncRNAs have similar value in keloids.
Circular RNAs
Due to their special 3′ end and 5′ end covalently linked structure, circular RNAs (circRNAs) can act as miRNA sponges and competitively inhibit miRNA binding to target gene mRNAs. circRNAs are expressed in a variety of physiological and pathological processes and have predictive significance for the screening and prognosis of a variety of tumors [169, 170]. Considering the high stability and low off-target ability of circRNAs, the design of artificial sponges aimed at miRNAs in specific diseases is a promising novel future direction for the advancement of targeted drugs.
circRNA_101238 is shown to be involved in the regulation of cyclins as a competitive endogenous RNA [143]. circCOL5A1 is upregulated in keloids and promotes fibroblast proliferation and collagen synthesis. RNA fluorescence in situ hybridization suggested that this effect was accomplished by adjusting the release of Epac1 by circCOL5A1 as a sponge of miR-7-5p [171]. Likewise, circNRIP1 is involved in the proliferation and apoptosis of tumor cells in breast and gastric cancers and is highly expressed in human keloids. circNRIP1 blocks the ubiquitination of FXR1, a key molecule of miR-503 maturation, by binding to it [172]. The elevation of circNRIP1 is ultimately accompanied by an increase in miR-503, which has been shown to escalate extracellular matrix deposition and promote cell division and differentiation (Table 5).
Table 5.
Tumor-associated circular RNAs abnormally expressed in keloid scars
| circRNA | circRNA expression | Potential value | Target miRNAs | Main associated tumors | References |
|---|---|---|---|---|---|
| circRNA COL5A1 | Up-regulated | Promoted proliferation, migration and ECM deposition Inhibited cell apoptosis |
miR-7-5p | Renal Cell Carcinoma | [171] |
| circRNA NRIP1 | Up-regulated | Promoted proliferation Increased expression of ECM-associated proteins Suppressed apoptosis |
miR-503-3p miR-503-5p |
Lung cancer Gastric cancer |
[172] |
| circRNA_0008259 | Down-regulated | Inhibited type I and III collagen expression | N/A | Gastric cancer | [174] |
miRNA microRNA, circRNA circular RNA, ECM extracellular matrix, N/A not available
Studies on circRNAs have been combined with biological information to predict potential functions and screen target miRNAs based on sequence information. A circRNA microarray analysis of keloids identified 76 significantly differentially expressed circRNAs and corresponding specifically bound miRNAs. For instance, circRNA_0043688 may have adsorption effects on miRNA-942-5p, miRNA-3177-3p and miRNA-5010-5p [173]. Until now, there have been few studies on circRNAs in keloids, and further experimental studies are necessary to evaluate circRNA biomolecules that are expected to be utilized in the clinical diagnosis and treatment of keloids [174].
DNA methylation
As an essential epigenetic process in eukaryotes, DNA methylation modifies chromatin structure and gene expression by establishing, maintaining and removing methyl groups [175]. Methylation of different components has dissimilar effects on gene expression, and DNA methylation of promoters constrains gene expression. Conversely, high methylation of the silencer is positively related to gene expression. Undisciplined DNA methyltransferase and hypermethylation of normal nonmethylated CpG islands are chief mechanisms of genomic DNA modification to induce tumors [176].
Of the 450,000 cytosine sites scanned in keloids, 37% of differentially methylated genes were hypermethylated, and 63% were hypomethylated, such as MMP3 and asporin [177, 178]. CDC2L1 promoters had a higher methylation rate, up to 50%, compared with 0% in normal skin tissue. This increased methylation rate is associated with a higher fibroblast growth rate and impedes the expression of the apoptosis-related protein cyclin-dependent kinase (CDK)11p58 [179]. Hypermethylation of the secreted frizzled-related protein 1 (SFRP1) promoter in keloids and epigenetic silencing of SFRP1 lead to mitigated inhibition of the Wnt/β-catenin signaling pathway [180], which participates in proliferation, invasion, fibrosis and EMT processes in a variety of tumors and keloids (Table 6).
Table 6.
Mechanism and clinical value of DNA methylation in keloids
| DNA methylation | Alteration | Potential value | Downstream gene or pathway | Main associated tumors | References |
|---|---|---|---|---|---|
| CDC2L1 | Hypermethylation | Reduced apoptosis | CDK11-p58 | Melanoma Neuroblastoma |
[179] |
| SFRP1 | Hypermethylation | Increased protein expression of α-SMA | Wnt/β-catenin pathway | Prostate cancer Breast cancer |
[180] |
| HOXA9 | Hypermethylation | A component of the tumorigenic phenotype of keloids | N/A | Leukemia Ovarian cancer |
[178] |
| MMP3 | Hypomethylation | Promoted proliferation | N/A | Esophageal cancer Melanoma |
[178] |
| ASPN | Hypomethylation | Collagen binding | N/A | Colon cancer Ductal breast carcinoma |
[178] |
α-SMΑ alpha-smooth muscle actin, N/A not available, MMP metalloprotease, CDC2L1 cell division cycle 2-like 1, SFRP1 secreted frizzled-related protein 1, HOXA9 homeobox A9, ASPN asporin, CDK11 cyclin-dependent kinase 11, N/A not available
DNA methylation can be detected directly in blood and body fluids, and diagnostic models based on the methylation levels of multiple genes can accurately and noninvasively screen for tumors [181, 182]. Moreover, due to the reversibility of DNA methylation, the hypothesis of curbing tumor growth by reorganizing the levels of DNA methylation without changing the gene sequence, reactivating tumor suppression genes or silencing oncogenes has been in the experimental stage of intervention in T-cell lymphoma, colon cancer and rectal cancer [183, 184]. Therefore, this approach is expected to become an auxiliary treatment for keloids when they mature.
Conclusions
Maturing molecular biology and burgeoning experimental detection technology have promoted the in-depth investigation of the pathological mechanism of keloids. Epigenetic signatures and liquid biopsy may have greater progression potential. Since the former do not require alterations in the gene sequence, their reversibility and tissue specificity make them more ideal therapeutic targets. Liquid biopsy is noninvasive and accurate, it is more suitable for the early screening of people with keloids. It is worth mentioning that serum levels of sHLA-E can be used as a predictive biomarker and potential therapeutic target for keloids due to its high sensitivity and specificity, but considering the ethnic specificity and genetic predisposition of keloids, more large-sample, multicenter studies are needed to determine the prevalence of sHLA-E. In addition, current studies on keloids mostly concentrate on a single molecule. Whether the sensitivity of diagnosis can be upgraded by integrating multiple biomarkers remains to be clarified in future studies.
