Abstract
Combined microautoradiography and fluorescence in situ hybridization (FISH) was used to investigate carbon metabolism in uncultured bacteria from the genus Achromatium. All of the Achromatium species identified in a freshwater sediment from Rydal Water, Cumbria, United Kingdom, which were distinguishable only by FISH, assimilated both [14C]bicarbonate and [14C]acetate. This extends previous findings that Achromatium spp. present at another location could only utilize organic carbon sources. Achromatium spp., therefore, probably exhibit a range of physiologies, i.e., facultative chemolithoautotrophy, mixotrophy, and chemoorganoheterotrophy, similar to other large sulfur bacteria (e.g., Beggiatoa spp.).
Achromatium oxaliferum is a large, morphologically conspicuous sulfur-oxidizing bacterium found principally in freshwater and brackish sedimentary environments (10, 11, 15, 20). Recent studies have shown that natural communities of this uncultured organism previously thought to be homogeneous in fact comprised a number of phylogenetically, morphologically, and ecologically distinct subpopulations (3, 5, 6, 11). The degree of identity observed in Achromatium-derived 16S rRNA sequences (<97.5%) was consistent with the occurrence of different species of Achromatium both within a single sediment community and at geographically separated locations (21). Fluorescence in situ hybridization (FISH) with combinations of fluorescently labeled oligonucleotide probes revealed that divergent sequences recovered from purified A. oxaliferum cells corresponded to genetically distinct Achromatium subpopulations. Upon analysis of their cell size distributions and population dynamics, these subpopulations also proved to be morphologically and ecologically distinct (6). However, considerable overlap between size classes has meant that whole-cell in situ hybridization provides the only reliable means for identifying and differentiating the coexisting Achromatium species present in any environment.
Microautoradiography was used to investigate carbon metabolism in Achromatium populations from different geographical locations (7). Uptake of 14C-labelled substrates demonstrated that Achromatium cells from freshwater sediments were able to assimilate 14C from bicarbonate, acetate, and protein hydrolysate; however, 14C-labeled glucose was not assimilated. The pattern of substrate uptake by Achromatium spp. was therefore similar to that of a number of other freshwater and marine sulfur-oxidizing bacteria. However, different patterns of radiolabeled bicarbonate uptake were noted in communities of Achromatium from different geographical locations and indicated that at least some of the Achromatium spp. present in one community (Rydal Water) possessed autotrophic potential, while those present in a second (Hell Kettles) did not. However, the pattern of organic and inorganic carbon uptake by Achromatium cells from the Rydal Water sediment suggested that physiological diversity also existed within this natural community. FISH indicated that approximately 98% of the Achromatium cells present in the Rydal Water sediment were potentially metabolically active; however, only 51.3 ± 2.2% of Achromatium cells assimilated carbon from bicarbonate (7). Furthermore, considerable variation in the assimilation of specific substrates was noted in the Rydal Water Achromatium community sampled at different times of the year (unpublished results). For example, the percentage of cells assimilating labeled bicarbonate was 78.7 ± 1.7% in a sample taken in August 1997 and 51.3 ± 2.2% in a sample taken in April 1998. This variation might be linked to temporal changes in the community structure already noted (6). With these results in mind and on the basis of the clearly defined differences in carbon substrate assimilation in Achromatium spp. from different geographical locations, it was tentatively suggested that phylogenetically distinct Achromatium species are also functionally different (7). For instance, within the Rydal Water community, there may be Achromatium species, like those from the Hell Kettles, which are chemoorganoheterotrophs or chemolithoheterotrophs, together with Achromatium species that are obligate chemolithoautotrophs, facultative chemolithoautotrophs, or true mixotrophs.
In this study microautoradiography and FISH were combined to investigate the carbon metabolism of individual Achromatium species within a complex community by simultaneously determining substrate uptake by, and the genetic identity of, individual cells. The approach is very similar to methods previously used to analyze the metabolic activities of specific bacterial communities in activated sludge, defined cell mixtures, and natural bacterioplankton populations (16, 17, 18). Here, the approach has been applied to a natural mixed community of bacteria from the genus Achromatium to identify differences in substrate assimilation by coexisting Achromatium spp. which can only be discriminated using whole-cell in situ hybridization.
