Abstract
ABO blood group is long known to be an influencing factor for the susceptibility to infectious diseases, and many studies have been describing associations between ABO blood types and COVID-19 infection and severity, with conflicting findings. This narrative review aims to summarize the literature regarding associations between the ABO blood group and COVID-19. Blood type O is mostly associated with lower rates of SARS-CoV-2 infection, while blood type A is frequently described as a risk factor. Although results regarding the risk of severe outcomes are more variable, blood type A is the most associated with COVID-19 severity and mortality, while many studies describe O blood type as a protective factor for the disease progression. Furthermore, genetic associations with both the risk of infection and disease severity have been reported for the ABO locus. Some underlying mechanisms have been hypothesized to explain the reported associations, with incipient experimental data. Three major hypotheses emerge: SARS-CoV-2 could carry ABO(H)-like structures in its envelope glycoproteins and would be asymmetrically transmitted due to a protective effect of the ABO antibodies, ABH antigens could facilitate SARS-CoV-2 interaction with the host’ cells, and the association of non-O blood types with higher risks of thromboembolic events could confer COVID-19 patients with blood type O a lower risk of severe outcomes. The hypothesized mechanisms would affect distinct aspects of the COVID-19 natural history, with distinct potential implications to the disease transmission and its management.
Keywords: ABO blood Group system, ABO blood Types, COVID-19, SARS-CoV-2, ABO antibodies, Glycobiology
1. Introduction
The Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2), the causing agent of COVID-19, emerged in late 2019 and rapidly spread worldwide, becoming a global health threat that, in one year, accounted for about 70 million cumulative cases [1]. Its high infectivity [2], with the occurrence of asymptomatic transmission [3], may have contributed to such a scenario. Mainly characterized by symptoms like fever, headache, cough, fatigue, breathing difficulties, anosmia, and ageusia; COVID-19 can either remain asymptomatic or develop to severe and critical manifestations, like dyspnea, hypoxia, acute respiratory distress syndrome, and multiorgan failure [4]. Age and co-morbidities, like diabetes and cardiovascular diseases, increase the risk of severe outcomes [5]. However, given the poorly understood pathogenesis of COVID-19 and the variability in clinical presentation and outcomes, there has been a search for other biological markers that could predict the susceptibility to SARS-CoV-2 infection and the risk of severe outcomes [4,6].
The ABO blood group, which comprises distinct carbohydrate epitopes at the surface of blood cells and other tissues, like the respiratory and gastrointestinal epithelium [7], has been considered as an influencing factor for the susceptibility to infectious diseases. For instance, associations between the ABO blood type and the risk or severity of infections have been described for the Human Immunodeficiency Virus (HIV) [8], hepatitis B [8,9], West Nile virus (WNV) [10], and Helicobacter pylori [11,12]. Blood group antigens can influence infections directly, acting as receptors or co-receptors for microorganisms and toxins; or indirectly, through the anti-blood group antibodies, which can be prompted by bacteria and enveloped viruses bearing blood group-like antigens [13].
After a first report [14], a wealth of studies have been investigating a possible association between the ABO blood group and SARS-CoV-2 infection, with conflicting findings. Some authors pointed to the ABO blood type as an influencing factor for SARS-CoV-2 infection [15,16] or even its severity [17] and mortality [18], while others failed to demonstrate such associations [19,20]. Generally, blood type A has been linked to higher risk of SARS-CoV-2 infection, and blood type O, to lower risk. It is noteworthy that a similar association was early described for SARS-CoV, in which blood type O was negatively associated with infection among a group of health workers [21].
Although these associations and their potential implications remain unclear; some underlying mechanisms have been hypothesized, like a protective effect of the ABO antibodies [22], the link between ABO blood type and cardiovascular risk [23], and a role for ABO(H) antigens in facilitating viral entry into target cells. Thus, this paper aims to review the associations between ABO blood group and COVID-19 and its possible underlying mechanisms.
2. ABO blood group system: an overview
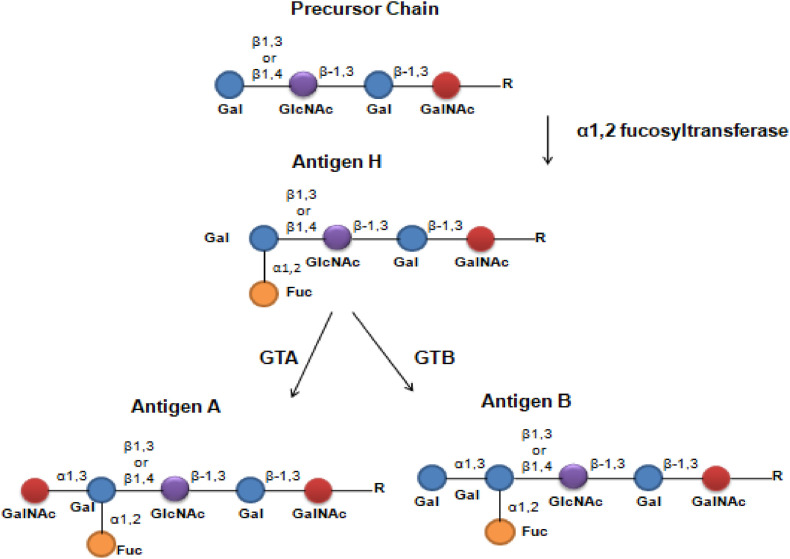
The ABO blood group system consists of carbohydrate antigens forming four main blood types: A, B, AB, and O. Blood types A and B are determined by the presence of the homonymous antigens, while blood types AB and O, by the presence of both or none, respectively [24]. Discovered in the Red Blood Cells (RBCs); the ABO antigens were later found in several tissues and secretions, such as saliva, intestinal mucosa, endothelium [13], and the respiratory tract epithelium [7]. As an autosomally inherited trait, the ABO blood type is determined by the polymorphism of the ABO gene, located at the 9q34.2 locus [6]. Alleles A and B are codominant over allele O and determine the expression of the homonymous antigens. Allele O, in turn, is recessive and does not determine the expression of any antigen. Like carbohydrates, antigens A and B are secondary products of the ABO gene, which primarily encodes specific enzymes that catalyze the addition of monosaccharide motifs to an oligosaccharide backbone anchored to glycoproteins or glycolipids through N- or O-linked glycosylation. Such process depends on antigen H, the acceptor of antigens A and B at the growing oligosaccharide chain [13,[25], [26], [27]].
Antigen H is synthesized through the addition of terminal fucose (Fucα1-2Gal) to lactosamine (GlcNac). On RBCs, platelets, and endothelium, this process is catalyzed by an α1,2-fucosyltransferase encoded by the FUT1 gene and forms a Fucα1-2Galβ1-4GlcNAc. FUT1 is independent of the ABO gene and its product uses insoluble oligosaccharides chains (type 2) as the acceptor structure for fucose, which generates insoluble antigen H. However, in epithelial cells, mucus, and secretions, antigen H is added to a soluble chain (type 1), which originates the secretor phenotype in a process catalyzed by an α1,2-fucosyltransferase encoded by the FUT 2 gene. The type 1 and type 2 chains differ in the link between galactoses: β1-3 and β1-4, respectively (Fig. 1 ) [7,27]. Although located at the same chromosome – 19q13 – FUT1 and FUT2 are independent genes with differential expression throughout the organism. The homozygous inheritance of FUT 1 null alleles produces the rare Bombay phenotype, on which the individuals are unable to synthesize antigen H and, thus, antigens A and B. In turn, the homozygous inheritance of FUT2 null alleles results in the inability to synthesize soluble antigen H and, therefore, in the non-secretor phenotype, characterized by the lack of ABO antigens in epithelial tissues and secretion. Unlike Bombay, the non-secretor phenotype is not rare and is found in about 20% of the western population [7].
Fig. 1.
- Sequential synthesis the ABO antigens. Gal: d-Galactose, GlcNAc: N-Acetylglucosamine, GalNAc: N-Acetyl-galactosamine, Fuc: l-fucose, R: Radical group, GTA: Glucosyltransferase A, GTB: Glucosyltransferase B, β1,3: Type 1 chain, β1,4: Type 2 chain. (Two-column fitting image).
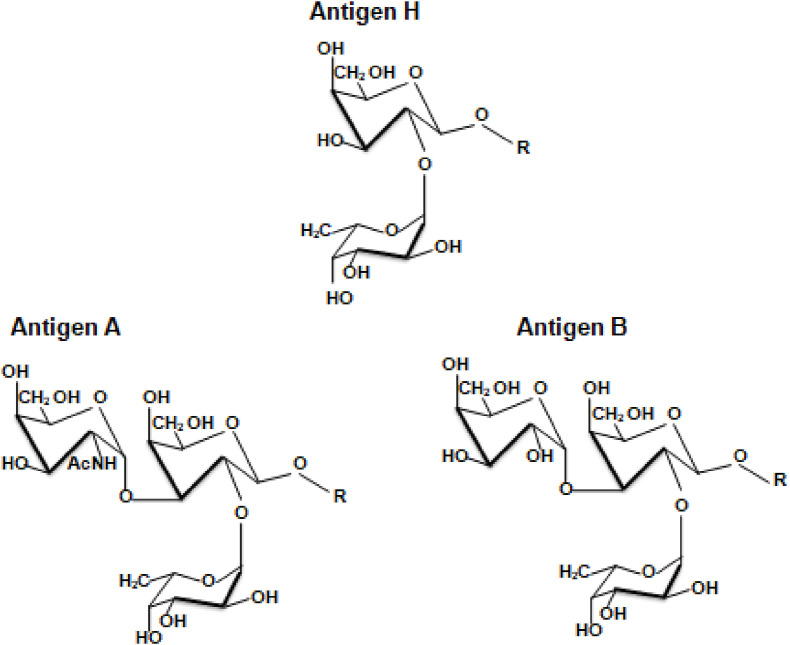
Once antigen H is formed, antigens A and B are synthesized by the glucosyltransferases A (GTA) and B (GTB), respectively encoded by the A and B alleles of the ABO gene (Fig. 1) [26]. The O allele, in turn, produces a truncated protein instead of a functional enzyme [28]. Antigen A is determined by a N-acetyl-galactosamine (GalNAc) motif, and antigen B, by d-Galactose (Gal), respectively characterized by acetamido (AcNH) and hydroxyl (OH) groups (Fig. 2 ). Both are added to antigen H through an α1-3 glycosidic bond, forming the GalNAcα1-3(Fucα1-2)Gal- and Galα1-3(Fucα1-2)Gal-chains, respectively [26,27,29]. The specificity of GTA and GTB relies on a size-based complementarity with the nucleotide-sugar donors Uridine diphosphate(UDP)-GalNAc and UDP-Gal, respectively [30].
Fig. 2.
- Structural representations of the ABO blood group antigens. (1.5-column fitting image).
Another component of the ABO blood group system is the ABO antibodies, which are specific antibodies directed toward the missing A/B antigen even in the absence of a previous alloimmunization. Therefore, A and O blood type individuals produce anti-B, B and O individuals produce anti-A, and AB individuals are devoid of both anti-A and anti-B [31]. The origin of ABO antibodies is still controversial, however, their synthesis seems to be mostly triggered by either microbiota or pathogenic microorganisms bearing ABO(H)-like antigens [32,33]. The ABO antibodies are able to induce complement-mediated lysis of RBCs and are mainly of IgM class [31].
