Abstract
Large populations (200 to 5,000 cells ml−1 in snowmelt) of bacteria were present in surface snow and firn from the south pole sampled in January 1999 and 2000. DNA isolated from this snow yielded ribosomal DNA sequences similar to those of several psychrophilic bacteria and a bacterium which aligns closely with members of the genus Deinococcus, an ionizing-radiation- and desiccation-resistant genus. We also obtained evidence of low rates of bacterial DNA and protein synthesis which indicates that the organisms were metabolizing at ambient subzero temperatures (−12 to −17°C).
There are no reports which document active metabolism of bacteria in the surface snow of the interior of the Antarctic continent. At the south pole, temperatures are extreme and austral winter air temperatures reach about −85°C, while in summer it can warm to about −13°C (mean monthly air temperature in December is −26°C). Bacteria have previously been cultured from samples taken from Antarctic ice cores (1), and deep cores from the accreted ice above subglacial Lake Vostok revealed a high diversity (24) of species that were reported to be metabolically active when warmed to 3°C (16). We report here bacterial populations and associated metabolic activity in surface (upper 20 cm) snow and firn collected at the south pole in the austral summer.
Sampling.
Snow samples were collected on 9 and 18 January 1999 and 10 January 2000 at the Amundsen-Scott South Pole Station. Care was taken to sample at the edge of the Clean Sector of the National Oceanic and Atmospheric Administration Clean Air Laboratory upwind of the Station (grid 115 to 120), so that contaminating bacteria from the human habitat would not be collected. Snow was sampled using sterile procedures, and Snowpak containers, which hold ca. 60 to 80 liters of snow, were returned frozen to the Crary laboratory at McMurdo Station for analysis within 24 h of collection.
Microscopy.
Microbes were concentrated by filtering 20 to 50 ml of snowmelt onto a 0.02-μm-pore-size Anodisc filter, and bacteria were stained with the DNA fluorescent dye SYBR green (22). Counts were done using a Zeiss Axioskop microscope at ×1,000. Scanning electron microscopy (SEM) was done on samples preserved in 4% glutaraldehyde–1.8% RNase-free sucrose–10 mM phosphate buffer (pH 7.4) and held at 1°C until processing for SEM.
DNA sequencing.
Melted snow (10 liters) from each snow sample was filtered through a 0.2-μm-pore-size Nalgene filtration unit on a clean bench. The sample retained on the membrane was then subjected to DNA purification using cetyltrimethylammonium bromide and chloroform according to a procedure as modified in reference 8. Briefly, the membrane was incubated with 1 ml of Tris-EDTA buffer containing lysozyme (1 mg/ml) at 37°C for 1 h with constant shaking (250 rpm). Subsequently, NaCl and sodium dodecyl sulfate were added at final concentrations of 0.7 M and 1%, respectively, and the mixture was incubated at 70°C for 15 min (gently hand shaken every 5 min). Next, the mixture was incubated with 1 mg of RNase per ml at 37°C for 30 min. Cetyltrimethylammonium bromide dissolved in 0.7 M NaCl was added at a final concentration of 1% (wt/vol), and the mixture was incubated at 56°C for 10 min. After a wash with an equal volume of chloroform, DNA in the aqueous phase was precipitated with an equal volume of isopropanol for 2 h or overnight at −20°C. To prevent contamination in any step of the procedure, a negative control was made by filtering 2 liters of the autoclaved distilled and deionized water used in the above-described experiments with the same filtration unit, and the membrane was subjected to the same procedure as the sample. After centrifugation and washing of the DNA pellet with 75% ethanol, the DNA was dissolved in autoclaved deionized and distilled water. Two microliters of the DNA preparation from both the sample and the negative control was used in a PCR to amplify the gene for the small subunit of ribosomal RNAs of prokaryotes (16S ribosomal DNAs (rDNAs). Primers were initially Bac1 (forward, 5′-AGA GTT TGA TCM TGG CTC AG-3′ [M is A or C]) and Bac2 (reverse, 5′-ACC TTG TTA CGA CTT CAC-3′) (15). In some cases, a secondary PCR was required and a nested primer was used, Bac5 (forward, 5′-ATG TGG TTT AAT TCG A-3′) (15). One microliter of the first PCR product from both the sample and the negative control was used as the template for the secondary PCR. On the 16S rDNA sequences from the south pole samples, we found regions that were universally conserved among all bacteria, including the Deinococcus-Thermus group. A universal primer was designed based on one of these regions and used in a PCR for samples from the second sampling season, Bac14 (forward; CGG GAG GCA GCA GTT AGG AAT). The PCR products were extracted from agarose gel using Spin X ultrafiltration units and cloned into a TA cloning kit (19). Ten to 12 clones were randomly selected for sequencing with an ABI Prizm automatic sequencer. The nucleotide sequences obtained were compared against those in the GenBank database using FASTA and aligned with representatives of major bacterial divisions using CLUSTAL W. Phylogenetic analysis was performed using PHYLIP (10).