Grab samples of sediment containing Achromatium cells were obtained from Rydal Water (4) and were used to prepare reconstituted sediment cores (7). All incubations were carried out in 10-ml glass vials (SH Scientific, Blyth, Northumberland, United Kingdom). Freshly collected sediment (5 ml) was added to each vial and covered with 5 ml of lake water. The reconstituted cores were preincubated for 1 day at room temperature before the addition of radiochemicals (7). In separate incubations 0.4 MBq (0.2 μmol) of radiochemical was added (sodium [14C]bicarbonate [2.0 GBq/mmol] or [1(2)-14C]acetic acid [2.07 GBq/mmol]). To ensure that all Achromatium cells in the sediment cores were exposed to the radiolabeled substrates, they were added by inserting a syringe needle vertically to the base of the reconstituted core and withdrawing the needle slowly while depressing the syringe plunger. This precluded the possibility that cells which did not incorporate the radiolabel failed to do so due to the diffusional limitation of substrate transport to cells deeper in the sediment core. Furthermore, preliminary experiments indicated that a similar proportion of cells assimilated radiolabel whether the radiolabeled substrate was added to the overlying water or injected throughout a sediment core. Therefore, diffusional limitation was not likely to be significant on the timescale of the experiments and did not result in false-negative signals. Since no account was taken of indigenous substrate concentrations, and hence their specific activities in sediment cores, we have not attempted to quantify the relative rates of accumulation of the radiolabeled-carbon sources in Achromatium cells. All radiochemicals were purchased from Amersham International (Little Chalfont, Buckinghamshire, United Kingdom). Control incubations containing a 2% (wt/vol) final concentration of buffered formaldehyde (pH 6.5, 0.1 M phosphate buffer) were also prepared. Incubations were performed in the dark at 20°C over a 3-h period to limit formation of 14CO2 from the degradation of acetate by heterotrophic bacteria (12). After incubation, Achromatium cells were crudely purified from sediments (4), decalcified using 0.1 M HCl (1 ml), and washed three times in filter sterilized water to remove residual radioactivity. Achromatium cells were fixed as described previously (10) prior to whole-cell in situ hybridization.
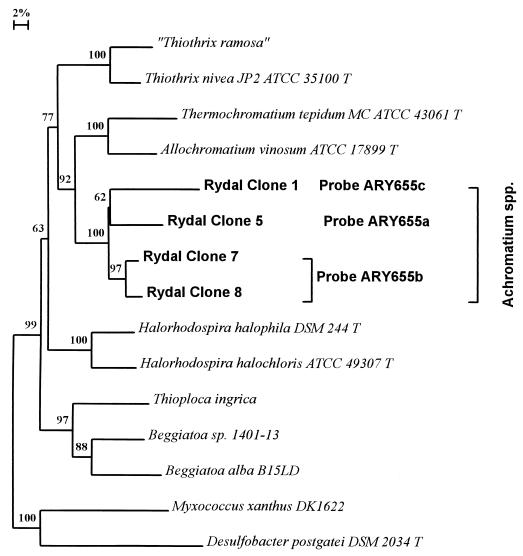
Fixed cells were pelleted (13,000 rpm, 1 min; MSE Microcentaur, Sanyo, United Kingdom) washed once in phosphate-buffered saline (PBS) and either resuspended in PBS for short-term storage at 4°C (no more than 2 days) or washed in sterile distilled water and used immediately. Cells were placed on gelatin-coated microscope slides and incubated for a few minutes at 50°C until just dry. Cells were hybridized in 19 μl of hybridization buffer (1, 10) containing 45% formamide at 42°C in an isotonic chamber for 2 to 3 h. Combinations of Achromatium-specific oligonucleotide probes ARY655a (5′-ACCCCCCTCTCTCGTACT-3′), ARY655b (5′-ACCCCCCTCTCTAGTACT-3′), and ARY655c (5′-ACCCCCCTCTATCGTACT-3′) (6) (1 μl) were added to hybridization reactions after 30 min of preincubation in hybridization buffer. The oligonucleotide probes used targeted the three deeper-branching