Abstract
Skeletal muscle fibers are malleable and undergo rapid remodeling in response to increased contractile activity (i.e., exercise) or prolonged periods of muscle inactivity (e.g., prolonged bedrest). Exploration of the cell signaling pathways regulating these skeletal muscle adaptations reveal that redox signaling pathways play a key role in the control of muscle remodeling during both exercise and prolonged muscle inactivity. In this regard, muscular exercise results in an acute increase in the production of reactive oxygen species (ROS) in the contracting fibers; however, this contraction-induced rise in ROS production rapidly declines when contractions cease. In contrast, prolonged muscle disuse results in a chronic elevation in ROS production within the inactive fibers. This difference in the temporal pattern of ROS production in muscle during exercise and muscle inactivity stimulates divergent cell-signaling pathways that activate both genomic and nongenomic mechanisms to promote muscle remodeling. This review examines the role that redox signaling plays in skeletal muscle adaptation in response to both prolonged muscle inactivity and endurance exercise training. We begin with a summary of the sites of ROS production in muscle fibers followed by a review of the cellular antioxidants that are responsible for regulation of ROS levels in the cell. We then discuss the specific redox-sensitive signaling pathways that promote skeletal muscle adaptation in response to both prolonged muscle inactivity and exercise. To stimulate future research, we close with a discussion of unanswered questions in this exciting field.
Keywords: Renin angiotensin system, Muscle atrophy, Mechanical ventilation, Diaphragm, Muscle wasting
Graphical abstract
1. Introduction
The discovery that contracting skeletal muscles produce free radicals was first reported >40 years ago. This seminal study also revealed that exhaustive exercise results in both oxidative damage and mitochondrial dysfunction in muscle fibers [1]. Paradoxically, it was later discovered that prolonged muscle inactivity (e.g., limb immobilization) also results in a chronic increase in the production of radicals and other reactive oxygen species (ROS); importantly, this amplified ROS production has been identified as a required contributor to the fiber atrophy that occurs during prolonged muscle disuse [2]. Collectively, these discoveries launched the field of muscle redox biology and stimulated research into the effects that ROS have on skeletal muscle structure and function.
Although it was widely believed in the 1980's that exercise-induced ROS production in skeletal muscle was damaging and potentially cytotoxic to muscle fibers, the consensus that ROS are important signaling molecules to promote muscle remodeling did not emerge until the early 2000's. Indeed, it is now established that muscle ROS production plays an important role in the control of signaling pathways that stimulate muscle adaptation in response to both exercise training and inactivity-induced muscle wasting. This review summarizes our current understanding of the role that redox signaling plays in skeletal muscle remodeling. The first segment of this report discusses the sites of ROS production in skeletal muscles during exercise and prolonged periods of muscle inactivity; this section also highlights the antioxidant networks that regulate ROS levels in muscle fibers. We then review key examples of important redox signaling targets and discuss how specific redox signaling pathways impact skeletal muscle in response to prolonged muscle inactivity and endurance exercise training.
2. ROS production in skeletal muscles
Numerous “reactive chemical species” exist, and several species exert profound effects on cellular function by causing molecular damage or contributing to redox signaling events. Reactive species are named by identifying their reactive atom (i.e., ROS or reactive nitrogen species (RNS)). Specifically, ROS are a group of molecules derived from molecular oxygen; these reactive species are formed from reduction-oxidation (redox) reactions or by electronic excitation [3]. The chemical reactivity of the various ROS with cellular targets is variable and spans several orders of magnitude across the ROS species [3]. It follows that the term “ROS” is not a specific chemical molecule and therefore, use of the term ROS does not refer to a specific reactive species. Nonetheless, because of the challenges in identifying the individual ROS in cells, the term ROS is used in redox biology to denote all reactive species [3]. Although the superoxide radical (O2-) is the parent of all ROS, hydrogen peroxide (H2O2) is accepted as the major ROS involved in redox signaling in cells [[4], [5], [6]]. Hence, a major focus of this review will be redox signaling pathways linked to H2O2. To date, a total 41 different enzymes are known to generate O2- or H2O2 [7]. An overview of the major sites of ROS production in muscle fibers during both exercise and prolonged muscle inactivity follows.
2.1. Sources of ROS production in contracting muscle
The major source of ROS production in contracting muscle has been debated for decades. As mentioned earlier, O2- is the parent species of all oxygen ROS produced in skeletal muscle and generation of these reactive species is increased during contractile activity [[8], [9], [10]]. Because this review is focused ROS-mediated redox signaling in muscle fibers, we will emphasize the putative sources of ROS in contracting muscle. Although multiple cell types are present in skeletal muscle (e.g., vascular smooth muscle cells, endothelial cells, etc.), muscle fibers are the main cell type found in skeletal muscle and this fact suggests that muscle fibers are the dominant source of ROS production during exercise [8]. Indeed, it established that both contracting myotubes [11] and mature (isolated) muscle fibers [12] produce ROS. In the next paragraphs we highlight key studies that provide insight into the primary locations of ROS production in contracting skeletal muscles.
During the 1980's and 1990's, mitochondria were postulated to be the major site of ROS production in contracting skeletal muscle [1,13]. This hypothesis emerged from early reports indicating that ∼2–5% of all oxygen consumed by mitochondria is used to form O2- [14,15]. Based on this assumption, it was predicted that the increased oxidative phosphorylation that occurs during muscular contractions results in a proportional surge in the production of O2-. Nonetheless, recent data do not support this concept. For example, contemporary work reveals that the upper estimate of the fraction of molecular oxygen that forms O2- in the mitochondria is on the order ∼0.15% and not the original prediction of 2–5% [16,17]. Moreover, compared to active state 3 respiration (i.e., exercise), mitochondria produce more O2- during state 4 (i.e., resting) respiration [16,18,19]. Specifically, skeletal muscle mitochondria produce 13–27 times more ROS during state 4 respiration than in state 3 [18,19]. Furthermore, assessments of the mitochondrial redox potential in contracting single muscle fibers reveal that ROS production does not increase in mitochondria during electrically stimulated muscle contractions [20]. Similar conclusions have reached using different experimental approaches [21]. Collectively, these findings reveal that mitochondria are not the dominant source of ROS production in contracting muscles [8,22,23].
The experimental evidence that mitochondrial are not a dominant source of ROS production in contracting muscle lead to investigations of other potential ROS producing pathways in muscle and several lines of evidence indicate that NAD(P)H oxidase enzymes play a key role in contraction-induced ROS generation in muscle fibers [20,21,[24], [25], [26]]. In this regard, skeletal muscles express three isoforms of NAD(P)H oxidases (Nox1, Nox2, and Nox4) and evidence indicates that Nox2, located in the sarcolemma, is a major source of ROS production in contracting skeletal muscles [27].
Although NAD(P)H oxidases are likely a primary site of ROS production in contracting muscle fibers, phospholipase A (PLA2) is another potential source of ROS in muscle fibers during exercise [28,29]. Indeed, ROS emission from contracting murine skeletal muscles is reduced when PLA2 activity is pharmacologically inhibited [28,29].
In summary, evidence supports the concept that NAD(P)H oxidases are a primary source of ROS in contracting skeletal muscles. Additional sites of ROS in contracting skeletal muscles include PLA2 and mitochondria sources. For more details about the sources of ROS in contracting skeletal muscles, the reader is referred to comprehensive reviews on this topic [[8], [22], [30]].
2.2. Sites of ROS production in skeletal muscle during prolonged inactivity
The fact that prolonged muscle disuse results in oxidative stress in muscle fibers was first reported in 1991 [2] and this milestone discovery has been confirmed in many studies and summarized in scientific reviews [[31], [32], [33]]. Several pre-clinical models of muscle inactivity have been employed to investigate the mechanisms responsible for disuse-induced oxidative stress in muscle fibers; these include rodent models of limb immobilization, hindlimb suspension, and prolonged mechanical ventilation resulting in diaphragmatic inactivity. The search for the sites of ROS production in muscle fibers during prolonged inactivity reveals that mitochondria are a dominant site of ROS generation in both immobilized hindlimb muscles [34,35] and in diaphragm muscle fibers during prolonged mechanical ventilation [19,[34], [35], [36], [37], [38], [39], [40], [41]]. Although both xanthine oxidase and NAD(P)H oxidases also produce ROS in inactive muscle fibers, these sources of ROS production likely play small roles in the total ROS production within chronically inactive fibers [19,38,40].
The important and unanswered question remains, why does prolonged skeletal muscle inactivity promote mitochondrial dysfunction resulting in significant increases in mitochondrial ROS production? Unfortunately, a definitive explanation to this question is not currently available. However, evidence indicates that several factors can contribute to inactivity-induced mitochondrial dysfunction including an imbalance in mitochondrial dynamics (i.e., fission/fusion), increased ROS production via activation of angiotensin 2 type 1 receptors on the sarcolemma, and an impairment in mitophagy. For more details see the following recent reviews [33,[42], [43], [44], [45], [46]].
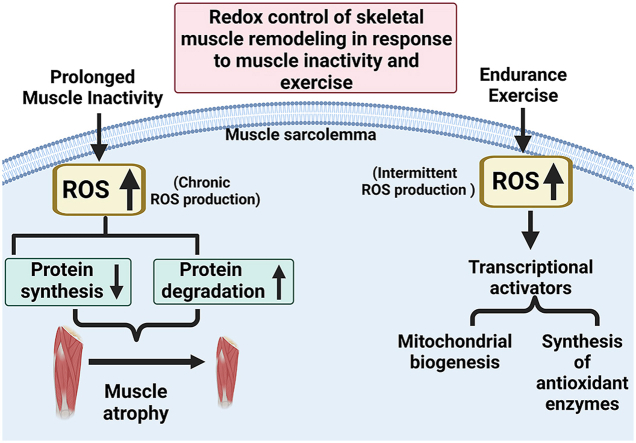
Notably, key differences exist in the temporal pattern of ROS production in muscle fibers during exercise and during prolonged periods of inactivity. Specifically, although muscular contractions promote an acute increase in ROS production, this contraction-induced rise in ROS emission returns to basal levels rapidly when contractions stop [20,47]. In contrast, prolonged periods of muscle inactivity results in a habitual increase in ROS production leading to protracted oxidative stress and disturbed redox signaling [18,34,35]. The impact of these temporal differences in ROS production on redox signaling pathways in skeletal muscle will be discussed later in this report.
3. Control of intra- and extracellular levels of H2O2
Cells regulate ROS levels using a variety of enzymatic and non-enzymatic antioxidants. Complete details of cellular antioxidant systems exceed the scope of this review and therefore, only a brief overview of key antioxidant enzymes is provided. For more information on cellular redox sinks the reader is referred to detailed reviews [9,48,49].
