Highlights
-
•
Virus-like particles (VLPs) related to wastewater treatment plant effluent quality.
-
•
VLP concentrations vary between wastewater treatment plants.
-
•
Effects of tertiary treatment on VLP concentrations varied.
-
•
Phage community composition differed between treatment plants.
-
•
Phage-host associations showed several connections to Comamonadaceae.
Keywords: Activated sludge, Bacteriophages, Metaviromics, Microbial ecology, Tertiary treatment
Abstract
With stringent effluent requirements and the implementation of new processes for micropollutant removal, it is increasingly important for wastewater treatment plants (WWTPs) to understand the factors affecting effluent quality. Phages (viruses infecting prokaryotes) are abundant in the biological treatment processes. They can contribute to organic carbon in the treated effluent both because they are organic in nature and occur in the effluent and because they cause lysis of microorganisms. Today very little is known about the effects of phages on effluent quality. The goal of this study was, therefore, to determine the relationship between phages and organic carbon in WWTP effluents. We also examined the diversity, taxonomy, and host-association of DNA phages using metagenomics. Effluent samples were collected from four WWTPs treating municipal wastewater. Significant differences in both organic carbon and virus-like particle concentrations were observed between the plants and there was a linear relationship between the two parameters. The phage communities were diverse with many members being taxonomically unclassified. Putative hosts were dominated by bacteria known to be abundant in activated sludge systems such as Comamonadaceae. The composition of phages differed between the WWTPs, suggesting that local conditions shape the communities. Overall, our findings suggest that the abundance and composition of phages are related to effluent quality. Thus, there is a need for further research clarifying the association between phage dynamics and WWTP function.
Introduction
Most municipal wastewater treatment plants (WWTPs) rely on biological processes. Both suspended growth processes, such as activated sludge, and biofilm processes, such a moving bed biofilm reactors and trickling filters, are commonly used. These biological reactors contain high numbers of microorganisms that treat the wastewater by removing organic compounds, nitrogen, and phosphorus. The prokaryotic communities in WWTPs have been investigated in many studies (e.g. Wu et al., 2019; Yang et al., 2011), but the viral communities are less explored. Phages, i.e., viruses that infect prokaryotes, are the most abundant living entities on Earth (Clokie et al., 2011). Their life cycle includes adsorption and injection of nucleic acids into the host, replication using the host cell's molecular machinery, and assembly of virus particles (virions), which are released into the environment to infect other cells. Lytic phages immediately start to replicate inside the host cell whereas lysogenic phages integrate into the host cell's genome and remain there until their lytic cycle is activated (Weinbauer, 2004). As phages are dependent on host cells to replicate, we can expect to find phages wherever prokaryotes are found (Dennehy and Abedon, 2021). Often, phages outnumber prokaryotic cells by an order of magnitude, although there is a large variation in the phage-to-prokaryote ratio between different environments (Parikka et al., 2017). Since there are high concentrations of prokaryotes in wastewater treatment processes, we can expect to find high concentrations of phages there as well (Shapiro and Kushmaro, 2011). Indeed, previous studies enumerating phage particles in WWTPs have found that biological treatment processes have high concentrations (Otawa et al., 2007). Typically, activated sludge and anaerobic digesters have higher phage particle concentrations than the influent and effluent wastewater (Brown et al., 2019; Wu and Liu, 2009). Brown et al. (2019) found higher concentrations in the effluent than the influent, suggesting that a production of phage particles in the biological reactors leads to a net increase in the wastewater as it moves through a WWTP.
Phages are believed to have a strong influence on the microbial community composition and ecosystem function in wastewater treatment processes, but we still know relatively little about their effects (Brown et al., 2019). There are examples where phages have been associated with changes in bioreactor performance. For example, Barr et al. (2010) hypothesized that phage infection of Candidatus Accumulibacter phosphatis was responsible for a decline in the performance of biological phosphorus removal in laboratory-scale sequencing batch reactors. Liu et al. (2017) found a decline of nitrifying bacteria corresponding to an increase of phages associated with these bacteria in bulking sludge.
Phage infections can potentially affect the concentration of organic compounds in effluent wastewater. Lysis of host cells leads to release of organic compounds (Middelboe et al., 1996). Some of these organics are immediately used as substrates by other microorganisms (Rosenberg et al., 2010) while others may be more recalcitrant and remain in the effluent. In other environments such as the oceans, phage-induced lysis of microorganisms is known to contribute to the pool of dissolved organic matter (Breitbart et al., 2018; Suttle, 2005). The phage particles themselves are also organic and although they are small (the capsid size of Caudovirales members is typically 45–185 nm in diameter Dion et al., 2020), high concentrations in the water could potentially make a noticeable contribution to the organic carbon concentration. Today, it is becoming increasingly important for WWTPs to reach low concentrations of organic carbon in their effluent. This is driven both by tougher regulations on effluent biochemical oxygen demand and by prospective implementation of new technologies, such as ozonation and activated carbon adsorption, for removal of micropollutants. For example, both the required ozone dose and the lifetime of activated carbon filters depend on the organic carbon concentration in the water (Boehler et al., 2012; Nöthe et al., 2009; Rizzo et al., 2019). A small increase in organic carbon concentration can thus lead to increased resource consumption and costs at the treatment plant. It is therefore important to understand the causes of variations in effluent organic carbon concentrations. Phage activity could be one important cause, which we currently know very little about. Thus, the goal of this study was to determine the relationship between phage particles and organic carbon concentrations in the effluent of WWTPs and to examine the phage communities using metagenomics.
Materials and methods
Wastewater treatment plants and sampling
For analysis of concentrations of organic carbon and phage particles, 24-h flow-proportional samples were collected from four Swedish WWTPs treating municipal wastewater (see process schemes in Fig. S1, supplementary material). For plant A, B, and C, samples were collected in May 2017. For plant D, samples were collected from June to December 2017. Samples were stored in plastic containers at a temperature of 4 °C until analysis.
Plant A has pre-precipitation in settling tanks followed by a high-loaded activated sludge process. The settled effluent from the activated sludge is partly recycled over nitrifying trickling filters to anoxic zones in the inlet of the activated sludge tanks. Another part of the secondary effluent flows though nitrifying and denitrifying moving bed biofilm reactors (MBBRs). Methanol is added for the post-denitrification step. The final effluent is filtered through 15-µm pore size disk filters before discharge. The average inflow to the WWTP was 4.4 m3 s−1 and the 7-days biochemical oxygen demand (BOD7) was 170 mg/L. The sludge age was 4.6 d, and the organic load to the activated sludge was 0.30 kg BOD7 kg−1 suspended solids d−1.