Achromatium lineages present in Rydal water (i.e., those exhibiting <97% 16S rRNA sequence identity; Fig. 1). ARY655b targeted two distinct 16S rRNA sequences which shared 97.3% sequence identity (clones 7 and 8). ARY655a and ARY655c probes targeted single Achromatium 16S rRNA sequences (clones 5 and 1, respectively). Two pairwise combinations of differentially labeled probes specific for each cluster were used to distinguish the three subpopulations as previously described (6). Hybridized cells were washed three times (20 μl of hybridization buffer, 15 min) at 42°C and washed once in filter-sterilized deionized water (Milli-Q50; Millipore, Molsheim, France). Finally, 10 μl of Citifluor antifadent (Citifluour, Canterbury, United Kingdom) was pipetted onto slides. Hybridized cells were viewed under a coverslip using an Olympus BH-2-RFCA microscope fitted with a high-pressure mercury vapor lamp and blue and green filter sets (Olympus BP545 and BP409). Photomicrographs were taken using Kodak Ektachrome Elite 400 film. Automatic exposure was used for phase-contrast micrographs and exposure times of 10 s (rhodamine fluorescence) and 30 s (fluorescein fluorescence) were used to obtain double-exposure epifluorescence micrographs. The position of photographed cells on slides was recorded using the microscope stage vernier scale. After phase-contrast and fluorescence micrographs were taken, the slides were carefully dipped in absolute ethanol to remove the coverslip and Citifluor antifadent. After air drying, the slides were dipped in Amersham LM-1 autoradiography emulsion (Amersham International). Slides were exposed in a light-proof box at 4°C for 2 weeks and developed and fixed using Kodak GBX developer and fixative-replenisher (Sigma, Poole, Dorset, United Kingdom). Cells were relocated on the slides, and phase-contrast micrographs were taken using automatic exposure. Examples of the results obtained are shown in Fig. 2. Cells were often washed from the slides, reducing the number of cells counted for each subpopulation.
FIG. 1.
Phylogenetic distance tree based on the comparative analysis of almost-full-length 16S rRNA gene sequences recovered from Achromatium cells purified from Rydal Water sediments (6, 10). The sequences targeted by the oligonucleotide probes used in this study are labeled. The scale bar denotes 2% sequence divergence, and the values at the nodes indicate the percentage of bootstrap trees that contained the cluster to the right of the node. Bootstrap values of <50 are not shown.
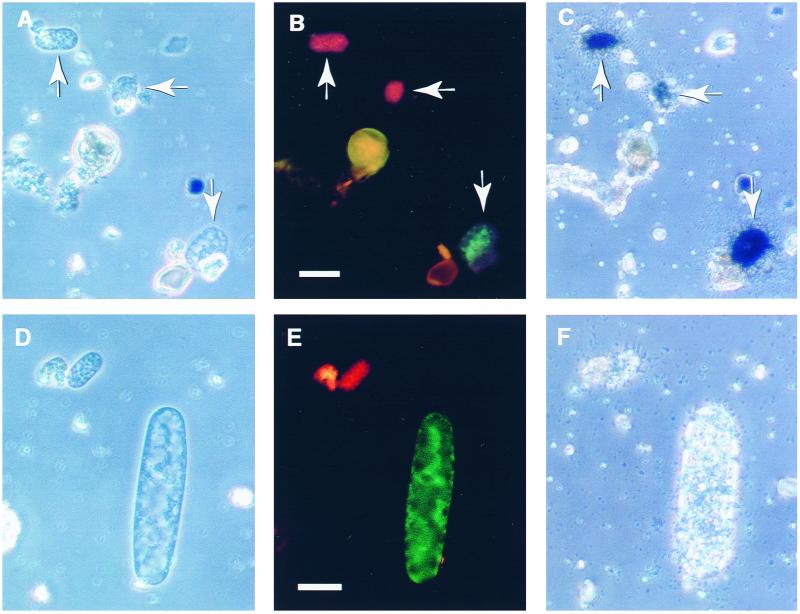
FIG. 2.
Combined whole-cell in situ hybridization and microautoradiography of Achromatium cells from Rydal Water sediments. Whole-cell hybridization was done using a fluorescein-labeled RY clone 1-specific probe (ARY655c) and a rhodamine-labeled RY clone 8-specific probe (ARY655b). The micrographs are phase-contrast (A and D), epifluorescence (B and E), and microautoradiographic (C and F) images of the same fields of view. The cells shown were purified from sediment cores treated with radiolabeled bicarbonate. A proportion of cells of all Achromatium spp. assimilated both bicarbonate and acetate (data not shown). Not all probe-positive cells assimilated labeled substrates (D to F). The scale bars represent 20 μm.