Abbreviations
- CCND1: CyclinD1; CDC2L1: Cell division cycle 2-like 1; CDK11: Cyclin-dependent kinase 11; circRNA: Circular RNA; ECM: Extracellular matrix; EGFR: Epidermal growth factor receptor; EMT: Endothelial–mesenchymal transition; FasL: Fas ligand; FGF2: Fibroblast growth factor 2; FOXF1: Forkhead box F1; HIF1AN: Hypoxia inducible factor 1 subunit alpha inhibitor; HOXA9:
Homeobox A9; HSP: Heat shock protein; IL-6: Interleukin 6; MMP-2: Matrix metallopeptidase 2; miRNA: MicroRNA; OPN: Osteopontin; PTEN: Phosphatase and tensin homolog; SFRP1: Secreted frizzled-related protein 1; α-SMΑ: α-Smooth muscle actin; TGFβ: Transforming growth factor beta; VEGF: Vascular endothelial growth factor; ZEB2: Zinc finger E-box binding homeobox 2; ZNF217: Zinc finger protein 217.
Authors’ contributions
XYJ and WYB conceived and wrote the manuscript. SMJ and HY reviewed the manuscript. SMJ, LH and LZY checked the manuscript. HY and CQ provided useful discussions. All authors contributed to the article and approved the submitted version.
Contributor Information
Yijun Xia, Department of Plastic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China.
Youbin Wang, Department of Plastic Surgery, Peking Union Medical College Hospital, Beijing, China.
Mengjie Shan, Department of Plastic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China.
Yan Hao, Department of Plastic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China.
Hao Liu, Department of Plastic Surgery, Peking Union Medical College Hospital, Beijing, China.
Qiao Chen, Department of Plastic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China.
Zhengyun Liang, Department of Plastic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China.
Funding
This study was supported by The National Natural Science Foundation of China (81871538) and Beijing Municipal Commission of Science and Technology (Z191100006619009).
Conflicts of interest
None declared.
References
- 1. Tan S, Khumalo N, Bayat A. Understanding keloid pathobiology from a quasi-neoplastic perspective: less of a scar and more of a chronic inflammatory disease with cancer-like tendencies. Front Immunol. 2019;10:1810. 10.3389/fimmu.2019.01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ogawa R. Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis. Int J Mol Sci. 2017;18:606. 10.3390/ijms18030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macarak EJ, Wermuth PJ, Rosenbloom J, Uitto J. Keloid disorder: fibroblast differentiation and gene expression profile in fibrotic skin diseases. Exp Dermatol. 2021;30:132–45. [DOI] [PubMed] [Google Scholar]
- 4. Al-Attar A, Mess S, Thomassen JM, Kauffman CL, Davison SP. Keloid pathogenesis and treatment. Plast Reconstr Surg. 2006;117:286–300. [DOI] [PubMed] [Google Scholar]
- 5. Park TH, Park JH, Tirgan MH, Halim AS, Chang CH. Clinical implications of single- versus multiple-site keloid disorder: a retrospective study in an Asian population. Ann Plast Surg. 2015;74:248–51. [DOI] [PubMed] [Google Scholar]
- 6. Ogawa R. Keloid and hypertrophic scarring may result from a mechanoreceptor or mechanosensitive nociceptor disorder. Med Hypotheses. 2008;71:493–500. [DOI] [PubMed] [Google Scholar]
- 7. Butler PD, Longaker MT, Yang GP. Current progress in keloid research and treatment. J Am Coll Surg. 2008;206:731–41. [DOI] [PubMed] [Google Scholar]
- 8. Arno AI, Amini-Nik S, Blit PH, Al-Shehab M, Belo C, Herer E, et al. Effect of human Wharton's jelly mesenchymal stem cell paracrine signaling on keloid fibroblasts. Stem Cells Transl Med. 2014;3:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hahn JM, McFarland KL, Combs KA, Supp DM. Partial epithelial-mesenchymal transition in keloid scars: regulation of keloid keratinocyte gene expression by transforming growth factor-β1. Burns Trauma. 2016;4:30. 10.1186/s41038-016-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lim KH, Itinteang T, Davis PF, Tan ST. Stem cells in keloid lesions: a review. Plast Reconstr Surg Glob Open. 2019;7:e2228. 10.1097/GOX.0000000000002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawarazaki A, Horinaka M, Yasuda S, Numajiri T, Nishino K, Sakai T. Sulforaphane suppresses cell growth and collagen expression of keloid fibroblasts. Wound Repair Regen. 2017;25:224–33. [DOI] [PubMed] [Google Scholar]
- 12. Jian X, Qu L, Wang Y, Zou Q, Zhao Q, Chen S, Gao X, Chen H, He C. Trichostatin A-induced miR-30a-5p regulates apoptosis and proliferation of keloid fibroblasts via targeting BCL2. Mol Med Rep. 2019;19:5251–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi K, Qiu X, Zheng W, Yan D, Peng W. MiR-203 regulates keloid fibroblast proliferation, invasion, and extracellular matrix expression by targeting EGR1 and FGF2. Biomed Pharmacother. 2018;108:1282–8. [DOI] [PubMed] [Google Scholar]
- 14. Wentzensen N, Clarke MA. Liquid biopsy for cancer detection: clinical and epidemiologic considerations. Clin Cancer Res. 2021;27:5733–5. [DOI] [PubMed] [Google Scholar]
- 15. Keup C, Suryaprakash V, Hauch S, Storbeck M, Hahn P, Sprenger-Haussels M, et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. Genome Med. 2021;13:85. 10.1186/s13073-021-00902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409–24. [DOI] [PubMed] [Google Scholar]
- 17. Xu H, Zhu Z, Hu J, Sun J, Wo Y, Wang X, et al. Downregulated cytotoxic CD8(+) T-cell identifies with the NKG2A-soluble HLA-E axis as a predictive biomarker and potential therapeutic target in keloids. Cell Mol Immunol. 2022;19:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen H, Chengalvala V, Hu H, Sun D. Tumor-derived exosomes: Nanovesicles made by cancer cells to promote cancer metastasis. Acta Pharm Sin B. 2021;11:2136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11:2783–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M, Jiang M, Meng J, Tao L. Exosomes: carriers of pro-fibrotic signals and therapeutic targets in fibrosis. Curr Pharm Des. 2019;25:4496–509. [DOI] [PubMed] [Google Scholar]
- 21. Qin XJ, Zhang JX, Wang RL. Exosomes as mediators and biomarkers in fibrosis. Biomark Med. 2020;14:697–712. [DOI] [PubMed] [Google Scholar]