The primary antioxidant enzymes involved in the elimination of ROS from cells include superoxide dismutase, glutathione peroxidases, peroxiredoxins, and catalase. Superoxide dismutase (SOD) exists in three isoforms and is responsible for dismutation of O2- to form H2O2 and oxygen [9]. Importantly, these SOD isoforms are found in divergent locations in the cell (i.e., mitochondria, cytosol, and extracellular spaces); this compartmentalization of SOD is key to rapid removal of O2- at the site of production [22].
Three types of antioxidant enzymes act as redox sinks to eliminate H2O2; these include peroxiredoxins (PRX), glutathione peroxidases (GPX), and catalase. PRX exists in six isoforms and this family of enzymes is arguably the most important peroxide scavenging enzymes [50,51]. PRX can reduce H2O2 via several reactions and most of these reactions result in oxidized PRX which is reduced by thioredoxin to restore its catalytic ability [50,51]. GPX also regulates H2O2 levels across different cellular compartments (e.g., cytosol and mitochondria) and several isoforms of GPX exist [52]. All forms of GPX convert H2O2 or organic hydroperoxides to water and alcohol [22]. Most GPX isoforms require a supply of electrons provided by reduced glutathione (GSH); this need for a continuous supply of GSH is met by oxidized GSH being reduced by glutathione reductase using NADPH to provide the reducing power [50,51]. Finally, catalase has one of highest turnover rates of all enzymes and converts H2O2 to water; similar to PRX and GPX, catalase exists across several cellular compartments [53]. Together, PRX, GPX, and catalase work cooperatively to remove H2O2 from cells.
Cellular levels of ROS often transition between steady state conditions and situations when ROS levels increase rapidly due changing metabolic circumstances. Under homeostatic conditions, H2O2 is maintained at steady-state levels; this steady-state environment results from both stringent control of ROS production and removal of ROS via redox sinks (e.g., antioxidant enzymes) [3]. Although the overall H2O2 concentration in cells is estimated to range between 1 and 10 nM, the levels of H2O2 varies markedly between the cytosol and various cellular organelles [3]. Moreover, compared to the intracellular H2O2 concentration, the extracellular level of H2O2 is substantially higher resulting in a steep gradient of H2O2 concentration (i.e., 100–500 fold) between the intracellular and extracellular spaces [3].
Movement of H2O2 across cell membranes occurs via two primary mechanisms. First, as a small uncharged molecule, H2O2 can cross cell membranes by passive diffusion; however, the rate of H2O2 diffusion across the membrane is relatively slow [3]. Secondly, H2O2 can also cross cell membranes via water channels (aquaporins) in the membrane; compared to simple diffusion, the rate of H2O2 transfer across the membrane via water channels is relatively rapid. Indeed, aquaporins facilitate the transfer of H2O2 across membranes and contribute to the establishment of the H2O2 gradient across cell membranes [54]. Moreover, evidence reveals that a key aquaporin (aquaporin-8; AQP8) contains a gating mechanism that is under redox control; this finding suggests that H2O2 is transported across membranes in a redox-regulated process [55].
Because mitochondria are a source of H2O2 production, the key question arises, can H2O2 cross the mitochondrial membrane? Unfortunately, a definitive answer to this question is not available as debate continues as to whether H2O2 can freely cross the mitochondrial membrane [3]. Nonetheless, when thioredoxin is inhibited within the mitochondrial intermembrane space, movement of H2O2 from the mitochondria to the cytosol does occur [56]. Moreover, although controversial, it has been suggested that mitochondrial membranes contain AQP8 that facilitates H2O2 movement from the mitochondrial to the cytosol [57]. If this is the case, H2O2 produced in the mitochondria could cross mitochondrial membranes and impact redox signaling in the cytosol. The next segment highlights several examples of prototypical ROS signaling targets.
4. Examples of redox signaling pathways in skeletal muscle
Redox signaling can influence muscle fiber structure and function in numerous ways. For example, changes in redox signaling can affect the function of numerous proteins leading to alterations in enzyme activity, membrane transport, and gene transcription [3]. Moreover, redox signaling can also impact gene expression by oxidative modification of mRNA [58]. In the following sections, we present several prototypical examples of redox signaling targets in skeletal muscles.
4.1. Nrf2/KEAP signaling
Nuclear factor erythroid 2-related factor (Nrf2) is a transcription factor recognized as a key regulator of the transcriptional response to oxidative stress [59]. Specifically, Nrf2 regulates both the basal and inducible expression of >200 genes including proteins involved in drug detoxication, numerous antioxidants, enzymes involved in carbohydrate metabolism, and NADPH regeneration enzymes [60,61]. In reference to antioxidant enzymes, Nrf2 regulates the expression of several isoforms of both glutathione peroxidase and peroxiredoxin as well as thioredoxin and glutathione reductase [60]. In the absence of oxidative stress, the levels of Nrf2 in the nucleus remain low due to the direct interaction of Nrf2 with the inhibitory protein Kelch-like ECH-associated protein 1 (KEAP1) [62,63]. KEAP1 inhibits Nrf2's movement into the nucleus in two ways. First, KEAP1 sequesters Nrf2 in the cytoplasm, preventing Nrf2 from migrating to the nucleus [61]. Second, KEAP1's interaction with Nrf2 in the cytoplasm targets Nrf2 for polyubiquitination and degradation by the ubiquitin-proteasome system (UPS) [61]. Thus, during homeostatic redox conditions, the low levels of Nrf2 in the nucleus maintain basal expression of antioxidant and detoxification enzymes. However, during periods of increased ROS production in cells, this electrophilic stress modifies redox-sensitive cysteine residues on KEAP1 that allows Nrf2 translocation to the nucleus to upregulate the expression of both antioxidant and detoxification genes [63].
4.2. NF-ĸB signaling
The transcriptional activating factor nuclear kappa B (NF-ĸB) includes a family of five transcriptional factors (p65, Rel B, c-Rel, p52, and p50) [64]. To become a transcriptional activator, two of these NF-ĸB family members must dimerize to acquire transcriptional competency [64]. Although all five of these NF-ĸB family members are expressed in skeletal muscle, evidence indicates that the p50-p65 heterodimer is responsible for much of the NF-ĸB activity in muscle [65]. NF-ĸB is subject to complex regulation and abundant evidence reveals that this regulation includes redox control. During homeostatic conditions, the nuclear localization sequence of NF-ĸB is bound to the inhibitory protein, IĸB; this prevents the dimerization of p50-p65 and therefore, averts NF-ĸB movement into the nucleus [66]. Acute oxidative stress is associated with increased NF-ĸB activation and the concomitant increase in gene expression [64].
Although an increase in oxidative stress in cells can promote NF-ĸB-mediated gene expression, extremely high levels of ROS production can diminish the ability of NF-ĸB to bind to DNA [64,67]. Specifically, oxidation of NF-ĸB dimers can directly inhibit NF-ĸB binding with DNA and therefore, redox signaling can both promote and inhibit NF-ĸB -mediated gene expression [67]. In regard to ROS-mediated NF-ĸB inhibition, a specific cysteine of p50 (cys-62) is sensitive to oxidation and this oxidation is often followed by S-glutathionylation; this is significant because glutathionated NF-ĸB has less transcriptional activity [68].
4.3. Regulation of FOXO transcription factors
The forkhead box protein O (FOXO) family of transcriptional factors regulates a wide range of cellular functions including apoptosis and proteolysis via its influence on both autophagy and the UPS [69]. Indeed, FOXO signaling is known to contribute to skeletal muscle wasting in a variety of conditions and this topic will be addressed in detail later. While the activation of FOXO transcriptional factors occurs via several pathways, oxidative stress can activate these transcriptional activators via posttranslational modifications such as phosphorylation or direct oxidation of cysteine in FOXO family members [[70], [71], [72]]. For example, exposure of myotubes to oxidative stress (i.e., H2O2) activates FOXO signaling (i.e., FOXO3a) resulting in the increased expression of key UPS proteins (e.g., muscle specific E3 ligases) and vital autophagy proteins (e.g., LC3) [73].
4.4. Regulation of cellular kinases
Many families of protein kinases exist in muscle fibers and several kinases are activated by oxidative stress [3,74]. A detailed discussion of the redox control of kinases exceeds the scope of this review. Nonetheless, this section will highlight the mammalian target of rapamycin complex 1 (mTORC1), mitogen-activated kinase (MAPK) family, and the AMP-activated kinase as examples of redox control of kinase activity.
mTORC1 activation plays an important role in both muscle protein synthesis and the control of muscle protein degradation via both autophagy and the ubiquitin-protease system [69]. mTORC1 can be phosphorylated and activated by Akt as well as several other phosphorylation pathways [75]. Activation of Akt/mTOR1 signaling regulates protein synthesis through numerous downstream effectors that control protein translation [76]. For instance, active mTORC1 phosphorylates both the eukaryotic initiation factor 4E-binding protein (4E-BP1) and ribosomal protein S6 kinase p70S6K1; collectively, this results in increased rates of translation and protein synthesis [77].
Studies investigating the impact of ROS on mTORC1 provide divergent results with some reports concluding that ROS depresses Akt/mTORC1 signaling in both muscle fibers and neurons [78,79] whereas other studies conclude that ROS activates Akt/mTORC1 in both cancer and embryonic kidney cells [[80], [81], [82]]. These contradictory findings likely result from differences in the cell types studied and the variances in ROS treatments used in the experiments. For example, a well-designed study using myocytes reveals that exposure to low levels of oxidants depresses Akt signaling whereas high levels of ROS activate Akt [83]. Hence, the impact of ROS on the activation of the Akt/mTORC1 pathways may depend upon the cellular level of oxidative stress.
MAP kinases are a family of ubiquitous proline-directed, protein-serine/threonine kinases that participate in a variety of signal transduction pathways [84]. All mammalian cells possess multiple MAPK signaling pathways but the three best-described MAPK family members are p38 MAPK, c-Jun N-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK) [85]. Although these three MAPK's are structurally similar, each MAPK has distinct functions. Importantly, each of these MAPKs is activated by oxidative stress and notably, MAPK family members contribute to both exercise-induced muscle adaptation and skeletal muscle atrophy [74,86]. Details about the role that MAPKs play in exercise-induced muscle adaptations and prolonged muscle inactivity is addressed later.