Fluorescent in situ hybridization.
A protocol previously reported (2) was modified. First, 250 ml of snowmelt was fixed with formaldehyde (final concentration, 3.7% [vol/vol]) at 4°C for 2 h. The fixed water sample was then filtered onto a 0.2-μm-pore-size Nucleopore membrane at a vacuum pressure of 5 lb/in2. Ethanol (50% [vol/vol] in sterilized double-distilled water) was applied to the membrane and incubated for 15 min. After the liquid was filtered, the cells retained on the membrane were transferred onto a slide by placing the membrane upside down on a poly-l-lysine-coated slide. The slide had been briefly rinsed with 70% ethanol to remove potentially contaminating bacteria before use. When the slide and the membrane became dry, the membrane was removed gently and a hydrophobic boundary was drawn with a PAP pen on the covered area on the slide (Energy Beam Sciences, Inc., Agawam, Mass.). The cells encircled within the boundary were then incubated with 50 μl of hybridization buffer (0.9 M HCl, 20 mM Tris [pH 7.5], 5 mM EDTA, 35% formamide, 0.01% sodium dodecyl sulfate) containing 5 ng of the oligonucleotide probe designed for eubacteria (Rhodamin-CGCGGTAATACGGAGGG) per μl. Incubation at 46°C lasted overnight. Next, the slide was washed in Oligo wash buffer (70 mM NaCl, 20 mM Tris · HCl [pH 7.4], 5 mM EDTA) at 48°C for 15 min. After a brief rinse in double-distilled water, the slide was mounted with Gel Mount. A negative control was made with a slide without samples being applied, and the slide was subjected to the same procedures as those described above.
DNA and protein synthesis.
Assays using about 3.8 μCi of either [methyl-3H]thymidine or [3H]leucine (6, 17) were conducted on minicores of snow in sterile 7-ml polystyrene round-bottom tubes (12 by 75 mm) with snap-top closures. Incubations were done in a walk-in cold room at temperatures between −12 and −17°C, and in an attempt to duplicate irradiance at the south pole, some tubes were exposed to 250 μmol of photons m−2 s−1 (photosynthetically active radiation [PAR]) of radiant energy from incandescent lamps (typical December and January PAR at the snow surface at the south pole ranges from 750 to 1,200 μmol of photons m−2 s−1). Incubation temperature was measured within a 7-ml tube that contained snow and was placed under the lighting. Two-hundred-microliter volumes of high-specific-activity thymidine (73 Ci/mmol) or leucine (64 Ci/mmol) was injected with a Hamilton microliter syringe in several line injections into each tube removed from the cold room on ice in the Radiation Laboratory. Injected samples were immediately returned to the cold room. Controls included (i) zero time-harvested samples (i.e., samples injected with radioisotope and immediately fixed with trichloroacetic acid [TCA]); (ii) samples incubated at −80°C; and (iii) samples amended with TCA at initiation and incubated over several hours. After termination of assays by addition of TCA to a final concentration of 5% and rapid melting of snow, assays were conducted according to standard procedures and included both filtration and centrifugation (29) protocols. Both a procedure to determine flux of isotopes into bulk macromolecules (TCA-ethanol rinse) and that to determine flux into DNA (NaOH digestion, phenol-chloroform rinse) were used (6).
Bacterial concentrations.