- 22. Lin F, Zeng Z, Song Y, Li L, Wu Z, Zhang X, et al. YBX-1 mediated sorting of miR-133 into hypoxia/reoxygenation-induced EPC-derived exosomes to increase fibroblast angiogenesis and MEndoT. Stem Cell Res Ther. 2019;10:263. 10.1186/s13287-019-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ringuette Goulet C, Bernard G, Tremblay S, Chabaud S, Bolduc S, Pouliot F. Exosomes induce fibroblast differentiation into cancer-associated fibroblasts through TGFβ Signaling. Molecular Cancer Research: MCR. 2018;16:1196–204. [DOI] [PubMed] [Google Scholar]
- 24. Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–30. [DOI] [PubMed] [Google Scholar]
- 25. Xie F, Wen G, Sun W, Jiang K, Chen T, Chen S, et al. Mechanical stress promotes angiogenesis through fibroblast exosomes. Biochem Biophys Res Commun. 2020;533:346–53. [DOI] [PubMed] [Google Scholar]
- 26. He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: biology and translational medicine. Theranostics. 2018;8:237–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sheridan C. Exosome cancer diagnostic reaches market. Nat Biotechnol. 2016;34:359–60. [DOI] [PubMed] [Google Scholar]
- 28. Li Q, Fang L, Chen J, Zhou S, Zhou K, Cheng F, et al. Exosomale microRNA-21 promotes keloid fibroblast proliferation and collagen production by inhibiting Smad7. J Burn Care Res. 2021;42:1266–74. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Zhang J, Lei Y, Lyu L, Zuo R, Chen T. MicroRNA-21 in skin fibrosis: potential for diagnosis and treatment. Mol Diagn Ther. 2017;21:633–42. [DOI] [PubMed] [Google Scholar]
- 30. Aheget H, Tristán-Manzano M, Mazini L, Cortijo-Gutierrez M, Galindo-Moreno P, Herrera C, Martin F, Marchal JA, Benabdellah K: Exosome: A new player in translational Nanomedicine. J Clin Med 2020;9:2380. 10.3390/jcm9082380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang L, Zhang Y, Liu T, Wang X, Wang H, Song H, et al. Exosomes derived from TSG-6 modified mesenchymal stromal cells attenuate scar formation during wound healing. Biochimie. 2020;177:40–9. [DOI] [PubMed] [Google Scholar]
- 32. Hu S, Li Z, Cores J, Huang K, Su T, Dinh P-U, et al. Needle-free injection of exosomes derived from human dermal fibroblast spheroids ameliorates skin Photoaging. ACS Nano. 2019;13:11273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marneros AG, Norris JEC, Watanabe S, Reichenberger E, Olsen BR. Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. J Invest Dermatol. 2004;122:1126–32. [DOI] [PubMed] [Google Scholar]
- 34. Roovers RC, Laeremans T, Huang L, De Taeye S, Verkleij AJ, Revets H, et al. Van Bergen en Henegouwen PMP: efficient inhibition of EGFR signaling and of tumour growth by antagonistic anti-EFGR Nanobodies. Cancer Immunology, Immunotherapy: CII. 2007;56:303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chin GS, Liu W, Steinbrech D, Hsu M, Levinson H, Longaker MT. Cellular signaling by tyrosine phosphorylation in keloid and normal human dermal fibroblasts. Plast Reconstr Surg. 2000;106:1532–40. [DOI] [PubMed] [Google Scholar]
- 36. Tsai C-H, Ogawa R. Keloid research: current status and future directions. Scars, Burns & Healing. 2019;5:2059513119868659. 10.1177/2059513119868659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dmytrzak A, Boroń A, Łoniewska B, Lewandowska K, Gorący I, Kaczmarczyk M, et al. Two functional genetic variants and predisposition to keloid scarring in Caucasians. Dermatol Res Pract. 2019;2019:6179063. 10.1155/2019/6179063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiao H, Dong P, Yan L, Yang Z, Lv X, Li Q, et al. TGF-β1 induces Polypyrimidine tract-binding protein to Alter fibroblasts proliferation and fibronectin deposition in keloid. Sci Rep. 2016;6:38033. 10.1038/srep38033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saed GM, Ladin D, Olson J, Han X, Hou Z, Fivenson D. Analysis of p53 gene mutations in keloids using polymerase chain reaction-based single-strand conformational polymorphism and DNA sequencing. Arch Dermatol. 1998;134:963–7. [DOI] [PubMed] [Google Scholar]
- 40. Liu Y-b, Gao J-h, Duan H-j, Liu X-j. Investigation of p53 gene mutations in keloids using PCR-SSCP. Chin J Burns. 2003;19:258–60. [PubMed] [Google Scholar]
- 41. Nyika DT, Khumalo N, Bayat A. Genetics and epigenetics of keloids. Adv Wound Care. 2022;11:192–201. [DOI] [PubMed] [Google Scholar]
- 42. Brown JJ, Ollier WER, Thomson W, Bayat A. Positive association of HLA-DRB1*15 with keloid disease in Caucasians. Int J Immunogenet. 2008;35:303–7. [DOI] [PubMed] [Google Scholar]
- 43. Lu W-S, Wang J-F, Yang S, Xiao F-L, Quan C, Cheng H, et al. Association of HLA-DQA1 and DQB1 alleles with keloids in Chinese Hans. J Dermatol Sci. 2008;52:108–17. [DOI] [PubMed] [Google Scholar]
- 44. Cao ZD, Liu W. Examining the pathogenesis and therapeutic strategy of keloids from the perspective of systemic inflammation. Chin J Burns. 2020;36:334–8. [DOI] [PubMed] [Google Scholar]
- 45. Ogawa R, Akaishi S. Endothelial dysfunction may play a key role in keloid and hypertrophic scar pathogenesis - keloids and hypertrophic scars may be vascular disorders. Med Hypotheses. 2016;96:51–60. [DOI] [PubMed] [Google Scholar]
- 46. Suarez E, Syed F, Alonso-Rasgado T, Bayat A. Identification of biomarkers involved in differential profiling of hypertrophic and keloid scars versus normal skin. Arch Dermatol Res. 2015;307:115–33. [DOI] [PubMed] [Google Scholar]
- 47. Sayah DN, Soo C, Shaw WW, Watson J, Messadi D, Longaker MT, et al. Downregulation of apoptosis-related genes in keloid tissues. J Surg Res. 1999;87:209–16. [DOI] [PubMed] [Google Scholar]
- 48. Teofoli P, Barduagni S, Ribuffo M, Campanella A, De Pita O, Puddu P. Expression of Bcl-2, p53, c-Jun and c-fos protooncogenes in keloids and hypertrophic scars. J Dermatol Sci. 1999;22:31–7. [DOI] [PubMed] [Google Scholar]
- 49. Hu Z, Lou L, Luo S. Experimental study of the expression of c-myc, c-fos and proto-oncogenes on hypertrophic and scars. Chin J Burns. 2002;18:165–7. [PubMed] [Google Scholar]