Another important kinase that contributes to maintaining cellular homeostasis and exercise-induced adaptations in skeletal muscles is AMP-activated kinase (AMPK) [87,88]. AMPK is responsible for phosphorylating down-stream substrates that regulate numerous cellular functions including control of glucose uptake and fatty acid oxidation in skeletal muscle [89]. Moreover, AMPK is also known for its regulation of mTORC1 [89]. Although the allosteric regulation of AMPK activity is under complex control, research indicates that AMPK activity is impacted by redox control; this redox regulation does not appear to be due to a direct influence of ROS on AMPK but is a secondary consequence of redox effects on other processes [90].
4.5. Regulation of calcium ion channels
Cellular ion channels are responsible for maintaining ionic homeostasis between the extracellular and intracellular spaces. Indeed, stringent ion homeostasis is critical for a variety of cellular functions. In particular, calcium ions (Ca2+) are vital second messengers required for the regulation of numerous signaling pathways [91]. Perturbations in the control of Ca2+ channels often have deleterious effects on cellular function, and it is confirmed that both Ca2+ channels and Ca2+ transporters are under redox control [69,92,93]. For example, the voltage-gated Ca2+ channel family contains five subgroups of channels and all are subject to redox regulation; these channels are widely expressed in many cell types and function in the control of muscle contraction, protease activation (e.g., calpain), gene expression, and other metabolic functions [92]. In reference to skeletal muscle, ROS mediated oxidation of the voltage-gated ryanodine receptor located on the sarcoplasmic reticulum (SR)) results in leakage of Ca2+ from the SR into the cytosol [94].
Because high cytosolic Ca2+ levels are toxic to cells, the plasma membrane is equipped with active transport Ca2+ pumps (i.e., plasma membrane Ca2+ ATPases) to transport Ca2+ across the cell membrane [95]. Notably, the ROS-mediated formation of the reactive aldehyde, 4-hydroxy-2,3-trans-nonenal, can inhibit the activity of these Ca2+ ATPases and hinder Ca2+ removal from the cell [96]. Therefore, oxidative stress can promote disturbances in intracellular Ca2+ homeostasis due to both increased Ca2+ conductance through voltage-gated Ca2+ channels and the diminished ability to remove Ca2+ from the cell [32,42,69,93]. Details about the role that disturbed redox homeostasis plays in muscle wasting follows.
5. Prolonged inactivity-induced atrophy of skeletal muscles: role of disturbed redox signaling
The size of skeletal muscle fibers is regulated by the balance between the rates of protein synthesis and degradation. Indeed, when the rates of protein breakdown exceed the rate of protein synthesis, muscles lose protein and fiber atrophy ensues. The first suggestion that oxidative stress contributes to disuse muscle atrophy was reported by Kondo and colleagues over 30 years ago [2]. However, this original account did not provide evidence of how disturbed redox signaling promotes muscle atrophy and details of the connection between oxidants and muscle wasting did not emerge until the past two decades. The next segments summarize the evidence that oxidative stress depresses protein synthesis and accelerates proteolysis.
5.1. Chronic oxidative stress inhibits anabolic signaling and decreases muscle protein synthesis
As discussed previously, significant differences exist in both the source and temporal pattern of ROS production in muscle fibers during prolonged periods of disuse compared to ROS emission during muscle contractions. Whereas acute exercise results in a temporary rise in ROS production that ceases when contractions stop, prolonged muscle inactivity results in a chronic elevation in ROS production within the inactive muscle fibers. Moreover, the primary source of ROS production in muscles differ between exercise-induced oxidant production and the ROS produced during chronic inactivity. Again, the increased ROS production within inactive fibers is primarily derived from mitochondria whereas NAD(P)H oxidase is the dominant source of ROS emission in contracting muscle fibers. Notably, these differences in both the sources and temporal pattern of ROS emissions result in diverse redox signaling responses in muscle fibers during prolonged periods of inactivity compared to acute exercise. A discussion of the impact of inactivity-induced disturbed redox signaling on muscle protein synthesis follows.
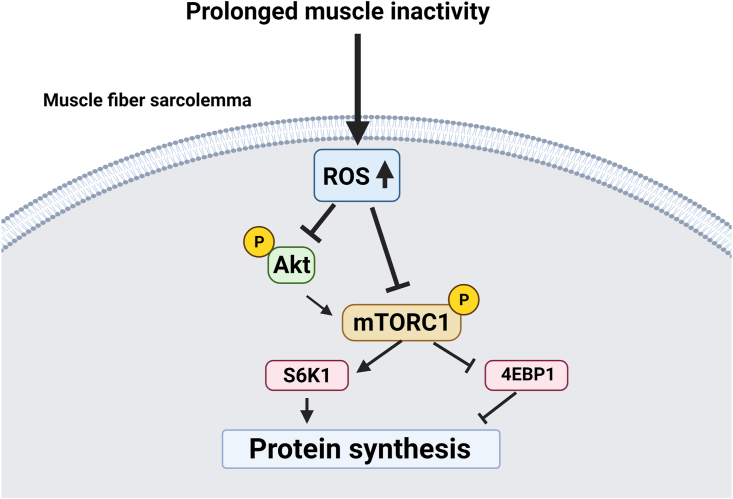
As introduced earlier, the major signaling pathway that regulates muscle protein synthesis is the protein kinase b (Akt)/mTORC 1 pathway. Stimulation of this pathway promotes protein synthesis whereas inhibition of this pathway depresses this process [69]. The impact of ROS on activation of the Akt/mTOR signaling pathway depends on both the specific ROS species produced and the levels of ROS production. For example, low levels of oxidants (e.g., H2O2) depress Akt signaling whereas high levels of ROS can activate Akt [83]. Therefore, does the level of ROS production in muscle fibers during prolonged inactivity promote a decrease in muscle protein synthesis? Two independent studies demonstrate that the answer to this question is yes. First, an investigation of cardiac myocytes isolated from the rat heart reveal that exposure of myocytes to pathophysiological levels of H2O2 leads to dephosphorylation of 4E-BP1 and a decrease in global protein synthesis [97]. Similarly, an in vivo study, using mitochondrial-targeted antioxidants to prevent inactivity-induced oxidative stress in diaphragm muscle concluded that oxidative stress plays a key role in inactivity-induced decreases in muscle protein synthesis [98]. Specifically, these experiments reveal that inactivity-induced oxidative stress in diaphragm muscle is associated with decreased levels of both phosphorylated Akt and mTORC1. Notably, prevention of the inactivity-induced oxidative stress in diaphragm fibers increased the levels of phosphorylated Akt, mTORC1, and 4E-BP1 and these increases were paralleled by higher rates of protein synthesis [98]. Together, these findings support the notion that oxidative stress depresses muscle protein synthesis by decreasing Akt/mTORC1 signaling and subsequently, mRNA translation (Fig. 1).
Fig. 1.
Illustration of the Akt/mTORC1 pathway leading to protein synthesis. Notice that increased production of ROS inhibits several steps in this process. See text for details.
5.2. Oxidants activate proteases and accelerate proteolysis in skeletal muscle fibers
Like most cells, skeletal muscles possess four major proteolytic systems (e.g., calpains, UPS, caspase-3, autophagy) and each of these proteases can be activated by oxidants produced in muscle fibers during prolonged periods of inactivity [32,99]. Details of the mechanism(s) linking oxidative stress to the activation of each of these proteolytic systems has been described in other reviews and therefore, only a short summary is provided here [32,69,99].
5.2.1. Oxidative stress-induced activation of calpains
Calpains are Ca2+-activated proteases that cleave >100 targeted proteins in skeletal muscles including oxidized contractile proteins (e.g., actin and myosin) [100]. Moreover, active calpains can also cleave structural sarcomeric proteins such as titin and nebulin along with a host of kinases and phosphatases [100]. While humans possess 15 different calpain genes, the two dominant calpains that contribute to inactivity-induced muscle wasting are calpain I and calpain II [93].
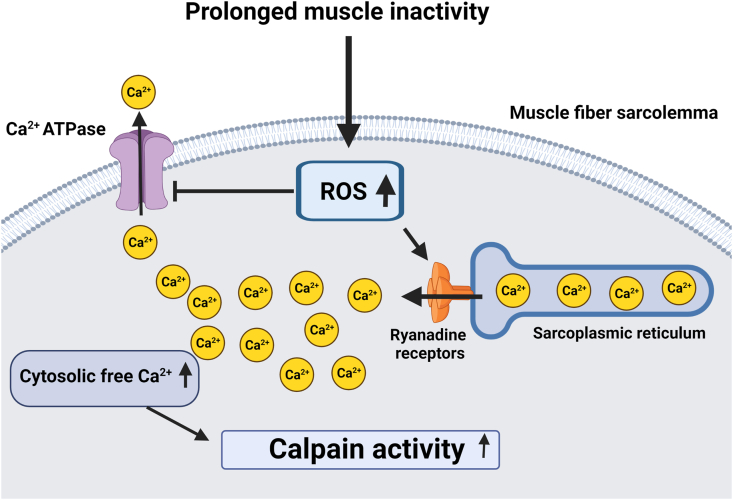
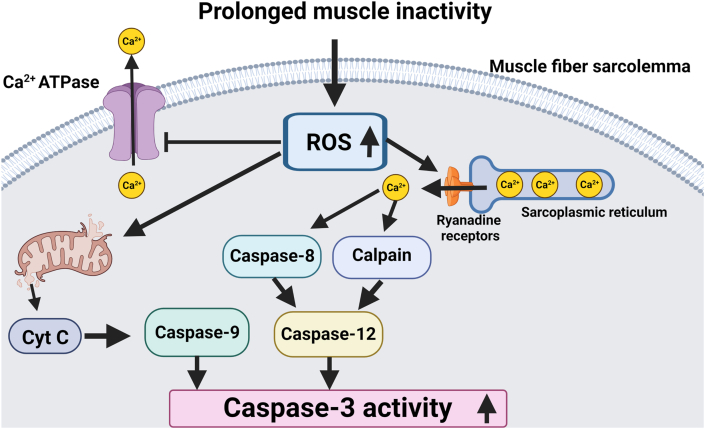
Abundant evidence demonstrates that oxidative stress activates calpains in skeletal muscles [32,93]. The key mechanism linking oxidative stress with calpain activation is the oxidant-directed increase in free Ca2+ in the cytosol [91,92,101]. Specifically, evidence indicates that ROS-mediated oxidation of the ryanodine receptor in skeletal muscle leads to leakage of Ca2+ from the SR into the cytosol [102,103]. Moreover, oxidative stress inhibits the activity of plasma membrane Ca2+ ATPases resulting in a diminished ability to remove Ca2+ from the fiber [96]. Therefore, an elevation in cellular ROS contributes to disturbances in cellular Ca2+ homeostasis due to the combination of Ca2+ leakage from the SR and a decreased ability to remove Ca2+ from the cytsol [32,42,69,93] (Fig. 2).
Fig. 2.