3. ABO blood group and link to infections
ABO and FUT2 are clearly polymorphic genes and their products are highly present on epithelial cells in contact with the external environment. They are maximally expressed in mucins, which are thought to have a protective role in binding to microorganisms and are found in the body's exposed surfaces, like in the lungs and the gastrointestinal tract. Such features are suggestive of a possible function for ABO antigens in the interaction with environmental pathogens [7,34]. In addition, phylogeny of the ABO determinants showed that their firstly appeared as tissular antigens [35].
The widespread distribution of carbohydrates on cells' surfaces and their structural diversity place them as informational molecules able to act in cell recognition [36]. Indeed, pathogenic microorganisms and toxins can require binding to specific carbohydrate antigens to trigger infection and disease [37]. This was observed for H. pylori, whose infection was shown to depend on the expression of the Lewis b (Leb) antigen in the gastric mucosa [38], and for the uropathogenic E. coli R45 strain, which binds to galactosyl-globoside - a precursor of the type 1 chain exposed at the membrane of non-secretor uroepithelial cells - and its sialylated derivate [39]. Furthermore carbohydrate antigens akin to that found on the surface of hosts’ cells can be synthesized by bacteria in a co-evolving process suggestive of molecular mimicry, in which pathogenic or symbiotic microorganisms mimic carbohydrate motifs found at the host species to evade their innate immune system [37]. Indeed, Georg et al. screened 282 gram-negative strains isolated from humans, of which 48% showed ABO blood group activity [40].
Likewise, enveloped viruses can also acquire carbohydrate antigens from their previous host as part of their structures and be asymmetrically transmitted due to a protective effect of the naturally occurring antibodies [41]. During their synthesis, the viral proteins are glycosylated in the Golgi apparatus and the endoplasmic reticulum of the host cells by their glycosylation enzymes [42]. Hence, viral glycoproteins can bear the glycosylation pattern of the cell in which they were synthesized. This was experimentally demonstrated for the Human T-cell Lymphotropic Virus (HTLV) iiib, which was neutralized by monoclonal anti-A antibodies after propagation in peripheral blood mononuclear cells (PBMC) from A-type individuals, but not after propagation in PBMC of B or O individuals [42]. A similar effect was observed for the Measles virus produced in cells transfected with cDNA encoding human A, B, or null “O” transferases. The viruses produced in A- or B-type cells were partially neutralized, in a complement-dependent manner, by human pre-immune serum containing anti-A or anti-B antibodies, respectively, while the viruses produced in type O cells were not neutralized [43]. Moreover, anti-A monoclonal antibodies and type O plasma inhibited the adhesion of transfected CHO cells co-expressing the A antigen and the Spike (S) protein of SARS-CoV to ACE2-expressing Vero cells [44].
The link between ABO polymorphism and infectious disease has been extensively studied and is detailed reviewed elsewhere [13]. Associations have been reported for instance, with HIV [8], hepatits B [9], West Nile Virus [10], and malaria [45]. Thus, the ABO polymorphism has been considered as an adaptive trait that could narrow the spread of infections within a given species. In turn, pathogenic microorganisms have been considered as a selective pressure generating diversity in the distribution of blood types among different populations, according to the exposition to distinct pathogens [37,46]. It is worth mentioning that associations between the ABO blood type and infectious diseases are most reliant on statistics, with few experimental data, and that illness may be multifactorial in nature, with the blood type constituting one of the multiple predisposing factors [34].
4. Associations between ABO blood types and COVID-19
4.1. Susceptibility to SARS-CoV-2 infection
Soon after the onset of the COVID-19 pandemic, an association between the ABO blood group and SARS-CoV-2 infection was firstly reported by Zhao et al., in which blood type A was significantly related to higher risk of COVID-19 infection in comparison to non-A blood types; while blood type O was significantly related to lower risk of COVID-19 infection compared to non-O blood types [14]. Subsequently, several studies have been reporting significant associations between blood type A and higher susceptibility to SARS-CoV-2 infection and/or blood type O and lower susceptibility [15,[47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58]]. A summary of the findings is described in Table 1 .
Table 1.
Studies on associations between ABO blood types and the risk of SARS-CoV-2 infection.
| Reference | Country | Study |
Cases |
Controls |
ABO |
Riska |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Designb | Definitionc | N | Definitiond | N | A | B | AB | O | ||
| Zhao et al. [14] | China | I | I | 2.173 | I | 27.080 | ↑ | ns | ns | ↓ |
| Li et al. [15] | China | II | II | 265 | I | 3.694 | ↑ | ns | ns | ↓ |
| Muñiz-Diaz et al. [18] | Spain | II | III | 854 | II | 75.870 | ↑ | ns | ns | ↓ |
| Franchini et al. [53] | Italy | III | III | 447 | II | 16.911 | ns | ns | ns | ↓ |
| Dzik et al. [19] | USA | II | II | 957 | III | 5.840 | ns | ns | ns | ns |
| Wu et al. [16] | China | II | II | 187 | III | 1.991 | ↑ | ns | ns | ↓ |
| Padhi et al. [69] | India | IV | IV | – | I | – | ns | ↑ | ns | ns |
| Khalil; Feghali; Hassoun [20] | Lebanon | II | II | 146 | III | 6.497 | ns | ns | ns | ns |
| Liu et al. [74] | International | I | I | 9.383 | IV | 54.218 | ↑ | ↑ | ns | ↓ |
| Fan et al. [52] | China | II | II | 105 | III | 103 | ↑ | ns | ns | ns |
| Solmaz; Araç [54] | Turkey | V | II | 1.667 | I | 127.091 | ↑ | ns | ns | ↓ |
| Barnkob et al. [47] | Denmark | II | I | 7.422 | I | 2.204.742 | ↑ | ns | ↑ | ↓ |
| Boudin et al. [76] | France | II | I | 1.279 | IV | 409 | ns | ns | ns | ns |
| Latz et al. [68] | US | II | I | 1.289 | V | 7.648 | ns | ↑ | ↑ | ↓ |
| Levi et al. [66] | Brazil | II | I | 2.037 | VI | 1.813.237 | ns | ns | ns | ns |
| Yalaoui et al. [55] | Tunisia | III | I | 51 | II or IV | 63.375 or 1.506 | ↑ | ns | ns | ns |
| Göker et al. [56] | Turkey | III | V | 186 | II | 1.881 | ↑ | ns | ns | ↓ |
| Almadhi et al. [59] | Bahrain | V | I | 2.334 | II | 4.985 | ns | ↑ | ↓ | ns |
| Lehrer; Rheinstein [65] | UK | II | I | 717 | I | 11.855 | ns | ns | ns | ns |
| Mullins et al. [67] | US | II | II | 227 | I | – | ns | ns | ns | ns |
| Negro et al. [78] | Italy | II | VI | 129 | VII | 38 | ↑ | ↓ | ns | ↓ |
| Ray et al. [70] | Canada | II | I | 7.071 | IV | 218.485 | ns | ↑ | ↑ | ↓ |
| El-Shitany et al. [51] | Saudi Arabia/Egypt | V | I | 726 | IV | 707 | ns | ns | ns | ↓ |
| Kibler et al. [77] | France | II | VII | 22 | VIII | 680 | ↑ | ns | ns | Ref |
| Leaf et al. [63],e | US | II | VIII | 2.033 | II | 3.100.000 | ↑ | ns | ns | ↓ |
| Niles et al. [64],e | US | II | I | 34.178 | IV | 242.358 | ns | ns | ns | ↓ |
| Al-Youha et al. [71] | Kuwait | II | I | 3.304 | I | 3.730.027 | ns | ↑ | ↑ | ↓ |
| Muñoz-Culla et al. [57] | Spain | II | VI | 412 | II | 17.796 | ↑ | ↑ | ↑ | ↓ |
| Kotila et al. [72] | Nigeria | II | I | 302 | II | 9.138 | ns | ↑ | ↑ | ↓ |
| Mahmud et al. [58] | Bangladesh | II | V | 438 | I | – | ↑ | ns | ns | ns |
| Schetelig et al. [48] | Germany | V | I | 6.257 | IV | 118.868 | ↑ | ns | ↑ | Ref |
| Ozkarafakili, Gareayaghi & Kara [60] | Turkey | II | II | 492 | II | 51.966 | ns | ns | ns | ns |
| Gamboa-Aguilar et al. [61] | Mexico | III | V | 2.416 | II | 5.000 | ↓ | ns | ↓ | ↑ |
| Enguita-Gérman et al. [49] | Spain | II | IX | 87.090 | – | – | ↑ | ns | ns | ↓ |
| Jawdat et al. [73] | Saudi Arabia | II | I | 373 | I | 9.939 | ns | ↑ | ↑ | ↓ |
| Kander et al. [75] | Sweden | II | X | 338 | IX | 29.174 | ns | ns | ↑ | ns |
| Gurung et al. [62] | Nepal | III | I | 1091 | II | 2.182 | ns | ns | ↑ | ns |
| Jukić et al. [50] | Croatia | II | VIII | 780 | II, V and X | 614.673 | ↑ | ns | ns | ↓ |
vs. other blood types. Upward arrow: increased risk, downward arrow: decreased risk; ns: non-significant = p˃0.05.
I: Meta-analysis, II: Cohort study, III: Case-control study, IV: Mixed method, V: Cross-sectional study.
I: SARS-CoV-2+ individuals (Mostly RT-PCR), II: COVID-19 hospitalized patients, III: Convalescent plasma-donors, IV: Officially reported cases, V: General COVID-19 patients, VI: Symptomatic cases, VII: COVID-19 patients underwent TAVR (Transaortic valvular replacement), VIII: Critically ill patients, IX: General population, X: COVID-19 patients admitted to ICU (Intensive care unit).
I: Local anthropological data, II: Blood donors, III: Non-COVID-19 hospitalized patients, IV: SARS-CoV-2- individuals, V: Individuals tested for SARS-CoV-2, VI: Patients in general, VII: Asymptomatic COVID-19 cases, VIII: Patients underwent TAVR, IX: Non-COVID-19 patients admitted to ICU, X: Pregnant women with ABO typing.
Adjusted for ethnicity.
As can be seen from Table 1, blood type A is frequently associated with higher risks of acquiring COVID-19, while blood type O is mostly described as a protective factor. Nonetheless, there are several disagreements, especially regarding the higher susceptibility. For instance, Muñiz-Diaz et al. evaluated the distribution of ABO blood types in two cohorts – convalescent plasma donors, with regular donors as the control group, and hospitalized COVID-19 patients, with general patients as the control group -. In the former, blood type A was significantly overrepresented compared to non-A blood types, while blood type O was significantly underrepresented compared to non-O types. However, no significant association was found in the later cohort [18].
Indeed, in a multiethnic cohort enrolling hospitalized COVID-19 patients - with general patients as the control group – no significant difference in ABO phenotype distribution was found [19]. In this study, the authors challenged the use of regular blood donors as the control group in studies concerning associations between ABO blood types and COVID-19 susceptibility. Blood type O individuals are preferentially recruited for blood donation and might be overrepresented among regular donors, which may have biased some previous results towards O-related protection. However, with a similar cohort choice, Wu et al. found significant associations between ABO blood group and the risk of SARS-CoV-2 infection, in which blood type A was associated with higher susceptibility to SARS-CoV-2 infection, and blood type O, to lower susceptibility [16]. Furthermore, studies using blood bank data do not always associate blood type O to lower risk of SARS-CoV-2 infection [[59], [60], [61], [62]].