- 50. De Felice B, Ciarmiello LF, Mondola P, Damiano S, Seru R, Argenziano C, et al. Differential p63 and p53 expression in human keloid fibroblasts and hypertrophic scar fibroblasts. DNA Cell Biol. 2007;26:541–7. [DOI] [PubMed] [Google Scholar]
- 51. Chen ZY, Yu XF, Huang JQ, Li DL. The mechanisms of β-catenin on keloid fibroblast cells proliferation and apoptosis. Eur Rev Med Pharmacol Sci. 2018;22:888–95. [DOI] [PubMed] [Google Scholar]
- 52. Igota S, Tosa M, Murakami M, Egawa S, Shimizu H, Hyakusoku H, et al. Identification and characterization of Wnt signaling pathway in keloid pathogenesis. Int J Med Sci. 2013;10:344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu D, Shang Y, Yuan J, Ding S, Luo S, Hao L. Wnt/β-catenin Signaling exacerbates keloid cell proliferation by regulating telomerase. Cell Physiol Biochem. 2016;39:2001–13. [DOI] [PubMed] [Google Scholar]
- 54. Hamburg-Shields E, DiNuoscio GJ, Mullin NK, Lafyatis R, Atit RP. Sustained β-catenin activity in dermal fibroblasts promotes fibrosis by up-regulating expression of extracellular matrix protein-coding genes. J Pathol. 2015;235:686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Canfield AE, Hadfield KD, Rock CF, Wylie EC, Wilkinson FL. HtrA1: a novel regulator of physiological and pathological matrix mineralization? Biochem Soc Trans. 2007;35:669–71. [DOI] [PubMed] [Google Scholar]
- 56. Yamawaki S, Naitoh M, Kubota H, Aya R, Katayama Y, Ishiko T, et al. HtrA1 is specifically up-regulated in active keloid lesions and stimulates keloid development. Int J Mol Sci. 2018;19:1275. 10.3390/ijms19051275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jiao H, Zhang T, Fan J, Xiao R. The superficial dermis may initiate keloid formation: histological analysis of the keloid dermis at different depths. Front Physiol. 2017;8:885. 10.3389/fphys.2017.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin JU, Kim SH, Kim H, Noh JY, Jin S, Park CO, et al. TSLP is a potential initiator of collagen synthesis and an activator of CXCR4/SDF-1 Axis in keloid pathogenesis. J Invest Dermatol. 2016;136:507–15. [DOI] [PubMed] [Google Scholar]
- 59. Campbell CA, Burdick MD, Strieter RM. Systemic Fibrocyte levels and keloid expression of the chemoattractant CXCL12 are upregulated compared with patients with normal scar. Ann Plast Surg. 2021;87:150–5. 10.1097/SAP.0000000000002929. [DOI] [PubMed] [Google Scholar]
- 60. Zhang M, Xu Y, Liu Y, Cheng Y, Zhao P, Liu H, et al. Chemokine-like factor 1 (CKLF-1) is overexpressed in keloid patients: a potential indicating factor for keloid-predisposed individuals. Medicine. 2016;95:e3082. 10.1097/MD.0000000000003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dong X, Zhang M, Chen Y, Li C, Wang Y, Jin X. A comparison expression analysis of CXCR4, CXCL9 and Caspase-9 in dermal vascular endothelial cells between keloids and normal skin on chemotaxis and apoptosis. J Plast Surg Hand Surg. 2022;56:93–102. [DOI] [PubMed] [Google Scholar]
- 62. Campbell CA, Burdick MD, Strieter RM. Systemic Fibrocyte levels and keloid expression of the chemoattractant CXCL12 are upregulated compared with patients with normal scar. Ann Plast Surg. 2021;87:150–5. [DOI] [PubMed] [Google Scholar]
- 63. Le Huu D, Matsushita T, Jin G, Hamaguchi Y, Hasegawa M, Takehara K, et al. IL-6 blockade attenuates the development of murine sclerodermatous chronic graft-versus-host disease. J Invest Dermatol. 2012;132:2752–61. [DOI] [PubMed] [Google Scholar]
- 64. Juhl P, Bondesen S, Hawkins CL, Karsdal MA, Bay-Jensen A-C, Davies MJ, et al. Dermal fibroblasts have different extracellular matrix profiles induced by TGF-β, PDGF and IL-6 in a model for skin fibrosis. Sci Rep. 2020;10:17300. 10.1038/s41598-020-74179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luckett LR, Gallucci RM. Interleukin-6 (IL-6) modulates migration and matrix metalloproteinase function in dermal fibroblasts from IL-6KO mice. Br J Dermatol. 2007;156:1163–71. [DOI] [PubMed] [Google Scholar]
- 66. Uchida G, Yoshimura K, Kitano Y, Okazaki M, Harii K. Tretinoin reverses upregulation of matrix metalloproteinase-13 in human keloid-derived fibroblasts. Exp Dermatol. 2003;12:35–42. [DOI] [PubMed] [Google Scholar]
- 67. Imaizumi R, Akasaka Y, Inomata N, Okada E, Ito K, Ishikawa Y, et al. Promoted activation of matrix metalloproteinase (MMP)-2 in keloid fibroblasts and increased expression of MMP-2 in collagen bundle regions: implications for mechanisms of keloid progression. Histopathology. 2009;54:722–30. [DOI] [PubMed] [Google Scholar]
- 68. Fujiwara M, Muragaki Y, Ooshima A. Keloid-derived fibroblasts show increased secretion of factors involved in collagen turnover and depend on matrix metalloproteinase for migration. Br J Dermatol. 2005;153:295–300. [DOI] [PubMed] [Google Scholar]
- 69. Gauglitz GG, Bureik D, Zwicker S, Ruzicka T, Wolf R. The antimicrobial peptides psoriasin (S100A7) and koebnerisin (S100A15) suppress extracellular matrix production and proliferation of human fibroblasts. Skin Pharmacol Physiol. 2015;28:115–23. [DOI] [PubMed] [Google Scholar]
- 70. Kuwatsuka Y, Murota H. Involvement of Periostin in skin function and the pathogenesis of skin diseases. Adv Exp Med Biol. 2019;1132:89–98. [DOI] [PubMed] [Google Scholar]
- 71. Supp DM, Hahn JM, Glaser K, McFarland KL, Boyce ST. Deep and superficial keloid fibroblasts contribute differentially to tissue phenotype in a novel in vivo model of keloid scar. Plast Reconstr Surg. 2012;129:1259–71. [DOI] [PubMed] [Google Scholar]
- 72. Subraman V, Thiyagarajan M, Malathi N, Rajan ST. OPN-revisited. J Clin Diagn Res. 2015;9:ZE10–3. 10.7860/JCDR/2015/12872.6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miragliotta V, Pirone A, Donadio E, Abramo F, Ricciardi MP, Theoret CL. Osteopontin expression in healing wounds of horses and in human keloids. Equine Vet J. 2016;48:72–7. [DOI] [PubMed] [Google Scholar]