Plant B has pre-precipitation in settling tanks followed by a high-loaded activated sludge process. The settled activated sludge effluent flows through a nitrifying trickling filter followed by post-denitrification in methanol-fed MBBRs. This is followed by dissolved air flotation with the possibility of adding precipitation chemicals. The average inflow to the WWTP was 1.2 m3 s−1 and the BOD7 was 225 mg/L. The sludge age was 2.9 d, and the organic load was 0.42 kg BOD7 kg−1 suspended solids d−1.
Plant C has primary settling followed by a low-loaded activated sludge process with nitrification and pre-denitrification. This is followed by chemical treatment and sedimentation. The final effluent is further treated in a pond system. The average inflow to the WWTP was 0.38 m3 s−1 and the BOD7 208 mg/L. The sludge age was 7–21 days, and the organic load was 0.058 kg BOD7 kg−1 suspended solids d−1.
Plant D has an activated sludge process with nitrification and pre-denitrification in parallel with an activated sludge process using sequencing batch reactors (SBRs) process. The effluent from both these processes go to chemical treatment with dosage of aluminum chloride and final settling. About 10% of the effluent was treated in a pilot-scale ultrafiltration (UF) unit, with a nominal pore size of 0.02 µm, and a granular activated carbon (GAC) filter unit, with an empty bed contact time (EBCT) of 19 min on average. The total average wastewater inflow was 0.20 m3 s−1 and the BOD7 was 290 mg/L. The sludge age was 9 d in the activated sludge and 14 d in the SBRs. The organic load was 0.87 kg BOD7 kg−1 suspended solids d−1, where 55% was treated in the activated sludge process and 45% in the SBRs.
Samples for analysis of organic carbon and phage particles were collected from the final effluent in all WWTPs. In plant C and D, samples were collected both after the conventional treatment processes and after tertiary treatment with ponds (plant C) or UF and GAC (plant D). Samples for metagenomics were collected from plants A, B, and C. Grab samples collected on three days a month apart were analyzed from plant A. One 24-h, flow-proportional sample each, was analyzed from plant B and C (after the conventional treatment processes).
Analytical methods
The collected samples were filtered through 0.45-µm pore size polyethylene sulfone filters before analysis. The organic carbon concentration of the filtered samples was analyzed with a TOC analyzer (TOC-V, Shimadzu). This fraction of the organic carbon content in water is conventionally referred to as dissolved organic carbon (DOC), although it contains a mix of truly dissolved and colloidal substances, such as phage particles. Therefore, we use the acronym DOC for the organic carbon concentrations measured in the study.
The concentration and size distribution of phage particles were analyzed using nanoparticle tracking analysis (NTA, Nanosight NS-300, Malvern). In NTA, a laser is used to visualize individual particles in a water sample and a video of the particles is recorded. The particle concentration is calculated based on the number of identified particles. The hydrodynamic diameters of the particles are calculated based on the Brownian motion observed in the videos. We determined phage concentrations and size distributions using at least three to five 30 s videos of each sample.
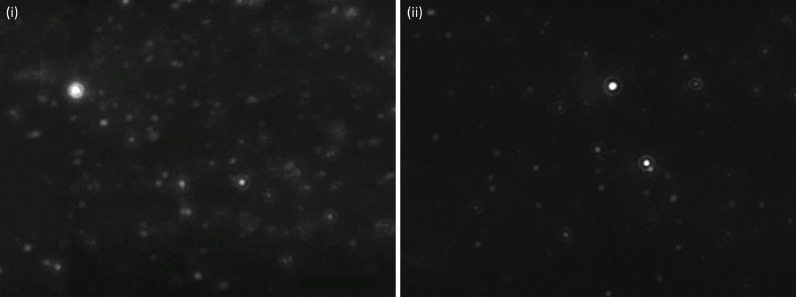
NTA has previously been used to measure phages and adenoviruses in water (Anderson et al., 2011; Kramberger et al., 2012). It has also been compared to other techniques such as the plaque assay, which is the standard way of analyzing concentrations of infectious phages and viruses. We also made this comparison using P1 phages (Text S1, supplementary material). The ratio between total virion concentrations measured with NTA and the infectious phage concentrations measured as plaque forming units (PFU) was 12 ± 8, which is in line with previous observations (Anderson et al., 2011; Du et al., 2010). We also confirmed that the observed particles in WWTP effluents were indeed phages by staining them with SYBR Gold. The SYBR Gold nucleic acid stain (concentrate in DMSO, ThermoFisher) was diluted 50 times in Tris-EDTA buffer (pH 7.4, Supelco) and 25 µL of the dilution was added to 10 mL effluent wastewater, resulting in a 20 000 times dilution of the original SYBR Gold stock solution. The mixture was heated at 80 °C for 10 min and allowed to cool to room temperature. Then, NTA was carried out with a blue laser (488 nm). The instrument was equipped with a 500 nm light filter, which can be inserted in the light path. Without the filter, all particles can be observed. With the filter in the light path, only particles stained with SYBR Gold can be observed. The stain rapidly bleached when exposed to the laser which meant only a few seconds long videos could be recorded, and hydrodynamic diameters could not be determined. Therefore, DNA staining was only carried out on a few samples to confirm that the observed particles were indeed phages (see Video S1, supplementary material) while quantification was done without SYBR Gold. Examples of still images of stained and unstained samples are shown in Fig. 1. We refer to the particles quantified with NTA as virus-like particles (VLP).
Fig. 1.
SYBR Gold-labelled particles (i) and non-labelled particles (ii) in effluent wastewater visualized using NTA.
To determine the contribution of VLP to the DOC concentration, a zinc hydroxide precipitation method was used to remove the fine particles from the water (Mamais et al., 1993). For the samples from plant D, this was also compared to measuring the DOC concentration in samples collected before and after the ultrafiltration step.
Metagenomics
The five samples for metagenomics were filtered through 0.2-µm pore size polyethylene sulfone filters. The VLP were concentrated using Amicon Ultra-15 centrifugal filters with a molecular weight cutoff of 100 kDa (Millipore). Approximately 50–100 ml sample volume was concentrated to 1 ml. To remove extracellular DNA, 10 µl (20 U) of DNase I (Invitrogen) was added to the 1 ml concentrate. It was incubated at room temperature for 15 min and thereafter the DNase was inactivated at 75 °C for 5 min. Phage DNA was then extracted from the samples using Norgen's Phage DNA Isolation Kit (Norgen Biotek) following the manufacturer's instructions. DNA fragmentation was performed using a Covaris E220 system targeting an insert size of 350–400 bp. Library preparation was carried out using the SMARTer ThruPLEX DNA-seq Kit (Takara). Paired-end 151 bp sequencing was done using a NovaSeq 6000 system, SP flow cell, and v1 sequencing chemistry (Illumina). The raw sequence reads are deposited at the European Nucleotide Archive under project number PRJEB50470.
