Abstract
The coronavirus disease 2019 (COVID-19) pandemic constitutes a global health emergency. Currently, there are no completely effective therapeutic medications for the management of this outbreak. The cytokine storm is a hyperinflammatory medical condition due to excessive and uncontrolled release of pro-inflammatory cytokines in patients suffering from severe COVID-19, leading to the development of acute respiratory distress syndrome (ARDS) and multiple organ dysfunction syndrome (MODS) and even mortality. Understanding the pathophysiology of COVID-19 can be helpful for the treatment of patients. Evidence suggests that the levels of tumor necrosis factor alpha (TNF-α) and interleukin (IL)-1 and IL-6 are dramatically different between mild and severe patients, so they may be important contributors to the cytokine storm. Several serum markers can be predictors for the cytokine storm. This review discusses the cytokines involved in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, focusing on interferons (IFNs) and ILs, and whether they can be used in COVID-19 treatment. Moreover, we highlight several microRNAs that are involved in these cytokines and their role in the cytokine storm caused by COVID-19.
Keywords: SARS-CoV-2, COVID-19, microRNAs, cytokines, interferons, interleukins, NF-κB, Janus kinases, STAT transcription factors
Graphical abstract
Our research indicated discrepancies in the reported results related to the impact of many cytokines on the treatment of COVID-19, and some studies reported that the disease phase is very important in the use of cytokines and will have different effects. Moreover, we suggest that cytokine-related microRNAs may be a promising target in treating COVID-19.
Introduction
The local outbreak of coronavirus disease 2019 (COVID-19) in December 2019 was recognized for the first time in Wuhan, China. The disease spread rapidly to all parts of the world and, on March 11, the World Health Organization (WHO) designated this outbreak as a global pandemic.1,2
The infectious disease COVID-19 is caused by a new coronavirus (CoV), called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The CoVs are a family of positive single-stranded (+ss) RNA viruses covered by an envelope, containing four genera based on nucleic acid and protein sequence: alpha CoVs, beta CoVs, gamma CoVs, and delta CoVs.2,3 Examples of beta-CoVs are Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), human coronavirus HKU1 (HCoV-HKU1), human coronavirus OC43 (HCoV-OC43), and human coronavirus 229E (HCoV-229E).3,4 From 2002 up to now, three global disease outbreaks have been shown to be caused by beta-CoVs that affect the airways, especially the lower tract, which were SARS-CoV in 2002, MERS-CoV in 2012, and SARS-CoV-2 from 2019. These infections resulted in multiple organ dysfunction syndrome (MODS), acute respiratory distress syndrome (ARDS), and even mortality.5,6
COVID-19 remains a health issue of great concern throughout the world. Fever, non-productive cough, fatigue, myalgia, rhinorrhea, pharyngalgia, diarrhea, dyspnea, and hypoxemia are the most frequently reported symptoms.7,8 Some cases have no or only mild symptoms, but others require hospitalization and intensive care. It has been found that pneumonia and respiratory failure are the leading causes of death, and death rates are very high in patients who require mechanical ventilation.9,10
Many studies have reported that the spike (S) protein of SARS-CoV-2 mediates its entry into the targeted human cells, such as airway epithelial cells, and olfactory receptor neurons. This may be the starting point of a process that will eventually lead to ARDS, MODS, and death. Viruses can multiply and spread rapidly to other target cells and organs. It is reported that the virus can affect a wide range of cells by direct attachment to the receptor or indirectly via cytokine production.11, 12, 13
Cytokines are cell signaling molecules composed of proteins, glycoproteins, or peptides. They are secreted by numerous cell types and play a crucial role in many biological functions by binding to cell surface receptors at particular concentrations. Cytokines exert either pro-inflammatory or anti-inflammatory effects, and inflammation is associated with an imbalance between anti-inflammatory and pro-inflammatory cytokines.14, 15, 16
In severe COVID-19, the virus activates both the innate and adaptive immune systems, resulting in uncontrolled inflammatory responses and overproduction of cytokines, finally producing a cytokine storm. A cytokine storm is a medical condition in which excessive and uncontrolled levels of pro-inflammatory cytokines are produced in response to different triggers, and then continue to be produced in a destructive auto-amplifying cycle typical of MOD.17,18 In COVID-19 pathogenesis, cells undergo apoptosis or necrosis induced directly or indirectly by cytokines. The cytokines can bind to death receptors or activate other cells such as natural killer (NK) cells. The breach of the blood-airway barrier caused by the cytokine storm leads to pulmonary edema and respiratory failure. It also permits the penetration of SARS-CoV-2 into the systemic circulation, where it can then cause systemic MOD.17,19,20 Contradictory results have been found regarding the modulation of cytokines to manage the disease. Some researchers believe that there are insufficient studies supporting the benefits of cytokine gene regulation in reducing the mortality rate in COVID-19.18,21
MicroRNAs (miRNAs, or miR) or short non-coding single-stranded RNAs, are able to regulate the expression of one-third of genes in human beings.22,23 It has been shown that miRNAs can affect many signaling pathways and regulate cytokine production. Cytokines can affect miRNA expression by inducing transcription factor miRNAs.23,24
Collectively, the cytokine profiles in COVID-19 are very complex, and the role of cytokine dysregulation induced by SARS-CoV-2 needs more clarification. Our goal in this article is to provide researchers and physicians a comprehensive overview of cytokine-related effects in COVID-19 up to the present.
Cytokines
Cytokines (in Greek, cyto means cell, and kinos means movement) are small proteins, glycoproteins, or signaling peptides produced and released by various cell types. They have several biological functions at various concentrations by attaching to specific cell receptors.14,25 Based on their biological effects, the cytokine superfamily can be divided into six main families: interleukins (ILs), interferons (IFNs), tumor necrosis factors (TNFs), hematopoietic growth factors, chemokines, and transforming growth factor (TGF) β (TGF-β). Cytokines can also be divided into pro-inflammatory or anti-inflammatory sub-classes.15,26
Cytokines can mediate cell-cell interactions and communication. They may act in an autocrine, paracrine, or endocrine manner, and can exhibit pleiotropic effects. Various cell types may secrete the same cytokine, or be affected by a single cytokine. Often, one cytokine acts on certain cells to produce additional cytokines, forming a cytokine cascade.15
There are numerous reports of abnormal cytokine concentrations in patients during COVID-19 infection. One critical process in severe COVID-19 patients is that excessive rates of inflammatory cytokine production and dysregulated gene expression in affected cells results in a cytokine storm. One of the leading reasons for multiple organ failure (MOF) and ARDS is the cytokine storm.27, 28, 29 Therefore, it is necessary to understand the cytokines involved in COVID-19 that have been studied so far.
IFNs
IFNs are a group of soluble glycoproteins. They provide the most critical first line of defense against pathogenic agents like viruses.30 According to their protein structure and receptors, IFNs are categorized into three classes: type I IFNs (IFN subtypes alpha, beta, epsilon, kappa, and omega), type II IFNs (IFN-γ), and type III IFNs ( IFN-λ).31,32
Type I IFNs
The type I IFNs in mammalian species include alpha (IFN-α), beta (IFN-β), kappa (IFN-κ), delta (IFN-δ), epsilon (IFN-ε), tau (IFN-τ), omega (IFN-ω), and zeta (IFN-ζ, also called limitin).25,33 They all have a specific type I IFN cell surface receptor (IFNAR) in target cells and act in an autocrine and/or paracrine manner. The Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway and nuclear factor κB NF-κB) pathway are mainly involved in type I IFN functions.34, 35, 36, 37, 38
When viral infection occurs and the body recognizes it, type I IFNs are produced. Different cell types can secrete type I IFNs. This stimulates a powerful antiviral defense response involving numerous IFN-stimulated genes (ISGs) designed to interfere with the replication and pathogenesis of the virus.30,33 Reportedly, the new CoV, SARS-CoV-2, is more sensitive to type I IFNs compared with SARS-CoV.39 Nevertheless, in COVID-19, it has been reported that the weak type I IFN response in patients suffering from severe hyperinflammation driven by NF-κB correlates with negligible clearance of the virus. Studies on the efficacy of type I IFN therapy in COVID-19 patients were disappointing.25,40
IFN-α
IFN-α is produced by a wide range of cells, such as respiratory epithelial cells, macrophages, and dendritic cells (DCs). It is known that many types of cells, especially peripheral blood B cells and monocytes, are IFN-α targets.41,42 Studies have shown a low level of IFN-α in the blood of hospitalized COVID-19 patients that go on to develop severe/life-threatening COVID-19.43 It has been shown that IFN-α plays a pivotal role in early-stage COVID-19. IFN-α can reduce the number of viruses, resulting in fewer symptoms and a shorter disease duration. IFN-α administration has been helpful in the management of some viral diseases like SARS. It can also induce angiotensin-converting enzyme (ACE) 2 as an IFN-stimulated gene (ISG) in human upper airway epithelial cells.43, 44, 45
A clinical trial found that IFN-α2b administration in the early phase of COVID-19 could decrease the mortality rate, while late use of IFN-α2b could actually increase mortality. Therefore, IFN-α2b administration in early-stage COVID-19 may produce promising results.46
IFN-β
IFN-β is secreted by pulmonary epithelial cells and monocyte-derived cells.41 IFN-β activates B cells, T cells, monocytes, macrophages, and DCs.47
Open reading frame (ORF) 6, ORF8, and nucleocapsid proteins related to SARS-CoV-2 are potent inhibitors of IFN-β and NF-κB-responsive gene promoters. In Sendai virus infection, these proteins inhibited the IFN-stimulated response element (ISRE), and ORF6 and ORF8 proteins after IFN-β treatment could block the ISRE.48 In critical COVID-19 patients, highly impaired production and response of IFN-β has been found, and the viral load remains high. In these patients, severe inflammatory responses have been observed.49
IFN-β can also induce useful antiviral activity via ISGs, but ISGs are significantly inhibited in SARS-CoV-2 infection. In early-stage COVID-19 infection, non-structural proteins (NSPs) and ORFs related to the virus block host IFNs and dysregulate the ISGs, while ISGs with pro-inflammatory potential are stimulated during the later phase of SARS-CoV-2 infection.50,51
It was previously reported that IFN-β-1b administration in severe COVID-19 patients was associated with positive clinical improvement and shorter duration of hospital stay without serious adverse effects in patients.52 IFN-β can block the overexpression of IL-6 and IL-8 and enhance immune response. Studies have reported that glucocorticoids could inhibit IFN-β signaling and many other cytokines in the human lungs, so care must be taken when using glucocorticoids in viral-induced ARDS.53,54
IFN-κ
IFN-κ affects innate immune-related cells and can be used to control systemic or local immune responses.55 It is produced by monocyte-derived macrophages, DCs, and keratinocytes. IFN-κ affects target cells such as monocytes and DCs. It has been shown that inhalation of IFN-κ plus Trefoil factor 2 (TFF2) can promote the repair of airway epithelial cells exposed to injury, and significantly improves symptoms such as cough in asthma patients. Use of this combination in COVID-19 treatment may significantly improve clinical symptoms, virus negativity, and computed tomography (CT) scan results, and reduce the duration of hospitalization.31,56,57
IFN-δ
IFN-δ is secreted by porcine blastocysts. It is reported that IFN-δ has immunomodulatory and antiviral activity via type I IFNs, but its antiviral activity is lower than IFN-α.58,59 Few relevant studies are available for the effects of IFN-δ.
IFN-ε
IFN-ε is expressed in the lung, brain, skin, intestinal tract, and reproductive tissues (uterus, cervix, vagina, and ovary). IFN-ε contributes to antiviral and antibacterial mucosal immunity. It has been reported that IFN-ε can suppress human immunodeficiency virus (HIV) replication.60, 61, 62
The IFN-ε ligand can bind to IFNAR1 and IFNAR2 and affect the JAK-STAT pathway. These pathways regulate mucosal immune responses resulting in protection from viral and bacterial invasion.63 In some mammalian species, such as pangolins, which are a potential natural reservoir of CoVs, IFN-ε is produced by epithelial cells and contributes to mucosal immunity in skin, lung, intestine, and reproductive tissues.64
Some COVID-19 patients reported changes in skin pigmentation, rash, and low spermatogenesis rate. IFN-ε is expressed in both tissues (skin and testes), so it is possible that COVID-19 infection is an androgen-mediated process.65, 66, 67 IFN-ε protects the reproductive system in women from HIV-1 and other viral infections, and is regulated by endogenous and exogenous hormones. Therefore IFN-ε may be an explanation of decreased mortality in women with SARS-CoV-2 infection.63,68
IFN-τ
IFN-τ is released by bovine blastocysts.69 Although IFN-τ is not expressed in humans, human and mouse cells, such as monocyte-derived macrophages, are able to respond to its administration. IFN-τ has antiviral and antitumor effects, and binds to IFN-α receptors. It induces intracellular signaling leading to antiviral cytokine production, such as IL-4 and IL-6.70 IFN-τ reduces the replication of various viruses, including human papillomavirus (HPV), feline immunodeficiency virus (FIV), and HIV.71 There is no report of its effect on SARS-CoV-2. It has been reported that administration of IFN-τ in a mouse allergy model decreased cell infiltration into lung tissue under inflammatory conditions.70
IFN-ω
DCs can produce IFN-ω in response to viral infection.72 Leukocytes and epithelial cells may be target cells for IFN-ω.73 It stimulates a signaling pathway similar to the IFN-α/β receptor (IFNAR). It has also been reported that the activation of phosphoinositide-3-kinase/protein kinase B (P13K/Akt) signaling is required for IFN-ω activity.74,75 It was shown that IFN-ω had an anti-SARS activity similar to IFN-β. IFN-ω administration reduced the severity of the disease and inhibited CoV replication in monkeys. A study reported that some patients with severe COVID-19-mediated pneumonia showed neutralizing immunoglobulin (Ig) G against IFN-ω.76
IFN-ζ
IFN-ζ is only found in mice, and is secreted by the salivary duct and bronchial epithelial cells. It has sequence homology with IFN-α, and shows similar antiviral and immunomodulatory effects. IFN-ζ has no or low lympho-myelo-suppressive activity. It has been suggested that IFN-α/βR could be a receptor for IFN-ζ. It was shown that the IRF-1 pathway in fibroblasts was involved with IFN-ζ activity and antiviral functions.77, 78, 79 To date, no research has been done on its effects in COVID-19.
Type II IFNs (IFN-γ)
IFN-γ is secreted by macrophages, T cells, and NK cells. The primary source of IFN-γ in rhinovirus infections was bronchial epithelial cells. Its receptors are expressed on immune cells like T cells and NK cells. Similar to type I IFN, IFN-γ can stimulate the JAK-STAT signaling pathway and the NF-κB pathway. IFN-γ can act through the activation of JAK1, JAK2, and STAT signaling pathways.80,81
IFN-γ contributes to the inhibition of acute inflammation, and the transition from innate to acquired immunity. It was reported that the interplay of IFN-γ with IL-6/sIL-6R signaling had positive effects on neutrophil recruitment and phagocytosis. Also, it was observed that patients suffering from severe COVID-19 exhibited a larger IL-6/IFN-γ ratio than moderate cases. IFN-γ may enhance the cytokine storm, leading to lung failure. By contrast, another study reported that IFN-γ production tended to be lower in the severe forms of COVID-19 than in the moderate cases.82,83 It seems that more studies are needed.
Type III IFNs (IFN-λ)
IFN-λ is secreted by epithelial cells, monocyte-derived macrophages, NK cells, dendritic cells (DCs), cytotoxic T cells, and regulatory T cells. There are four sub-classes of IFN-λ: IFN-λ1 (or IL-29), IFN-λ2 (or IL-28A), IFN-λ3 (or IL-28B), and IFN-λ4. These can bind to a complex resulting in IL-10R2 and IL-28RA formation.84, 85, 86, 87 IFN-λ has the highest expression level among all the IFNs, and is secreted in mucosal barriers (like the respiratory tract) in response to viral infections. The target cells of IFN-λ3 include keratinocytes, neutrophils, macrophages, DCs, endothelial cells (ECs), and respiratory epithelial cells.85,88
IFN-λ can delay or prolong the JAK/STAT signaling pathway. Moreover, it can also initiate the mitogen-activated protein kinases (MAPK) and PI3-kinase pathways.35,37
IFN-λ or IFN-γ can bind to the type III IFN receptor (IFNLR). These receptors are mainly found on epithelial cells of many organ systems, including the respiratory, gastrointestinal, and reproductive system, and some myeloid cells. Type II alveolar epithelial cells produce IFN-λ to protect respiratory tract epithelium by the antiviral activity of this IFN. IFN-λ could be a promising therapeutic agent to reduce viral pathogenesis and the cytokine storm to prevent pneumonia and ARDS caused by COVID-19.89, 90, 91
It was reported that MERS-CoV protein functions as an IFN antagonist and suppresses IFN-λ production. During infection by SARS-CoV-2, IFN-λ reduced the viral load and inflammatory responses but had no effect on mortality rates.92,93
Hematopoietic growth factors
Granulocyte colony-stimulating factors
Granulocyte colony-stimulating factors (G-CSFs) are glycoproteins that stimulate the production of granulocytes. They promote survival and proliferation of neutrophil precursors, and the function of mature neutrophils. G-CSFs can increase the production of ILs such as IL-10, IL-8, IL-6, and IL-1 and soluble tumor necrosis factor receptors (sTNF-Rs), and they also decrease the concentrations of IFN-λ, TNF-α, and granulocyte-macrophage colony-stimulating factor (GM-CSF).89 In the severe form of COVID-19, the concentrations of IL-2, IL-6, IL-10, and IFN-γ were elevated. Therefore, G-CSFs have been suggested to play a role in disease severity by facilitating the formation of the cytokine storm.90
Furthermore, G-CSFs may increase the oxidative burst within alveolar neutrophils and macrophages, which could increase the risk of respiratory failure during G-CSF administration. Cancer patients often receive G-CSFs to improve neutropenia caused by their cancer treatment. Therefore, the administration of G-CSFs in cancer patients with COVID-19 should be treated with caution.91,94
GM-CSFs
GM-CSFs are important myelopoietic growth factors secreted by various cells such as epithelial cells and leukocytes. GM-CSF is an important pro-inflammatory cytokine stimulating both innate and adaptive immunity. There were increased numbers of GM-CSF-expressing leukocytes in the blood of COVID-19 patients. It was also reported that levels of GM-CSF were increased in COVID-19 patients compared with non-infected individuals.3,95
GM-CSF is probably implicated in the occurrence of the cytokine storm. It stimulates IL-1 and IL-6 production and targets macrophages and neutrophils. IL-1 and IL-6 play an important role in the cytokine storm, and they may also be aggravated by the cytokine storm itself.96
In COVID-19 patients suffering from life-threatening pneumonia, GM-CSF inhibition resulted in more rapid clinical improvement in symptoms and a further decrease in inflammation.97 By contrast, another study reported that GM-CSF had an effective role in macrophage homeostasis and pathogen clearance in the lungs.98 Due to these contradictory results, further research is needed to evaluate the effects of GM-CSF in COVID-19.