Other studies enrolling multiethnic cohorts in the U.S reported no significant association between ABO blood group and the risk of acquiring COVID-19 [63] or significantly higher odds for blood type O [64]. Nonetheless, after adjusting for ethnicity, blood type A was significantly associated with a higher risk of SARS-CoV-2 infection [63], and blood type O, with lower risk [63,64], among white non-Hispanic participants, but not in the Hispanic and black non-Hispanic groups. Indeed, a more recent study conducted in Mexico described a significantly higher risk of SARS-CoV-2 infection among blood type O-individuals, while blood type A was significantly related to lower risk [61]. Ethnicity constitutes, therefore, a potential confounding factor in studies concerning ABO blood types and the risk of SARS-CoV-2 infection and should be considered in the interpretation of their results.
While several studies failed to reproduce significant associations between ABO blood-type and SARS-CoV-2 infection [20,60,[65], [66], [67]], others reached significant associations for blood types other than A and O. Blood type B was related to significantly higher risk of SARS-CoV-2 infection in some studies [59,[68], [69], [70], [71], [72], [73]]; and a meta-analysis indicated that blood type B is associated with higher risk of SARS-CoV-2 infection in Asian, but not in European cohorts [74]. AB blood type, in turn, has been significantly related to either higher [47,62,68,[70], [71], [72], [73],75] or lower [59,61] risk of acquiring COVID-19.
Moreover, some authors studied the association between ABO blood types and the risk of SARS-CoV-2 infection in more specific backgrounds. Boudin et al. found no significant difference in ABO phenotype distribution between infected and uninfected crewmembers exposed to SARS-CoV-2 during an outbreak in an aircraft carrier [76]. Conversely, blood type A was significantly associated with higher risk of developing COVID-19 among patients who underwent transcatheter aortic valve replacement (TAVR) [77]. Furthermore, after no association between ABO phenotype and SARS-CoV-2 positivity was found, Negro et al. sought to investigate whether ABO blood type correlates with the symptomatic disease in SARS-CoV-2-positive individuals. A significant positive association was found between blood type A and the development of symptomatic COVID-19, while blood types B and O were significantly related to lower risks of developing symptomatic infection [78].
Although studies concerning ABO blood types and the risk of SARS-CoV-2 infection are highly heterogeneous, we were unable to find any trend in susceptibility according to the study design or the type and size of the case and control groups. However, among the studies summarized here, a dichotomy between blood types A – higher risk – and O –lower risk – was more clearly observed in European and East Asian settings, while the involvement of blood types B or AB has been mostly observed in Middle Eastern, Indian, and North American settings (Table 1). Data from Africa and Latin America are scarce, but it is noteworthy that in both the settings, blood type A was not associated yet with higher susceptibility to SARS-CoV-2 infection [61,66,72] and, in the case of Latin America, blood type O was not associated yet with lower susceptibility [61,66].
4.2. COVID-19 severity and outcomes
In addition to the risk of SARS-CoV-2 infection or symptomatic disease, the ABO blood group has also been associated with severe outcomes. Severity is usually defined as hospitalization, admission to Intensive Care Unit (ICU), mechanical ventilation requirement, or decease. The association of ABO blood types with COVID-19 severity is generally performed by comparing the blood type distribution between distinct subgroups of patients. Studies on this subject were mostly designed to uncover associations for both the risk of infection and severity, with fewer studies designed to evaluate associations for severity only. A summary of the findings is described in Table 2 .
Table 2.
- Studies on associations between ABO blood types and the risk of severe COVID-19 outcomes.
| Reference | Country | Study Designb | Cases |
Type of Association | ABO Riska |
B | AB | O | |
|---|---|---|---|---|---|---|---|---|---|
| Definitionc | N | A | |||||||
| Muñiz-Diaz et al. [18] | Spain | I | I | 965 | Mortality | ↑ | ns | ns | ↓ |
| Dzik et al. [19] | USA | I | II | 957 | Mortality | ns | ns | ns | ns |
| Padhi et al. [69] | India | II | III | – | Mortality | ns | ↑ | ns | ↓ |
| Khalil; Feghali; Hassoun [20] | Lebanon | I | IV | 146 | Severity and mortality | ns | ns | ns | ns |
| Solmaz; Araç [54] | Turkey | III | II | 1.667 | Admission to ICUd and death | ns | ns | ns | ns |
| Barnkob et al. [47] | Denmark | I | V | 7.422 | Hospitalization and death | ns | ns | ns | ns |
| Latz et al. [68] | US | I | IV | 1289 | Hospitalization, admission to ICU, intubation, and death | ns | ns | ns | ns |
| Almadhi et al. [59] | Bahrain | III | IV | 2.334 | Admission to ICU | ns | ns | ns | ns |
| Lehrer; Rheinstein [65] | UK | I | V | 717 | Mortality | ns | ns | ns | ns |
| Mullins et al. [67] | US | I | II | 227 | Severity, admission to ICU, intubation, dialysis, hospital or ICU LOSe, and death | ns | ns | ns | ns |
| Negro et al. [78] | Italy | I | V | 167 | Severity and mortality | ns | ns | ns | ns |
| Ray et al. [70] | Canada | I | V and VI | 218.485 | Severity and mortality | ns | ns | ns | ↓ |
| El-Shitany et al. [51] | Saudi Arabia/Egypt | III | IV | 726 | Oxygen saturation, artificial respiration, myalgia, and recovery time | ↓ | ↓ | ns | ↑ |
| Garibaldi et al. [85] | Brazil | I | VII | 72 | Severity and risk of death (SOFAf and SAPS-3g) | ↑ | ns | ns | ↓ |
| Hoiland et al. [86] | Canada | I | VIII | 95 | Mechanical ventilation, CRRTh, and ICU LOS | ↑ | ↓ | ↑ | ↓ |
| Kibler et al. [77] | France | I | IX | 22 | Hospitalization and death | ↑ | ns | ns | ns |
| Sardu et al. [87] | Italy | I | X | 164 | Cardiac injury and death | ns | ns | ns | ↓ |
| Solhpour et al. [17] | Iran | I | XI | 93 | Severity | ↑ | ns | ns | ↓ |
| Valenti et al. [80] | Italy | I | VII | 505 | Mechanical ventilation and admission to ICU | ns | ns | ns | ref |
| Göker et al. [56] | Turkey | IV | IV | 186 | Admission to ICU, intubation, and death | ns | ns | ns | ns |
| Al-Youha et al. [71] | Kuwait | I | V | 3.304 | Respiratory support, admission to ICU, and death | ns | ns | ns | ns |
| Badedi et al. [79] | Saudi Arabia | I | V | 404 | Symptom presentation, hospital admission, disease duration and severity | ns | ns | ns | ns |
| Muñoz-Culla et al. [57] | Spain | I | VII, XII, XIII | 213 | Severity | ns | ns | ns | ns |
| Garg et al. [84] | India | I | IV | 383 | Severity | ↑ | ↑ | ↓ | ↓ |
| Szymanski et al. [88] | US | I | IV | 4.968 | Mortality | ↑ | ns | ns | ref |
| Ozkarafakili, Gareayaghi & Kara [60] | Turkey | I | II | 492 | Admission to ICU and death | ns | ns | ns | ns |
| Gamboa-Aguilar et al. [61] | Mexico | IV | IV | 2.416 | Hospitalization and death | ns | ns | ns | ns |
| Enguita-Gérman [49] | Spain | I | V | 6.771 | Hospitalization and death | ns | ns | ns | ns |
| Jawadat et al. [73] | Saudi Arabia | I | V | 373 | Hospitalization and admission to ICU | ns | ns | ns | ns |
| Nafakhi et al. [89] | Iraq | V | VII and XIII | 169 | Respiratory support, admission to ICU, death, and long-term symptoms | ns | ↑ | ↑ | ref |
| Gursoy & Avci [81] | Turkey | I | II | 969 | Hspital LOS, admission to ICU, ICU LOS, and death | ns | ns | ns | ns |
| Bokhary et al. [82] | Saudi Arabia | I | II | 316 | Admission to ICU and death | ns | ns | ns | ns |
| Jelinek et al. [83] | United Arab Emirates | III | VII | 193 | Severity | ns | ns | ns | ↓ |
| Alabsi et al. [90] | Saudi Arabia | III | XIV | 100 | Chronic chemosensitive dysfunction | – | – | – | ↑ |
Risk of severity and death vs. other blood types. Upward arrow: increased risk, downward arrow: decreased risk, ns: non-significant = p˃0.05.
I: Cohort study, II: Mixed method, III: Cross-sectional study, IV: Case-control study, V: Longitudinal observational study.
I: COVID-19 patients transfused during hospitalization, II: Hospitalized COVID-19 patients, III: Officially reported death rates, IV: General COVID-19 patients, V: SARS-CoV-2-positive individuals (Mostly RT-PCR), VI: SARS-CoV-2-negative individuals, VII: Severe COVID-19 patients, VIII: Patients admitted to intensive care unit (ICU), IX: COVID-19 patients underwent TAVIR, X: Hypertensive COVID-19 patients, XI: COVID-19 patients with severe hypoxia, XII: light COVID-19, XIII: Mild COVID-19, XIV: Patients recovered from COVID-19.
Intensive Care Unit.
Length of Stay.
Sequencial Organ Failure Assessment.
Simplified Acute Physiology Score 3.
Continuous Renal Replacement Therapy.
As can be seen from Table 2, results regarding severity and outcomes are more variable than those regarding the risk of infection. Several studies that reported associations between ABO blood type and the risk of SARS-CoV-2 infection failed to describe any association for severity [47,49,54,56,57,59,61,68,71,73,78] or reached distinct associations [51,70]. Other studies also reported no association between ABO blood types and severe COVID-19 outcomes but in accordance with the finds reported for the risk of infection [19,20,60,65,67]. Further studies also failed to report any association between the ABO blood group and severe COVID-19 outcomes [[79], [80], [81], [82]].
Nonetheless, blood type A has been the most associated with severe outcomes, while blood type O frequently appears as a protective factor against the disease progression [17,18,69,70,77,[83], [84], [85], [86], [87], [88]]. For instance, although failing to describe a significant difference in ABO phenotype distribution between hospitalized COVID-19 patients and the control group, Muñiz-Diaz et al. described a significant positive association between blood type A and mortality, and a significant negative association for blood type O [18]. Blood type A was significantly associated with hospitalization and mortality among COVID-19 patients who underwent TAVR [77] and to severe hypoxia among young COVID-19 patients without co-morbidities [17]. Blood type O, in turn, was significantly associated with lower risks of cardiac injury and death, and with lower pro-thrombotic indexes, among hypertensive COVID-19 patients [87].
Enrolling COVID-19 patients admitted to ICU, Hoiland et al. described significantly higher odds of requiring mechanical ventilation and Continuous Renal Replacement Therapy (CRRT), as well as a higher ICU length of stay (LOS), for A and AB groups; while B and O groups were negatively associated with these outcomes [86]. Conversely, El-Shitany et al. reported an association between blood type O and unfavorable outcomes, such as lower oxygen saturation and mechanical ventilation requirement, while blood type A was linked to lower odds of requiring artificial ventilation, and blood type B, to shorter recovery periods [51]. Significant associations with severity [84] and mortality [69,89] were also reported for blood type B, while blood type AB has been linked to either favorable [84], or unfavorable [89] outcomes. More recently, some studies sought to investigate whether ABO blood types are associated with long-term symptoms presentation after COVID-19 recovery. Blood type AB was significantly associated with long-term palpitation and dizziness [89], and blood type O, with subjective loss of smell [90].