The raw sequence reads were quality filtered using fastp v0.20.0 (Chen et al., 2018). The coverage was normalized to a target depth of 100 and a minimum depth of 2 using BBNorm (v38.61, https://sourceforge.net/projects/bbmap/). Then, contigs were assembled individually from each sample using Spades v3.15.3 with the setting –metaviral (Antipov et al., 2020). CheckV v0.8.1 was used to determine whether the contigs represented viral genomes (Nayfach et al., 2021). All contigs with at least one viral gene and more than twice as many viral genes than host genes were retained. The relative abundances of the contigs in each sample were determined by mapping the quality filtered reads to the phage contigs using CoverM (https://github.com/wwood/CoverM) with bwa-mem v0.7.17 (Vasimuddin et al., 2019) as mapper. For a contig to be considered present in a sample, at least 80% of its nucleotides had to have a coverage exceeding one.
Taxonomic classification of the phage contigs was done using PhagCN with default settings (Shang et al., 2021). Potential hosts of the phages were identified using two methods. In the first method, we mapped the phage contigs against the CRISPR spacer database of CrisprOpenDB (Dion et al., 2021). In the second, we used ANVIO v7 (Eren et al., 2021) to taxonomically classify tRNA sequences in the phage contigs. In ANVIO, tRNAScan-SE is used for identification of tRNA sequences (Chan and Lowe 2019). The results from the two methods were combined and the phage-host associations were visualized using Circos (Krzywinski et al., 2009).
The presence and function of auxiliary metabolic genes in the phage genomes was determined in ANVIO using the function anvi-estimate-metabolism. Genes were predicted using Prodigal v2.6.3 (Hyatt et al., 2010). Gene functions were determined by matching the genes to KEGG orthologs using the KOfam database (Aramaki et al., 2020). The genes were then categorized into metabolic pathways as defined by KEGG modules (Muto et al., 2013).
Statistical methods
To determine differences in VLP concentration, DOC concentrations, and particle sizes between WWTPs, Levene's test for equal variances was first carried out. Then, a one-way ANOVA was done to determine if statistically significant differences existed. Finally, Tukey's Honest Significant Difference (HSD) post-hoc test was done for pairwise comparisons. The Levene and ANOVA tests were done using Scipy (Virtanen et al., 2020) and Tukey's HSD was done using Statsmodels (Seabold and Perktold, 2010). Correlation between parameters was examined using ordinary linear regression in Statsmodels.
Diversity was calculated as the Hill number of diversity order 1 (1D) (Jost, 2006). This diversity index weighs each phage contig according to its relative abundance in the sample and the index can therefore be interpreted as the number of phage contigs that are “common” in the sample. Pielou's evenness index (Pielou, 1966) was used to quantify the distribution of phage contigs detected in a sample. The index ranges from 0 to 1 and a high value means that all detected contigs have similar relative abundance in the sample (i.e. high evenness). The difference in phage composition between samples was calculated as Hill-based dissimilarity of diversity order 1. This index ranges from 0 to 1 and can be interpreted as the fraction of “common” phage contigs not shared between pairs of samples. Diversity indices and principal coordinate analysis (PCoA) were calculated using qdiv (Modin et al., 2020).
Results
The concentrations of VLP and DOC were different in the four WWTPs
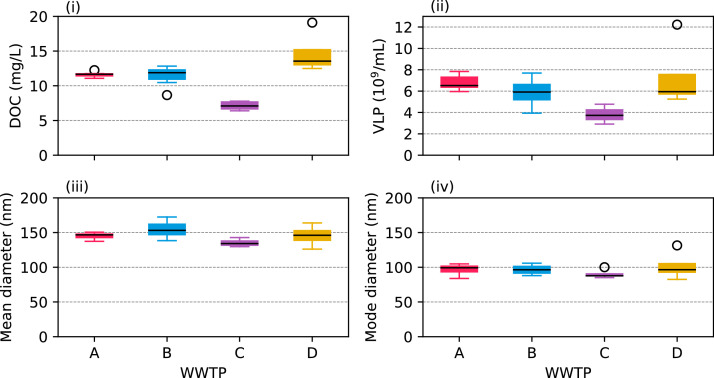
In Fig. 2, the concentrations of DOC and VLP, and the sizes of the VLP in the four WWTPs are shown. There were significantly different concentrations of effluent DOC between WWTPs (p = 4∙10−6, ANOVA). These ranged from 7.1 ± 0.6 mg L−1 (average±standard deviation, n = 7) for plant C to 14.7 ± 3.0 mg L−1 (n = 4) for plant D. Post-hoc pairwise comparisons showed significant differences (p < 0.02, Tukey's HSD) between all plants except A and B. For VLP, there were also significant differences (p = 0.01, ANOVA), with concentration ranging from 3.8(±0.7)∙109 VLP mL−1 in plant C to 7.3(±3.3)∙109 VLP mL−1 in plant D. Pairwise comparisons showed that plant C had significantly lower concentrations than plant A and D (p < 0.02, Tukey's HSD). No significant difference in the mean and mode of the VLP diameters between the WWTPs was observed. The VLP size distribution is also shown in Fig. S2 (supplementary material). The mode of the size distribution was between 90±5 nm in plant C and 102±22 nm in plant D.
Fig. 2.
Concentration of DOC (i) and VLP (ii) in the four WWTPs, and the mean (iii) and mode (iv) of the hydrodynamic diameters of the VLPs. The boxes show the 1st, 2nd, and 3rd quartiles. The whiskers extend 1.5 times the interquartile range and measured values outside this range are shown as circles. The data is based on samples collected after secondary treatment in each WWTP (Fig. S1, supplementary material).
There was a linear correlation between VLP and DOC concentrations
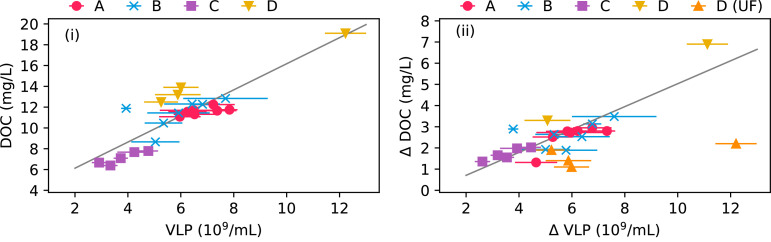
Using linear regression, the correlation between DOC and VLP concentrations was investigated (Fig. 3i). There was a significant correlation (R2adj = 0.73, p < 0.001) and the slope of the regression line was 1.25(±0.33)·10−12 mg DOC VLP−1 (95% confidence interval). When linear regression was carried out on the data from each plant individually, positive correlations could be observed for all plants with Pearson's r ranging from 0.54 to 0.99. The correlations were statistically significant for plants C and D (p < 0.05) (Fig. S3, supplementary material).
Fig. 3.