Our central hypothesis was that different coexisting Achromatium spp. would exhibit different patterns of carbon source (bicarbonate and acetate) assimilation. Based on their uptake of [14C]bicarbonate (clone 1, 40.0 ± 21.5% positive [n = 20]; clone 5, 47.6 ± 21.6% positive [n = 21]; clones 7 and 8, 82.4 ± 18.3% positive [n = 17]) and [14C]acetate (clone 1, 53.3 ± 25.3% positive [n = 15]; clone 5, 71.4 ± 33.6% positive [n = 7]; clones 7 and 8, 58.8 ± 23.4% positive [n = 17]) (data represent the mean number of positive cells ± 1.96 × the standard error), all Achromatium species identified by our oligonucleotide probes possessed autotrophic potential and were also capable of assimilating acetate. Thus, our data refuted our original hypothesis and ecological differentiation of coexisting Achromatium spp. is not a result of the use of acetate by some species and bicarbonate by others. Nevertheless, this study cannot be viewed as definitive since the sample sizes were relatively small. A more extensive study with larger sample sizes may permit smaller differences between populations to be resolved.
Although the uptake of radiolabeled bicarbonate by Achromatium cells suggests that they are potentially autotrophic, this evidence is not conclusive. Heterotrophic bacteria can incorporate CO2 via alternative enzymatic pathways (e.g., pyruvate carboxylase), but in such cases inorganic carbon contributes little to cell growth. Several strands of evidence support the notion that some Achromatium spp. fix inorganic carbon by an autotrophic pathway. First, Achromatium is closely related to other morphologically conspicuous sulfur bacteria, such as Beggiatoa spp., for which obligate and facultative chemolithoautotrophy have been confirmed (8). Second, and more compellingly, a homologue of the RuBisCO large subunit gene (rbcL) has been detected in DNA extracts from pure preparations of Achromatium cells from Rydal Water which showed bicarbonate uptake but was never detected in DNA extracted from Achromatium cells purified from Hell Kettles sediments, where [14C]bicarbonate uptake was never observed (11). While this evidence is circumstantial (we have not directly detected the rbcL gene within Achromatium cells or looked for expression of the gene), it does suggest that the bicarbonate fixation observed in Achromatium spp. is via an autotrophic pathway. Furthermore, if inorganic carbon assimilation was associated with a heterotrophic pathway, one would expect that all Achromatium spp. whether heterotrophic, autotrophic, or mixotrophic, would assimilate inorganic carbon by a similar pathway. However, we know that Achromatium cells from Hell Kettles sediments are almost certainly heterotrophs, yet they do not assimilate inorganic carbon (11).
Although particular care was taken in these experiments to ensure that cells at different depths in the sediment were exposed to similar concentrations of radiolabeled compounds, a proportion of probe-positive cells from each subpopulation did not assimilate substrate. This inconsistency may be explained by a functional diversity that is manifest at a higher degree of phylogenetic resolution than our rRNA-targeted probes were designed to identify. Indeed, probe ARY655b targets two Achromatium-derived sequences (clones 7 and 8) that show >2.5% sequence divergence and accordingly could be classified as separate species (21). However, we have no information about the relative representation of the two sequence types within the clone 7 and 8 subpopulation or even whether they represent two distinct genotypes or heterogeneous rRNA operons in a single genotype (6). It is also possible that our other probes, which target a region of the 16S rRNA that is relatively conserved in Achromatium spp., may also fail to detect some similar but distinct genotypes present in the sediment. The use of multiple signature probes or probes targeting less-conserved regions of the 16S rRNA would potentially resolve this issue. However, if one were to assume that the Achromatium subpopulations identified by FISH were not functionally homogeneous, then the functional diversity (carbon substrate utilization) in a single subpopulation would actually be greater than has been found to exist between subpopulations (Fig. 2).