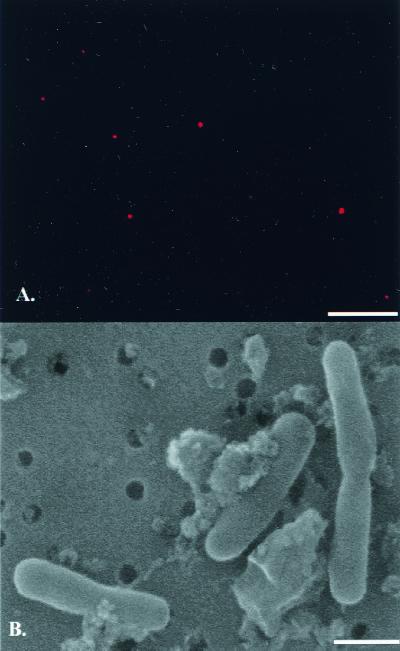
Direct counts by epifluorescence microscopy revealed population densities from about 200 to 5,000 cells ml of snowmelt−1 (mean ± standard error, 3,140 ± 771 cells ml−1, n = 18) for both years sampled. This value compares to similar densities of bacteria in surface snows from the Ross Ice Shelf and typical concentrations of about 106 bacteria ml of surface seawater−1 near McMurdo Station (12). In situ hybridization revealed the presence of eubacteria in the snowmelt from the south pole (Fig. 1A), while the negative control indicated no contamination. Examination of snowmelt using SEM indicated the presence of coccoid and rod-shaped bacteria, some of which appeared to be dividing (Fig. 1B).
FIG. 1.
Micrographs of fluorescent in situ hybridization (A) and SEM of bacteria from south pole snow. Bar = 10 μm (A) and 1.0 μm (B).
DNA.
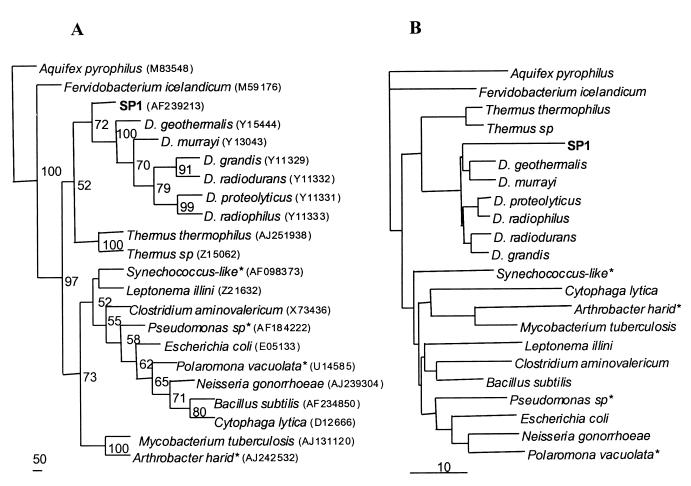
Amplification and sequencing of 16S rDNAs from DNAs isolated from bacteria in snow samples indicate the presence of bacteria that align most closely with the genus Deinococcus. The 16S rDNA sequences of these microbes were about 86 to 87% identical to those of Deinococcus. species (closest to D. grandis and D. geothermalis), the closest relatives found from GenBank databases. This genus has also been observed in soils of the Antarctic Dry Valleys that lie near the coast of Antarctica (28). We also cloned and sequenced one sample from Taylor Valley and found a sequence closest to Deinococcus sp. and D. grandis (84% identity). Phylogenetic analysis comparing our samples with representatives from the major 11 bacterial divisions, including some Antarctic and Arctic bacteria, showed that these south pole snow microbes are clustered with the Deinococcus-Thermus group (Fig. 2) and possibly make up a new genus. As reported before, Deinococcus and Thermus are closely related (3). The 16S rRNA cloned from south pole snow samples also contained bacteria from psychrophilic or unidentified taxa. FASTA comparison showed that these sequences (∼1.2 kb cloned, >800 bp sequenced and matched with existing sequences) were most closely related to Alcaligenes sp. (clone Spd17), Cytophaga sp. (clones Spd1_8 and Spd2B_4), and Bacteroides sp. (Spd1_12). Based on the number of clones randomly selected for sequencing, Deinococcus-like organisms accounted for about 10 to 20% of the total snow bacteria.