(i) Correlation between concentrations of DOC and VLP. (ii) Correlation between changes in concentrations of VLP and DOC after flocculation with zinc hydroxide. In samples D (UF), ultrafiltration was used to remove particles from the water. Error bars show standard deviations of multiple VLP analyses (n = 3 or n = 4).
VLP contributed to the DOC concentrations
The possible contribution of VLP to the DOC was investigated by removing particles from the water using a flocculation method and then measure the DOC and VLP concentrations again (Fig. 3ii). The reduction in DOC concentration after flocculation was between 22 ± 4% (average±standard deviation, n = 7) in plant A and 30 ± 6% (n = 2) in plant D. The reduction in VLP concentration ranged from 88 ± 5% in plant A to 99 ± 1% in plant B (n = 7). The reduction in DOC was correlated with the reduction in VLP (R2adj = 0.73, p < 0.001) and the slope of the regression line was 0.54(±0.15)·10−12 mg DOC VLP−1 (95% confidence interval). There was also a linear correlation for all plants individually with Pearson's r ranging from 0.45 to 0.93, although the correlations were only significant for plants A and C (Fig. S4, supplementary material).
As plant D was equipped with a UF unit with a cutoff of 0.02 µm, this provided an alternative way of analyzing the contribution of VLP to DOC. The DOC concentrations dropped from 14.7 ± 2.6 to 13.0 ± 2.4 mg L−1, corresponding to a reduction of 11.3 ± 2.6%. The VLP concentrations dropped from 7.3±(2.8)·109 to 0.022±(0.004)·109 VLP ml−1, corresponding to a reduction of 99.7 ± 0.1% (n = 4). For these four samples, no statistically significant linear correlation could be determined but the ratio between changes in DOC and VLP was 0.24±(0.07)·10−12 mg DOC VLP−1 (average±standard deviation, n = 4).
Tertiary treatment had varying effects on the DOC and VLP concentrations
Plant C was equipped with a pond system as tertiary treatment before the effluent was discharged. As water flowed through the ponds, the DOC concentration decreased by 9 ± 11%, from 7.1 ± 0.5 mg L−1 to 6.4 ± 0.4 mg L−1 (average±standard deviation, n = 5). The VLP concentration, on the other hand, decreased by 75 ± 4%, from 3.8(±0.7)∙109 VLP mL−1 to 0.96(±0.23)∙109 VLP mL−1. There was no change in the size distribution of VLPs (Fig. S5, supplementary material).
Plant D was equipped with a UF unit followed by GAC filter unit. As mentioned above, the UF removed 99.7 ± 0.1% of the VLP and 11.3 ± 2.6% of the DOC. The following GAC filtration led to a decrease in the DOC concentration by 14±9%, from 13.0 ± 2.4 mg L−1 to 11.0 ± 1.3 mg L−1. The VLP concentration increased across the GAC filter from 0.02(±0.004)∙109 VLP mL−1 to 0.14(±0.04)∙109 VLP mL−1.
Phage communities from different WWTPs had different composition
The bioinformatics resulted in 366 phage contigs out of which 106 were classified as complete phage genomes while 43, 73, and 143 were classified a high-, medium-, and low-quality genomes, respectively, by CheckV.
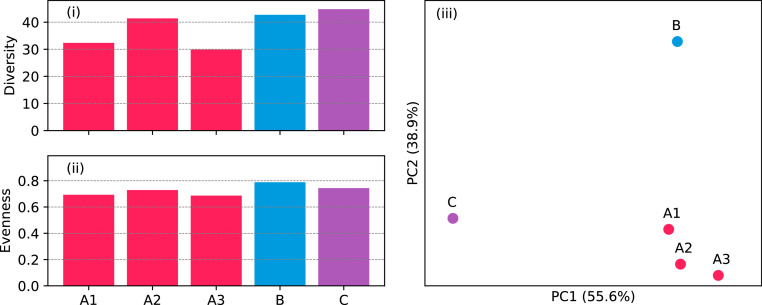
The phage communities had similar alpha diversity and evenness (Fig. 4i,ii). For beta diversity, the three samples from plant A clustered, suggesting they had similar composition in comparison to the samples from the other two WWTPs (Fig. 4iii).
Fig. 4.
(i) Phage diversity measured as Hill number of diversity order 1 (1D. (ii) Evenness measured as Pielou's index. (iii) Principal coordinate analysis based on Hill-based dissimilarities of order 1.
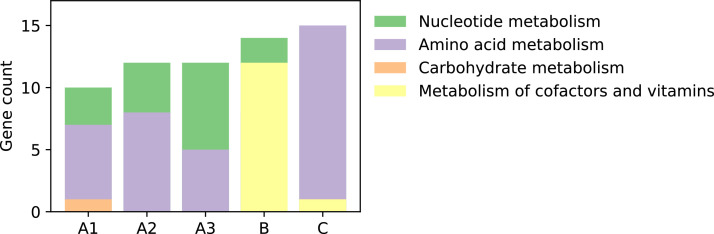
A difference in composition between samples from different plants was also seen in the analysis of auxiliary metabolic genes in the viral contigs (Fig. 5). These are genes that encode various metabolic functions in the host cells (Breitbart et al., 2018). The sample from plant A had an even distribution between genes associated with nucleotide- and amino acid metabolism. In plant B, metabolism of co-factors and vitamins dominated while amino acid metabolism dominated in plant C.
Fig. 5.
Auxiliary metabolic genes detected in the metaviromes. The gene functions are categorized based on KEGG modules.