Studies regarding associations between ABO blood types and COVID-19 severity and outcomes are even more heterogeneous than those concerning the risk of infection. The studies evaluated distinct outcomes and some of them had a unique design, which makes it more difficult to establish trends. Among the studies summarized here (Table 2), the majority did not reach statistical significance, and we were unable to identify any trend in the outcomes according to the size of the enrolled groups. However, it is possible to observe a trend towards significance when the enrolled group consisted of critically ill COVID-19 patients – like those requiring ICU admission [86] or blood transfusion [18] and those presenting severe hypoxia [17] - or patients with cardiovascular co-morbidities [77,87]. Taken together, in these groups of patients, blood type A was the most associated with unfavorable outcomes and blood type O constituted a protective factor. Conversely, in broader enrolled groups, like general hospitalized COVID-19 patients or individuals with a positive test, there was a trend towards non-significant results (Table 2).
4.3. Genetic link with susceptibility and severity
Several studies have been designed to uncover single nucleotide polymorphisms (SNPs) that could predict COVID-19 susceptibility or severity, and associations have been reported for the ABO locus. In a meta-analysis of two Genome-Wide Association Studies (GWAS) enrolling COVID-19 patients with respiratory failure, Ellinghaus et al. analyzed 8.582.968 SNPs and described a cross-replicating association with the Reference SNP (rs)657152 at locus 9q34, which corresponds to the ABO gene [6]. Other GWAS also reported associations for the ABO gene. An association with rs9411378 at chromosome 9q34.2 was identified by Shelton et al. in contrasting SARS-CoV-2-positive and –negative individuals, while no association with this SNP was identified when comparing severity phenotypes [91]. In this study, blood type O was shown to be protective regarding susceptibility, while blood types A, B, and AB did not differ from each other. An association with rs91280525 for susceptibility was replicated in a meta-analyses of previous GWAS [92]. Nonetheless, a GWAS enrolling critically ill COVID-19 patients failed to identify any association with the ABO locus [93].
More recently, Vargas-Alarcón et al. evaluated the allele distribution of six SNPs located at the ABO gene among COVID-19 patients and healthy hospital staff members. Two SNPs located at the coding region - rs8176740 and rs512770 – were found to be associated with the risk of developing COVID-19, with the risk alleles being related to O blood type [94]. The rs8176740 polymorphism was also associated with high platelet blood levels [94]. In contrast, after finding significantly lower levels of blood type O among critical COVID-19 patients (vs. non-critical patients), Jelinek et al. performed a candidate gene approach based on previous GWAS data and examined 240 SNPs in the ABO gene for association with critical COVID-19 presentation. 8 SNPs located at the ABO 3'untranslated region were significantly associated with disease severity, with significantly lower frequencies of the risk alleles in non-critical patients with blood type O [83].
Furthermore, some studies sought to genotype the ABO and/or FUT2 genes in COVID-19 patients and control groups. AB-related alleles were significantly associated with SARS-CoV-2 infection and O-related alleles were significantly less represented among the patients’ group, while no association was described for FUT2 [95]. Conversely, a study found no significant difference in the distribution of the rs8176719 deletion allele – the main O blood type-determinant – between patients and controls nor between severe and non-severe patients [96], while the rs601338 G˃A variant – the main non-secretor determinant – was associated with lower risks of ICU admission, mechanical ventilation and decease, especially in blood type A-individuals [80]. Another study described associations of the A/B-related rs8176747 with higher susceptibility to COVID-19 infection and the O-related rs8176719 with lower susceptibility, failing to describe any association with the FUT2-related rs1047781. Nonetheless, when comparing ABO secretor individuals vs. O and non-secretors, significant results were obtained, with the former group being more prone to COVID-19 onset [97].
Data from previous GWAS have been integrated to protein (p) or expression (e) quantitative trait loci (QTL) and colocalization analysis in Summary-Based Mendelian Randomization studies. In a multi-omics-based study, a protein-increasing allele of the ABO gene (rs505922) co-localized with susceptibility and severity-associated loci in both lungs and blood; and increased levels of blood ABO proteins was associated with higher risk of COVID-19 and its severity [98]. Likewise, in a Mendelian Randomization study based on previous GWAS, the blood level of ABO proteins was associated with hospitalization, respiratory support requirement, and decease [99]. Further QTL-based studies have also been reporting associations between the expression of ABO proteins (rs505922) and COVID-19 severity phenotypes [100,101]; however, pQTL for rs505922 was also associated with the levels of several other proteins [100,101], which suggests potential pleiotropic effects. Interestingly, among the proteins associated with rs505922, CD209 antigen was recently described as a receptor for SARS-CoV-2 S protein in lung and kidney epithelium and endothelial tissue [102]. Moreover, genetically predicted ABO levels were significantly associated with COVID-19 clinical phenotypes driven by the circulatory system, such as deep vein thrombosis, pulmonary heart disease, and acute pulmonary infarction [103]. Genetic associations between the ABO locus and COVID-19 are summarized in Table 3 .
Table 3.
- Genetic associations of the ABO locus with COVID-19 infection and severity.
| Reference | Country | Study Design | Sample size (n) | SNP | Outcomes |
|---|---|---|---|---|---|
| Ellinghaus et al. [6] | Italy/Spain | COVID-19 with RFa vs. blood donors, and non-COVID-19 controls | 1.610 cases vs.2.205 controls | rs657152 | Severe COVID-19 with respiratory failure |
| Pairo-Castineira et al. [93] | UK | Critically ill COVID-19 vs. matched controls | 2.244 cases vs.1.157.277 controls | – | None |
| Cordero et al. [98] | International | Integrative genomics | – | rs505922 | Susceptibility and severity |
| Valenti et al. [80] | Italy | Non-O vs. O COVID-19 with RF | Non-O 256 vs.O 153 | rs601338 (FUT2) | Mechanical ventilation, ICUa admission, and mortality |
| Matzhold et al. [95] | Austria | Hospitalized COVID-19 patients vs. blood donors | 336 patients vs. 250.298 controls | Several SNPs corresponding to variants of ABO alleles, FUT-2 rs601338, FUT3 | Suscetibility to infection, hospitalization, ICU admission, mechanical ventilation, and mortality |
| Coto et al. [96] | Spain | Hospitalized COVID-19 patients vs. matched healthy controls | 318 patients vs. 350 controls | rs8176719 | Susceptibility and severity |
| Zhou et al. [100] | International | Hospitalized COVID-19 patients vs. healthy controls. | 14.134 cases vs.1.2 million controls | rs505922 | Severity |
| Zhu et al. [101] | International | Hospitalized COVID-19 patients vs. healthy controls. | 6.492 patients vs.1.012.809 controls | rs505922 | Susceptibility, hospitalization and severity. |
| Pathak et al. [103] | International | COVID-19 hospitalized patients vs.controls | 7.885 cases vs. 961.804 controls | – | Severity |
| CHGI [92] | International (multiethnic) | COVID-19 patients vs. matched controls | 49.562 cases vs. 2.000.000 controls | rs91280525 | Susceptibility and severity |
| Shelton et al. [91] | US/UK | SARS-CoV-2 positive vs. controls | 12.972 cases vs. 101.268 controls | rs9411378 | Susceptibility |
| Vargas-Alarcón et al. [94] | Mexico | COVID-19 patients vs. healthy hospital staff members | 415 cases vs. 288 controls | rs8176740 and rs512770 | Susceptibility and platelets concentration |
| Jelinek et al. [83] | United Arab Emirates | Critical vs. non-critical COVID-19 patients | 193 cases vs. 453 controls | rs199969472, rs34266669, rs76700116, rs7849280, rs34039247, rs10901251, rs9411475, and rs13291798 | Severity |
| Nishida et al. [97] | Japan | COVID-19 patients vs. healthy controls | 461 cases vs. 1.193 controls | rs8176719 and rs8176747 (ABO) and rs1047781 (FUT2) | Susceptibility |
| Palmos et al. [99] | International | Mendelian Randomiation | – | Several SNPs related to the ABO gene. | Severity |
Intensive Care Unit.
5. Hypothesis linking ABO blood type and COVID-19: underlying mechanisms and their contribution to the interpretation of reported associations
Since an association between the ABO blood group and COVID-19 was firstly reported, several underlying mechanisms have been hypothesized [22,23]. The proposed hypotheses are most reliant on what is already known for other enveloped viruses, including SARS-CoV [21,44], or in the previous associations between ABO blood group and other COVID-19 risk factors and co-morbidities [23,104]. The hypothesized mechanisms could affect either the risk of infection or the disease severity and outcomes, with distinct potential implications to the transmission dynamics or disease management.
5.1. ABO antibodies and host's glycosylation pattern
Upon entry in the human body, SARS-CoV-2 can infect and replicate in the upper respiratory tract [105,106], several cell types of the bronchial epithelium, and the alveolar type 1 pneumocytes [107], as well as in the enterocytes [108]. Epithelial cells of both the human airways and digestive tract are known to express host factors used by SARS-CoV-2, like ACE2 and TMPRSS2 [[109], [110], [111]], in addition to A/B transferases in secretor individuals [7,112].
During the replication cycle of SARS-CoV-2, viral RNA is synthesized within double-membrane vesicles that bud from the host endoplasmic reticulum following extensive remodeling [113], and the structural proteins assemble to the viral genome at the reticulum-Golgi intermediate compartment (ERGIC) to form progeny virions, which are released from the cell surface by exocytose [114]. The passage of virions through the secretory pathway raises questions concerning the processing of the viral surface glycoproteins [115], due to the presence of glycosylation enzymes at this pathway [116].
Indeed, the SARS-CoV-2 spike (S) protein is a highly glycosylated trimer, with 22 canonical N-linked glycosylation sites per protomer [117]. Glycomics-oriented studies revealed a high occupancy of N-linked sequons and the presence of O-linked glycosylation sites with low occupancy in recombinant S protein of SARS-CoV-2 produced in either mammalian [115,[117], [118], [119]] or insect [120] cell lines. In addition, mass spectrometry approaches showed that the glycosylation profile of the S protein is highly heterogeneous and cell type-dependent. In recombinant S protein produced in insect Sf9 cells, a predominance of less processed glycans, like oligomannose, was found [120]; while a more complex glycan processing, like fucosylation, galactosylation, and sialylation, was shown to take place in mammalian HEK 293 cells [115,117,119]. Nonetheless, these glycans do not fully represent those found at the envelope glycoproteins of virions emitted from the human respiratory or digestive tract and cannot be decorated with ABH antigens, since Hek293 cells do not synthesize ABH transferases [25].
Aiming to uncover whether the S protein can be decorated with ABH antigens when synthesized in ABO- and FUT2-expressing cells, Deleers et al. produced recombinant S protein in transfected CHO cells expressing either the A or B transferase and the FUT2 gene. This approach showed that the S protein can carry ABH antigens according to the glycosyltransferase repertoire of the previous host cell [121]. Hence, ABO antibodies could constitute a barrier to SARS-CoV-2 infection according to the rules of blood transfusion. When transmitted in ABO-incompatible situations, SARS-CoV-2 could be partially neutralized by anti-A or anti-B antibodies, which can explain the lower incidence of COVID-19 in type O individuals, who display both anti-A and anti-B. Nonetheless, such protection would only be initial as, once the infection is established, the virions would carry the host's ABO profile and the autologous ABO antibodies would no longer be effective [122]. Therefore, this possible underlying mechanism can only explain associations between the ABO blood type and the risk of SARS-CoV-2 infection and does not apply to the reported associations concerning severity and outcomes.