FIG. 2.
Phylogenetic trees based on the 16S rDNA sequence. (A) Maximum-parsimonious analysis performed using PHYLIP with 100 replicates of bootstrap. Included in this analysis were two identical south pole sequences (SP1) that showed highest similarity to the Deinococcus group, representatives from the 11 major bacterial divisions, and some polar bacteria (denoted with asterisks). Bootstrap values are shown at the nodes. Scale bar = 50 steps. (B) Neighbor-joining analysis carried out with PHYLIP. The scale bar indicates a 10% substitution. GenBank accession numbers for these bacteria are given in parentheses following the species names in panel A.
DNA and protein synthesis.
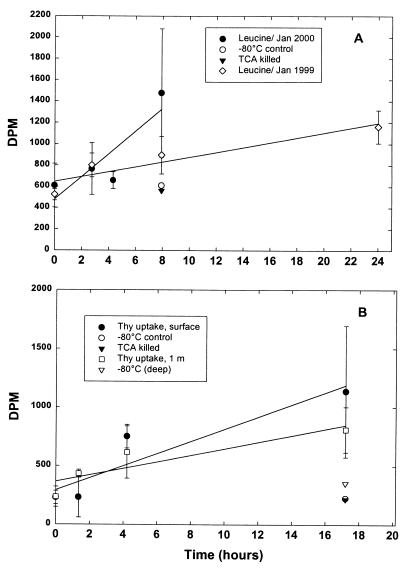
Remarkably, we also detected low levels of apparent DNA and protein synthesis in minicores of snow incubated with tritiated ([3H]thymidine and [3H]leucine, respectively) precursors of these macromolecules incubated at temperatures between −12° and −17°C (Fig. 3) (29). Over the course of 2 to 18 h, radioactive counts recovered in the macromolecular fraction generally increased by 1.5- to 5-fold, relative to counts in zero time-harvested and control samples incubated at −80°C or with TCA from the outset. Positive results were noted for 16 of 18 leucine experiments and 12 of 18 thymidine experiments.
FIG. 3.
Time courses of incorporation of [3H]leucine and [methyl-3H]thymidine into protein and DNA in south pole surface snows. First-order regressions are indicated. (A) The slope is equivalent to a rate of 0.31 ± 0.12 (n = 3) pmol of leucine liter of snowmelt−1 h−1 for the sample taken in January of 2000 and 0.09 ± 0.03 pmol liter of leucine of snowmelt−1 h−1 for the sample taken in January of 1999. (B) Results are for two thymidine-uptake experiments in January 2000 using samples from the very surface and snows 1-m deep. For the surface samples, the rate of thymidine incorporation was 0.13 pmol liter of snowmelt−1 h−1, and for the 1-m samples, the rate was 0.07 ± 0.02 pmol liter of snowmelt−1 h−1. Experimental samples from 1999 were incubated at −12°C, while those from 2000 were incubated at −15 to −17°C. Results from additional controls (incubated at 80°C or amended with TCA at zero time) from January 2000 are also indicated.
Deinococcus is a remarkable bacterium in that it possesses the ability to withstand very high doses of ionizing radiation; it was originally isolated from cans of meat irradiated for sterilization (14). It has been posited that since there are virtually no terrestrial environments that generate radiation doses intolerable to this organism, this ability must have been selected for as a by-product of other DNA damage mechanisms. Extreme desiccation and UV flux can also cause breakage of DNA, and it is thought that Deinococcus may have evolved in a very arid environment (20). The Antarctic snow environment in the interior of the continent is extremely dry, and the level of liquid water is vanishingly low (see below). The Antarctic interior also receives high fluxes of UV radiation during the austral summer.