TNFs
TNFs are pro-inflammatory cytokines significantly implicated in the cytokine storm. Levels of TNFs are high in COVID-19, and it has been suggested that early levels of TNFs can predict the mortality risk. Anti-TNF administration reduces the generation of other pro-inflammatory cytokines (like IL-1 and IL-6) in patients with rheumatoid arthritis. Thus, blocking TNF in individuals with COVID-19 is a potential immunomodulatory treatment by decreasing the production of other pathogenic cytokines. A major cause of lung failure in COVID-19 infection is capillary leakage, resulting from pro-inflammatory cytokines like TNF, IL-1, and IL-6. It has been suggested that anti-TNF therapy might reduce inflammation-mediated vessel leakage in COVID-19, but this requires further study.99,100
TGF-β
TGF-β is well-known as a multifunctional immunoregulatory cytokine. It belongs to the TGF family. TGF-β is a potent immunosuppressive factor in the body that inhibits the immune response but accelerates healing. TGF-β may also reduce the number of lymphocytes in the peripheral circulation by inhibiting proliferation and differentiation.101 TGF signaling is activated via both canonical and non-canonical signaling pathways. A nucleocapsid protein interacting with Smad3 activates the canonical pathway, while a papain-like protein activates the non-canonical pathway, upregulating TGF-β. Apoptosis, inflammation, and fibrosis are all triggered by this activation, leading to lung failure and tissue loss. According to recent research, the untimely production of TGF-β is a feature of severe COVID-19 and may impair NK cell activity and early viral control.
Furthermore, SARS-CoV-2 has been shown to cause a TGF-β-related immune response that is not directed toward itself.102, 103, 104, 105 TGF-β inhibits the anti-oxidant system, activates the plasminogen activator inhibitor 1 (PAI-1) and nuclear factor-kappa-light-chain-enhancer of activated B cells, and downregulates the cystic fibrosis transmembrane conductance regulator (CFTR) via internalizing the epithelial sodium channel (ENaC) (NF-κB). These alterations result in inflammation, lung damage, and coagulopathy.106 Because TGF-β plays such an essential role in the initiation and development of the disease, several pieces of research have been conducted to target it for COVID-19 therapy.107, 108, 109, 110
TGF induction has been demonstrated to contribute considerably to the short- and long-term effects of COVID-19, while ACE inhibition affects TGF receptors, resulting in slight blunting of TGF-β receptor activity and effects.111,112 TGF- β1 levels in the blood were considerably higher throughout the early and middle phases of COVID-19 and were associated with SARS-CoV-2-specific IgA levels. As a result, the TGF-β1-IgA axis may play a key role in COVID-19 pathogenesis.113
Chemokines
Chemokines aid in the fight against viral infections by attracting innate and adaptive immune cells to infection sites and boosting their cytotoxic action and production of antiviral mediators. On the other hand, some viruses employ chemokines to avoid the immune system. For example, certain large DNA viruses, such as herpesviruses, create chemokine-like molecules that disrupt signaling and the immune response, resulting in viral multiplication and persistence.114 Chemokines are further divided into four subtypes based on their main amino acid sequence: CC, CXC, CX3C, and XC. Atypical chemokine receptors (aCKRs) and conventional chemokine receptors (cCKRs) are the two types of chemokine receptors found on the surface of cells. Chemokine receptors may be found on a wide range of cells, including leukocytes, airway epithelial cells, mesenchymal cells, and ECs.115,116
Chemokines can help leukocytes to migrate, multiply, and degranulate. They can also increase cytokine production. Several chemokines have been found to have direct antibacterial activity.115,117 They have antiviral effects through several mechanisms. Chemokines direct cells from the innate and adaptive immune system to infection sites. Here they boost antiviral activity by promoting the generation of antiviral mediators.118 Compared with SARS and MERS, the high transmissibility and low fatality rate of SARS-CoV-2 might be attributed to distinct chemokine profiles, requiring more investigation.
CCL-2, CCL-7 (MCP-3), CXCL-9 (MIG), CXCL-10, and CCL-3 serum levels were shown to be higher in COVID-19 patients with clinical symptoms, according to Chi et al.119 COVID-19 patients have been shown to have higher levels of chemokines such as CCL3, CCL5, CXCL10, CCL19, CCL20, while CCL5 remained unchanged. When compared with healthy controls, Huang et al. found that the chemokines CXCL-8 (IL-8), CXCL-10 (IP-10), CCL-2 (MCP-1), CCL-3 (M1α), and CCL-4 (MIP-1β) had already reached high plasma levels, as well as elevated levels of several inflammatory cytokines (IL-IL-1α, IL-7, IL-9, IL-10, G-CSF, FGF, GM-CSF, PDGF, IFN-γ, TNF-α, and vascular endothelial growth factors (VEGFs).95
According to reports, T cells, NK cells, monocytes, macrophages, and neutrophils are the primary immune cells that invade the lungs in patients with COVID-19. CCL2, CCL7, and CCL8 are abundant in infiltrating macrophages at first.120, 121, 122 Corticosteroids have been found to reduce chemokines such as CXCL10 and CXCL8 that directly contribute to the pathogenesis of SARS-CoV-2.123 CCL2, CXCL10, and CXCL8 are the most commonly found chemokines associated with COVID-19 severity. Future research should to be focused on these cytokines. Furthermore, COVID-19 patients have higher CXCL-1, CXCL-2, and CXCL-6 expression than healthy people.124
ILs
ILs are expressed by many types of cells, especially leukocytes. They play both pro-inflammatory and anti-inflammatory roles. ILs are involved in intercellular communication, activation, and migration of immune cells and cytokine production. They also mediate the proliferation, maturation, and adhesion of immune cells. There is a growing list of identified ILs, and approximately 40 different types of ILs are now known.125,126 Studies have observed abnormal levels of ILs in COVID-19 patients, such as IL-17, IL-13, IL-12, IL-10, IL-7, IL-6, IL-4, IL-2, and IL-1.95,127 The following is an overview of COVID-19-related ILs.
IL-1
The prototypic pro-inflammatory cytokine IL-1 can be either IL-1α or IL-1β, whose bioactivities are indistinguishable. Some studies reported that IL-1α is produced by damaged ECs, epithelial cells, and myeloid cells. In comparison, IL-1β is released by infiltrating neutrophils and some monocyte-derived cells.128,129
The SARS-CoV-2 present in the lower respiratory tract can cause SARS associated with excessive pro-inflammatory cytokine levels like IL-1β. Anakinra is an antagonist for the IL-1 receptor capable of blocking IL-1α and IL-1β activity, and is used to manage auto-inflammatory conditions. One study found that using an IL-1 blocker ameliorated clinical symptoms in 72% of patients with COVID-19 and managed the ARDS.130,131
IL-2
In 1976, IL-2 was described as a T cell growth factor (TCGF) during basic immunology research in human tumors. It can exert a pleiotropic effect on immunity and is routinely prescribed to manage some human cancers.132 It has been suggested that administration of IL-2 may be an effective therapy by increasing lymphocyte numbers in COVID-19 patients.133
IL-2 can stimulate all T cells, including T regulatory, T helper, and NK cells. These are vital antiviral cells essential for SARS-CoV-2 clearance. They can restrict the severe form of COVID-19-mediated cytokine storm. Therefore IL-2 has been suggested to be a therapeutic agent to control COVID-19.134,135
IL-4
The anti-inflammatory cytokine IL-4 has a specific receptor (IL-4R). IL-4 can suppress the cytotoxic activity of macrophages and reduce nitric oxide (NO) production. Patients with COVID-19 have shown an elevated level of IL-4. IL-4 induces T cell differentiation into T helper 2 cells. These cells subsequently release additional IL-4 from other T cells in a positive-feedback loop. T helper 2 cells were found to be present in high amounts in COVID-19 patients.136, 137, 138, 139
IL-6
The anti-inflammatory and pro-inflammatory cytokine IL-6 is expressed in various cells like ECs, T cells, and macrophages. The functions of IL-6 include differentiation of B cells resulting in IgM, IgE, and IgA production; regulation of T cell activation; and T cell differentiation.134,135
COVID-19 infection increases the expression level of IL-6, especially in patients with lung damage and pulmonary inflammation. Overproduction of IL-6 is more significant compared with other cytokines in the severe form of COVID-19. IL-6 has a pivotal role in the occurrence of cytokine storm in COVID-19.140,141
Elevated IL-6 levels are correlated with a poor prognosis of COVID-19 patients. IL-6 is a key mediator governing the generation of T helper 17 cells.30 Therefore, the excessive IL-6 concentration in COVID-19 patients may be a reason for the increased number of activated T helper 17 cells seen in these individuals. Moreover, it has been reported that excessive IL-6 signaling leads to an increase in effector T cells along with increased vessel permeability. Therefore, inhibition of IL-6 could be helpful in COVID-19 treatment, which may be due to its ability to inhibit the cytokine storm.122,142,143
IL-7
The pleiotropic cytokine IL-7 is an important mediator for lymphocyte survival and proliferation. Lymphopenia and T cell depletion in the spleen and other organs can occur in severe COVID-19 patients, which reduces the ability to fight against viruses. IL-7 administration can be safe for those with life-threatening COVID-19, and can restore the lymphocytes to a usual number, appearing to reverse a pathologic characteristic of COVID-19. The IL-7-mediated restoration of lymphocytes boosted the antiviral activity and reduced the viral load.143, 144, 145
IL-8
The pro-inflammatory cytokine IL-8 plays a crucial role in neutrophil chemotaxis and activation during inflammation. IL-8 levels increase in SARS-CoV-2, suggesting that IL-8 may contribute to neutrophilia in COVID-19 pathophysiology. It has been reported that IL-8 and IL-6 are closely correlated with COVID-19 severity (moderate to end-organ damage). The IL-8 concentration can make a distinct difference between severe progression and recovery in SARS-CoV-2 infection, and IL-8 could be a more appropriate marker to determine the status of COVID-19 compared with IL-6. Moreover, IL-8 could be a potential target in the treatment of COVID-19.99,146,147
IL-10
IL-10 is usually considered to be an anti-inflammatory or immunosuppressive cytokine. It has been called cytokine synthesis inhibitory factor (CSIF).140
A particular property of the COVID-19-induced cytokine storm compared with that found in SARS-CoV infection is a significant increase in IL-10 in critically ill patients. A high concentration of IL-10 may be a predictor of a poor prognosis in COVID-19 cases. Moreover, the elevated concentration of IL-10 occurs earlier in the course of the infection compared with IL-6, and it was shown that IL-10 levels in serum were dramatically higher in COVID-19 patients admitted to the intensive care unit (ICU).95,141,148
Some studies have suggested that IL-10 acts as an anti-inflammatory cytokine by a negative feedback mechanism. At the same time, other clinical evidence has suggested that early elevation of IL-10 may have a detrimental role in COVID-19 intensity.149 Therefore more studies are needed on the role of IL-10.
IL-17
IL-17 is an important mediator in innate immunity and an essential link between T cells and neutrophils.150 Moreover, in the ARDS inflammatory process, IL-17A (the most studied subtype of IL-17) acts on macrophages, resulting in increased expression of NO synthase, IL-1, IL-6, TNFα, and chemokines, which makes the disease worse. Moreover, alveolar endothelial and epithelial cells release IL-17, leading to the production of IL-6, GM-CSF, IL-1β, TNFα, TGF-β, and G-CSF from various cell types.151,152
It has been reported that IL-17 is involved in the hyperinflammatory state in COVID-19. The upregulation of IL-17A in the cytokine storm is produced by T helper 17 cells, and is mainly responsible for ARDS. Therefore, the possible therapeutic use of IL-17 inhibitors in COVID-19 has been suggested.153,154
IL-21
Activated CD4+ T cells, NK T cells, and T helper 17 cells all release IL-21, which belongs to the γ-chain cytokine family. Both innate and adaptive immune responses are governed by IL-21. IL-21 promotes IgG1 and IgG3 class switching in humans while suppressing IgE class switching. Dendritic cells are activated by IL-21, which causes NK cells to release more IFN-γ, boosting their cytotoxic activity. IL-21 suppresses T helper 1 cell differentiation and promotes CD8+ T cell proliferation. IL-21 regulates the expression of CD27 and CD28, which is important for virus-specific effector CD8+ T cells.
In COVID-19 patients, measurement of serum IL-21 shows a high predictive value for disease development. High levels of IL-6 and IL-21 in the blood at admission are independent risk factors for clinical deterioration.155,156 IL-21 has also been demonstrated to reduce the generation of IL-6 and TNF-α, lowering the inflammatory proteins implicated in the cytokine storm.157 T follicular helper cell-induced IL-21 modulates germinal center B cell differentiation, which is critical for forming effective virus-neutralizing antibodies, similar to other viral infections. When the B cells in the germinal center of patients with acute COVID-19 were analyzed, IgG2-producing B cells responsive to type 1 IFN were dominant at the beginning of ICU hospitalization; however, as the disease progressed, the number of IgG1- and IgA1-producing B cells responsive to IL-21 and TGF-β increased, and ultimately the number of IgA2-producing B cells responsive to TGF-β increased. TGF-β produces lung fibrosis, the most preventable consequence of SARS-CoV-2 infection in the chronic post-inflammatory phase. In acute COVID-19, IL-21 and TGF-β may be out of balance, thus exacerbating the pathological state.156,158 IL-21 and IL-15 combined together promote a more effective immune response than either IL alone. As a result, a clinical trial exploring the combination of IL-15 and IL-21 for COVID-19 patients is recommended.159 However, anticancer clinical trials of IL-21 have been unsatisfactory, and it has been suggested that its clinical use is problematic.156 However, some research suggests that high circulating levels of IL-21 are linked to illness severity in COVID-19 patients.160
IL-23
IL-23 belongs to the family IL-12 cytokine as a pro-inflammatory heterodimeric cytokine mainly secreted by the skin, intestinal mucosa, lung-activated macrophages, and DCs. IL-23 triggers IFN-γ expression and T cell proliferation, as well as IFN-α responsiveness to eliminate hepatitis C virus (HCV). It also enhances the resistance of the host to viruses by the IL-23/IL-17 axis. IL-23 triggers the production of suppressor of cytokine signaling 1 (SOCS1) and impairs T cell function in HIV infection. Biologics that target IL-23 are helpful for psoriasis treatment.161,162
It has been shown that low levels of IL-23 may lead to deficient immunity in the mucosal barrier, and an enhanced risk of respiratory infections.161 A study found that psoriasis patients treated with biologics had a higher risk for SARS-CoV-2 infection and hospital admission. However, there was no difference in mortality rate or mechanical ventilation in the ICU. At the same time, a case report study showed an improvement in SARS-CoV-2 infection after administration of guselkumab, an IL-23 antagonist.163,164
IL-27
IL-27 is a pleiotropic cytokine and forms heterodimeric complexes of EBI3 and p28 subunits. IL-27 is involved in T helper 1 differentiation and acts on various cells, such as monocyte-derived, NK, T, and B cells. It is secreted by diverse cell types, especially activated macrophages and DCs. IL-27 signaling downregulates IL-4 and reduces mucin production. IL-27 can also induce anti-inflammatory responses by stimulating IL-10 production. It has been suggested that low levels of IL-27 could be a promising biomarker for COVID-19 prognosis.164, 165, 166
IL-33
IL-33 belongs to the family of IL-1 cytokine with pleiotropic functions. IL-33 is produced by barrier tissues, and in the lungs damaged alveolar epithelial cells release IL-33.167 Reportedly, the production of IL-33 is associated with SARS-CoV-2 infection. IL-33 may be involved in pulmonary fibrosis by stimulating various cytokines. Anti-IL-33 therapy might be promising in COVID-19 management, and is presently under investigation.168
Cytokine storm
The role of the cytokine storm is illustrated in Figures 1 and 2. The cytokine storm, also called hypercytokinemia, is mostly an uncontrolled hyperimmune response to infection. This term was first introduced in 1993 with regard to tissue graft rejection. Since then, the term cytokine storm has been more often used in viral infections, such as influenza viruses, SARS, and MERS. Studies have found that in SARS, high concentrations of pro-inflammatory cytokines, such as TNF-α, IL-6, IL-1β, IL-12, IFN-γ, IP10, and MCP1, were associated with lung damage.169,170 The cytokine storm has also been observed in some COVID-19 patients. Extraordinarily high levels of inflammatory cytokines were found in life-threatening and severe forms of COVID-19, resulting in pulmonary inflammation, lung failure, and MODS. It was also reported that the levels of IL-1, TNF-α, and IL-6 could clearly differentiate mild cases from severe forms of COVID-19.95
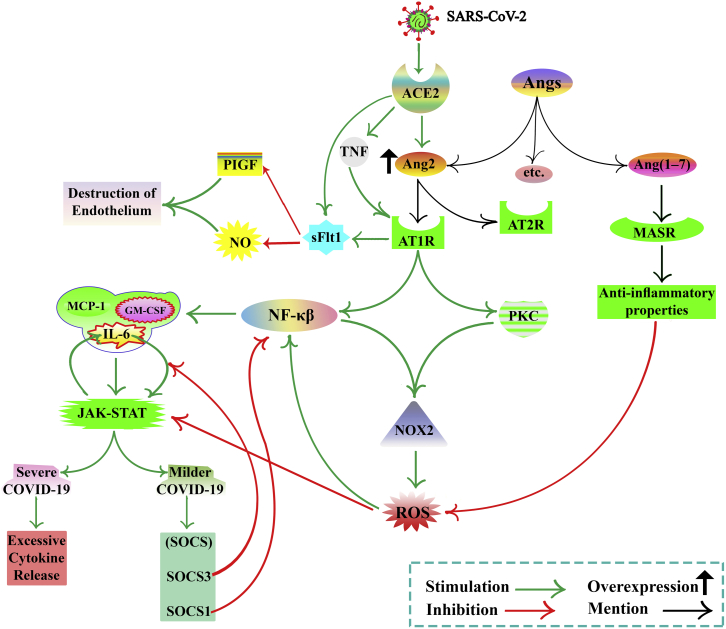
Figure 1.