- 74. Shevtsov M, Multhoff G. Heat shock protein-peptide and HSP-based immunotherapies for the treatment of cancer. Front Immunol. 2016;7:171. 10.3389/fimmu.2016.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Calderwood SK, Gong J. Heat shock proteins promote cancer: It's a protection racket. Trends Biochem Sci. 2016;41:311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Totan S, Echo A, Yuksel E. Heat shock proteins modulate keloid formation. Eplasty. 2011;11:e21. [PMC free article] [PubMed] [Google Scholar]
- 77. Chen JJ, Zhao S, Cen Y, Liu XX, Yu R, Wu DM. Effect of heat shock protein 47 on collagen accumulation in keloid fibroblast cells. Br J Dermatol. 2007;156:1188–95. [DOI] [PubMed] [Google Scholar]
- 78. Leite de Oliveira R, Hamm A, Mazzone M. Growing tumor vessels: more than one way to skin a cat - implications for angiogenesis targeted cancer therapies. Mol Asp Med. 2011;32:71–87. [DOI] [PubMed] [Google Scholar]
- 79. Cho WC, Jour G, Aung PP. Role of angiogenesis in melanoma progression: update on key angiogenic mechanisms and other associated components. Semin Cancer Biol. 2019;59:175–86. [DOI] [PubMed] [Google Scholar]
- 80. Tanaka R, Umeyama Y, Hagiwara H, Ito-Hirano R, Fujimura S, Mizuno H, et al. Keloid patients have higher peripheral blood endothelial progenitor cell counts and CD34(+) cells with normal vasculogenic and angiogenic function that overexpress vascular endothelial growth factor and interleukin-8. Int J Dermatol. 2019;58:1398–405. [DOI] [PubMed] [Google Scholar]
- 81. Tanaka R, Umeyama Y, Hagiwara H, Ito-Hirano R, Fujimura S, Mizuno H, et al. Keloid patients have higher peripheral blood endothelial progenitor cell counts and CD34 cells with normal vasculogenic and angiogenic function that overexpress vascular endothelial growth factor and interleukin-8. Int J Dermatol. 2019;58:1398–405. [DOI] [PubMed] [Google Scholar]
- 82. Syed F, Bayat A. Notch signaling pathway in keloid disease: enhanced fibroblast activity in a Jagged-1 peptide-dependent manner in lesional vs. extralesional fibroblasts. Wound Repair Regen. 2012;20:688–706. [DOI] [PubMed] [Google Scholar]
- 83. Syed F, Bayat A. Notch signaling pathway in keloid disease: enhanced fibroblast activity in a Jagged-1 peptide-dependent manner in lesional vs. extralesional fibroblasts. Wound Repair Regen. 2012;20:688–706. [DOI] [PubMed] [Google Scholar]
- 84. Jiang D-y, Fu X-b, Chen W, Sun T-z. Relationship of overexpression of angiogenesis factors and their receptors with invasive growth of keloid. Chinese Journal of Plastic Surgery. 2004;20:128–31. [PubMed] [Google Scholar]
- 85. Jiang DY, Fu XB, Chen W, Sun TZ. Relationship of overexpression of angiogenesis factors and their receptors with invasive growth of keloid. Zhonghua Zheng Xing Wai Ke Za Zhi. 2004;20:128–31. [PubMed] [Google Scholar]
- 86. Wu WS, Wang FS, Yang KD, Huang CC, Kuo YR. Dexamethasone induction of keloid regression through effective suppression of VEGF expression and keloid fibroblast proliferation. J Invest Dermatol. 2006;126:1264–71. [DOI] [PubMed] [Google Scholar]
- 87. Ogawa R, Akaishi S. Endothelial dysfunction may play a key role in keloid and hypertrophic scar pathogenesis - keloids and hypertrophic scars may be vascular disorders. Med Hypotheses. 2016;96:51–60. [DOI] [PubMed] [Google Scholar]
- 88. Ong CT, Khoo YT, Tan EK, Mukhopadhyay A, Do DV, Han HC, et al. Epithelial-mesenchymal interactions in keloid pathogenesis modulate vascular endothelial growth factor expression and secretion. J Pathol. 2007;211:95–108. [DOI] [PubMed] [Google Scholar]
- 89. Jumper N, Paus R, Bayat A. Functional histopathology of keloid disease. Histol Histopathol. 2015;30:1033–57. [DOI] [PubMed] [Google Scholar]
- 90. Kurokawa N, Ueda K, Tsuji M. Study of microvascular structure in keloid and hypertrophic scars: density of microvessels and the efficacy of three-dimensional vascular imaging. J Plast Surg Hand Surg. 2010;44:272–7. [DOI] [PubMed] [Google Scholar]
- 91. Li Q, Qin Z, Nie F, Bi H, Zhao R, Pan B, et al. Metabolic reprogramming in keloid fibroblasts: aerobic glycolysis and a novel therapeutic strategy. Biochem Biophys Res Commun. 2018;496:641–7. [DOI] [PubMed] [Google Scholar]
- 92. Vincent AS, Phan TT, Mukhopadhyay A, Lim HY, Halliwell B, Wong KP. Human skin keloid fibroblasts display bioenergetics of cancer cells. J Invest Dermatol. 2008;128:702–9. [DOI] [PubMed] [Google Scholar]
- 93. Lei R, Li J, Liu F, Li W, Zhang S, Wang Y, et al. HIF-1α promotes the keloid development through the activation of TGF-β/Smad and TLR4/MyD88/NF-κB pathways. Cell Cycle. 2019;18:3239–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lin X, Wang Y, Jiang Y, Xu M, Pang Q, Sun J, et al. Sumoylation enhances the activity of the TGF-β/SMAD and HIF-1 signaling pathways in keloids. Life Sci. 2020;255:117859. 10.1016/j.lfs.2020.117859. [DOI] [PubMed] [Google Scholar]
- 95. Hong KH, Yoo SA, Kang SS, Choi JJ, Kim WU, Cho CS. Hypoxia induces expression of connective tissue growth factor in scleroderma skin fibroblasts. Clin Exp Immunol. 2006;146:362–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Persa OD, Niessen CM. Epithelial polarity limits EMT. Nat Cell Biol. 2019;21:299–300. [DOI] [PubMed] [Google Scholar]
- 97. Venhuizen JH, Jacobs FJC, Span PN, Zegers MM. P120 and E-cadherin: double-edged swords in tumor metastasis. Semin Cancer Biol. 2020;60:107–20. [DOI] [PubMed] [Google Scholar]
- 98. Do DV, Ong CT, Khoo YT, Carbone A, Lim CP, Wang S, et al. Interleukin-18 system plays an important role in keloid pathogenesis via epithelial-mesenchymal interactions. Br J Dermatol. 2012;166:1275–88. 10.1111/j.1365-2133.2011.10721.x. [DOI] [PubMed] [Google Scholar]
- 99. Yan L, Cao R, Liu Y, Wang L, Pan B, Lv X, et al. MiR-21-5p links epithelial-mesenchymal transition phenotype with stem-like cell signatures via AKT Signaling in keloid keratinocytes. Sci Rep. 2016;6:28281. 10.1038/srep28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yan L, Wang LZ, Xiao R, Cao R, Pan B, Lv XY, et al. Inhibition of microRNA-21-5p reduces keloid fibroblast autophagy and migration by targeting PTEN after electron beam irradiation. Lab Investig. 2020;100:387–99. 10.1038/s41374-019-0323-9. [DOI] [PubMed] [Google Scholar]