The ABO antibodies hypothesis has some implications for the dynamics of COVID-19 transmission. Firstly, the reduced risk of viral infections in case of ABO-incompatible transmission does not constitute individual protection, but rather collective protection that could delay the spread of newly emerging viruses and slow down the epidemics, in addition to the required behavioral changes [44]. Nonetheless, as the epidemic progresses, more and more ABO-compatible transmission might occur, and the final number of cases may not be reduced to a great extent [25]. This could explain, for instance, the lack of association reported by Boudin et al. [76], in which the study participants consisted of crewmembers in close enough contact within an aircraft carrier that may had facilitate ABO-compatible transmission and reduced any O-related protection.
Moreover, the protection provided by ABO antibodies in ABO-incompatible transmission would critically depend on the relative frequency of ABO phenotypes in a given population. Such protection would be more effective in populations with low coefficients of variation between the ABO phenotypes and is expected to be inefficient in populations with high coefficients of variation, in which more ABO-compatible transmission may occur [25]. Importantly, although blood type O is likely more protective to SARS-CoV-2 infection due to the presence of both anti-A and anti-B antibodies, this advantage seems to disappear in populations in which this blood type is highly predominant, like in some Central and South American countries [25].
Following this rationale, differences in ABO phenotypes distribution between distinct cohorts can explain some interstudy discrepancies. For instance, the frequency of the B phenotype is increased in Asia at the expense of both A and O phenotypes, which, together, are largely predominant in Western Europe, with the Middle East displaying an intermediary distribution [25]. Such difference could explain the association between blood type B and the increased risk of SARS-CoV-2 infection reported by some Indian or Middle Eastern studies [59,69,71,73]. Likewise, the lack of association between ABO blood type and the risk of SARS-CoV-2 infection [19,63] or a significant positive association with blood type O [64] in U.S multiethnic cohorts before adjusting for ethnicity can be explained by the fact that ethnic groups among who blood type O is predominant, like Hispanics and black non-Hispanics, have been disproportionally affected by the COVID-19 pandemic in the U.S [19,64]. In addition, the association between blood type O and the risk of COVID-19 onset described in a Mexican study can be explained by the high prevalence of this blood type in the Mexican population [61].
More recently, Boukhari et al. sought to investigate the impact of ABO compatibility on a secondary attack rate among the spouses of hospital staff members who were symptomatic for COVID-19. The risk of symptomatic COVID-19 transmission was significantly lower for ABO-incompatible couples, while ABO compatibility was associated with higher transmissibility regardless of the ABO blood type of the partner. This supports that all blood types are equally susceptible to COVID-19, and the protection provided by ABO-incompatibility may be due to ABO antibodies [123].
Furthermore, one should consider the variability of the ABO antibodies titer. The levels of ABO antibodies decrease with improving hygiene and tend to be low in developed countries [124], which may pose a challenge to its protective effect against viral infections and constitutes a potential confounding factor in association studies. Indeed, a study enrolling COVID-19 patients and asymptomatic controls showed that, for both A, B, and O blood types, the patients’ group displayed significantly lower IgM agglutination scores compared to the control group [121]. In this way, some authors have been suggesting the promotion of anti-glycan antibodies through dedicated microbiota to enhance the natural anti-viral protection and slow down the spread of newly emergent viruses [121,125].
5.2. S protein priming and SARS-CoV-2-host cells interaction
The S protein of SARS-CoV-2 harbors a multibasic S1/S2 cleavage site, which constitutes a target for furin, a proprotein convertase. The furin-mediated S1/S2 cleavage generates two subunits – S1 and S2 – and is pivotal for the S protein's activation. S1 binds to cellular receptors, while S2 is further cleaved by TMPRSS2 and promotes virus-cell membrane fusion. The furin cleavage site was shown to be required for SARS-CoV-2 infection of the human lung cells Calu-3 and for the S protein-mediated cell-cell fusion, which promotes a systemic viral spread [126]. Although no correlation was yet reported for the furin levels and ABO polymorphism, an association between blood type O and lower levels of the furin-related proprotein convertase subtilisin-kexin type 9 (PCSK9) was previously reported [127]. Therefore, Abdelmassih et al. hypothesized that furin levels may follow the same pattern as PCSK9 and be decreased in blood type O individuals, which might reduce S protein priming and SARS-CoV-2 entry into host cells [128].
S1, in turn, is subdivided into the S1A and S1B domains. The former corresponds to the amino-terminal portion of the polypeptide chain, namely NTD (from N-terminal domain), and, in some coronaviruses, interacts with glycolipids and glycoproteins displaying terminal sialic acid motifs at the host cells’ surface [129]. The latter constitutes the Receptor-Binding Domain (RBD) and binds to the ACE2 receptor [130]. In silico studies based on molecular docking simulations [131] and electronic density mapping surface [132] predict the existence of a sialic acid-binding pocket in the SARS-CoV-2 NTD similar to that found in MERS-CoV, which might confer to SARS-CoV-2 the ability to infect human cells in a dual manner, resulting in a higher diffusion speed [132].
ABH antigens are known to modulate the clustering of sialylated glycans at the cell's surface through carbohydrate-carbohydrate interactions and to regulate their interaction with cognate-binding proteins. A model was proposed in which antigen A is peripherally located at the sialylated glycan clusters, allowing for continuous clusters with increased sialic acids concentrations. Conversely, antigen B is mostly located at the center of the cluster, which may pose a steric hindrance for sialic acids-binding proteins, while in type O cells, the clusters are small and lack H antigens [133]. Following this rationale, it was hypothesized that ABO blood group may correlate with COVID-19 susceptibility and severity through carbohydrate-carbohydrate interactions between ABH antigens and sialylated glycans at the cells surface. According to such hypothesis, antigen A maximizes the interaction of SARS-CoV-2 with the target cells by promoting the clustering of sialylated glycans and increasing the likelihood of NTD biding [134].
Moreover, in a glycan microarray with ABH antigens, the SARS-CoV-2 RBD bound preferentially to the type I structure of antigen A, like the SARS-CoV RBD, which suggests a possible role for antigen A in coronaviruses entry into respiratory or digestive tract cells [48]. A similar result was described by Wu et al., who also demonstrated sequence similarities between the SARS-CoV-2 RBD and galectins, a family of carbohydrate-binding proteins [135]. This could explain, for instance, the genetic association reported by Valenti et al. between non-secretor status and lower risk of severe outcomes among blood type A-individuals [80]. Conversely, Boukhari et al. failed to detect any preferential biding of recombinant RBD to both A type I saccharide and lung tissue from A and O donors, as well as to salivary mucin from secretor-phenotype saliva [123]. Furthermore, although individuals with blood type A1 display more than fourfold higher A epitope density compared to those with A2 subtype, comparable rates of infection between A1 and A2 participants were reported by Schetelig et al., which suggests that the A antigen itself does not facilitate viral entry [48].
Hypotheses linking the ABO blood type to S protein priming or SARS-CoV-2 cellular entry remain largely elusive and need further studies. These hypotheses could explain both the higher SARS-CoV-2 infection rates and higher disease severity reported for blood type A, as well as the lower rates of infection and lower disease severity reported for blood type O. Unlike the ABO antibodies hypothesis, the possible role of ABO antigens in regulating virus-cell interaction may be mostly reliant on the antigen itself and may not depend on the ABO coefficient of variation at a population level. In addition, the possible pleiotropy effect of the ABO gene (rs505922) over the CD209 receptor gene [101] could also be considered into the framework of these hypotheses.
5.3. ABO blood group and cardiovascular risk
Associations between non-O blood types and increased risk of cardiovascular disorders are well documented and are detailed reviewed elsewhere [[136], [137], [138]]. Associations have been reported, for instance, with venous thromboembolism, myocardial infarction, peripheral vascular disease, cerebrovascular stroke, and pulmonary embolism. A possible explanation relies on the ∼30% lower levels of von Willebrand factor (vWF) and coagulation factor VIII (FVIII) in blood type O individuals [139,140]. vWF mediates platelet aggregation and adhesion and can be released in response to inflammatory stimuli, like infections. It also plays a role in hemostasis by complexing with FVIII and stabilizing it at the circulation [141]. As a glycoprotein, vWF carries ABO(H)-like structures in Asp-linked sugar chains [142], which are likely to mediate its homeostasis. Indeed, glycosylation is likely to mediate the clearance of vWF, which is largely performed by lectin receptors [143,144]. A and B antigens were shown to protect vWF from proteolysis by the ADAMTS13 metalloprotease [145]. Furthermore, the ABO blood group indirectly modulates the circulating levels of FVIII by affecting the clearance of the vWF/FVIII complex, accounting for about ∼30% of its variation [140].
Likewise, some adhesion molecules, like the intercellular cell adhesion molecule 1 (ICAM1) and P-selectin, are protected by antigen A from enzymatic cleavage, prompting the attachment of leucocytes to them at the vascular wall, which increases inflammation and decreases circulation [23]. In addition, a study enrolling COVID-19 hypertensive patients reported significantly higher values of pro-thrombotic indexes - vWF, FVIII, D-dimer, and activated pro-thrombin time - in non-O vs O patients, which was consistent with the higher rates of cardiac injury and death in the former group [87]. Blood type O individuals are therefore less prone to thromboembolic events that constitute relevant outcomes in COVID-19 disease, especially among ICU-admitted patients [146,147].
The link between ABO blood group and cardiovascular disorders does not affect the susceptibility to SARS-CoV-2 infection and cannot explain the reported associations between ABO blood type and the risk of infection. However, it constitutes a possible underlying mechanism for the lower risk of severe outcomes associated with blood type O and, at a lesser extent, the higher risk related to blood type A in some studies (Table 2), especially among patients with cardiovascular co-morbidities [77,87]. Into the framework of this hypothesis, the reported associations rely on constitutive tracts related to ABO blood type, which is in line with a study that reported associations between genetically-driven ABO expression levels and COVID-19 clinical phenotypes driven by the circulatory system [103]. Following this rationale, a risk stratification for cardiovascular complications in COVID-19 patients based on the ABO phenotype has been suggested, as well as the introduction of early anticoagulant therapy in non-O hypertensive patients [87].
6. Conclusions
A literature survey of epidemiological studies suggests that the ABO blood type may be an influencing factor for both SARS-CoV-2 infection and COVID-19 severity. Although the findings are conflicting and quite variable, blood type O is mostly associated with lower risk of SARS-CoV-2 infection, while blood type A, with higher risk. Blood types B and AB were also associated with higher risk of infection in some studies. Although the majority of the studies regarding COVID-19 severity and outcomes failed to report any association with the ABO blood group, associations with lower risk of severe outcomes have been frequently described for blood type O, while blood type A has been frequently described as a risk factor for severe outcomes. Moreover, genetic links with both susceptibility to infection and severity have been reported for the ABO locus.
Driven by the previous knowledge about ABO blood group biology and its role in the pathogenesis of infectious diseases, some underlying mechanisms that could explain the reported associations emerge; like a protective effect of the ABO antibodies against viral infections, a role for ABO(H) antigens in facilitating viral entry into host cells, and the association between non-O blood types and higher risk of thromboembolic events. These underlying mechanisms may affect different phases of the natural history of COVID-19, with distinct implications to the SARS-CoV-2 transmission dynamics and to COVID-19 outcomes. Nonetheless, there are little experimental data on this subject and more studies are needed to elucidate the mechanisms underlying the reported associations, which is pivotal for a deeper understanding of the relationship between the ABO blood group and COVID-19 and whether or not it can be translated to disease prevention or management strategies.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Code availability
Not applicable.