COVID-19 and cytokine storm, focusing on Ang2 concentration
The concentrations of TNF-α, IL-6, and IL-10 are the most important mediator in cytokine storm formation. IL-6 receptors classify into two groups, mIL-6R and sIL-6R. During virus infection, host cells can express PRRs. PRRs detect PAMPs. ACE2 receptor acts as a PRR. ACE2 cleaves Ang2. ACE2 binds to viral S protein, so Ang2 concentrations increase. Ang2 has two types of receptors, AT1R and AT2R. The Ang2/AT1R complex activates PKC and NF-κB, leading to NOX2 activation and cytokine production. ROS production mediated by NOX2 and ROS activates NF-κB. NF-κB enhances the expression of IL-6, GM-CSF, MCP-1, etc. IL-6 induces the activation of NOX. IL-6, GM-CSF, and MCP-1 activate JAK-STAT signaling, resulting in an elevated level of SOCS, but, in severe disease, it leads to excessive cytokine production. SOCS blocks the JAK-STAT signaling. SOCS may provide a novel therapy for the treatment of COVID-19. The JAK-STAT pathway activates by ROS. IL-6 and JAK-STAT signaling pathway interaction can be defined as positive feedback. SOCS3 disrupts this vicious cycle by inhibiting IL-6 signaling. SOCS-1 inhibits NF-κB activation. NF-κB increases ACE2 expression. sFLT1 production is not clear (by binding the Ang2 to AT1 or directly induced by SARS-CoV-2 infection or upregulation of AT1 Receptor by TNF). sFLT1 inhibits PlGF, a VEGF, and impairs NO production, resulting in endothelial damage. Ang-(1–7) binds to the MASR and causes inhibition of ROS and anti-inflammatory properties. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; Ang2, angiotensin2; IL, interleukin; TNF-α, tumor necrosis factor alpha; PRRs, pattern recognition receptors; ; PAMPs, pathogen-associated molecular patterns; ACE2, angiotensin-converting enzyme 2; AT1R, angiotensin II type 1 receptor; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NOX, nicotinamide adenine dinucleotide phosphate oxidase; AT2R, angiotensin II type 2 receptor; GM-CSF, granulocyte-macrophage colony-stimulating factor; PKC, protein kinase C; ROS, reactive oxygen species; MCP-1, monocyte chemoattractant protein-1; SOCS, suppressor of cytokine signaling; JAK-STAT, Janus kinase-signal transducer and activator of transcription; sFLT1, soluble Fms-like tyrosine kinase-1; MASR, Mas oncogene receptor; gp130, glycoprotein 130; PlGF, placenta growth factor; NO, nitric oxide; sIL-6R, soluble IL-6 receptor; VEGF, vascular endothelial growth factor.
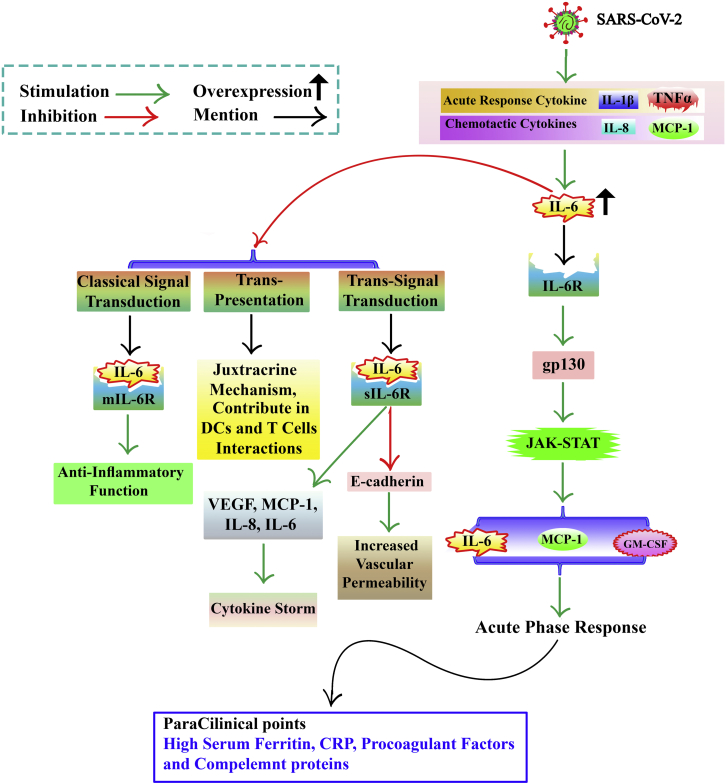
Figure 2.
COVID-19 and cytokine storm, focusing on IL-6
IL-1b and TNF, as acute-response cytokines, and MCP-1 and IL-8, chemotactic cytokines, increase hypercytokinemia, which elevates IL-6. The IL-6/IL-6R acts on gp130 to increase IL-6, MCP-1, and GM-CSF by activating the JAK-STAT pathway. IL-6, GM-CSF, and MCP-1 may activate an acute-phase response indicated by a high serum ferritin, CRP, and pro-coagulant factors in paraclinical tests. Three IL-6 signal transductions of trans-presentation, trans-signal transduction, and classical signal transduction. IL-6/mIL-6R complex contributes to the classical signal transduction mode, which mediates anti-inflammatory function. IL-6 trans-signaling is more related to inflammatory processes. In this signaling, IL-6 binds to the sIL-6R. This leads to the production of VEGF, MCP-1, IL-8, IL-6, and E-cadherin. Expression in ECs is reduced. This increases vascular permeability and exacerbates the cytokine storm. IL-6 trans-presentation signaling pathway is a juxtracrine mechanism that contributes to dendritic and T cell interactions. mIL-6R, membrane-bound form of Interleukin-6 Receptor; CRP, C-reactive protein.
The concentrations of IL-6, TNF-α, and IL-10 were reported to be related to the severity of SARS-CoV-2 infection. Raised concentrations of IL-1β, IFN-γ, MCP1, and IP10 in ICU COVID-19 patients might stimulate the activation of T helper 1 cells to further increase inflammation.95,170
IL-6 is one of the most important mediators in the cytokine storm. Various cells like T cells, ECs, fibroblasts, and monocyte-derived cells secrete IL-6, which then acts on B cells, T cells, and granulocytes.134 IL-6 receptors can be classified into two groups: soluble IL-6 receptor (sIL-6R) and membrane-bound IL-6 receptor (mIL-6R). mIL-6R is expressed by only some of the target cells of IL-6. sIL-6R functions as a transporter of IL-6 to various parts of the body, and is generated by mIL-6R cleavage mediated by a disintegrin and metallopeptidase domain 10 (ADAM10) and ADAM17. It has also been reported that sIL-6R can be produced by alternative splicing of IL-6R mRNA.134
During virus infection, host cells can express pattern recognition receptors (PRRs) on their surface. PRRs detect molecules related to pathogens (bacteria, fungi, and viruses) that are different from host molecules, called pathogen-associated molecular patterns (PAMPs). The ACE2 receptor recognized by SARS-CoV-2 serves as a PRR expressed in many types of cells, such as pulmonary alveolar cells and ECs in blood vessels.171
ACE2 is a zinc metalloprotease enzyme that cleaves angiotensin II (Ang2). Peptides that are produced as a result of Ang2 cleavage may activate signaling pathways that counteract Ang2 signaling. ACE2 binds to the viral S protein so its activity is reduced; therefore, Ang2 concentrations may be significantly higher in infected lungs.172
Ang2 type 1 receptor (AT1R) is considered an important Ang2 receptor. The Ang2/AT1R complex can activate several molecules and signaling pathways in ECs, such as protein kinase C (PKC) and NF-κB. NF-κB acts as a transcription factor leading to NADPH oxidase (NOX) 2 activation, cytokine production, etc. Reactive oxygen species (ROS) production in ECs is increased by NOX2, and the ROS can activate the NF-κB signaling pathway.173,174
Activation of NF-κB enhances the expression of many cytokines, such as IL-1, TNF-α, IL-6, IL-2, IL-12, GM-CSF, IL-8, MCP-1, and MIP-1. Moreover, IL-6 can induce the activation of NOX, resulting in oxidative stress.167,175,176
The main source of MCP-1, GM-CSF, and IL-6 are fibroblasts, ECs, epithelial cells, etc. These cytokines may affect macrophages, neutrophils, and other immune cells via the JAK-STAT signaling pathway resulting in increased levels of suppressor of cytokine signaling (SOCS), and excessive cytokine production in severe COVID-19.177, 178, 179, 180
The SOCS family function as intracellular checkpoint inhibitors. SOCS regulates JAK-STAT signaling in a negative feedback loop. STATs enhance transcription of the SOCS genes, while SOCS proteins bind to JAKs to inhibit the JAK/STAT pathway. The JAK/STAT pathway is critically involved in the transmission of extracellular cytokine signals into the nucleus.181 Therefore, SOCS may be a target for the treatment of COVID-19, using SOCS mimetics or stabilizers. The JAK-STAT pathway can be inhibited by SOCS, and activated by ROS.182,183
IL-6 activates STAT3, and to a lesser extent STAT1. The JAK-STAT pathway enhances the generation of IL-6 in a positive-feedback loop. SOCS3 could disrupt this vicious cycle in severe COVID-19 by inhibiting IL-6 signaling.184,185
SOCS can also interfere with NF-κB activity. It has been reported that, in infected cells, inhibition of SOCS1 increases the IκB protein concentration, and dramatically increases NF-κB activation. NF-κB can modulate ACE2 expression, and one study reported that ACE2 mRNA could be suppressed by NF-κB inhibitors, such as pyrrolidine dithiocarbamate (PDTC).186,187
Since SARS-CoV-2 can bind to ACE2, NF-κB inhibitors have been suggested to be beneficial in COVID-19. This binding also decreases the level of ACE2, resulting in increased Ang2 levels. The increased Ang2 binds to its receptors (AT1 and AT2) and directly impairs endothelial function.188
Soluble fms-like tyrosine kinase-1 (sFLT1) is a soluble inhibitor of VEGFs. It acts as an endothelial decoy receptor produced by binding of Ang2 to AT1 in response to hypoxia, only in ECs or monocytes. It was shown that the level of sFLT-1 was considerably higher in COVID-19 patients with pneumonia.189, 190, 191
It has not yet been determined whether sFLT-1 production is directly triggered by SARS-CoV-2 infection or stimulated in response to increased Ang2 concentrations. There is also a hypothesis that COVID-associated inflammatory mediators directly or indirectly cause the production of sFLT-1; for example, TNF can induce AT1 receptor upregulation.191,192
sFLT1 has many functions, such as inhibition of VEGFs, such as placental growth factor (PlGF); impairment of NO production resulting in endothelial damage; and sensitizing ECs to Ang2 resulting in the formation of a positive-feedback loop. It has also been reported that the Ang2/AT1 complex induces cytokine production and ROS generation via the NF-κB pathway in macrophages.177,193,194
In addition to Ang2, there are other biologically active members of the angiotensin peptide family. Some studies have been performed on the role of Ang-(1–7) in COVID-19 disease. However, no study has been done on other types of angiotensins in SARS-CoV-2 infection. Ang-(1–7) binds to the MAS1 oncogene receptor (MASR), a G protein-coupled receptor. This inhibits ROS production and vasodilation, and has anti-inflammatory properties. Therefore Ang-(1–7) may be beneficial in COVID-19.195
It has been reported that the levels of TNF and IL-1 (acute-response cytokines) as well as MCP-1 and IL-8 (chemotactic cytokines) increase initially in hypercytokinemia, resulting in a sustained increases in IL-6 concentration. The IL-6/IL-6R axis acts on transmembrane glycoprotein 130 (gp130) to increase the level of IL-6 in a positive-feedback loop. GM-CSF and MCP-1 activate the JAK-STAT pathway to prolong the inflammatory process.178,179
IL-6 and other cytokines activate an acute-phase response. Acute-phase proteins are stimulated by IL-6 and secreted from the liver, including C-reactive protein (CRP), ferritin, pro-coagulant factors like fibrinogen, serum amyloid A (SAA), haptoglobin, and a1-antichymotrypsin. At the same time, the levels of fibronectin, albumin, and transferrin are reduced due to IL-6 activation. High serum ferritin levels, CRP, and pro-coagulant factors can be measured by paraclinical tests.180
GM-CSF and possibly IL-6 are secreted by the pathogenic IFN-ɣ+ GM-CSF+ T helper 1 cells to increase inflammatory CD14+CD16+ monocyte responses by increasing the concentration of IL-6 in COVID-19 patients. Moreover, monocytes secrete TNF-α, IL-6, and IL-1b.3
IL-6 employs a highly complex signaling pathway, making it difficult to know exactly how IL-6 signaling exerts its biological effects. There are three main IL-6 signal transduction mechanisms: trans-presentation, trans-signal transduction, and classical signal transduction. IL-6 receptor signaling has protective roles in normal individuals, such as regulating various metabolic processes and tissue repair.196
It has been reported that the IL-6/mIL-6R complex contributes to the classical signal transduction process, which mediates the anti-inflammatory activity of IL-6. This complex causes gp130 dimerization, since gp130 is expressed by all cells. Studies have suggested that IL-6 can bind to both forms of the receptor and subsequently interact with gp130. Activated gp130 initiates intracellular signaling pathways that finally alter gene expression. IL-6 signal transduction by gp130 activation in cells that do not express mIL-6R can also be triggered by sIL-6R binding. Therefore, sIL-6R increases the target cells that can respond to IL-6 and accounts for its pleiotropic functions.196, 197, 198
IL-6 trans-signaling is more related to pro-inflammatory processes. In this signaling, IL-6 binds to sIL-6R, and then a gp130 dimer complex is formed. Next, there is activation of IL-6-sIL-6R-JAK-STAT3 signaling. Finally, this causes the production of VEGF, IL-8, MCP-1, and additional amounts of IL-6. Moreover, the production of E-cadherin in ECs is reduced, thereby increasing vascular permeability and exacerbating the cytokine storm.143,198,199
The IL-6 signaling pathway of trans-presentation is a juxtracrine mechanism that contributes to DC and T cell interactions. It may lead to the stimulation of pathogenic T helper 17 cell production, inhibition of T regulatory cells, and inflammation.199
The COVID-19-induced cytokine storm mechanism is complex, and many molecules and cells in various cascades and feedback loops are involved in creating it or eliminating it. Therefore, there is a need for further research to reach a more solid understanding of this condition, because the early diagnosis of cytokine storms is important to decrease the mortality rate of COVID-19.