Illustration of the impact of increased ROS production on cellular levels of free Ca+2 and calpain activation. See text for details.
5.2.2. Oxidative stress-induced activation of the UPS
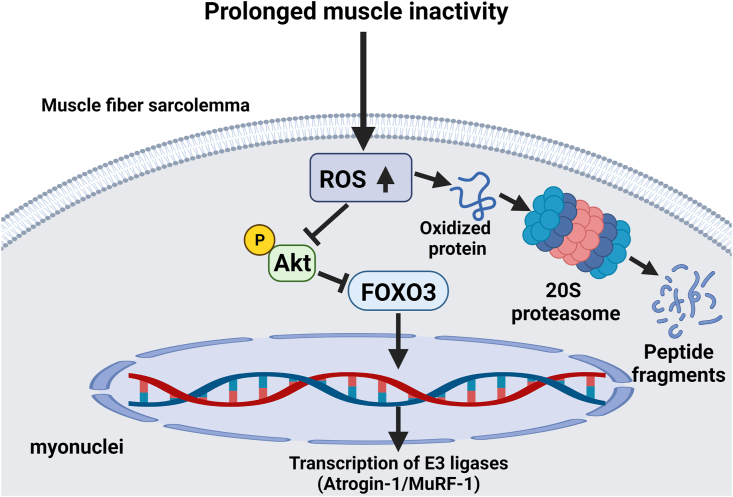
The total UPS complex (26S) is comprised of a core proteasome subunit (20S) that is regulated by two complexes connected to each end of the 20S subunit; notably, this proteasome subunit is responsible for protein breakdown [[104], [105], [106]]. The UPS becomes active when damaged proteins are tagged for degradation by the covalent binding of ubiquitin to a protein [[104], [105], [106]]. This binding of ubiquitin to the damaged molecule involves several reactions including a final stage whereby specialized protein ligases (E3 ligases) identify and tag specific protein substrates with ubiquitin [31]. Notably, oxidized proteins can also be degraded by the 20S protease without a ubiquitin tag [[104], [105], [106]].
Oxidative stress can influence UPS-mediated proteolysis in several ways. First, ROS promotes the increased expression of muscle specific E3 ligases including atrogin-1 and muscle ring finger1 [104,105]. Second, while oxidative stress has been reported to allosterically lower 26S proteasome activity, the 20S proteasome is less susceptible to downregulation by ROS [104,105]. Lastly, oxidized proteins are more susceptible for degradation by the UPS [104,105]. Hence, abundant evidence supports the concept that increased ROS production in cells accelerates protein degradation via the UPS (Fig. 3).
Fig. 3.
Illustration of the influence of ROS on the ubiquitin-proteasome system of proteolysis in skeletal muscle fibers during prolonged periods of inactivity. See text for details.
5.2.3. Oxidative stress-induced activation of caspase-3
Caspase-3 is a member of the cysteine-aspartic acid protease family that normally exists in an inactive (proenzyme) form. Activation of caspase-3 degrades numerous cellular proteins and plays an active role in apoptosis [101]. In regard to caspase-3 and muscle atrophy, it is recognized that active caspase-3 contributes to inactivity-induced muscle atrophy by degrading actomyosin complexes [107].
It is well-documented that oxidative stress activates caspase-3 in skeletal muscle fibers [19,41,108] and that oxidative stress is responsible for the activation of caspase-3 in skeletal muscle during prolonged periods of inactivity [34,35,109]. Further, ROS-mediated oxidation of myofibrillar proteins increases the susceptibility of both actin and myosin to degradation by caspase-3 [107]. Cooperatively, these findings corroborate that ROS accelerates proteolysis in skeletal muscle fibers via the activation of caspase-3 (Fig. 4).
Fig. 4.
Increased ROS production in muscle fibers can activate caspase-3 via multiple signaling pathways. See text for details.
5.2.4. Oxidative stress-induced activation of autophagy
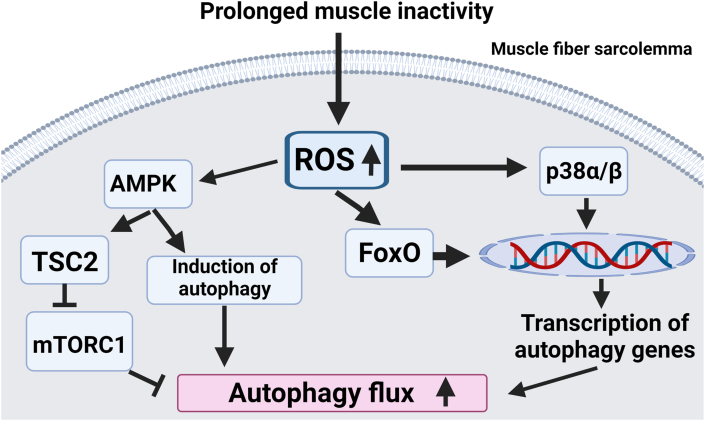
Autophagy is a lysosomal proteolytic pathway for the degradation of damaged cytosolic proteins and organelles in numerous cells including skeletal muscle fibers [110]. The delivery of damaged proteins and organelles to lysosomes occurs in three different ways (i.e., microautophagy, chaperone-mediated autophagy, macroautophagy). Our focus will be macroautophagy, hereafter referred to as “autophagy”. Abundant evidence indicates that prolonged muscle disuse increases the expression of autophagy genes, activates lysosomal proteases (i.e., cathepsin B, D, and L) and accelerates autophagic breakdown of muscle proteins [35,[111], [112], [113]]. Studies also document that autophagy is accelerated during disuse muscle atrophy [114]. For example, evidence indicates that enhanced autophagy is obligatory for mechanical ventilation-induced diaphragmatic atrophy [113]. Together, these studies confirm that autophagy contributes to the proteolysis associated with prolonged muscle inactivity.
Numerous cell culture studies demonstrate that oxidative stress stimulates autophagy in myotubes [35,112,113,115]. For example, direct exposure of C2C12 myotubes to H2O2 activates autophagy [73,116,117]. Indeed, oxidative stress promotes autophagy in several ways. First, increased ROS production in cells can activate AMPK leading to suppression of mTORC1 activation via the tuberous sclerosis complex 2; together, these signaling events activate autophagy as active mTORC1 inhibits the induction of autophagy [115]. Furthermore, ROS can stimulate autophagy by promoting the expression of key autophagy genes. Precisely, exposing cells to H2O2 increases the expression of several autophagy genes including LC3 and Beclin-1; the importance of this increased expression of autophagy genes is supported by the observation that oxidative stress is linked to the increased formation of autophagosomes (reviewed in Ref. [115]). An important signaling pathway connected to ROS-mediated expression of autophagy genes involves activation of the mitogen-activated kinase, p38 alpha/beta. Indeed, activation of p38 elevates the expression of important autophagy-related genes (e.g., Atg7) [73]. Collectively, these factors provide a mechanistic connection between oxidative stress and accelerated autophagy in myotubes.
In addition to the aforementioned in vitro studies, in vivo studies provide added support that oxidative stress promotes autophagy in skeletal muscles during prolonged muscle inactivity. For example, inhibition of inactivity-induced oxidative stress in skeletal muscles blocks the activation of FoxO signaling; this is important because active FoxO elevates the expression of several autophagy-related proteins and promotes autophagy in muscle fibers [32]. Together, these in vivo and in vitro experiments provide robust evidence that oxidative stress plays a key role in activating autophagy in skeletal muscles during periods of prolonged inactivity (Fig. 5).
Fig. 5.
Illustration of the impact of increased ROS production on autophagy in skeletal muscle fibers exposed to prolonged periods of inactivity. Note that increased ROS production accelerates autophagy flux via multiple signaling pathways. See text for details.
6. Endurance exercise-induced adaptation in skeletal muscles: role of ROS
Skeletal muscle is a plastic tissue capable of significant adaptation in response to endurance exercise training. Notably, exercise-induced muscle adaptations occur rapidly as demonstrated by the observation that as few as 5 consecutive days of exercise results in significant changes in muscle fibers [118]. Research during the past three decades has provided key information about the signaling pathways that trigger exercise-induced adaptations in skeletal muscle fibers. In this regard, substantial evidence reveals that oxidants contribute to exercise-induced muscle adaptions. The next segments review the data indicating that increased ROS production plays a key role in exercise-mediated changes in skeletal muscle. We then highlight two key redox-sensitive signaling pathways that contribute to the endurance exercise-induced changes in the biochemical make-up of skeletal muscles.
6.1. Evidence that increased ROS production contributes to endurance exercise-induced muscle adaptation
Davies and colleagues were the first to postulate that contraction-induced ROS production is a primary stimulus to promote muscle adaptation to exercise training [1]. Since this early report, many studies support the notion that increased ROS production is a key stimulus for exercise-induced adaptations in skeletal muscle fibers. For example, abundant evidence confirms that numerous redox-sensitive genes exist in skeletal muscle. Indeed, in vitro studies indicate that exposure of myotubes to oxidants (e.g., hydrogen peroxide) results in increased expression of many genes, including key antioxidant enzymes along with genes regulated by the transcriptional coactivator, peroxisome proliferator-activated receptor-γ coactivator-1 protein-α (PGC-1α) [116,[119], [120], [121]]. It is noteworthy that several in vitro studies have employed non-physiological levels of H2O2 and therefore, the outcomes of these studies may not reflect in vivo occurrences. Nonetheless, the evaluable evidence indicates that these oxidant-mediated increases in gene expression are due to altered redox signaling [116]. Similarly, it has also been shown that ROS production is also required for the contraction-induced gene expression associated with PGC-1α activation in primary rat muscle cells [122]. Together, these studies provide proof of concept that oxidants are capable of significantly altering in vitro gene expression in myotubes.
Importantly, numerous in vivo studies have concluded that contraction-induced ROS production plays a required role in endurance exercise-induced adaptations in skeletal muscle in vivo. The universal approach in these studies is to treat animals or humans with antioxidants to scavenge ROS and abolish the exercise-induced redox signaling effects. The first in vivo studies to demonstrate that antioxidant supplementation suppresses endurance exercise-induced muscle adaptations were preclinical studies. Specifically, in the early 2000's, two independent studies concluded that antioxidant supplement suppressed exercise-induced expression of heat shock protein 72 in rodent skeletal muscles 72 [123,124]. These findings were later supported by human studies revealing that antioxidant supplementation with relatively high doses of vitamin E (400 IU/day) and vitamin C (1 g/day) blunt endurance exercise-induced increases in both antioxidant enzymes and key markers of mitochondrial biogenesis [[125], [126], [127], [128]]. Nonetheless, this topic is not without controversy as studies using lower doses of antioxidants have often reported no negative effects of vitamin E and vitamin C supplementation on skeletal muscle adaptation to exercise [[129], [130], [131]]. However, a close examination of both preclinical and human studies reveals that most studies supplementing with high doses of both vitamin E and vitamin C conclude that exercise-induced ROS production is a prerequisite for optimal training-induced adaptations in skeletal muscle. The next sections highlight the role that redox signaling plays in endurance exercise-induced increases in both mitochondrial biogenesis and the expression of antioxidant enzymes.