- 101. Lim CP, Phan TT, Lim IJ, Cao X. Stat3 contributes to keloid pathogenesis via promoting collagen production, cell proliferation and migration. Oncogene. 2006;25:5416–25. [DOI] [PubMed] [Google Scholar]
- 102. Lee WJ, Park JH, Shin JU, Noh H, Lew DH, Yang WI, et al. Endothelial-to-mesenchymal transition induced by Wnt 3a in keloid pathogenesis. Wound Repair Regen. 2015;23:435–42. [DOI] [PubMed] [Google Scholar]
- 103. Ma X, Chen J, Xu B, Long X, Qin H, Zhao RC, et al. Keloid-derived keratinocytes acquire a fibroblast-like appearance and an enhanced invasive capacity in a hypoxic microenvironment in vitro. Int J Mol Med. 2015;35:1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang M, Liu S, Guan E, Liu H, Dong X, Hao Y, et al. Hyperbaric oxygen therapy can ameliorate the EMT phenomenon in keloid tissue. Medicine (Baltimore). 2018;97:e11529. 10.1097/md.0000000000011529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Macarak EJ, Wermuth PJ, Rosenbloom J, Uitto J. Keloid disorder: fibroblast differentiation and gene expression profile in fibrotic skin diseases. Exp Dermatol. 2021;30:132–45. [DOI] [PubMed] [Google Scholar]
- 106. Qu M, Song N, Chai G, Wu X, Liu W. Pathological niche environment transforms dermal stem cells to keloid stem cells: a hypothesis of keloid formation and development. Med Hypotheses. 2013;81:807–12. [DOI] [PubMed] [Google Scholar]
- 107. Zhang YX, Liu LP, Li M, Huang JL, Xu H, Chen XD, et al. Development of individualized induced pluripotent stem cells from fibroblasts of keloid lesions in patients. Transplant Proc. 2018;50:2868–71. [DOI] [PubMed] [Google Scholar]
- 108. Simandi Z, Horvath A, Wright LC, Cuaranta-Monroy I, De Luca I, Karolyi K, et al. OCT4 acts as an integrator of pluripotency and signal-induced differentiation. Mol Cell. 2016;63:647–61. [DOI] [PubMed] [Google Scholar]
- 109. Zhang Q, Yamaza T, Kelly AP, Shi S, Wang S, Brown J, et al. Tumor-like stem cells derived from human keloid are governed by the inflammatory niche driven by IL-17/IL-6 axis. PLoS One. 2009;4:e7798. 10.1371/journal.pone.0007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lee SY, Kim EK, Seo HB, Choi JW, Yoo JH, Jung KA, et al. IL-17 induced stromal cell-derived Factor-1 and Profibrotic factor in keloid-derived skin fibroblasts via the STAT3 pathway. Inflammation. 2020;43:664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bagabir RA, Syed F, Shenjere P, Paus R, Bayat A. Identification of a potential molecular diagnostic biomarker in keloid disease: Syndecan-1 (CD138) is overexpressed in keloid scar tissue. J Invest Dermatol. 2016;136:2319–23. [DOI] [PubMed] [Google Scholar]
- 112. Couchman JR. Syndecan-1 (CD138), carcinomas and EMT. Int J Mol Sci. 2021;22:4227. 10.3390/ijms22084227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Cui J, Jin S, Jin C, Jin Z. Syndecan-1 regulates extracellular matrix expression in keloid fibroblasts via TGF-β1/Smad and MAPK signaling pathways. Life Sci. 2020;254:117326. 10.1016/j.lfs.2020.117326. [DOI] [PubMed] [Google Scholar]
- 114. Stevenson AW, Melton PE, Moses EK, Wallace HJ, Wood FM, Rea S, et al. A Methylome and transcriptome analysis of normal human scar cells reveals a role for FOXF2 in scar maintenance. J Invest Dermatol. 2021;142:1489–98. [DOI] [PubMed] [Google Scholar]
- 115. Shin JU, Lee WJ, Tran T-N, Jung I, Lee JH. Hsp70 knockdown by siRNA decreased collagen production in keloid fibroblasts. Yonsei Med J. 2015;56:1619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yun IS, Lee MH, Rah DK, Lew DH, Park J-C, Lee WJ. Heat shock protein 90 inhibitor (17-AAG) induces apoptosis and decreases cell migration/motility of keloid fibroblasts. Plast Reconstr Surg. 2015;136:44e–53. 10.1097/PRS.0000000000001362. [DOI] [PubMed] [Google Scholar]
- 117. Tan KT, McGrouther DA, Day AJ, Milner CM, Bayat A. Characterization of hyaluronan and TSG-6 in skin scarring: differential distribution in keloid scars, normal scars and unscarred skin. J Eur Acad Dermatol Venereol. 2011;25:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li X-Y, Weng X-J, Li X-J, Tian X-Y. TSG-6 inhibits the growth of keloid fibroblasts via mediating the TGF-β1/Smad Signaling pathway. J Invest Surg. 2021;34:947–56. [DOI] [PubMed] [Google Scholar]
- 119. Li X, Chen Z, Li X, Wang H. In vitro analysis of the role of tumor necrosis factor-stimulated gene-6 in keloid. Mol Med Rep. 2019;19:919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zhang Q, Oh CK, Messadi DV, Duong HS, Kelly AP, Soo C, et al. Hypoxia-induced HIF-1 alpha accumulation is augmented in a co-culture of keloid fibroblasts and human mast cells: involvement of ERK1/2 and PI-3K/Akt. Exp Cell Res. 2006;312:145–55. [DOI] [PubMed] [Google Scholar]
- 121. Song KX, Liu S, Zhang MZ, Liang WZ, Liu H, Dong XH, et al. Hyperbaric oxygen therapy improves the effect of keloid surgery and radiotherapy by reducing the recurrence rate. J Zhejiang Univ Sci B. 2018;19:853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Long F, Si L, Long X, Yang B, Wang X, Zhang F. 2ME2 increase radiation-induced apoptosis of keloid fibroblasts by targeting HIF-1α in vitro. Australas J Dermatol. 2016;57:e32–8. 10.1111/ajd.12340. [DOI] [PubMed] [Google Scholar]
- 123. Si L, Zhang M, Guan E, Han Q, Liu Y, Long X, et al. Resveratrol inhibits proliferation and promotes apoptosis of keloid fibroblasts by targeting HIF-1α. J Plast Surg Hand Surg. 2020;54:290–6. [DOI] [PubMed] [Google Scholar]
- 124. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sandiford OA, Moore CA, Du J, Boulad M, Gergues M, Eltouky H, et al. Human aging and cancer: role of miRNA in tumor microenvironment. Adv Exp Med Biol. 2018;1056:137–52. [DOI] [PubMed] [Google Scholar]
- 126. Xu M, Sun J, Yu Y, Pang Q, Lin X, Barakat M, et al. TM4SF1 involves in miR-1-3p/miR-214-5p-mediated inhibition of the migration and proliferation in keloid by regulating AKT/ERK signaling. Life Sci. 2020;254:117746. 10.1016/j.lfs.2020.117746. [DOI] [PubMed] [Google Scholar]