Molecular mechanisms of cytokine production in SARS-CoV-2
During SARS-CoV-2 infection, the immune response is divided into a protective phase based on immunological defense against the virus, and a second phase marked by severe inflammation. Because the virus inhibits the host immune response in the first stage and causes an inflammatory storm in the second stage, some SARS-CoV-2-infected patients become seriously ill. DCs, neutrophils, macrophages, and NK cells are the first line of defense in the immune, and influence the type and severity of the response. The SARS-CoV-2 cytokine storm is fueled by these cells and other tissue-resident cells, including epithelial cells and ECs. COVID-19 patients have high levels of pro-inflammatory cytokines and chemokines such as IL-1β, IL-2, IL-6, IL-7, IL-10, IFN-γ, TNF-α, G-CSF, CCL2, and CXCL10. These inflammatory chemokines and cytokines then recruit additional innate immune cells from the peripheral tissues and activate adaptive immune cells (CD4+ and CD8+ T cells) to produce long-lasting inflammatory cytokines like IL-2, IFN-γ, and TNF-α, which cause myelopoiesis, leading to rapid granulopoiesis, and aggravate lung epithelial damage. Furthermore, macrophage activation is triggered by the overexpression of systemic cytokines, including IL-2, IFN-γ, GM-CSF, and TNF-α. These cytokines eventually result in ARDS. As a result, SARS-CoV-2 infection causes an overproduction of pro-inflammatory cytokines, often resulting in severe consequences.200,201
The heterodimeric inhibitory receptor CD94/NK group 2 member A (NKG2A) is expressed by NK cells. NKG2A can bind to peptide-loaded non-classical human leukocyte antigen (HLA) class I molecules (HLA-E) to reduce NK cell toxicity and cytokine release. NKG2A expression was found to be high in COVID-19 patients in some studies.202,203 Due to SP1 intracellular expression in lung epithelial cells, NK cells displayed elevated NKG2A/CD94 inhibitory receptor levels, according to a recent study.204
One recently discovered beta CoV protein is SARS-CoV-2 ORF8. ORF8 is a hypervariable gene that allows the virus to adapt to the human host more easily.205 ORF8 was identified as the most critical viral protein that was inhibited by type I IFN (IFN-β) and NF-κB-responsive promoters. ORF8 also influenced intracellular immunity and growth pathways through IFN-I signaling and MAPKs.206
After recognizing a wide range of PAMPs via PRRs such as toll-like receptors (TLRs), macrophages generate pro-inflammatory cytokines. The SARS-CoV-2 spike protein S1 component substantially promotes IL-6 and IL-1β production in murine and human macrophages by activating TLR4 signaling via the JNK and NF-κB pathways, according to Shirato et al.207,208 In macrophages, the P44/42 MAPK and Akt pathways produce pro-inflammatory molecules. Infection with SARS-CoV-2 activated the JAK-STAT pathway via CD147, resulting in increased production of cyclophilin A (CyPA), which then bound to CD147 and activated the MAPK pathway. PAMPs initiate various signaling cascades in the innate immune system, leading to the transcriptional activation of type I and type III IFNs.31,209
NSPs from SARS-CoV-2 have a role in immune regulation
The CoV NSPs are essential components of the viral replication mechanism. They aid viral RNA transcription and replication while evading the natural host defenses.210 The (alpha-beta)-CoV genome is a non-segmented, single-strand, positive-sense RNA that encodes for six primary ORFs. The first two-thirds of viral RNA contains two overlapping reading frames, ORF1a and ORF1b, which encode large polyproteins that are then cleaved into 16 mature NSPs. These are thought to play essential roles in regulating cellular pathways and adjusting the cellular environment for optimized virus replication while attempting to avoid immunity.195,211
The virus capacity to avoid and inhibit the host innate immune responses is one of the most critical drivers of viral infection. Both alpha and beta-CoV nsp1 proteins are likely to promote more effective, coordinated immune suppression. Nsp1 proteins have been demonstrated to interfere with multiple phases of the immune response.195 Different strains of (alpha-beta)-CoVs have been shown to suppress IFN-I with variable degrees of efficiency. Nsp1 and nsp6 effectively suppressed IFN-I via blocking STAT1 and STAT2 phosphorylation.212 IFN-β-expression was measured after Nsp1 transfection into HEK293T cells, and activity from a reporter plasmid expressing an IFN transcription factor in a study to examine the immune response effects of SARS-CoV nsp1. Nsp1 can suppress the activation of NF-κB, IRF3, IRF7, and the transcription factor c-Jun, according to the findings.213,214 SARS-CoV-2 nsp9 and nsp10 target NKRF (NF-κB repressor) to increase IL-6/IL-8 production, according to one study using peripheral blood mononuclear cells (PBMCs).215 Nsp3 is a multi-domain transmembrane protein containing an ADP-ribose phosphatase domain (ADRP/MacroD) that is thought to disrupt the host immune response by removing ADP-ribose from ADP-ribosylated proteins or RNA. The PLPro/deubiquitinase domain cleaves viral polyprotein and inhibits the host innate immune response.216, 217, 218 Furthermore, the papain-like proteinase nsp3 has been shown to reduce protective immunity by suppressing innate immunity genes and preventing the host immunological response.219
Another study found that SARS-CoV-2 nsp6 could disrupt lysosome acidification in lung epithelial cells by interacting with the vacuolar-type H+-ATPase component ATP6AP1, resulting in blockage of autophagic flux, pyroptosis, and activation of NLRP3 inflammasomes.220 It has been found that nsp6 can suppress IFN regulation by interacting with TANK binding kinase 1 (TBK1). Orf6 binds importin karyopherin α 2 (KPNA2) to restrict IRF3 nuclear translocation. Nsp13 binds and prevents TBK1 phosphorylation, while Orf13 binds and blocks IRF3 nuclear translocation.221 Another study found that nsp8 and nsp9 both bind to 7SL RNA in the signal recognition particle after infection, and prevent protein trafficking to the cell membrane. The IFN response to viral infection is suppressed when these essential cellular processes are disrupted.222 Xu et al. recently discovered that NSP12 might activate RIPK1 and cause a cytokine storm leading to death.223
miRNAs involved with IFNs and ILs
miRNAs are a class of short (20–25 or 21–23 bases) single-stranded oligonucleotides, acting as non-coding RNAs (ncRNAs). It has been reported that the expression of one-third of all human genes can be regulated by miRNAs.22,23
miRNAs are produced from pri-miRNAs that then form pre-miRNAs with a stem-loop structure. Endonucleases process the pre-miRNAs to form mature miRNA duplexes. The miRNA duplex is finally loaded into the Argonaute protein, resulting in the formation of a mature RNA interference silencing complex (RISC).224 The mature single-stranded miRNA regulates gene expression in a post-transcriptional manner mediated by binding to sites within the 5′- or 3′-untranslated region of complementary messenger RNAs.22 Moreover, miRNAs may be involved in cell-to-cell signaling by entering into neighboring cells and affecting the expression of their mRNAs.225
miRNAs can affect many signaling pathways and regulate cytokine production. Moreover, cytokines are capable of altering miRNA expression by triggering transcription factor miRNAs. It is known that the miRNAs expression profile is specific to the type of cell.226,227
We have provided a list of cytokine-related miRNAs involved in COVID-19 pathogenesis. Understanding the miRNAs that regulate cytokines or are regulated by them can help us to better understand their role in the pathogenesis and clinical treatment of COVID-19, since miRNA expression is specific to the type of cell.228 We suggest that the expression profile of these cellular miRNAs in COVID-19 patients be examined to help identify the major cells involved in SARS-CoV-2 infection.
There have been numerous reports about IFN-related miRNAs (Table 1). Within the type 1 IFNs family, only IFN-α and IFN-β have been studied in depth. It was reported that IFN-α was regulated by miR-466l, miR-22, and miR-122,229, 230, 231 while miR-146a, miR-26a, miR-34, and Let-7b can regulate IFN-β.232,233 In addition, it was observed that IFN-α regulates the production of miR-130a/301, miR-203, and miR-122.234, 235, 236 Many types of miRNAs are known to be regulated by IFN-β. These are miR-155, miR-29a, miR-26a, miR-34a, Let-7b, miR-21, and miR-122.233,234,237, 238, 239 Moreover, it was shown that IFN-τ (not found in humans) can regulate bta-miR-204 in bovine endometrial epithelial cells.240
Table 1.
IFNs
| IFNs | Subtype | Cell source | Target cells | Common pathway | Known functions | COVID-19 | Regulated miRNA | Regulatory miRNAs | References |
|---|---|---|---|---|---|---|---|---|---|
| IFN-I | IFN-α | pulmonary epithelial cells, DCs, macrophages | many cell types, B cells, and monocytes | NF-κB, JAK-STAT (MAPK, PI3-kinase) | induces ACE2 as an ISG in human upper airway epithelial cells | reduced the number of viruses, resulting in relief of symptoms, leading to shorter disease duration | miR-130a/301, miR-203, miR-122 | miR-466l, miR-22, miR-122 | 34, 35, 36, 37, 38,41, 42, 43, 44, 45, 46,156, 157, 158,161, 162, 163,241 |
| IFN-β | pulmonary epithelial cells, DCs, macrophages | immune cells (B cells, T cells), monocytes, macrophages, DCs | NF-κB, JAK-STAT (MAPK, PI3-kinase) | effective antiviral action via ISGs | IFN-β-1b administration in severe COVID-19 had positive effects on clinical improvement and duration of hospital stay without serious adverse effects in patients | miR-155, miR-29a, miR-26a, miR-34a, Let-7b, miR-21, miR-122 | miR-146a miR-26a miR-34, Let-7b |
34, 35, 36, 37, 38,41,47, 48, 49, 50, 51, 52, 53, 54,159, 160, 161,164, 165, 166 | |
| IFN-κ | macrophages, monocytes, DCs, keratinocytes | monocytes, dendritic cells | NF-κB, JAK-STAT (MAPK, PI3-kinase) | influence innate immune system cells. Improved symptoms such as cough in patients with asthma | IFN-κ plus TFF2 could significantly enhance clinical improvement | not reported | not reported | 31,34, 35, 36, 37, 38,55, 56, 57 | |
| IFN-δ | porcine blastocysts | not reported | not reported | antiviral and immunomodulatory activity. Lower antiviral activity than IFN-α | not reported | not reported | not reported | 58,59 | |
| IFN-ε | lung, brain, skin tissue, intestinal system, reproductive tissues (Uterus, Cervix, Vagina, Ovary) | macrophages | NF-κB, JAK-STAT (MAPK, PI3-kinase) | mucosal immunity against viral and bacterial infections. Suppression of HIV replication. Protection of reproductive system against viral infections | may be explanation for lower mortality rate in women with SARS-CoV-2 infection than men | not reported | not reported | 34, 35, 36, 37, 38,60, 61, 62, 63, 64, 65, 66, 67, 68 | |
| IFN-τ | bovine blastocysts, endometrial cells | can affect human macrophages | JAK-STAT (bovine) | reduced inflammatory cell infiltration into lung tissue in mouse model of allergy. Antiviral activity. Antiproliferative effects | not reported | bta-miR-204 (bovine endometrial epithelial cells) | not reported | 69, 70, 71,242,243 | |
| IFN-ω | dendritic cells | leukocytes, epithelial cells | NF-κB, JAK-STAT (MAPK, PI3-kinase, P13K/Akt) signaling) | antiviral effects | anti-SARS activity similar to IFN-β. Useful in severe COVID-19 patients with pneumonia | not reported | not reported | 34, 35, 36, 37, 38,72, 73, 74, 75, 76 | |
| IFN-ζ | in mice bronchial epithelial cells, salivary duct cells |
IFN-α/βR-expressing cells | not exactly known (IRF-1 pathway?) | antiviral and immunomodulatory effects | not reported | not reported | not reported | 77,79 | |
| IFN-II | – | bronchial epithelial cells, NK cells, T cells, macrophages | T cells, NK cells | NF-κB, JAK-STAT (MAPK, PI3-kinase) | inhibited acute inflammation (inhibited innate/acquired immunity transition) | expression of IFN tends to be lower in severe COVID-19 than mild cases | miR-29a, miR-155, miR-520b | miR-29, miR-181a | 80, 81, 82, 83,168,243, 244, 245, 246, 247 |
| IFN-III | IFN-λ1 (or IL-29), IFN-λ2 (or IL-28A), IFN-λ3 (or IL-28B), IFN-λ4 | epithelial cells, macrophages, DCs, cytotoxic T cells, NK cells, regulatory T cells | keratinocytes, neutrophils, macrophages, DCs, ECs, respiratory epithelial cells | JAK-STAT (MAPK, PI3-kinase) | reduced systemic inflammation | reduced viral load and inflammatory responses | miR-15a | miR-548, miR-29 | 35,37,84, 85, 86, 87, 88,92,93,169,248, 249, 250, 251, 252 |
Some IFN-II- and IFN-III-related miRNAs have also been identified. It was observed that miR-29 and miRNA181a can regulate IFN-II,240,253 while miRNA-548 and miR-29 affect IFN-III.254,255 It was also reported that IFN-II affects some miRNAs, miR-29a, miR-155, and miR-520b,256, 257, 258, 259 while IFN-III can regulate miR-15a.260
In addition, the effects that ILs and miRNAs have on each other have been investigated (Table 2). The expression of many COVID-19-related ILs can be altered by miRNAs. For instance, IL-1 is affected by miRNA-146a;261 IL-2 by miRNA-221-3p;262 and IL-4 by miRNA-221-3p, miR-210, miR-524-5p, and miR-340/429.263, 264, 265, 266 IL-6 is affected by mmu-miR-7578, miRNA-136-5p, miRNA-146a, miRNA-30b, and miR-365;267, 268, 269, 270, 271 IL-8 by miRNA-146a, miR-520b, miR-155, miR-106a, and miR-16;272, 273, 274, 275, 276 IL-10 by miRNA 27a-3p, hsa-miR-106a, and hsa-miR-106a;277, 278, 279 IL-17 by miR-30a and miR-136;280,281 IL-23 by miRNA-155;282 and IL-33 by RNA-200, miR-524-5p, and miR-378a-3p.283, 284, 285
Table 2.
ILs
| IL type | COVID-19 | Regulated miRNA | Regulatory miRNA | References |
|---|---|---|---|---|
| IL-1 | IL-1 inhibitor improved clinical symptoms in 72% of patients with ARDS Excessive expression of pro-inflammatory cytokines like IL-1β in lower respiratory tract cells |
miR-155 | miR-146a | 118,119,170,194 |
| IL-2 | administration of IL-2 may be effective by increasing lymphocyte numbers in critically ill patients Il-2 can stimulate T cells, including T helper, T regulatory, and NK cells, which are essential for viral clearance, and limiting the severe cytokine storm |
not reported | miR-221-3p | 121,122,171 |
| IL-4 | patients have elevated IL-4 | miR-124, miR-142-5p, miR-130a-3p | miR-221-3p, miR-210, miR-524-5p, miR-340/429 | 126,167,172, 173, 174,177,286 |
| IL-6 | high IL-6 concentrations have been reported correlated with patient pulmonary inflammation and lung damage elevated level IL-6 is correlated with poor prognosis inhibition of IL-6 can be effective in blocking cytokine storm |
miR-15a/-16, miR-1275 | mmu-miR-7578, miR-136-5p, miR-146a, miR-30b, miR-365 | 129,130,175,176,178,179,287, 288, 289 |
| IL-7 | IL-7 administration was safe in life-threatening COVID-19. Restored lymphocytes to a normal count, reversed COVID-19 pathology | miR-6852 | – | 132,133,180,290 |
| IL-8 | IL-8 is a more appropriate marker than IL-6 potential therapeutic target |
mir-200 | miR-146a, miR-520b, miR-155, miR-106a, miR-16 | 99,136,137,181, 182, 183, 184,196,291 |
| IL-10 | IL-10 acts as an anti-inflammatory cytokine by a negative feedback mechanism. Other clinical evidence suggested that early IL-10 elevation may play a pathological role | miR-155, miR-375 | miR-27a-3p, miR-106a, miR-106a | 134,185, 186, 187,196, 197, 198 |
| IL-17 | IL-17 is related to hyperinflammatory state and cytokine storm upregulation of IL-17A produced by increased T helper 17 cells is partly responsible for ARDS Possible therapeutic use of IL-17 inhibitors |
miR-873, miR-155-5p, miR-497 | miR-30a, miR-136 | 141, 142, 143,188,189,198,199,292 |
| IL-21 | in acute COVID-19, TGF-β and IL-21 may be out of balance, exacerbating disease | miR-663b, miR-29 | miR-155, miR-30b, miR-423-5p | 244, 245, 246, 247,251,252 |
| IL-23 | IL-23 antagonist improved clinical symptoms | miR-25 | miR-155 | 147,190,199 |
| IL-27 | IL-27 can induce anti-inflammatory effects by stimulating IL-10 production. Low levels of IL-27 may be a prognostic marker for COVID-19 | miR-935 | – | 140,147,148,200 |
| IL-33 | IL-33 production was increased | miR-320 | miR-200, miR-524-5p, miR-378a-3p | 150,191, 192, 193,201 |
It has also been demonstrated that ILs can regulate the concentration of many miRNAs. For instance, IL-1 regulates miRNA-155;293 IL-4 regulates miR-124, miR-142-5p, and miR-130a-3p;294,295 IL-6 regulates miRNA-15a/-16 and miR-1275;296,297 IL-7 regulates hsa-miR-6852;298 IL-8 regulates mir-200;299 IL-10 regulates miR-155 and miR-375;299, 300, 301 IL-17 regulates miR-873 and miR-155-5p;301,302 IL-23 regulates miRNA-25;302 IL-27 regulates miR-935;303 and IL-33 regulates miR-320.304
There is a wide range of cytokine-related miRNAs that it is not possible to cover completely in this article. We suggest that future research should concentrate on those miRNAs that are associated with all COVID-19-related cytokines.