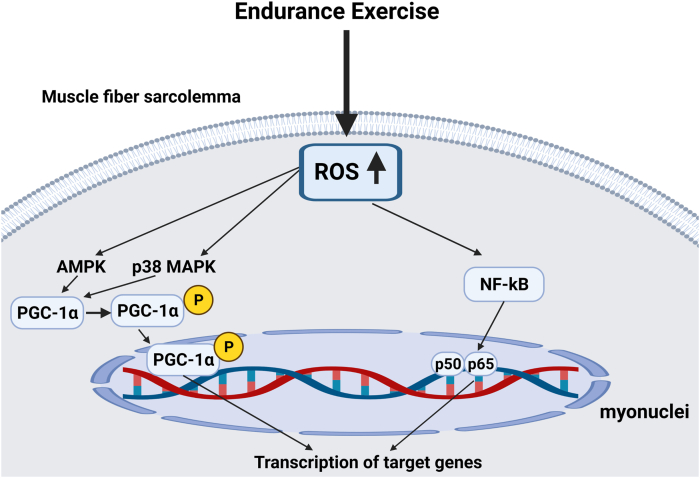
6.2. Role of redox signaling in exercise-induced mitochondrial biogenesis in skeletal muscle
A hallmark of endurance exercise training is an increase in mitochondrial volume within the exercised skeletal muscle fibers. Indeed, it is well-established that a bout of endurance exercise results in significant increases in mitochondrial biogenesis. In this regard, PGC-1α is considered the master regulator of mitochondrial biogenesis [132,133]. PGC-1α is regulated at both the transcriptional and post-translational levels [134]; a detailed discussion of the regulation of PGC-1α is beyond the scope of this review and therefore, we will focus on redox sensitive factors that regulate PGC-1α. Briefly, transcriptional activation of PGC-1α is regulated by numerous signals including up-stream signals from AMPK and p38 MAPK [132,135,136] (Fig. 6). As discussed earlier, both kinases are upregulated by increased ROS production and redox sensitive processes [[137], [138], [139]]. Further, numerous studies conclude that treatment of both animals and humans with antioxidants dampens the exercise-induced increase in mitochondrial biogenesis in skeletal muscles. Additional evidence that exercise-induced mitochondrial biogenesis is a redox-sensitive process is provided by the finding that allopurinol, a xanthine oxidase inhibitor, decreases exercise-induced ROS production in skeletal muscles and blunts exercise-induced PGC-1α gene expression and mitochondrial biogenesis [140]. Together, these results corroborate that exercise-induced increases in ROS production provide a key stimulus to trigger mitochondrial biogenesis.
Fig. 6.
Schematic diagram illustration two key signaling pathways involved in exercise-induced ROS-mediated gene expression in skeletal muscle fibers. See text for details.
6.3. Role of redox signaling in promoting exercise-increased increases in antioxidant enzymes within skeletal muscle
It is established that endurance exercise training elevates the levels of numerous antioxidant enzymes in skeletal muscle [141]. Further, the magnitude of this exercise-induced increase in antioxidants follows an exercise/intensity dose-response curve and antioxidant enzymes increase in only those muscles engaged in the exercise activity [[142], [143], [144]]. This exercise-induced increase in the expression of antioxidant enzymes is due, in part, to redox-regulation of distinct, but interrelated signaling pathways. In particular, exercise-induced increases in antioxidant enzymes occur due activation of PGC-1α, Nrf2, and NF-ĸB [132]. Again, these signaling pathways are activated by several factors including increases in ROS and activation of select redox sensitive kinases (e.g., AMPK, p38 MAPK) [116].
Active PGC-1α regulates the expression of mitochondrial defense in muscle fibers by increasing the levels of manganese SOD (SOD2), catalase, peroxiredoxin 3, peroxiredoxin 5, and thioredoxin [134]. Further, the redox sensitive transcription factor NF-ĸB also contributes to increased SOD2 expression [145]. Finally, one of the major pathways for exercise-induced expression of antioxidant enzymes is the Keap1-Nrf2 pathway. Although the exercise-induced increases in muscle antioxidant enzymes occurs due to a cooperative interaction between PGC-1α, Nrf2, and NF-ĸB signaling, evidence reveals that Nrf2 plays a required role in this process. For example, experiments using Nrf2 knockout mice indicate that Nrf2 is essential for exercise-induced increases in gene expression for SOD1, SOD2, and catalase [146]. Nonetheless, at present, the details of how PGC-1α and NF-ĸB interact with Nrf2 to promote increased antioxidant gene expression remain obscure.
7. Future directions
The discovery that the production of ROS increases in both contracting skeletal muscle fibers and in muscle fibers exposed to prolonged periods of inactivity ushered in the exciting field of research in skeletal muscle redox biology. In the 30–40 years following these landmark discoveries, much has been learned about the role that redox signaling plays in skeletal muscle adaptation in response to exercise training and during prolonged periods of muscle inactivity. However, several key questions remain unanswered. For example, while it is clear that mitochondria are a dominant site of ROS production in muscle fibers exposed to prolonged inactivity, the mechanism(s) that trigger mitochondrial damage and this increase in ROS production remain ambiguous.
Further, while H2O2 can cross plasma membranes of numerous cells via water channels (aquaporins), the abundance of these channels in the sarcolemma of muscle fibers remains unknown. This is an important topic that needs experimental attention.
To date, the long-standing query of whether H2O2 freely crosses mitochondrial membranes remains unresolved. An improved understanding of both the mechanisms and kinetics of H2O2 transfer across mitochondrial membranes is essential to better understand the role that increased mitochondrial ROS emission plays in redox signaling in muscle fibers and other cell types.
Finally, as noted earlier, exercise-induced increases in antioxidant enzymes within muscle fibers are driven by interactions between PGC-1α, Nrf2, and NF-ĸB signaling. However, the precise details of these interactions remain unclear. Therefore, additional studies are needed to better understand the interaction between PGC-1α, Nrf2, and NF-ĸB in regulating the exercise-induced increase in the expression of antioxidant enzymes in skeletal muscle.
Author contributions
Conceptualization, Scott Powers and Matthew Schrager; Review of literature, Scott Powers and Matthew Schrager; Writing – original draft, Scott Powers; Writing – review & editing, Scott Powers and Matthew Schrager.
Submission declaration and verification
This work has not been published previously and is not under consideration for publication elsewhere. Moreover, both authors fully approve of the contents of this report.
Declaration of competing interest
The authors (Scott Powers and Matthew Schrager) do not have a conflict of interest to report.
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R21AR063956 to SKP). Figures created with BioRender.com.
References
- 1.Davies K.J., Packer L., Brooks G.A. Exercise bioenergetics following sprint training. Arch. Biochem. Biophys. 1982;215:260–265. doi: 10.1016/0003-9861(82)90303-4. [DOI] [PubMed] [Google Scholar]
- 2.Kondo H., Miura M., Itokawa Y. Oxidative stress in skeletal muscle atrophied by immobilization. Acta Physiol. Scand. 1991;142:527–528. doi: 10.1111/j.1748-1716.1991.tb09191.x. [DOI] [PubMed] [Google Scholar]
- 3.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 4.Forman H.J., Maiorino M., Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J. Biol. Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winterbourn C.C. Biological production, detection, and fate of hydrogen peroxide. Antioxidants Redox Signal. 2018;29:541–551. doi: 10.1089/ars.2017.7425. [DOI] [PubMed] [Google Scholar]
- 7.Go Y.M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84:227–245. doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson M.J., Vasilaki A., McArdle A. Cellular mechanisms underlying oxidative stress in human exercise. Free Radic. Biol. Med. 2016;98:13–17. doi: 10.1016/j.freeradbiomed.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 9.Powers S.K., Ji L.L., Kavazis A.N., Jackson M.J. Reactive oxygen species: impact on skeletal muscle. Compr. Physiol. 2011;1:941–969. doi: 10.1002/cphy.c100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powers S.K., Nelson W.B., Hudson M.B. Exercise-induced oxidative stress in humans: cause and consequences. Free Radic. Biol. Med. 2011;51:942–950. doi: 10.1016/j.freeradbiomed.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Pattwell D.M., McArdle A., Morgan J.E., Patridge T.A., Jackson M.J. Release of reactive oxygen and nitrogen species from contracting skeletal muscle cells. Free Radic. Biol. Med. 2004;37:1064–1072. doi: 10.1016/j.freeradbiomed.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Palomero J., Vasilaki A., Pye D., McArdle A., Jackson M.J. Aging increases the oxidation of dichlorohydrofluorescein in single isolated skeletal muscle fibers at rest, but not during contractions. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;305:R351–R358. doi: 10.1152/ajpregu.00530.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koren A., Sauber C., Sentjurc M., Schara M. Free radicals in tetanic activity of isolated skeletal muscle. Comp. Biochem. Physiol. B. 1983;74:633–635. doi: 10.1016/0305-0491(83)90241-9. [DOI] [PubMed] [Google Scholar]
- 14.Boveris A., Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loschen G., Azzi A., Richter C., Flohe L. Superoxide radicals as precursors of mitochondrial hydrogen peroxide. FEBS Lett. 1974;42:68–72. doi: 10.1016/0014-5793(74)80281-4. [DOI] [PubMed] [Google Scholar]
- 16.Anderson E.J., Neufer P.D. Type II skeletal myofibers possess unique properties that potentiate mitochondrial H(2)O(2) generation. Am. J. Physiol. Cell Physiol. 2006;290:C844–C851. doi: 10.1152/ajpcell.00402.2005. [DOI] [PubMed] [Google Scholar]