CRediT authorship contribution statement
Eric Pereira: Writing – original draft, Formal analysis, Data curation, Conceptualization. Stela Felipe: Writing – review & editing, Conceptualization. Raquel de Freitas: Writing – review & editing. Valdevane Araújo: Data curation, Conceptualization. Paula Soares: Data curation, Conceptualization. Jannison Ribeiro: Supervision, Data curation, Conceptualization. Luiz Henrique dos Santos: Conceptualization. Juliana Osório Alves: Writing – review & editing. Natália Canabrava: Conceptualization. Mauricio van Tilburg: Supervision, Conceptualization. Maria Izabel Guedes: Supervision, Conceptualization. Vânia Ceccatto: Writing – review & editing, Supervision, Data curation, Conceptualization.
Declaration of competing interest
The authors have no financial or non-financial competing interests to disclose.
Data availability
No data was used for the research described in the article.
References
- 1.World Health Organization . 2020. COVID-19 Weekly Epidemiological Update.https://www.who.int/publications/m/item/weekly-epidemiological-update---15-december-2020 [Internet] Available from: [Google Scholar]
- 2.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han D., Li R., Han Y., Zhang R., Li J. Covid-19: insight into the asymptomatic sars-cov-2 infection and transmission. Int. J. Biol. Sci. 2020;16:2803–2811. doi: 10.7150/ijbs.48991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velavan T.P., Pallerla S.R., Rüter J., Augustin Y., Kremsner P.G., Krishna S., et al. Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine. 2021:72. doi: 10.1016/j.ebiom.2021.103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA, J. Am. Med. Assoc. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 6.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., et al. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/nejmoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marionneau S., Cailleau-Thomas A., Rocher J., Le Moullac-Vaidye B., Ruvoën N., Clément M., et al. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie. 2001;83:565–573. doi: 10.1016/S0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- 8.Batool Z., Durrani S.H., Tariq S. Association of abo and Rh blood group types to hepatitis B, hepatitis C, hiv and Syphilis infection, A five Year’ experience in healthy blood donors in A tertiary care hospital. J. Ayub Med. Coll. Abbottabad. 2017;29:90–92. [PubMed] [Google Scholar]
- 9.Jing W., Zhao S., Liu J., Liu M. ABO blood groups and hepatitis B virus infection: a systematic review and meta-analysis. BMJ Open. 2020;10 doi: 10.1136/bmjopen-2019-034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaidarova Z., Bravo M.D., Kamel H.T., Custer B.S., Busch M.P., Lanteri M.C. Blood group A and D negativity are associated with symptomatic West Nile virus infection. Transfusion. 2016;56:1699–1706. doi: 10.1111/trf.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Mattos L.C., Cintra J.R., Sanches F.E., Alves Da Silva R.D.C.M., Ruiz M.A., Moreira H.W. ABO, Lewis, secretor and non-secretor phenotypes in patients infected or uninfected by the Helicobacter pylori bacillus. Sao Paulo Med. J. 2002;120:55–58. doi: 10.1590/s1516-31802002000200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins L.C., de Oliveira Corvelo T.C., Oti H.T., do Socorro Pompeu Loiola R., Aguiar D.C.F., dos Santos Barile K.A., et al. ABH and Lewis antigen distributions in blood, saliva and gastric and H pylori infection in gastric ulcer patients. World J. Gastroenterol. 2006;12:1120–1124. doi: 10.3748/wjg.v12.i7.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooling L. Blood groups in infection and host susceptibility. Clin. Microbiol. Rev. 2015;28:801–870. doi: 10.1128/CMR.00109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X., et al. Relationship between the ABO blood group and the COVID-19 susceptibility. Clin. Infect. Dis. 2021;73:328–331. doi: 10.1093/cid/ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Wang X., Chen J., Cai Y., Deng A., Yang M. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br. J. Haematol. 2020;190:24–27. doi: 10.1111/bjh.16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Feng Z., Li P., Yu Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin Chim Acta. Netherlands. 2020;509:220–223. doi: 10.1016/j.cca.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solhpour A., Jafari A., Pourhoseingholi M., Soltani F. Corona Covid-19 virus and severe hypoxia in young patients without underlying disease: high prevalence rate with blood group A. Trends Anaesth Crit Care. 2020;34:63–64. doi: 10.1016/j.tacc.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñiz-Diaz E., Llopis J., Parra R., Roig I., Ferrer G., Grifols J., et al. Relationship between the ABO blood group and COVID-19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfus. Italy. 2021;19:54–63. doi: 10.2450/2020.0256-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzik S., Eliason K., Morris E.B., Kaufman R.M., North C.M. COVID-19 and ABO blood groups. Transfusion. 2020;60:1883–1884. doi: 10.1111/trf.15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil A., Feghali R., Hassoun M. The Lebanese COVID-19 cohort; A challenge for the ABO blood group system. Front. Med. 2020;7:1–7. doi: 10.3389/fmed.2020.585341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y., Cheng G., Chui C.H., Lau F.Y., Al E. ABO blood group and susceptibility to severe acute respiratory syndrome to. J. Am. Med. Assoc. 2005;293:1450–1451. doi: 10.3122/jabfm.19.6.648. [DOI] [PubMed] [Google Scholar]
- 22.Gérard C., Maggipinto G., Minon J.M. COVID-19 and ABO blood group: another viewpoint. Br. J. Haematol. 2020;190:e93–e94. doi: 10.1111/bjh.16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X. ABO blood group predisposes to COVID-19 severity and cardiovascular diseases. Eur. J. Prev. Cardiol. 2020;27:1436–1437. doi: 10.1177/2047487320922370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean L. In: Blood Groups and Red Cell Antigens. Dean L., editor. National Center for Biotechnology Information; Bethesda: 2005. The ABO blood group; pp. 31–39. [Google Scholar]
- 25.Le Pendu J., Breiman A., Rocher J., Dion M., Ruvoën-Clouet N. ABO blood types and COVID-19: Spurious, Anecdotal, or truly important relationships? A reasoned review of Available data. Viruses. 2021;13:1–20. doi: 10.1016/b978-0-08-022449-7.50017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins W.M. In: Biochemistry of Human Genetics. Wolstenholme C., O'Connor C., editors. Little, Brown and Company; Boston: 1959. Some genetical aspects of the biosynthesis of human blood group substances; pp. 217–238. [Google Scholar]
- 27.Yamamoto F. Review: ABO blood group system--ABH oligosaccharide antigens, anti-A and anti-B, A and B glycosyltransferases, and ABO genes. Immunohematol. 2004;20:3–22. [PubMed] [Google Scholar]
- 28.Yamamoto F.I., Clausen H., White T., Marken J., Hakomori S.I. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 29.Watkins W.M. The ABO blood group system: historical background. Transfus. Med. 2001;11:243–265. doi: 10.1046/j.1365-3148.2001.00321.x. [DOI] [PubMed] [Google Scholar]
- 30.Patenaude S.I., Seto N.O.L., Borisova S.N., Szpacenko A., Marcus S.L., Palcic M.M., et al. The structural basis for specificity in human abo(h) blood group biosynthesis. Nat. Struct. Biol. 2002;9:685–690. doi: 10.1038/nsb832. [DOI] [PubMed] [Google Scholar]
- 31.Branch D.R. Anti-A and anti-B: what are they and where do they come from? Transfusion. 2015;55:S74–S79. doi: 10.1111/trf.13087. [DOI] [PubMed] [Google Scholar]
- 32.Springer G.F., Horton R.E. Blood group isoantibody stimulation in man by feeding blood group-active bacteria. J. Clin. Invest. 1969;48:1280–1291. doi: 10.1172/JCI106094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bello-Gil D., Audebert C., Olivera-Ardid S., Pérez-Cruz M., Even G., Khasbiullina N., et al. The formation of glycan-specific natural antibodies repertoire in Galt-KO mice is determined by gut microbiota. Front. Immunol. 2019;10:1–13. doi: 10.3389/fimmu.2019.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwell P. Blood group antigens: molecules seeking a function? Glycoconj. J. 1997;14:159–173. doi: 10.1023/A:1018581503164. [DOI] [PubMed] [Google Scholar]
- 35.Oriol R., Le Pendu J., Mollicone R. Genetics of ABO, H, Lewis, X and related antigens. Vox Sang. 1986;51:161–171. doi: 10.1111/j.1423-0410.1986.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 36.Geisel J., Steuer M.K., Ko H.L., Beuth J. The role of ABO blood groups in infections induced by Staphylococcus saprophyticus and Pseudomonas aeruginosa. Zentralblatt fur Bakteriol. 1995;282:427–430. doi: 10.1016/S0934-8840(11)80714-0. [DOI] [PubMed] [Google Scholar]
- 37.Gagneux P., Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- 38.Borén T., Falk P., Roth K.A., Larson G., Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262 doi: 10.1016/s0887-7963(05)80040-9. 1892–5. [DOI] [PubMed] [Google Scholar]
- 39.Stapleton A., Nudelman E., Clausen H., Hakomori S.I., Stamm W.E. Binding of uropathogenic escherichia coli R45 to glycolipids extracted from vaginal epithelial cells is dependent on histo-blood group secretor status. J. Clin. Invest. 1992;90:965–972. doi: 10.1172/JCI115973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georg F., Springer M.D., Williamson P., Brandes W. Blood group Activity of gram-negative bacteria. J. Exp. Med. 1961;113:1077–1093. doi: 10.1046/j.1537-2995.1988.28388219149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seymour R.M., Allan M.J., Pomiankowski A., Gustafsson K. Evolution of human ABO polymorphism by two complementary selective pressures. Proc. R Soc. B Biol. Sci. 2004;271:1065–1072. doi: 10.1098/rspb.2004.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arendrup M., Hansen J.-E.S., Clausen H., Nielsen C., Mathiesen L.R., Nielsen J.O. Antibody to histo-blood group A antigen neutralizes HIV produced by lymphocytes from blood group A donors but not from blood group B or O donors. AIDS. 1991;5:441–444. doi: 10.1097/00002030-199104000-00014. [DOI] [PubMed] [Google Scholar]
- 43.Preece A.F., Strahan K.M., Devitt J., Yamamoto F.I., Gustafsson K. Expression of ABO or related antigenic carbohydrates on viral envelopes leads to neutralization in the presence of serum containing specific natural antibodies and complement. Blood. 2002;99:2477–2482. doi: 10.1182/blood.V99.7.2477. [DOI] [PubMed] [Google Scholar]