Conclusion
The COVID-19-mediated cytokine storm involves an imbalance between anti-inflammatory cytokines and pro-inflammatory cytokines, leading to an overproduction of pro-inflammatory cytokines. This causes serious problems, including tissue damage, ARDS, MOD, and death. There are some clinical studies about the early diagnosis of cytokine storms. This is essential to decrease mortality. There are some discrepancies in published studies about the effects of various cytokines on the treatment of COVID-19, and some studies have suggested that the disease phase is very important in the clinical use of cytokines and that they will have different effects. Moreover, we suggest that cytokine-related miRNAs may be a promising new target for the treatment of COVID-19. Based on the data, Ang(1–7), MASR, SOCSs, NF-kβ inhibitors, and JAK-STAT inhibitors should be further investigated as potential therapeutic agents.
Availability of data and material
The primary data for this study are available from the authors on request.
Acknowledgments
The authors wish to thank all physicians, nurses, technicians, pharmacists, administrators, and all other frontline heroes for their plights and heroic efforts. All people around the world have been struck by their mind-blowing dedication. They are the front line, endangering themselves and their families for all of our benefits. They try not to be discouraged in these challenging conditions. They are now being asked to work even if exposed to SARS-CoV-2 infection, and they never say no. Some of them will no longer see their families. We dedicate this article to all healthcare workers and managers who lost their lives and who are now fighting COVID-19. M.R.H. was supported by US NIH grants R01AI050875 and R21AI121700.
Author contributions
H.M. and B.R. contributed in conception, design, statistical analysis, and drafting of the manuscript. M.H.J., F.Z.R., and S.M.R.H. contributed in data collection and manuscript drafting. M.R.H. critically revised the manuscript. All authors approved the final version for submission.
Declaration of interests
M.R.H. declares the following potential conflicts of interest. Scientific advisory boards for Transdermal Cap Inc (Cleveland, OH), Hologenix Inc (Santa Monica, CA), Vielight (Toronto, Canada), and JOOVV Inc (Minneapolis St Paul MN); consulting for USHIO Corp (Japan) and Sanofi-Aventis Deutschland GmbH (Frankfurt am Main, Germany). The other authors declare no competing interests.
Contributor Information
Bahman Rashidi, Email: b_rashidi@med.mui.ac.ir.
Seyed Mohammad Reza Hashemian, Email: smrhashemian@sbmu.ac.ir.
Hamed Mirzaei, Email: mirzaei-h@kaums.ac.ir, h.mirzaei2002@gmail.com.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry J.D., Jones S., Drebot M.A., Andonov A., Sabara M., Yuan X.Y., Weingartl H., Fernando L., Marszal P., Gren J., et al. Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods. 2004;120:87–96. doi: 10.1016/j.jviromet.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B., Zhou X., Qiu Y., Feng F., Feng J., Jia Y., Zhu H., Hu K., Liu J., Liu Z. Clinical characteristics of 82 death cases with COVID-19. MedRxiv. 2020 doi: 10.1101/2020.02.26.20028191. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cakir Edis E. Chronic pulmonary diseases and COVID-19. Turk. Thorac. J. 2020;21:345–349. doi: 10.5152/turkthoracj.2020.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Yu Y., Xu J., Shu H., Xia J., Wu Y., Zhang L., Yu Z., Fang M., Yu T., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/s2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. Soc. 2020;55:2000607. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z., Mcgoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. J. Am. Med. Assoc. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro dos Santos Miggiolaro A.F., da Silva Motta Junior J., Busatta Vaz de Paula C., Nagashima S., Alessandra Scaranello Malaquias M., Baena Carstens L., N Moreno-Amaral A., Pellegrino Baena C., De Noronha L. Covid-19 cytokine storm in pulmonary tissue: anatomopathological and immunohistochemical findings. Respir. Med. Case. Rep. 2020;31:101292. doi: 10.1016/j.rmcr.2020.101292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bienvenu J., Monneret G., Fabien N., Revillard J.P. The clinical usefulness of the measurement of cytokines. Clin. Chem. Lab. Med. 2000;38:267–285. doi: 10.1515/CCLM.2000.040. [DOI] [PubMed] [Google Scholar]
- 15.Xie W.-R., Deng H., Li H., Bowen T., Strong J., Zhang J.-M. Robust increase of cutaneous sensitivity, cytokine production and sympathetic sprouting in rats with localized inflammatory irritation of the spinal ganglia. Neuroscience. 2006;142:809–822. doi: 10.1016/j.neuroscience.2006.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J.-M., An J. Cytokines, inflammation and pain. Int. Anesthesiol. Clin. 2007;45:27–37. doi: 10.1097/aia.0b013e318034194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarzi-Puttini P., Giorgi V., Sirotti S., Marotto D., Ardizzone S., Rizzardini G., Antinori S., Galli M. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin. Exp. Rheumatol. 2020;38:337–342. doi: 10.55563/clinexprheumatol/xcdary. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robba C., Battaglini D., Pelosi P., Rocco P.R.M. Multiple organ dysfunction in SARS-CoV-2: MODS-CoV-2. Expet Rev. Respir. Med. 2020;14:865–868. doi: 10.1080/17476348.2020.1778470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Virgiliis F., Di Giovanni S. Lung innervation in the eye of a cytokine storm: neuroimmune interactions and COVID-19. Nat. Rev. Neurol. 2020;16:645–652. doi: 10.1038/s41582-020-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Shukla G.C., Singh J., Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 24.Kin K., Miyagawa S., Fukushima S., Shirakawa Y., Torikai K., Shimamura K., Daimon T., Kawahara Y., Kuratani T., Sawa Y. Tissue-and plasma-specific micro RNA signatures for atherosclerotic abdominal aortic aneurysm. J. Am. Heart Assoc. 2012;1:e000745. doi: 10.1161/jaha.112.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartee E., Mcfadden G. Cytokine synergy: an underappreciated contributor to innate anti-viral immunity. Cytokine. 2013;63:237–240. doi: 10.1016/j.cyto.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krejsek J. In: Klinická imunologie. Krejsek J., Kopecky O., editors. 2004. Imunitní systém jako informační soustava; pp. 41–52. [Google Scholar]
- 27.Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39:517–528. doi: 10.1007/s00281-017-0639-8. [DOI] [PubMed] [Google Scholar]
- 28.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/mmbr.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X., Wang T., Cai D., Hu Z., Chen J., Zhi L., Wei H., Zhang Z., Qiu Y., Wang J., et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38–42. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hündgen M., Von Eick H.I. Grundlagen und Anwendung in Klinik und Praxis. Med. Monatsschr. Pharm. 1991;14:164–173. [PubMed] [Google Scholar]
- 33.Hardy M.P., Owczarek C.M., Jermiin L.S., Ejdebäck M., Hertzog P.J. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Platanias L.C. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 35.Donnelly R.P., Kotenko S.V. Interferon-lambda: a new addition to an old family. J. Interferon. Cytokine. Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dumoutier L., Lejeune D., Hor S., Fickenscher H., Renauld J.-C. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT) 1, STAT2 and STAT3. Biochem. J. 2003;370:391–396. doi: 10.1042/bj20021935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z., Hamming O.J., Ank N., Paludan S.R., Nielsen A.L., Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 2007;81:7749–7758. doi: 10.1128/jvi.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Z., Wei L., Murti A., Pfeffer S.R., Fan M., Yang C.H., Pfeffer L.M. Non-conventional signal transduction by type 1 interferons: the NF-κB pathway. J. Cell. Biochem. 2007;102:1087–1094. doi: 10.1002/jcb.21535. [DOI] [PubMed] [Google Scholar]
- 39.Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Menachery V.D., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. bioRxiv. 2020 doi: 10.1101/2020.03.07.982264. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mckechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell. Host. Microbe. 2020;27:863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi O., Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065x.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogue S.L., Preston B.T., Stalder J., Bebbington C.R., Cardarelli P.M. The receptor for type I IFNs is highly expressed on peripheral blood B cells and monocytes and mediates a distinct profile of differentiation and activation of these cells. J. Interferon. Cytokine. Res. 2004;24:131–139. doi: 10.1089/107999004322813372. [DOI] [PubMed] [Google Scholar]
- 43.Commission, RBNH. National Administration of Traditional Chinese Medicine on March 3 Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7) Chin. Med. J. 2020;133:1087–1095. doi: 10.1097/CM9.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Tawfiq J.A., Momattin H., Dib J., Memish Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int. J. Infect. Dis. 2014;20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell. Host. Microbe. 2020;28:455–464.e2. doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (α/β) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–335. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 48.Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., Ge X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus. Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park A., Iwasaki A. Type I and type III interferons–induction, signaling, evasion, and application to combat COVID-19. Cell. Host. Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rahmani H., Davoudi-Monfared E., Nourian A., Khalili H., Hajizadeh N., Jalalabadi N.Z., Fazeli M.R., Ghazaeian M., Yekaninejad M.S. Interferon β-1b in treatment of severe COVID-19: a randomized clinical trial. Int. Immunopharmacol. 2020;88:106903. doi: 10.1016/j.intimp.2020.106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung I.F.-N., Lung K.-C., Tso E.Y.-K., Liu R., Chung T.W.-H., Chu M.-Y., Ng Y.-Y., Lo J., Chan J., Tam A.R., et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/s0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jalkanen J., Pettilä V., Huttunen T., Hollmén M., Jalkanen S. Glucocorticoids inhibit type I IFN beta signaling and the upregulation of CD73 in human lung. Intensive Care. Med. 2020;46:1937–1940. doi: 10.1007/s00134-020-06086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nardelli B., Zaritskaya L., Semenuk M., Cho Y.H., Lafleur D.W., Shah D., Ullrich S., Girolomoni G., Albanesi C., Moore P.A. Regulatory effect of IFN-κ, a novel type I IFN, on cytokine production by cells of the innate immune system. J. Immunol. 2002;169:4822–4830. doi: 10.4049/jimmunol.169.9.4822. [DOI] [PubMed] [Google Scholar]
- 56.Fu W., Liu Y., Xia L., Li M., Song Z., Hu H., Yang Z., Wang L., Cheng X., Wang M., et al. A clinical pilot study on the safety and efficacy of aerosol inhalation treatment of IFN-κ plus TFF2 in patients with moderate COVID-19. EClinicalMedicine. 2020;25:100478. doi: 10.1016/j.eclinm.2020.100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu W., Liu Y., Liu L., Hu H., Cheng X., Liu P., Song Z., Zha L., Bai S., Xu T., et al. An open-label, randomized trial of the combination of IFN-κ plus TFF2 with standard care in the treatment of patients with moderate COVID-19. EClinicalMedicine. 2020;27:100547. doi: 10.1016/j.eclinm.2020.100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chon T.W., Bixler S. Interferon-τ: current applications and potential in antiviral therapy. J. Interferon. Cytokine. Res. 2010;30:477–485. doi: 10.1089/jir.2009.0089. [DOI] [PubMed] [Google Scholar]
- 59.Sang Y., Rowland R.R.R., Hesse R.A., Blecha F. Differential expression and activity of the porcine type I interferon family. Physiol. Genomics. 2010;42:248–258. doi: 10.1152/physiolgenomics.00198.2009. [DOI] [PubMed] [Google Scholar]
- 60.Zwarthoff E.C., Mooren A.T.A., Trapman J. Organization, structure and expression of murine interferon alpha genes. Nucleic. Acids. Res. 1985;13:791–804. doi: 10.1093/nar/13.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demers A., Kang G., Ma F., Lu W., Yuan Z., Li Y., Lewis M., Kraiselburd E.N., Montaner L., Li Q. The mucosal expression pattern of interferon-ϵ in rhesus macaques. J. Leukoc. Biol. 2014;96:1101–1107. doi: 10.1189/jlb.3a0214-088rrr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tasker C., Subbian S., Gao P., Couret J., Levine C., Ghanny S., Soteropoulos P., Zhao X., Landau N., Lu W., Chang T.L. IFN-ε protects primary macrophages against HIV infection. JCI Insight. 2016;1:e88255. doi: 10.1172/jci.insight.88255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marks Z.R., Campbell N., Deweerd N.A., Lim S.S., Gearing L.J., Bourke N.M., Hertzog P.J. Properties and functions of the novel type i interferon epsilon. Semin. Immunol. 2019;43:101328. doi: 10.1016/j.smim.2019.101328. [DOI] [PubMed] [Google Scholar]
- 64.Choo S.W., Rayko M., Tan T.K., Hari R., Komissarov A., Wee W.Y., Yurchenko A.A., Kliver S., Tamazian G., Antunes A., et al. Pangolin genomes and the evolution of mammalian scales and immunity. Genome. Res. 2016;26:1312–1322. doi: 10.1101/gr.203521.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z., Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9:920. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.EstébAnez A., Pérez-Santiago L., Silva E., Silva E., Guillen-Climent S., Guillen-Climent S., García-Vázquez A., Ramón M.D., EstebAnez A., GArciA-Vazquez A. Cutaneous manifestations in COVID-19: a new contribution. J. Eur. Acad. Dermatol. Venereol. 2020;34:e250–e251. doi: 10.1111/jdv.16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wambier C.G., Goren A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is likely to be androgen mediated. J. Am. Acad. Dermatol. 2020;83:308–309. doi: 10.1016/j.jaad.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fung K.Y., Mangan N.E., Cumming H., Horvat J.C., Mayall J.R., Stifter S.A., De Weerd N., Roisman L.C., Rossjohn J., Robertson S.A., et al. Interferon-ε protects the female reproductive tract from viral and bacterial infection. Science. 2013;339:1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubisch H.M., Larson M.A., Kiesling D.O. Control of interferon-τ secretion by in vitro-derived bovine blastocysts during extended culture and outgrowth formation. Mol. Reprod. Dev. 2001;58:390–397. doi: 10.1002/1098-2795(20010401)58:4<390::aid-mrd6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 70.Mujtaba M.G., Villarete L., Johnson H.M. IFN-τ inhibits IgE production in a murine model of allergy and in an IgE-producing human myeloma cell line. J. Allergy. Clin. Immunol. 1999;104:1037–1044. doi: 10.1016/s0091-6749(99)70086-2. [DOI] [PubMed] [Google Scholar]
- 71.Pontzer C.H., Yamamoto J.K., Bazer F.W., Ott T.L., Johnson H.M. Potent anti-feline immunodeficiency virus and anti-human immunodeficiency virus effect of IFN-tau. J. Immunol. 1997;158:4351–4357. [PubMed] [Google Scholar]
- 72.Shibabaw T., Molla M.D., Teferi B., Ayelign B. Role of IFN and complements system: innate immunity in SARS-CoV-2. J. Inflamm. Res. 2020;13:507–518. doi: 10.2147/jir.s267280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrat F.J., Crow M.K., Ivashkiv L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019;20:1574–1583. doi: 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adolf G.R., Frühbeis B., Hauptmann R., Kalsner I., Maurer-Fogy I., Ostermann E., Patzelt E., Schwendenwein R., Sommergruber W., Zöphel A. Human interferon ω1: isolation of the gene, expression in Chinese hamster ovary cells and characterization of the recombinant protein. Biochim. Biophys. Acta. 1991;1089:167–174. doi: 10.1016/0167-4781(91)90004-6. [DOI] [PubMed] [Google Scholar]
- 75.Seo Y., Kim M., Choi M., Kim S., Park K., Oh I., Chung S., Suh H., Hong S., Park S. Possible role of phosphoinositide-3-kinase in Mx1 protein translation and antiviral activity of interferon-omega-stimulated HeLa cells. Pharmacology. 2011;87:224–231. doi: 10.1159/000324536. [DOI] [PubMed] [Google Scholar]
- 76.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oritani K., Hirota S., Nakagawa T., Takahashi I., Kawamoto S.-I., Yamada M., Ishida N., Kadoya T., Tomiyama Y., Kincade P.W., Matsuzawa Y. T lymphocytes constitutively produce an interferonlike cytokine limitin characterized as a heat-and acid-stable and heparin-binding glycoprotein. Blood. 2003;101:178–185. doi: 10.1182/blood-2002-01-0045. [DOI] [PubMed] [Google Scholar]
- 78.Oritani K., Medina K.L., Tomiyama Y., Ishikawa J., Okajima Y., Ogawa M., Yokota T., Aoyama K., Takahashi I., Kincade P.W., Matsuzawa Y. Limitin: an interferon-like cytokine that preferentially influences B-lymphocyte precursors. Nat. Med. 2000;6:659–666. doi: 10.1038/76233. [DOI] [PubMed] [Google Scholar]
- 79.Oritani K., Tomiyama Y. Interferon-3/Limitin: novel type I interferon that displays a narrow range of biological activity. Int. J. Hematol. 2004;80:325–331. doi: 10.1532/ijh97.04087. [DOI] [PubMed] [Google Scholar]