- 127. Huang Y, Wang Y, Lin L, Wang P, Jiang L, Liu J, et al. Overexpression of miR-133a-3p inhibits fibrosis and proliferation of keloid fibroblasts by regulating IRF5 to inhibit the TGF-β/Smad2 pathway. Mol Cell Probes. 2020;52:101563. 10.1016/j.mcp.2020.101563. [DOI] [PubMed] [Google Scholar]
- 128. Feng J, Xue S, Pang Q, Rang Z, Cui F. miR-141-3p inhibits fibroblast proliferation and migration by targeting GAB1 in keloids. Biochem Biophys Res Commun. 2017;490:302–8. [DOI] [PubMed] [Google Scholar]
- 129. Wang R, Bai Z, Wen X, Du H, Zhou L, Tang Z, et al. MiR-152-3p regulates cell proliferation, invasion and extracellular matrix expression through by targeting FOXF1 in keloid fibroblasts. Life Sci. 2019;234:116779. 10.1016/j.lfs.2019.116779. [DOI] [PubMed] [Google Scholar]
- 130. Zhu W, Wu X, Yang B, Yao X, Cui X, Xu P, et al. miR-188-5p regulates proliferation and invasion via PI3K/Akt/MMP-2/9 signaling in keloids. Acta Biochim Biophys Sin. 2019;51:185–96. [DOI] [PubMed] [Google Scholar]
- 131. Xu Z, Guo B, Chang P, Hui Q, Li W, Tao K. The differential expression of miRNAs and a preliminary study on the mechanism of miR-194-3p in keloids. Biomed Res Int. 2019;2019:8214923. 10.1155/2019/8214923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Xu Q, Jiang S. miR-194-5p serves a suppressive role in human keloid fibroblasts via targeting NR2F2. Mol Med Rep. 2021;2357. 10.3892/mmr.2020.11695. [DOI] [PubMed] [Google Scholar]
- 133. Kashiyama K, Mitsutake N, Matsuse M, Ogi T, Saenko VA, Ujifuku K, et al. miR-196a downregulation increases the expression of type I and III collagens in keloid fibroblasts. J Invest Dermatol. 2012;132:1597–604. [DOI] [PubMed] [Google Scholar]
- 134. Yang J, Deng P, Qi Y, Feng X, Wen H, Chen F. NEAT1 knockdown inhibits keloid fibroblast progression by miR-196b-5p/FGF2 Axis. J Surg Res. 2021;259:261–70. [DOI] [PubMed] [Google Scholar]
- 135. Shi K, Qiu X, Zheng W, Yan D, Peng W. MiR-203 regulates keloid fibroblast proliferation, invasion, and extracellular matrix expression by targeting EGR1 and FGF2. Biomed Pharmacother. 2018;108:1282–8. [DOI] [PubMed] [Google Scholar]
- 136. Yuan W, Sun H, Yu L. Long non-coding RNA LINC01116 accelerates the progression of keloid formation by regulating miR-203/SMAD5 axis. Burns. 2021;47:665–75. [DOI] [PubMed] [Google Scholar]
- 137. Tang H, Chen Q, Yu W, Zhao T. MiR-4328 inhibits proliferation, metastasis and induces apoptosis in keloid fibroblasts by targeting BCL2 expression. Open Life Sci. 2020;15:638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Liu P, Hu Y, Xia L, Du M, Hu Z. miR-4417 suppresses keloid fibrosis growth by inhibiting CyclinD1. J Biosci. 2020;45:47. [PubMed] [Google Scholar]
- 139. Wu ZY, Lu L, Liang J, Guo XR, Zhang PH, Luo SJ. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genetics and Molecular Research: GMR. 2014;13:2727–38. [DOI] [PubMed] [Google Scholar]
- 140. El-Kott AF, Shati AA, Ali Al-Kahtani M, Alharbi SA. The apoptotic effect of resveratrol in ovarian cancer cells is associated with downregulation of galectin-3 and stimulating miR-424-3p transcription. J Food Biochem. 2019;43:e13072. 10.1111/jfbc.13072. [DOI] [PubMed] [Google Scholar]
- 141. Zhang J, Xu D, Li N, Li Y, He Y, Hu X, et al. Downregulation of microRNA-31 inhibits proliferation and induces apoptosis by targeting in human keloid. Oncotarget. 2017;8:74623–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Rang Z, Wang Z-Y, Pang Q-Y, Wang Y-W, Yang G, Cui F. MiR-181a targets PHLPP2 to augment AKT Signaling and regulate proliferation and apoptosis in human keloid fibroblasts. Cell Physiol Biochem. 2016;40:796–806. [DOI] [PubMed] [Google Scholar]
- 143. Yang D, Li M, Du N. Effects of the circ_101238/miR-138-5p/CDK6 axis on proliferation and apoptosis keloid fibroblasts. Exp Ther Med. 2020;20:1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wu W, Xie F, Zhang Y, Wang X, Xia L, Wu X, et al. A novel regulatory function for miR-217 targetedly suppressing fibronectin expression in keloid fibrogenesis. Int J Clin Exp Pathol. 2018;11:1866–77. [PMC free article] [PubMed] [Google Scholar]
- 145. Wang Z, Feng C, Song K, Qi Z, Huang W, Wang Y. lncRNA-H19/miR-29a axis affected the viability and apoptosis of keloid fibroblasts through acting upon COL1A1 signaling. J Cell Biochem. 2020;121:4364–76. [DOI] [PubMed] [Google Scholar]
- 146. Wu D, Zhou J, Tan M, Zhou Y. LINC01116 regulates proliferation, migration, and apoptosis of keloid fibroblasts by the TGF-β1/SMAD3 signaling via targeting miR-3141. Anal Biochem. 2021;627:114249. 10.1016/j.ab.2021.114249. [DOI] [PubMed] [Google Scholar]
- 147. Lv W, Ren Y, Wu M, Luo X, Yu J, Zhang Q, et al. Identifying miRNA modules associated with progression of keloids through weighted gene co-expression network analysis and experimental validation in vitro. Burns: J Int Soc Burn Injur. 2021;47:1359–72. [DOI] [PubMed] [Google Scholar]
- 148. Chao L, Hua-Yu Z, Wen-Dong B, Mei S, Bin X, Da-Hai H, et al. miR-96 promotes collagen deposition in keloids by targeting Smad7. Exp Ther Med. 2019;17:773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Su X, Ma Y, Wang Q, Gao Y. LncRNA HOXA11-AS aggravates keloid progression by the regulation of HOXA11-AS-miR-205-5p-FOXM1 pathway. J Surg Res. 2021;259:284–95. [DOI] [PubMed] [Google Scholar]
- 150. Yao X, Cui X, Wu X, Xu P, Zhu W, Chen X, et al. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-β1/Smad3 signaling pathway. Biochem Biophys Res Commun. 2018;495:713–20. [DOI] [PubMed] [Google Scholar]
- 151. Pang Q, Wang Y, Xu M, Xu J, Xu S, Shen Y, et al. MicroRNA-152-5p inhibits proliferation and migration and promotes apoptosis by regulating expression of Smad3 in human keloid fibroblasts. BMB Rep. 2019;52:202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhang Y, Guo B, Hui Q, Li W, Chang P, Tao K. Downregulation of miR-637 promotes proliferation and metastasis by targeting Smad3 in keloids. Mol Med Rep. 2018;18:1628–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zhu H-Y, Bai W-D, Li C, Zheng Z, Guan H, Liu J-Q, et al. Knockdown of lncRNA-ATB suppresses autocrine secretion of TGF-β2 by targeting ZNF217 via miR-200c in keloid fibroblasts. Sci Rep. 2016;6:24728. 10.1038/srep24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Liu Y, Ren L, Liu W, Xiao Z. MiR-21 regulates the apoptosis of keloid fibroblasts by caspase-8 and the mitochondria-mediated apoptotic signaling pathway via targeting FasL. Biochem Cell Biol. 2018;96:548–55. [DOI] [PubMed] [Google Scholar]