- 44.Guillon P., Clément M., Sébille V., Rivain J.G., Chou C.F., Ruvoën-Clouet N., et al. Inhibition of the interaction between the SARS-CoV Spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008;18:1085–1093. doi: 10.1093/glycob/cwn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe J.A., Handel I.G., Thera M.A., Deans A.M., Lyke K.E., Koné A., et al. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc. Natl. Acad. Sci. USA. 2007;104:17471–17476. doi: 10.1073/pnas.0705390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ségurel L., Gao Z., Przeworski M. Ancestry runs deeper than blood: the evolutionary history of ABO points to cryptic variation of functional importance. Bioessays. 2013;35:862–867. doi: 10.1002/bies.201300030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnkob M.B., Pottegård A., Støvring H., Haunstrup T.M., Homburg K., Larsen R., et al. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4:4990–4993. doi: 10.1182/BLOODADVANCES.2020002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schetelig J., Baldauf H., Wendler S., Heidenreich F., Real R., Kolditz M., et al. Blood group A epitopes do not facilitate entry of SARS-CoV-2. J. Intern. Med. 2021;290:223–226. doi: 10.1111/joim.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enguita-Gérman M., Librero J., Leache L., Gutiérrez-Valencia M., Tamayo I., Jericó C., et al. Role of the AB0 blood group in COVID-19 infection and complications: a population-based study. Transfus. Apher. Sci. 2022 doi: 10.1016/j.transci.2022.103357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jukić I., Hećimović A., Vuk T., Vinković M., Kereš T., Lampalo M., et al. Prevalence of ABO and RhD blood group phenotypes in the Croatian population and in patients with severe COVID-19 in Croatia. Blood Transfus. 2022 doi: 10.2450/2022.0311-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Shitany N.A., El-Hamamsy M., Alahmadi A.A., Eid B.G., Neamatallah T., Almukadi H.S., et al. The impact of abo blood grouping on covid-19 vulnerability and seriousness: a retrospective cross-sectional controlled study among the Arab community. Int. J. Environ. Res. Publ. Health. 2021;18:1–19. doi: 10.3390/ijerph18010276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Q., Zhang W., Li B., Li D.-J., Zhang J., Zhao F. Association between ABO blood group system and COVID-19 susceptibility in Wuhan. Front. Cell. Infect. Microbiol. 2020;10:1–10. doi: 10.3389/fcimb.2020.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franchini M., Glingani C., Del Fante C., Capuzzo M., Di Stasi V., Rastrelli G., et al. The protective effect of O blood type against SARS-CoV-2 infection. Vox Sang. 2021;116:249–250. doi: 10.1111/vox.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solmaz İ., Araç S. ABO blood groups in COVID-19 patients; Cross-sectional study. Int. J. Clin. Pract. 2021;75 doi: 10.1111/ijcp.13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yalaoui S., Fakhfakh R., Tritar F., Chaouch N., Mestiri T., Besbes M., et al. Groupes sanguins ABO et risque d ’ atteinte par le covid-19. Tunis. Med. 2020;98:888–891. [PubMed] [Google Scholar]
- 56.Göker H., Aladağ-Karakulak E., Demiroğlu H., Ayaz C.M., Büyükaşik Y., İnkaya A.C., et al. The effects of blood group types on the risk of COVID-19 infection and its clinical outcome. Turk. J. Med. Sci. 2020;50:679–683. doi: 10.3906/sag-2005-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muñoz-Culla M., Roncancio-Clavijo A., Martinez B., Gorostidi-Aicua M., Piñeiro L., Azkune A., et al. O group is a protective factor for COVID19 in Basque population. PLoS One. 2021;16:3–6. doi: 10.1371/journal.pone.0249494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahmud R., Rassel M.A., Monayem F.B., Sayeed S.K.J.B., Islam M.S., Islam M.M., et al. Association of ABO blood groups with presentation and outcomes of confirmed SARS CoV-2 infection: a prospective study in the largest COVID-19 dedicated hospital in Bangladesh. PLoS One. 2021;16:1–10. doi: 10.1371/journal.pone.0249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almadhi M.A., Abdulrahman A., Alawadhi A., Rabaan A.A., Atkin S., AlQahtani M. The effect of ABO blood group and antibody class on the risk of COVID-19 infection and severity of clinical outcomes. Sci. Rep. 2021;11:19–23. doi: 10.1038/s41598-021-84810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozkarafakili M.A., Gareayaghi N., Kara Z.M.Y. Relationship between ABO blood types and COVID-19 severity. Med. Bull. Sisli Hosp. 2022;56:1–20. doi: 10.14744/SEMB.2021.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamboa-aguilar J., Zamorano-montaño Á.C., Enríquez-osorio A., Torres-cubillas W., López-arroyo J.L., Mata A., et al. Abo blood group, Atherothrombotic comorbidities, and COVID-19: a case-control study of their association in the Mexican population. Arch. Med. Res. 2022;53:100–108. doi: 10.1016/j.arcmed.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurung S., Mahotra N.B., Shrestha L., Sherpali A., Joshi S.P., Shrestha G., et al. Association of ABO blood group with susceptibility to SARS-CoV-2 infection in Rupandehi district of Nepal. Sage Open Med. 2022;10:1–5. doi: 10.1177/20503121221095413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leaf R.K., Al-Samkari H., Brenner S.K., Gupta S., Leaf D.E. ABO phenotype and death in critically ill patients with COVID-19. Br. J. Haematol. 2020;190:204–208. doi: 10.1111/bjh.16984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niles J.K., Karnes H.E., Dlott J.S., Kaufman H.W. Association of ABO/Rh with SARS-CoV-2 positivity: the role of race and ethnicity in a female cohort. Am. J. Hematol. 2021;96:1–3. doi: 10.1002/ajh.26019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehrer S., Rheinstein P.H. ABO blood groups, COVID-19 infection and mortality. Blood Cells. Mol. Dis. 2021;89:1–4. doi: 10.1016/j.bcmd.2021.102571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levi J.E., Telles P.R., Scrivani H., Campana G. Lack of association between ABO blood groups and susceptibility to SARS-CoV-2 infection. Vox Sang. 2021;116:251–252. doi: 10.1111/vox.13015. [DOI] [PubMed] [Google Scholar]
- 67.Mullins J., Al-Tarbsheh A.H., Chieng H., Chaukiyal P., Ghalib S., Jain E., et al. The association of ABO blood type with the risk and severity of COVID-19 infection. Am. J. Blood Res. 2021;11:53–58. [PMC free article] [PubMed] [Google Scholar]
- 68.Latz C.A., DeCarlo C., Boitano L., Png C.Y.M., Patell R., Conrad M.F., et al. Blood type and outcomes in patients with COVID-19. Ann. Hematol. Annals of Hematology. 2020;99:2113–2118. doi: 10.1007/s00277-020-04169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padhi S., Suvankar S., Dash D., Panda V.K., Pati A., Panigrahi J., et al. ABO blood group system is associated with COVID-19 mortality: an epidemiological investigation in the Indian population. Transfus. Clin. Biol. 2020;27:253–258. doi: 10.1016/j.tracli.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray J.G., Schull M.J., Vermeulen M.J., Park A.L. Association between ABO and Rh blood groups and SARS-CoV-2 infection or severe COVID-19 illness : a population-based cohort study. Ann. Intern. Med. 2021;174:308–315. doi: 10.7326/M20-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Youha S.A., Alduaij W., Al-Serri A., Almazeedi S.M., Al-Haddad M., Jamal M.H., et al. The impact of ABO blood groups on clinical outcomes and susceptibility to COVID-19: a retrospective study in an unselected population. Transfusion. 2021;61:1631–1641. doi: 10.1111/trf.16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kotila T.R., Alonge T.O., Fowotade A., Famuyiwa O.I., Akinbile A.S. Association of the ABO blood group with SARS-CoV-2 infection in a community with low infection rate. Vox Sang. 2021;116:910–915. doi: 10.1111/vox.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jawdat D., Hajeer A., Massadeh S., Aljawini N., Abedalthagafi M.S., Alaamery M. Correlation between ABO blood group phenotype and the risk of COVID-19 infection and severity of disease in a Saudi Arabian cohort. J. Epidemiol. Glob. Health. 2022;12:85–91. doi: 10.1007/s44197-021-00023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu N., Zhang T., Ma L., Zhang H., Wang H., Wei W., et al. The impact of ABO blood group on COVID-19 infection risk and mortality: a systematic review and meta-analysis. Blood Rev. 2021;48 doi: 10.1016/j.blre.2020.100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kander T., Bjurström M.F., Frigyesi A., Jöud M., Nilsson C.U. ABO and RhD blood group are not associated with mortality and morbidity in critically ill patients; a multicentre observational study of 29 512 patients. BMC Anesthesiol. 2022;22:1–8. doi: 10.1186/s12871-022-01626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boudin L., Janvier F., Bylickei O., Dutasta F. ABO blood groups are not associated with the risk of acquiring SARS-CoV-2 infection in young adults. Haematologica. 2020;105:2841–2843. doi: 10.3810/psm.1999.02.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kibler M., Carmona A., Marchandot B., Matsushita K., Trimaille A., Kanso M., et al. Risk and severity of COVID-19 and ABO blood group in transcatheter aortic valve patients. J. Clin. Med. 2020;9:1–12. doi: 10.1101/2020.06.13.20130211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Negro P., Congedo M., Zizza A., Guido M., Sacquegna G., Pulito G., et al. Role of ABO blood system in COVID-19: findings from a southern Italian study. Transfus. Med. 2021;1–5 doi: 10.1111/tme.12797. [DOI] [PubMed] [Google Scholar]
- 79.Badedi M., Alnami A., Darraj H., Alrajhi A., Mutawwam D.A., Somaily M., et al. Clinical characteristics and ABO blood groups in COVID-19 patients, Saudi Arabia. Medicine. 2021;100:1–6. doi: 10.1097/MD.0000000000026738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valenti L., Villa S., Baselli G., Temporiti R., Bandera A., Scudeller L., et al. Association of ABO blood group and secretor phenotype with severe COVID-19. Transfusion. 2020;60:3067–3070. doi: 10.1111/trf.16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gursoy V., Avci S. Effect of ABO blood groups on length of hospital stay according to age in Covid-19 patients. Hematol Transfus Cell Ther. 2022;44:7–12. doi: 10.1016/j.htct.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bokhary D.H., Bokhary N.H., Seadawi L.E., Moafa A.M., Khairallah H.H., Bakhsh A.A. Variation in COVID-19 disease severity and clinical outcomes between different ABO blood groups. Cureus. 2022;12 doi: 10.3389/fmicb.2021.799519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jelinek H.F., Mousa M., Alkaabi N., Alefishat E., Daw Elbait G., Kannout H., et al. Allelic variants within the ABO blood group phenotype confer protection against critical COVID-19 hospital presentation. Front. Med. 2022;8 doi: 10.3389/fmed.2021.759648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garg I., Srivastava S., Dogra V., Bargotya M., Bhattar S., Gupta U., et al. Potential association of COVID-19 and ABO blood group: an Indian study. Microb. Pathog. 2021;158:1–7. doi: 10.1016/j.micpath.2021.105008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garibaldi P.M.M., Oliveira L.C., da Fonseca B.A., Martins M.A., Miranda C.H., Almado C.E.L., et al. Histo-blood group A is a risk factor for severe COVID-19. Transfus. Med. 2021;1–4 doi: 10.1111/tme.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoiland R.L., Fergusson N.A., Mitra A.R., Griesdale D.E.G., Devine D.V., Stukas S., et al. The association of ABO blood group with indices of disease severity and multiorgan dysfunction in COVID-19. Blood Adv. 2020;4:4981–4989. doi: 10.1182/BLOODADVANCES.2020002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sardu C., Marfella R., Maggi P., Messina V., Cirillo P., Codella V., et al. Implications of AB0 blood group in hypertensive patients with covid-19. BMC Cardiovasc. Disord. 2020;20:1–10. doi: 10.1186/s12872-020-01658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szymanski J., Mohrmann L., Carter J., Nelson R., Chekuri S., Assa A., et al. ABO blood type association with SARS-CoV-2 infection mortality: a single-center population in New York City. Transfusion. 2021;61:1064–1070. doi: 10.1111/trf.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nafakhi A., Rabeea I.S., Al-Darraji R., Nafakhi H., Mechi A., Al-Khalidi A., et al. Association of ABO blood group with in-hospital adverse outcome and long term persistent symptoms of COVID-19 infection : a single-center longitudinal observational study. Heal Sci Reports. 2022;5:1–6. doi: 10.1002/hsr2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alabsi R.A.M., Sandeepa N.C., Misfer R.T., Alraqdi M.M., Hamdi M.I.M. Correlation between post-COVID-19, chemosensitive function, blood group, and oral health-related quality of life. Int. J. Dentristy. 2022. 2022:1–8. doi: 10.1155/2022/8715777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Sonmez T., Coker D., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53:801–808. doi: 10.1038/s41588-021-00854-7. [DOI] [PubMed] [Google Scholar]