- 17.St-Pierre J., Buckingham J.A., Roebuck S.J., Brand M.D. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 18.Kavazis A.N., Talbert E.E., Smuder A.J., Hudson M.B., Nelson W.B., Powers S.K. Mechanical ventilation induces diaphragmatic mitochondrial dysfunction and increased oxidant production. Free Radic. Biol. Med. 2009;46:842–850. doi: 10.1016/j.freeradbiomed.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powers S.K., Hudson M.B., Nelson W.B., Talbert E.E., Min K., Szeto H.H., Kavazis A.N., Smuder A.J. Mitochondria-targeted antioxidants protect against mechanical ventilation-induced diaphragm weakness. Crit. Care Med. 2011;39:1749–1759. doi: 10.1097/CCM.0b013e3182190b62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaelson L.P., Shi G., Ward C.W., Rodney G.G. Mitochondrial redox potential during contraction in single intact muscle fibers. Muscle Nerve. 2010;42:522–529. doi: 10.1002/mus.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakellariou G.K., Vasilaki A., Palomero J., Kayani A., Zibrik L., McArdle A., Jackson M.J. Studies of mitochondrial and nonmitochondrial sources implicate nicotinamide adenine dinucleotide phosphate oxidase(s) in the increased skeletal muscle superoxide generation that occurs during contractile activity. Antioxidants Redox Signal. 2013;18:603–621. doi: 10.1089/ars.2012.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers S.K., Jackson M.J. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol. Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakellariou G.K., Jackson M.J., Vasilaki A. Redefining the major contributors to superoxide production in contracting skeletal muscle. The role of NAD(P)H oxidases. Free Radic. Res. 2014;48:12–29. doi: 10.3109/10715762.2013.830718. [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Vegas A., Campos C.A., Contreras-Ferrat A., Casas M., Buvinic S., Jaimovich E., Espinosa A. ROS production via P2Y1-PKC-NOX2 is triggered by extracellular ATP after electrical stimulation of skeletal muscle cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinosa A., Leiva A., Pena M., Muller M., Debandi A., Hidalgo C., Carrasco M.A., Jaimovich E. Myotube depolarization generates reactive oxygen species through NAD(P)H oxidase; ROS-elicited Ca2+ stimulates ERK, CREB, early genes. J. Cell. Physiol. 2006;209:379–388. doi: 10.1002/jcp.20745. [DOI] [PubMed] [Google Scholar]
- 26.Xia R., Webb J.A., Gnall L.L., Cutler K., Abramson J.J. Skeletal muscle sarcoplasmic reticulum contains a NADH-dependent oxidase that generates superoxide. Am. J. Physiol. Cell Physiol. 2003;285:C215–C221. doi: 10.1152/ajpcell.00034.2002. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira L.F., Laitano O. Regulation of NADPH oxidases in skeletal muscle. Free Radic. Biol. Med. 2016;98:18–28. doi: 10.1016/j.freeradbiomed.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong M.C., Arbogast S., Guo Z., Mathenia J., Su W., Reid M.B. Calcium-independent phospholipase A2 modulates cytosolic oxidant activity and contractile function in murine skeletal muscle cells. J. Appl. Physiol. 1985;100:399–405. doi: 10.1152/japplphysiol.00873.2005. 2006. [DOI] [PubMed] [Google Scholar]
- 29.Nethery D., Stofan D., Callahan L., DiMarco A., Supinski G. Formation of reactive oxygen species by the contracting diaphragm is PLA(2) dependent. J. Appl. Physiol. 1985;87:792–800. doi: 10.1152/jappl.1999.87.2.792. 1999. [DOI] [PubMed] [Google Scholar]
- 30.Nemes R., Koltai E., Taylor A.W., Suzuki K., Gyori F., Radak Z. Reactive oxygen and nitrogen species regulate key metabolic, anabolic, and catabolic pathways in skeletal muscle. Antioxidants. 2018;7 doi: 10.3390/antiox7070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers S.K., Kavazis A.N., McClung J.M. Oxidative stress and disuse muscle atrophy. J. Appl. Physiol. 2007;102:2389–2397. doi: 10.1152/japplphysiol.01202.2006. 1985. [DOI] [PubMed] [Google Scholar]
- 32.Powers S.K., Ozdemir M., Hyatt H. Redox control of proteolysis during inactivity-induced skeletal muscle atrophy. Antioxidants Redox Signal. 2020 doi: 10.1089/ars.2019.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powers S.K., Wiggs M.P., Duarte J.A., Zergeroglu A.M., Demirel H.A. Mitochondrial signaling contributes to disuse muscle atrophy. Am. J. Physiol. Endocrinol. Metab. 2012;303:E31–E39. doi: 10.1152/ajpendo.00609.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min K., Smuder A.J., Kwon O.S., Kavazis A.N., Szeto H.H., Powers S.K. Mitochondrial-targeted antioxidants protect skeletal muscle against immobilization-induced muscle atrophy. J. Appl. Physiol. 1985;(111):1459–1466. doi: 10.1152/japplphysiol.00591.2011. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talbert E.E., Smuder A.J., Min K., Kwon O.S., Szeto H.H., Powers S.K. Immobilization-induced activation of key proteolytic systems in skeletal muscles is prevented by a mitochondria-targeted antioxidant. J. Appl. Physiol. 1985;115:529–538. doi: 10.1152/japplphysiol.00471.2013. 2013. [DOI] [PubMed] [Google Scholar]
- 36.Falk D.J., Deruisseau K.C., Van Gammeren D.L., Deering M.A., Kavazis A.N., Powers S.K. Mechanical ventilation promotes redox status alterations in the diaphragm. J. Appl. Physiol. 1985;101:1017–1024. doi: 10.1152/japplphysiol.00104.2006. 2006. [DOI] [PubMed] [Google Scholar]
- 37.Falk D.J., Kavazis A.N., Whidden M.A., Smuder A.J., McClung J.M., Hudson M.B., Powers S.K. Mechanical ventilation-induced oxidative stress in the diaphragm: role of heme oxygenase-1. Chest. 2011;139:816–824. doi: 10.1378/chest.09-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClung J.M., Van Gammeren D., Whidden M.A., Falk D.J., Kavazis A.N., Hudson M.B., Gayan-Ramirez G., Decramer M., DeRuisseau K.C., Powers S.K. Apocynin attenuates diaphragm oxidative stress and protease activation during prolonged mechanical ventilation. Crit. Care Med. 2009;37:1373–1379. doi: 10.1097/CCM.0b013e31819cef63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanely R.A., Zergeroglu M.A., Lennon S.L., Sugiura T., Yimlamai T., Enns D., Belcastro A., Powers S.K. Mechanical ventilation-induced diaphragmatic atrophy is associated with oxidative injury and increased proteolytic activity. Am. J. Respir. Crit. Care Med. 2002;166:1369–1374. doi: 10.1164/rccm.200202-088OC. [DOI] [PubMed] [Google Scholar]
- 40.Whidden M.A., McClung J.M., Falk D.J., Hudson M.B., Smuder A.J., Nelson W.B., Powers S.K. Xanthine oxidase contributes to mechanical ventilation-induced diaphragmatic oxidative stress and contractile dysfunction. J. Appl. Physiol. 1985;106:385–394. doi: 10.1152/japplphysiol.91106.2008. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whidden M.A., Smuder A.J., Wu M., Hudson M.B., Nelson W.B., Powers S.K. Oxidative stress is required for mechanical ventilation-induced protease activation in the diaphragm. J. Appl. Physiol. 1985;108:1376–1382. doi: 10.1152/japplphysiol.00098.2010. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyatt H., Deminice R., Yoshihara T., Powers S.K. Mitochondrial dysfunction induces muscle atrophy during prolonged inactivity: a review of the causes and effects. Arch. Biochem. Biophys. 2019;662:49–60. doi: 10.1016/j.abb.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powers S.K., Morton A.B., Hyatt H., Hinkley M.J. The renin-angiotensin system and skeletal muscle. Exerc. Sport Sci. Rev. 2018;46:205–214. doi: 10.1249/JES.0000000000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romanello V., Sandri M. Mitochondrial biogenesis and fragmentation as regulators of protein degradation in striated muscles. J. Mol. Cell. Cardiol. 2013;55:64–72. doi: 10.1016/j.yjmcc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Romanello V., Sandri M. Mitochondrial quality control and muscle mass maintenance. Front. Physiol. 2015;6:422. doi: 10.3389/fphys.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romanello V., Sandri M. The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass. Cell. Mol. Life Sci. 2021;78:1305–1328. doi: 10.1007/s00018-020-03662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid M.B., Haack K.E., Franchek K.M., Valberg P.A., Kobzik L., West M.S. Reactive oxygen in skeletal muscle. I. Intracellular oxidant kinetics and fatigue in vitro. J. Appl. Physiol. 1985;73:1797–1804. doi: 10.1152/jappl.1992.73.5.1797. 1992. [DOI] [PubMed] [Google Scholar]
- 48.He L., He T., Farrar S., Ji L., Liu T., Ma X. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 2017;44:532–553. doi: 10.1159/000485089. [DOI] [PubMed] [Google Scholar]
- 49.Powers S.K., Deminice R., Ozdemir M., Yoshihara T., Bomkamp M.P., Hyatt H. Exercise-induced oxidative stress: friend or foe? J. Sport Health Sci. 2020 doi: 10.1016/j.jshs.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhee S.G., Kil I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 52.Brigelius-Flohe R., Flohe L. Regulatory phenomena in the glutathione peroxidase superfamily. Antioxidants Redox Signal. 2020;33:498–516. doi: 10.1089/ars.2019.7905. [DOI] [PubMed] [Google Scholar]
- 53.Chance B., Sies H., Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 54.Henzler T., Steudle E. Transport and metabolic degradation of hydrogen peroxide in Chara corallina: model calculations and measurements with the pressure probe suggest transport of H(2)O(2) across water channels. J. Exp. Bot. 2000;51:2053–2066. doi: 10.1093/jexbot/51.353.2053. [DOI] [PubMed] [Google Scholar]