There have been previous records of microbial metabolism at subzero temperatures. For example, net photosynthetic activity (CO2 exchange) has been detected in Antarctic lichens at −11 and −17°C (18, 26). Bacterial cultures have been established from Siberian permafrost at a mean temperature of −10°C, and because of the age of the permafrost (103 to 106 years), it was argued that the bacteria must have maintained some metabolic activity (27). Viable bacteria have also been isolated from Antarctic snow and exhibited growth in culture at −7°C (30). In Antarctica, in several McMurdo Dry Valley lakes, cyanobacteria and bacteria can be metabolically active in spite of being within a 3- to 6-m lake ice cover due to austral summer radiation-induced heating of small particles (23).
Liquid water is generally thought necessary for life, and some liquid water exists in snow at temperatures below freezing. For example, even at −10°C in Siberian permafrost, 0.5 to 3% of water is unfrozen (9). Furthermore, the thickness of the quasiliquid water layer on the surface of an ice crystal has been calculated to be about 50 nm at a temperature of −10°C (4). Recent data indicate that there is some uncertainty as to the thickness of this layer. It is thought that pristine ice may have less of a layer but that natural ice, as is found in Antarctica, has contaminants which may alter the thickness of the quasiliquid layer. Furthermore, psychrophilic microorganisms have made several adaptations to life at cold temperatures. For some, soluble carbohydrates and polyols serve as cryoprotectants (21) and increased amounts of unsaturated fatty acids in membranes enhance their fluidity (14), and there are enzymes in psychrophils which are adapted to work at low temperatures (13).
A possible source of energy for these bacteria in south pole snow is allochthonous input of marine microbes and other particulate organic matter in snowfall. Some cloud condensation nuclei over the Ross Ice Shelf in Antarctica have been shown to be of biogenic origin and consist partially of plankton from the Southern Ocean (25). The south pole receives sea salt aerosols that are transported within about two days from the Antarctic coast to the south pole (7). These sea salts serve as cloud condensation nuclei, and presumably some carry marine microbes and other particulate organic matter.
Antarctica, the second smallest continent, has an area of 14 by 106 km2, and only 2% of the surface is terrestrial while the remainder is snow and ice. If our observations of bacterial activity and density are representative of the surface snow over the continent, then this significantly extends the range of life on earth in both a physical and a physiological sense. Furthermore, these observations have relevance for the search for life on other planets, such as Mars, which has a polar ice cap.
Nucleotide sequence accession number.
The nucleotide sequences of the Taylor Valley sample were assigned GenBank accession numbers AF239213 and AF239800, respectively.
Acknowledgments
We thank Pilar Heredia for technical help; Paul Sullivan and Eivind Jensen for aid in snow sampling; and Roland Psenner, Birgit Sattler, Anders Hansen, Edward DeLong, Andrei Chistoserdov, Allison Murray, and John Battista for advice.
Our research was supported by NSF's LEXEN program.
REFERENCES
- 1.Abyzov S S. Microorganisms in the Antarctic ice. In: Friedman E I, editor. Antarctic microbiology. New York, N.Y: Wiley-Liss; 1993. pp. 265–295. [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson A W, Nordan H C, Cain R F, Parrish G, Duggan D. Studies on a radio-resistant micrococcus. 1. Isolation, morphology, cultural characteristics, and resistance to gamma-radiation. Food Technol. 1956;10:575–577. [Google Scholar]
- 4.Anderson D M. Ice nucleation and the substrate-ice interface. Nature. 1967;216:563–566. [Google Scholar]
- 5.Battista J R. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 6.Bell R T. Estimating production of heterotrophic bacterioplankton via incorporation of tritiated thymidine. In: Kemp P F, Sherr B F, Sherr E B, Cole J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Press; 1993. pp. 495–503. [Google Scholar]