- 92.CHGI Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-021-03767-x. [DOI] [PubMed] [Google Scholar]
- 94.Vargas-Alarcón G., Ramírez-Bello J., Posadas-Sánchez R., Rojas-Velasco G., López-Reyes A., Martínez-Gómez L., et al. The rs8176740 T/A and rs512770 T/C genetic variants of the ABO gene increased the risk of COVID-19, as well as the plasma concentration platelets. Biomolecules. 2022;12:1–11. doi: 10.3390/biom12040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matzhold E.M., Berghold A., Bemelmans M.K.B., Banfi C., Stelzl E., Kessler H.H., et al. Lewis and ABO histo-blood types and the secretor status of patients hospitalized with COVID-19 implicate a role for ABO antibodies in susceptibility to infection with SARS-CoV-2. Transfusion. 2021;61:2736–2745. doi: 10.1111/trf.16567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coto E., Albaiceta G.M., Clemente M.G., Gómez J. Lack of association between SNPsrs8176719 (O blood group) and COVID-19: data from Spanish age matched patients and controls. Transfusion. 2021;61:654–656. doi: 10.1111/trf.16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nishida N., Sugiyama M., Kawai Y., Naka I., Iwamoto N., Suzuki T., et al. Genetic association of IL17 and the importance of ABO blood group antigens in saliva to COVID-19. Sci. Rep. 2022;12:1–12. doi: 10.1038/s41598-022-07856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hernández Cordero A.I., Li X., Milne S., Yang C.X., Bossé Y., Joubert P., et al. Multi-omics highlights ABO plasma protein as a causal risk factor for COVID-19. Hum. Genet. 2021;140:969–979. doi: 10.1007/s00439-021-02264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palmos A.B., Millischer V., Menon D.K., Nicholson T.R., Taams L.S., Michael B., et al. Proteome-wide Mendelian randomization identifies causal links between blood proteins and severe COVID-19. PLoS Genet. 2022;18:1–23. doi: 10.1371/journal.pgen.1010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou S., Butler-Laporte G., Nakanishi T., Morrison D.R., Afilalo J., Afilalo M., et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med. 2021;27:659–667. doi: 10.1038/s41591-021-01281-1. [DOI] [PubMed] [Google Scholar]
- 101.Zhu J., Wu C., Wu L. Associations between genetically predicted protein levels and COVID-19 severity. J. Infect. Dis. 2021;223:19–22. doi: 10.1093/infdis/jiaa660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amraei R., Yin W., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Cent. Sci. 2021;7:1156–1165. doi: 10.1021/acscentsci.0c01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pathak G.A., Singh K., Miller-Fleming T.W., Wendt F.R., Ehsan N., Hou K., et al. Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-24824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koster T., Blann A., Briët E., Vandenbroucke J., Rosendaal F. Role of clotting factor VIII in effect of von Willebrand factor occurrence of deep-vein thrombosis. Lancet. 1995;345:152–155. doi: 10.1016/s0140-6736(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 105.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/nejmc2000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 107.Hui K.P.Y., Cheung M.C., Perera R.A.P.M., Ng K.C., Bui C.H.T., Ho J.C.W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., et al. TMPRSS2 and TMPRSS4 mediate SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;3582:1–14. doi: 10.1101/2020.04.21.054015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., et al. SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:1–15. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ravn V., Dabelsteen E. Tissue distribution of histo-blood group antigens. Apmis. 2000;108:1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x. [DOI] [PubMed] [Google Scholar]
- 113.Bouhaddou M., Memon D., Meyer B., White K., Rezelj V. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Haan C.A.M., Rottier P.J.M. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1101/2020.06.25.172403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanda M., Morrison L., Goldman R. N-and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021;93 doi: 10.1021/acs.analchem.0c03173. 2003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun Z., Ren K., Zhang X., Chen J., Jiang Z., Jiang J., et al. Mass spectrometry analysis of newly emerging coronavirus HCoV-19 spike protein and human ACE2 reveals camouflaging glycans and unique post-translational modifications. Engineering. 2021;7:1441–1451. doi: 10.1016/j.eng.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deleers M., Breiman A., Daubie V., Maggetto C., Barreau I., Besse T., et al. Covid-19 and blood groups: ABO antibody levels may also matter. Int. J. Infect. Dis. 2021;104:242–249. doi: 10.1016/j.ijid.2020.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zaidi F.Z., Zaidi A.R.Z., Abdullah S.M., Zaidi S.Z.A. COVID-19 and the ABO blood group connection. Transfus. Apher. Sci. 2020;59:1–2. doi: 10.1016/j.transci.2020.102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boukhari R., Breiman A., Jazat J., Ruvoën-Clouet N., Martinez S., Damais-Cepitelli A., et al. ABO blood group incompatibility protects against SARS-CoV-2 transmission. Front. Microbiol. 2022;12 doi: 10.3389/fmicb.2021.799519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mazda T., Yabe R., NaThalang O., Thammavong T., Tadokoro K. Differences in ABO antibody levels among blood donors: a comparison between past and present Japanese, Laotian, and Thai populations. Immunohematol. 2007;23:38–41. doi: 10.21307/immunohematology-2019-261. [DOI] [PubMed] [Google Scholar]
- 125.Breiman A., Ruvën-Clouet N., Le Pendu J. Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLoS Pathog. 2020;16:8–11. doi: 10.1371/journal.ppat.1008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li S., Xu R.X., Guo Y.L., Zhang Y., Zhu C.G., Sun J., et al. ABO blood group in relation to plasma lipids and proprotein convertase subtilisin/kexin type 9. Nutr. Metabol. Cardiovasc. Dis. 2015;25:411–417. doi: 10.1016/j.numecd.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 128.AbdelMassih A., Mahrous R., Taha A., Saud A., Osman A., Kamel B., et al. The potential use of ABO blood group system for risk stratification of COVID-19. Med. Hypotheses. 2020;145:1–6. doi: 10.1016/j.mehy.2020.110343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alejandra Tortorici M., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Renhong Y., Yuanyuan Z., Yaning L., Lu X., Yingying G., Qiang Z. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Awasthi M., Gulati S., Sarkar D.P., Tiwari S., Kateriya S., Ranjan P., et al. The sialoside-binding pocket of SARS-CoV-2 spike glycoprotein structurally resembles MERS-CoV. Viruses. 2020;12:1–10. doi: 10.3390/v12090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Milanetti E., Miotto M., Di Rienzo L., Nagaraj M., Monti M., Golbek T.W., et al. In-silico evidence for a two receptor based strategy of SARS-CoV-2. Front. Mol. Biosci. 2021;8:1–11. doi: 10.3389/fmolb.2021.690655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cohen M., Hurtado-Ziola N., Varki A. ABO blood group glycans modulate sialic acid recognition on erythrocytes. Blood. 2009;114:3668–3676. doi: 10.1182/blood-2009-06-227041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silva-filho J.C., de Melo C.G.F., de Oliveira J.L. The influence of ABO blood groups on COVID-19 susceptibility and severity: a molecular hypothesis based on carbohydrate-carbohydrate interactions. Med. Hypotheses. 2020;144:1–6. doi: 10.1016/j.mehy.2020.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu S.C., Arthur C.M., Wang J., Verkerke H., Josephson C.D., Kalman D., et al. The SARS-CoV-2 receptor-binding domain preferentially recognizes blood group A. Blood Adv. 2021;5:1305–1309. doi: 10.1182/bloodadvances.2020003259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stowell S.R., Stowell C.P. Biologic roles of the ABH and Lewis histo-blood group antigens part II: thrombosis, cardiovascular disease and metabolism. Vox Sang. 2019;114:535–552. doi: 10.1111/vox.12786. [DOI] [PubMed] [Google Scholar]
- 137.Wu O., Bayoumi N., Vickers M.A., Clark P. ABO(H) blood groups and vascular disease: a systematic review and meta-analysis. Thromb. Haemostasis. 2007;6:62–69. doi: 10.1160/TH08-05-0302. [DOI] [PubMed] [Google Scholar]
- 138.Dentali F., Sironi A.P., Ageno W., Turato S., Bonfanti C., Frattini F., et al. Non-O blood type is the commonest genetic risk factor for VTE: results from a meta-analysis of the literature. Semin. Thromb. Hemost. 2012;38:535–547. doi: 10.1055/s-0032-1315758. [DOI] [PubMed] [Google Scholar]
- 139.Yamamoto F., Cid E., Yamamoto M., Blancher A. ABO research in the modern Era of Genomics. Transfus. Med. Rev. 2012;26:103–118. doi: 10.1016/j.tmrv.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 140.Kamphuisen P., Eikenboom J., Bertina R. Elevated factor VIII levels and risk of venous thrombosis. Arterioscler. Thromb. Vasc. Biol. 2001;21:731–738. doi: 10.1111/j.1365-2141.2012.09134.x. [DOI] [PubMed] [Google Scholar]
- 141.Kiouptsi K., Reinhardt C. In: Vertabrate and Invertebrate Respiratory Proteins, Lipoproteins and Other Body Fluid Proteins. Hoeger U., Harris J.R., editors. Springer; Cham: 2020. Physiological roles of the von Willebrand factor-factor VIII interaction; pp. 437–464. [Google Scholar]
- 142.Matsui T., Fujimura Y., Nishida S., Titani K. Human plasma α2-macroglobulin and von Willebrand factor possess covalently linked ABO(H) blood group antigens in subjects with corresponding ABO phenotype. Blood. 1993;82:663–668. doi: 10.1182/blood.v82.2.663.bloodjournal822663. [DOI] [PubMed] [Google Scholar]
- 143.Grewal P.K., Uchiyama S., Ditto D., Varki N., Le D.T., Nizet V., et al. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat. Med. 2008;14:648–655. doi: 10.1038/nm1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rydz N., Swystun L.L., Notley C., Paterson A.D., Riches J.J., Sponagle K., et al. The C-type lectin receptor CLEC4M binds, internalizes, and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood. 2013;121:5228–5237. doi: 10.1182/blood-2012-10-457507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bowen D.J. An influence of ABO blood group on the rate of proteolysis of von Willebrand factor by ADAMTS13. J. Thromb. Haemostasis. 2003;1:33–40. doi: 10.1046/j.1538-7836.2003.00007.x. [DOI] [PubMed] [Google Scholar]
- 146.Tan B.K., Mainbourg S., Friggeri A., Bertoletti L., Douplat M., Dargaud Y., et al. Arterial and venous thromboembolism in COVID-19: a study-level meta-analysis. Thorax. 2021;76:970–979. doi: 10.1136/thoraxjnl-2020-215383. [DOI] [PubMed] [Google Scholar]
- 147.Mai V., Kim B., Mainbourg S., Potus F., Cucherat M., Lega J.C., et al. Venous thromboembolism in COVID-19 compared to non-COVID-19 cohorts: a systematic review with meta-analysis. Vasc. Pharmacol. 2021;139:1–8. doi: 10.1016/j.vph.2021.106882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.