- 80.Lin F.-C., Young H.A. Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014;25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Darwich L., Coma G., Peña R., Bellido R., Blanco E.J.J., Este J.A., Borras F.E., Clotet B., Ruiz L., Rosell A., et al. Secretion of interferon-γ by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. 2009;126:386–393. doi: 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mcloughlin R.M., Witowski J., Robson R.L., Wilkinson T.S., Hurst S.M., Williams A.S., Williams J.D., Rose-John S., Jones S.A., Topley N. Interplay between IFN-γ and IL-6 signaling governs neutrophil trafficking and apoptosis during acute inflammation. J. Clin. Invest. 2003;112:598–607. doi: 10.1172/jci17129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen G., Di Wu W.G., Guo W., Yong Cao D.H., Hongwu Wang T.W., Huang D., Zhang H.C., Haijing Yu X.Z., Zhang S.W., Jianxin Song T.C., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kindler E., Thiel V., Weber F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus. Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mora-Arias T., Amezcua-Guerra L.M. Type III Interferons (lambda Interferons) in rheumatic autoimmune diseases. Arch. Immunol. Ther. Exp. 2020;68:1. doi: 10.1007/s00005-019-00564-3. [DOI] [PubMed] [Google Scholar]
- 86.Kotenko S.V., Gallagher G., Baurin V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 87.Wang B.X., Fish E.N. Global virus outbreaks: interferons as 1st responders. Semin. Immunol. 2019;43:101300. doi: 10.1016/j.smim.2019.101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou J.-H., Wang Y.-N., Chang Q.-Y., Ma P., Hu Y., Cao X. Type III interferons in viral infection and antiviral immunity. Cell. Physiol. Biochem. 2018;51:173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 89.Hartung T., Docke W., Gantner F., Krieger G., Sauer A., Stevens P., Volk H.D., Wendel A. Effect of granulocyte colony-stimulating factor treatment on ex vivo blood cytokine response in human volunteers. Blood. 1995;85:2482–2489. [PubMed] [Google Scholar]
- 90.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karlin L., Darmon M., Thiéry G., Ciroldi M., De Miranda S., Lefebvre A., Schlemmer B., Azoulay E., Lefebvre A. Respiratory status deterioration during G-CSF-induced neutropenia recovery. Bone. Marrow. Transplant. 2005;36:245–250. doi: 10.1038/sj.bmt.1705037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang C.-Y., Liu H.M., Chang M.-F., Chang S.C. Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J. Virol. 2020;94 doi: 10.1128/jvi.00099-20. e00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye Q., Wang B., Mao J. Cytokine storm in COVID-19 and treatment. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alkan A., Uncu A., Taşkıran I., Tanrıverdi Ö. Double-edged sword: granulocyte colony stimulating factors in cancer patients during the COVID-19 era. Clinic. 2020;75:e2033. doi: 10.6061/clinics/2020/e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Griffin J.D., Cannistra S.A., Demetri G.D., Ernst T.J., Kanakura Y., Sullivan R. The biology of GM-CSF: regulation of production and interaction with its receptor. Int. J. Cell. Clon. 1990;8:35–45. doi: 10.1002/stem.5530080705. [DOI] [PubMed] [Google Scholar]
- 97.Temesgen Z., Assi M., Shweta F., Vergidis P., Rizza S.A., Bauer P.R., Pickering B.W., Razonable R.R., Libertin C.R., Burger C.D. GM-CSF neutralization with Lenzilumab in severe COVID-19 pneumonia: a case-cohort study. Mayo. Clin. Proc. 2020;95:2382–2394. doi: 10.1016/j.mayocp.2020.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamilton J.A. GM-CSF in inflammation. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190945. e20190945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robinson P.C., Richards D., Tanner H.L., Feldmann M. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2:e653–e655. doi: 10.1016/s2665-9913(20)30309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen W.-X., Luo R.-C., Wang J.-Q., Chen Z.-S. Features of cytokine storm identified by distinguishing clinical manifestations in COVID-19. Front. Public Health. 2021;9:671788. doi: 10.3389/fpubh.2021.671788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beller A., Kruglov A., Durek P., Von Goetze V., Werner K., Heinz G.A., Ninnemann J., Lehmann K., Maier R., Hoffmann U., et al. Specific microbiota enhances intestinal IgA levels by inducing TGF-β in T follicular helper cells of Peyer's patches in mice. Eur. J. Immunol. 2020;50:783–794. doi: 10.1002/eji.201948474. [DOI] [PubMed] [Google Scholar]
- 103.Zhao X., Nicholls J.M., Chen Y.-G. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-β signaling. J. Biol. Chem. 2008;283:3272–3280. doi: 10.1074/jbc.m708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carlson F.R., Jr., Bosukonda D., Keck P.C., Carlson W.D. Multiorgan damage in patients with COVID-19: is the TGF-β/BMP pathway the missing link? JACC Basic Transl. Sci. 2020;5:1145–1148. doi: 10.1016/j.jacbts.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Witkowski M., Tizian C., Ferreira-Gomes M., Niemeyer D., Jones T.C., Heinrich F., Frischbutter S., Angermair S., Hohnstein T., Mattiola I., et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature. 2021;600:295–301. doi: 10.1038/s41586-021-04142-6. [DOI] [PubMed] [Google Scholar]
- 106.Hamidi S.H., Kadamboor Veethil S., Hamidi S.H. Role of pirfenidone in TGF-β pathways and other inflammatory pathways in acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a theoretical perspective. Pharmacol. Rep. 2021;73:712–727. doi: 10.1007/s43440-021-00255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen W. A potential treatment of COVID-19 with TGF-β blockade. Int. J. Biol. Sci. 2020;16:1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huntington K.E., Carlsen L., So E.-Y., Piesche M., Liang O., El-Deiry W. Integrin/TGF-Beta1 inhibitor GLPG-0187 blocks SARS-CoV-2 Delta and Omicron pseudovirus infection of airway epithelial cells which could attenuate disease severity. medRxiv. 2022 doi: 10.3390/ph15050618. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen W. A potential treatment of COVID-19 with TGF-β blockade. Int. J. Biol. Sci. 2020;16:1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trieu V., Saund S., Rahate P., Barge V., Nalk S., Windlass H., Uckun F. Targeting TGF-b pathway with COVID-19 drug candidate ARTIVeda/PulmoHeal accelerates recovery from mild-moderate COVID-19. medRxiv. 2021 Preprint at. [Google Scholar]
- 111.Leeming D.J., Genovese F., Sand J.M.B., Rasmussen D.G.K., Christiansen C., Jenkins G., Maher T.M., Vestbo J., Karsdal M.A. Can biomarkers of extracellular matrix remodelling and wound healing be used to identify high risk patients infected with SARS-CoV-2?: lessons learned from pulmonary fibrosis. Respir. Res. 2021;22:38. doi: 10.1186/s12931-020-01590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kang N.S., Yim H.E., Bae I.S., Choi J.H., Choi B.M., Yoo K.H., Hong Y.S., Lee J.W., Kim S.K. ACE inhibition modulates transforming growth factor-β receptors in the young rat. Pediatr. Nephrol. 2003;18:865–871. doi: 10.1007/s00467-003-1220-3. [DOI] [PubMed] [Google Scholar]
- 113.Wang E.Y., Chen H., Sun B.Q., Wang H., Qu H.Q., Liu Y., Sun X.Z., Qu J., Fang Z.F., Tian L., et al. Serum levels of the IgA isotype switch factor TGF-β1 are elevated in patients with COVID-19. FEBS. Lett. 2021;595:1819–1824. doi: 10.1002/1873-3468.14104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khalil B.A., Elemam N.M., Maghazachi A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021;19:976–988. doi: 10.1016/j.csbj.2021.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nomiyama H., Osada N., Yoshie O. Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history. Gene. Cell. 2013;18:1–16. doi: 10.1111/gtc.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bachelerie F., Ben-Baruch A., Burkhardt A.M., Combadiere C., Farber J.M., Graham G.J., Horuk R., Sparre-Ulrich A.H., Locati M., Luster A.D., et al. International union of basic and clinical pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wolf M., Moser B. Antimicrobial activities of chemokines: not just a side-effect? Front. Immunol. 2012;3:213. doi: 10.3389/fimmu.2012.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Melchjorsen J., Sørensen L.N., Paludan S.R. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J. Leukoc. Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chi Y., Ge Y., Wu B., Zhang W., Wu T., Wen T., Liu J., Guo X., Huang C., Jiao Y., et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J. Infect. Dis. 2020;222:746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hue S., Beldi-Ferchiou A., Bendib I., Surenaud M., Fourati S., Frapard T., Rivoal S., Razazi K., Carteaux G., Delfau-Larue M.-H., et al. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am. J. Respir. Crit. Care. Med. 2020;202:1509–1519. doi: 10.1164/rccm.202005-1885oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Proudfoot A.E.I. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2002;2:106–115. doi: 10.1038/nri722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/s2213-2600(20)30076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cinatl J., Michaelis M., Morgenstern B., Doerr H.W. High-dose hydrocortisone reduces expression of the pro-inflammatory chemokines CXCL8 and CXCL10 in SARS coronavirus-infected intestinal cells. Int. J. Mol. Med. 2005;15:323–327. doi: 10.3892/ijmm.15.2.323. [DOI] [PubMed] [Google Scholar]
- 124.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vaillant A.J., Qurie A. StatPearls [Internet]; 2019. Interleukin. [Google Scholar]
- 126.Akdis M., Burgler S., Crameri R., Eiwegger T., Fujita H., Gomez E., Klunker S., Meyer N., O’mahony L., Palomares O., et al. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J. Allergy. Clin. Immunol. 2011;127:701–721.e70. doi: 10.1016/j.jaci.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 127.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D. Novel coronavirus (2019-nCoV) infections trigger an exaggerated cytokine response aggravating lung injury. Preprin at ChinaXiv. 2019 [Google Scholar]
- 128.Miura R. 19F-NMR study on the interaction of fluorobenzoate with porcine kidney D-amino acid oxidase. J. Biochem. 1989;105:318–322. doi: 10.1093/oxfordjournals.jbchem.a122660. [DOI] [PubMed] [Google Scholar]
- 129.Dinarello C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/s2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Conti P., Ronconi G., Caraffa A., Gallenga C., Ross R., Frydas I., Kritas S., CArAffA A. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 132.Morgan D.A., Ruscetti F.W., Gallo R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science. 1976;193:1007–1008. doi: 10.1126/science.181845. [DOI] [PubMed] [Google Scholar]
- 133.Zhu M.-E., Wang Q., Zhou S., Wang B., Ke L., He P. Recombinant interleukin-2 stimulates lymphocyte recovery in patients with severe COVID-19. Exp. Ther. Med. 2021;21:227. doi: 10.3892/etm.2021.9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Narazaki M., Kishimoto T. The two-faced cytokine IL-6 in host defense and diseases. Int. J. Mol. Sci. 2018;19:3528. doi: 10.3390/ijms19113528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qian S., Gao Z., Cao R., Yang K., Cui Y., Li S., Meng X., He Q., Li Z. Transmissible gastroenteritis virus infection up-regulates fcrn expression via nucleocapsid protein and secretion of tgf-β in porcine intestinal epithelial cells. Front. Microbiol. 2019;10:3085. doi: 10.3389/fmicb.2019.03085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gregory G.D., Raju S.S., Winandy S., Brown M.A. Mast cell IL-4 expression is regulated by Ikaros and influences encephalitogenic Th1 responses in EAE. J. Clin. Invest. 2006;116:1327–1336. doi: 10.1172/jci27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Min B., Prout M., Hu-Li J., Zhu J., Jankovic D., Morgan E.S., Urban J.F., Jr., Dvorak A.M., Finkelman F.D., Legros G., Paul W.E. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J. Exp. Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Patton E.A., La Flamme A.C., Pedras-Vasoncelos J.A., Pearce E.J. Central role for interleukin-4 in regulating nitric oxide-mediated inhibition of T-cell proliferation and gamma interferon production in schistosomiasis. Infect. Immun. 2002;70:177–184. doi: 10.1128/iai.70.1.177-184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eskdale J., Kube D., Tesch H., Gallagher G. Mapping of the human IL10 gene and further characterization of the 5’flanking sequence. Immunogenetics. 1997;46:120–128. doi: 10.1007/s002510050250. [DOI] [PubMed] [Google Scholar]
- 141.Han H., Ma Q., Li C., Liu R., Zhao L., Wang W., Zhang P., Liu X., Gao G., Liu F., et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microb. Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanaka T., Narazaki M., Kishimoto T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy. 2016;8:959–970. doi: 10.2217/imt-2016-0020. [DOI] [PubMed] [Google Scholar]
- 144.Mackall C.L., Fry T.J., Gress R.E. Harnessing the biology of IL-7 for therapeutic application. Nat. Rev. Immunol. 2011;11:330–342. doi: 10.1038/nri2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barata J.T., Durum S.K., Seddon B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 2019;20:1584–1593. doi: 10.1038/s41590-019-0479-x. [DOI] [PubMed] [Google Scholar]
- 146.Li L., Li J., Gao M., Fan H., Wang Y., Xu X., Chen C., Liu J., Kim J., Aliyari R., Zhang J., Jin Y., Li X., Ma F., Shi M., Cheng G., Yang H. Interleukin-8 as a biomarker for disease prognosis of coronavirus disease-2019 patients. Front. Immunol. 2020;11:602395. doi: 10.3389/fimmu.2020.602395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baggiolini M., WAlz A., Kunkel S., WAlz A. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J. Clin. Invest. 1989;84:1045–1049. doi: 10.1172/jci114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., Xu B., Dai Y., Li X., Zhang C., et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5:139834. doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lu L., Zhang H., Zhan M., Jiang J., Yin H., Dauphars D.J., Li S.-Y., Li Y., He Y.-W. Preventing mortality in COVID-19 patients: which cytokine to target in a raging storm? Front. Cell Dev. Biol. 2020;8:677. doi: 10.3389/fcell.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Miossec P., Kolls J.K. Targeting IL-17 and TH 17 cells in chronic inflammation. Nat. Rev. Drug. Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 151.Righetti R.F., Dos Santos T.M., Camargo L.D.N., Aristóteles L.R.C.R.B., Fukuzaki S., de Souza F.C.R., Santana F.P.R., de Agrela M.V.R., Cruz M.M., Alonso-Vale M.I.C., et al. Protective effects of anti-IL17 on acute lung injury induced by LPS in mice. Front. Pharmacol. 2018;9:1021. doi: 10.3389/fphar.2018.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Blois S., Tometten M., Kandil J., Hagen E., Klapp B.F., Margni R.A., Arck P.C. Intercellular adhesion molecule-1/LFA-1 cross talk is a proximate mediator capable of disrupting immune integration and tolerance mechanism at the feto-maternal interface in murine pregnancies. J. Immunol. 2005;174:1820–1829. doi: 10.4049/jimmunol.174.4.1820. [DOI] [PubMed] [Google Scholar]
- 153.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pacha O., Sallman M.A., Evans S.E. COVID-19: a case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Acet Ozturk N.A., Ursavas A., Gorek Di̇lektasli A., Demi̇rdogen E., Coskun N.F., Coskun N.F., Ediger D., Yoyen-Ermi̇s D., Yoyen Ermis D., Karaca M., et al. Interleukin-21: a potential biomarker for diagnosis and predicting prognosis in COVID-19 patients. Turk. J. Med. Sci. 2021;51:2274–2284. doi: 10.3906/sag-2102-24. [DOI] [PubMed] [Google Scholar]
- 156.Asao H. Interleukin-21 in viral infections. Int. J. Mol. Sci. 2021;22:9521. doi: 10.3390/ijms22179521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wilz S.W. 2021. The Variable Immune Response to SARS-CoV-2 Infection and Potential Treatment with Combination IL-15 and IL-21. [Google Scholar]