- 7.Bodhaine B A, Deluisi J J, Harris J M, Houmere P, Bauman S. Aerosol measurements at the South Pole. Tellus. 1986;38B:223–235. [Google Scholar]
- 8.Doyle J J, Doyle J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 9.Ershov E D. Obshchaja geokriologija (general geocryology). Moscow, Russia: Nedra; 1990. [Google Scholar]
- 10.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:146–164. [Google Scholar]
- 11.Ferreira A C, Nobre M F, Rainey F A, Silvia M T, Wait R, Burghardt J, Chung A P P, daCosta M S. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int J Syst Bacteriol. 1997;47:939–947. doi: 10.1099/00207713-47-4-939. [DOI] [PubMed] [Google Scholar]
- 12.Garrison D L, Sullivan C W, Ackley S F. Sea ice microbial communities in Antarctica. Bioscience. 1986;36:243–250. [Google Scholar]
- 13.Gerday C, Aittaleb M, Arpigny J L, Baise E, Chessa J P, Garsoux G, Petrescu I, Feller G. Psychrophilic enzymes: a thermodynamic challenge. Biochim Biophys Acta. 1997;1342:119–131. doi: 10.1016/s0167-4838(97)00093-9. [DOI] [PubMed] [Google Scholar]
- 14.Gounot A M. Psychrophilic and psychrotrophic microorganisms. Experimentia. 1986;42:1192–1197. doi: 10.1007/BF01946390. [DOI] [PubMed] [Google Scholar]
- 15.Janson S, Bergman B, Carpenter E J, Giovannoni S J, Vergin K. Genetic analysis of natural populations of the marine diazotrophic cyanobacterium Trichodesmium. FEMS Microbiol Ecol. 1999;30:57–65. [Google Scholar]
- 16.Karl D M, Bird D F, Björkman K, Houlihan T, Shackelford R, Tupas L. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science. 1999;286:2144–2147. doi: 10.1126/science.286.5447.2144. [DOI] [PubMed] [Google Scholar]
- 17.Kirchman D L. Leucine incorporation as a measure of biomass production by heterotrophic bacteria. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Boca Raton, Fla: Lewis Press; 1993. pp. 509–517. [Google Scholar]
- 18.Lang O L. Der CO2-Gaswechsel von Flechten bei tiefen Temperturen. Planta. 1996;64:1–19. [Google Scholar]
- 19.Lin S, Carpenter E J. A PSTTLRE-form of cdc2-like gene in the marine microalga Dunaliella tertiolecta. Gene. 1999;239:39–48. doi: 10.1016/s0378-1119(99)00383-2. [DOI] [PubMed] [Google Scholar]
- 20.Mattimore V, Battista J R. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1998;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteil P O, Cowan D A. Possible role of soluble carbohydrates and polyols as cryoprotectants in Antarctic plants. In: Heywood R B, editor. Proceedings of University Research in Antarctica. Cambridge, United Kingdom: British Antarctic Survey; 1993. pp. 119–125. [Google Scholar]
- 22.Noble R T, Fuhrman J. Use of SYBR Green I for rapid epifluorescence of marine viruses and bacteria. Aquat Microbiol Ecol. 1992;14:113–118. [Google Scholar]
- 23.Priscu J C, et al. Perennial Antarctic Lake ice: an oasis for life in a polar desert. Science. 1998;280:2095–2098. doi: 10.1126/science.280.5372.2095. [DOI] [PubMed] [Google Scholar]
- 24.Priscu J C, et al. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science. 1999;286:2141–2144. doi: 10.1126/science.286.5447.2141. [DOI] [PubMed] [Google Scholar]
- 25.Saxena V K. Evidence of biogenic nuclei involvement in Antarctic coastal clouds. J Phys Chem. 1983;87:4130–4134. [Google Scholar]
- 26.Schroeter B, Green T G A, Kappen L, Seppelt R D. Carbon dioxide exchange at subzero temperatures: field measurements on Umbilicarina aprina in Antarctica. Cryptogam Bot. 1994;4:233–241. [Google Scholar]
- 27.Shi T, Reever R H, Gilchinsky D A, Friedman E I. Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb Ecol. 1997;33:169–179. doi: 10.1007/s002489900019. [DOI] [PubMed] [Google Scholar]
- 28.Siebert J, Hirsch P. Characterization of 15 selected coccal bacteria isolated from Antarctic rock and soil samples from the McMurdo-Dry Valleys (South-Victoria Land) Polar Biol. 1988;9:37–44. doi: 10.1007/BF00441762. [DOI] [PubMed] [Google Scholar]
- 29.Smith D C, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- 30.Straka R P, Stokes J L. Psychrophilic bacteria from Antarctica. J Bacteriol. 1960;80:622–625. doi: 10.1128/jb.80.5.622-625.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]