Putative viral hosts were diverse
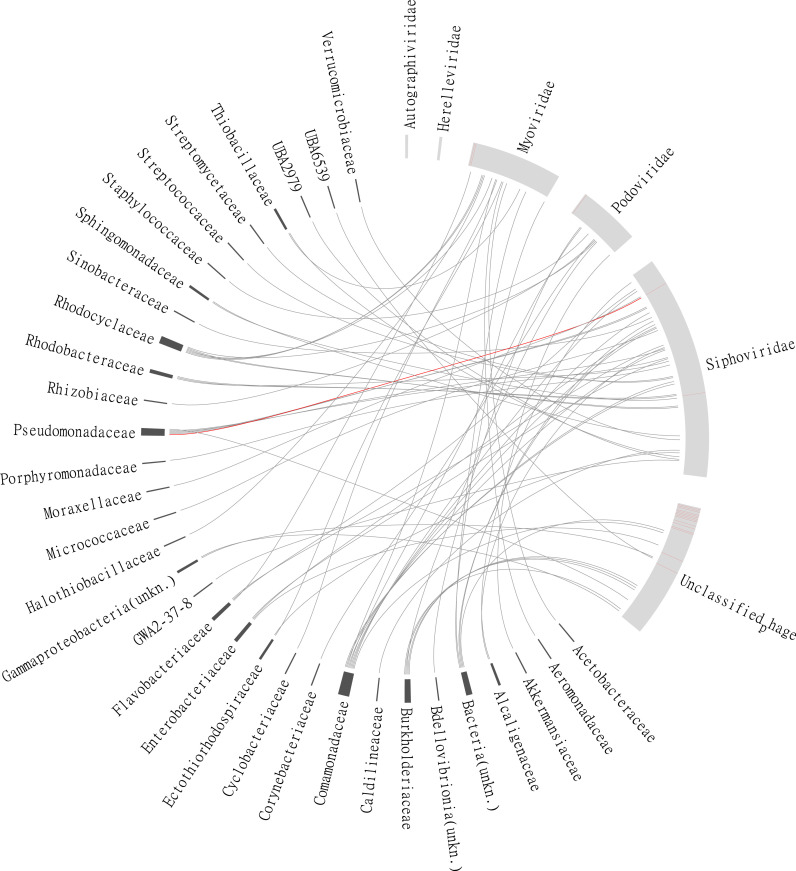
Putative associations between phages and hosts are shown in Fig. 6. Among phage classifications, Siphoviridae, Myoviridae, and Podoviridae were the most common. Many contigs were also unclassified phages. Out of the 366 phage contigs, 70 could be linked to a putative host. Comamonadaceae, Pseudomonadaceae, Rhodocyclaceae, and Burkholderiaceae were the most common host affiliations represented by 9, 6, 6, and 5 contigs, respectively. There were 20 phage contigs that could be detected in all five samples. Most of the contigs in this core community were unclassified and only one, a Siphoviridae, had a host link to a Pseudomonadaceae sp. The most common host affiliation that could be observed in all five samples were Comamonadaceae, Pseudomonadaceae, Flavobacteriaceae, Enterobacteriaceae, and Rhodobacteraceae (Fig. S6, supplementary material).
Fig. 6.
Links between phages (light gray bars) and putative host microorganisms (dark gray bars). The size of the bars in the outer ring is proportional to the number of phage-contigs with the specified taxonomic affiliation. Red bars and links refer to contigs belonging to a core phage community (i.e., contigs that are present in all five samples). The abbreviation unkn after the host name means that host was unclassified at the family level, but had a higher-level classification.
Discussion
Can we predict phage concentrations in WWTP effluents?
Phage concentrations and the phage-to-prokaryote ratio is highly variable in different environments (Parikka et al., 2017) and fluctuates over time (Brown et al., 2019). There is also an order of magnitude difference in phage concentrations observed in different process streams and WWTPs. In activated sludge, 107 to 109 particles mL−1 have been measured (Brown et al., 2019; Otawa et al., 2007; Wu and Liu, 2009), and in anaerobic digesters 1010 particles mL−1 were observed (Wu and Liu, 2009). The effluent concentrations are typically lower. Wu and Liu (2009) measured a concentration of 108 particles ml−1 in the effluent from a Singaporean WWTP and Brown et al. (2019) measured concentrations of around 108 to 109 particles ml−1 from an English WWTP. In this study, the VLP concentration measurements in the effluent from the four plants varied by a factor of 5. Despite the variability, there are indications that WWTP process design and operation could be a predictor of phage concentrations. Phage production can be linked to prokaryotic productivity (Parikka et al., 2017; Zimmerman et al., 2020). This has, for example, been observed in marine environments where lytic viral infections and phage production was higher when the prokaryotic productivity was high during the summer season, while lysogeny was more prevalent during the winter season (Payet and Suttle, 2013). A WWTP is a highly productive environment, and we can therefore expect high phage concentrations. However, the productivity (i.e., microbial growth rate) depends on the process configuration. We observed that the activated sludge plants with high organic loading rate (A, B, and D) had higher VLP concentrations than plant C (Fig. 2). This could be explained by faster microbial growth and lower fraction of inert material in high-loaded systems. In plant C, we can see that the VLP concentration dropped by 75 ± 4% after the effluent passed through a pond system. The microbial activity and density were likely much lower in the ponds than in the activated sludge system, leading to the decline in VLP concentration. In the tertiary treatment of plant D, we could also observe a link between microbial activity and VLP concentration. After VLPs were removed by the UF unit, an increase in VLP concentration could be observed following the GAC filtration unit, which might be explained by biological activity. Organic compounds are degraded by a biofilm forming on the activated carbon surface (Acevedo Alonso et al., 2021). The biofilm growth in the GAC could thus explain the increase in phages in the water. Many other factors at WWPTs, such as the type of activated sludge process (SBR or continuous flow) and the presence of chemical treatment, could also affect the VLP concentration as well as the relationship between VLP and DOC, and should be investigated in further work.
Can we predict phage community composition in WWTP effluents?
There were clear differences in phage composition between the WWTPs. The beta diversity analysis suggested that each WWTP had its own distinct phage community. Despite being samples taken a month apart, the three phage communities from plant A clearly grouped and were different from the phage communities in plant B and C (Fig. 4ii). Phage communities are shaped by interactions with the hosts (Liu et al., 2021). The Red Queen hypothesis (Van Valen, 2014) suggests a continuous arms race between phages and their hosts (Rohwer and Segall, 2015). When the host evolves improved defense systems against a phage, the phage counteracts this by evolving improved ways of infecting the host. A genome study of Candidatus Accumulibacter species in separate bioreactors found that the species mainly differed in genomic regions related to phage defense, suggesting that local arms races with phages likely drives speciation of both the host and the phage (Kunin et al., 2008). Speciation caused by arms races between bacteria and phages would lead to different species evolving in different locations. This would explain how geographically separate WWTPs evolve distinct phage communities, which could be observed in our study. Differences phage community composition could lead to different relationships between phage concentrations and DOC at different WWTPs.
Despite having different composition, several of the host associations could be seen in samples from all the WWTPs (Fig. S6, supplementary material). Among the identified hosts (Fig. 6), many of the taxa are known to belong to the core microbiome of activated sludge. These include, for example, Comamonadaceae and Rhodocyclaceae (Wu et al., 2019).
What are the practical consequences of phage activity for WWTPs?
Phage activity affects WWTPs in several ways. In the effluent, the phage particles themselves contribute to the DOC concentration. The zinc hydroxide precipitation we used to remove phages from the water suggested that they accounted for 22–30% of the DOC and that a phage contains 0.54(±0.15)·10−12 mg carbon. This is likely an overestimation as other organic compounds than phages could have been removed from the water in the flocculation process. A more realistic estimate was obtained from plant D, which used ultrafiltration with a 0.02 µm cutoff. Although this could also have removed non-phage organic particles, the number of such particles in the investigated size range was likely small in comparison to the number of phages, as indicated by SYBR Gold staining during NTA (supplementary video). The measurements across the UF unit in plant D suggested that phages made up 11.3 ± 2.6% of the DOC in the effluent and that a phage contains 0.24(±0.07)·10−12 mg carbon. This is similar to previous estimates of the carbon content of phage particles, which range from 0.055·10−12 to 0.2·10−12 mg (Steward et al., 2007; Wilhelm and Suttle, 1999). Jover et al. (2014) established a relationship between the elemental content of a phage and its capsid size. Based on the size measurements in Fig. 2, we estimate that in an effluent containing 1010 phages per ml, the phages would contribute with 0.72 mg L−1 DOC, 0.29 mg L−1 total nitrogen, and 0.11 mg L−1 total phosphorus (Text S2, supplementary material). Especially the phosphorus content may be relevant for WWTPs, which often have strict discharge limits on total phosphorus.