It was not possible to determine the uptake of more than one radiolabeled substrate at a time by individual cells. Therefore, the probability that Achromatium cells from the different subpopulations utilized both bicarbonate and acetate was determined statistically. If the combined percentages of radiolabeled cells in the bicarbonate and acetate uptake experiments can be shown statistically to exceed 100%, then at least a fraction of the cells assimilated both substrates. For example, 82.4 ± 18.3% (P = 0.05) clone 7 and 8 cells were positive for bicarbonate uptake and 58.8 ± 23.4% (P = 0.05) cells were positive for acetate uptake. When the data from bicarbonate and acetate uptake are grouped, a total of 141.2 ± 29.6% (range, 111.6 to 170.8%; P = 0.05) were positive for radiolabeled substrate uptake. Since all values in the range exceed 100%, a proportion of the cells were capable of assimilating both substrates and were thus probably mixotrophs. However, the data for clone 1 (range, 45.8 to 132.9%; P = 0.05) and clone 5 cells (range, 79.4 to 158.6%; P = 0.05) are equivocal, and mixotrophy cannot be confirmed in these subpopulations. Furthermore, it cannot be confirmed that mixotrophy occurred in all cells in the clone 7 and 8 subpopulation. As a consequence, the pattern of substrate uptake observed, i.e., the presence of probe-positive cells that did not assimilate substrate, may indicate that these cells are facultative chemolithoautotrophs. Facultative chemolithoautotrophs can obtain all of their cellular carbon from inorganic sources and derive energy and reducing power from the oxidation of reduced sulfur species. Under different environmental conditions, they can grow mixotrophically or heterotrophically (19). Thus, in any population of a single Achromatium sp. inhabiting a heterogeneous sedimentary environment there may be cells expressing different metabolic pathways. Facultative autotrophy of this nature has been demonstrated in the sulfur-oxidizing marine Beggiatoa sp. strain MS-81-6 (8). This isolate was capable of growth as a chemolithoautotroph but could also use a range of simple organic carbon sources for energy generation and biosynthesis. When provided with organic carbon, ribulose-1,5-bisphosphate carboxylase/oxygenase activity in this organism was downregulated in comparison to strictly chemolithoautotrophically grown cells (8).
Rather than facultative metabolism, an alternative explanation for the observed pattern of substrate uptake is that Achromatium cells which have retained detectable numbers of ribosomes, and hence gave a positive probe hybridization signal, were metabolically inactive. Previous studies have shown that while there was a strong correlation between oligonucleotide probe binding and uptake of radiolabeled substrates in both pure cultures and natural bacterial communities (13, 18), a proportion of probe-positive cells in a community of marine bacterioplankton did not assimilate radiolabeled amino acids (<10%) (13, 18). However, in the Rydal Water Achromatium community the proportion of probe-positive but apparently metabolically inactive Achromatium cells ranged from 12 to 60%. On balance, this unexpectedly high proportion of probe-positive but putatively inactive cells suggests that cells exhibiting facultative metabolism is a more likely explanation for this observation. However, the data presently do not unequivocally prove this since cells which have been specifically inhibited and are thus metabolically inactive can retain high levels of ribosomes detectable using FISH (22) and the possibility exists that Achromatium cells that have been inactivated during sampling and preparation of the sediment cores may retain significant numbers of ribosomes.
Knowledge of the physiology of Achromatium spp. now encompasses heterotrophy and mixotrophy/facultative chemolithoautotrophy, but no evidence of obligate chemolithoautotrophy has been obtained. This is comparable with the known metabolic features of freshwater Beggiatoa spp., which are not known to be capable of chemolithoautotrophic growth.
The use of in situ hybridization in combination with microautoradiography has increased our knowledge of the ecological roles of coexisting Achromatium species. Although it has been previously suggested that ecological differentiation of coexisting Achromatium species might be based on chemolithoautotrophy/mixotrophy versus chemoorganoheterotrophy (which occurs in Achromatium cells from a different geographical location [7]), the data presented here have indicated that this is not the case. However, while there is considerable metabolic overlap with respect to the known carbon metabolism in coexisting Achromatium species from Rydal Water sediments; they have been shown previously to be ecologically distinct (6). With this in mind, the data presented here suggest that different Achromatium spp. utilize similar carbon sources but do so optimally under different conditions. For example, diffusion-limited, larger cells might occupy a substrate-rich niche, while smaller cells might be favored by lower substrate concentrations (14). Interestingly, size differentiation is a characteristic frequently associated with resource partitioning and niche differentiation in functionally similar but coexisting and competing species (2). Alternatively, the ecological differentiation of coexisting Achromatium spp. may be based on differential utilization of carbon substrates not tested here, and future studies should explore this possibility. Furthermore, perturbation experiments combined with the methodology used here may permit optimal conditions for selection of different subpopulations to be identified and help elucidate the nature of competition between different species, i.e., do they occupy completely separate ecological niches or do they competitively coexist? (9). By manipulation of sediment microcosms with respect to redox conditions and substrate type and availability it may be possible to elicit different patterns of carbon substrate utilization in different Achromatium species and to thus deduce the metabolic repertoire expressed by Achromatium spp. Since location within the sediment may play a crucial role in controlling the metabolism of Achromatium cells, depth-resolved analyses to determine carbon substrate utilization in individual subpopulations found in different sediment redox zones would also be informative. In a wider context, studies of this nature may provide a model for determining the ecological principles that govern the distribution, abundance, and evolutionary change within more-complex bacterial communities.