- 55.Medrano-Fernandez I., Bestetti S., Bertolotti M., Bienert G.P., Bottino C., Laforenza U., Rubartelli A., Sitia R. Stress regulates aquaporin-8 permeability to impact cell growth and survival. Antioxidants Redox Signal. 2016;24:1031–1044. doi: 10.1089/ars.2016.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pak V.V., Ezerina D., Lyublinskaya O.G., Pedre B., Tyurin-Kuzmin P.A., Mishina N.M., Thauvin M., Young D., Wahni K., Martinez Gache S.A., et al. Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metabol. 2020;31:642–653 e646. doi: 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamma G., Valenti G., Grossini E., Donnini S., Marino A., Marinelli R.A., Calamita G. Aquaporin membrane channels in oxidative stress, cell signaling, and aging: recent advances and research trends. Oxid. Med. Cell. Longev. 2018 doi: 10.1155/2018/1501847. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong Q., Lin C.L. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell. Mol. Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sykiotis G.P., Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 2010 doi: 10.1126/scisignal.3112re3. 3, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Lacher S.E., Lee J.S., Wang X., Campbell M.R., Bell D.A., Slattery M. Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radic. Biol. Med. 2015;88:452–465. doi: 10.1016/j.freeradbiomed.2015.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji L.L. Antioxidant signaling in skeletal muscle: a brief review. Exp. Gerontol. 2007;42:582–593. doi: 10.1016/j.exger.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Jackman R.W., Kandarian S.C. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- 66.Jackman R.W., Cornwell E.W., Wu C.L., Kandarian S.C. Nuclear factor-kappaB signalling and transcriptional regulation in skeletal muscle atrophy. Exp. Physiol. 2013;98:19–24. doi: 10.1113/expphysiol.2011.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lingappan K. NF-kappaB in oxidative stress. Curr. Opin. Toxicol. 2018;7:81–86. doi: 10.1016/j.cotox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morgan M.J., Liu Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powers S.K., Morton A.B., Ahn B., Smuder A.J. Redox control of skeletal muscle atrophy. Free Radic. Biol. Med. 2016;98:208–217. doi: 10.1016/j.freeradbiomed.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eijkelenboom A., Burgering B.M. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 71.Eijkelenboom A., Mokry M., Smits L.M., Nieuwenhuis E.E., Burgering B.M. FOXO3 selectively amplifies enhancer activity to establish target gene regulation. Cell Rep. 2013;5:1664–1678. doi: 10.1016/j.celrep.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 72.Klotz L.O., Steinbrenner H. Cellular adaptation to xenobiotics: interplay between xenosensors, reactive oxygen species and FOXO transcription factors. Redox Biol. 2017;13:646–654. doi: 10.1016/j.redox.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McClung J.M., Judge A.R., Powers S.K., Yan Z. p38 MAPK links oxidative stress to autophagy-related gene expression in cachectic muscle wasting. Am. J. Physiol. Cell Physiol. 2010;298:C542–C549. doi: 10.1152/ajpcell.00192.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Powers S.K., Duarte J., Kavazis A.N., Talbert E.E. Reactive oxygen species are signalling molecules for skeletal muscle adaptation. Exp. Physiol. 2010;95:1–9. doi: 10.1113/expphysiol.2009.050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White J.P., Puppa M.J., Gao S., Sato S., Welle S.L., Carson J.A. Muscle mTORC1 suppression by IL-6 during cancer cachexia: a role for AMPK. Am. J. Physiol. Endocrinol. Metab. 2013;304 doi: 10.1152/ajpendo.00410.2012. E1042-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hornberger T.A. Mechanotransduction and the regulation of mTORC1 signaling in skeletal muscle. Int. J. Biochem. Cell Biol. 2011;43:1267–1276. doi: 10.1016/j.biocel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodman C.A. Role of mTORC1 in mechanically induced increases in translation and skeletal muscle mass. J. Appl. Physiol. 1985;127:581–590. doi: 10.1152/japplphysiol.01011.2018. 2019. [DOI] [PubMed] [Google Scholar]
- 78.Berdichevsky A., Guarente L., Bose A. Acute oxidative stress can reverse insulin resistance by inactivation of cytoplasmic JNK. J. Biol. Chem. 2010;285:21581–21589. doi: 10.1074/jbc.M109.093633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Durgadoss L., Nidadavolu P., Valli R.K., Saeed U., Mishra M., Seth P., Ravindranath V. Redox modification of Akt mediated by the dopaminergic neurotoxin MPTP, in mouse midbrain, leads to down-regulation of pAkt. Faseb. J. 2012;26:1473–1483. doi: 10.1096/fj.11-194100. [DOI] [PubMed] [Google Scholar]
- 80.Mackey A.M., Sanvicens N., Groeger G., Doonan F., Wallace D., Cotter T.G. Redox survival signalling in retina-derived 661W cells. Cell Death Differ. 2008;15:1291–1303. doi: 10.1038/cdd.2008.43. [DOI] [PubMed] [Google Scholar]
- 81.Sadidi M., Lentz S.I., Feldman E.L. Hydrogen peroxide-induced Akt phosphorylation regulates Bax activation. Biochimie. 2009;91:577–585. doi: 10.1016/j.biochi.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarbassov D.D., Sabatini D.M. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 2005;280:39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]
- 83.Tan P.L., Shavlakadze T., Grounds M.D., Arthur P.G. Differential thiol oxidation of the signaling proteins Akt, PTEN or PP2A determines whether Akt phosphorylation is enhanced or inhibited by oxidative stress in C2C12 myotubes derived from skeletal muscle. Int. J. Biochem. Cell Biol. 2015;62:72–79. doi: 10.1016/j.biocel.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 84.Pearson G., Robinson F., Beers Gibson T., Xu B.E., Karandikar M., Berman K., Cobb M.H. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 85.Cuschieri J., Maier R.V. Mitogen-activated protein kinase (MAPK) Crit. Care Med. 2005;33:S417–S419. doi: 10.1097/01.ccm.0000191714.39495.a6. [DOI] [PubMed] [Google Scholar]
- 86.Powers S.K., Talbert E.E., Adhihetty P.J. Reactive oxygen and nitrogen species as intracellular signals in skeletal muscle. J. Physiol. 2011;589:2129–2138. doi: 10.1113/jphysiol.2010.201327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S., Song P., Zou M.H. AMP-activated protein kinase, stress responses and cardiovascular diseases. Clin. Sci. (Lond.) 2012;122:555–573. doi: 10.1042/CS20110625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kjobsted R., Hingst J.R., Fentz J., Foretz M., Sanz M.N., Pehmoller C., Shum M., Marette A., Mounier R., Treebak J.T., et al. AMPK in skeletal muscle function and metabolism. Faseb. J. 2018;32:1741–1777. doi: 10.1096/fj.201700442R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hinchy E.C., Gruszczyk A.V., Willows R., Navaratnam N., Hall A.R., Bates G., Bright T.P., Krieg T., Carling D., Murphy M.P. Mitochondria-derived ROS activate AMP-activated protein kinase (AMPK) indirectly. J. Biol. Chem. 2018;293:17208–17217. doi: 10.1074/jbc.RA118.002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bogeski I., Niemeyer B.A. Redox regulation of ion channels. Antioxidants Redox Signal. 2014;21:859–862. doi: 10.1089/ars.2014.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bogeski I., Kappl R., Kummerow C., Gulaboski R., Hoth M., Niemeyer B.A. Redox regulation of calcium ion channels: chemical and physiological aspects. Cell Calcium. 2011;50:407–423. doi: 10.1016/j.ceca.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 93.Hyatt H.W., Powers S.K. The role of calpains in skeletal muscle remodeling with exercise and inactivity-induced atrophy. Int. J. Sports Med. 2020 doi: 10.1055/a-1199-7662. [DOI] [PubMed] [Google Scholar]
- 94.Andersson D.C., Betzenhauser M.J., Reiken S., Meli A.C., Umanskaya A., Xie W., Shiomi T., Zalk R., Lacampagne A., Marks A.R. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metabol. 2011;14:196–207. doi: 10.1016/j.cmet.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.DeSantiago J., Batlle D., Khilnani M., Dedhia S., Kulczyk J., Duque R., Ruiz J., Pena-Rasgado C., Rasgado-Flores H. Ca2+/H+ exchange via the plasma membrane Ca2+ ATPase in skeletal muscle. Front. Biosci. 2007;12:4641–4660. doi: 10.2741/2414. [DOI] [PubMed] [Google Scholar]
- 96.Siems W., Capuozzo E., Lucano A., Salerno C., Crifo C. High sensitivity of plasma membrane ion transport ATPases from human neutrophils towards 4-hydroxy-2,3-trans-nonenal. Life Sci. 2003;73:2583–2590. doi: 10.1016/s0024-3205(03)00661-1. [DOI] [PubMed] [Google Scholar]
- 97.Pham F.H., Sugden P.H., Clerk A. Regulation of protein kinase B and 4E-BP1 by oxidative stress in cardiac myocytes. Circ. Res. 2000;86:1252–1258. doi: 10.1161/01.res.86.12.1252. [DOI] [PubMed] [Google Scholar]
- 98.Hudson M.B., Smuder A.J., Nelson W.B., Wiggs M.P., Shimkus K.L., Fluckey J.D., Szeto H.H., Powers S.K. Partial support ventilation and mitochondrial-targeted antioxidants protect against ventilator-induced decreases in diaphragm muscle protein synthesis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Powers S.K., Smuder A.J., Criswell D.S. Mechanistic links between oxidative stress and disuse muscle atrophy. Antioxidants Redox Signal. 2011;15:2519–2528. doi: 10.1089/ars.2011.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goll D.E., Thompson V.F., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 101.Powers S.K., Kavazis A.N., DeRuisseau K.C. Mechanisms of disuse muscle atrophy: role of oxidative stress. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R337–R344. doi: 10.1152/ajpregu.00469.2004. [DOI] [PubMed] [Google Scholar]
- 102.Matecki S., Dridi H., Jung B., Saint N., Reiken S.R., Scheuermann V., Mrozek S., Santulli G., Umanskaya A., Petrof B.J., et al. Leaky ryanodine receptors contribute to diaphragmatic weakness during mechanical ventilation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:9069–9074. doi: 10.1073/pnas.1609707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matecki S., Jung B., Saint N., Scheuermann V., Jaber S., Lacampagne A. Respiratory muscle contractile inactivity induced by mechanical ventilation in piglets leads to leaky ryanodine receptors and diaphragm weakness. J. Muscle Res. Cell Motil. 2017;38:17–24. doi: 10.1007/s10974-017-9464-x. [DOI] [PubMed] [Google Scholar]
- 104.Korovila I., Hugo M., Castro J.P., Weber D., Hohn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lefaki M., Papaevgeniou N., Chondrogianni N. Redox regulation of proteasome function. Redox Biol. 2017;13:452–458. doi: 10.1016/j.redox.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shang F., Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Smuder A.J., Kavazis A.N., Hudson M.B., Nelson W.B., Powers S.K. Oxidation enhances myofibrillar protein degradation via calpain and caspase-3. Free Radic. Biol. Med. 2010;49:1152–1160. doi: 10.1016/j.freeradbiomed.2010.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jang Y.C., Rodriguez K., Lustgarten M.S., Muller F.L., Bhattacharya A., Pierce A., Choi J.J., Lee N.H., Chaudhuri A., Richardson A.G., et al. Superoxide-mediated oxidative stress accelerates skeletal muscle atrophy by synchronous activation of proteolytic systems. Geroscience. 2020;42:1579–1591. doi: 10.1007/s11357-020-00200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Talbert E.E., Smuder A.J., Min K., Kwon O.S., Powers S.K. Calpain and caspase-3 play required roles in immobilization-induced limb muscle atrophy. J. Appl. Physiol. 1985;(114):1482–1489. doi: 10.1152/japplphysiol.00925.2012. 2013. [DOI] [PubMed] [Google Scholar]