The linear correlation between DOC and VLP concentrations observed in this study (Fig. 3i) cannot be solely explained by the carbon content of the phages themselves. Phage activity can also contribute to effluent DOC concentrations by causing lysis of prokaryotic cells in the biological treatment processes. The term “viral shunt” has been used to describe the recycling of nutrients that occurs when viral lysis of host cells results in the release of cellular debris, which can be taken up by other prokaryotic cells (Zimmerman et al., 2020). A fraction of the cellular debris is likely recalcitrant (Jiao et al., 2010) and remains in the WWTP effluent. High levels of lytic infection would thus both result in high concentrations of phage particles and organic carbon derived from cellular debris. Phage activity can also lead to changes in the microbial composition of biological treatment process and indirectly affect treatment performance. The viral shunt will cause a decline in the population infected by the phage and growth of the co-existing microorganisms able to use the released nutrients, which affects the balance between different populations. Such phage-driven changes in the relative abundance of different taxa were previously reported for a membrane bioreactor (Shapiro et al., 2010). The arms race between phages and their hosts also leads to diversification, and the evolution of phage resistance in the host can be associated with changes in metabolism, for example, the ability to metabolize organic compounds (Middelboe et al., 2009). This could theoretically have consequences for the performance of wastewater treatment processes. Phages can also contribute to horizontal gene transfer (Touchon et al., 2017) and affect the metabolism of the microorganisms they infect. For example, marine phages were shown to carry auxiliary metabolic genes for several nitrogen conversion processes (Gazitúa et al., 2021). Nitrogen conversions are important in wastewater treatment and the role of phages for these processes should be explored further. In our data set, we could see that the distribution of auxiliary metabolic genes where different in phage communities from different WWTPs, indicating that the effects of phages on microbial metabolism could differ depending on the WWTP.
Both the concentration and the composition of DOC in the effluent from biological treatment has effects on the performance of tertiary treatment technologies. The effectiveness of advanced oxidation processes is reduced by presence of DOC (Dantas et al., 2012) and both the concentration and oxidation state of DOC were shown to determine the removal efficiency of micropollutants during ozonation (Ekblad et al., 2019). The sorption efficiency of activated carbon for micropollutant removal is negatively affected by increasing DOC concentration (Boehler et al., 2012). Disinfection by chlorination is used as the final treatment step at WWTPs around the world. The presence of DOC would increase the chlorine consumption and lead to the formation of toxic byproducts (Summers et al., 2020). Managing the DOC content in WWTP effluents is, therefore, an important challenge. Phage activity is important for the function of biological treatment processes in WWTPs and, as shown in this study, there is a relationship between DOC and phage concentrations.
Conclusions
Based on samples from four Swedish WWTPs, a linear correlation between DOC- and VLP concentrations could be established. The contribution of the VLP to the DOC concentration is in the order of one mg per liter. In plant D equipped with UF, this corresponded to 11.3 ± 2.6% of the total DOC in the water. Since the fraction of the DOC that can be attributed to carbon in VLPs was quite small, there most be another explanation for the observed linear correlation between DOC and VLP. Phage infection of bacterial cells would also cause an increase in DOC concentration in the wastewater because of organic compounds released by the lysed host cells. High concentrations of VLP could indicate high rates of infections and many lysed host cells, which could explain the linear correlation between DOC and VLP.
The phage diversity was high and many phage contigs were taxonomically unidentified. The diversity of putative hosts was also high and corresponded to bacterial families frequently found in activated sludge systems. Differences in phage composition between treatment plants and a possible relationship between organic loading rate and phage concentration and composition suggest that it may be possible to control phage activity by the design and operation of WWTPs. This warrants further investigations because even small changes in effluent quality caused by phage activity can have consequences for tertiary treatment processes for micropollutants removal and disinfection.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Funding was provided by the Swedish Research Council for Sustainable Development (FORMAS, project 2012–1433) and the Swedish Water and Wastewater Association (Svenskt Vatten) through VA Teknik Södra. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. Part of the bioinformatic work was enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council through grant agreement no. 2018–05973. Open access funding was provided by Chalmers University of Technology.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.wroa.2022.100146.