- 155. Hou Z, Fan F, Liu P. BTXA regulates the epithelial-mesenchymal transition and autophagy of keloid fibroblasts via modulating miR-1587/miR-2392 targeted ZEB2. Biosci Rep. 2019;39:BSR20190679. 10.1042/BSR20190679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23:205–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Li J, Zhang X, Liu C. The computational approaches of lncRNA identification based on coding potential: status quo and challenges. Comput Struct Biotechnol J. 2020;18:3666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, et al. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci. 2017;18:2659. 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139:269–80. [DOI] [PubMed] [Google Scholar]
- 160. McCabe EM, Rasmussen TP. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 2021;75:38–48. [DOI] [PubMed] [Google Scholar]
- 161. Zhang J, Le TD, Liu L, Li J. Inferring and analyzing module-specific lncRNA-mRNA causal regulatory networks in human cancer. Brief Bioinform. 2019;20:1403–19. [DOI] [PubMed] [Google Scholar]
- 162. Deng Y, Xu Y, Xu S, Zhang Y, Han B, Liu Z, et al. Secondary data mining of GEO database for long non-coding RNA and competing endogenous RNA network in keloid-prone individuals. Aging. 2020;12:25076–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Xu L, Sun N, Li G, Liu L. LncRNA H19 promotes keloid formation through targeting the miR-769-5p/EIF3A pathway. Mol Cell Biochem. 2021;476:1477–87. [DOI] [PubMed] [Google Scholar]
- 164. Wan J, He X-L, Jian Q-C, Fan Z-F, Shi Y, Luo L-F. LINC00937 suppresses keloid fibroblast proliferation and extracellular matrix deposition by targeting the miR-28-5p/MC1R axis. Histol Histopathol. 2021;36:995–1005. [DOI] [PubMed] [Google Scholar]
- 165. Jin J, Jia Z-H, Luo X-H, Zhai H-F. Long non-coding RNA HOXA11-AS accelerates the progression of keloid formation via miR-124-3p/TGFβR1 axis. Cell Cycle. 2020;19:218–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Jin J, Zhai H-F, Jia Z-H, Luo X-H. Long non-coding RNA HOXA11-AS induces type I collagen synthesis to stimulate keloid formation via sponging miR-124-3p and activation of Smad5 signaling. Am J Physiol Cell Physiol. 2019;317:C1001–10. 10.1152/ajpcell.00319.2018. [DOI] [PubMed] [Google Scholar]
- 167. Duan X, Wu Y, Zhang Z, Lu Z. Identification and analysis of dysregulated lncRNA and associated ceRNA in the pathogenesis of keloid. Ann Transl Med. 2020;8:222. 10.21037/atm.2020.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhao X, Jie X, Gao Y-K, Nie B, Jiang H. Long non-coding RNA CACNA1G-AS1 promotes proliferation and invasion and inhibits apoptosis by regulating expression of miR-205 in human keloid fibroblasts. Biosci Rep. 2020;40:BSR20192839. 10.1042/BSR20192839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. [DOI] [PubMed] [Google Scholar]
- 171. Lv W, Liu S, Zhang Q, Hu W, Wu Y, Ren Y. Circular RNA CircCOL5A1 sponges the MiR-7-5p/Epac1 Axis to promote the progression of keloids through regulating PI3K/Akt Signaling pathway. Front Cell Dev Biol. 2021;9:626027. 10.3389/fcell.2021.626027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wang B, Yin H, Zhang H, Wang T. circNRIP1 facilitates keloid progression via FXR1-mediated upregulation of miR-503-3p and miR-503-5p. Int J Mol Med. 2021;47:70. 10.3892/ijmm.2021.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Shi J, Yao S, Chen P, Yang Y, Qian M, Han Y, et al. The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol Biol Rep. 2020;47:201–9. [DOI] [PubMed] [Google Scholar]
- 174. Zhang Z, Yu K, Liu O, Xiong Y, Yang X, Wang S, et al. Expression profile and bioinformatics analyses of circular RNAs in keloid and normal dermal fibroblasts. Exp Cell Res. 2020;388:111799. 10.1016/j.yexcr.2019.111799. [DOI] [PubMed] [Google Scholar]
- 175. Chater-Diehl E, Goodman SJ, Cytrynbaum C, Turinsky AL, Choufani S, Weksberg R. Anatomy of DNA methylation signatures: emerging insights and applications. Am J Hum Genet. 2021;108:1359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wang R, Xu J. Genomic DNA methylation and histone methylation. Yi Chuan. 2014;36:191–9. [PubMed] [Google Scholar]
- 177. Jones LR, Young W, Divine G, Datta I, Chen KM, Ozog D, et al. Genome-wide scan for methylation profiles in keloids. Dis Markers. 2015;2015:943176. 10.1155/2015/943176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, et al. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhang G, Guan Q, Chen G, Qian F, Liang J. DNA methylation of the CDC2L1 gene promoter region decreases the expression of the CDK11p58 protein and reduces apoptosis in keloid fibroblasts. Arch Dermatol Res. 2018;310:107–15. [DOI] [PubMed] [Google Scholar]
- 180. Liu J, Zhu H, Wang H, Li J, Han F, Liu Y, et al. Methylation of secreted frizzled-related protein 1 (SFRP1) promoter downregulates Wnt/β-catenin activity in keloids. J Mol Histol. 2018;49:185–93. [DOI] [PubMed] [Google Scholar]
- 181. Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep. 2015;2:145–54. [DOI] [PubMed] [Google Scholar]
- 182. Raut JR, Guan Z, Schrotz-King P, Brenner H: Whole-blood DNA methylation markers for risk stratification in colorectal cancer screening: : a systematic review. Cancers (Basel) 2019;11:912. 10.3390/cancers11070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cowan LA, Talwar S, Yang AS. Will DNA methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics. 2010;2:71–86. [DOI] [PubMed] [Google Scholar]
- 184. Guzzetta AA, Pisanic Ii TR, Sharma P, Yi JM, Stark A, Wang TH, et al. The promise of methylation on beads for cancer detection and treatment. Expert Rev Mol Diagn. 2014;14:845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]