- 110.Kirkin V., Rogov V.V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 111.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metabol. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 112.Smuder A.J., Sollanek K.J., Min K., Nelson W.B., Powers S.K. Inhibition of forkhead boxO-specific transcription prevents mechanical ventilation-induced diaphragm dysfunction. Crit. Care Med. 2015;43:e133–142. doi: 10.1097/CCM.0000000000000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smuder A.J., Sollanek K.J., Nelson W.B., Min K., Talbert E.E., Kavazis A.N., Hudson M.B., Sandri M., Szeto H.H., Powers S.K. Crosstalk between autophagy and oxidative stress regulates proteolysis in the diaphragm during mechanical ventilation. Free Radic. Biol. Med. 2018;115:179–190. doi: 10.1016/j.freeradbiomed.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Franco-Romero A., Sandri M. Role of autophagy in muscle disease. Mol. Aspect. Med. 2021;82 doi: 10.1016/j.mam.2021.101041. [DOI] [PubMed] [Google Scholar]
- 115.Navarro-Yepes J., Burns M., Anandhan A., Khalimonchuk O., del Razo L.M., Quintanilla-Vega B., Pappa A., Panayiotidis M.I., Franco R. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxidants Redox Signal. 2014;21:66–85. doi: 10.1089/ars.2014.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Irrcher I., Ljubicic V., Hood D.A. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am. J. Physiol. Cell Physiol. 2009;296:C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 117.Rahman M., Mofarrahi M., Kristof A.S., Nkengfac B., Harel S., Hussain S.N. Reactive oxygen species regulation of autophagy in skeletal muscles. Antioxidants Redox Signal. 2014;20:443–459. doi: 10.1089/ars.2013.5410. [DOI] [PubMed] [Google Scholar]
- 118.Vincent H.K., Powers S.K., Stewart D.J., Demirel H.A., Shanely R.A., Naito H. Short-term exercise training improves diaphragm antioxidant capacity and endurance. Eur. J. Appl. Physiol. 2000;81:67–74. doi: 10.1007/PL00013799. [DOI] [PubMed] [Google Scholar]
- 119.Franco A.A., Odom R.S., Rando T.A. Regulation of antioxidant enzyme gene expression in response to oxidative stress and during differentiation of mouse skeletal muscle. Free Radic. Biol. Med. 1999;27:1122–1132. doi: 10.1016/s0891-5849(99)00166-5. [DOI] [PubMed] [Google Scholar]
- 120.McArdle F., Spiers S., Aldemir H., Vasilaki A., Beaver A., Iwanejko L., McArdle A., Jackson M.J. Preconditioning of skeletal muscle against contraction-induced damage: the role of adaptations to oxidants in mice. J. Physiol. 2004;561:233–244. doi: 10.1113/jphysiol.2004.069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McClung J.M., Judge A.R., Talbert E.E., Powers S.K. Calpain-1 is required for hydrogen peroxide-induced myotube atrophy. Am. J. Physiol. Cell Physiol. 2009;296:C363–C371. doi: 10.1152/ajpcell.00497.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Silveira L.R., Pilegaard H., Kusuhara K., Curi R., Hellsten Y. The contraction induced increase in gene expression of peroxisome proliferator-activated receptor (PPAR)-gamma coactivator 1alpha (PGC-1alpha), mitochondrial uncoupling protein 3 (UCP3) and hexokinase II (HKII) in primary rat skeletal muscle cells is dependent on reactive oxygen species. Biochim. Biophys. Acta. 2006;1763:969–976. doi: 10.1016/j.bbamcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 123.Hamilton K.L., Staib J.L., Phillips T., Hess A., Lennon S.L., Powers S.K. Exercise, antioxidants, and HSP72: protection against myocardial ischemia/reperfusion. Free Radic. Biol. Med. 2003;34:800–809. doi: 10.1016/s0891-5849(02)01431-4. [DOI] [PubMed] [Google Scholar]
- 124.Jackson M.J., Khassaf M., Vasilaki A., McArdle F., McArdle A. Vitamin E and the oxidative stress of exercise. Ann. N. Y. Acad. Sci. 2004;1031:158–168. doi: 10.1196/annals.1331.015. [DOI] [PubMed] [Google Scholar]
- 125.Gomez-Cabrera M.C., Domenech E., Romagnoli M., Arduini A., Borras C., Pallardo F.V., Sastre J., Vina J. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am. J. Clin. Nutr. 2008;87:142–149. doi: 10.1093/ajcn/87.1.142. [DOI] [PubMed] [Google Scholar]
- 126.Morrison D., Hughes J., Della Gatta P.A., Mason S., Lamon S., Russell A.P., Wadley G.D. Vitamin C and E supplementation prevents some of the cellular adaptations to endurance-training in humans. Free Radic. Biol. Med. 2015;89:852–862. doi: 10.1016/j.freeradbiomed.2015.10.412. [DOI] [PubMed] [Google Scholar]
- 127.Paulsen G., Cumming K.T., Holden G., Hallen J., Ronnestad B.R., Sveen O., Skaug A., Paur I., Bastani N.E., Ostgaard H.N., et al. Vitamin C and E supplementation hampers cellular adaptation to endurance training in humans: a double-blind, randomised, controlled trial. J. Physiol. 2014;592:1887–1901. doi: 10.1113/jphysiol.2013.267419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ristow M., Zarse K., Oberbach A., Kloting N., Birringer M., Kiehntopf M., Stumvoll M., Kahn C.R., Bluher M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yfanti C., Akerstrom T., Nielsen S., Nielsen A.R., Mounier R., Mortensen O.H., Lykkesfeldt J., Rose A.J., Fischer C.P., Pedersen B.K. Antioxidant supplementation does not alter endurance training adaptation. Med. Sci. Sports Exerc. 2010;42:1388–1395. doi: 10.1249/MSS.0b013e3181cd76be. [DOI] [PubMed] [Google Scholar]
- 130.Yfanti C., Fischer C.P., Nielsen S., Akerstrom T., Nielsen A.R., Veskoukis A.S., Kouretas D., Lykkesfeldt J., Pilegaard H., Pedersen B.K. Role of vitamin C and E supplementation on IL-6 in response to training. J. Appl. Physiol. 1985;112:990–1000. doi: 10.1152/japplphysiol.01027.2010. 2012. [DOI] [PubMed] [Google Scholar]
- 131.Yfanti C., Nielsen A.R., Akerstrom T., Nielsen S., Rose A.J., Richter E.A., Lykkesfeldt J., Fischer C.P., Pedersen B.K. Effect of antioxidant supplementation on insulin sensitivity in response to endurance exercise training. Am. J. Physiol. Endocrinol. Metab. 2011;300:E761–E770. doi: 10.1152/ajpendo.00207.2010. [DOI] [PubMed] [Google Scholar]
- 132.Mason S.A., Trewin A.J., Parker L., Wadley G.D. Antioxidant supplements and endurance exercise: current evidence and mechanistic insights. Redox Biol. 2020;35 doi: 10.1016/j.redox.2020.101471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fernandez-Marcos P.J., Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rius-Perez S., Torres-Cuevas I., Millan I., Ortega A.L., Perez S. PGC-1alpha, inflammation, and oxidative stress: an integrative view in metabolism. Oxid. Med. Cell. Longev. 2020 doi: 10.1155/2020/1452696. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Akimoto T., Pohnert S.C., Li P., Zhang M., Gumbs C., Rosenberg P.B., Williams R.S., Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J. Biol. Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 136.Wu Z., Huang X., Feng Y., Handschin C., Feng Y., Gullicksen P.S., Bare O., Labow M., Spiegelman B., Stevenson S.C. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miranda S., Foncea R., Guerrero J., Leighton F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochem. Biophys. Res. Commun. 1999;258:44–49. doi: 10.1006/bbrc.1999.0580. [DOI] [PubMed] [Google Scholar]
- 138.Piantadosi C.A., Suliman H.B. Redox regulation of mitochondrial biogenesis. Free Radic. Biol. Med. 2012;53:2043–2053. doi: 10.1016/j.freeradbiomed.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Trewin A.J., Bahr L.L., Almast A., Berry B.J., Wei A.Y., Foster T.H., Wojtovich A.P. Mitochondrial reactive oxygen species generated at the complex-II matrix or intermembrane space microdomain have distinct effects on redox signaling and stress sensitivity in Caenorhabditis elegans. Antioxidants Redox Signal. 2019;31:594–607. doi: 10.1089/ars.2018.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kang C., O'Moore K.M., Dickman J.R., Ji L.L. Exercise activation of muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha signaling is redox sensitive. Free Radic. Biol. Med. 2009;47:1394–1400. doi: 10.1016/j.freeradbiomed.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 141.Powers S.K., Ji L.L., Leeuwenburgh C. Exercise training-induced alterations in skeletal muscle antioxidant capacity: a brief review. Med. Sci. Sports Exerc. 1999;31:987–997. doi: 10.1097/00005768-199907000-00011. [DOI] [PubMed] [Google Scholar]
- 142.Powers S.K., Criswell D., Lawler J., Ji L.L., Martin D., Herb R.A., Dudley G. Influence of exercise and fiber type on antioxidant enzyme activity in rat skeletal muscle. Am. J. Physiol. 1994;266:R375–R380. doi: 10.1152/ajpregu.1994.266.2.R375. [DOI] [PubMed] [Google Scholar]
- 143.Powers S.K., Criswell D., Lawler J., Martin D., Ji L.L., Herb R.A., Dudley G. Regional training-induced alterations in diaphragmatic oxidative and antioxidant enzymes. Respir. Physiol. 1994;95:227–237. doi: 10.1016/0034-5687(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 144.Powers S.K., Criswell D., Lawler J., Martin D., Lieu F.K., Ji L.L., Herb R.A. Rigorous exercise training increases superoxide dismutase activity in ventricular myocardium. Am. J. Physiol. 1993;265:H2094–H2098. doi: 10.1152/ajpheart.1993.265.6.H2094. [DOI] [PubMed] [Google Scholar]
- 145.Ji L.L., Gomez-Cabrera M.C., Vina J. Role of nuclear factor kappaB and mitogen-activated protein kinase signaling in exercise-induced antioxidant enzyme adaptation. Appl. Physiol. Nutr. Metabol. 2007;32:930–935. doi: 10.1139/H07-098. [DOI] [PubMed] [Google Scholar]
- 146.Merry T.L., Ristow M. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J. Physiol. 2016;594:5195–5207. doi: 10.1113/JP271957. [DOI] [PMC free article] [PubMed] [Google Scholar]