Appendix. Supplementary materials
References
- Acevedo Alonso V., Kaiser T., Babist R., Fundneider T., Lackner S. A multi-component model for granular activated carbon filters combining biofilm and adsorption kinetics. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117079. [DOI] [PubMed] [Google Scholar]
- Anderson B., Rashid M.H., Carter C., Pasternack G., Rajanna C., Revazishvili T., Dean T., Senecal A., Sulakvelidze A. Enumeration of bacteriophage particles: comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage. 2011;1(2):86–93. doi: 10.4161/bact.1.2.15456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov D., Raiko M., Lapidus A., Pevzner P.A. MetaviralSPAdes: assembly of viruses from metagenomic data. Bioinformatics. 2020;36(14):4126–4129. doi: 10.1093/bioinformatics/btaa490. [DOI] [PubMed] [Google Scholar]
- Aramaki T., Blanc-Mathieu R., Endo H., Ohkubo K., Kanehisa M., Goto S., Ogata H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36(7):2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J.J., Slater F.R., Fukushima T., Bond P.L. Evidence for bacteriophage activity causing community and performance changes in a phosphorus-removal activated sludge. FEMS Microbiol. Ecol. 2010;74(3):631–642. doi: 10.1111/j.1574-6941.2010.00967.x. [DOI] [PubMed] [Google Scholar]
- Boehler M., Zwickenpflug B., Hollender J., Ternes T., Joss A., Siegrist H. Removal of micropollutants in municipal wastewater treatment plants by powder-activated carbon. Water Sci. Technol. 2012;66(10):2115–2121. doi: 10.2166/wst.2012.353. [DOI] [PubMed] [Google Scholar]
- Breitbart M., Bonnain C., Malki K., Sawaya N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018;3(7):754–766. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- Brown M.R., Baptista J.C., Lunn M., Swan D.L., Smith S.J., Davenport R.J., Allen B.D., Sloan W.T., Curtis T.P. Coupled virus - bacteria interactions and ecosystem function in an engineered microbial system. Water Res. 2019;152:264–273. doi: 10.1016/j.watres.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie M.R.J., Millard A.D., Letarov A.V., Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas R.F., Dominguez V., Cruz A., Sans C., Esplugas S. Application of advanced oxidation for the removal of micropollutants in secondary effluents. J. Water Reuse Desalin. 2012;2(2):121–126. [Google Scholar]
- Dennehy J.J., Abedon S.T. Harper D.R., Abedon S.T., Burrowes B.H., McConville M.L., editors. Bacteriophages: Biology, Technology, Therapy. 2021:253–294. [Google Scholar]
- Dion M.B., Oechslin F., Moineau S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020;18(3):125–138. doi: 10.1038/s41579-019-0311-5. [DOI] [PubMed] [Google Scholar]
- Dion M.B., Plante P.L., Zufferey E., Shah S.A., Corbeil J., Moineau S. Streamlining CRISPR spacer-based bacterial host predictions to decipher the viral dark matter. Nucleic. Acids Res. 2021;49(6):3127–3138. doi: 10.1093/nar/gkab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Kendall K., Morris S., Sweet C. Measuring number-concentrations of nanoparticles and viruses in liquids on-line. J. Chem. Technol. Biotechnol. 2010;85(9):1223–1228. [Google Scholar]
- Ekblad M., Falås P., El-Taliawy H., Nilsson F., Bester K., Hagman M., Cimbritz M. Is dissolved COD a suitable design parameter for ozone oxidation of organic micropollutants in wastewater? Sci. Total Environ. 2019;658:449–456. doi: 10.1016/j.scitotenv.2018.12.085. [DOI] [PubMed] [Google Scholar]
- Eren A.M., Kiefl E., Shaiber A., Veseli I., Miller S.E., Schechter M.S., Fink I., Pan J.N., Yousef M., Fogarty E.C., Trigodet F., Watson A.R., Esen Ö.C., Moore R.M., Clayssen Q., Lee M.D., Kivenson V., Graham E.D., Merrill B.D., Karkman A., Blankenberg D., Eppley J.M., Sjödin A., Scott J.J., Vázquez-Campos X., McKay L.J., McDaniel E.A., Stevens S.L.R., Anderson R.E., Fuessel J., Fernandez-Guerra A., Maignien L., Delmont T.O., Willis A.D. Community-led, integrated, reproducible multi-omics with anvi'o. Nat. Microbiol. 2021;6(1):3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazitúa M.C., Vik D.R., Roux S., Gregory A.C., Bolduc B., Widner B., Mulholland M.R., Hallam S.J., Ulloa O., Sullivan M.B. Potential virus-mediated nitrogen cycling in oxygen-depleted oceanic waters. ISME J. 2021;15(4):981–998. doi: 10.1038/s41396-020-00825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11(1):119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao N., Herndl G.J., Hansell D.A., Benner R., Kattner G., Wilhelm S.W., Kirchman D.L., Weinbauer M.G., Luo T., Chen F., Azam F. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010;8(8):593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113(2):363–375. [Google Scholar]
- Jover L.F., Effler T.C., Buchan A., Wilhelm S.W., Weitz J.S. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 2014;12(7):519–528. doi: 10.1038/nrmicro3289. [DOI] [PubMed] [Google Scholar]
- Kramberger P., Ciringer M., Štrancar A., Peterka M. Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol. J. 2012;9(1):265. doi: 10.1186/1743-422X-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., He S., Warnecke F., Peterson S.B., Garcia Martin H., Haynes M., Ivanova N., Blackall L.L., Breitbart M., Rohwer F., McMahon K.D., Hugenholtz P. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18(2):293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Li Z., Han G., Cun S., Yang M., Liu X. Bacteriophage ecology in biological wastewater treatment systems. Appl. Microbiol. Biotechnol. 2021;105(13):5299–5307. doi: 10.1007/s00253-021-11414-8. [DOI] [PubMed] [Google Scholar]
- Liu R., Qi R., Wang J., Zhang Y., Liu X., Rossetti S., Tandoi V., Yang M. Phage-host associations in a full-scale activated sludge plant during sludge bulking. Appl. Microbiol. Biotechnol. 2017;101(16):6495–6504. doi: 10.1007/s00253-017-8429-8. [DOI] [PubMed] [Google Scholar]
- Mamais D., Jenkins D., Pitt P. A rapid physical-chemical method for the determination of readily biodegradable soluble COD in municipal wastewater. Water Res. 1993;27(1):195–197. [Google Scholar]
- Middelboe M., Holmfeldt K., Riemann L., Nybroe O., Haaber J. Bacteriophages drive strain diversification in a marine Flavobacterium: implications for phage resistance and physiological properties. Environ. Microbiol. 2009;11(8):1971–1982. doi: 10.1111/j.1462-2920.2009.01920.x. [DOI] [PubMed] [Google Scholar]
- Middelboe M., Jorgensen N., Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 1996;62(6):1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modin O., Liébana R., Saheb-Alam S., Wilén B.M., Suarez C., Hermansson M., Persson F. Hill-based dissimilarity indices and null models for analysis of microbial community assembly. Microbiome. 2020;8(1):132. doi: 10.1186/s40168-020-00909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Kotera M., Tokimatsu T., Nakagawa Z., Goto S., Kanehisa M. Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J. Chem. Inf. Model. 2013;53(3):613–622. doi: 10.1021/ci3005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S., Camargo A.P., Schulz F., Eloe-Fadrosh E., Roux S., Kyrpides N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021;39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthe T., Fahlenkamp H., von Sonntag C. Ozonation of wastewater: rate of ozone consumption and hydroxyl radical yield. Environ. Sci. Technol. 2009;43(15):5990–5995. doi: 10.1021/es900825f. [DOI] [PubMed] [Google Scholar]
- Otawa K., Lee S.H., Yamazoe A., Onuki M., Satoh H., Mino T. Abundance, diversity, and dynamics of viruses on microorganisms in activated sludge processes. Microb. Ecol. 2007;53(1):143–152. doi: 10.1007/s00248-006-9150-9. [DOI] [PubMed] [Google Scholar]
- Parikka K.J., Le Romancer M., Wauters N., Jacquet S. Deciphering the virus-to-prokaryote ratio (VPR): insights into virus–host relationships in a variety of ecosystems. Biol. Rev. 2017;92(2):1081–1100. doi: 10.1111/brv.12271. [DOI] [PubMed] [Google Scholar]
- Payet J.P., Suttle C.A. To kill or not to kill: the balance between lytic and lysogenic viral infection is driven by trophic status. Limnol. Oceanogr. 2013;58(2):465–474. [Google Scholar]
- Pielou E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. [Google Scholar]
- Rizzo L., Malato S., Antakyali D., Beretsou V.G., Đolić M.B., Gernjak W., Heath E., Ivancev-Tumbas I., Karaolia P., Lado Ribeiro A.R., Mascolo G., McArdell C.S., Schaar H., Silva A.M.T., Fatta-Kassinos D. Consolidated vs new advanced treatment methods for the removal of contaminants of emerging concern from urban wastewater. Sci. Total Environ. 2019;655:986–1008. doi: 10.1016/j.scitotenv.2018.11.265. [DOI] [PubMed] [Google Scholar]
- Rohwer F., Segall A.M. A century of phage lessons. Nature. 2015;528(7580):46–47. doi: 10.1038/528046a. [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Bittan-Banin G., Sharon G., Shon A., Hershko G., Levy I., Ron E.Z. The phage-driven microbial loop in petroleum bioremediation. Microb. Biotechnol. 2010;3(4):467–472. doi: 10.1111/j.1751-7915.2010.00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold, S. and Perktold, J. (2010) statsmodels: econometric and statistical modeling with python.