- 158.Ferreira-Gomes M., Kruglov A., Durek P., Heinrich F., Tizian C., Heinz G.A., Pascual-Reguant A., Du W., Mothes R., Fan C., et al. SARS-CoV-2 in severe COVID-19 induces a TGF-β-dominated chronic immune response that does not target itself. Nat. Commun. 2021;12:1961. doi: 10.1038/s41467-021-22210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wilz S.W. A clinical trial of IL-15 and IL-21 combination therapy for COVID-19 is warranted. Cytokine. Growth. Factor. Rev. 2021;58:49–50. doi: 10.1016/j.cytogfr.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Khan M., Mathew B.J., Gupta P., Garg G., Khadanga S., Vyas A.K., Singh A.K. Gut dysbiosis and IL-21 response in patients with severe COVID-19. Microorganisms. 2021;9:1292. doi: 10.3390/microorganisms9061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mckenzie B.S., Kastelein R.A., Cua D.J. Understanding the IL-23–IL-17 immune pathway. Trends. Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 162.Garg A., Rawat P., Spector S.A. Interleukin 23 produced by myeloid dendritic cells contributes to T-cell dysfunction in HIV type 1 infection by inducing SOCS1 expression. J. Infect. Dis. 2015;211:755–768. doi: 10.1093/infdis/jiu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Benhadou F., Del Marmol V. Improvement of SARS-CoV-2 symptoms following Guselkumab injection in a psoriatic patient. J. Eur. Acad. Dermatol. Venereol. 2020;34:e363–e364. doi: 10.1111/jdv.16590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Damiani G., Pacifico A., Bragazzi N.L., Malagoli P. Biologics increase the risk of SARS-CoV-2 infection and hospitalization, but not ICU admission and death: real-life data from a large cohort during red-zone declaration. Dermatol. Ther. 2020;33:e13475. doi: 10.1111/dth.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tamayo-Velasco A., Martinez-Paz P., Peñarrubia M.J., De La Fuente I., Pérez-González S., Fernández I., Dueñas C., Gómez-Sánchez E., Lorenzo-López M., Gómez-Pesquera E. HGF, IL-1α and IL-27 are robust biomarkers in early severity stratification of COVID-19 patients. J. Clin. Med. 2021;10:2017. doi: 10.3390/jcm10092017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang Q., Lenardo M.J., Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stanczak M.A., Sanin D.E., Apostolova P., Nerz G., Lampaki D., Hofmann M., Steinmann D., Krohn-Grimberghe M., Thimme R., Mittler G., et al. IL-33 expression in response to SARS-CoV-2 correlates with seropositivity in COVID-19 convalescent individuals. Nat. Commun. 2021;12:2133. doi: 10.1038/s41467-021-22449-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., Lit L.C.W., Hui D.S.C., Chan M.H.M., Chung S.S.C., Sung J.J.Y. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang J., Tecson K.M., Mccullough P.A. Endothelial dysfunction contributes to COVID-19-associated vascular inflammation and coagulopathy. Rev. Cardiovasc. Med. 2020;21:315. doi: 10.31083/j.rcm.2020.03.126. [DOI] [PubMed] [Google Scholar]
- 172.Li X., Molina-Molina M., Abdul-Hafez A., Ramirez J., Serrano-Mollar A., Xaubet A., Uhal B.D. Extravascular sources of lung angiotensin peptide synthesis in idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006;291:L887–L895. doi: 10.1152/ajplung.00432.2005. [DOI] [PubMed] [Google Scholar]
- 173.Forrester S.J., Booz G.W., Sigmund C.D., Coffman T.M., Kawai T., Rizzo V., Scalia R., Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol. Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang M.T., Lokuta A.J., Farrell E.F., Wolff M.R., Haworth R.A., Valdivia H.H. Abnormal Ca2+ release, but normal ryanodine receptors, in canine and human heart failure. Circ. Res. 2002;91:1015–1022. doi: 10.1161/01.res.0000043663.08689.05. [DOI] [PubMed] [Google Scholar]
- 175.Li Q., Verma I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 176.Ali M.I., Chen X., Didion S.P. Heterozygous eNOS deficiency is associated with oxidative stress and endothelial dysfunction in diet-induced obesity. Phys. Rep. 2015;3:e12630. doi: 10.14814/phy2.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nickenig G., Harrison D.G. The AT1-type angiotensin receptor in oxidative stress and atherogenesis: part I: oxidative stress and atherogenesis. Circulation. 2002;105:393–396. doi: 10.1161/hc0302.102618. [DOI] [PubMed] [Google Scholar]
- 178.Bhaskar S., Sinha A., Banach M., Mittoo S., Weissert R., Kass J.S., Rajagopal S., Pai A.R., Kutty S. Cytokine storm in COVID-19—immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper. Front. Immunol. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Panigrahy D., Gilligan M.M., Huang S., Gartung A., Cortés-Puch I., Sime P.J., Phipps R.P., Serhan C.N., Hammock B.D. Inflammation resolution: a dual-pronged approach to averting cytokine storms in COVID-19? Cancer. Metastasis. Rev. 2020;39:337–340. doi: 10.1007/s10555-020-09889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.r700016200. [DOI] [PubMed] [Google Scholar]
- 182.Durham G.A., Williams J.J., Nasim M.T., Palmer T.M. Targeting SOCS proteins to control JAK-STAT signalling in disease. Trends. Pharmacol. Sci. 2019;40:298–308. doi: 10.1016/j.tips.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 183.Simon A.R., Rai U., Fanburg B.L., Cochran B.H. Activation of the JAK-STAT pathway by reactive oxygen species. Am. J. Physiol. Cell. Physiol. 1998;275:C1640–C1652. doi: 10.1152/ajpcell.1998.275.6.c1640. [DOI] [PubMed] [Google Scholar]
- 184.Babon J.J., Varghese L.N., Nicola N.A. Inhibition of IL-6 family cytokines by SOCS3. Semin. Immunol. 2014;26:13–19. doi: 10.1016/j.smim.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee C., Lim H.-K., Sakong J., Lee Y.-S., Kim J.-R., Baek S.-H. Janus kinase-signal transducer and activator of transcription mediates phosphatidic acid-induced interleukin (IL)-1β and IL-6 production. Mol. Pharmacol. 2006;69:1041–1047. doi: 10.1124/mol.105.018481. [DOI] [PubMed] [Google Scholar]
- 186.Wei H., Wang S., Chen Q., Chen Y., Chi X., Zhang L., Huang S., Gao G.F., Chen J.-L. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog. 2014;10:e1003845. doi: 10.1371/journal.ppat.1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lee M.C., Chen Y.-K., Hsu Y.-J., Lin B.-R. Zinc supplement augments the suppressive effects of repurposed drugs of NF-kappa B inhibitor on ACE2 expression in human lung cell lines in vitro. bioRxiv. 2021 doi: 10.1101/2021.01.27.428372. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dimmeler S., Rippmann V., Weiland U., Haendeler J., Zeiher A.M. Angiotensin II induces apoptosis of human endothelial cells: protective effect of nitric oxide. Circ. Res. 1997;81:970–976. doi: 10.1161/01.res.81.6.970. [DOI] [PubMed] [Google Scholar]
- 189.Zhou C.C., Ahmad S., Mi T., Xia L., Abbasi S., Hewett P.W., Sun C., Ahmed A., Kellems R.E., Xia Y. Angiotensin II induces soluble fms-Like tyrosine kinase-1 release via calcineurin signaling pathway in pregnancy. Circ. Res. 2007;100:88–95. doi: 10.1161/01.res.0000254703.11154.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Giardini V., Carrer A., Casati M., Contro E., Vergani P., Gambacorti-Passerini C. Increased sFLT1/PlGF ratio in COVID-19: a novel link to Angiotensin II-mediated endothelial dysfunction. Am. J. Hematol. 2020;95:E188–E191. doi: 10.1002/ajh.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.De Lemos J.A., Mcguire D.K., Drazner M.H. B-type natriuretic peptide in cardiovascular disease. Lancet. 2003;362:316–322. doi: 10.1016/s0140-6736(03)13976-1. [DOI] [PubMed] [Google Scholar]
- 192.Peng J., Gurantz D., Tran V., Cowling R.T., Greenberg B.H. Tumor necrosis factor-α–induced AT1 receptor upregulation enhances angiotensin II–mediated cardiac fibroblast responses that favor fibrosis. Circ. Res. 2002;91:1119–1126. doi: 10.1161/01.res.0000047090.08299.d5. [DOI] [PubMed] [Google Scholar]
- 193.Murphy S.R., Cockrell K. Regulation of soluble fms-like tyrosine kinase-1 production in response to placental ischemia/hypoxia: role of angiotensin II. Physiol. Rep. 2015;3:e12310. doi: 10.14814/phy2.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guo F., Chen X.-L., Wang F., Liang X., Sun Y.-X., Wang Y.-J. Role of angiotensin II type 1 receptor in angiotensin II-induced cytokine production in macrophages. J. Interferon. Cytokine. Res. 2011;31:351–361. doi: 10.1089/jir.2010.0073. [DOI] [PubMed] [Google Scholar]
- 195.Yuan S., Balaji S., Lomakin I.B., Xiong Y. Coronavirus Nsp1: immune response suppression and protein expression inhibition. Front. Microbiol. 2021;12:752214. doi: 10.3389/fmicb.2021.752214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Uciechowski P., Dempke W.C. Interleukin-6: a masterplayer in the cytokine network. Oncology. 2020;98:131–137. doi: 10.1159/000505099. [DOI] [PubMed] [Google Scholar]
- 197.Baran P., Hansen S., Waetzig G.H., Akbarzadeh M., Lamertz L., Huber H.J., Ahmadian M.R., Moll J.M., Scheller J. The balance of interleukin (IL)-6, IL-6· soluble IL-6 receptor (sIL-6R), and IL-6· sIL-6R· sgp130 complexes allows simultaneous classic and trans-signaling. J. Biol. Chem. 2018;293:6762–6775. doi: 10.1074/jbc.ra117.001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. The pro-and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 199.Heink S., Yogev N., Garbers C., Herwerth M., Aly L., Gasperi C., Husterer V., Croxford A.L., Möller-Hackbarth K., Bartsch H.S., et al. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH 17 cells. Nat. Immunol. 2017;18:74–85. doi: 10.1038/ni.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gu W., Gan H., Ma Y., Xu L., Cheng Z.J., Li B., Zhang X., Jiang W., Sun J., Sun B., Hao C. The molecular mechanism of SARS-CoV-2 evading host antiviral innate immunity. Virol. J. 2022;19:49. doi: 10.1186/s12985-022-01783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Targeted Ther. 2021;6:255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Houchins J.P., Lanier L.L., Niemi E.C., Phillips J.H., Ryan J.C. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J. Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- 203.Nguyen S., Beziat V., Dhedin N., Kuentz M., Vernant J.P., Debre P., Vieillard V. HLA-E upregulation on IFN-γ-activated AML blasts impairs CD94/NKG2A-dependent NK cytolysis after haplo-mismatched hematopoietic SCT. Bone. Marrow. Transplant. 2009;43:693–699. doi: 10.1038/bmt.2008.380. [DOI] [PubMed] [Google Scholar]
- 204.Bortolotti D., Gentili V., Rizzo S., Rotola A., Rizzo R. SARS-CoV-2 spike 1 protein controls natural killer cell activation via the HLA-E/NKG2A pathway. Cells. 2020;9:1975. doi: 10.3390/cells9091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zinzula L. Lost in deletion: the enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021;538:116–124. doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Díaz J., Valcarcel A., Bensussen A., Álvarez-Buylla E.R. SARS-CoV-2 ORF8 protein: pathogenic and therapeutic implications. Front. Genet. 2021;1517 doi: 10.3389/fgene.2021.693227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shirato K., Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7:e06187. doi: 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shirato K., Takanari J., Kizaki T. Standardized extract of Asparagus officinalis stem attenuates SARS-CoV-2 spike protein-induced IL-6 and IL-1β production by suppressing p44/42 MAPK and Akt phosphorylation in murine primary macrophages. Molecules. 2021;26:6189. doi: 10.3390/molecules26206189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.García-Sastre A. Ten strategies of interferon evasion by viruses. Cell. Host. Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Qiu Y., Xu K. Functional studies of the coronavirus nonstructural proteins. STEMedicine. 2020;1:e39. doi: 10.37175/stemedicine.v1i2.39. [DOI] [Google Scholar]
- 211.Pizzato M., Baraldi C., Boscato Sopetto G., Finozzi D., Gentile C., Gentile M.D., Marconi R., Paladino D., Raoss A., Riedmiller I., et al. SARS-CoV-2 and the host cell: a tale of interactions. Front. Virol. 2022 doi: 10.3389/fviro.2021.815388. [DOI] [Google Scholar]
- 212.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/jvi.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Li J., Guo M., Tian X., Liu C., Wang X., Yang X., Wu P., Xiao Z., Qu Y., Yin Y. Virus-host interactome and proteomic survey of PMBCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.03.31.019216. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Rohaim M.A., El Naggar R.F., Clayton E., Munir M. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 2021;150:104641. doi: 10.1016/j.micpath.2020.104641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Michalska K., Kim Y., Jedrzejczak R., Maltseva N.I., Stols L., Endres M., Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ. 2020;7:814–824. doi: 10.1107/s2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Russo L.C., Tomasin R., Matos I.A., Manucci A.C., Sowa S.T., Dale K., Caldecott K.W., Lehtiö L., Schechtman D., Meotti F.C., et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021;297:101041. doi: 10.1016/j.jbc.2021.101041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7 doi: 10.1128/mbio.01721-16. e01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sun X., Liu Y., Huang Z., Xu W., Hu W., Yi L., Liu Z., Chan H., Zeng J., Liu X., et al. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell. Death. Differ. 2022;29:1240–1254. doi: 10.1038/s41418-021-00916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Thomas S. Mapping the nonstructural transmembrane proteins of severe acute respiratory syndrome coronavirus 2. J. Comput. Biol. 2021;28:909–921. doi: 10.1089/cmb.2020.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Xu G., Li Y., Zhang S., Peng H., Wang Y., Li D., Jin T., He Z., Tong Y., Qi C., et al. SARS-CoV-2 promotes RIPK1 activation to facilitate viral propagation. Cell. Res. 2021;31:1230–1243. doi: 10.1038/s41422-021-00578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bartel D.P. Metazoan micrornas. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chen X., Liang H., Zhang J., Zen K., Zhang C.-Y. Secreted microRNAs: a new form of intercellular communication. Trends. Cell. Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 226.De Gonzalo-Calvo D., Iglesias-Gutiérrez E., Llorente-Cortés V. Epigenetic biomarkers and cardiovascular disease: circulating microRNAs. Rev. Esp. Cardiol. (Engl. Ed.) 2017;70:763–769. doi: 10.1016/j.rec.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 227.Suárez Y., Wang C., Manes T.D., Pober J.S. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J. Immunol. 2010;184:21–25. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jansen F., Schäfer L., Wang H., Schmitz T., Flender A., Schueler R., Hammerstingl C., Nickenig G., Sinning J.M., Werner N. Kinetics of circulating micro RNA s in response to cardiac stress in patients with coronary artery disease. J. Am. Heart Assoc. 2017;6:e005270. doi: 10.1161/jaha.116.005270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li Y., Fan X., He X., Sun H., Zou Z., Yuan H., Xu H., Wang C., Shi X. MicroRNA-466l inhibits antiviral innate immune response by targeting interferon-alpha. Cell. Mol. Immunol. 2012;9:497–502. doi: 10.1038/cmi.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yoshikawa T., Takata A., Otsuka M., Kishikawa T., Kojima K., Yoshida H., Koike K. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci. Rep. 2012;2:637. doi: 10.1038/srep00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kadmon C.S., Landers C.T., Li H.S., Watowich S.S., Rodriguez A., King K.Y. MicroRNA-22 controls interferon alpha production and erythroid maturation in response to infectious stress in mice. Exp. Hematol. 2017;56:7–15. doi: 10.1016/j.exphem.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hou J., Wang P., Lin L., Liu X., Ma F., An H., Wang Z., Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 233.Witwer K.W., Sisk J.M., Gama L., Clements J.E. MicroRNA regulation of IFN-β protein expression: rapid and sensitive modulation of the innate immune response. J. Immunol. 2010;184:2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li C., Wang Y., Wang S., Wu B., Hao J., Fan H., Ju Y., Ding Y., Chen L., Chu X., et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J. Virol. 2013;87:2193–2205. doi: 10.1128/jvi.02831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhang X., Daucher M., Armistead D., Russell R., Kottilil S. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS One. 2013;8:e55733. doi: 10.1371/journal.pone.0055733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Buggele W.A., Horvath C.M. MicroRNA profiling of Sendai virus-infected A549 cells identifies miR-203 as an interferon-inducible regulator of IFIT1/ISG56. J. Virol. 2013;87:9260–9270. doi: 10.1128/jvi.01064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Reinsbach S., Nazarov P.V., Philippidou D., Schmitt M., Wienecke-Baldacchino A., Muller A., Vallar L., Behrmann I., Kreis S. Dynamic regulation of microRNA expression following interferon-γ-induced gene transcription. RNA Biol. 2012;9:978–989. doi: 10.4161/rna.20494. [DOI] [PubMed] [Google Scholar]
- 238.Nathans R., Chu C.-Y., Serquina A.K., Lu C.-C., Cao H., Rana T.M. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ohno M., Natsume A., Kondo Y., Iwamizu H., Motomura K., Toda H., Ito M., Kato T., Wakabayashi T. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol. Cancer Res. 2009;7:2022–2030. doi: 10.1158/1541-7786.mcr-09-0319. [DOI] [PubMed] [Google Scholar]