Acknowledgments
This work was supported by the IFE and the FBA. Financial support was provided by the Natural Environmental Research Council (grant GR3/9148 to I.M.H., R.W.P., and J.G.J. and a studentship, GT4/95/235/F, to R.H.).
REFERENCES
- 1.Amann R, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begon M, Harper J L, Townsend C R. Ecology: individuals, populations and communities. 3rd ed. Oxford, England: Blackwell Science, Ltd.; 1996. [Google Scholar]
- 3.Glöckner F O, Babenzien H-D, Wulf J, Amann R. Phylogeny and diversity of Achromatium oxaliferum. Syst Appl Microbiol. 1999;22:28–38. doi: 10.1016/S0723-2020(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 4.Gray N D, Pickup R W, Jones J G, Head I M. Ecophysiological evidence that Achromatium oxaliferum is responsible for the oxidation of reduced sulfur species to sulfate in a freshwater sediment. Appl Environ Microbiol. 1997;63:1905–1910. doi: 10.1128/aem.63.5.1905-1910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray N D, Head I M. New insights on old bacteria: diversity and function of morphologically conspicuous sulfur bacteria in aquatic systems. Hydrobiologia. 1999;401:97–112. [Google Scholar]
- 6.Gray N D, Howarth R, Rowan A, Pickup R W, Jones J G, Head I M. Natural communities of Achromatium oxaliferum comprise genetically morphologically and ecologically distinct sub-populations. Appl Environ Microbiol. 1999;65:5089–5099. doi: 10.1128/aem.65.11.5089-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray N D, Howarth R, Pickup R W, Jones J G, Head I M. Substrate utilisation by the uncultured bacteria from the genus Achromatium determined by the use of microautoradiography. Appl Environ Microbiol. 1999;65:5100–5106. doi: 10.1128/aem.65.11.5100-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagen K D, Nelson D C. Organic carbon utilization by obligately and facultatively autotrophic Beggiatoa strains in homogeneous and gradient cultures. Appl Environ Microbiol. 1996;62:947–953. doi: 10.1128/aem.62.3.947-953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 10.Head I M, Gray N D, Clarke K J, Pickup R W, Jones J G. The phylogenetic position and ultrastructure of the uncultured bacterium Achromatium oxaliferum. Microbiology. 1996;142:2341–2354. doi: 10.1099/00221287-142-9-2341. [DOI] [PubMed] [Google Scholar]
- 11.Head I M, Gray N D, Howarth R, Pickup R W, Jones J G. Achromatium oxaliferum: understanding the unmistakable Adv. Microb Ecol. 1999;16:1–40. [Google Scholar]
- 12.Hoppe H-G. Determination and properties of actively metabolizing heterotrophic bacteria in the sea, investigated by means of micro-autoradiography. Mar Biol. 1976;36:291–302. [Google Scholar]
- 13.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch A L. What size should a bacterium be? A question of scale. Annu Rev Microbiol. 1996;50:317–348. doi: 10.1146/annurev.micro.50.1.317. [DOI] [PubMed] [Google Scholar]
- 15.La Rivière J W M, Schmidt K. Morphologically conspicuous sulfur-oxidizing eubacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 3934–3947. [Google Scholar]
- 16.Lee N P H, Nielsen, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen P H, Andreasen K, Lee N, Wagner M. Use of microautoradiography and fluorescent in situ hybridisation for characterization of microbial activity in activated sludge. Wat Sci Technol. 1999;39:1–9. [Google Scholar]
- 18.Ouverney C C, Fuhrman J A. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson L A, Kuenen J G. The colorless sulfur bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 385–413. [Google Scholar]
- 20.Schewiakoff W. Über einen neuen bakterienähnlichen Organismus des Sßwassers. Heidelberg, Germany: Habilitationsschrift; 1893. [Google Scholar]
- 21.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 22.Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. In situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol. 1995;18:251–264. [Google Scholar]