- Shang J., Jiang J., Sun Y. Bacteriophage classification for assembled contigs using graph convolutional network. Bioinformatics. 2021;37(Supplement_1):i25–i33. doi: 10.1093/bioinformatics/btab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro O.H., Kushmaro A. Bacteriophage ecology in environmental biotechnology processes. Curr. Opin. Biotechnol. 2011;22(3):449–455. doi: 10.1016/j.copbio.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Shapiro O.H., Kushmaro A., Brenner A. Bacteriophage predation regulates microbial abundance and diversity in a full-scale bioreactor treating industrial wastewater. ISME J. 2010;4(3):327–336. doi: 10.1038/ismej.2009.118. [DOI] [PubMed] [Google Scholar]
- Steward G.F., Fandino L.B., Hollibaugh J.T., Whitledge T.E., Azam F. Microbial biomass and viral infections of heterotrophic prokaryotes in the sub-surface layer of the central Arctic Ocean. Deep Sea Res. Part I. 2007;54(10):1744–1757. [Google Scholar]
- Summers R.S., Shiokari S.T., Johnson S., Peterson E., Yu Y., Cook S. Reuse treatment with ozonation, biofiltration, and activated carbon adsorption for total organic carbon control and disinfection byproduct regulation compliance. AWWA Water Sci. 2020;2(5):e1190. [Google Scholar]
- Suttle C.A. Viruses in the sea. Nature. 2005;437(7057):356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Touchon M., Moura de Sousa J.A., Rocha E.P.C. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017;38:66–73. doi: 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- Van Valen L. Foundations of Macroecology: Classic Papers With Commentaries. University of Chicago Press; 2014. Foundations of Macroecology: Classic Papers With Commentaries; pp. 284–314. Felisa, A.S., John, L.G. and James, H.B. (eds) [Google Scholar]
- Vasimuddin, M., Misra, S., Li, H. and Aluru, S. (2019) Efficient architecture-aware acceleration of BWA-MEM for multicore systems, pp. 314–324.
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., van der Walt S.J., Brett M., Wilson J., Millman K.J., Mayorov N., Nelson A.R.J., Jones E., Kern R., Larson E., Carey C.J., Polat I., Feng Y., Moore E.W., VanderPlas J., Laxalde D., Perktold J., Cimrman R., Henriksen I., Quintero E.A., Harris C.R., Archibald A.M., Ribeiro A.H., Pedregosa F., van Mulbregt P., SciPy 1.0 Contributors SciPy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004;28(2):127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wilhelm S.W., Suttle C.A. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience. 1999;49(10):781–788. [Google Scholar]
- Wu L., Ning D., Zhang B., Li Y., Zhang P., Shan X., Zhang Q., Brown M.R., Li Z., Van Nostrand J.D., Ling F., Xiao N., Zhang Y., Vierheilig J., Wells G.F., Yang Y., Deng Y., Tu Q., Wang A., Acevedo D., Agullo-Barcelo M., Alvarez P.J.J., Alvarez-Cohen L., Andersen G.L., de Araujo J.C., Boehnke K.F., Bond P., Bott C.B., Bovio P., Brewster R.K., Bux F., Cabezas A., Cabrol L., Chen S., Criddle C.S., Deng Y., Etchebehere C., Ford A., Frigon D., Sanabria J., Griffin J.S., Gu A.Z., Habagil M., Hale L., Hardeman S.D., Harmon M., Horn H., Hu Z., Jauffur S., Johnson D.R., Keller J., Keucken A., Kumari S., Leal C.D., Lebrun L.A., Lee J., Lee M., Lee Z.M.P., Li Y., Li Z., Li M., Li X., Ling F., Liu Y., Luthy R.G., Mendonça-Hagler L.C., de Menezes F.G.R., Meyers A.J., Mohebbi A., Nielsen P.H., Ning D., Oehmen A., Palmer A., Parameswaran P., Park J., Patsch D., Reginatto V., de los Reyes F.L., Rittmann B.E., Noyola A., Rossetti S., Shan X., Sidhu J., Sloan W.T., Smith K., de Sousa O.V., Stahl D.A., Stephens K., Tian R., Tiedje J.M., Tooker N.B., Tu Q., Van Nostrand J.D., De los Cobos Vasconcelos D., Vierheilig J., Wagner M., Wakelin S., Wang A., Wang B., Weaver J.E., Wells G.F., West S., Wilmes P., Woo S.-.G., Wu L., Wu J.-.H., Wu L., Xi C., Xiao N., Xu M., Yan T., Yang Y., Yang M., Young M., Yue H., Zhang B., Zhang P., Zhang Q., Zhang Y., Zhang T., Zhang Q., Zhang W., Zhang Y., Zhou H., Zhou J., Wen X., Curtis T.P., He Q., He Z., Brown M.R., Zhang T., He Z., Keller J., Nielsen P.H., Alvarez P.J.J., Criddle C.S., Wagner M., Tiedje J.M., He Q., Curtis T.P., Stahl D.A., Alvarez-Cohen L., Rittmann B.E., Wen X., Zhou J., Global Water Microbiome Consortium Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 2019;4(7):1183–1195. doi: 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- Wu Q., Liu W.T. Determination of virus abundance, diversity and distribution in a municipal wastewater treatment plant. Water Res. 2009;43(4):1101–1109. doi: 10.1016/j.watres.2008.11.039. [DOI] [PubMed] [Google Scholar]
- Yang C., Zhang W., Liu R., Li Q., Li B., Wang S., Song C., Qiao C., Mulchandani A. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ. Sci. Technol. 2011;45(17):7408–7415. doi: 10.1021/es2010545. [DOI] [PubMed] [Google Scholar]
- Zimmerman A.E., Howard-Varona C., Needham D.M., John S.G., Worden A.Z., Sullivan M.B., Waldbauer J.R., Coleman M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020;18(1):21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.