- 240.Wang X., Yuan T., Yin N., Ma X., Yang Y., Yang J., Shaukat A., Deng G. Interferon-τ regulates the expression and function of bovine leukocyte antigen by downregulating bta-miR-204. Exp. Ther. Med. 2021;21:594. doi: 10.3892/etm.2021.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Contoli M., Papi A., Tomassetti L., Rizzo P., Vieceli Dalla Sega F., Fortini F., Torsani F., Morandi L., Ronzoni L., Zucchetti O., et al. Blood interferon-α levels and severity, outcomes, and inflammatory profiles in hospitalized COVID-19 patients. Front. Immunol. 2021;12:648004. doi: 10.3389/fimmu.2021.648004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Rogez C., Martin M., Dereuddre-Bosquet N., Martal J., Dormont D., Clayette P. Anti-human immunodeficiency virus activity of tau interferon in human macrophages: involvement of cellular factors and β-chemokines. J. Virol. 2003;77:12914–12920. doi: 10.1128/jvi.77.23.12914-12920.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gabryelska A., Kuna P., Antczak A., Białasiewicz P., Panek M. IL-33 mediated inflammation in chronic respiratory diseases—understanding the role of the member of IL-1 superfamily. Front. Immunol. 2019;10:692. doi: 10.3389/fimmu.2019.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rasmussen T.K., Andersen T., Bak R.O., Yiu G., Sørensen C.M., Stengaard-Pedersen K., Mikkelsen J.G., Utz P.J., Holm C.K., Deleuran B. Overexpression of microRNA-155 increases IL-21 mediated STAT3 signaling and IL-21 production in systemic lupus erythematosus. Arthritis Res. Ther. 2015;17:154. doi: 10.1186/s13075-015-0660-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Adoro S., Cubillos-Ruiz J.R., Chen X., Deruaz M., Vrbanac V.D., Song M., Park S., Murooka T.T., Dudek T.E., Luster A.D., et al. IL-21 induces antiviral microRNA-29 in CD4 T cells to limit HIV-1 infection. Nat. Commun. 2015;6:7562. doi: 10.1038/ncomms8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ortega P.A., Saulle I., Mercurio V., Ibba S.V., Lori E.M., Fenizia C., Masetti M., Trabattoni D., Caputo S.L., Vichi F., et al. Interleukin 21 (IL-21)/microRNA-29 (miR-29) axis is associated with natural resistance to HIV-1 infection. Aids. 2018;32:2453–2461. doi: 10.1097/qad.0000000000001938. [DOI] [PubMed] [Google Scholar]
- 247.Wang M., Guo J., Zhao Y.-Q., Wang J.-P. IL-21 mediates microRNA-423-5p/claudin-5 signal pathway and intestinal barrier function in inflammatory bowel disease. Aging. 2020;12:16099–16110. doi: 10.18632/aging.103566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kotenko S.V., Rivera A., Parker D., Durbin J.E. Type III IFNs: beyond antiviral protection. Semin. Immunol. 2019;43:101303. doi: 10.1016/j.smim.2019.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Andreakos E., Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 2020;12:e12465. doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Prasun P. Letter to the editor: COVID-19, mitochondria, and interferon. J. Interferon. Cytokine. Res. 2020;40:466–467. doi: 10.1089/jir.2020.0112. [DOI] [PubMed] [Google Scholar]
- 251.Wang T., Yang L., Yuan M., Farber C.R., Spolski R., Leonard W.J., Ganta V.C., Annex B.H. MicroRNA-30b is both necessary and sufficient for interleukin-21 receptor-mediated angiogenesis in experimental peripheral arterial disease. Int. J. Mol. Sci. 2021;23:271. doi: 10.3390/ijms23010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.De Cecco L., Capaia M., Zupo S., Cutrona G., Matis S., Brizzolara A., Orengo A.M., Croce M., Marchesi E., Ferrarini M., et al. Interleukin 21 controls mRNA and microRNA expression in CD40-activated chronic lymphocytic leukemia cells. PLoS One. 2015;10:e0134706. doi: 10.1371/journal.pone.0134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Amado T., Amorim A., Enguita F.J., Romero P.V., Inácio D., De Miranda M.P., Winter S.J., Simas J.P., Krueger A., Schmolka N., et al. MicroRNA-181a regulates IFN-γ expression in effector CD8+ T cell differentiation. J. Mol. Med. 2020;98:309–320. doi: 10.1007/s00109-019-01865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fang J., Hao Q., Liu L., Li Y., Wu J., Huo X., Zhu Y. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J. Virol. 2012;86:1010–1020. doi: 10.1128/jvi.06169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Li Y., Xie J., Xu X., Wang J., Ao F., Wan Y., Zhu Y. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-λ1. Protein Cell. 2013;4:130–141. doi: 10.1007/s13238-012-2081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Schmitt M.J., Philippidou D., Reinsbach S.E., Margue C., Wienecke-Baldacchino A., Nashan D., Behrmann I., Kreis S. Interferon-γ-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun. Signal. 2012;10:41. doi: 10.1186/1478-811x-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., Hua M., Li N., Yao H., Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 258.Kim J.-H., Jou I., Joe E.-H. Suppression of miR-155 expression in IFN-γ-treated astrocytes and microglia by DJ-1: a possible mechanism for maintaining SOCS1 expression. Exp. Neurobiol. 2014;23:148–154. doi: 10.5607/en.2014.23.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yadav D., Ngolab J., Lim R.S.-H., Krishnamurthy S., Bui J.D. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-γ-induced microRNA. J. Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yuan Y., Kasar S., Underbayev C., Vollenweider D., Salerno E., Kotenko S.V., Raveche E. Role of microRNA-15a in autoantibody production in interferon-augmented murine model of lupus. Mol. Immunol. 2012;52:61–70. doi: 10.1016/j.molimm.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Perry M.M., Moschos S.A., Williams A.E., Shepherd N.J., Larner-Svensson H.M., Lindsay M.A. Rapid changes in microRNA-146a expression negatively regulate the IL-1β-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xu H., Yao Y., Smith L.P., Nair V. MicroRNA-26a-mediated regulation of interleukin-2 expression in transformed avian lymphocyte lines. Cancer. Cell. Int. 2010;10:15. doi: 10.1186/1475-2867-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhou Y., Yang Q., Xu H., Zhang J., Deng H., Gao H., Yang J., Zhao D., Liu F. miRNA-221-3p enhances the secretion of interleukin-4 in mast cells through the phosphatase and tensin homolog/p38/nuclear factor-kappaB pathway. PLoS One. 2016;11:e0148821. doi: 10.1371/journal.pone.0148821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kopriva S.E., Chiasson V.L., Mitchell B.M., Chatterjee P. TLR3-induced placental miR-210 down-regulates the STAT6/interleukin-4 pathway. PLoS One. 2013;8:e67760. doi: 10.1371/journal.pone.0067760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Xie N., Jia Z., Li L. miR-320a upregulation contributes to the development of preeclampsia by inhibiting the growth and invasion of trophoblast cells by targeting interleukin 4. Mol. Med. Rep. 2019;20:3256–3264. doi: 10.3892/mmr.2019.10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim E.S., Choi Y.E., Hwang S.J., Han Y.-H., Park M.-J., Bae I.H. IL-4, a direct target of miR-340/429, is involved in radiation-induced aggressive tumor behavior in human carcinoma cells. Oncotarget. 2016;7:86836–86856. doi: 10.18632/oncotarget.13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhang J., Xie S., Ma W., Teng Y., Tian Y., Huang X., Zhang Y. A newly identified microRNA, mmu-miR-7578, functions as a negative regulator on inflammatory cytokines tumor necrosis factor-α and interleukin-6 via targeting Egr1 in vivo. J. Biol. Chem. 2013;288:4310–4320. doi: 10.1074/jbc.m112.351197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ou M., Hao S., Chen J., Zhao S., Cui S., Tu J. Downregulation of interleukin-6 and C-reactive protein underlies a novel inhibitory role of microRNA-136-5p in acute lower extremity deep vein thrombosis. Aging (Albany NY) 2020;12:21076–21090. doi: 10.18632/aging.103140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yang Z., Peng Y., Yang S. MicroRNA-146a regulates the transformation from liver fibrosis to cirrhosis in patients with hepatitis B via interleukin-6. Exp. Ther. Med. 2019;17:4670–4676. doi: 10.3892/etm.2019.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang N., Zhang Q., Yang W., Miao L., Wang N., Wei S., Ge J., Li X., Wu J. Decreased expression of microRNA-30b promotes the development of pulpitis by upregulating the expression of interleukin-6 receptor. Exp. Ther. Med. 2019;17:3233–3238. doi: 10.3892/etm.2019.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Xu Z., Xiao S.-B., Xu P., Xie Q., Cao L., Wang D., Luo R., Zhong Y., Chen H.-C., Fang L.-R. miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-κB. J. Biol. Chem. 2011;286:21401–21412. doi: 10.1074/jbc.m110.198630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Perng D.-W., Yang D.-M., Hsiao Y.-H., Lo T., Lee O.K.-S., Wu M.-T., Wu Y.-C., Lee Y.-C. miRNA-146a expression positively regulates tumor necrosis factor-α-induced interleukin-8 production in mesenchymal stem cells and differentiated lung epithelial-like cells. Tissue. Eng. 2012;18:2259–2267. doi: 10.1089/ten.tea.2011.0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Hu N., Zhang J., Cui W., Kong G., Zhang S., Yue L., Bai X., Zhang Z., Zhang W., Zhang X., Ye L. miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J. Biol. Chem. 2011;286:13714–13722. doi: 10.1074/jbc.m110.204131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bhattacharyya S., Balakathiresan N.S., Dalgard C., Gutti U., Armistead D., Jozwik C., Srivastava M., Pollard H.B., Biswas R. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J. Biol. Chem. 2011;286:11604–11615. doi: 10.1074/jbc.m110.198390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhou R., Li X., Hu G., Gong A.-Y., Drescher K.M., Chen X.-M. miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One. 2012;7:e30772. doi: 10.1371/journal.pone.0030772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zhang J., Shao N., Yang X., Xie C., Shi Y., Lin Y. Interleukin-8 promotes epithelial-to-mesenchymal transition via downregulation of mir-200 family in breast cancer cells. Technol. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820979672. 153303382097967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Quinn S.R., Mangan N.E., Caffrey B.E., Gantier M.P., Williams B.R., Hertzog P.J., Mccoy C.E., O'neill L.A. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J. Biol. Chem. 2014;289:4316–4325. doi: 10.1074/jbc.m113.522730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hussain T., Zhao D., Shah S.Z.A., Wang J., Yue R., Liao Y., Sabir N., Yang L., Zhou X. MicroRNA 27a-3p regulates antimicrobial responses of murine macrophages infected by Mycobacterium avium subspecies paratuberculosis by targeting interleukin-10 and TGF-β-activated protein kinase 1 binding protein 2. Front. Immunol. 2017;8:1915. doi: 10.3389/fimmu.2017.01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sharma A., Kumar M., Aich J., Hariharan M., Brahmachari S.K., Agrawal A., Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc. Natl. Acad. Sci. USA. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhao M., Sun D., Guan Y., Wang Z., Sang D., Liu M., Pu Y., Fang X., Wang D., Huang A., et al. Disulfiram and diphenhydramine hydrochloride upregulate miR-30a to suppress IL-17-associated autoimmune inflammation. J. Neurosci. 2016;36:9253–9266. doi: 10.1523/jneurosci.4587-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wan L., Zhao Q., Niu G., Xiang T., Ding C., Wang S. Plasma miR-136 can be used to screen patients with knee osteoarthritis from healthy controls by targeting IL-17. Exp. Ther. Med. 2018;16:3419–3424. doi: 10.3892/etm.2018.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Podsiad A., Standiford T.J., Ballinger M.N., Eakin R., Park P., Kunkel S.L., Moore B.B., Bhan U. MicroRNA-155 regulates host immune response to postviral bacterial pneumonia via IL-23/IL-17 pathway. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2016;310:L465–L475. doi: 10.1152/ajplung.00224.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tang X., Wu F., Fan J., Jin Y., Wang J., Yang G. Posttranscriptional regulation of interleukin-33 expression by microRNA-200 in bronchial asthma. Mol. Ther. 2018;26:1808–1817. doi: 10.1016/j.ymthe.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Chen L., Wang G., Qiao X., Wang X., Liu J., Niu X., Zhong M. Downregulated miR-524-5p participates in the tumor microenvironment of ameloblastoma by targeting the Interleukin-33 (IL-33)/suppression of tumorigenicity 2 (ST2) Axis. Med. Sci. Monit. 2020;26:e921863. doi: 10.12659/msm.921863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Dubois-Camacho K., Diaz-Jimenez D., De La Fuente M., Quera R., Simian D., Martínez M., Landskron G., Olivares-Morales M., Cidlowski J.A., Xu X., et al. Inhibition of miR-378a-3p by inflammation enhances IL-33 levels: a novel mechanism of alarmin modulation in ulcerative colitis. Front. Immunol. 2019;10:2449. doi: 10.3389/fimmu.2019.02449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Alifano M., Alifano P., Forgez P., Iannelli A. Renin-angiotensin system at the heart of COVID-19 pandemic. Biochimie. Clin. 2020;174:30–33. doi: 10.1016/j.biochi.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., Lambrecht B.N. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Tesch G.H., Schwarting A., Kinoshita K., Lan H.Y., Rollins B.J., Kelley V.R. Monocyte chemoattractant protein-1 promotes macrophage-mediated tubular injury, but not glomerular injury, in nephrotoxic serum nephritis. J. Clin. Invest. 1999;103:73–80. doi: 10.1172/jci4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Mauer J., Chaurasia B., Goldau J., Vogt M.C., Ruud J., Nguyen K.D., Theurich S., Hausen A.C., Schmitz J., Brönneke H.S., et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat. Immunol. 2014;15:423–430. doi: 10.1038/ni.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lenardo M.J. lnterleukin-2 programs mouse αβ T lymphocytes for apoptosis. Nature. 1991;353:858–861. doi: 10.1038/353858a0. [DOI] [PubMed] [Google Scholar]
- 291.Dranoff G., Jaffee E., Lazenby A., Golumbek P., Levitsky H., Brose K., Jackson V., Hamada H., Pardoll D., Mulligan R.C., LAzenby A. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc. Natl. Acad. Sci. USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Arts N., Cané S., Hennequart M., Lamy J., Bommer G., Van Den Eynde B., De Plaen E. microRNA-155, induced by interleukin-1ß, represses the expression of microphthalmia-associated transcription factor (MITF-M) in melanoma cells. PLoS One. 2015;10:e0122517. doi: 10.1371/journal.pone.0122517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Veremeyko T., Siddiqui S., Sotnikov I., Yung A., Ponomarev E.D. IL-4/IL-13-dependent and independent expression of miR-124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS One. 2013;8:e81774. doi: 10.1371/journal.pone.0081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Su S., Zhao Q., He C., Huang D., Liu J., Chen F., Chen J., Liao J.-Y., Cui X., Zeng Y., et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun. 2015;6:8523. doi: 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hao M., Zhang L., An G., Sui W., Yu Z., Zou D., Xu Y., Chang H., Qiu L. Suppressing miRNA-15a/-16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J. Hematol. Oncol. 2011;4:37. doi: 10.1186/1756-8722-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhou Y.F., Fu Z.Y., Chen X.H., Cui Y., Ji C.B., Guo X.R. Tumor necrosis factor-α and interleukin-6 suppress microRNA-1275 transcription in human adipocytes through nuclear factor-κB. Mol. Med. Rep. 2017;16:5965–5971. doi: 10.3892/mmr.2017.7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Poudyal D., Herman A., Adelsberger J.W., Yang J., Hu X., Chen Q., Bosche M., Sherman B.T., Imamichi T. A novel microRNA, hsa-miR-6852 differentially regulated by Interleukin-27 induces necrosis in cervical cancer cells by downregulating the FoxM1 expression. Sci. Rep. 2018;8:900. doi: 10.1038/s41598-018-19259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Hong Z., Hong H., Liu J., Zheng X., Huang M., Li C., Xia J. miR-106a is downregulated in peripheral blood mononuclear cells of chronic hepatitis B and associated with enhanced levels of interleukin-8. Mediators. Inflamm. 2015:629862. doi: 10.1155/2015/629862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Cui B., Liu W., Wang X., Chen Y., Du Q., Zhao X., Zhang H., Liu S.-L., Tong D., Huang Y. Brucella Omp25 upregulates miR-155, miR-21-5p, and miR-23b to inhibit interleukin-12 production via modulation of programmed death-1 signaling in human monocyte/macrophages. Front. Immunol. 2017;8:708. doi: 10.3389/fimmu.2017.00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Garikipati V.N.S., Krishnamurthy P., Verma S.K., Khan M., Abramova T., Mackie A.R., Qin G., Benedict C., Nickoloff E., Johnson J., et al. Negative regulation of miR-375 by interleukin-10 enhances bone marrow-derived progenitor cell-mediated myocardial repair and function after myocardial infarction. Stem. Cells. 2015;33:3519–3529. doi: 10.1002/stem.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ksiazek-Winiarek D., Szpakowski P., Turniak M., Szemraj J., Glabinski A. IL-17 exerts anti-apoptotic effect via miR-155-5p downregulation in experimental autoimmune encephalomyelitis. J. Mol. Neurosci. 2017;63:320–332. doi: 10.1007/s12031-017-0981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mei Z., Chen S., Chen C., Xiao B., Li F., Wang Y., Tao Z. Interleukin-23 facilitates thyroid cancer cell migration and invasion by inhibiting SOCS4 expression via MicroRNA-25. PLoS One. 2015;10:e0139456. doi: 10.1371/journal.pone.0139456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Lopetuso L.R., De Salvo C., Pastorelli L., Rana N., Senkfor H.N., Petito V., Di Martino L., Scaldaferri F., Gasbarrini A., Cominelli F., et al. IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proc. Natl. Acad. Sci. USA. 2018;115:E9362–E9370. doi: 10.1073/pnas.1803613115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The primary data for this study are available from the authors on request.